Summary
The Hsp90 family of heat shock proteins is an abundantly expressed and highly conserved family of ATP-dependent molecular chaperones. Hsp90 facilitates remodeling and activation of hundreds of proteins. In this study we developed a screen to identify Hsp90 defective mutants in E. coli. The mutations obtained define a region incorporating residues from the middle and C-terminal domains of E. coli Hsp90. The mutant proteins are defective in chaperone activity and client binding in vitro. We constructed homologous mutations in S. cerevisiae Hsp82 and identified several that caused defects in chaperone activity in vivo and in vitro. However, the Hsp82 mutant proteins were less severely defective in client binding to a model substrate than the corresponding E. coli mutant proteins. Our results identify a region in Hsp90 important for client binding in E. coli Hsp90 and suggest an evolutionary divergence in the mechanism of client interaction by bacterial and yeast Hsp90.
Keywords: HtpG, DnaK, Hsp70, DnaJ, GrpE, CbpA, Hsp40, L2, Δ131Δ, GR, Sti1, Aha1, Ste11, v-Src
Introduction
Heat shock protein 90, Hsp90, is a highly conserved and abundant ATP-dependent molecular chaperone (Johnson, 2012; Taipale et al., 2010; Wandinger et al., 2008). It is essential in eukaryotes, where it participates in the remodeling and activation of more than 200 client proteins (Johnson, 2012; Taipale et al., 2010; Wandinger et al., 2008). Cytosolic eukaryotic Hsp90 proteins have numerous cochaperones to assist in remodeling the diverse set of client proteins (Johnson, 2012; Taipale et al., 2010). Some cochaperones regulate the ATPase activity of Hsp90, while others promote Hsp90 interaction with specific client proteins.
Hsp90 proteins are highly conserved from bacteria to eukaryotes. For example, Escherichia coli Hsp90, the product of the htpG gene and referred to as Hsp90Ec, shares approximately 55% sequence similarity with human Hsp90. In E. coli, Hsp90Ec is a very abundant protein under normal growth conditions and its expression is further induced during heat stress (Neidhardt et al., 1984). While it is not essential for growth, cells deleted for Hsp90Ec grow more slowly at high temperatures (Bardwell and Craig, 1988) and exhibit a slight increase in aggregated proteins upon heat-stress (Thomas and Baneyx, 2000), compared to wild type cells. Although proteomic studies suggest there are many Hsp90 client proteins in E. coli (Arifuzzaman et al., 2006; Butland et al., 2005), only one E. coli protein, ribosomal protein L2, has been confirmed to interact with Hsp90Ec (Motojima-Miyazaki et al., 2010). The physiological significance of the interaction between L2 and Hsp90Ec is unknown. Additionally, recent genetic studies have shown that Hsp90Ec is a positive modulator of the CRISPR system, a system that provides adaptive immunity from phage and horizontally transferred DNA and RNA (Yosef et al., 2011). In this system, Hsp90Ec acts by maintaining adequate levels of a component of the CRISPR system, Cas3.
Hsp90 functions as a homodimer with each protomer consisting of three domains: an N-terminal domain (N-domain), which is involved in ATP binding and hydrolysis, a middle domain (M-domain) and a C-terminal domain (C-domain), which is required for dimerization (Krukenberg et al., 2011). In addition, eukaryotic Hsp90 proteins contain a charged linker region of approximately 50 amino acids connecting the N- and M-domains, and a C-terminal region of about 35 amino acids that is involved in the binding of several cochaperones.
Under nucleotide-free conditions apo-Hsp90Ec crystallized in a ‘V’-shaped conformation (Shiau et al., 2006). Multiple conformational rearrangements have been reported for Hsp90 underscoring the flexibility of the protein (Graf et al., 2009; Krukenberg et al., 2008; Krukenberg et al., 2009; Ratzke et al., 2010). Nucleotide binding induces further conformational transitions causing the dimer to adopt a closed conformation (Ali et al., 2006), seemingly poised for ATP hydrolysis. The different conformational states of Hsp90 are conserved across species, although the equilibrium between conformational states is variable (Southworth and Agard, 2008). Substrate binding also induces conformational changes in Hsp90. This was demonstrated recently using the model substrate Δ131Δ, a natively unfolded fragment of the staphylococcal nuclease. Hsp90Ec interacts directly with Δ131Δ, causing Hsp90Ec to contract, and in the presence of an ATP analog, Δ131Δ binds to the closed conformation of Hsp90Ec with higher affinity (Street et al., 2011). Further evidence for conformational changes in Hsp90 promoted by client binding comes from studies showing that several Hsp90-interacting proteins, including Δ131Δ, L2 and the ligand-binding domain of the glucocorticoid receptor stimulate ATP hydrolysis by Hsp90 (McLaughlin et al., 2002; Motojima-Miyazaki et al., 2010; Street et al., 2011).
In contrast to eukaryotes that possess a large number of Hsp90 cochaperones (Johnson, 2012; Taipale et al., 2010), prokaryotes appear to lack homologs of the eukaryotic cochaperones with the exception of DnaK, the Hsp70 homolog. We recently demonstrated that Hsp90Ec acts in collaboration with the DnaK system, comprised of DnaK, DnaJ or CbpA, and GrpE, to reactivate several heat- or chemically-inactivated proteins in vitro (Genest et al., 2011). Importantly, reactivation requires the ATPase activity of Hsp90Ec demonstrating that the ATP-dependent chaperone function of Hsp90Ec is required for protein remodeling reactions (Genest et al., 2011).
We report here that a conserved region of Hsp90 containing residues from the M- and C-terminal domains is required for client protein interactions. We identified these residues through the use of a novel screen that exploits toxic effects associated with overexpression of Hsp90Ec in E. coli. We found that the residues identified in our screen are important for protein remodeling activity and client protein binding. Moreover, we constructed homologous mutations in yeast Hsp90 and tested them for chaperone activity in vivo. Several of these residues are important for Hsp90 function in yeast. However, in vitro the yeast mutant proteins are less defective for client binding to a model substrate than the E. coli mutant proteins.
Results
Identification of mutations in Hsp90Ec that abrogate the toxic effects of Hsp90Ec overexpression
To identify regions of Hsp90Ec that are important for activity, we developed a screen to select for Hsp90Ec defective mutants. The phenotypes associated with cells lacking Hsp90Ec, which are slower growth at high temperatures (Bardwell and Craig, 1988) and a slight increase in protein aggregation upon heat-stress (Thomas and Baneyx, 2000), are not suitable for the selection of Hsp90Ec defective mutants. Therefore we looked for conditions where the overexpression of Hsp90Ec was detrimental to cell growth.
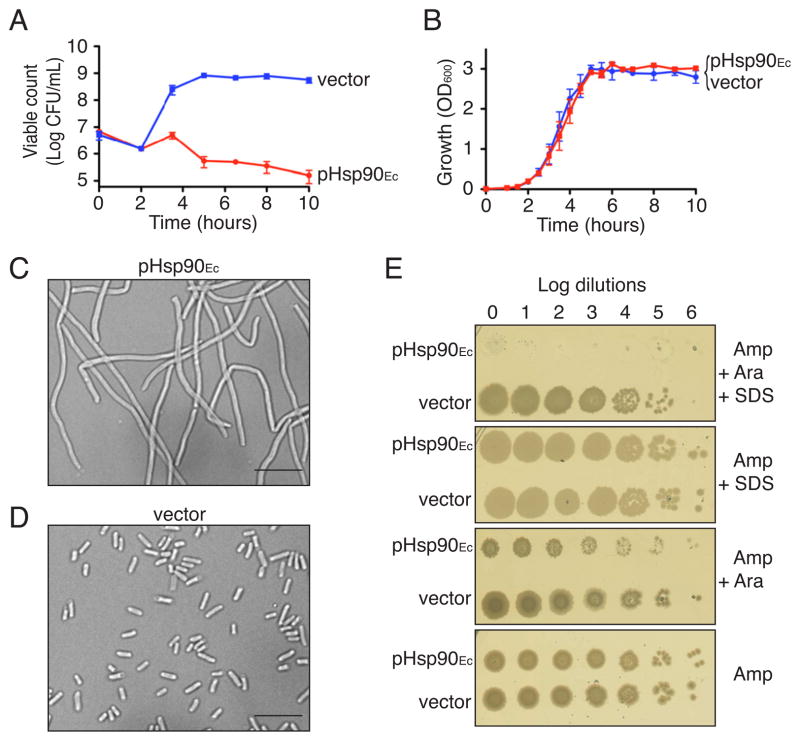
The gene encoding Hsp90Ec, htpG, was cloned into an expression plasmid under the control of the arabinose-inducible promoter. After induction, the cells carrying the plasmid, pHsp90Ec, expressed a high level of Hsp90Ec (Figure S1A). We examined the growth phenotypes of Hsp90Ec-overexpressing cells in liquid media and observed a dramatic decrease in colony forming units compared to cells carrying the empty vector (Figure 1A). However, we detected no significant difference in growth rates of the two strains as measured by O.D.600 (Figure 1B). Since a decrease in colony forming units without an accompanying decrease in O.D.600 can be indicative of cell filamentation, we examined the cells by microscopy. The cells overproducing Hsp90Ec appeared filamentous in the presence of arabinose (Figure 1C). Cells carrying the pHsp90Ec expression plasmid grown in the absence of arabinose and cells carrying the empty vector grown in the presence or absence of arabinose appeared as small rods, characteristic of wild type E. coli (Figure 1D and S1B).
Figure 1. Hsp90Ec overproduction leads to decreased cell survival, filamentation and SDS sensitivity in E. coli.
(A) E. coli MG1655 cells harboring the Hsp90Ec overexpression plasmid (pHsp90Ec) or vector were grown at 37°C in the presence of 0.2% arabinose to induce Hsp90Ec production. The number of viable cells was determined as described in Supplemental Information and plotted as colony forming units (CFU) per mL.
(B) Cells described in (A) were incubated at 37°C in the presence of 0.2% arabinose and growth was followed by measuring OD600.
(C–D) Cells expressing Hsp90Ec from pHsp90Ec (C) or carrying the empty vector
(D) were grown overnight at 37°C in the presence of 0.2% arabinose and visualized by DIC microscopy. Size bars are 10 μm.
(E) Cells expressing Hsp90Ec from pHsp90Ec or carrying the empty vector were grown without arabinose to early stationary phase and aliquots of 10-fold serial dilutions were spotted on LB-ampicillin plates (Amp) containing 0.2% arabinose (Ara) and 1% SDS as indicated. Plates were incubated overnight at 37°C.
In (A) and (B), data from three replicates are presented as mean ± SEM.
See also Figure S1.
By testing the Hsp90Ec overexpressing cells for growth under various conditions, we discovered that cells carrying the pHsp90Ec expression plasmid were unable to grow on agar plates containing 1% SDS and arabinose, whereas cells carrying the empty vector grew well under identical conditions (Figure 1E). Cells containing either the pHsp90Ec plasmid or the empty vector grew similarly in the presence of SDS, in the presence of arabinose, or in the absence of both SDS and arabinose (Figure 1E). We tested several other growth conditions, including growth at high temperature and growth on media containing Tween 20, Triton X-100, NP-40 or high salt, but did not observe growth defects caused by overexpression of Hsp90Ec. Our results suggest that high levels of Hsp90Ec impair cell division and impart sensitivity to SDS, however the mechanisms leading to these defects are not understood. Identical results were observed when the experiments were carried out using an E. coli host with a deletion of htpG, indicating that Hsp90Ec expressed from the chromosome did not affect the overexpression phenotypes (Figure S1C and S1D).
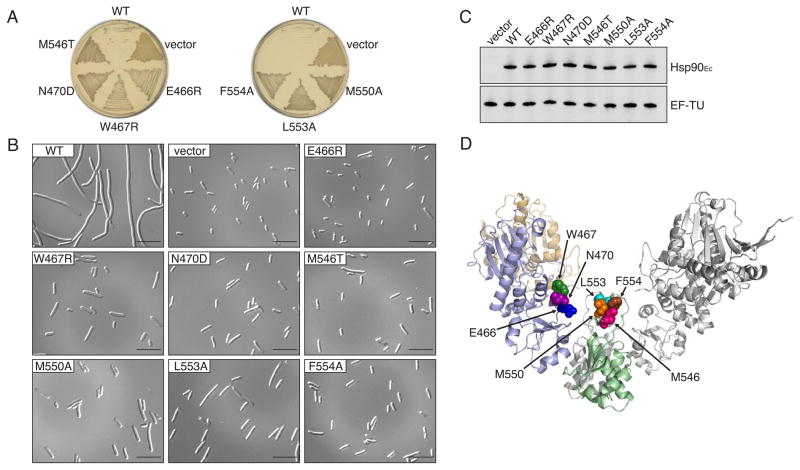
We used the detergent-sensitive phenotype of the Hsp90Ec-overproducing strain as a tool to select for Hsp90Ec mutants that lacked the overexpression phenotype. We performed random mutagenesis on plasmid-encoded htpG and selected cells able to grow on plates containing arabinose and 1% SDS. Eleven mutants were isolated using this screen and the mutant proteins were purified (Table S1). Eight mutants were not analyzed further since the gel filtration elution and/or trypsin digestion profiles of the purified proteins deviated from those of wild type. The other three Hsp90Ec mutants, W467R, N470D and M546T, exhibited physical properties indistinguishable from the wild type (Figure S2A, S2B). E. coli ΔhtpG cells expressing high levels of Hsp90Ec W467R, N470D or M546T grew on plates containing SDS and arabinose (Figure 2A) and did not form long filaments (Figure 2B). The Hsp90Ec mutant proteins were produced at similar high levels as wild type Hsp90Ec (Figure 2C). Two of these mutants, W467R and N470D, have amino acid substitutions in the middle domain, while M546T has a substitution in the C-terminal domain (Figure 2D). Based on the crystal structure of Hsp90Ec (Harris et al., 2004; Shiau et al., 2006), residues W467, N470 and M546 are spatially close to each other and on the surface of the protein.
Figure 2. Hsp90Ec mutations in the middle and C-terminal domains block the overproduction phenotypes.
(A) E. coli MG1655ΔhtpG cells overexpressing plasmid-encoded Hsp90Ec wild type (WT) or the indicated mutants were incubated at 37°C overnight on LB-ampicillin plates containing 0.2% arabinose and 1% SDS.
(B) Strains used in (A) were grown overnight at 37°C in the presence of 0.2% arabinose and analyzed by DIC microscopy. Size bars are 10 μm.
(C) Strains used in (A) were grown overnight at 37°C in the presence of 0.2% arabinose and protein expression determined by Western blot analysis using Hsp90Ec antibody. Detection of EF-TU was used as a loading control.
(D) Model of the Hsp90Ec dimer made from the X-ray structures of Hsp90Ec in the apo form (pdb: 2ioq) and the C-terminal domain of Hsp90Ec (pdb: 1sf8) visualized using PYMOL (www.pymol.org). In one protomer, the N-terminal domain is colored tan, the middle domain is light blue and the C-terminal domain is green. Residues that were mutated are represented as CPK models.
To obtain additional mutants, we performed site-directed mutagenesis on pHsp90Ec, targeting 16 surface exposed residues in close proximity to W467, N470 and M546 (Table S1). Overexpression of nine of these Hsp90Ec mutant proteins did not cause SDS sensitivity or extensive cellular filamentation (Table S1, Figure 2A, 2B). Hsp90Ec E466R and M550A were chosen for further investigation because these residues are conserved in E. coli Hsp90Ec, Saccharomyces cerevisiae Hsp82 and human Hsp90. Two mutants with substitutions in non-conserved residues, L553A and F554A, were also further characterized. Hsp90Ec E466R, M550A, L553A and F554A were purified and their physical properties were like the wild type protein (Figure S2A, S2B). The mutant proteins were expressed at high levels similar to wild type protein following induction (Figure 2C).
Altogether, we identified seven residues in Hsp90Ec that are in close proximity to one another and are important for the Hsp90Ec function(s) causing the overexpression phenotypes associated with wild type Hsp90Ec. Three residues are located in the middle domain (E466, W467 and N470) and four residues are in the C-terminal domain (M546, M550, L553, and F554) (Figure 2D).
Hsp90Ec mutants are defective in chaperone activity in vitro
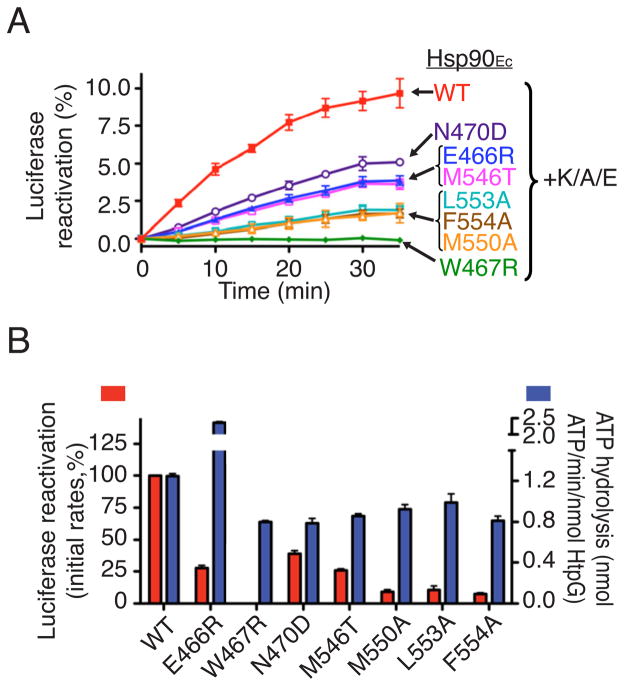
To determine if the Hsp90Ec mutant proteins that we isolated were defective for chaperone activity, we tested the mutant proteins in an in vitro chaperone assay. We monitored the reactivation of heat-inactivated luciferase in collaboration with the DnaK system, comprised of DnaK, CbpA and GrpE (Genest et al., 2011). In vitro luciferase reactivation depends on Hsp90Ec and the DnaK system, requires ATP hydrolysis by Hsp90Ec and is inhibited by geldanamycin (Genest et al., 2011). In this reaction, Hsp90Ec and the DnaK chaperone system reactivate only the soluble fraction of the denatured luciferase that corresponds to about 20% of the total luciferase (Genest et al., 2011). When we analyzed our mutant proteins, we found that Hsp90Ec W467R was unable to reactivate luciferase with the DnaK system (Figure 3A, 3B). The other Hsp90Ec mutant proteins, E466R, N470D, M546T, M550A, L553A and F554A, were partially defective for luciferase reactivation compared to wild type, with rates of reactivation between 10% and 40% of the wild type (Figure 3A, 3B). Like wild type Hsp90Ec, the mutant proteins were unable to reactivate luciferase in the absence of the DnaK system (Figure S3). Thus all of the Hsp90Ec mutant proteins exhibit defective ATP-dependent protein remodeling activity in vitro, supporting the use of our in vivo overexpression screen to identify Hsp90Ec defective mutants.
Figure 3. The Hsp90Ec mutant proteins exhibit defective chaperone activity in vitro.
(A) Reactivation of heat-denatured luciferase by Hsp90Ec wild type or mutants in conjunction with DnaK, CbpA and GrpE (K/A/E) was measured over time. The value obtained with K/A/E alone was subtracted. It was previously shown that only the soluble fraction of the denatured luciferase is reactivated by Hsp90Ec and the DnaK system and that this soluble fraction corresponds to about 20% of the total luciferase (Genest et al., 2011). Thus Hsp90Ec wild type in combination with the DnaK system reactivates about 50% of the soluble inactive luciferase in this experiment.
(B) Initial linear rates of luciferase reactivation (red) by Hsp90Ec wild type or mutants in the presence of K/A/E were calculated from Figure 3A and the rate of wild type reactivation set to 100%. The ATPase activity of Hsp90Ec wild type or mutants was measured and is represented in blue.
In (A) and (B), data from three replicates are presented as mean ± SEM.
See also Figure S3.
Since the ATPase activity of Hsp90Ec is required for protein remodeling in vitro (Genest et al., 2011), we tested if our chaperone-defective mutant proteins hydrolyze ATP. We observed that all of the mutants hydrolyze ATP at rates between 65 and 80% of wild type, except E466R, which hydrolyzes ATP ~2-fold faster than wild type (Figure 3B). Importantly, the variation in the rates of ATP hydrolysis did not correlate with changes in chaperone activity (Figure 3B). These results imply that the mutations impaired an activity other than ATP hydrolysis, which is important for protein remodeling by Hsp90Ec.
Hsp90Ec chaperone-defective mutants are impaired in their ability to bind substrates
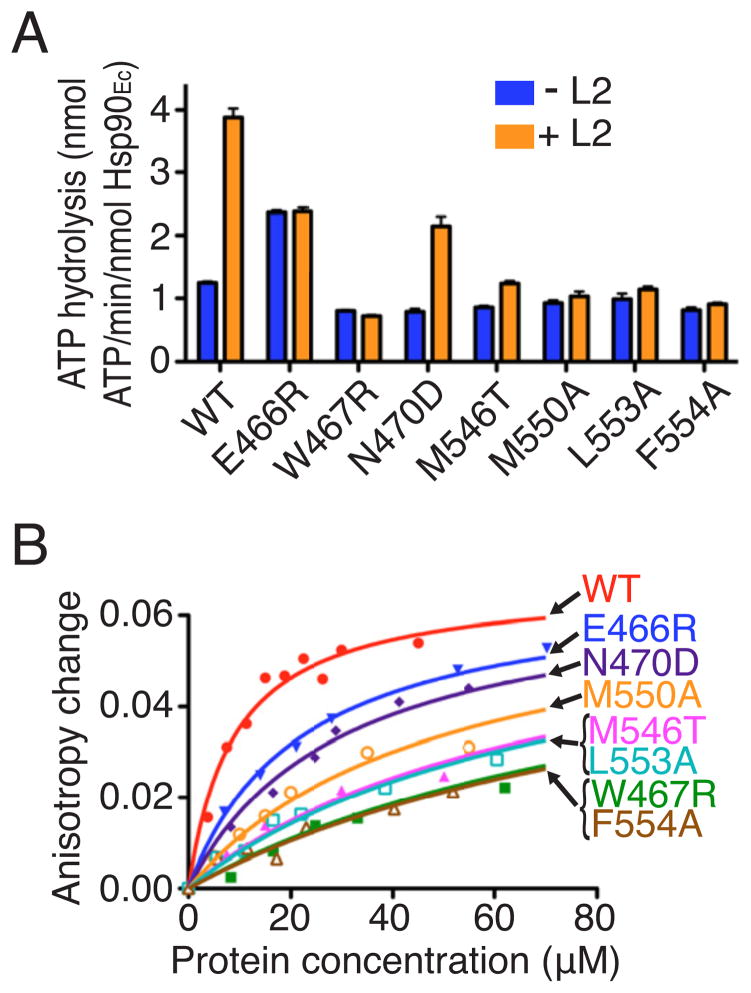
Since our Hsp90Ec mutants were defective in chaperone activity but not significantly impaired in ATPase activity, we wanted to test if they were defective in client binding. Client binding is an essential step in the pathway of protein remodeling by Hsp90Ec but does not require ATP hydrolysis (Motojima- Miyazaki et al., 2010; Street et al., 2011). To probe for Hsp90Ec-client interaction, we used ribosomal protein L2, which has been shown to interact with Hsp90Ec in the absence of ATP (Motojima-Miyazaki et al., 2010). In addition, the ATPase activity of Hsp90Ec is stimulated in the presence of L2 (Motojima-Miyazaki et al., 2010) (Figure 4A). Our results show that in contrast to the ATPase activity of wild type Hsp90Ec, the ATPase activity of Hsp90Ec E466R, W467R, M546T, M550A, L553A and F554A was not stimulated by L2 (Figure 4A). Hsp90Ec N470D ATPase activity was stimulated by L2 comparably to wild type, consistent with the observation that Hsp90Ec N470D exhibited higher chaperone activity than the other mutants (Figure 4A, 3A). Similar results were obtained when the ATPase activity of the Hsp90Ec mutant proteins was measured in the presence of another Hsp90Ec model client protein, a fragment of Staphylococcal nuclease referred to as Δ131Δ (Figure S4). Together these results suggest that the Hsp90Ec mutants are defective in chaperone activity because of impaired client binding.
Figure 4. Hsp90Ec mutants are defective in client binding.
(A) The ATPase activity of Hsp90Ec wild type or mutants with or without L2 was measured. Data from 3 replicates are presented as mean ± SEM.
(B) Binding of Hsp90Ec wild type or mutants to IAEDANS-labeled Δ131Δ was measured by fluorescence anisotropy. Binding curves are the average of two independent measurements.
See also Figure S4.
We next tested directly if the Hsp90Ec mutants bind client proteins more weakly than the wild type Hsp90Ec. We measured the fluorescence anisotropy of an IAEDANS-labeled Δ131Δ variant in the presence of Hsp90Ec or mutant Hsp90Ec. We observed that all seven of the mutant Hsp90Ec proteins bound Δ131Δ more weakly than wild type (Figure 4B). The affinity constants were not calculated because binding of this model substrate to the mutant proteins was weak and saturation was not reached. Among the Hsp90Ec mutants, W467R and F554A were most defective in Δ131Δ binding and N470D and E466R were least defective. Consistent with the relative client binding activity, Hsp90Ec W467R is the most defective in luciferase reactivation whereas N470D is the least defective (Figure 3A). Residue W467 of Hsp90Ec has been implicated in the interaction with Δ131Δ in a recent study (Street et al., 2012), which demonstrated that an A substitution at position 467 resulted in reduced affinity for Δ131Δ.
Altogether, these results show that Hsp90Ec E466R, W467R, N470D, M546T, M550A, L553A and F554A are defective for chaperone function and are impaired in their ability to interact with client proteins. They suggest that the chaperone activity is defective because of weak substrate binding. The results imply that the region of Hsp90Ec defined by these mutations is important for interaction with multiple client proteins.
Hsp82 mutants homologous to the Hsp90Ec mutants are minimally defective in Δ131Δ binding but several exhibit physiological defects
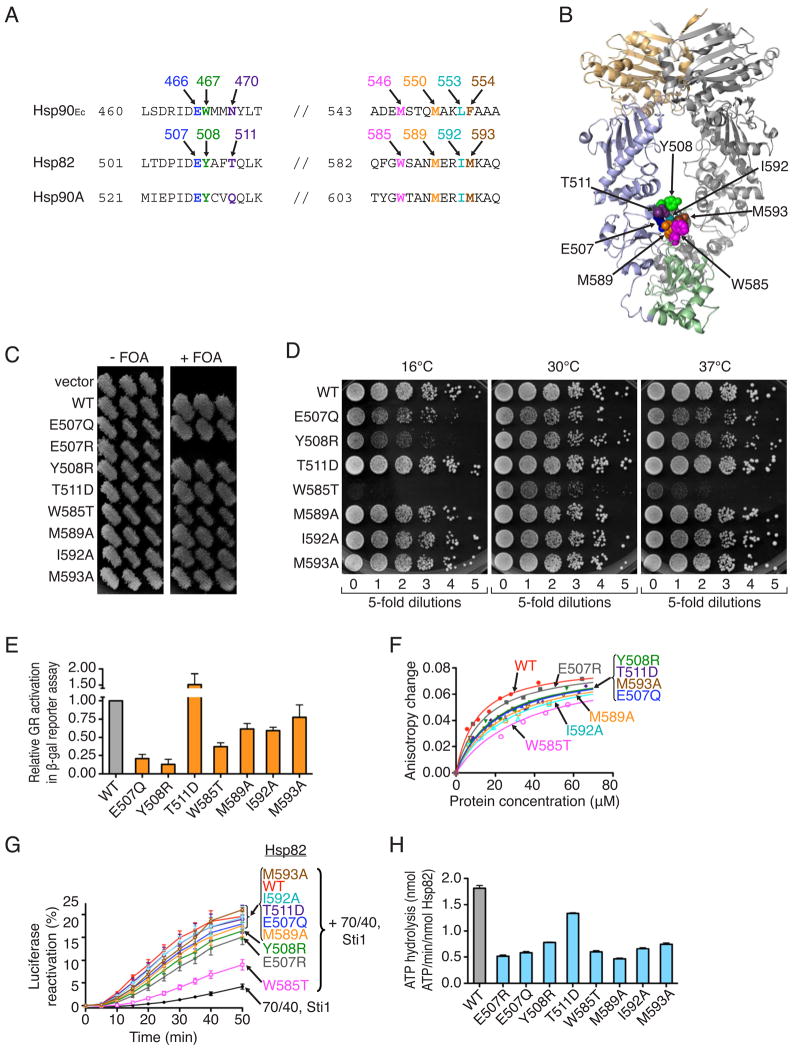
Since the Hsp90Ec mutant proteins were severely defective in client binding, we wanted to know if the same mutations might affect the function of eukaryotic Hsp90 and, in particular, if they also affect client binding. To explore this possibility, mutants homologous to our seven Hsp90Ec mutants were constructed in S. cerevisiae Hsp82 (Figure 5A). The mutated residues were close to each other on the model of the crystal structure of Hsp82 (Figure 5B) as they were on the model of Hsp90Ec (Figure 2D). However, since Hsp82 and Hsp90Ec were crystallized in different conformations, it is not possible to know with certainty if the comparable amino acids will be in similar structural environments in the two proteins.
Figure 5. Several amino acids residues important for Hsp90Ec function are also important for yeast Hsp82 function.
(A) Alignment of E. coli Hsp90Ec, S. cerevisiae Hsp82 and human Hsp90A showing mutated residues.
(B) Model of the Hsp82 dimer made from the X-ray structure of Hsp82 in the closed conformation (pdb: 2cg9) visualized using PYMOL (www.pymol.org). In one protomer, the N-terminal domain is colored tan, the middle domain is light blue and the C-terminal domain is green. Residues that were mutated are represented as CPK models.
(C) Ability of Hsp82 mutants to support growth was assessed via a plasmid shuffle assay as described in Supplemental Information. S. cerevisiae G612 lacks chromosomal copies of HSC82 and HSP82 and is supported by wild type HSP82 on a URA3 plasmid. Strains with HSP82 alleles on LEU2 plasmids that support viability grow on FOA, which prevents growth of cells carrying the URA3-based HSP82 plasmid. Three individual transformants are shown.
(D) Overnight cultures of strains expressing Hsp82 wild type or mutants that supported growth were diluted to equal density, and aliquots of five-fold serial dilutions were spotted onto YPAD plates and incubated for three days at the indicated temperatures. The cells expressing the Hsp82 defective mutants looked normal by microscopy at the permissive temperature.
(E) Glucocorticoid receptor maturation. Strains expressing Hsp82 wild type or mutants that supported growth were transformed with plasmids encoding GR and a downstream LacZ reporter (Louvion et al., 1996). After growth in the presence of deoxycorticosteroid, β-galactosidase activity was measured. Values are averages of three independent measurements reported as relative to wild type. Error bars indicate standard deviations.
(F) Binding of Hsp82 wild type or mutants to IAEDANS-labeled Δ131Δ was measured by fluorescence anisotropy. Binding curves are the average of two independent measurements.
(G) Reactivation of heat-denatured luciferase by Hsp82 wild type or mutants in conjunction with Hsp70 and Ydj1 (70/40), and Sti1 was measured over time. Data from three replicates are presented as mean ± SEM.
(H) The ATPase activity of Hsp82 wild type or mutants was measured. Data from three replicates are presented as mean ± SEM.
See also Figure S5.
We tested if the mutant proteins were able to support yeast growth as the sole source of Hsp90 using a plasmid shuffle method. Plasmids expressing Hsp82 mutant proteins were transformed into a yeast strain in which the only source of wild type Hsp82 was provided by another plasmid. When negative selection was applied, we observed that the strain expressing Hsp82 E507R was non-viable in the absence of the wild type Hsp82 plasmid (Figure 5C). The other mutants, Hsp82 Y508R, T511D, W585T, M589A, I592A and M593A, supported growth as the only source of Hsp82 (Figure 5C). We also constructed a plasmid expressing Hsp82 E507Q, since substitution of the homologous residue in Hsp90Ec, E466, with glutamine caused less severe defects than its substitution with arginine (Table S1). Hsp82 E507Q supported growth as the only source of Hsp82 (Figure 5C). Under these conditions the wild type and mutant Hsp82 proteins were expressed at similar levels (Figure S5A and S5B).
We next monitored the growth of Hsp82 mutant cells at various temperatures to determine if the mutations caused temperature sensitive growth defects under stress conditions. We found that cells expressing Hsp82 W585T as the sole Hsp82 source were unable to grow at 16°C and grew very poorly at 30°C and 37°C compared to wild type (Figure 5D and S5C). Cells expressing Hsp82 E507Q or Y508R also grew poorly at 16°C, but were able to grow at 30°C and 37°C (Figure 5D). Growth of cells expressing the other mutants was similar to wild type at all three temperatures. Thus one of the seven residues in the region that is important for client binding by Hsp90Ec is essential for the in vivo activity of Hsp82. Two others are important for growth under stress conditions.
We also assessed the ability of the Hsp82 mutants to perform specific protein remodeling activities in vivo. For example, maturation of the mammalian glucocorticoid receptor, GR, by the mutants was monitored (Picard, 2006). We observed that GR activity in cells expressing Hsp82 E507Q, Y508R or W585T was 20, 12 and 30%, respectively, compared to cells expressing wild type Hsp82 (Figure 5E). GR activity was less severely affected in cells expressing Hsp82 M589A, I592A and M593A. The Hsp82 mutants were also tested for their ability to activate the oncogenic tyrosine kinase v-Src (Mollapour et al., 2010; Nathan and Lindquist, 1995) and the yeast Ste11 kinase (Louvion et al., 1998) (Figure S5D and S5E). In both assays, W585T was more defective than the others. Taken together the results show that of the seven residues shown to be important for client binding by Hsp90Ec three are important for the in vivo chaperone function of Hsp82, E507, Y508 and W585.
Importantly, we tested if the Hsp82 mutant proteins were defective in client binding. We observed that all of the Hsp82 mutants bound Δ131Δ more weakly than wild type Hsp82, as measured by fluorescence anisotropy, with Hsp82 W585T being the most defective (Figure 5F). In contrast to the E. coli Hsp90Ec mutants, none of the Hsp82 mutant proteins were severely defective for Δ131Δ binding. These results suggest that the mechanism of client binding by Hsp90Ec and Hsp82 may differ. However, it is possible that a difference in the position of the mutated residues in Hsp82 compared to Hsp90Ec might account for the very mild client binding defects of the yeast Hsp82 mutants.
We tested several other properties of the mutant proteins but were unable to correlate the in vivo defects with in vitro defects. For example, the Hsp82 mutant proteins were slightly defective in luciferase reactivation in combination with Hsp70, Hsp40 and the Sti1 cochaperone; Hsp82 W585T was the most defective (Figure 5G). In addition, all of the mutants exhibited decreased rates of ATP hydrolysis (Figure 5H). Moreover, the ATPase activity of all of the mutants was inhibited by the Sti1 cochaperone (Figure S5F) (Prodromou et al., 1999) and all of the mutants, with the exception of E507R, were stimulated by the Aha1 cochaperone (Figure S5G) (Panaretou et al., 2002). Altogether these results demonstrate that three of the Hsp82 mutants that are homologous to the Hsp90Ec client binding mutants have impaired activity in vivo. However, the underlying causes of the in vivo defects are not known. The results showing that the Hsp90Ec mutants exhibit large defects in Δ131Δ binding and the homologous Hsp82 mutants exhibit modest defects suggest that the mechanisms by which Hsp90Ec and Hsp82 interact with substrates have diverged through evolution.
Discussion
We identified a region of Hsp90Ec that is important for ATP-dependent protein remodeling by Hsp90Ec in vitro and, more specifically, for client protein binding. The client-binding region of Hsp90Ec we identified is comprised of residues in the middle and C-terminal domains that are on the surface of Hsp90Ec at the base of the cleft formed by the two protomers of a dimer (Figure 2D). This region may be responsible for interaction with multiple client proteins, since the Hsp90Ec mutants identified in our screen are defective in binding or activating a structurally diverse set of model substrates, including Δ131Δ, L2, and denatured luciferase. Our study does not exclude the possibility that there are other client binding sites in Hsp90Ec in addition to the region identified here.
We found that mutations in this region of Hsp82 affect viability and chaperone activity in vivo but only marginally affect client protein binding in vitro using a model substrate. Consistent with this difference between client binding by Hsp90Ec and Hsp82, interactions of eukaryotic Hsp90 with client proteins are known to be complex (Didenko et al., 2012; Li et al., 2012). Different client proteins have been shown to bind to separate domains of Hsp90, suggesting that the binding process may vary with the substrate and with cochaperones. For example, the tumor suppressor protein p53 interacts with all three domains of Hsp90 in a dynamic fashion (Hagn et al., 2011; Park et al., 2011). Another structural study of Hsp90 in complex with the kinase Cdk4 and the cochaperone Cdc37 showed that Cdk4 interacts with both the N- and M-domains of Hsp90 (Vaughan et al., 2006). In addition, the interaction between Hsp82 and GR has been shown to involve residues in the C-terminal domain of Hsp82 (Fang et al., 2006). Recently, Wu et al. reported that Toll-like receptors and integrins interact with the C-terminal domain of GRP94, the endoplasmic reticulum Hsp90 (Wu et al., 2012). Interestingly, the region of GRP94 identified in that study involves residues homologous to the C-terminal domain residues in Hsp90Ec identified in our study, M546, M550, L553 and F554.
Although several genetic studies have identified mutants of Hsp82 (Bohen and Yamamoto, 1993; Nathan and Lindquist, 1995), this study is the first that uses a genetic tool to select for defective mutants of Hsp90 in E. coli. The molecular basis for the Hsp90Ec overproduction phenotypes remains unknown, but the mutants characterized were defective for ATP-dependent chaperone activity in vitro, demonstrating that this screen can be used to identify chaperone- defective Hsp90Ec mutants. Moreover, from studies exploring the conformational changes in Hsp90Ec upon binding Δ131Δ, Street et al. predicted and then confirmed by site-directed mutagenesis that residue W467 is important for binding to Δ131Δ (Street et al., 2012). Using our in vivo selection approach, we identified the same residue as essential for the Hsp90Ec overexpression phenotypes, further supporting the use of this phenotype to identify defective mutants. By testing known Hsp90Ec mutants that are defective in ATP binding (D80N) or hydrolysis (E34A) (Genest et al., 2011; Motojima-Miyazaki et al., 2010), we found that the ATPase activity of Hsp90Ec is not required for the in vivo overexpression phenotype (Table S1). These results imply that an Hsp90Ec ATPase-independent activity within the pathway of client activation is involved in this phenotype. Client binding by Hsp90Ec is independent of ATP hydrolysis (Motojima-Miyazaki et al., 2010; Street et al., 2011). Since the defective mutants obtained by our screen were altered for client binding (Figures 4A, 4B and S4), one speculation could be that overproduction of Hsp90Ec could sequester one or more client proteins and thereby block a normal function of the client protein. However, our screen was not saturated and therefore mutants with defects in Hsp90Ec activities other than client binding may also abrogate the in vivo overexpression phenotype.
Our results do not exclude the possibility that the mutations we identified could also affect long-range communication between the different domains of Hsp90, although we have shown that the physical properties of the wild type and mutant proteins were indistinguishable (Figures S2, S5H and S5I). Indeed, the differences in ATPase activity we measured between the mutant and wild type proteins are not understood and could result from altered communication between domains. Retzlaff and colleagues have identified a single residue in the C-terminal domain of Hsp90 that, when mutated, affects the conformational equilibrium of the protein, thus altering ATPase activity, association of both the N- and C-terminal domains, and chaperone activity in vivo and in vitro (Retzlaff et al., 2009). In addition, other mutations in the M-domain of Hsp90 lead to variations in the rates of ATPase activity (Hawle et al., 2006; Meyer et al., 2003).
In summary, we identified a region of Hsp90Ec that is involved in client protein binding. Similar mutations in this region of yeast Hsp82 only slightly reduce interaction with a model substrate, suggesting that the mechanism of client binding may have diverged through evolution. Further work is needed to better understand the mechanism of client protein activation by the very important Hsp90 family of proteins.
Experimental Procedures
E. coli strains and plasmids
E. coli strains MG1655 and MG1655ΔhtpG (Supplemental Information) were used. Cells were grown in Luria Broth at 37°C and, when necessary, 50 μg/mL ampicillin was added to maintain plasmid selection. Cells were imaged by differential interference contrast (DIC) microscopy as described in Supplemental Information.
Plasmid pHsp90Ec was constructed by digesting pET-HtpG (Genest et al., 2011) with XbaI/HindIII restriction enzymes and the resulting htpG containing DNA fragment cloned into similarly digested pBAD18. Single substitution mutations of Hsp90Ec were introduced into pHsp90Ec and pET-HtpG (Genest et al., 2011) and single substitutions of Hsp82 were introduced into pRSETA-Hsp82 (Prodromou et al., 2000) with the QuikChange mutagenesis system (Stratagene). All mutations were verified by DNA sequencing. The Hsp90Ec random mutagenesis library was constructed as described in Supplemental Information. Plasmids allowing the overproduction of Sti1 and Aha1 were constructed as described in Supplemental Information.
Proteins and in vitro assays
Hsp90Ec wild type and mutant proteins (Genest et al., 2011), Hsp82 wild type and mutant proteins (Supplemental Information), DnaK (Skowyra and Wickner, 1993), CbpA (Ueguchi et al., 1994), GrpE (Skowyra and Wickner, 1993), L2 (Motojima-Miyazaki et al., 2010), human Hsp70 (Miot et al., 2011), Ydj1 (Miot et al., 2011), Aha1 (Supplemental Information), Sti1 (Supplemental Information) and Δ131Δ (Street et al., 2011) were isolated as described. Hsp90Ec wild type and mutant proteins exhibited similar gel filtration chromatograms and trypsin digestion profiles (Figure S2A and S2B) as did Hsp82 and Hsp82 mutant proteins (Figure S5H, S5I). Concentrations given are for Hsp90Ec, Hsp82, CbpA, Ydj1 and GrpE dimers and DnaK, L2, Sti1, Aha1, Δ131Δ and luciferase monomers.
Luciferase reactivation was performed as described (Genest et al., 2011) using 80 nM luciferase, 0.75 μM DnaK, 0.15 μM CbpA, 0.05 μM GrpE and 0.5 μM Hsp90Ec wild type or mutant. DnaK, CbpA and GrpE were pre-incubated 20 minutes with heat-denatured luciferase before the addition of Hsp90Ec wild type or mutant. Luciferase reactivation with Hsp82 was performed similarly using 2.0 μM human Hsp70, 0.6 μM Ydj1, 0.5 μM Sti1 and 0.2 μM Hsp82. Heat denatured luciferase was added directly to chaperones without a pre-incubation step.
ATPase assays for Hsp90Ec were performed at 37°C in 25 mM Hepes, pH 7.5, 200 mM KCl, 5 mM DTT, 5 mM MgCl2, and 2 mM ATP using a pyruvate kinase/lactate dehydrogenase enzyme-coupled assay as described (Graf et al., 2009) with 1 μM Hsp90Ec wild type or mutant and 1 μM L2, where indicated. ATPase measurements for Hsp82 were performed at 37°C with the same enzyme-coupled assay using 1.25 μM Hsp82 and a similar buffer containing 20 mM KCl. Where indicated, 2.0 μM Aha1 was added. To monitor effects of Sti1, 2.0 μM Hsp82 and 2.2 μM Sti1 were used in a similar buffer containing 100 mM KCl.
Δ131Δ binding was carried out as described (Street et al., 2011). Briefly, 500 nM IAEDANS-labeled Δ131Δ was incubated for 30 minutes with varying concentrations of Hsp90Ec wild type or mutant, or Hsp82 wild type or mutant. Fluorescence anisotropy was measured on a temperature controlled Jobin Horiba fluorometer using excitation/emission wavelengths of 340/480 nm, 7 nm slits, and 0.3 s integration time. Data were fit to a noncooperative one site specific binding model using GraphPad Prism Version 5.0d.
Yeast strains, plasmids, media, growth conditions and assays
All yeast genetic manipulations were performed using derivatives of strain G612 (hsc82::KanMX hsp82::KanMX; (Jones et al., 2004)), which is isogenic to strain 779-6A (Jung and Masison, 2001). Yeast plasmid construction is detailed in Supplemental Information. Yeast cells were grown in rich medium (YPAD) or in synthetic media supplemented with appropriate nutrients for plasmid selection and containing 0.7% yeast nitrogen base with 2% glucose (SD), 2% raffinose (SRaf), or 2% galactose plus 2% raffinose (SGal). Plasmid shuffle assay (Supplemental Information) and GR activation assay (Louvion et al., 1996) were performed as described. Additional details are in Supplemental Information.
Supplementary Material
Figure S1, related to Figure 1: Hsp90Ec overproduction in E. coli.
Figure S2, related to Figure 2: Physical properties of Hsp90Ec mutant proteins
Figure S3, related to Figure 3: Hsp90Ec mutants do not reactivate heat- denatured luciferase in the absence of the DnaK chaperone system.
Figure S4, related to Figure 4: Stimulation of the ATPase activity of Hsp90Ec wild type and mutants by the Δ131Δ fragment of staphylococcal nuclease.
Figure S5, related to Figure 5: Characterization of yeast Hsp82 mutants.
Table S1, related to Figure 2: Hsp90Ec mutants studied in this work
Highlights.
Using a genetic screen, we selected for defective mutants of E. coli Hsp90
The mutants are impaired in ATP-dependent chaperone activity
The mutations define a client-binding region of E. coli Hsp90
Mutation of homologous residues in yeast Hsp90 causes in vivo and in vitro defects
Acknowledgments
We thank Jill Johnson (University of Idaho) for the Ste11ΔN plasmid, Len Neckers (NCI) for yeast plasmids, and Shannon Doyle and Danielle Johnston for critical reading of the manuscript and helpful discussions. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, National Institute of Diabetes and Digestive and Kidney Diseases and Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interests
Author Contributions: O. G., J. R. H., T. O. S., J. C. and M. R. designed experiments, performed experiments, interpreted data and wrote paper; S. W., D. A. and D. M. designed experiments, interpreted data, and wrote paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Huang HC, Hirai A, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell JC, Craig EA. Ancient heat shock gene is dispensable. J Bacteriol. 1988;170:2977–2983. doi: 10.1128/jb.170.7.2977-2983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohen SP, Yamamoto KR. Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc Natl Acad Sci USA. 1993;90:11424–11428. doi: 10.1073/pnas.90.23.11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Didenko T, Duarte AM, Karagoz GE, Rudiger SG. Hsp90 structure and function studied by NMR spectroscopy. Biochim Biophys Acta. 2012;1823:636–647. doi: 10.1016/j.bbamcr.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Fang L, Ricketson D, Getubig L, Darimont B. Unliganded and hormone-bound glucocorticoid receptors interact with distinct hydrophobic sites in the Hsp90 C-terminal domain. Proc Natl Acad Sci USA. 2006;103:18487–18492. doi: 10.1073/pnas.0609163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O, Hoskins JR, Camberg JL, Doyle SM, Wickner S. Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc Natl Acad Sci USA. 2011;108:8206–8211. doi: 10.1073/pnas.1104703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf C, Stankiewicz M, Kramer G, Mayer MP. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 2009;28:602–613. doi: 10.1038/emboj.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagn F, Lagleder S, Retzlaff M, Rohrberg J, Demmer O, Richter K, Buchner J, Kessler H. Structural analysis of the interaction between Hsp90 and the tumor suppressor protein p53. Nat Struct Mol Biol. 2011;18:1086–1093. doi: 10.1038/nsmb.2114. [DOI] [PubMed] [Google Scholar]
- Harris SF, Shiau AK, Agard DA. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure. 2004;12:1087–1097. doi: 10.1016/j.str.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Hawle P, Siepmann M, Harst A, Siderius M, Reusch HP, Obermann WM. The middle domain of Hsp90 acts as a discriminator between different types of client proteins. Mol Cell Biol. 2006;26:8385–8395. doi: 10.1128/MCB.02188-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim Biophys Acta. 2012;1823:607–613. doi: 10.1016/j.bbamcr.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Jones G, Song Y, Chung S, Masison DC. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol. 2004;24:3928–3937. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA. Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg KA, Southworth DR, Street TO, Agard DA. pH-dependent conformational changes in bacterial Hsp90 reveal a Grp94-like conformation at pH 6 that is highly active in suppression of citrate synthase aggregation. J Mol Biol. 2009;390:278–291. doi: 10.1016/j.jmb.2009.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Q Rev Biophys. 2011:1–27. doi: 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Louvion JF, Abbas-Terki T, Picard D. Hsp90 is required for pheromone signaling in yeast. Mol Biol Cell. 1998;9:3071–3083. doi: 10.1091/mbc.9.11.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvion JF, Warth R, Picard D. Two eukaryote-specific regions of Hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc Natl Acad Sci USA. 1996;93:13937–13942. doi: 10.1073/pnas.93.24.13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin SH, Smith HW, Jackson SE. Stimulation of the weak ATPase activity of human hsp90 by a client protein. J Mol Biol. 2002;315:787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Miot M, Reidy M, Doyle SM, Hoskins JR, Johnston DM, Genest O, Vitery MC, Masison DC, Wickner S. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci USA. 2011;108:6915–6920. doi: 10.1073/pnas.1102828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Tsutsumi S, Donnelly AC, Beebe K, Tokita MJ, Lee MJ, Lee S, Morra G, Bourboulia D, Scroggins BT, et al. Swe1Wee1-dependent tyrosine phosphorylation of Hsp90 regulates distinct facets of chaperone function. Mol Cell. 2010;37:333–343. doi: 10.1016/j.molcel.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motojima-Miyazaki Y, Yoshida M, Motojima F. Ribosomal protein L2 associates with E. coli HtpG and activates its ATPase activity. Biochem Biophys Res Commun. 2010;400:241–245. doi: 10.1016/j.bbrc.2010.08.047. [DOI] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, VanBogelen RA, Vaughn V. The genetics and regulation of heat-shock proteins. Annu Rev Genet. 1984;18:295–329. doi: 10.1146/annurev.ge.18.120184.001455. [DOI] [PubMed] [Google Scholar]
- Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- Park SJ, Borin BN, Martinez-Yamout MA, Dyson HJ. The client protein p53 adopts a molten globule-like state in the presence of Hsp90. Nat Struct Mol Biol. 2011;18:537–541. doi: 10.1038/nsmb.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D. Chaperoning steroid hormone action. Trends Endocrinol Metab. 2006;17:229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Prodromou C, Panaretou B, Chohan S, Siligardi G, O’Brien R, Ladbury JE, Roe SM, Piper PW, Pearl LH. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Siligardi G, O’Brien R, Woolfson DN, Regan L, Panaretou B, Ladbury JE, Piper PW, Pearl LH. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzke C, Mickler M, Hellenkamp B, Buchner J, Hugel T. Dynamics of heat shock protein 90 C-terminal dimerization is an important part of its conformational cycle. Proc Natl Acad Sci USA. 2010;107:16101–16106. doi: 10.1073/pnas.1000916107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzlaff M, Stahl M, Eberl HC, Lagleder S, Beck J, Kessler H, Buchner J. Hsp90 is regulated by a switch point in the C-terminal domain. EMBO Rep. 2009;10:1147–1153. doi: 10.1038/embor.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Wickner S. The interplay of the GrpE heat shock protein and Mg2+ in RepA monomerization by DnaJ and DnaK. J Biol Chem. 1993;268:25296–25301. [PubMed] [Google Scholar]
- Southworth DR, Agard DA. Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol Cell. 2008;32:631–640. doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street TO, Lavery LA, Agard DA. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol Cell. 2011;42:96–105. doi: 10.1016/j.molcel.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street TO, Lavery LA, Verba KA, Lee CT, Mayer MP, Agard DA. Cross-monomer substrate contacts reposition the hsp90 N-terminal domain and prime the chaperone activity. J Mol Biol. 2012;415:3–15. doi: 10.1016/j.jmb.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Thomas JG, Baneyx F. ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol Microbiol. 2000;36:1360–1370. doi: 10.1046/j.1365-2958.2000.01951.x. [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Kakeda M, Yamada H, Mizuno T. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:1054–1058. doi: 10.1073/pnas.91.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- Wu S, Hong F, Gewirth D, Guo B, Liu B, Li Z. The molecular chaperone gp96/GRP94 interacts with Toll-like receptors and integrins via its C-terminal hydrophobic domain. J Biol Chem. 2012;287:6735–6742. doi: 10.1074/jbc.M111.309526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc Natl Acad Sci USA. 2011;108:20136–20141. doi: 10.1073/pnas.1113519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1: Hsp90Ec overproduction in E. coli.
Figure S2, related to Figure 2: Physical properties of Hsp90Ec mutant proteins
Figure S3, related to Figure 3: Hsp90Ec mutants do not reactivate heat- denatured luciferase in the absence of the DnaK chaperone system.
Figure S4, related to Figure 4: Stimulation of the ATPase activity of Hsp90Ec wild type and mutants by the Δ131Δ fragment of staphylococcal nuclease.
Figure S5, related to Figure 5: Characterization of yeast Hsp82 mutants.
Table S1, related to Figure 2: Hsp90Ec mutants studied in this work