Abstract
Stem cells are essential for development and tissue maintenance and display molecular markers and functions distinct from those of differentiated cell types in a given tissue. Malignant cells that exhibit stem cell-like activities have been detected in many types of cancers and have been implicated in cancer recurrence and drug resistance. Normal stem cells and cancer stem cells have striking commonalities, including shared cell surface markers and signal transduction pathways responsible for regulating quiescence vs. proliferation, self-renewal, pluripotency and differentiation. As the search continues for markers that distinguish between stem cells, progenitor cells and cancer stem cells, growing evidence suggests that a unique chromatin-associated protein called DEK may confer stem cell-like qualities. Here, we briefly describe current knowledge regarding stem and progenitor cells. We then focus on new findings that implicate DEK as a regulator of stem and progenitor cell qualities, potentially through its unusual functions in the regulation of local or global chromatin organization.
Keywords: chromatin organization, stem cells, cancer stem cells, DEK, heterochromatin
Introduction
Stem cells are responsible for creating and maintaining the various tissues and organs within the body. These include multipotent embryonic stem cells from the inner cell mass of a blastocyst, which are responsible for creating the new organism, and pluripotent adult stem cells, which maintain tissues after birth by replacing old or damaged cells. Through asymmetric division, stem cells can give rise to progenitor cells. Progenitor cells, or transit amplifying cells, differ in that they are partially committed in lineage and have limited or no self-renewal potential, but they often are more proliferative and migratory in response to growth factors and cytokines, resulting in significant contributions to the bulk of the tissue, particularly in response to injury.1,2 In 2006 and 2007, an additional type of stem cell was reported in murine and human cells, respectively: induced pluripotent stem cells (iPSCs). These artificially derived stem cells originated from adult somatic cells and were generated by introducing four key transcription factors, Oct4, Sox2, Klf4 and c-Myc (OSKM),3-5 or variations thereof. The common features of these stem cells are their (1) pluripotency, or the ability to generate many cell types in the mature organism, and (2) capacity for self-renewal and multi-lineage differentiation via symmetrical or asymmetrical cell division.
These properties of stem cells are essential for organismal survival; however, if these cells undergo genetic damage, or if mutated cells acquire the above stem cell-like qualities, then they can potentially lead to cancer. By analogy, “cancer stem cells” (CSCs) or “tumor-initiating cells” can create the diverse sets of non-tumorigenic cancer cells that form the bulk of a tumor. First described in acute myeloid leukemia and later in several (but not all) types of solid tumors, cancer stem cells can recreate a tumor from as few as 100 cells, whereas the remainder of the cancer cells from the same patient or animal cannot form tumors in xenograft studies.6,7 Although the origin of CSCs is under debate, it is clear that their existence holds significant biological and clinical importance.
Molecular drivers of stem and progenitor cell viability and pluripotency are poorly understood; therefore, it is increasingly important to identify factors that are essential for stem cell maintenance. Ideally, protein targets for new cancer therapies would be differentially expressed between normal stem and progenitor cells and cancer stem cells in order to control the drivers of tumorigenesis without permanently damaging the normal stem cell compartment required for tissue homeostasis. Additionally, a better general understanding of the molecular biology of both normal and cancer stem cells might open up new avenues for treatment strategies. Here, we briefly discuss the different types of stem and progenitor cells and some of the molecular pathways currently known to be required for “stem-ness.” For this review, we will briefly describe the Notch, NFκB and Wnt/β-catenin pathways and the p53 family in the context of stem cell biology; detailed reviews on the roles of each of these pathways for stem cell pluripotency and self-renewal already exist.8-11 Then, we focus on the chromatin-bound DEK protein, its function in chromatin organization and its ability to regulate the activity of several molecular pathways involved in stem and progenitor cell biology. We also summarize growing evidence implicating DEK activities in progenitor and cancer stem cell proliferation. Finally, DEK depletion has been suggested as a novel therapeutic method for selectively targeting cancer cells, and we will address the challenges of this approach with regards to maintaining a healthy, normal stem and progenitor cell compartment.
Normal and Cancer Stem Cells
Several types of normal multipotent cells have been reported, including embryonic stem cells (ESC) from the inner cell mass of a blastocyst, adult stem and progenitor cells in mature tissues and iPSCs derived from somatic cells. The existence of adult stem and progenitor cells have been demonstrated in several tissues, including the hematopoietic cells of the bone marrow and umbilical cord blood; epithelial tissues such as skin, intestine and the mammary gland; mesenchymal tissues, including adipose, lung and dental pulp; neuronal stem cells, particularly in the hippocampal region of the brain, and many more.12-19 Stem and progenitor cells reside in specialized, isolated compartments within the tissues called niches. The stem cell niche creates a microenvironment that supports stem cell self-renewal and inhibits differentiation through interactions with differentiated cells, extracellular matrix, blood vessels and controlled exposure to soluble factors.19-21 Embryonic stem cells and iPSCs are considered pluripotent, because they can differentiate into many different tissue types and have unlimited self-renewal ability. However, adult stem cells and progenitor cells are multipotent, limited to differentiate into only certain cell types based on their tissue of origin, with progenitor cells being more limited than the stem cells. The difference between adult stem cells and progenitor cells is still a topic of controversy, depending upon the tissue in question. However, it is commonly thought that adult stem cells have unlimited self-renewal capacity, whereas progenitor cells have limited self-renewal ability and are further along in the differentiation process.2 An additional complicating factor is that several tissues, including intestine and skin, have multiple distinct stem cell populations that proliferate at different rates and in response to different environmental cues.1,22,23
The cancer stem cell hypothesis postulates that tumor formation is initiated and advances due to aberrant stem-like cells, also referred to as tumor-initiating cells. The origin of cancer stem cells is still hotly debated, but there are two prevailing theories. The first line of thought is that cancer stem cells originate from normal, tissue-specific adult stem or progenitor cells or stem cell niches. This hypothesis is supported by recently published lineage tracing experiments that marked normal cells in the stem cell compartments of the colon, skin and brain and followed their progeny through the transformation process.24-26 However, this does not rule out a second line of thought that differentiated cells can aberrantly acquire stem cell-like qualities. A recent report has also implicated changes in the stem cell niche or microenvironment for initiating a “switch” from a normal stem cell to a cancer stem cell. Chen et al. discovered that growing GFP-labeled mouse iPS cells in conditioned media from one of four murine carcinoma cells, or co-cultured with murine mammary carcinoma cells, led to the acquisition of cancer stem cell phenotypes, including the ability to grow in sphere culture and form undifferentiated carcinomas in vivo.27 In any of the above scenarios, key signaling pathways used by stem cells to regulate self-renewal, proliferation and differentiation are interrupted or deregulated, and their activities culminate in the creation of a tumor initiating cell. The tumor initiating cell(s) can then generate additional tumor cells, which form the bulk of the tumor.28,29
The presence of cancer stem cells is of significant clinical concern. These cells have been implicated in tumor recurrence, due in part to their ability to acquire easily chemotherapeutic resistance, and therefore may be the main contributors to disease-specific mortality. Recurrence is further complicated by tumor heterogeneity and the presence of bulk tumor cells with different sensitivities to varieties of chemotherapies.11,30-32 It is also possible that cancer stem cells may be the source of metastases.29,30 Therefore, to improve cancer survival, the focus should be on identifying novel cancer therapies that target both cancer stem cells and bulk tumor cells in order to prevent metastasis and recurrence.
Stem cell markers
Molecular markers are used routinely to identify and prospectively isolate stem cells from a mixed population of cells in the tumor or tissue. Descriptions of these markers have been extensively reviewed in other manuscripts.11,29,33-35 However, these molecules do not uniformly identify stem cells in all tissues. Many adult stem cells express distinctive combinations of these markers that vary from tissue to tissue. Briefly, there are several categories of stem cell markers. The expression of unique transcription factors is common, including Oct4, Nanog, Sox2, p63, etc., which, in turn, regulate the expression of a myriad of genes involved in self-renewal and differentiation.3,5 Detoxifying proteins are also expressed in normal and cancer stem cells originating from many tissue types. These include the ABCG family of proteins, which can be assessed by identifying the “side population” – a population of cells that actively pumps out fluorescent dyes such as Hoechst 33342.36-39 A second detoxifying enzyme, aldehyde dehydrogenase 1 (ALDH1), demonstrates increased activity in some types of stem cells and can be measured using fluorescent substrates for this enzyme.40 One of the most commonly used classes of markers is cell surface antigens, which differ between hematological and solid tissues. Normal hematopoietic stem cells can by enriched by isolating the lin-/Sca1+/c-kit+ (LSK phenotype), with enhanced detection using the “signaling lymphocyte activation molecule” (SLAM) family of proteins, while lineage specific progenitor cells display other unique cell surface marker combinations.41 Additional cell surface glycoproteins CD34 and CD38 can be used to refine the LSK population into a smaller pool from which to identify hematopoietic stem cells, although the usage of these molecules differs between mice and humans.13 Non-hematologic stem cells can be identified by the expression of different combinations of cell surface antigens, depending on their tissue of origin, including CD133, CD44, epithelial specific antigen (ESA/Epcam) and the lack of other antigens, including CD24.6,11 These cell surface markers often change as stem cells give rise to progenitor cells, which eventually results in the generation of terminally differentiated cells. Stem cells also exhibit increased activation of signaling pathways that inhibit differentiation and regulate self-renewal, such as Wnt and Notch. DNA label retention is a classical tool to demonstrate asymmetric cell division, conservative DNA replication as opposed to semi-conservative replication, where the self-renewing stem cell retains the original DNA strands and passes on the newly synthesized DNA strand to progenitor daughter cells.42,43 Another marker associated with self-renewing stem cells that is related to DNA maintenance is the expression of telomerase, which prevents telomere shortening during successive rounds of cell division, allowing the cell to delay or escape the Hayflick limit and replicate indefinitely.44,45
Finally, functional studies are ultimately used to identify the stem cell population. For example, non-adherent conditions ex vivo are used to enrich for and study stem cell populations from several tissues, such as sphere-forming assays for neural and mammary epithelial stem cells and embryoid body formation for human ESCs.12,46,47 Other functional assays, including tissue reconstitution, teratoma formation and serial transplantation, are used in animal models. However, in vitro techniques such as long-term culture-initiating cell (LTC-IC) and colony-forming cell assays also are frequently employed in the study of hematopoietic stem and progenitor cells and ESCs.47,48
A complicating problem is that many of these markers are shared between normal and cancer stem cells, thus it is difficult to study both the origin of cancer stem cells and to develop targeted therapies to cancer stem cells that preserve normal stem cells. One unique property of cancer stem cells is enhanced tumorigenicity using orthotopic xenografts in non-obese diabetic/severe combined immune deficient (NOD/SCID) mice in limiting dilutions, accompanied by tumor formation following serial transplantation, compared with other bulk tumor cells.49 However, there are concerns regarding this method including the physiological relevance of xenotransplantation.33 Likewise, the role of the immune system is lost in this model; this methodology may thus either underestimate or overestimate tumor-initiating cell frequency, as the immune system can clearly regulate cancer stem cell fitness.33,50 Leukemic stem cells also have a unique CD34+/CD38- cell surface antigen signature; however, a reliable signature distinguishing normal stem cells from cancer stem cells of solid tumor malignancies has yet to be discovered.7
Molecular pathways regulating stem cells
The microenvironment of the stem cell niche limits differentiation by regulating the activity of several signaling pathways. These pathways have been implicated in controlling diverse stem cell relevant processes, such as quiescience vs. proliferation, pluripotency vs. differentiation and self-renewal vs. exhaustion. Among the most important pathways for stem cell maintenance are Notch, NFκB, Wnt/β-catenin and TGFβ/SMAD signaling pathways, along with growth factor-mediated stimulation of both the MAPK/ERK and PI3K/AKT pathways and the activity of p53 family members to regulate proliferation and genome maintenance.8-10,51 The activities of these pathways are modulated throughout the progression from stem to progenitor to terminally differentiated cells in order to regulate gene expression and function of the cells within the tissue.
The Notch signaling pathway communicates information between adjacent cells in a given tissue. This pathway is activated by the interaction between one of four Notch receptors and one of their membrane-tethered ligands, Delta, Serrata or Lag2. This results in the proteolytic cleavage of Notch by γ-secretases and metalloproteinases. The cleaved fragment, Notch intracellular domain (NCID), translocates to the nucleus, where it functions as a transcriptional co-activator for proteins such as suppressor of hairless (SuH). Notch target genes, which include the hairy/enhancer of split (HES) family members such as Hes(1–7) and Hey(1–3), CCND1 (cyclin D1), c-myc and others, promote stem cell proliferation and direct the timely regulation of differentiation and cell fate decisions throughout the course of development in a cell type-specific manner.11,52,53 New evidence, such as that recently published in pancreatic precursor cells in zebrafish, also indicates that Notch function may be dose-dependent in order to regulate proliferation rates and differentiation.54 Likewise, due to its varied roles in proliferation and differentiation, this molecule has been described as both a tumor suppressor and an oncogene, depending on the tissue of interest.53 Recent work has focused on modulating Notch pathway activity to target bulk tumor and cancer stem cells while maintaining the health of the normal adult stem cell population. For example, Ninov et al. recently showed an upregulation in Notch signaling molecules in sphere cultures of tumor cells compared with normal murine mammary stem cells. Furthermore, treatment with the γ-secretase inhibitor MRK-003 could irreversibly inhibit tumor initiating cell proliferation and survival but had reversible effects on normal mammospheres, thus permitting normal stem cell survival.55 Additional studies have also examined the importance of Notch signaling in the cancer stem cell population. Notch inhibition was also prominent in glioblastoma neurospheres, whose growth was attenuated upon treatment with all-trans retinoic acid, an agent typically used to induce differentiation.56 Finally, Notch inhibition with γ-secretase can obstruct, and possibly eliminate, the leukemia-initiating cells in a mouse model of T-cell acute lymphoblastic leukemia (T-ALL).57
A second prominent stem cell-associated signaling mechanism is the NFκB pathway, which regulates the expression of genes involved in proliferation, differentiation, inflammation and immune responses. Five NFκB transcription factor family members—cRel, RelA/p65, RelB, p52, and p50—can homo- and heterodimerize to mediate changes in gene transcription. Typically, these proteins are bound to a member of the IκB inhibitory molecule family and inactivated until a stimulatory signal (such as infections, oxidative stress, or TNFα) is received by the cell. In canonical signaling, the IκB molecule is phosphorylated and degraded in response to stimuli, and p65/RelA is phosphorylated by a number of different mechanisms, most notably AKT, p38, protein kinase A (PKA) and protein kinase C (PKC), allowing it to translocate to the nucleus. Non-canonical NFκB signaling results in the formation of the active RelB:p52 dimer.8,58 The role(s) of the NFκB pathway in stem cell biology is just now being elucidated. Currently, there is conflicting evidence as to the role of NFκB in human embryonic stem cells and adult cells. Using p65 inhibitors, Armstrong et al. showed that the inhibition of NFκB signaling promotes differentiation of hESCs.59 However, Yang et al. have shown that the opposite may be true. Chemical or RNAi-mediated inhibition of canonical NFκB signaling actually promoted a transcriptional profile reminiscent of pluripotency and upregulated the expression of canonical NFκB pathway members during differentiation. However, it was the inhibition of non-canonical NFκB signaling that promoted the expression of genes associated with differentiation.58 In support of this, Zhang et al. reported that canonical NFκB signaling was associated with neural differentiation and asymmetrical division in neuronal stem cells.60
In comparison, the Wnt/β-catenin pathway has been extensively studied in the context of hESC, adult stem cells and iPS cells. The canonical Wnt/β-catenin pathway is activated when Wnt ligands bind to the Frizzled and LRP5/6 receptors, resulting in the activation of Dishevelled. Dishevelled then inhibits the APC/Axin/GSK3β complex, allowing the stabilization, accumulation and nuclear translocation of β-catenin. In the nucleus, β-catenin binds to TCF/LEF transcription factors to regulate the expression of numerous target genes involved in proliferation, self-renewal, pluripotency/differentiation and migration. Wnt signaling is critical for maintaining homeostasis in epithelial tissues, wherein cells are constantly lost and replaced. In intestinal epithelia, Wnt activity stimulates the proliferation of stem cells in the intestinal crypt. Wnt/β-catenin signaling is also crucial for the development and proper function of other epithelial tissues that contain stem cells, such as the mammary gland.61,62 In human and mouse ESCs, active Wnt/β-catenin signaling can maintain the cells in an undifferentiated state via the expression of Rex1, Nanog and Oct4.63 Furthermore, pathway activation dramatically enhances the efficiency of cell reprogramming in cell fusion experiments and c-myc, a gene upregulated by β-catenin and other signaling pathways, is one of the four classical transcription factors sufficient to induce reprogramming in iPSCs.3,64
The roles of p53 family members
The p53 family consists of three related proteins—p53, p63 and p73—that share many functions including transactivation of overlapping gene targets and the ability to induce apoptosis. All three family members have been implicated in stem cell biology, although p63 and p73 are more tissue-specific with regards to their functions, as determined by studies using knockout mice. p63 and p73 each have two independent promoters and undergo extensive alternative splicing in order to create approximately six different transcripts each, with three variants (α, β, and γ) of a full-length TA (transactivation domain) isoform and three variants of a ΔN isoform that lacks the N-terminal transactivation domain.65 The TA and ΔN isoforms tend to have opposing functions, with the TA isoforms being most reminiscent of p53 and exhibiting tumor suppressive activity, whereas the ΔN isoforms promote proliferation and are often characterized as oncogenic.66
Specifically, p73 was required for proper central nervous system development. Knockout mice exhibited cortical loss accompanied by hypoplasia of the hippocampus and caudal cortex along with hydrocephalus.67 The p73 gene locus also was required for the maintenance of the neuronal stem cell population and for self-renewal of neural stem cells in neurosphere cultures from whole brain tissue. However, the mechanism that results in this hypoplasia is not well defined. Gonazalez-Cano et al. showed that loss of the p73 locus resulted in increased apoptosis of neuronal stem cells and premature differentiation, whereas Talos et al. described a decrease in proliferation and increase in senescence, via deregulation of the Sox and Notch pathways, along with impaired differentiation.67,68 The differences may be due to the genetic background of the mice and/or that Gonzalez-Cano studied neuronal stem cells from the olfactory bulb while Talos cultured whole brain tissue. Results from Talos et al. also were further supported in another report by Agostini et al.69
Compared with p73, the roles of p63 and p53 in stem cells are better understood. In brief, the generation of two versions of p63 locus knockout mice resulted in premature death due to a dramatic loss or underdevelopment of stratified epithelial tissues, including skin, breast, prostate and urothelia.70,71 This observation helped shape our view of p63 as one of the first proteins associated with stem cell maintenance.71 The ΔNp63α isoform is the most commonly expressed isoform in the basal layer of stratified epithelia and is expressed in cells destined to differentiate into other cell types of the epithelium, including skin, breast, cervix, urogenital tract, prostate, thymus and olfactory neuroepithelium. It is required for cell proliferation and for maintaining the undifferentiated state of basal epithelial stem cells by suppressing Notch signaling and promoting the expression of Wnt-responsive genes.72 However, the relationship between p63 and Notch signaling is complex, as each protein can regulate the other and p63 can be both an agonist and an antagonist for Notch signaling, depending on the cellular context.73 The ΔNp63α isoform also has been implicated in cancer stem cell maintenance in tumors of epithelial origin. In both MCF7 breast cancer cells and head and neck squamous cell carcinomas (HNSCCs), ΔNp63α overexpression caused increased expression of the cancer stem cell marker CD44.74,75 Additionally, Du et al. demonstrated that ΔNp63α overexpression in MCF7 cells increased the size of the CD44+CD24- cancer stem cell population with a corresponding increase in mammosphere growth, enhanced tumor formation efficiency in the mammary fat pad and drug resistance to doxorubicin.75
Substantially more work has analyzed the role of p53 and its ability to monitor genome integrity in different stem cell populations. It is critically important that stem cells protect genome integrity and minimize the accumulation of mutations so that their differentiating daughter cells can perpetuate cellular identity. Several reports revealed that both murine and human ESCs are hypersensitive to DNA damage caused by irradiation and genotoxic chemicals, undergoing substantial apoptosis as a way to eliminate potentially mutant ESCs and to protect the integrity of the stem cell population.76 This is in direct opposition to cancer stem cells which are typically resistant to these insults. Furthermore, surviving stem cells must repair DNA damage efficiently and properly. Mouse embryonic stem cells prefer the “error-free” homologous recombination pathway to repair DNA damage, whereas somatic cells predominantly use the “error-prone” non-homologous end-joining pathway.77 Independent of the DNA repair pathway in use, p53 ultimately directs cell survival following DNA damage. In agreement with these findings, p53 was found to limit iPS cell formation and protect genome integrity in these stem cells. Marión et al. reported that p53-null mouse embryonic fibroblasts (MEFs) have increased reprogramming efficiency and significantly decreased apoptosis during reprogramming, despite the presence of extensive DNA damage, persistent activation of DNA damage response and chromosomal abnormalities.78 p53 also limits the reprogramming of human embryonic fibroblasts and primary keratinocytes.79 An additional role for p53 in stem cells is regulating differentiation. There is evidence that p53 both promotes and inhibits the differentiation of stem and progenitor cells, and this may be either tissue-specific or dependent upon the presence of DNA damaging agents in the environment.76,80 In human ESCs stimulated to differentiate with the addition of retinoic acid, p53 can promote differentiation by activating the expression of miR-34a and miR-145, which then repress the expression of stem cell factors OCT4, KLF4, SOX2 and LIN28A.81 In contrast, p53 can inhibit differentiation in mouse ESCs, but not in neural progenitors or MEFs, in response to genotoxic stress by activating the expression of multiple Wnt ligands. It is hypothesized that this Wnt-mediated anti-differentiation program may be used to stimulate the proliferation of undamaged or minimally damaged cells, while severely damaged cells undergo apoptosis to maintain a healthy stem cell population.82 The requirement of p53 for tumor suppression and genome stability is well-documented, so it is probable that it is required to prevent transformation of stem cells into cancer stem cells through similar mechanisms. An additional role for p53 in regulating self-renewal and cell division polarity of normal and cancer stem cells also has been characterized. For example, stem cells from pre-malignant mouse mammary glands of p53-null mice exhibited elevated rates of symmetric divisions, which would be expected to increase the number of cells in the (cancer) stem cell population.83 As future work will define the relationship between normal and cancer stem cells, and the possibility that mutations or stem cell niche problems cause the transformation of normal stem cells into cancer stem cells, relevant activities of the “master regulator” p53 are bound to emerge.
Chromatin organization and epigenetic properties of stem cells
Besides the important stem cell signaling pathways outlined above, stem cells exhibit distinct epigenetic features and present with a unique global chromatin organization. Chromatin, the highly dynamic packaging polymer of DNA in every eukaryotic nucleus, represents the context for the regulation of gene expression and has vital roles in cell fate determination. Chromatin is composed of arrayed repeats of the basic building block, the nucleosome, which contains two copies of each of the core histones (H2A, H2B, H3 and H4, or function-specific variants thereof) and 146 bp of DNA. Chromatin is a major determinant in regulating virtually all DNA-dependent processes and represents a major target for response to environmental change. Global and local cell-specific higher-order chromatin structure is tightly regulated in stem and differentiated cells. Major mechanisms of epigenetic regulation are represented by posttranslational modifications (PTM) of histones, DNA methylation, chromatin remodeling, exchange of histone variants, non-coding RNAs directing heterochromatin formation and selective binding of non-histone proteins, all of which are subject to intimate and highly organized crosstalk.84-86 Two major structural variants of chromatin are represented by the “open” (transcriptionally active) euchromatin and the “closed” (transcriptionally inactive) heterochromatin, with multiple intermediate forms recently reported.87 Deregulation of the establishment, maintenance or coordinated transition between these chromatin states can affect the balance between hetero and euchromatin and can thus trigger the pathogenesis of a variety of human diseases, including carcinogenesis.
One major form of epigenetic regulation is through the control of chromatin conformation as determined by post-translational modifications of histone variants. Compared with differentiated cells, stem cells exhibit a remarkably “open” chromatin conformation, as exemplified by a higher ratio of euchromatin to heterochromatin.88 In line with this, increased abundance of active epigenetic marks (e.g., histone H3 trimethylated on lysine 4 (H3K4me3), hyperacetylation of histones H3 and H4) is observed, a finding which at least in part explains the hypertranscriptional activity in stem cells. Furthermore, many so-called “poised genes” maintained in a bivalent conformation carry simultaneously repressive (histone H3 trimethylated on lysine 27, H3K27Me3) and active marks (H3K4me3). Depending upon the differentiation stimulus, either one of these marks can be erased, resulting in transcriptional activation or repression, respectively. On the other hand, heterochromatin, as exemplified by the repressive marks H3K9me3 and H3K27me3, is under-represented in stem cells, yet increases upon cellular differentiation.
A second type of epigenetic regulation occurs through differential cytosine methylation, typically at CpG islands, in promoters to repress gene expression. In H1 human embryonic stem cells, nearly 25% of cytosine methylation in the DNA occurred outside of the CpG context; this non-CpG methylation disappeared during induced differentiation and also was absent in IMR-90 fetal fibroblasts.89 Furthermore, these differentially methylated regions between stem cells and differentiated cells were often proximal to genes associated with pluripotency and differentiation.
Outside of post-translational modifications of histones and DNA methylation, chromatin or nucleosomal structure itself, the interaction and spacing of histones and DNA, can be remodeled or organized in order to direct proper gene expression and facilitate the maintenance of the open euchromatin state. On one hand, there are four families of chromatin remodeling proteins, CHD (chromodomain helicase DNA-binding), ISWI (imitation switch), INO80 (inositol-requiring 80) and SWI/SNF (switch/sucrose nonfermentable), and members of each of these families are essential for the timely orchestration of differentiation by regulating euchromatin abundance and gene expression.88 On the other hand, the network of so-called linker proteins has been further proposed to regulate accessibility of nucleosomal DNA.90 These factors compete for binding to the entry and exit site of nucleosomal DNA and can keep this “gate” either locked, ajar or allow for “breathing.” Whereas factors like HMG1/2 and PARP-1 positively influence transcription by keeping nucleosomal DNA ajar, MeCP2 or HP1 lock the nucleosomal gate, thereby negatively influencing transcription.90 Due to direct competition for the same binding site in the nucleosome and depending on the environmental stimulus, these factors can be exchanged by distinct mechanisms, which further enable the fine-tuning of local and global chromatin structure.
Although euchromatin is predominant in stem cells, the architectural factors involved in heterochromatin formation (e.g., heterochromatin protein 1, HP1), are still expressed albeit undergo substantial relocalization during terminal differentiation. In particular, variant HP1α diffusely stains the nuclei in hES cells but re-localizes to large foci during all-trans retinoic acid-induced differentiation and in neighboring mouse embryonic fibroblast feeder cells.91 Collectively, DNA and protein modifications are tightly controlled to regulate chromatin topology in stem cells and these factors are required to reverse the presence of heterochromatin during reprogramming to form iPSCs. However, the molecular mechanisms that trigger these chromatin modifications, and the identity of the proteins involved, are poorly understood.
DEK Regulates Chromatin Structure and Function in Normal and Transformed Cells
Cellular and molecular functions of the DEK oncogene
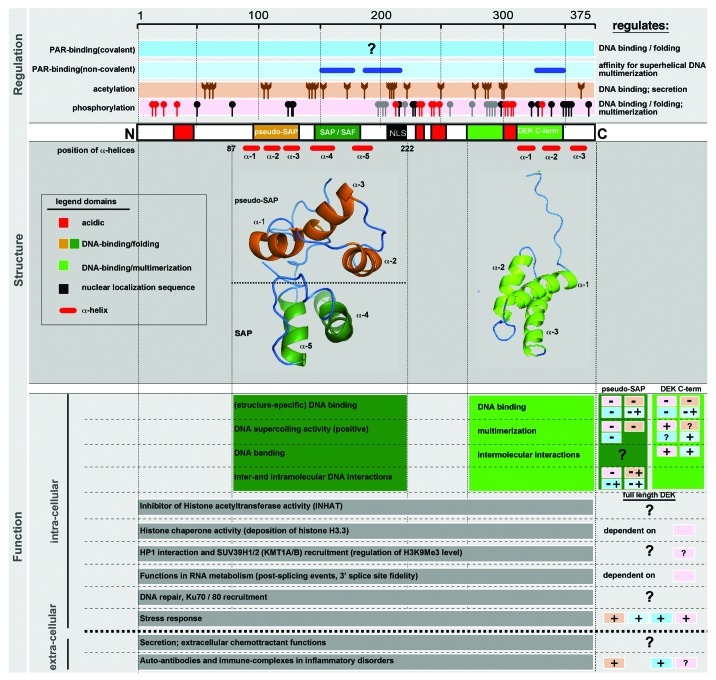
DEK is a unique non-histone chromosomal factor of 43 kDa (375 amino acids) with no known enzymatic activity (Fig. 1).92 This molecule is the only representative of its own protein class, as no paralogs have been identified thus far. However, DEK is highly conserved through evolution, present in almost all higher eukaryotes, but is not found in lower eukaryotes such as yeast or C. elegans. This suggests that DEK functions represent an evolutionary requirement as genome organization and regulation became more complex throughout evolution. DEK is a basic protein (pI = 8.69), yet harbors four discrete long patches of aspartic/glutamic acid residues that span its sequence. Of note, besides the domain between residues 149 to 183, which shows homology to the putative SAF/SAP DNA binding motif, no further homologies to other proteins can be identified.93,94 The SAF/SAP motif is a highly conserved 35-residue DNA-binding motif that is the founding feature of a diverse group of chromatin-associated proteins involved in transcription, DNA repair, RNA processing, chromosomal organization and apoptotic chromatin degradation.93 Whereas in most SAF/SAP box containing proteins, this motif is located either in the far N- or C terminus, the SAF/SAP box of DEK is located centrally within the molecule. Another unusual feature of the SAF/SAP box motif in DEK is that it is accompanied by a pseudo-SAP box, which is located N-terminally of the SAF/SAP box. This pseudo-SAP box exhibits a strikingly similar structure to the SAF/SAP box, yet shows no sequence homology, thus making this combination unique among SAF/SAP box containing proteins (discussed below). A series of mutational and structural in vitro studies highlighted the pseudo-SAP-SAP/SAF box region of DEK as the central and dominant DNA-binding hub, which is responsible for the signature DNA binding and folding features of DEK. These studies showed that the pseudo-SAP-SAF/SAP box, fragment 87–187, conveys preferential binding of DEK to unusual DNA structures, including supercoiled, cruciform/four-way junction or distorted DNA.95,96 They also described its DNA-bending activity, its ability to form large nucleo-protein complexes through inter- and intramolecular interactions and, finally, its ability to introduce constrained positive supercoils into closed circular DNA, later referred to as DNA folding activity.97 Intriguingly, this DNA folding activity of DEK appears to be independent of energy consumption, thus might be transmitted through structural changes to DNA upon DEK binding and multimerization and could further impact on overall chromatin structure.
Figure 1. Structure, function, and regulation of the DEK oncogene. Shown is the linear sequence of DEK with DNA binding domains (yellow, pseudo-SAP-box; dark green, SAP-box; light green, C-terminal DNA binding domain; red lines, position of α-helices, as revealed by NMR) and other functional features [red, acidic regions; black, putative nuclear localization signal (NLS, not yet confirmed experimentally)] indicated. Upper panel (Regulation): Depicted are known post-translational modifications of DEK. Dark blue box, covalent poly(ADP-ribosyl)ation, where the precise attachment sites are not known; light blue box, non-covalent attachment of poly(ADP-ribose), with the responsible regions indicated by blue lines; brown box, the position of mapped acetylation sites are indicated; red box, the position of mapped phosphorylation sites are indicated. Red sites were mapped both in vivo and in vitro; black sites were mapped only in vivo and gray sites mapped only in vitro; implications for regulation are indicated on the right side.106,160 Middle panel (Structure): The NMR structures of DEK 78–222 (pseudo-SAP/ SAP box) and the C-terminal DNA binding domain (DEK 310–375) are pictured as ribbon structures. Positions of α-helices are indicated in relation to the primary structure. Lower panel (Function): Intra- and extracellular functions of DEK are described, and their respective regulation by post-translational modifications, if known, is specified on the right side. The dark (pseudo-SAP-SAF/SAP box) and light green (DEK C-term) boxes indicate the specific functions of the individual DNA binding regions of DEK, as revealed mostly by in vitro studies. For all other functions the responsible region(s) within DEK are not known. The boxes on the right side indicate the regulation by post-translational modifications of functions mediated by either the pseudo-SAP-SAF/SAP box (dark green) or the C-terminal DNA binding domains (light green), using the identical color coding scheme from the top panel [dark blue, covalent poly(ADP-ribosyl)ation; light blue, non-covalent attachment of poly(ADP-ribose); brown, acetylation; red, phosphorylation]. For the regulation of all other indicated DEK functions, where no specific region within DEK can be assigned, the color-coding as above is used. (+) indicates a positive effect, (-) indicates a negative effect, and (?) indicates that regulation and effect are unknown.
Available structural information stems from the NMR solution structure of two DEK fragments 78–208 (pseudo-SAP and SAF/SAP box) and 309–375.98-100 DEK78–208 adopts a distinct globular five-helix bundle structure99 that has not been reported for other proteins.101 This helical bundle consists of the pseudo-SAP (α−helices 1–3) and SAF/SAP folds (α−helices 4 and 5) related by a plane of symmetry that efficiently binds DNA (Fig. 1).96 The SAF/SAP helix-extended loop-helix homology motif spans helices α4 and α5 of the C-terminal half. The N-terminal half, spanning (α−helices 1–3), superimposes well with the SAF/SAP motif of PIAS1, which binds double-stranded DNA (dsDNA) and might be a binding site for p53.102 Chemical shift mapping experiments have confirmed the binding of DEK78–208 to dsDNA and showed that this binding occurs on the first three-helices of the pseudo-SAP box and on the second helix (α5) of the SAF/SAP motif, which also appears to be involved in protein-protein interactions.99 Taken together, these data suggest that DEK78–208 has two DNA-binding domains, which might also interact with other proteins. The DEK309–375 region folds into a three-helix bundle with structural homology to the winged-helix motif,103 although the mechanism through which this region of DEK binds dsDNA is different compared with the winged-helix motif.104 Interestingly, this C-terminal region of DEK has been shown to partially rescue the DNA damage sensitivity phenotype of fibroblasts isolated from patients with ataxia telangiectasia.105
Together, the data derived from structural and in vitro studies suggest that the DEK protein has three DNA-binding sites but their biological significance and the relationship between DEK structure and function is still poorly understood. There is quite some knowledge regarding the regulation of DEK DNA binding activity in vitro and in the cellular environment. DEK is heavily phosphorylated by casein kinase 2 (CK2) and potentially other kinases, which reduces its overall binding to DNA and enhances its multimerization.106,107 To date, a striking number of 42 phosphorylation sites have been mapped on DEK by multiple approaches, suggesting phosphorylation as a major mechanism in regulating DEK functions (Fig. 1). Additionally, functions for acetylation and non-covalent as well as covalent attachment of poly(ADP-ribose) moieties to DEK, mostly under stress conditions, have been identified as critical regulators of its DNA/chromatin binding, localization and biological activity.108-110 Even though formally only shown for a few scenarios, these modifications are believed to be pivotal in integrating and regulating its various functions, controlling its subcellular localization and directing its interaction with other partners or structures. They may additionally play a central part in human disease phenotypes. In support of this notion, acetylation of DEK was associated with the formation of autoantibodies in juvenile idiopathic arthritis, and phosphorylation and poly(ADP-ribosyl)ation have been linked to increased programmed cell death.108,111,112
However, DEK is highly phosphorylated in normally cycling cells and still remains tightly bound to chromatin; only in vitro did phosphorylation decrease DNA affinity.106 Perhaps phosphorylation weakens the affinity of individual DEK molecules to DNA, but its subsequent oligomerization increases the collective affinity. Since many environmental stimuli utilize differential phosphorylation to relay signals within the cell, it is possible that differential phosphorylation of DEK, and the resulting changes in oligomerization and DNA binding, result in subtle but functionally significant changes in chromatin architecture and DNA accessibility in response to damage, stress or mitogenic signals. Additionally, DNA binding of DEK can also be influenced by acetylation, as Cleary et al. have shown that DEK, acetylated in the N-terminus by P/CAF, is driven into interchromatin granule clusters (IGCs), thereby exhibiting reduced DNA binding abilities.113 However, there are also many regions of the DEK protein with unknown function, especially the highly acidic regions. Interestingly, a recent study has shown that the acidic DEK fragment 78–222 can be taken up by neighboring cells in vitro and in vivo.114 This has many biological implications, as reports have shown DEK is not solely nuclear but also can be secreted into the extracellular space, where it functions as a chemotactic factor and autoantigen in the pathogenesis of juvenile arthritis.108,115 Perhaps in addition to being a chromatin-binding protein, DEK is also an intercellular signaling molecule, secreted by a certain population of cells and subsequently taken up by neighboring cells. It is becoming increasingly clear that there are many varied DEK activities, and continued structure-function studies are needed to explore these in different cell types and disease models.
In general, DEK expression is high in proliferating cells, in part because it is an E2F target gene that is upregulated following Rb inhibition.116,117 Yet expression levels vary in different tissues and are substantially decreased or fully absent in differentiated cells. Whether or not the two known alternative splice variants of human DEK (isoform 1 and 2) are subject to altered expression during development or differentiation also is not clear at the current time. Isoform 2 lacks amino acids 49–82, a region shown to be necessary for the positive DNA supercoiling activity of DEK. This would suggest negative DNA folding activities for isoform 2, and even though purely speculative, could have severe effects on overall chromatin organization. Furthermore, no loss-of-function SNPs are known to date.
In the cellular environment, the majority of DEK localizes to the nucleus, where a subfraction of approximately 10% is found associated with RNA of unknown nature, in agreement with functions of DEK in RNA metabolism.118,119 However, the vast majority of DEK associates with chromatin.119 By using biochemical chromatin fractionation protocols and other techniques, DEK was shown to reside in hetero- as well as in euchromatin, which is in agreement with functional roles for DEK in transcriptional activation and repression.92,119-122 Furthermore, extensive biochemical studies identified specific DNA/chromatin/histone binding features and DNA-folding activities for DEK that generally result in the compaction of chromatin in vitro. These distinct biochemical DNA binding features (e.g., preferential binding to unusual DNA structures) classified DEK as a chromatin architectural protein, similar to HMGB1, and therefore to the class of linker-proteins.92,95-97,99,106,107,109,111,112,115,119,123 How these DNA/chromatin binding features of DEK impact upon its biological function in the cellular context is a topic of ongoing studies.
Despite significant knowledge derived from its biochemistry and its association with multiple cellular functions outlined above, a principle biological function of DEK remains elusive. Cytoplasmic activities for DEK are currently uncharacterized, but secreted forms of DEK that have been found in synovial fluid were implicated in triggering autoimmune events associated with juvenile idiopathic arthritis and found in the urine of over 80% of bladder cancer patients.108,115,124 The nuclear fraction of DEK carries out numerous activities associated with DNA binding and chromatin organization. However, recent reports might have uncovered principal biological modes of action for DEK that could account for its diverse, cellular functions. One report demonstrated that DEK is a Su(var), a positive regulator of heterochromatin, acting through heterochromatin protein 1α (HP1α) that is required for heterochromatin integrity.125 Another report identified histone chaperone activities for DEK that strictly depended on its interaction with and phosphorylation by CK2, which resulted in gene activation.126 As previously mentioned, in cell-free assays DEK could introduce constrained positive supercoils into DNA and facilitate the ligation of linear DNA molecules in vitro.95,97 Most studies indicate that DEK binding to nucleic acids is not sequence-specific but rather has a preference for super-coiled and cruciform structures.95 However, DEK also has been shown to exhibit DNA sequence-specificity in certain scenarios, such as binding to the peri-ets (pets) site in the HIV-2 promoter.127 These roles in chromatin topology and nucleic acid binding have led to functional associations of DEK with nuclear processes, including mRNA splicing, transcriptional regulation, DNA replication and DNA damage repair.118,128-133 With regard to the latter, DEK was necessary for timely Ku70/80 heterodimer formation following DNA damage and for the subsequent activation of DNA-PK for proper non-homologous end joining (NHEJ)-mediated DNA damage repair.130 This role in DNA damage repair supports findings that DEK promotes resistance to genotoxic drugs such as doxorubicin and cisplatin, many of which are used in the clinic to treat cancer patients.111,134 Collectively, current biochemical and functional data on DEK lead us to speculate that its primary function is to bind and change the topology of nucleic acids. This would result in the compaction and/or coiling of nucleic acids, altering the accessibility of these molecules and their orientation in three-dimensional space and possibly encouraging the formation of unique tertiary structures that can subsequently result in altered nucleosomal structure, promote DNA replication and repair and RNA splicing. Furthermore, the binding of DEK to nucleic acids may result in the recruitment of proteins required for essential processes such as chromatin organization and DNA repair (e.g., HP1α and Ku70/80, respectively). Through cis- and trans-acting mechanisms, perturbed DEK expression levels could thus dramatically alter nucleic acid structures and globally impact processes such as transcription, mRNA splicing, DNA replication and DNA repair, which could, in turn, activate or repress multiple signaling pathways.
With a plethora of activities in maintaining genome integrity and regulating gene expression, it is not surprising that de-regulated DEK expression can be detrimental to cell survival. Numerous studies using DEK specific RNAi have shown a significant increase in cellular apoptosis or senescence through p53-dependent and -independent mechanisms. Relevant mechanisms include p53 stabilization and the transcriptional repression of myeloid cell leukemia 1 (MCL-1) protein in several types of cancer.134-137 These molecular mechanisms of DEK-mediated apoptosis and senescence inhibition are tissue specific; for instance, in melanoma cells, DEK inhibition did not induce p53 dependent apoptosis, but rather caused downregulation of MCL-1.134 Furthermore, the cytotoxic response to DEK loss was significantly more pronounced in rapidly proliferating cancer cells compared with normal, differentiated cells, suggesting that dividing cancer cells are dependent upon high levels of DEK expression.137,138 The reason for the biological requirement of DEK in proliferating compared with non-proliferating cells is not understood. In addition, DEK suppressed quiescence and senescence in muscle cells, basal keratinocytes and cancer cells; furthermore, non-proliferating differentiated cells demonstrate severely diminished or absent DEK expression.117,139,140 Interestingly, studies in transgenic Drosophila indicate that aberrantly high levels of DEK, at least in normal differentiated cells which typically do not express DEK, may also be detrimental. Overexpression of human DEK in the fly eye caused a rough-eye phenotype due to caspase-9- and -3-mediated apoptosis and was accompanied by histone H3 and H4 hypoacetylation.141 It is clear that DEK overexpression in normal epithelia promotes transformation, but overexpression above physiological levels, especially in cancer cells, also is difficult to accomplish (data not shown).136,138,142 It remains unknown whether this is due to the degradation of excessive DEK mRNA or protein, or apoptosis of the overexpressing cell population. Together, the data indicate that DEK expression levels must be tightly controlled in a limited population of cells in order to balance proliferation and tissue integrity, likely due to its role(s) in regulating chromatin organization and (nucleosomal) DNA accessibility. Since DEK expression seems limited to proliferating cells and is absent, or very low, in differentiated cells, it is possible that DEK is expressed in stem or progenitor cells within tissues. Therefore, any potential therapeutic targeting of DEK expression, either for cancer or juvenile arthritis, must proceed with caution, so that the relevant cell populations are treated while normal tissue integrity is preserved.
The molecular connection between DEK and stem cells
The DEK oncogene regulates the activity of numerous molecules and signal transduction pathways, many of which are implicated in tumorigenesis and in stem/progenitor cell maintenance and survival. First, DEK was found to interact with RelA/p65 and repress its transcriptional activity in the NFκB pathway. Specifically, DEK expression blocked transcription of RelA/p65 reporter genes. Furthermore, chromatin immunoprecipitation and quantitative RT-PCR experiments revealed that TNFα treatment resulted in the removal of inhibitory DEK from NFκB regulated promoters, specifically from the cIAP2 and IL-8 genes, and subsequent transcriptional upregulation of these target genes in HeLa cells. In addition, NFκB reporter and target genes were significantly upregulated in mouse embryonic fibroblasts (MEFs) isolated from Dek-knockout mice compared with wild type mice.133 A later report confirmed the ability of DEK to regulate RelA/p65 activity by showing that DEK knockdown could relieve the transcriptional repression of the p65-regulated gene 1-cys Prx (1-cys peroxiredoxin).143 A recent report by Liu et al. also demonstrated that DEK depletion in Caski cervical cancer cells resulted in phosphorylation of ΙκBα and the nuclear translocation of p65 with increased DNA target sequence binding.140 In summary, the evidence indicates that DEK can repress NFκB signaling in numerous cell types, which may, in turn, suppress differentiation.
Recently, we reported that the loss of DEK in both human breast cancer cells and MEFs decreased the levels of activated β-catenin in the nucleus and significantly reduced β-catenin-driven transcription using a TCF/LEF responsive luciferase reporter assay.136 The addition of a constitutively active S37A β-catenin expression construct could rescue, at least partially, the associated defects in β-catenin activity and associated cellular invasion. We postulated that the DEK oncogene promotes β-catenin activation and nuclear translocation in multiple species, and in both cancer and normal cells.136 Further support for DEK as a regulator of the Wnt/β-catenin signaling was published by Shibata et al., where DEK was shown to upregulate the expression of Wnt10b, a canonical Wnt ligand which can activate the Wnt/β-catenin pathway, in neuroendrocrine carcinomas of the lung.142 Similar upregulation of Wnt10b was observed in human breast cancer cells using quantitative RT-PCR (our unpublished data). A feedback loop may exist as DEK has several GSK3β consensus sites, which, when phosphorylated, drive DEK ubiquitination and targeting for degradation by the Fbxw7 subunit-containing SCF (Skp1/Cullin/F-box protein) E3 ubiquitin ligase complex. GSK3β functions in a similar manner to signal β-catenin degradation and can phosphorylate Notch to inhibit its transcriptional activity.106,144,145 The exact mechanism(s) through which DEK positively regulates the Wnt/β-catenin pathway remains undefined.
A tentative association between DEK and Notch signaling also has been identified. In intestinal epithelia, loss of Fbxw7, the above component of the SCF (Skp1/Cullin/F-box protein) E3 ubiquitin ligase complex, caused increased Notch, Notch target gene (i.e., Hes 1) and Dek expression.144 Using cDNA microarrays from lung carcinoma cells expressing either non-targeting shRNA or DEK shRNA, Shibata et al. also identified Notch pathway members Hes1 and NeuroD1 as downregulated in DEKsh cells.142 The same study also identified a decrease in ABCA1 transporter, a member of the ABCG family of proteins responsible for drug resistance in stem cells, in DEKsh lung carcinoma cells.142 In other cell types, including MDA-MB-468 breast cancer cells and near diploid immortalized keratinocytes that form skin (NIKS cells), high DEK expression resulted in increased levels of the p53 family member, ΔNp63α, which is thought to be an epithelial stem cell marker that can regulate Notch signaling.136,146
As mentioned above, the depletion of DEK by RNAi stimulates p53-dependent and -independent apoptosis. A p53-independent role for DEK in controlling apoptosis has been identified in metastatic melanoma. Here, depletion of DEK by shRNA expression resulted in a substantial reduction of MCL-1 level, an important pro-survival member of the Bcl-2 family.134 Interestingly, rather than influencing MCL-1 protein stability, which represents a common mechanism of MCL-1 activation in cancer, DEK controls MCL-1 at the transcriptional level through unknown mechanisms. In hematopoietic stem cells and progenitor cells, expression of MCL-1 is tightly controlled by several cytokines and growth factor signaling pathways and plays essential roles in survival. MCL-1 is required for survival and self-renewal of HSCs, but expression decreases substantially during differentiation into multipotent progenitor cells.147,148 Whether or not DEK is involved in regulating MCL-1 during HSC differentiation remains unclear. There is additional evidence that DEK can regulate the expression of p53 family members. In HeLa cells, DEK overexpression was found to inhibit p53 protein stability, which resulted in decreased expression of p53 target genes using luciferase reporter assays, including Maspin, p21 and Bax. Conversely, DEK depletion by RNAi resulted in p53 stabilization and upregulated the expression of some p53 targets.137 Furthermore, DEK depletion was found to cause a modest increase in activated p53 phosphorylated on residue Ser15 in U2OS cells.130 Interestingly, DEK expression is also regulated by p53 activity: it was significantly downregulated when primary B-chronic lymphocytic leukemia cells were treated with Nutlin-3, a non-genotoxic activator of p53 signaling.149 Taken together, DEK and p53 may function in a feedback loop to regulate survival.
An additional link between DEK and DNA repair, namely its requirement for Ku70/80 dimerization at the sites of DNA damage, leads us to speculate that DEK may be involved in telomere maintenance in stem cells: Ku70/80 dimers bind to hTERT and the RNA component (hTR) of telomerase to enhance enzyme activity.150,151 Interestingly, in a recent screen Drosophila Dek was identified as a novel factor that could bind to telomere-associated sequence (TAS) repeats.152
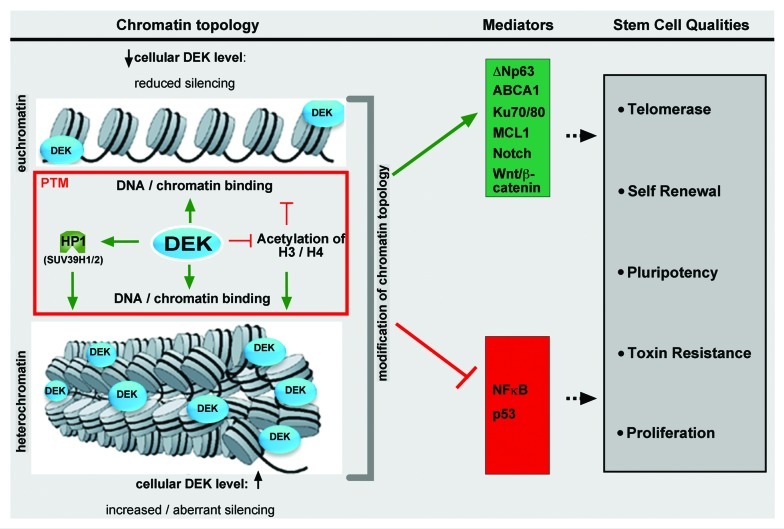
Although definitive studies have not been performed to elucidate the molecular functions of DEK specifically in stem and progenitor cells, growing evidence that changes in DEK expression impact several crucial pathways is suggestive that DEK, too, may be fundamentally important for the viability and function of the stem and/or progenitor cell populations. Combined, DEK either regulates or is regulated by several signaling pathways and transcription factors implicated in stem cell proliferation, differentiation and self-renewal, potentially through its roles in regulating chromatin topology (Fig. 2). Furthermore, its well-documented role as an oncogene indicates that DEK also may promote cancer stem cell creation or maintenance.
Figure 2. Schematic and simplified depiction of the role(s) of DEK in chromatin topology (left), and the speculative downstream effects on the transcription or functional regulation of specific mediators (middle). These mediators then may ultimately promote cellular phenotypes associated with stem and/or progenitor cells (right).
Evidence supporting a role for DEK in maintaining the stem cell population
Numerous recent reports have indicated that DEK controls normal adult stem and/or progenitor cell proliferation and self-renewal and supports cancer stem cell maintenance. Contrary to the well-established positive correlation between DEK expression and cell proliferation, Dek-knockout mice were shown to have more hematopoietic progenitor cells, based on colony-forming assays, and greater numbers of proliferating progenitors in the bone marrow and spleen when compared with wild type mice. In addition, recombinant human DEK inhibited the colony-forming unit-granulocyte macrophage (CFU-GM), burst forming unit-erythroid (BFU-E) and colony-forming unit-granulocyte erythroid macrophage megakaryocyte (CFU-GEMM) populations from wild type mice and human cord blood in a dose-dependent manner.153 It was not determined if decreased colony numbers were due to impaired proliferation or increased apoptosis. However, we have anecdotally observed that it is difficult to express DEK above physiological levels and, as previously mentioned, studies in Drosophila also indicate aberrantly high human DEK induces apoptosis. This suggests that DEK levels may have to be tightly controlled, such that too much is toxic, and, at least in undifferentiated or cancer-derived cells from solid tissues, too little may also be detrimental. It is important to note that by assessing repopulation following bone marrow transplantation in primary and secondary recipient mice, Broxmeyer et al. also reported that Dek is necessary for long-term engraftment of transplanted hematopoietic stem cells and perhaps their self-renewal capacity. Bone marrow from Dek-knockout mice was deficient in a competitive engraftment assay compared with bone marrow cells from wild type mice.153 Collectively, this suggests that murine Dek may have distinct roles in regulating proliferation and/or survival in hematopoietic stem cells vs. progenitor cells.
A growing amount of evidence points to a role for Dek in non-hematopoietic stem cell biology. For example, there are Dek-positive staining cells at the bottom of the murine intestinal crypt, which is the location of intestinal stem cells, Paneth cell progenitors and mature Paneth cells; however, co-staining was not performed to determine which cell type expressed Dek.144 Of interest, Dek was either absent or minimally expressed in the differentiated cells of the intestinal villi.144 In addition, in normal human epidermis, DEK expression is limited to rare cells in the basal compartment, which harbor epithelial stem and progenitor cells. Forced DEK overexpression led to hyperplasia and increased numbers of ΔNp63-positive epithelial cells in the basal compartment using an organotypic epithelial raft model system.146 The most compelling evidence to date that Dek is crucial for murine stem and/or progenitor cell biology was recently reported by Cheung et al. in the context of muscle stem cells required for muscle homeostasis and regeneration after injury.139 Specifically, Dek expression was absent in quiescent muscle satellite (stem) cells but was highly upregulated in activated satellite cells that are responsible for muscle regeneration.139 The expression of Dek was necessary for activated satellite cell proliferation and proliferative expansion of transit-amplifying myogenic progenitor cells.139 Furthermore, the regulation of Dek expression was controlled by the expression of miR-489, a microRNA that is highly expressed in quiescent satellite cells but is sharply downregulated upon muscle satellite cell activation as a result of stimulation by events such as tissue injury.139 Interestingly, Dek expression was asymmetric during mitosis following satellite cell activation. It was present only in the proliferating progenitor daughter cell destined for differentiation and was completely absent from the self-renewing stem cell which inherited the original DNA template.139
Interestingly, in hematopoietic stem cells, Dek expression appears to support “stemness” and lowers progenitor cell numbers whereas in muscle satellite cells it antagonizes “stemness” and supports proliferation of progenitor cells. There are a few explanations as to why this may occur. First, the manuscript describing the importance of Dek in murine hematopoietic stem and progenitor cells primarily analyzed the colony-forming ability on methylcellulose ex vivo, a system which may not be entirely representative of the stem and progenitor cell environment in vivo. This is partially supported by the absence of a difference in total nucleated cellularity in the bone marrow or spleen between wild type and Dek-knockout mice. Additional flow cytometric analyses to enumerate or enrich for stem and progenitor populations will be important to reconcile these observations. Furthermore, it is also possible that the recombinant human DEK, which inhibited colony formation, may not have undergone the extensive post-translational modifications present on endogenous murine and human DEK (Fig. 1), thus possibly perturbing its function. However, it is entirely plausible that there may be specific inherent molecular differences in hematopoietic vs. solid tissues that may account for the observed distinct, perhaps even opposing, Dek functions. This possibility may be further supported by the vastly different gene expression profiles and alternative mRNA splicing patterns observed in hematopoietic cells, both from whole blood and isolated CD34+ populations, when compared with 11 other solid tissues.154 Upstream regulators of Dek expression and post-translational modification (and, therefore, function) are likely to be significantly different between blood and solid tissues. Substantial additional research is required to elucidate the functions of Dek in these unique stem and progenitor cell populations.
In addition to growing evidence of functional activities for DEK in normal stem and progenitor cells, novel findings point toward DEK promoting transformation of normal stem cells and maintaining the cancer stem cell population. Oancea et al. reported that Dek is responsible, in part, for the leukemogenic transformation of hematopoietic stem cells.155 A rare but clinically severe type of acute myeloid leukemia is caused by the t(6;9) chromosomal translocation that creates a fusion gene between DEK and the CAN nuclear pore complex protein.156 This finding led to the original discovery of DEK, and the fusion protein maintains all but the C-terminal region of the DEK oncogene and is under control of the DEK promoter. The DEK-CAN fusion protein was able to induce transformation of Sca1+/c-Kit+/lin-/Flk2- long-term repopulating hematopoietic stem cells, but not Sca1+/c-Kit+/lin-/Flk2+ cells, and caused leukemia with 100% penetrance in lethally irradiated mice.155 DEK expression has also been implicated in the maintenance of the cancer stem cell population in both neuroendocrine carcinomas of the lung and in breast cancer. Shibata et al. examined the oncogenic roles of DEK in lung cancer and noted that targeting DEK expression with shRNA resulted in significantly decreased mRNA levels of four stem cell-related genes, including Aldh1a1, Ascl1, Pou5f1 (a.k.a., Oct3/4) and Foxo15.142 Finally, we have previously reported that, in MCF7 breast cancer cells, DEK overexpression increased the size of the side population, as determined by Hoechst dye exclusion, increased the percentage of CD44+ cells and doubled the number of mammospheres that formed in non-adherent culture, suggesting that DEK promotes cancer stem cell proliferation (data not shown).136 Likewise, DEK depletion by shRNA reduced the size of the side population, or the number of cells able to efflux Hoechst 33342, and resulted in fewer mammospheres.136
As previously mentioned, DEK is a Su(var) and histone chaperone, interacting with histones (in particular trimethylated H3K9 (H3K9me3)), HP1α, and casein kinase 2 to establish or maintain heterochromatin. Additional reports in human cells and Drosophila have indicated that DEK can inhibit histone acetyltransferases p300 and PCAF, resulting in histone H3 and H4 hypoacetylation, which are known contributors to heterochromatin formation.141,157 However, Hu et al. and Sawatsubashi et al. also published that DEK is found on euchromatin in humans and Drosophila, respectively.126,158 It is currently unclear whether DEK is required for heterochromatin establishment, the balance between heterochromatin and euchromatin or overall chromatin stability. As described earlier, stem cells have significantly more euchromatin than heterochromatin, and this ratio changes as daughter cells undergo differentiation. Furthermore, cancer cells, which almost universally overexpress DEK, often demonstrate heterochromatin instability and dramatic de-regulation of the epigenome.159 Given the different expression patterns of DEK in proliferating cancer vs. non-proliferating differentiated normal cells, as well as proliferating progenitors vs. quiescent stem cells, and the ability to influence the activity of signal transduction pathways involved in differentiation, it is possible that DEK is important for the regulation of chromatin organization and the ratio of heterochromatin to euchromatin as stem cells undergo asymmetric division and daughter cells begin to undergo differentiation. When this process is deregulated, such as through aberrantly high DEK protein levels or expression in a population normally devoid of it, resulting effects on global chromatin organization, DNA repair, and gene transcription might promote the transformation of proliferating stem and progenitor cells and, ultimately, carcinogenesis.
Conclusions
The collective body of evidence suggests that DEK is absent in quiescent stem cells but is expressed in response to environmental cues that induce stem and progenitor cell proliferation, at least in solid tissues. We hypothesize that DEK expression promotes proliferation and possibly directs early events in differentiation via its role in regulating nucleic acid structure. By directing nucleic acid topology, DEK may promote (hetero)chromatin organization, transcription, DNA replication, DNA repair and alternative mRNA splicing that could significantly alter the activity of multiple signal transduction pathways that direct the course of differentiation . How these processes relate to, and even regulate, each other remains to be determined. Furthermore, the fact that a secreted form of DEK can function as an extracellular chemo-attractant factor, and that fragments of the DEK protein can freely enter cells may have interesting implications for direct communication between progenitor cells producing DEK and neighboring cells receiving and responding to DEK in the stem cell niche.114,115 In addition, DEK is likely to support the maintenance of the cancer stem cell population and, at least when present as a fusion protein with CAN, can induce the transformation of normal stem cells. DEK expression appears to be absent in fully differentiated, non-proliferative cells, such as the suprabasal layers of the epidermis.146 Therefore, DEK expression may be a progenitor cell marker in multiple tissues, expressed during progenitor cell creation during asymmetric division of stem cells and in subsequent proliferating transit amplifying cells. The de-regulation of DEK, allowing its persistent expression, may then promote tumorigenesis and continued proliferation of the cancer stem cell. Mechanistically, this may be accomplished via (1) promoting proliferation and survival, (2) modulation of signal transduction pathways involved in differentiation, migration and self-renewal, (3) global changes in transcription, DNA repair and replication via changes in chromatin organization, (4) resistance to chemotherapeutic drugs or (5) a combination of any of these processes. An enticing hypothesis is that DEK expression, which may be limited to normal proliferating progenitor cells and both bulk and stem cells in cancers, could be ideally suited as a novel target for cancer therapy whose inhibition might eliminate cancer stem cells but retain the normal quiescent adult pluripotent stem cell population for tissue maintenance after disease remission. This hypothesis may be reflected by the reported normal phenotype of DEK-knockout mice, together with their relative protection from chemically induced papilloma formation in the skin compared with wild type controls.138 However, some DEK activities may be tissue-specific, as contradictory functions, particularly regarding proliferation, have been noted in hematopoietic cells compared with epithelial tissues. Future work should focus on defining these different tissue-specific functions of DEK and the molecular mechanism(s) by which this chromatin organizer regulates progenitor and cancer stem cell biology.
Acknowledgments
L.M.P.V. was supported by Kirschstein National Research Service Award (NRSA) F32CA139931 from the National Cancer Institute of the National Institutes of Health. Additional funding for L.M.P.V. was provided by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under award number K12HD051953, the Building Interdisciplinary Research Careers in Women’s Health (BIRCWH) Research Scholars mentored career development award sponsored by The Center for Clinical and Translational Science and Training at the University of Cincinnati. Research in the laboratory of F.K. is supported by the START Program of the Faculty of Medicine, RWTH Aachen and by the German Research Foundation (DFG). N.N. is supported by Public Health Service Grant CA115611–01. Research in the laboratory of S.I.W. is funded by Public Health Service grant R01 CA116316 and Department of Defense award W81XWH-12–1-0194. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/23121
References
- 1.Grompe M. Tissue stem cells: new tools and functional diversity. Cell Stem Cell. 2012;10:685–9. doi: 10.1016/j.stem.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seaberg RM, van der Kooy D. Stem and progenitor cells: the premature desertion of rigorous definitions. Trends Neurosci. 2003;26:125–31. doi: 10.1016/S0166-2236(03)00031-6. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 6.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 8.Dreesen O, Brivanlou AH. Signaling pathways in cancer and embryonic stem cells. Stem Cell Rev. 2007;3:7–17. doi: 10.1007/s12015-007-0004-8. [DOI] [PubMed] [Google Scholar]
- 9.Okita K, Yamanaka S. Intracellular signaling pathways regulating pluripotency of embryonic stem cells. Curr Stem Cell Res Ther. 2006;1:103–11. doi: 10.2174/157488806775269061. [DOI] [PubMed] [Google Scholar]
- 10.Sanges D, Cosma MP. Reprogramming cell fate to pluripotency: the decision-making signalling pathways. Int J Dev Biol. 2010;54:1575–87. doi: 10.1387/ijdb.103190ds. [DOI] [PubMed] [Google Scholar]
- 11.Natarajan TG, FitzGerald KT. Markers in normal and cancer stem cells. Cancer Biomark. 2007;3:211–31. doi: 10.3233/cbm-2007-34-506. [DOI] [PubMed] [Google Scholar]
- 12.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–5. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 14.Woodward WA, Chen MS, Behbod F, Rosen JM. On mammary stem cells. J Cell Sci. 2005;118:3585–94. doi: 10.1242/jcs.02532. [DOI] [PubMed] [Google Scholar]
- 15.Liu S, Dontu G, Wicha MS. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res. 2005;7:86–95. doi: 10.1186/bcr1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–60. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 17.Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair--current views. Stem Cells. 2007;25:2896–902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- 18.McKay R. Stem cells in the central nervous system. Science. 1997;276:66–71. doi: 10.1126/science.276.5309.66. [DOI] [PubMed] [Google Scholar]
- 19.Braun KM, Prowse DM. Distinct epidermal stem cell compartments are maintained by independent niche microenvironments. Stem Cell Rev. 2006;2:221–31. doi: 10.1007/s12015-006-0050-7. [DOI] [PubMed] [Google Scholar]
- 20.Cabarcas SM, Mathews LA, Farrar WL. The cancer stem cell niche--there goes the neighborhood? Int J Cancer. 2011;129:2315–27. doi: 10.1002/ijc.26312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9:11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- 22.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–4. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckert RL, Adhikary G, Balasubramanian S, Rorke EA, Vemuri MC, Boucher SE, et al. Biochemistry of epidermal stem cells. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagen.2012.07.002. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–30. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–5. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Kasai T, Li Y, Sugii Y, Jin G, Okada M, et al. A model of cancer stem cells derived from mouse induced pluripotent stem cells. PLoS One. 2012;7:e33544. doi: 10.1371/journal.pone.0033544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–82. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 29.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dontu G, Liu S, Wicha MS. Stem cells in mammary development and carcinogenesis: implications for prevention and treatment. Stem Cell Rev. 2005;1:207–13. doi: 10.1385/SCR:1:3:207. [DOI] [PubMed] [Google Scholar]
- 32.Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–85. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 33.Sengupta A, Cancelas JA. Cancer stem cells: a stride towards cancer cure? J Cell Physiol. 2010;225:7–14. doi: 10.1002/jcp.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schatton T, Frank NY, Frank MH. Identification and targeting of cancer stem cells. Bioessays. 2009;31:1038–49. doi: 10.1002/bies.200900058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K, et al. Cancer stem cell markers in common cancers - therapeutic implications. Trends Mol Med. 2008;14:450–60. doi: 10.1016/j.molmed.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Alvi AJ, Clayton H, Joshi C, Enver T, Ashworth A, Vivanco MM, et al. Functional and molecular characterisation of mammary side population cells. Breast Cancer Res. 2003;5:R1–8. doi: 10.1186/bcr563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asakura A, Rudnicki MA. Side population cells from diverse adult tissues are capable of in vitro hematopoietic differentiation. Exp Hematol. 2002;30:1339–45. doi: 10.1016/S0301-472X(02)00954-2. [DOI] [PubMed] [Google Scholar]
- 38.Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101:14228–33. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, Ludeman SM, et al. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci U S A. 1999;96:9118–23. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin KK, Goodell MA. Detection of hematopoietic stem cells by flow cytometry. Methods Cell Biol. 2011;103:21–30. doi: 10.1016/B978-0-12-385493-3.00002-4. [DOI] [PubMed] [Google Scholar]
- 42.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–8. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 43.Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 2005;132:681–7. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 44.Sharma HW, Sokoloski JA, Perez JR, Maltese JY, Sartorelli AC, Stein CA, et al. Differentiation of immortal cells inhibits telomerase activity. Proc Natl Acad Sci U S A. 1995;92:12343–6. doi: 10.1073/pnas.92.26.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrison SJ, Prowse KR, Ho P, Weissman IL. Telomerase activity in hematopoietic cells is associated with self-renewal potential. Immunity. 1996;5:207–16. doi: 10.1016/S1074-7613(00)80316-7. [DOI] [PubMed] [Google Scholar]
- 46.Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 47.O’Connor MD, Kardel MD, Eaves CJ. Functional assays for human embryonic stem cell pluripotency. Methods Mol Biol. 2011;690:67–80. doi: 10.1007/978-1-60761-962-8_4. [DOI] [PubMed] [Google Scholar]
- 48.Perry JM, Li L. Functional assays for hematopoietic stem cell self-renewal. Methods Mol Biol. 2010;636:45–54. doi: 10.1007/978-1-60761-691-7_3. [DOI] [PubMed] [Google Scholar]
- 49.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 50.Santisteban M, Reiman JM, Asiedu MK, Behrens MD, Nassar A, Kalli KR, et al. Immune-induced epithelial to mesenchymal transition in vivo generates breast cancer stem cells. Cancer Res. 2009;69:2887–95. doi: 10.1158/0008-5472.CAN-08-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lowry WE, Richter L. Signaling in adult stem cells. Front Biosci. 2007;12:3911–27. doi: 10.2741/2360. [DOI] [PubMed] [Google Scholar]
- 52.Wang MM. Notch signaling and Notch signaling modifiers. Int J Biochem Cell Biol. 2011;43:1550–62. doi: 10.1016/j.biocel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 54.Ninov N, Borius M, Stainier DY. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139:1557–67. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kondratyev M, Kreso A, Hallett RM, Girgis-Gabardo A, Barcelon ME, Ilieva D, et al. Gamma-secretase inhibitors target tumor-initiating cells in a mouse model of ERBB2 breast cancer. Oncogene. 2012;31:93–103. doi: 10.1038/onc.2011.212. [DOI] [PubMed] [Google Scholar]
- 56.Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin CR, et al. Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene. 2011;30:3454–67. doi: 10.1038/onc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tatarek J, Cullion K, Ashworth T, Gerstein R, Aster JC, Kelliher MA. Notch1 inhibition targets the leukemia-initiating cells in a Tal1/Lmo2 mouse model of T-ALL. Blood. 2011;118:1579–90. doi: 10.1182/blood-2010-08-300343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang C, Atkinson SP, Vilella F, Lloret M, Armstrong L, Mann DA, et al. Opposing putative roles for canonical and noncanonical NFκB signaling on the survival, proliferation, and differentiation potential of human embryonic stem cells. Stem Cells. 2010;28:1970–80. doi: 10.1002/stem.528. [DOI] [PubMed] [Google Scholar]
- 59.Armstrong L, Hughes O, Yung S, Hyslop L, Stewart R, Wappler I, et al. The role of PI3K/AKT, MAPK/ERK and NFkappabeta signalling in the maintenance of human embryonic stem cell pluripotency and viability highlighted by transcriptional profiling and functional analysis. Hum Mol Genet. 2006;15:1894–913. doi: 10.1093/hmg/ddl112. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Liu J, Yao S, Li F, Xin L, Lai M, et al. Nuclear factor kappa B signaling initiates early differentiation of neural stem cells. Stem Cells. 2012;30:510–24. doi: 10.1002/stem.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–40. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–77. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 64.Lluis F, Pedone E, Pepe S, Cosma MP. Periodic activation of Wnt/beta-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell. 2008;3:493–507. doi: 10.1016/j.stem.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 65.Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol. 2000;1:199–207. doi: 10.1038/35043127. [DOI] [PubMed] [Google Scholar]
- 66.Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–83. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 67.Talos F, Abraham A, Vaseva AV, Holembowski L, Tsirka SE, Scheel A, et al. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ. 2010;17:1816–29. doi: 10.1038/cdd.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzalez-Cano L, Herreros-Villanueva M, Fernandez-Alonso R, Ayuso-Sacido A, Meyer G, Garcia-Verdugo JM, et al. p73 deficiency results in impaired self renewal and premature neuronal differentiation of mouse neural progenitors independently of p53. Cell Death Dis. 2010;1:e109. doi: 10.1038/cddis.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agostini M, Tucci P, Chen H, Knight RA, Bano D, Nicotera P, et al. p73 regulates maintenance of neural stem cell. Biochem Biophys Res Commun. 2010;403:13–7. doi: 10.1016/j.bbrc.2010.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 71.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 72.Nekulova M, Holcakova J, Coates P, Vojtesek B. The role of p63 in cancer, stem cells and cancer stem cells. Cell Mol Biol Lett. 2011;16:296–327. doi: 10.2478/s11658-011-0009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dotto GP. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer. 2009;9:587–95. doi: 10.1038/nrc2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boldrup L, Coates PJ, Gu X, Nylander K. DeltaNp63 isoforms regulate CD44 and keratins 4, 6, 14 and 19 in squamous cell carcinoma of head and neck. J Pathol. 2007;213:384–91. doi: 10.1002/path.2237. [DOI] [PubMed] [Google Scholar]
- 75.Du Z, Li J, Wang L, Bian C, Wang Q, Liao L, et al. Overexpression of ΔNp63α induces a stem cell phenotype in MCF7 breast carcinoma cell line through the Notch pathway. Cancer Sci. 2010;101:2417–24. doi: 10.1111/j.1349-7006.2010.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tichy ED. Mechanisms maintaining genomic integrity in embryonic stem cells and induced pluripotent stem cells. Exp Biol Med (Maywood) 2011;236:987–96. doi: 10.1258/ebm.2011.011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tichy ED, Pillai R, Deng L, Liang L, Tischfield J, Schwemberger SJ, et al. Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev. 2010;19:1699–711. doi: 10.1089/scd.2010.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–53. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–4. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Menendez S, Camus S, Izpisua Belmonte JC. p53: guardian of reprogramming. Cell Cycle. 2010;9:3887–91. doi: 10.4161/cc.9.19.13301. [DOI] [PubMed] [Google Scholar]
- 81.Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, et al. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012;10:e1001268. doi: 10.1371/journal.pbio.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee KH, Li M, Michalowski AM, Zhang X, Liao H, Chen L, et al. A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci U S A. 2010;107:69–74. doi: 10.1073/pnas.0909734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–95. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 84.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 85.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–69. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 87.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–24. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zlatanova J, Seebart C, Tomschik M. The linker-protein network: control of nucleosomal DNA accessibility. Trends Biochem Sci. 2008;33:247–53. doi: 10.1016/j.tibs.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 91.Bártová E, Krejcí J, Harnicarová A, Kozubek S. Differentiation of human embryonic stem cells induces condensation of chromosome territories and formation of heterochromatin protein 1 foci. Differentiation. 2008;76:24–32. doi: 10.1111/j.1432-0436.2007.00192.x. [DOI] [PubMed] [Google Scholar]
- 92.Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein--an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 93.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–4. doi: 10.1016/S0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 94.Kipp M, Göhring F, Ostendorp T, van Drunen CM, van Driel R, Przybylski M, et al. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol Cell Biol. 2000;20:7480–9. doi: 10.1128/MCB.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Waldmann T, Baack M, Richter N, Gruss C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–10. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Böhm F, Kappes F, Scholten I, Richter N, Matsuo H, Knippers R, et al. The SAF-box domain of chromatin protein DEK. Nucleic Acids Res. 2005;33:1101–10. doi: 10.1093/nar/gki258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waldmann T, Eckerich C, Baack M, Gruss C. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J Biol Chem. 2002;277:24988–94. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- 98.Devany M, Kotharu NP, Matsuo H. Expression and isotopic labeling of structural domains of the human protein DEK. Protein Expr Purif. 2005;40:244–7. doi: 10.1016/j.pep.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 99.Devany M, Kappes F, Chen KM, Markovitz DM, Matsuo H. Solution NMR structure of the N-terminal domain of the human DEK protein. Protein Sci. 2008;17:205–15. doi: 10.1110/ps.073244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Devany M, Matsuo H. NMR resonance assignments for the DNA-supercoiling domain of the human protein DEK. J Biomol NMR. 2005;31:65. doi: 10.1007/s10858-004-6889-5. [DOI] [PubMed] [Google Scholar]
- 101.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–7. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 102.Okubo S, Hara F, Tsuchida Y, Shimotakahara S, Suzuki S, Hatanaka H, et al. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J Biol Chem. 2004;279:31455–61. doi: 10.1074/jbc.M403561200. [DOI] [PubMed] [Google Scholar]
- 103.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–6. doi: 10.1016/S0959-440X(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 104.Devany M, Kotharu NP, Matsuo H. Solution NMR structure of the C-terminal domain of the human protein DEK. Protein Sci. 2004;13:2252–9. doi: 10.1110/ps.04797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meyn MS, Lu-Kuo JM, Herzing LB. Expression cloning of multiple human cDNAs that complement the phenotypic defects of ataxia-telangiectasia group D fibroblasts. Am J Hum Genet. 1993;53:1206–16. [PMC free article] [PubMed] [Google Scholar]
- 106.Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004;24:6011–20. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kappes F, Scholten I, Richter N, Gruss C, Waldmann T. Functional domains of the ubiquitous chromatin protein DEK. Mol Cell Biol. 2004;24:6000–10. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mor-Vaknin N, Kappes F, Dick AE, Legendre M, Damoc C, Teitz-Tennenbaum S, et al. DEK in the synovium of patients with juvenile idiopathic arthritis: characterization of DEK antibodies and posttranslational modification of the DEK autoantigen. Arthritis Rheum. 2011;63:556–67. doi: 10.1002/art.30138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fahrer J, Popp O, Malanga M, Beneke S, Markovitz DM, Ferrando-May E, et al. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 2010;49:7119–30. doi: 10.1021/bi1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14:548–55. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- 111.Kappes F, Fahrer J, Khodadoust MS, Tabbert A, Strasser C, Mor-Vaknin N, et al. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28:3245–57. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tabbert A, Kappes F, Knippers R, Kellermann J, Lottspeich F, Ferrando-May E. Hypophosphorylation of the architectural chromatin protein DEK in death-receptor-induced apoptosis revealed by the isotope coded protein label proteomic platform. Proteomics. 2006;6:5758–72. doi: 10.1002/pmic.200600197. [DOI] [PubMed] [Google Scholar]
- 113.Cleary J, Sitwala KV, Khodadoust MS, Kwok RP, Mor-Vaknin N, Cebrat M, et al. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280:31760–7. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- 114.Cronican JJ, Beier KT, Davis TN, Tseng JC, Li W, Thompson DB, et al. A class of human proteins that deliver functional proteins into mammalian cells in vitro and in vivo. Chem Biol. 2011;18:833–8. doi: 10.1016/j.chembiol.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mor-Vaknin N, Punturieri A, Sitwala K, Faulkner N, Legendre M, Khodadoust MS, et al. The DEK nuclear autoantigen is a secreted chemotactic factor. Mol Cell Biol. 2006;26:9484–96. doi: 10.1128/MCB.01030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carro MS, Spiga FM, Quarto M, Di Ninni V, Volorio S, Alcalay M, et al. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5:1202–7. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- 117.Wise-Draper TM, Allen HV, Thobe MN, Jones EE, Habash KB, Münger K, et al. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J Virol. 2005;79:14309–17. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Soares LM, Zanier K, Mackereth C, Sattler M, Valcárcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–5. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 119.Kappes F, Burger K, Baack M, Fackelmayer FO, Gruss C. Subcellular localization of the human proto-oncogene protein DEK. J Biol Chem. 2001;276:26317–23. doi: 10.1074/jbc.M100162200. [DOI] [PubMed] [Google Scholar]
- 120.Takata H, Nishijima H, Ogura S, Sakaguchi T, Bubulya PA, Mochizuki T, et al. Proteome analysis of human nuclear insoluble fractions. Genes Cells. 2009;14:975–90. doi: 10.1111/j.1365-2443.2009.01324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu HG, Illges H, Gruss C, Knippers R. Distribution of the chromatin protein DEK distinguishes active and inactive CD21/CR2 gene in pre- and mature B lymphocytes. Int Immunol. 2005;17:789–96. doi: 10.1093/intimm/dxh261. [DOI] [PubMed] [Google Scholar]
- 122.Torrente MP, Zee BM, Young NL, Baliban RC, LeRoy G, Floudas CA, et al. Proteomic interrogation of human chromatin. PLoS One. 2011;6:e24747. doi: 10.1371/journal.pone.0024747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brázda V, Laister RC, Jagelská EB, Arrowsmith C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol Biol. 2011;12:33. doi: 10.1186/1471-2199-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Datta A, Adelson ME, Mogilevkin Y, Mordechai E, Sidi AA, Trama JP. Oncoprotein DEK as a tissue and urinary biomarker for bladder cancer. BMC Cancer. 2011;11:234. doi: 10.1186/1471-2407-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kappes F, Waldmann T, Mathew V, Yu J, Zhang L, Khodadoust MS, et al. The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 2011;25:673–8. doi: 10.1101/gad.2036411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24:159–70. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fu GK, Grosveld G, Markovitz DM. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc Natl Acad Sci U S A. 1997;94:1811–5. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Alexiadis V, Waldmann T, Andersen J, Mann M, Knippers R, Gruss C. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 2000;14:1308–12. [PMC free article] [PubMed] [Google Scholar]
- 129.Campillos M, García MA, Valdivieso F, Vázquez J. Transcriptional activation by AP-2alpha is modulated by the oncogene DEK. Nucleic Acids Res. 2003;31:1571–5. doi: 10.1093/nar/gkg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kavanaugh GM, Wise-Draper TM, Morreale RJ, Morrison MA, Gole B, Schwemberger S, et al. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res. 2011;39:7465–76. doi: 10.1093/nar/gkr454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–97. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McGarvey T, Rosonina E, McCracken S, Li Q, Arnaout R, Mientjes E, et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J Cell Biol. 2000;150:309–20. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sammons M, Wan SS, Vogel NL, Mientjes EJ, Grosveld G, Ashburner BP. Negative regulation of the RelA/p65 transactivation function by the product of the DEK proto-oncogene. J Biol Chem. 2006;281:26802–12. doi: 10.1074/jbc.M600915200. [DOI] [PubMed] [Google Scholar]
- 134.Khodadoust MS, Verhaegen M, Kappes F, Riveiro-Falkenbach E, Cigudosa JC, Kim DS, et al. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69:6405–13. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim DW, Chae JI, Kim JY, Pak JH, Koo DB, Bahk YY, et al. Proteomic analysis of apoptosis related proteins regulated by proto-oncogene protein DEK. J Cell Biochem. 2009;106:1048–59. doi: 10.1002/jcb.22083. [DOI] [PubMed] [Google Scholar]
- 136.Privette Vinnedge LM, McClaine R, Wagh PK, Wikenheiser-Brokamp KA, Waltz SE, Wells SI. The human DEK oncogene stimulates β-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene. 2011;30:2741–52. doi: 10.1038/onc.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H, Wells SI. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26:7506–19. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wise-Draper TM, Mintz-Cole RA, Morris TA, Simpson DS, Wikenheiser-Brokamp KA, Currier MA, et al. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009;69:1792–9. doi: 10.1158/0008-5472.CAN-08-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–8. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu K, Feng T, Liu J, Zhong M, Zhang S. Silencing of the DEK gene induces apoptosis and senescence in CaSki cervical carcinoma cells via the up-regulation of NFκB p65. Biosci Rep. 2012;32:323–32. doi: 10.1042/BSR20100141. [DOI] [PubMed] [Google Scholar]
- 141.Lee KS, Kim DW, Kim JY, Choo JK, Yu K, Seo SB. Caspase-dependent apoptosis induction by targeted expression of DEK in Drosophila involves histone acetylation inhibition. J Cell Biochem. 2008;103:1283–93. doi: 10.1002/jcb.21511. [DOI] [PubMed] [Google Scholar]
- 142.Shibata T, Kokubu A, Miyamoto M, Hosoda F, Gotoh M, Tsuta K, et al. DEK oncoprotein regulates transcriptional modifiers and sustains tumor initiation activity in high-grade neuroendocrine carcinoma of the lung. Oncogene. 2010;29:4671–81. doi: 10.1038/onc.2010.217. [DOI] [PubMed] [Google Scholar]
- 143.Kim DW, Kim JY, Choi S, Rhee S, Hahn Y, Seo SB. Transcriptional regulation of 1-cys peroxiredoxin by the proto-oncogene protein DEK. Mol Med Report. 2010;3:877–81. doi: 10.3892/mmr.2010.346. [DOI] [PubMed] [Google Scholar]
- 144.Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Espinosa L, Inglés-Esteve J, Aguilera C, Bigas A. Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J Biol Chem. 2003;278:32227–35. doi: 10.1074/jbc.M304001200. [DOI] [PubMed] [Google Scholar]
- 146.Wise-Draper TM, Morreale RJ, Morris TA, Mintz-Cole RA, Hoskins EE, Balsitis SJ, et al. DEK proto-oncogene expression interferes with the normal epithelial differentiation program. Am J Pathol. 2009;174:71–81. doi: 10.2353/ajpath.2009.080330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Opferman JT, Iwasaki H, Ong CC, Suh H, Mizuno S, Akashi K, et al. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 2005;307:1101–4. doi: 10.1126/science.1106114. [DOI] [PubMed] [Google Scholar]
- 148.Campbell CJ, Lee JB, Levadoux-Martin M, Wynder T, Xenocostas A, Leber B, et al. The human stem cell hierarchy is defined by a functional dependence on Mcl-1 for self-renewal capacity. Blood. 2010;116:1433–42. doi: 10.1182/blood-2009-12-258095. [DOI] [PubMed] [Google Scholar]
- 149.Secchiero P, Voltan R, di Iasio MG, Melloni E, Tiribelli M, Zauli G. The oncogene DEK promotes leukemic cell survival and is downregulated by both Nutlin-3 and chlorambucil in B-chronic lymphocytic leukemic cells. Clin Cancer Res. 2010;16:1824–33. doi: 10.1158/1078-0432.CCR-09-3031. [DOI] [PubMed] [Google Scholar]
- 150.Chai W, Ford LP, Lenertz L, Wright WE, Shay JW. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J Biol Chem. 2002;277:47242–7. doi: 10.1074/jbc.M208542200. [DOI] [PubMed] [Google Scholar]
- 151.Ting NS, Yu Y, Pohorelic B, Lees-Miller SP, Beattie TL. Human Ku70/80 interacts directly with hTR, the RNA component of human telomerase. Nucleic Acids Res. 2005;33:2090–8. doi: 10.1093/nar/gki342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Antão JM, Mason JM, Déjardin J, Kingston RE. Protein landscape at Drosophila melanogaster telomere-associated sequence repeats. Mol Cell Biol. 2012;32:2170–82. doi: 10.1128/MCB.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Broxmeyer HE, Kappes F, Mor-Vaknin N, Legendre M, Kinzfogl J, Cooper S, et al. DEK Regulates Hematopoietic Stem Engraftment and Progenitor Cell Proliferation. Stem Cells Dev. 2011;21:1449–54. doi: 10.1089/scd.2011.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tondeur S, Pangault C, Le Carrour T, Lannay Y, Benmahdi R, Cubizolle A, et al. Expression map of the human exome in CD34+ cells and blood cells: increased alternative splicing in cell motility and immune response genes. PLoS One. 2010;5:e8990. doi: 10.1371/journal.pone.0008990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Oancea C, Rüster B, Henschler R, Puccetti E, Ruthardt M. The t(6;9) associated DEK/CAN fusion protein targets a population of long-term repopulating hematopoietic stem cells for leukemogenic transformation. Leukemia. 2010;24:1910–9. doi: 10.1038/leu.2010.180. [DOI] [PubMed] [Google Scholar]
- 156.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, et al. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12:1687–97. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ko SI, Lee IS, Kim JY, Kim SM, Kim DW, Lee KS, et al. Regulation of histone acetyltransferase activity of p300 and PCAF by proto-oncogene protein DEK. FEBS Lett. 2006;580:3217–22. doi: 10.1016/j.febslet.2006.04.081. [DOI] [PubMed] [Google Scholar]
- 158.Hu HG, Scholten I, Gruss C, Knippers R. The distribution of the DEK protein in mammalian chromatin. Biochem Biophys Res Commun. 2007;358:1008–14. doi: 10.1016/j.bbrc.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 159.Carone DM, Lawrence JB. Heterochromatin instability in cancer: From the Barr body to satellites and the nuclear periphery. Semin Cancer Biol. 2012 doi: 10.1016/j.semcancer.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]