Abstract
The DNA damage response (DDR) pathways play critical roles in protecting the genome from DNA damage. Abrogation of DDR often results in elevated genomic instability and cellular sensitivity to DNA damaging agents. Many proteins involved in DDR are subjected to precise regulation at multiple levels, such as transcriptional control and posttranslational modifications, in response to DNA damage. MicroRNAs (miRNAs) are a class of small non-coding RNAs that negatively regulate gene expression at the post-transcriptional level. The expression levels of some miRNAs change in response to DNA damage. Some miRNAs, such as miR-24, 138, 96 and 182, have been implicated in DDR and/or DNA repair and affect cellular sensitivity to DNA damaging agents. In this review, we summarize recent findings related to the emerging roles of miRNAs in regulating DDR and DNA repair and discuss their potential in cancer therapy.
Keywords: microRNA, DNA damage response, DNA repair, chemotherapy, radiotherapy, DNA damaging agents
MicroRNAs (miRNAs) are a class of noncoding, endogenous, short (usually 21–25 nt) and single-stranded RNA molecules. MiRNA genes are located in both intragenic and intergenic regions throughout the genome, where they are transcribed independently or together with their host genes.1 MiRNAs represent a new class of post-transcriptional regulators of gene expression and play important roles in many biological processes and disease phenotypes.2 Particularly, the expression of miRNAs is responsive to various endogenous and exogenous stimuli, such as treatment with anticancer DNA damaging agents. Cells with defects in miRNA biogenesis machinery have abnormal cell cycle checkpoints and DNA repair.3,4 Some miRNAs, such as miR-16, miR-24, miR-138 and the miR-183-96-182 cluster, have been implicated in DNA damage response (DDR) and DNA repair.3,5-8 These findings suggest an important role for miRNAs in DDR and DNA repair pathways. Understanding the connection between miRNAs and DDR/DNA repair will advance our knowledge of cancer development and exploitation of miRNAs for cancer treatment. There are several excellent reviews about miRNAs and DDR/DNA repair.9-11 Here, we provide up-to-date summaries of miRNA-DDR interactions, propose our models for the roles of miRNAs in DDR and discuss the therapeutic potential of the miRNAs that regulate DDR/DNA repair.
DNA Damage Response and DNA Repair
To preserve genomic stability, cells have developed elaborate response pathways to handle DNA lesions that are continuously caused by genotoxic agents arising from various sources.12 This DDR is a signaling network initiated by lesion recognition and amplified by multiple mediator signaling proteins, which eventually activate downstream effectors to modulate cell fate by manipulating gene expression, cell cycle control, DNA repair, apoptosis and senescence.12 Activation of cell cycle checkpoints at either G1/S, intra-S or G2/M gives cells time to employ DNA repair machineries to resolve DNA lesions. Multiple DNA repair pathways have evolved to resolve various DNA lesions. These pathways include single-strand break repair, homologous recombination repair, non-homologous end-joining repair, nucleotide excision repair, mismatch repair, translesion DNA synthesis, the Fanconi anemia pathway and others. DNA damage levels that exceed the repair capacity of cells lead to the activation of apoptosis or senescence.12
Three of the phosphoinositide 3-kinases (PI3K)-like protein kinases, ATM, ATR and DNA-PKcs, play a central role in controlling the cellular response to DNA damage.13 ATM, activated after detection of DSBs, phosphorylates numerous substrates, including Chk2, p53 and BRCA1, and plays central roles in regulating cell cycle checkpoint activation and DNA repair.14 ATR, which is activated upon detection of single-stranded DNA molecules present at DNA lesions, including DSBs and stalled replication forks, phosphorylates Chk1 and other substrates to control cell cycle checkpoint and genomic stability.15,16 DNA-PKcs, activated upon induction of DSBs, phosphorylates itself and other substrates and plays an important role in DSB repair through non-homologous end joining.14
Tumor cells with defects in DDR or DNA repair are hypersensitive to certain types of DNA damaging and chemotherapeutic agents. For example, ATM-deficient cells are hypersensitive to ionizing radiation (IR).17 ATR-deficient cells are hypersensitive to multiple DNA damaging agents, including topoisomerase inhibitors, UV radiation and DNA crosslinking agents.18 Tumor cells with defective homologous recombination, such as breast and ovarian cancer cells with BRCA1 or BRCA2 deficiency, are hypersensitive to poly(ADP-ribose) polymerase (PARP) inhibitors,19 which promote DSB formation during replication due to unrepaired single-strand DNA breaks.20 Homologous recombination-deficient tumor cells are also sensitive to interstrand DNA crosslinking agents, such as cisplatin and carboplatin.21 Therefore, targeting DDR or DNA repair pathways is an important strategy for developing novel anticancer therapeutic agents and for chemosensitizing cancer cells to conventional chemotherapy.22
Biogenesis and Function of microRNAs
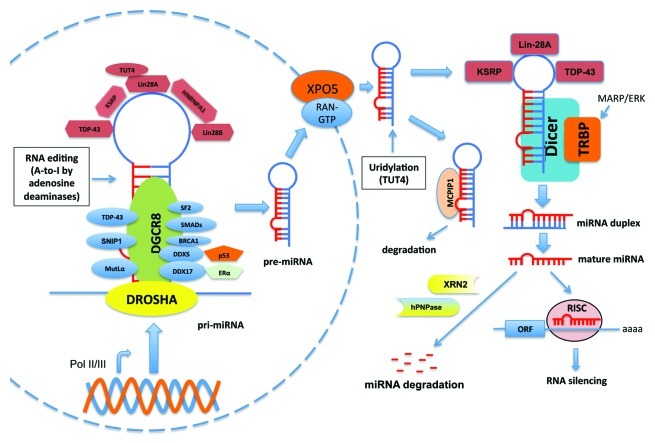
Maturation of miRNAs is a multistep process (Fig. 1). After being transcribed by RNA Pol II or Pol III in the nucleus,1,23 the “stem-loop”-like primary-miRNAs (pri-miRNAs) are recognized and processed by the microprocessor complex that contains the RNase III enzyme, Drosha and its dsRNA-binding partner, DGCR8/Pasha.1 The ~70 nucleotide excised hairpins (pre-miRNAs) are subsequently delivered to the cytosol by the exportin-5 (XPO5)-containing nuclear export protein complex.24 Further processing of pre-miRNAs by the cytosolic RNase III enzyme, Dicer, with its dsRNA-binding partner TARBP2 (TRBP), generates miRNA duplexes.25,26 Each strand of the duplex can be subsequently loaded into Argonaute (Ago) proteins to assemble the RNA-induced silencing complex (RISC),1 depending on the thermodynamic stability at the 5′ or 3′ ends of miRNA duplexes.27,28 The miRNA serves as a specificity factor for RISC by promoting the association of RISC with target mRNAs through base-paring interactions with complementary sites, mainly in the 3′-UTR of target mRNAs and sometimes in coding regions or 5′-UTR.29,30 This leads to translational repression of the transcript, mRNA degradation or both.31 Interestingly, let-7 can bind to the 3′ end of its own primary transcript and provide a feedback loop, suggesting that miRNAs can also regulate the expression of noncoding RNAs.32 In addition to the canonical miRNA biogenesis pathway, minor pathways that bypass the requirement of the enzymatic activities of either Drosha33,34 or Dicer35-37 have also been identified.
Figure 1. Canonical miRNA biogenesis and its regulation by interacting partners. MiRNAs genes are transcribed by RNA polymerases II or III (Pol II/III) into pri-miRNAs from intergenic or intronic regions. The pri-miRNAs are recognized and processed by the DROSHA-DGCR8 complex to generate pre-miRNAs, which are subsequently released into cytosol by the XPO5-containing nuclear export receptor complex. The cytosolic pre-miRNAs will be further digested by the Dicer-TRBP complex to produce miRNA duplexes, which are unwinded and incorporated into the RISC complex for target recognition and silencing. Single-stranded miRNA can be decayed by the 5′-3′ exoribonuclease XRN2 or 3′-5′ exoribonuclease human polynucleotide phosphrylase (hPNPase). The unprocessed pre-miRNAs can also bind to MCPIP1, leading to degradation of pre-miRNAs. The biogenesis of miRNA is subjected to regulation by many proteins that bind to either pri-miRNAs or pre-miRNAs (refer to text for details).
MiRNA biogenesis is subjected to regulation by various mechanisms (Fig. 1). Some RNA binding proteins (such as DDX5, DDX17, SMADs, SNIP1, SF2 and TDP-43) bind to the stem regions of pri-miRNAs and mediate the interaction between the Drosha-DGCR8 complex and a subset of pri-miRNAs, leading to efficient processing.38-43 The NF-90-NF-45 complex prevents access of the Drosha-DGCR8 complex to pri-miRNAs, causing a reduction of a subset of mature miRNAs.44 Several RNA-binding proteins (such as TDP-43, Lin-28, KSRP) bind to the terminal loops of some pri-miRNAs and/or pre-miRNAs and promote or limit their processing.42 Binding of other proteins (such as p53, ERα) to these RNA binding proteins can modulate their regulatory roles in miRNA processing.45,46 MCPIP1 can antagonize Dicer function to prevent the maturation of pre-miRNAs.47 Furthermore, miRNA biogenesis is also associated with RNA-editing pathways.48 For example, adenosine deaminases cause A-to-I editing of about 16% of human pri-miRNAs,49-51 which can block the maturation of pri- or pre-miRNAs and/or change target specificity.51,52 Deep sequencing of miRNA precursors also revealed extensive 3′ end uridylation and other modifications of pre-miRNAs.53 In particular, Lin28 interacts with some miRNA precursors, including those of let-7, miR-107, miR-143 and miR-200c, to facilitate their terminal uridylation by TUT4 uridylyl transferase and subsequent degradation in stem cells.54,55
The activity of the components of miRNA biogenesis machinery can be stimulated by post-translational modifications. For example, TRBP is phosphorylated by MAPK/ERK pathway upon growth factor stimulation, and this phosphorylation specifically regulates the expression of a subset of miRNAs.56 Also, phosphorylation of Drosha is required for its nuclear localization.57 These post-translational regulations of miRNA biogenesis proteins, together with the transcriptional control of pri-miRNAs, provide precise control over miRNA production under different physiological and pathological conditions, thereby relaying important roles in biological process and disease development. Furthermore, cellular miRNA levels also reflect the balance of biogenesis and turnover. Active miRNA degradation can be mediated by the 5′-3′ exoribonuclease XRN258 or the 3′-5′ exoribonuclease, human polynucleotide phosphorylase (hPNPase).59,60
Modulation of microRNA Expression in DNA Damage Response
Expression of miRNAs is altered in response to DNA damage
Multiple normal cell models, such as human endothelial cells,61 human lymphoblast IM9 cells,62,63 human thyroid cells64 and mouse embryonic fibroblasts,65 have been used to study IR-induced miRNA expression changes in various conditions. However, the lack of consistency among these cell lines indicates that the regulation of some miRNAs after DNA damage is cell-type specific. This is further supported by multiple studies in tumor cells. For example, Shin et al. identified a subset of miRNAs that were responsive to IR in A549 non-small cell lung cancer cells, but not in HCT116 colon carcinoma cells.66,67 Niemoeller et al. found that only a few miRNAs are commonly upregulated in all six glioma and head and neck squamous cell carcinoma cell lines examined upon IR treatment.68 Induction of miR-138 after IR treatment was observed in multiples cell lines, such as a HPV E6/E7-transformed lung epithelial cell line CRL-274169 and some glioma and head neck squamous cell carcinoma cell lines,68 but not in IM9 B-cell lyphoblastoma cells,63,66 A549 non-small cell lung cancer cells63,66 or U2OS osteosarcoma cells.6
In addition to IR, treatment with other DNA damaging agents also modulates miRNA expression. For example, UV radiation alters the expression of miRNAs in both HeLa cells and normal human fibroblasts (NHFs).3 Hydrogen peroxide (H2O2) treatment induces a subset of miRNAs, such as the miR-16 family, the miR-106b family and the miR-183 cluster, in NHFs.70 In another study, exposure to IR, H2O2 or etoposide causes differential miRNA expression in NHFs. In this study, some miRNAs can be modulated by two or all three types of DNA damage, while exposure to H2O2 and etoposide can modulate the expression of a unique set of miRNAs, respectively.71 Furthermore, cisplatin or 5-fluorouracil treatment, also elicits a unique miRNA response in two esophageal carcinoma cell lines (one adenocarcinoma and one squamous cell carcinoma).72 Thus, different DNA damaging agents can trigger different miRNA responses.
Mechanisms underlying DNA damage-induced miRNA response
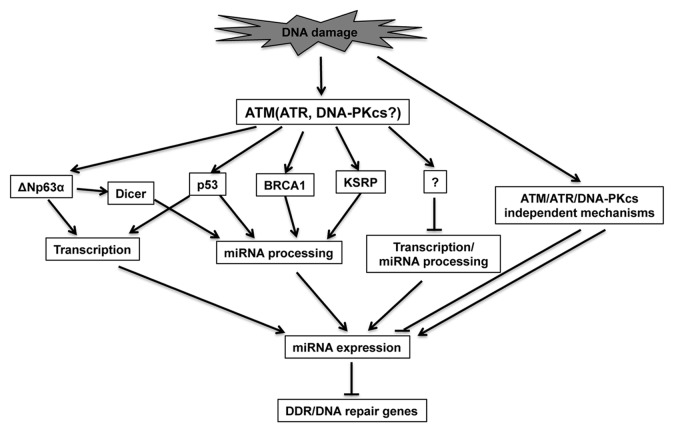
Regulation of miRNA expression can occur at both transcriptional and post-transcriptional levels in response to DNA damage.73 ATM, a key kinase that regulates DNA damage response, also plays a central role in miRNA expression after DNA damage, as loss of ATM prevents the alteration of a large set of miRNAs after DNA damage (Fig. 2).65
Figure 2. A proposed model for the roles of miRNAs in DDR. DNA damage triggers the activation of PI3K-like kinases, such as ATM. ATM phosphorylates a subset of substrates (i.e., p53, KSRP, BRCA1 and ΔNp63α), which activate the transcription of some miRNA genes or enhance their processing through regulating the interaction between miRNA processing complex and pri- or pre-miRNAs. Activation of PI3K-like kinases may also suppress the transcription or processing of some miRNAs through unknown mechanisms. Activation of PI3K-like kinase-independent pathways may also contribute to miRNA response upon DNA damage. The differentially expressed miRNAs may modulate DDR through regulating the expression of DDR/DNA repair genes or their regulatory factors.
ATM regulates miRNA expression through several different mechanisms. One mechanism is the ATM-dependent phosphorylation of KH-type splicing regulatory protein (KSRP).65 KSRP is an interacting factor of both Drosha and Dicer and regulates the maturation of a subset of miRNAs by recognizing the GGG triplets in their terminal loop region.74 ATM-mediated phosphorylation of KSRP at S132, S274 and S670, significantly strengthens the interaction between KSRP and pri-miRNAs and also increases KSRP activity in miRNA processing.65 Whether phosphorylation of KSRP at those sites is important for DDR remains to be determined. Since ATR and ATM share many of the same substrates, it will be interesting to determine whether KSRP is also phosphorylated by ATR in response to UV or replication stress, and whether KSRP phosphorylation plays a role in UV (or replication stress)-induced miRNA alteration.
Interestingly, while all of the 29 KSRP-dependent miRNAs are induced in an ATM-dependent manner in response to neocarzinostain (a radiomimetic agent) treatment,74 an additional 42 miRNAs are induced in an ATM-dependent but KSRP-independent manner.65
One possible KSRP-independent mechanism of ATM-mediated miRNA regulation is through p53. In response to DNA damage, ATM activates p53 by multiple mechanisms, such as Chk2-dependent phosphorylation of p53.75,76 p53 regulates miRNA expression both by transcriptional regulation and post-transcriptional regulation. For example, p53 promotes the processing of a subset of pri-miRNAs through interaction with the DDX5 RNA helicase45 and upregulates the transcription of pri-miR-34a-c after DNA damage.77 Seven of the 10 miRNAs that were induced by doxorubicin in a p53-dependent manner are also induced by DNA damage in an ATM-dependent manner in MEFs,45,65 suggesting that p53-mediated regulation is a mechanism by which ATM controls miRNA expression. However, only three of the seven miRNAs (miR-15a, miR-16 and miR-26b) are induced in a KSRP-dependent manner. This suggests that p53 and KSRP can regulate pri-miRNA processing differently.
Other mechanisms also contribute to ATM-dependent miRNA induction after DNA damage. ATM-dependent phosphorylation of ΔNp63α causes alteration of miRNA expression, both by transcriptionally modulating miRNA expression and by upregulating Dicer to stimulate miRNA maturation after cisplatin treatment.78-80 BRCA1, activated by ATM upon DNA damage, facilitates the processing of some pri-miRNAs through direct interaction with Drosha, DDX5 and pri-miRNAs.81 However, these mechanisms cannot explain the ATM-dependent reduction of some miRNAs. Thus, there are likely additional ATM substrates that are relevant to ATM-dependent reduction of those miRNAs. Identification of such substrates is important for our understanding of the mechanisms of DNA damage-induced alteration of miRNA expression.
A subset of miRNAs responds to DNA damage in an ATM-independent, but DNA-PKcs or ATR-dependent, manner.82,83 Although ATR and DNA-PKcs may share some substrates with ATM, they may differentially regulate other substrates and the expression of some miRNAs. Supporting this notion, Chaudhry et al. examined the expression of miRNAs in DNA-PKcs-deficient (M059J) and -proficient (M059K) glioma cells derived from the same patient.82 They found that miR-17-3p, miR-17-5p, miR-19a/b, miR-142-3p and miR-142-5p are upregulated in both cell lines, whereas miR-15a, miR-16, miR-143, miR-155 and miR-21 are upregulated only in DNA-PKcs-proficient cells, suggesting that their upregulation is dependent upon DNA-PKcs.82 In another study, miR-709 is induced in mouse testes possibly through ATR-dependent upregulation of Rfx-1, the host gene of miR-709.83
Kinases other than ATM, ATR and DNA-PKcs, such as MAPKs, are also activated in response to DNA damage.84 Their roles in DNA damage-induced miRNA response are far from being understood. Interestingly, Ago2 is phosphorylated at S387 by p38 MAPK, and this phosphorylation promotes the localization of the RISC complex to P-bodies.85 Phosphorylation of Ago2 at site Y529 by an unidentified kinase may also prevent small RNA binding.86 TRBP is phosphorylated by MAPK/ERK, and this phosphorylation modulates miRNA production by stabilizing the Dicer/TRBP complex.56 Whether these phosphorylation events can be induced by UV or other types of DNA damage needs to be tested.
Furthermore, DNA damage may modulate the expression of miRNA biogenesis genes. As mentioned above, cisplatin can stimulate the transcription of Dicer in a ΔNp63α-dependent manner.80 It is also possible that the enzymatic activities of Drosha and Dicer may be altered upon DNA damage by post-translational modifications of themselves or their binding partners, such as DGCR8, TRBP, DDX5 and DDX17. Interestingly, DDX17 is phosphorylated at two SQ sites (ATM/ATR consensus sites),87 though the functional relevance of this phosphorylation in DNA damage-induced miRNA response needs to be studied.
Functions of miRNAs in DNA Damage Response
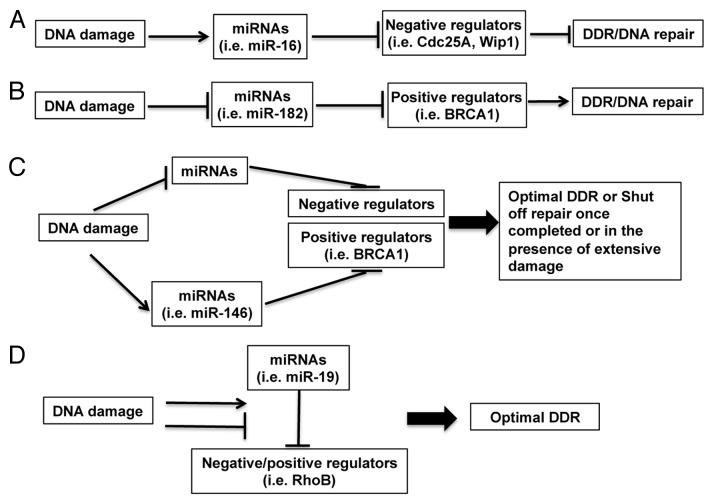
Although expression of a lot of miRNAs is altered in response to DNA damage, the roles for these miRNAs in DDR are still poorly understood. The conceptual models of the function of DDR-regulated miRNAs are shown in Figure 3. In Model A, upregulated miRNAs in DDR promote DDR by suppressing the negative regulators of DDR. In Model B, downregulated miRNAs in DDR release their inhibitory roles on DDR by targeting positive regulators of DDR. In Model C, upregulated miRNAs in DDR target positive regulators of DDR, or downregulated miRNAs target negative regulators of DDR. This results in fine-tuning DDR, or shutting off DNA repair upon the resolution of DNA damage or in the case of extensive and irreparable DNA damage. In Model D, DNA damage changes the interaction between unaltered microRNAs and the positive or negative regulators of DDR/DNA repair. Recent studies have shed some light on DDR-regulated miRNAs’ functions by identifying their targets relevant to cell cycle checkpoints and DNA repair (Table 1), both of which cooperate to prevent propagation of damaged genetic materials.
Figure 3. Conceptual models of the functions of miRNAs in DDR. (A) Upregulated miRNAs in DDR may promote DDR by suppressing the negative regulators of DDR; (B) downregulated miRNAs in DDR may release their inhibitory roles on DDR by targeting positive regulators of DDR; (C) upregulated miRNAs in DDR may target positive regulators of DDR or downregulated miRNAs may target negative regulators of DDR to reach an optimal DDR or to shut off DNA repair once completed or in the case of extensive damage; (D) DNA damage may change the interaction between unaltered microRNAs and the positive or negative regulators of DDR/DNA repair. Examples for miRNAs and their targeting DDR regulators are shown in brackets.
Table 1. MicroRNAs that directly regulate the expression of DDR genes.
| DDR genes | Function in DDR | miRNAs | References |
|---|---|---|---|
| ATM |
DNA repair |
miR-421, miR-100, miR-101 |
99
-
101
|
| H2AX |
DNA repair |
miR-24, miR-138 |
6
,
7
|
| BRCA1 |
DNA repair |
miR-182, miR-146a, miR-146b-5p |
8
,
102
|
| RAD23B |
DNA repair |
miR-373 |
134
|
| RAD51 |
DNA repair |
miR-96 |
5
|
| RAD52 |
DNA repair |
miR-210, miR-373 |
134
|
| REV1 |
DNA repair |
miR-96 |
5
|
| REV3L |
DNA repair |
miR-25, miR-32 |
135
|
| MSH2 |
DNA repair |
miR-21, miR-155 |
103
,
114
|
| MLH1 |
DNA repair |
miR-155 |
103
|
| FANCG |
DNA repair |
miR-23a |
136
|
| p53 |
cell cycle checkpoint, apoptosis |
miR-125b, miR-504, miR-25, miR-30d |
137
-
139
|
| Wee1 |
cell cycle checkpoint |
miR-128a, miR-155, miR-516–3p, miR-195 |
140
,
141
|
| Wip1 |
cell cycle checkpoint |
miR-16 |
88
|
| Cdc25A |
cell cycle checkpoint |
miR-16, miR-21, miR-322, miR-424, miR-503, miR-449a/b |
3
,
89
,
142
,
143
|
| c-myc |
cell cycle checkpoint |
miR-145, let-7 |
144
,
145
|
| p21 |
cell cycle |
miR-17, miR-106a/b |
96
,
146
|
| p27 |
cell cycle |
miR-221/222 |
147
|
| E2F |
cell cycle |
miR-17–92, miR-20a, miR-34a, let-7b |
92
|
| CDK2 | cell cycle | miR-302, miR-372, miR-885–5p | 148 - 150 |
In line with Model A, a number of studies find that some DNA damage-inducible miRNAs target the negative regulators of cell cycle checkpoint. The miR-16 family miRNAs, such as miR-16 and miR-15a/b, are induced by multiple DNA damage agents. Cdc25A and Wip1 are critical phosphatases that regulate DNA damage-induced cell cycle checkpoint. Induction of miR-16 leads to cell cycle checkpoint activation by suppressing Cdc25A and Wip1.3,88 MiR-21, induced upon IR irradiation, represses Cdc25A and modulates cell cycle checkpoint activation.89 The miR-34 family miRNAs (miR-34a, 34b and 34c) are induced by DNA damage in a p53-dependent manner. Induction of miR-34a can regulate the G1/S checkpoint by modulating multiple genes (E2F, cyclinE2, CDK4 and CDK6),77,90 whereas miR-34c regulates the S-phase checkpoint by targeting c-myc.91 The let-7 family miRNAs are also induced by DNA damage. Overexpression of let-7a induces G1/S cell cycle arrest by targeting E2F2 and cyclin D2 in prostate cancer cells.92 Overexpression of let-7b arrests primary fibroblasts at G2/M by direct repression of Cdc34 (a subunit of SCF E3 ligase complex), which leads to stabilization of Wee1 kinase.93 Overexpression of let-7b also downregulates the expression of cyclins D1/D3/A and Cdk4 in melanoma cells.94 Furthermore, miR-24 can activate G1/S cell cycle checkpoint mainly by targeting E2F2.95
In agreement with Model B, downregulation of some miRNAs is required for efficient cell cycle checkpoint activation. For example, miR-106b is downregulated in IR-irradiated LNCaP prostate cancer cells, and this downregulation is required for p21-mediated G2/M cell cycle arrest.96 Some of the miR-17 family miRNAs (three paralog clusters: miR-17–92, miR-106a-363 and miR-106b-25) are involved in cell cycle control. Inhibition of miR-17 or miR-20a leads to an E2F1-associated DDR and G1 checkpoint activation.97 MiR-17-5p acts specifically at the G1/S boundary by targeting more than 20 genes involved in the G1/S transition.98 Furthermore, downregulation of some miRNAs allows rapid accumulation of DNA repair proteins following DNA damage. Among these miRNAs, miR-421, miR-101 and miR-100 suppress the expression of ATM.99-101 MiR-101 also targets DNA-PKcs,101 and miR-138 and miR-24 both target histone H2AX.6,7 The miR-183-96-182 polycistronic miRNA cluster, which is rapidly downregulated in response to IR, may affect DNA repair through miR-182-mediated suppression of BRCA18 or miR-96-mediated suppression of RAD51 and REV1.5
There are also some miRNAs that are upregulated in response to DNA damage and target DNA repair or DDR genes. These miRNAs may impose a threshold on the activation of DNA repair or DDR, thereby buffering the cellular system for optimal activation of DNA repair and DDR (Fig. 3, Model C). For example, miR-146a and miR-146b-5p, which target BRCA1,102 are induced in response to DSBs.65 MiR-155, induced upon DNA damage, targets MLH1 and MSH2 and negatively regulates mismatch repair.103 In addition, the MLH1-PMS2 heterodimer facilitates the processing of mir-422a, which, in turn, suppresses MLH1 through base pairing with the MLH1 3′-untranslated region.104 These miRNAs may provide an opportunity for cells to choose DNA repair pathways in certain cell types or provide a way for cells to shut down DNA repair machinery after extensive damage. Moreover, the induction of Dicer expression is in concert with the induction of let-7 and miR-103/107 miRNAs after DNA damage, both of which can target Dicer.105-108 This suggests that a feedback control by let-7 and miR-103/107 may limit the extent of Dicer activity to reach an optimal activation of DDR.
Some DNA damage non-responsive miRNAs may also regulate DDR (Fig. 3, model D). For example, although miR-19 level was not changed after UV irradiation, UV prevents the binding of miR-19 to its target site in the 3′UTR of RhoB by dissociating the AU-rich element binding protein HuR from an adjacent binding site, thereby protecting the cell from apoptosis.109 This observation suggests that DNA damage may alter the function of miRNA in DDR through modulating the environment for efficient miRNA-target interaction. Further work is needed to evaluate the contribution of this type of regulation in DDR.
Thus, many DDR and DNA repair genes are subjected to precise control at the post-transcriptional level by miRNAs. Some of these genes are targeted by multiple miRNAs, while some miRNAs can also target multiple DDR/DNA repair genes. Many miRNAs are altered mildly in DNA damage, and the small change in expression of individual miRNA may not produce a significant effect on DDR. However, the collective effects of multiple miRNAs that target the same gene or a common pathway (such as cell cycle checkpoint or homologous recombination repair) may be important for an effective cellular response to DNA damage.
Interestingly, a new class of small non-coding RNAs, other than miRNAs, is also important for DDR. Wei et al. identified that a new class of Dicer-processed small non-coding RNAs, called DSB-induced small RNAs (diRNAs), can be generated at DNA damage sites in both Arabidopsis and human cells.4 These diRNAs are important for the activation of homologous recombination repair, possibly by directing certain chromatin modifications.4 Francia et al. further demonstrated that site-specific DICER and DROSHA RNA products, but not miRNAs, are required for efficient foci formation of several DDR proteins, including phospho-ATM, 53BP1, MDC1 and ATM/ATR substrates (recognized by phospho S/TQ antibody).110 However, this does not rule out the possibility that some specific miRNAs are required for efficient recruitment and accumulation of certain DDR proteins in the presence of substantial DNA damage.
MiRNAs Serve as Therapeutic Agents or Targets for Improving Cancer Treatment with DNA Damaging Agents
Because of the key role of DDR and DNA repair in cancer treatment with DNA damaging agents, targeting DDR and DNA repair is becoming a very attractive strategy to overcome chemo-/radio-resistance.22 In particular, miRNAs are promising agents for improving the efficacy of conventional cancer therapy with DNA damaging agents, attributing to their active involvement in DDR and their ability to target DDR components to control cellular response to DNA damaging agents.
For example, inhibition of ATM by miR-101, miR-100 and miR-421 or inhibition of DNA-PKcs by miR-101 causes increased cellular sensitivity to IR.99-101 Inhibition of H2AX expression by either miR-24 or miR-138 promotes cellular sensitivity to IR and/or genotoxic agents.6,7 Several other IR-responsive miRNAs, such as miR-521, miR-127, let-7g, miR-125a and miR-189, can also modulate radiosensitivity by targeting DDR genes,61,111 although the mechanisms are less clear. Furthermore, miR-34a negatively regulates cellular resistance to 5-FU by regulating Sirt1 and E2F3 in DLD-1 colon cancer cells,112 while miR-21 promotes resistance to 5-FU by suppressing a mismatch repair gene, MSH2, in colon cancer cells.113,114 MiR-143 and miR-145, whose expression is induced by DNA damage through p53, target MDM2 and, thus, provide positive feedback to strengthen the function of p53 in mediating DNA damage-induced cell death.115
To identify miRNAs that modulate chemosensitivity by inhibiting homologous recombination DNA repair, we have developed a cell-based high-throughput screen assay. We transfected individual miRNA mimics into a human osteosarcoma cell line U2OS, irradiated cells with IR and monitored RAD51 foci formation using automated fluorescence microscopy (Huang et al., manuscript in preparation). By screening the human miRNA mimic library, we identified a set of miRNAs that inhibit IR-induced RAD51 foci formation, including miR-96.5 MiR-96 targets both RAD51 and REV1, an error-prone Y-family DNA polymerase required for translesion synthesis across the unhooked interstrand crosslinks.116,117 Overexpression of miR-96 promotes cellular hypersensitivity to both cisplatin and a PARP inhibitor, AZD2281.5 Interestingly, Moskwa et al. reported that overexpression of miR-182, another member of the miR-183-96-182 cluster, inhibits BRCA1 and induces hypersensitivity to PARP inhibitors in vitro and in a xenograft model.8 However, we did not observe significant effects of miR-182 or miR-183 on chemosensitivity to cisplatin or AZD2281,5 suggesting a potential cell type-specific miRNA-target interaction.
Given the fact that some miRNAs can target multiple genes involved in DDR, modulating endogenous miRNA expression may be a promising way to overcome chemoresistance in cancer treatment. Overexpression of miRNA can be achieved by ectopic expression of mature miRNAs or their precursors. Suppression of endogenous miRNAs can be reached with anti-miR oligonucleotides or miRNA sponges, which express a transgene-containing multiple tandem binding sites for endogenous miRNA.118 MiRNA-masking antisense oligonucleotides can also be used to protect particular targets of miRNAs.119
Multiple technologies have been developed to achieve systemic delivery of miRNA mimics or anti-miRs. They include application of chemically modified oligonucleotides, adenoviral or lentiviral-based delivery and nanoparticle-based delivery.120-124 Chemical modification of oligonucleotides, such as adding sugar and phosphate modifications into the active and passenger strands of duplex miRNA mimics or anti-miRs, improves their activities and stability.125,126 Recent studies with modified nanoparticles also provide some potential selective targeting tools for delivering small RNAs in vivo.127,128 Substantial studies provide solid evidence for the use of miRNAs as therapeutic agents or tools in cancer therapy, but the side effects of miRNA therapy also need much attention. This is important since each miRNA may target numerous transcripts and high expression of miRNA mimics may interfere with the endogenous miRNAs or siRNAs by saturating the RISC complex. Therefore, the safety of miRNA formulations needs to be extensively evaluated in disease models.
Conclusions and Future Directions
In summary, miRNA expression is modulated in response to DNA damage at either the transcriptional or post-transcriptional level. Some of these miRNAs can regulate the expression of a wide range of DDR/DNA repair genes and modulate cellular sensitivity to DNA damaging agents. This level of regulation occurs at intervals between rapidly induced post-translational modifications and late-initiating transcriptional control,129 thus having fundamental roles on the integrity of DDR. However, many questions remain to be addressed for a more complete understanding of the importance of miRNA response in DDR.
First, we need to evaluate whether DNA damage triggers post-translational modifications (i.e., phosphorylation) of canonical miRNA biogenesis proteins to regulate miRNA maturation. For example, phosphorylation of Drosha at S300 or S302 was shown to be necessary for its nuclear retention and pri-miRNA processing,57 but analyses of other proteins involved in the miRNA biogenesis are incomplete. It is possible that these phosphorylation events are mediated by ATM, other PI3K-like kinases, their downstream kinases or completely different kinases.
Second, it is important to elucidate a mechanism by which some miRNAs are downregulated in DDR, since downregulation of certain miRNAs may release their inhibitory roles on cell cycle checkpoint control or DNA repair, allowing efficient DDR.
Third, it should be tested whether miRNAs can be edited to modulate their stability and/or activities in DDR.
Lastly, it will be interesting to investigate whether DNA damage triggers secretion of certain miRNAs that are taken up by surrounding cells to modulate the activation of DDR and cellular sensitivity to DNA damaging agents. Recent studies found that miRNAs can be secreted into serum by different mechanisms and taken up by adjacent cells.130 A significant number of miRNAs were found to be rapidly upregulated in the blood of radiotherapy-treated mice or patients.131-133 Whether these secreted miRNAs play roles in DDR needs to be studied.
Fully addressing these questions will help to understand the functions of miRNAs in DDR and provide solid evidence for designing novel strategies for cancer treatment.
Acknowledgments
We appreciate all the members of Taniguchi lab for helpful discussion. We thank Dr. Muneesh Tewari, Dr. Kiranjit Dhillon, Jen-wei Huang and Philamer Calses for critical reading of the manuscript. This work was supported by Howard Hughes Medical Institute, the NIH/ NHLBI (R21 HL092978 to T.T.), the NIH/ NCI (R01 CA125636 to T.T.), Fanconi Anemia Research Fund (to T.T.) and the NIH (P30 DK56465 to Y.W. and T.T.). Y.W. is a research fellow supported by Canadian Institute of Health Research.
Glossary
Abbreviations:
- miRNA
microRNA
- pri-miRNA
primary miRNA
- pre-miRNA
precursor miRNA
- DDR
DNA damage response
- DSBs
double-strand breaks
- PARP
poly(ADP-ribose) polymerase
- IR
ionizing radiation
- NHFs
normal human fibroblasts
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/23051
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pothof J, Verkaik NS, van IJcken W, Wiemer EA, Ta VT, van der Horst GT, et al. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009;28:2090–9. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, et al. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–12. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Huang JW, Calses P, Kemp CJ, Taniguchi T. MiR-96 downregulates REV1 and RAD51 to promote cellular sensitivity to cisplatin and PARP inhibition. Cancer Res. 2012;72:4037–46. doi: 10.1158/0008-5472.CAN-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, et al. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol Cancer Res. 2011;9:1100–11. doi: 10.1158/1541-7786.MCR-11-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009;16:492–8. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41:210–20. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landau DA, Slack FJ. MicroRNAs in mutagenesis, genomic instability, and DNA repair. Semin Oncol. 2011;38:743–51. doi: 10.1053/j.seminoncol.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu H, Gatti RA. MicroRNAs: new players in the DNA damage response. J Mol Cell Biol. 2011;3:151–8. doi: 10.1093/jmcb/mjq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Lu X. Posttranscriptional regulation of miRNAs in the DNA damage response. RNA Biol. 2011;8:960–3. doi: 10.4161/rna.8.6.17337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lempiäinen H, Halazonetis TD. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 2009;28:3067–73. doi: 10.1038/emboj.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bensimon A, Aebersold R, Shiloh Y. Beyond ATM: the protein kinase landscape of the DNA damage response. FEBS Lett. 2011;585:1625–39. doi: 10.1016/j.febslet.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21:879–85. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- 16.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kühne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Löbrich M. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 2004;64:500–8. doi: 10.1158/0008-5472.CAN-03-2384. [DOI] [PubMed] [Google Scholar]
- 18.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 20.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 21.Dhillon KK, Swisher EM, Taniguchi T. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci. 2011;102:663–9. doi: 10.1111/j.1349-7006.2010.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 23.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 24.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 25.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/S0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 28.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 29.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 31.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–26. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature. 2012;486:541–4. doi: 10.1038/nature11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–85. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–40. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci USA. 2010;107:15163–8. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–8. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–11. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 39.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–84. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA. 2008;105:10073–8. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci USA. 2012;109:3347–52. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, et al. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, et al. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29:3754–69. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 46.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, et al. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–7. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44:424–36. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 48.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–54. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, et al. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–80. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–7. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–40. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newman MA, Mani V, Hammond SM. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA. 2011;17:1795–803. doi: 10.1261/rna.2713611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 55.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 56.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–22. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang X, Zhang Y, Tucker L, Ramratnam B. Phosphorylation of the RNase III enzyme Drosha at Serine300 or Serine302 is required for its nuclear localization. Nucleic Acids Res. 2010;38:6610–9. doi: 10.1093/nar/gkq547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–9. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 59.Das SK, Sokhi UK, Bhutia SK, Azab B, Su ZZ, Sarkar D, et al. Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proc Natl Acad Sci USA. 2010;107:11948–53. doi: 10.1073/pnas.0914143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pawlicki JM, Steitz JA. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2010;20:52–61. doi: 10.1016/j.tcb.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner-Ecker M, Schwager C, Wirkner U, Abdollahi A, Huber PE. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat Oncol. 2010;5:25. doi: 10.1186/1748-717X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cha HJ, Seong KM, Bae S, Jung JH, Kim CS, Yang KH, et al. Identification of specific microRNAs responding to low and high dose gamma-irradiation in the human lymphoblast line IM9. Oncol Rep. 2009;22:863–8. [PubMed] [Google Scholar]
- 63.Cha HJ, Shin S, Yoo H, Lee EM, Bae S, Yang KH, et al. Identification of ionizing radiation-responsive microRNAs in the IM9 human B lymphoblastic cell line. Int J Oncol. 2009;34:1661–8. doi: 10.3892/ijo_00000297. [DOI] [PubMed] [Google Scholar]
- 64.Nikiforova MN, Gandhi M, Kelly L, Nikiforov YE. MicroRNA dysregulation in human thyroid cells following exposure to ionizing radiation. Thyroid. 2011;21:261–6. doi: 10.1089/thy.2010.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X, Wan G, Berger FG, He X, Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell. 2011;41:371–83. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK, Jin HO, et al. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int J Oncol. 2009;35:81–6. [PubMed] [Google Scholar]
- 67.Shin S, Cha HJ, Lee EM, Jung JH, Lee SJ, Park IC, et al. MicroRNAs are significantly influenced by p53 and radiation in HCT116 human colon carcinoma cells. Int J Oncol. 2009;34:1645–52. doi: 10.3892/ijo_00000295. [DOI] [PubMed] [Google Scholar]
- 68.Niemoeller OM, Niyazi M, Corradini S, Zehentmayr F, Li M, Lauber K, et al. MicroRNA expression profiles in human cancer cells after ionizing radiation. Radiat Oncol. 2011;6:29. doi: 10.1186/1748-717X-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–6. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev. 2009;130:731–41. doi: 10.1016/j.mad.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, Degraff W, et al. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One. 2009;4:e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hummel R, Wang T, Watson DI, Michael MZ, Van der Hoek M, Haier J, et al. Chemotherapy-induced modification of microRNA expression in esophageal cancer. Oncol Rep. 2011;26:1011–7. doi: 10.3892/or.2011.1381. [DOI] [PubMed] [Google Scholar]
- 73.Wan G, Mathur R, Hu X, Zhang X, Lu X. miRNA response to DNA damage. Trends Biochem Sci. 2011;36:478–84. doi: 10.1016/j.tibs.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–88. [PMC free article] [PubMed] [Google Scholar]
- 76.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–7. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 77.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang Y, Guerrero-Preston R, Ratovitski EA. Phospho-ΔNp63α-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle. 2012;11:1247–59. doi: 10.4161/cc.11.6.19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang Y, Chuang AY, Ratovitski EA. Phospho-ΔNp63α/miR-885-3p axis in tumor cell life and cell death upon cisplatin exposure. Cell Cycle. 2011;10:3938–47. doi: 10.4161/cc.10.22.18107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang Y, Chuang A, Hao H, Talbot C, Sen T, Trink B, et al. Phospho-ΔNp63α is a key regulator of the cisplatin-induced microRNAome in cancer cells. Cell Death Differ. 2011;18:1220–30. doi: 10.1038/cdd.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol. 2012;197:201–8. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaudhry MA, Sachdeva H, Omaruddin RA. Radiation-induced micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol. 2010;29:553–61. doi: 10.1089/dna.2009.0978. [DOI] [PubMed] [Google Scholar]
- 83.Tamminga J, Kathiria P, Koturbash I, Kovalchuk O. DNA damage-induced upregulation of miR-709 in the germline downregulates BORIS to counteract aberrant DNA hypomethylation. Cell Cycle. 2008;7:3731–6. doi: 10.4161/cc.7.23.7186. [DOI] [PubMed] [Google Scholar]
- 84.Dent P, Yacoub A, Fisher PB, Hagan MP, Grant S. MAPK pathways in radiation responses. Oncogene. 2003;22:5885–96. doi: 10.1038/sj.onc.1206701. [DOI] [PubMed] [Google Scholar]
- 85.Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413:429–36. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- 86.Rüdel S, Wang Y, Lenobel R, Körner R, Hsiao HH, Urlaub H, et al. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011;39:2330–43. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, et al. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70:7176–86. doi: 10.1158/0008-5472.CAN-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Oliveira PE, Zhang L, Wang Z, Lazo JS. Hypoxia-mediated regulation of Cdc25A phosphatase by p21 and miR-21. Cell Cycle. 2009;8:3157–64. doi: 10.4161/cc.8.19.9704. [DOI] [PubMed] [Google Scholar]
- 90.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 91.Cannell IG, Kong YW, Johnston SJ, Chen ML, Collins HM, Dobbyn HC, et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci USA. 2010;107:5375–80. doi: 10.1073/pnas.0910015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dong Q, Meng P, Wang T, Qin W, Qin W, Wang F, et al. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One. 2010;5:e10147. doi: 10.1371/journal.pone.0010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Legesse-Miller A, Elemento O, Pfau SJ, Forman JJ, Tavazoie S, Coller HA. let-7 Overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J Biol Chem. 2009;284:6605–9. doi: 10.1074/jbc.C900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–57. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 95.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol Cell. 2009;35:610–25. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li B, Shi XB, Nori D, Chao CK, Chen AM, Valicenti R, et al. Down-regulation of microRNA 106b is involved in p21-mediated cell cycle arrest in response to radiation in prostate cancer cells. Prostate. 2011;71:567–74. doi: 10.1002/pros.21272. [DOI] [PubMed] [Google Scholar]
- 97.Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2009;28:140–5. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest AR, et al. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ng WL, Yan D, Zhang X, Mo YY, Wang Y. Over-expression of miR-100 is responsible for the low-expression of ATM in the human glioma cell line: M059J. DNA Repair (Amst) 2010;9:1170–5. doi: 10.1016/j.dnarep.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 100.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci USA. 2010;107:1506–11. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One. 2010;5:e11397. doi: 10.1371/journal.pone.0011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011;3:279–90. doi: 10.1002/emmm.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA. 2010;107:6982–7. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mao G, Lee S, Ortega J, Gu L, Li GM. Modulation of microRNA processing by mismatch repair protein MutLα. Cell Res. 2012;22:973–85. doi: 10.1038/cr.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 106.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA. 2008;105:14879–84. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jakymiw A, Patel RS, Deming N, Bhattacharyya I, Shah P, Lamont RJ, et al. Overexpression of dicer as a result of reduced let-7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer. 2010;49:549–59. doi: 10.1002/gcc.20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–7. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 109.Glorian V, Maillot G, Polès S, Iacovoni JS, Favre G, Vagner S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 2011;18:1692–701. doi: 10.1038/cdd.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–5. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Josson S, Sung SY, Lao K, Chung LW, Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68:1599–606. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Akao Y, Noguchi S, Iio A, Kojima K, Takagi T, Naoe T. Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 2011;300:197–204. doi: 10.1016/j.canlet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 113.Yu Y, Wang Y, Ren X, Tsuyada A, Li A, Liu LJ, et al. Context-dependent bidirectional regulation of the MutS homolog 2 by transforming growth factor β contributes to chemoresistance in breast cancer cells. Mol Cancer Res. 2010;8:1633–42. doi: 10.1158/1541-7786.MCR-10-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci USA. 2010;107:21098–103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, Chen WT. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2012 doi: 10.1038/onc.2012.28. In press. [DOI] [PubMed] [Google Scholar]
- 116.Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Guéranger Q, et al. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol. 2010;30:1217–30. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ando Y, Maida Y, Morinaga A, Burroughs AM, Kimura R, Chiba J, et al. Two-step cleavage of hairpin RNA with 5′ overhangs by human DICER. BMC Mol Biol. 2011;12:6. doi: 10.1186/1471-2199-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–50. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–4. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 120.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 121.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 123.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–7. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci USA. 2005;102:12177–82. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Melo SA, Kalluri R. Molecular pathways: microRNAs as cancer therapeutics. Clin Cancer Res. 2012;18:4234–9. doi: 10.1158/1078-0432.CCR-11-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18:1121–6. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7:389–93. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arima H, Yoshimatsu A, Ikeda H, Ohyama A, Motoyama K, Higashi T, et al. Folate-PEG-appended dendrimer conjugate with α-cyclodextrin as a novel cancer cell-selective siRNA delivery carrier. Mol Pharm. 2012;9:2591–604. doi: 10.1021/mp300188f. [DOI] [PubMed] [Google Scholar]
- 129.Pothof J, Verkaik NS, Hoeijmakers JH, van Gent DC. MicroRNA responses and stress granule formation modulate the DNA damage response. Cell Cycle. 2009;8:3462–8. doi: 10.4161/cc.8.21.9835. [DOI] [PubMed] [Google Scholar]
- 130.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–32. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 131.Cui W, Ma J, Wang Y, Biswal S. Plasma miRNA as biomarkers for assessment of total-body radiation exposure dosimetry. PLoS One. 2011;6:e22988. doi: 10.1371/journal.pone.0022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Templin T, Paul S, Amundson SA, Young EF, Barker CA, Wolden SL, et al. Radiation-induced micro-RNA expression changes in peripheral blood cells of radiotherapy patients. Int J Radiat Oncol Biol Phys. 2011;80:549–57. doi: 10.1016/j.ijrobp.2010.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Templin T, Amundson SA, Brenner DJ, Smilenov LB. Whole mouse blood microRNA as biomarkers for exposure to γ-rays and (56)Fe ion. Int J Radiat Biol. 2011;87:653–62. doi: 10.3109/09553002.2010.549537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–9. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang S, Chen H, Zhao X, Cao J, Tong J, Lu J, et al. REV3L 3′UTR 460 T>C polymorphism in microRNA target sites contributes to lung cancer susceptibility. Oncogene. 2012 doi: 10.1038/onc.2012.32. in press. [DOI] [PubMed] [Google Scholar]
- 136.Tsai YS, Lin CS, Chiang SL, Lee CH, Lee KW, Ko YC. Areca nut induces miR-23a and inhibits repair of DNA double-strand breaks by targeting FANCG. Toxicol Sci. 2011;123:480–90. doi: 10.1093/toxsci/kfr182. [DOI] [PubMed] [Google Scholar]
- 137.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–76. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS, et al. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell. 2010;38:689–99. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kumar M, Lu Z, Takwi AA, Chen W, Callander NS, Ramos KS, et al. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene. 2011;30:843–53. doi: 10.1038/onc.2010.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Butz H, Likó I, Czirják S, Igaz P, Khan MM, Zivkovic V, et al. Down-regulation of Wee1 kinase by a specific subset of microRNA in human sporadic pituitary adenomas. J Clin Endocrinol Metab. 2010;95:E181–91. doi: 10.1210/jc.2010-0581. [DOI] [PubMed] [Google Scholar]
- 141.He JF, Luo YM, Wan XH, Jiang D. Biogenesis of MiRNA-195 and its role in biogenesis, the cell cycle, and apoptosis. J Biochem Mol Toxicol. 2011;25:404–8. doi: 10.1002/jbt.20396. [DOI] [PubMed] [Google Scholar]
- 142.Sarkar S, Dey BK, Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell. 2010;21:2138–49. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang X, Feng M, Jiang X, Wu Z, Li Z, Aau M, et al. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23:2388–93. doi: 10.1101/gad.1819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–8. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106:3207–12. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wong P, Iwasaki M, Somervaille TC, Ficara F, Carico C, Arnold C, et al. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70:3833–42. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tian RQ, Wang XH, Hou LJ, Jia WH, Yang Q, Li YX, et al. MicroRNA-372 is down-regulated and targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human cervical cancer, which may contribute to tumorigenesis. J Biol Chem. 2011;286:25556–63. doi: 10.1074/jbc.M111.221564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Afanasyeva EA, Mestdagh P, Kumps C, Vandesompele J, Ehemann V, Theissen J, et al. MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ. 2011;18:974–84. doi: 10.1038/cdd.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lin SL, Chang DC, Ying SY, Leu D, Wu DT. MicroRNA miR-302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. Cancer Res. 2010;70:9473–82. doi: 10.1158/0008-5472.CAN-10-2746. [DOI] [PubMed] [Google Scholar]