Abstract
The development of activation/dissociation techniques such as ultraviolet photodissociation (UVPD), infrared multiphoton dissociation (IRMPD) and electron transfer dissociation (ETD) as alternatives to collision induced dissociation (CID) has extended the range of strategies for characterizing biologically relevant molecules. Here, we describe a comprehensive comparison of CID, IRMPD, UVPD, ETD and hybrid processes termed ETcaD and ET-IRMPD (and analogous hybrid methods in the negative mode NETcaD and NET-IRMPD) for generating sequence specific-fragment ions and allowing adduction sites to be pinpointed for DNA/cisplatin adducts. Among the six MS/MS methods, the numerous products generated by the IRMPD and UVPD techniques resulted in the most specific and extensive backbone cleavages. We conclude that IRMPD and UVPD methods generally offer the best characteristics for pinpointing the cisplatin adduction sites in the fragment-rich spectra.
Keywords: tandem mass spectrometry, cisplatin, oligodeoxynucleotides, photodissociation, electron transfer dissociation
Introduction
DNA is a primary target for drug interaction as it is the key platform for replication, transcription and translation, cellular processes critical for protein synthesis and cell growth.[1-4] DNA-binding drugs have been reported to function via inhibition of DNA-dependent enzymes and restriction of protein-DNA interactions and consequently the activity of transcription factors, the latter leading to alteration of gene expression.[5-8] Both covalent and non-covalent modes are observed for DNA-drug binding. A number of anticancer drugs are based on a covalent binding modality owing to the fact that the formation of stable covalent adducts can lead to errors in the replication process that induce apoptosis in cancer cells. An understanding of the mechanism by which covalently binding drugs interact with DNA and the structures of the resulting adducts assists the design of agents that can bind with higher selectivity to DNA target sequences.
Cisplatin remains the most widely used anticancer drug in chemotherapy today.[9-11] It is particularly useful for the treatment of testicular, bladder, cervical, and ovarian cancers, melanoma, non-small cell lung cancer, small cell lung cancer, lymphomas and myelomas.[12-15] Within the cellular environment, inactive cis-[PtCl2(NH3)2] is hydrolyzed to the presumed active species cis-[PtCl(OH2)(NH3)2]+, a product which more readily coordinates to DNA. The hydrolyzed product preferentially binds to guanine N7 (the most electron rich site), first as a monoadduct, then subsequently forming a bifunctional adduct.[16-18] The main products are 1,2-intrastrand cross-links involving adjacent bases, primarily as cis-[Pt(d(GpG))(NH3)2]+ (60-65% abundance) or cis-[Pt(d(ApG))(NH3)2]+ (20-25% abundance) species.[19-21] Products arising from 1,3-intrastrand or G N7 – G N7 interstrand crosslinks involving non-adjacent guanines are known to be far less abundant. The adducts, especially the 1,2-intrastrand cross-links of GpG, bend the DNA, widen the minor groove, and partially unwind the helix.[22, 23] Characterization of these DNA/cisplatin adducts remains a challenging task.
Tandem mass spectrometry (MS/MS) has proven to be a versatile and sensitive strategy for elucidating the sequences of modified nucleic acids and the sites of modification. MS/MS is well-established for the characterization of oligonucleotides[24] and has been used with increasing success to obtain structural information for a variety of modified oligonucleotides, including cisplatin adducts.[25-30] In some of the earliest work, FAB tandem mass spectrometry was employed to characterize cisplatin adducts for both positive and negative ions, with loss of NH3 from cisplatin and N-glycosidic cleavage being the dominant pathways.[31, 32] Fragmentation of the cisplatin–5’-CACGTG-3’ adduct was reported by Iannitti-Tito et al. in 2000,[29] with the adducts also exhibiting the favorable loss of the amine groups and N-glycosidic cleavage. Egger et al. determined the binding kinetics and the location of the binding site of cisplatin (at GG and GTG sites) within double-stranded DNA oligonucleotides by FT-ICR MS/MS via CID and IRMPD.[33] Nyakas and co-worker reported that the binding of cisplatin substantially enhanced cleavage of the 3’-C-O bond next to a GG site in oligodeoxyribonucleotides, resulting in a notable w ion at the expense of other fragmentation pathways.[34] Recently, the same group investigated the gas-phase behavior of platinated RNA and rationalized the fragmentation pathways.[35]
To date, no single MS/MS method has established itself as the universal method for characterization of all types of biomolecules, and thus there continues to be widespread effort to develop, implement, and evaluate alternative ion activation methods, especially for biological molecules that possess key modifications (like peptides with post-translational modifications or oligonucleotide adducts). Collision-induced dissociation (CID) is by far the most common method employed for characterization of oligonucleotides[24, 36, 37] and adducts[28, 29, 33-35, 38-42]. Upon collisional activation, the loss of a base is typically a dominant fragmentation pathway with subsequent fragmentation along the phosphodiester backbone, leading to complementary w and (a - base) ions, according to the commonly adopted nomenclature shown in Supplemental Figure 1.[27, 36, 37, 39, 43-46] Infrared multiphoton dissociation (IRMPD) has also been evaluated and results in similar fragmentation pathways as CID, as demonstrated for sequencing oligonucleotides[47] and for analysis of DNA/drug complexes.[40-42, 48] Ultraviolet photodissociation (UVPD) has recently been implemented for sequencing oligonucleotides and mapping their modifications.[49] UVPD at 193 nm results in efficient charge reduction of the multiply charged oligodeoxynucleotide anions via electron detachment, in addition to extensive backbone cleavages to yield sequence ions of relatively low abundance. Electron-based activation methods in which energy deposition occurs through the capture of an energetic electron (electron capture dissociation, ECD)[50] or through the detachment of an electron (electron detachment dissociation, EDD)[51] or through the transfer of an electron from a donor molecule (electron transfer dissociation, ETD)[52], have recently been applied for sequencing oligonucleotides.[53-56] Electron activation of oligonucleotides generates exclusively charge-reduced oligonucleotide radical ions with limited backbone cleavages. More recently, our group reported the utility of hybrid tandem mass spectrometry (MS/MS) techniques combining electron transfer (ET) and CID, IRMPD, or UVPD for the characterization of a series of oligonucleotides, oligoribonucleotides, and several specific nucleic acid modifications, in which several advantages over traditional MS/MS methods were highlighted.[57] These hybrid MS/MS methods enabled rich fragmentation of modified oligodeoxynucleotides and oligoribonucleotides, with many product ions retaining the modifications and therefore affording characterization of their locations.
In an effort to develop ESI-MS/MS as an efficient screening tool for DNA/drug adducts, we have evaluated the capability of several MS/MS techniques, including CID, IRMPD (10.6 μm), ETD, NETD or UVPD (193 nm) and hybrid processes termed ETcaD, ET-IRMPD or ET-UVPD, (and analogous hybrid methods in the negative mode NETcaD, NET-IRMPD, and NET-UVPD) for generating sequence specific fragment ions and allowing adduction sites to be pinpointed.57 The hybrid methods are explored because they allow more explicit interrogation of odd electron precursors produced by electron attachment or detachment reactions. The results presented here provide a comparison of the fragmentation of platinated oligonucleotides by an array of different MS/MS techniques in both positive and negative ion modes. The strategy is also demonstrated for separation and identification of isobaric products via a capillary LC/MS/MS method.
Experimental
Materials
Single strand oligodeoxynucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA) on the 1 μmol scale and used without further purification. A summary of the single strands studied is shown in Table 1. Stock solutions were prepared in water at approximately 1 mM and the exact concentration was determined using a Nanodrop ND-1000 spectrophotometer at 260 nm (Wilmington, DE, USA) with the extinction coefficients provided by the manufacturer. Each oligodeoxynucleotide was diluted to approximately 10 μmol/L in a solution of 25 mmol/L ammonium acetate directly prior to ESI-MS analysis. Cis-Diamminedichloroplatinum (II) (cDDP) was purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA).
Table 1.
DNA oligonucleotide sequences used in this study.
| DNA name | Primary structure | MW(Da) | No. of guanines |
|---|---|---|---|
| SS1 | CCC CCC GGC CCC CC | 4066.7 | 2 |
| SS2 | CGG CCC CCC CCC CC | 4066.7 | 2 |
| SS3 | CCC CCC GCG CCC CC | 4066.7 | 2 |
| SS4 | CCC CCC GGG CCC CC | 4106.7 | 3 |
| SS5 | CGG CCC GGC CCC CC | 4146.7 | 4 |
| SS6 | CCC CCC GCC CCC CC | 4026.6 | 1 |
Cross-linking reaction of cisplatin
After the cDDP solution (500 μmol/L) was incubated with water for 24 h at 37 °C to form the reactive species [Pt(NH3)2(H2O)2], it was mixed with the corresponding oligonucleotide in a molar ratio of 3 : 1 (120 μmol/L : 40 μmol/L) and then incubated with ammonium acetate (90 mmol/L) for another 2 hours at 37 °C. Prior to mass spectrometric analysis, the reaction mixture was diluted with water to yield a final concentration of approximately 10 μmol/L.
Mass spectrometry
Analytical solutions were directly infused into a Bruker Daltonics HCTUltra quadrupole ion trap mass spectrometer (Bremen, Germany) or a Thermo Velos Pro linear ion trap mass spectrometer (San Jose, CA) at 1.5-3.0 μL/min using a Cole-Parmer syringe pump (Vernon Hills, USA). For the Bruker ion trap mass spectrometer, the ion charge control (ICC) target was set to 75,000 with a maximum accumulation time of 100 ms. The ion source parameters were set as follows: dry temperature 100 °C, nebulizer pressure 12 psi, and dry gas flow 6 L/min. The mass spectrometer was operated in both positive and negative ESI modes. Mass spectra were obtained with a Thermo Velos Pro with the heated capillary normally set at 90 °C. Typical conditions utilized a source voltage of –2.5 to –3 kV, nitrogen sheath gas set at 20–25 arbitrary units, each consisting of 30 microscans. Solutions of oligonucleotide or drug/oligonucleotide were admitted by direct infusion with a Hamilton syringe pump (Holliston, MA) at 3–5 μL/min.
Typical CID conditions consisted of an isolation width of 4.0 m/z and a fragmentation amplitude of 0.4-0.8 V. The fragmentation amplitude was adjusted so that there was survival of approximately 20% of the precursor for spectra acquired on the Bruker ion trap instrument. IRMPD was achieved using a Synrad (Mukilteo, WA) 50 W CO2 laser at a wavelength of 10.6 μm. Typical IRMPD conditions include an isolation width of 4.0 m/z, a q value of 0.1 with the laser operating at a power level of 9-10%. The irradiation time was adjusted to vary the amount of fragmentation observed, typically between 50 and 150 ms. UVPD was performed using a Coherent Excistar XS 500 ArF excimer laser (Munich, Germany), with a repetition rate of 500 Hz, and a pulse width of 5 ns. Typical UVPD conditions consisted of a pulse energy of 2-3 mJ/pulse and 2-3 laser pulses for spectra acquired on both the Bruker ion trap and the Thermo Velos Pro instrument. For hybrid MS/MS experiments, ETD was performed for typically 50-150 ms to maximize the abundance of the charge reduced radical species. In the NETD experiments described here, the oligonucleotide anion was first transferred to the ion trap (injection time 20-50 ms) and was mass isolated. Then, a given charge state of the oligonucleotide anion was mass selected and subjected to an ion-ion reaction with the fluoranthene radical cation (typically 100-200 ms). IRMPD and UVPD were carried out on the charge reduced radical species in a similar manner to the even electron precursors.
LC/MS
For the LCMS experiments a Dionex UltiMate 3000 capillary liquid chromatograph (Sunnyvale, CA, USA) was coupled to a Bruker Daltonics HCT Ultra quadrupole ion trap mass spectrometer with an electrospray (ESI) ion source. Separation was performed on an Agilent Zorbax Extend-C18 column (150 mm × 0.3 mm ID, 3.5 μm particles) at 25 °C. The injection volume was 5 μL. The mobile phases used in this study were solvent A: 10 mM ammonium acetate, solvent B: 50 % acetonitrile and 50 % 10 mM ammonium acetate. The gradient consisted of a linear gradient from 1% to 15% B over 40 min, hold at 99% B for 10 min, then equilibration back to 1% B for 1.5 min, all at a flow rate of 4 μL min–1. The injection cycle lasted for about 45 s, which includes the time required by the data acquisition program and the autosampler.
Results and Discussion
For the present study, the fragmentation patterns of cisplatin-modified oligodeoxynucleotides (ODNs) obtained by several activation techniques, including CID, IRMPD, ETD, UVPD (for both positively and negatively charged ODNs) and hybrid MS/MS methods including ETcaD, ET-IRMPD and ET-UVPD (for positively charged ODNs) were evaluated. MS/MS spectra were also acquired for the corresponding unmodified ODNs as benchmark data. The primary objective was to evaluate the analytical utility of this array of MS/MS methods in terms of mapping the cisplatin binding sites while simultaneously sequencing the ODNs. The production of high-abundance sequence ions covering the backbone of a oligodeoxynucleotide is vital for any MS/MS strategy that will be applied to the analysis of sites of modification, especially those sites in the middle of the chain which are typically more challenging to assign. Moreover, the production of a large array of fragment ions, which confers greater confidence in pinpointing the specific locations of modifications, is balanced by the dispersion of the ion signal over a larger number of dissociation channels, a factor which reduces sensitivity.
The ODNs were examined in both the positive and negative modes to allow the most systematic comparison of MS/MS spectra. Although the phosphodiester backbone is acidic and readily deprotonated to generate abundant negative ions, the ODNs also undergo efficient protonation of the nucleobases, thus making both polarities feasible for analysis of modified species. Incubation of cisplatin with the ODNs resulted primarily in formation of products consistent with intrastrand crosslinks (based on m/z values) as well as very low abundance interstrand crosslinks (typically less than 1% abundance). The low abundance of the interstrand crosslinks prohibited extensive characterization, and thus this study focused on the intrastrand crosslinks. The fragmentation patterns described in the following sections were assigned manually based on the nomenclature of McLuckey,[36, 37] as illustrated in Supplemental Figure 1. Internal fragments result from a double backbone cleavage. They have a phosphate at their 5′ end and a furan at the 3′ terminal. In some cases there are several isobaric assignments possible for a particular product ion in the MS/MS spectra, and in these cases just one reasonable assignment is labeled to prevent excessive congestion of the spectra. For example, for the symmetrical sequences it is impossible to distinguish between certain types of fragment ions (i.e., w versus d and a versus z) based on m/z alone because of the isobaric masses. The series of sequences used herein were cytosine-rich sequences containing a single or multiple guanine nucleobases in order to evaluate the impact of the position of the guanine sites on the reactivity with cisplatin.
MS/MS characterization of unmodified ODNs
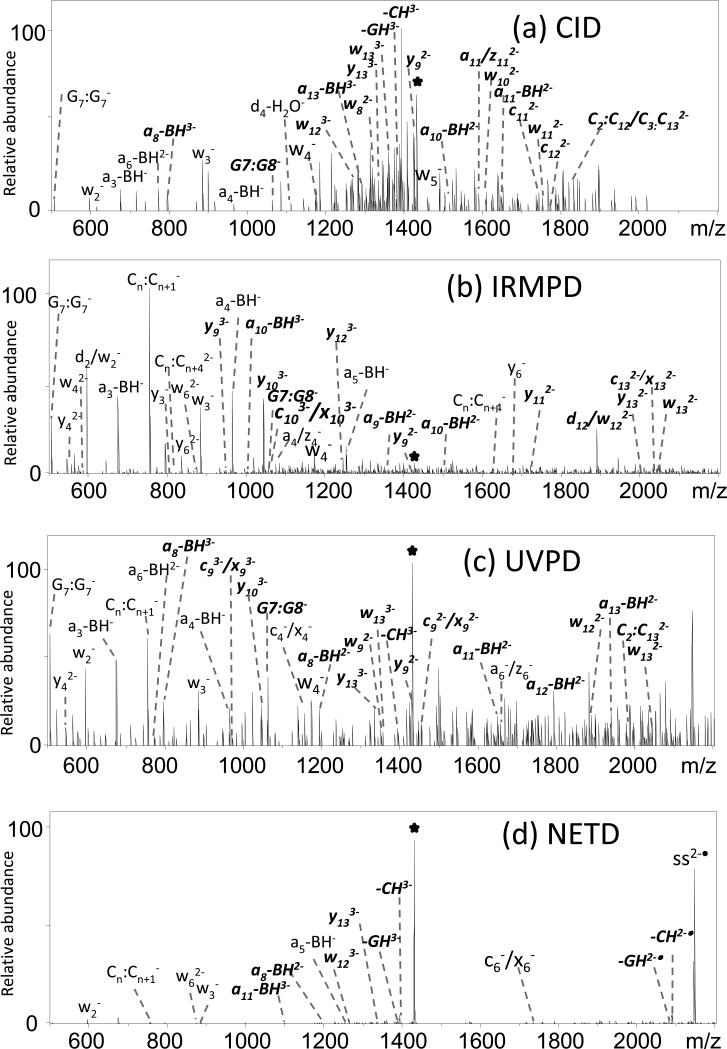
Upon electrospray ionization in the negative mode, the mass spectrum of each single strand alone is dominated by ions of the 3- and 4- charge states. The triply deprotonated and triply charged radical ions (generated by electron detachment from the quadruply deprotonated species) were analyzed for each ODN. The general fragmentation trends are consistent for each of the six ODNs, and thus only the results for the one sequence (ss1) are presented here, with representative spectra shown in Supplemental Figure 2 for the 3- charge state. The MS/MS spectra for the other ODNs are provided in Supplemental Figures 3-7. The congestion of the MS/MS spectra increases with the length of the oligodeoxynucleotides, thus prohibiting labeling all of the fragment ions, but the spectra are reproducible. The types of fragments observed upon CID of unmodified ODNs are consistent with those reported previously in which the loss of base without backbone cleavage is dominant. This base loss pathway provides no sequence information. The base loss products may subsequently dissociate into the complementary a – B and w sequence ions as well as some internal fragments denoted by Bx:By (in which the Bx and By denote the positions of the nucleobases that are included in the internal product) in Supplemental Figure 2a. IRMPD is a stepwise thermal heating process similar to CID,[58] although typically more fragments of lower m/z are observed due to alleviation of the low mass cutoff upon activation in an ion trap mass spectrometer. Nucleic acids undergo extremely efficient absorption of IR photons (10.6 μm) due to the extensive phosphodiester backbone, thus leading to high conversion of the ODN precursors into fragment ions.[59] As shown in Supplemental Figure 2b, IRMPD results in the production of numerous a - B ions, w ions, and internal fragment ions, similar to CID, but the uninformative base loss ions are minimized by their sequential dissociation into a - B and w ions. The x, y, z, c, and d series (corresponding to fragment ions arising from different backbone cleavage points, see Supplemental Figure 1) are notably absent from the CID and IRMPD spectra.
In contrast to CID and IRMPD, 193-nm UV photodissociation yielded significantly different fragmentation patterns (Supplemental Figure 2c). UVPD of ODN anions results in an extensive series of product ions arising from various backbone cleavages, as exemplified by the array of ions shown in Supplemental Figure 2c. A near complete series of a - B and w ions are observed as well as some a, b, c, x, y, and z ions. Internal ions are numerous, but base loss ions are low in abundance. Overall, UVPD generated the most diverse series of sequence ions of all the MS/MS techniques evaluated. The fourth activation method, NETD, causes efficient charge reduction of the deprotonated ODNs via electron detachment in addition to limited backbone cleavages to yield sequence ions of inconsequential abundance (Supplemental Figure 2d).
The charge-reduced ions produced by NETD were isolated and subjected to a second stage of activation, either CID or IRMPD (resulting in the hybrid MS/MS methods termed ETcaD or ET-IRMPD). NETcaD results in a modest decrease in uninformative base loss ions and an increase in w ions. The NETcaD and NET-IRMPD spectra produced product ion distributions that were similar to those observed upon CID or IRMPD. In addition to the w and a-B ion types, the NETcaD and NET-IRMPD spectra also contain some a, y, and z ions. The NETcaD and NET-IRMPD method promote greater diversity in the fragmentation pathways compared to corresponding non-hybrid activation methods. As expected, the hybrid activation methods, NETcaD and NET-IRMPD, combined the efficient charge-reduction, radical ion formation capabilities of electron transfer activation with the high efficiency of CID or IRMPD. They afford a compelling strategy for elucidation of sites of modifications of nucleic acids, but the overall efficiency of the two-stage process is rather low.
In the positive ion mode, ions in the 2+ and 3+ charge states are dominant for the size of the ODNS examined in this study, and the relative abundance of the 3+ charge state is much lower than that of the 2+ charge state. All of six activation methods, including CID, UVPD, IRMPD, ETD and hybrid MS/MS methods, resulted in similar dissociation trends for the positive ions as those described for the negative ions, although the sequence coverage was lower overall. The MS/MS spectra based on six activation methods for sequence ss1 created for the 3+ positive ions are shown in Supplemental Figure 8.
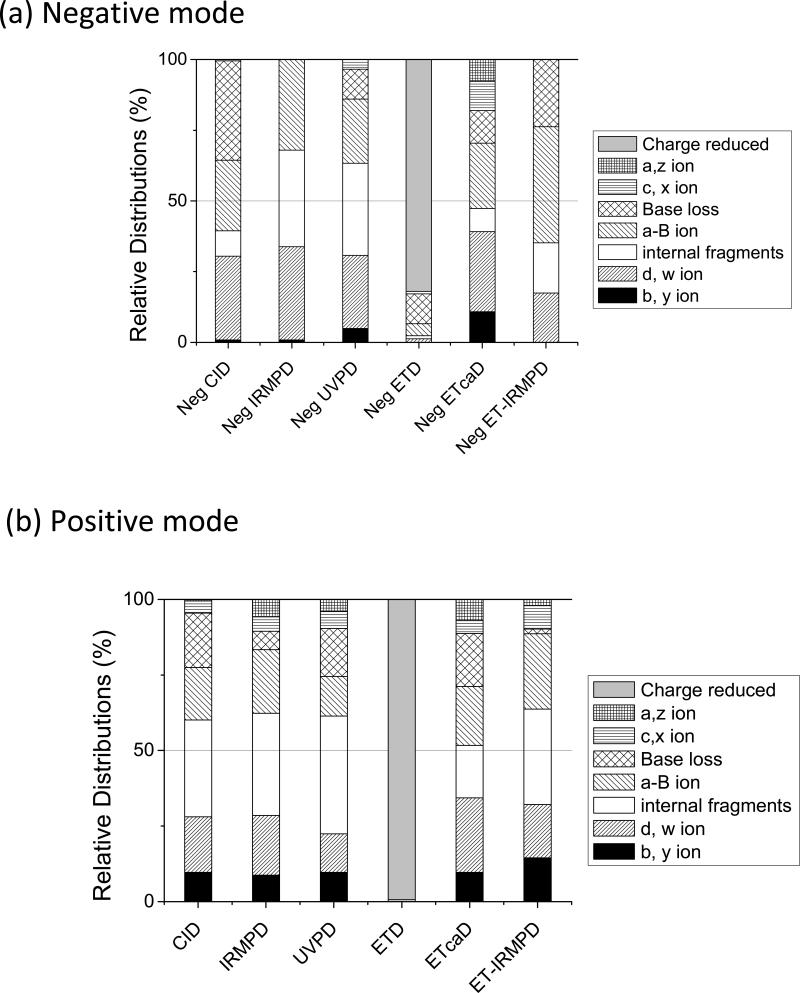
For comparative purposes, the distributions of the various types of product ions obtained for the negative and positive modes, i.e. charged-reduced, a/z, c/x, base loss, a – B, d/w, b/y, and internal fragments, are summarized in bar graph form for one ODN, ss1, in Figure 2. The types of fragments observed upon CID of unmodified ODNs in the positive ion mode are dominated by the loss of a nucleobase without subsequent backbone cleavage (Supplemental Figure 8a). The overall abundances of other types of diagnostic fragment ions are lower compared to the results in the negative mode, and more internal ions are observed in the positive mode. In addition to the w and a-B ion types, the CID spectra also result in an increase in a/z and c/x ions.
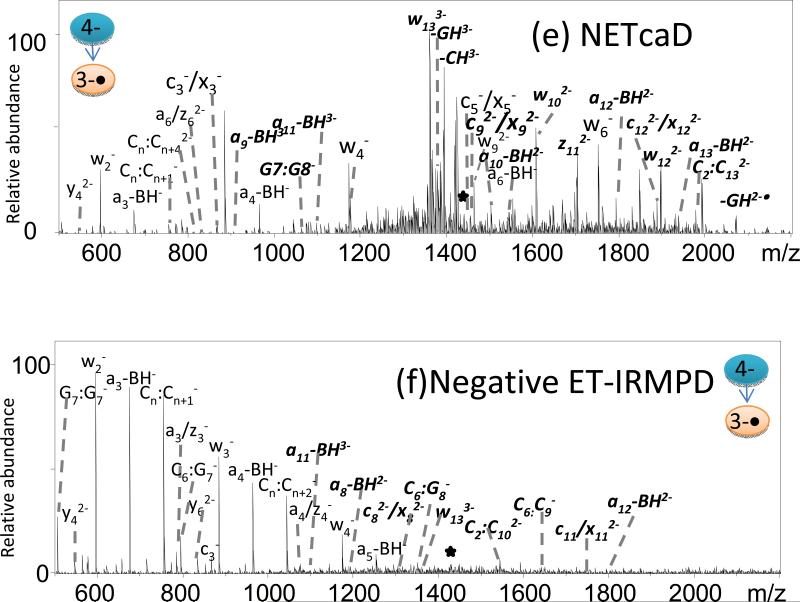
Figure 2.
MS/MS spectra of ss1/cisplatin adduct by (a) CID (3-), (b) IRMPD (3-), (c) 193-nm UVPD (3-), (d) neg ETD (3-), (e) neg ETcaD (3-•), and (f) neg ET-IRMPD (3-•). Precursor ions are noted with a star. Bolded, italicized product ions retain the modification.
UVPD of ODN cations results in a near complete series of a - B and w ions as well as some a, b, c, x, y, and z ions (Supplemental Figure 8c). Internal ions are numerous, but base loss ions are low in abundance. ETD causes efficient charge reduction of the deprotonated ODNs and limited backbone cleavages to yield low abundances of sequence ions (Supplemental Figure 8d). The ETcaD and ET-IRMPD spectra produced product ion distributions that were quite similar to those observed upon CID or IRMPD. In addition to the w and a-B ion types, the ETcaD spectra also contain some a/z ions, but with a lower abundance of internal ions than CID. The base loss ions and a/z ions in the ET-IRMPD spectra are lower in abundance than those observed upon IRMPD. In general, the types and distributions of fragment ions produced by each MS/MS method vary depending on the ion polarity and type of ion activation method. In terms of most consistent spectral quality and production of an array of diagnostic sequence ions, IRMPD exhibited the best performance.
MS/MS characterization of cisplatin-modified ODNs
ESI-MS/MS spectra of the cisplatin modified oligonucleotides were recorded in both the positive and negative ion modes. The signal-to-noise ratios were lower in the positive ion mode, owing to adduct formation between the oligonucleotides and sodium as well as potassium ions, thus leading to additional peaks in the spectra and division of the ion current among more species. Therefore, the discussion of the MS/MS results will focus mainly on the negatively charged ions.
The cisplatin-modified ODNs generally maintain the same charge state pattern as observed for the unmodified ODNs (mainly 3- and 4-). CID, IRMPD, UVPD, NETcaD, NET-IRMPD, and NET-UVPD were used to characterize the cisplatin-modified ODNs. Examples of the resulting spectra are shown in Figure 2 for cisplatin-modified ss1. Other MS/MS spectra and summaries of fragment ion distributions are provided as Supplemental Information (Supplemental Figures 9-14). The CID mass spectrum of the platinated-ODN displayed the same dominant product ions as observed for the unmodified ODN, as seen in Figure 2a, with most products stemming from base losses, in addition to some a - B and w ions. A few internal fragments ions were also detected by CID; however, these product ions were not as abundant as those in the IRMPD and UVPD mass spectra. Among the platinated fragment ions, the a - B ions, w ions and internal ions comprise the most abundant products in the MS/MS spectrum for all the activation methods. The most important diagnostic fragment, [G7:G8 + Pt]-, is observed in low abundance in Figure 2a. These internal ions allow the site of modification to be assigned with confidence in terms of the types of nucleobases that are crosslinked. The cisplatin modification is frequently retained by the a - B ions, w ions, and internal ions, which corresponds to the observation that the a - B ions, w ions and internal ions comprise the most abundant products in the MS/MS spectrum for almost all the activation methods, especially IRMPD and UVPD.
Uninformative labile base losses for both unmodified ODNs and the cisplatin/ODN crosslinks remain very dominant upon CID (Figure 2a), thus highlighting one of the shortcomings of conventional CID. Among the six activation methods, IRMPD and UVPD consistently produced lower abundances of neutral base loss ions for both unmodified and modified ODNs, either due to the conversion of these ions to other products (in the case of IRMPD) or due to the initial higher energy deposition of UVPD which allows access to other fragmentation pathways. As exemplified in Figure 2b, IRMPD resulted in similar fragmentation patterns as CID but with different relative abundances of ions, also with the additional formation of c/x ions. There is only one pair of guanine bases in the sequence of ss1. Thus, for the sequence of ss1 the detection of the internal ion [G7:G8 + Pt]- is an important diagnostic product which is observed in greater abundance in Figures 2b and 2c than in 2a. As shown in Figure 2c, UVPD at 193 nm yields even more b/y, c/x ions and fewer neutral base loss ions for the ODN/cisplatin adducts than for unmodified ODNs, and many of the informative ions retain the cisplatin modification. UVPD and IRMPD methods produce more internal ions with high abundances compared to other MS/MS methods. These short fragments are helpful for pinpointing the modification sites because they localize the modification to specific stretches of the ODN.
NETD of the ss1/cisplatin adduct also results in charge reduction by electron transfer as the primary reaction pathway, in addition to low abundance (<5%) sequence ions (Figure 2d). The NETD product ion is shifted by 1 Da compared to the ions formed directly by ESI, which confirms that charge reduction occurs via electron transfer. The lack of significant fragment ions upon NETD alone emphasizes the importance of considering the hybrid methods, such as NETcaD and NET-IRMPD, to convert the charge-reduced precursors into diagnostic products.
Analogous to the results observed for the ss1 DNA strand described above, the CID and NETcaD (Figure 2e) spectra resulted in similar ion distributions for platinated ss1. As an example, the charge-reduced species, [ss1 - 4H]3-•, produced upon electron detachment of the multi-deprotonated oligonucleotide (4-) was subjected to CID. The resulting NETcaD spectrum (Fig. 2e) displays an extensive series of abundant w, a, b, c, d, x, y, z, and a - B ions, in addition to internal fragments. Base loss also constitutes approximately 25% of the total ion abundance in the NETcaD spectrum. The relative abundance of the base loss product is decreased significantly compared to that observed in the CID spectrum. The observation of most important diagnostic fragment, [G7:G8 + Pt]-, assists in pinpointing the crosslinked nucleotides. The other fragment ions gave more information to confirm the location of modification.
In the context of determining the cisplatin adduction sites, the NET-IRMPD spectrum (Figure 2f) is overly congested with some ions un-assigned due to the low signal-to-noise ratio, making it difficult to confirm the site of adduction. Compared to unmodified ODNs, modified ODNs produced more c/x ions and b/y ions by NET-IRMPD. Unfortunately, in this case it is not possible to pinpoint the modification.
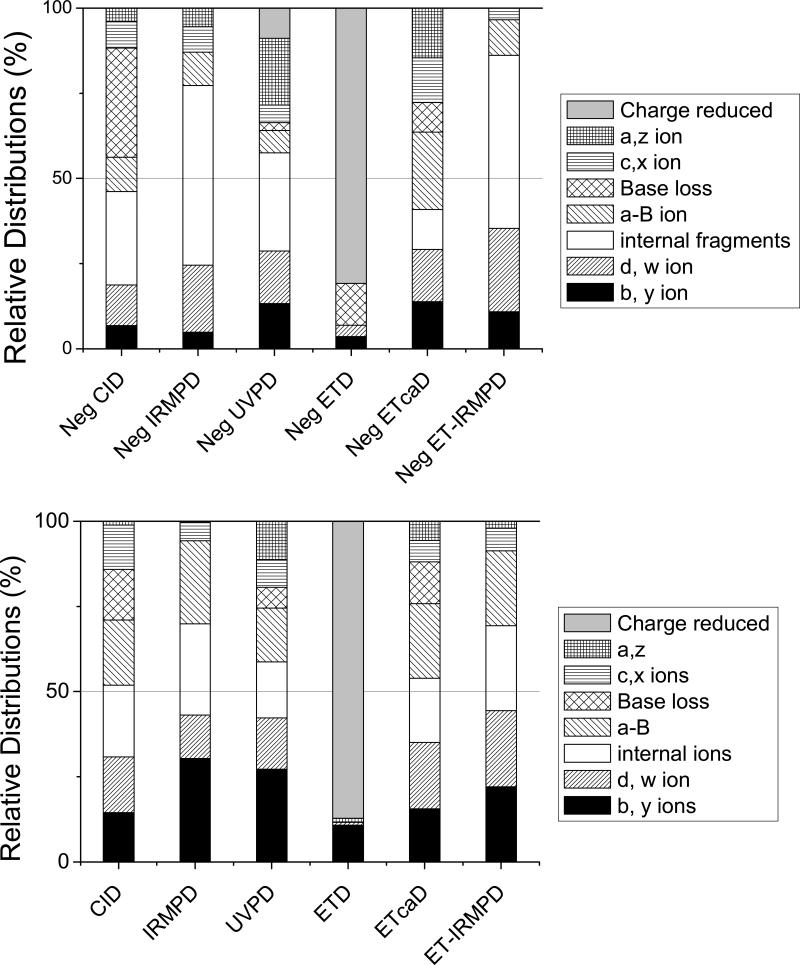
The MS/MS spectra based on six activation methods for platinated ss1 created for the 3+ positive ions are shown in Supplemental Figure 14. MS/MS experiments of ODN cations results in an extensive series of product ions arising from various backbone cleavages, as exemplified by the array of ions shown in Figure 3, and a consistent decrease of internal ions and an increase of b/y ions. Figure 3 shows the relative distributions of each type of fragment ion for platinated ss1 resulting from the six activation techniques in both negative mode and positive modes. These distributions for the ODN/cisplatin adducts show that the diversity of sequence ions differed slightly from that of unmodified ODNs, with greater preference for c/x and b/y ions for all MS/MS methods studied.
Figure 3.
Relative distributions of all ion types upon CID, IRMPD, UVPD, ETD, ETcaD, ETIRMPD for (a) the 3- (or 3-•) charge state and (b) 3+ (or 3+•) charge state of platinated ss1. The relative portion of each category of ion is obtained by dividing the summed abundance for all fragments in a particular category by the total abundance of all fragment ions.
The short Pt-free fragments (e.g., w2–, a4-BH-), and even the short internal fragments (e.g. G2:G3–) and the much more specific and interesting Pt-containing fragments are equally important for binding characterization. Briefly, the Pt-free fragments reveal which part of the ODNs sequence does not contain the Pt modification, while the Pt-containing fragments indicate which part of ODNs sequence contain the Pt modification. Among the six MS/MS methods, CID of platinum-modified ODNs produces more of the longer fragments, such as w13, a12-BH, C2:C13 etc. These very long fragments are less helpful for the identification of modification sites because the crosslinking site can not be readily extracted without more elaborate MSn strategies.In contrast, IRMPD and UVPD produce a larger array of fragment ions containing the platinum-modification.
Among the six activation techniques, four of them, including NETcaD, CID, IRMPD and UVPD, produce the specific internal ion [G7:G8+Pt]-, which is the most diagnostic ion. This fragment ion allows location of the modification site easily due to specificity of the ss1 sequence. Interestingly, for each MS/MS methods, certain ions, including mainly the a - B ions and w ions, generated by cleavage of the bonds between the platinum-modified GG base pair are lower abundance in the spectra. For ODNs with more random sequences, in order to confirm the cisplatin binding site unambiguously, all fragments ion in the MS/MS spectra must be considered. Based on the tabulation of sequence ions (both modified and unmodified ones) produced by the six MS/MS methods for the array of ODNs, IRMPD and UVPD generally offers the best characteristics for pinpointing the cisplatin adduction sites due to the superior quality of the spectra, with good signal abundances in the fragment-rich spectra.
MS/MS differentiation of cisplatin adducts
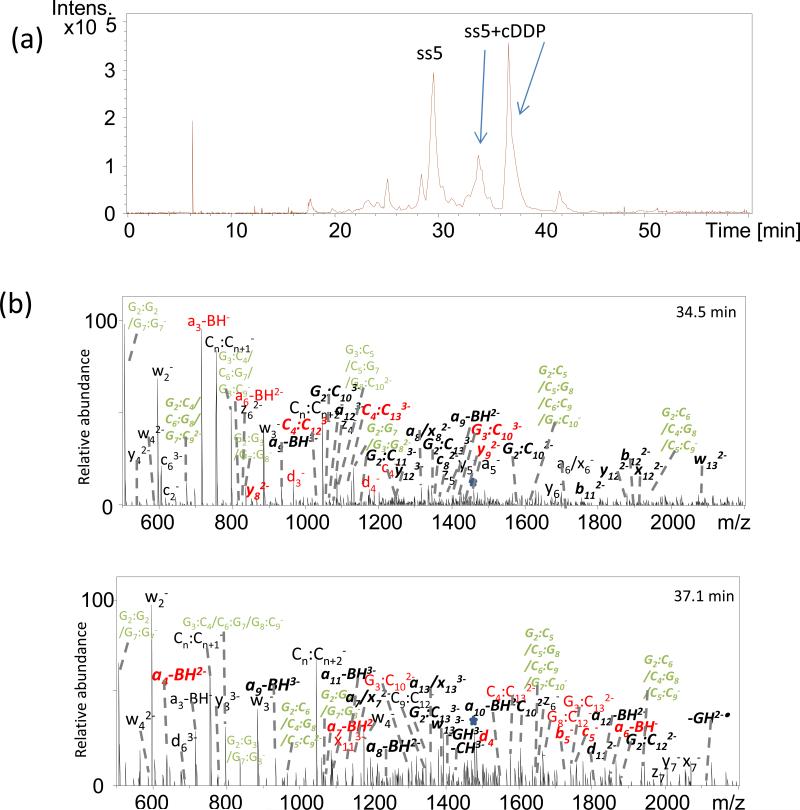
Cisplatin prefers to bind covalently to N7 purine nitrogen atoms (in GG, GNG, or AG segments), generating intrastrand cross-links[19-21]. The reaction of cisplatin with single strand ODN ss5, which bears two pairs of GG segments, results in the formation of two isobaric products which cannot be discriminated due to their same m/z values. To establish the feasibility of using the LC-MS/MS method to pinpoint the modification of DNA, the characteristic fragmentation of the ss5/cisplatin adducts were elucidated. Based on the comparison of all six methods, IRMPD in negative mode was used because it provided the best sequence coverage and variety of ions.
Displayed in Fig. 4 are the IRMPD mass spectra generated from the [ss5 + cDDP]3- ions (m/z 1457) of platinated oligonucleotide ss5 acquired via the LCMS strategy. The IRMPD spectra are dominated by abundant a – B, w and internal fragments ions. Fragment ions derived from cleavage of the oligonucleotide backbone, x, y, z, a, b, c, d ions are either absent or present at less than 10% relative ion abundance. The formation of complementary modified and unmodified a - B, w, and internal fragments gives the exact cisplatin binding site. In Fig. 4, the fragments marked in red are very important for localizing the Pt modification. The fragments marked in green alone cannot be assigned confidently due to the same m/z value for several possible structures. However, when the platinum modification is located based on the observation of fragments ions in red, the structure of fragments in green can be assigned. The two eluting products at 34.5 minutes and 37.1 minutes were thus identified as the G7G8 adduct and G2G3 adducts, respectively.
Figure 4.
(a) Chromatographic profile elution of two ss5/cisplatin adducts; (b) LCMS/MS spectra for products at 34.5 min and 37.1 min. Precursor ions are noted with a star. Bolded, italicized product ions retain the modification. The fragments in red are important for pinpointing the site of the Pt modification. The fragments in green cannot be assigned confidently due to m/z values that fit several isomeric sequences.
Conclusion
Our experiments were aimed at evaluating the relative merits of several MS/MS techniques for generating sequence specific fragment ions and allowing adduction sites to be pinpointed. IRMPD and UVPD proved more proficient for locating the platinum modification site in the negative mode. The types of fragments observed upon CID are dominated by base loss products as well as a – B, w and internal fragments. IRMPD and UVPD consistently produced lower abundances of neutral base loss ions. IRMPD resulted in similar fragmentation patterns as CID but with different relative abundances of ions, also with the additional formation of c/x ions. UVPD at 193 nm resulted in extensive fragmentation with the generation of a – B , w, internal fragments, b/y, and c/x ions. IRMPD and UVPD led to increased fragment ion diversity with many of the informative ions retaining the cisplatin modification, a factor which allowed the modification sites of multiply modified oligonucleotides to be ascertained. ETD and NETD caused efficient charge reduction of ODNs in addition to limited backbone cleavages to yield sequence ions of low abundance. The ETcaD and ET-IRMPD spectra produced product ion distributions that were quite similar to those observed upon CID or IRMPD (but with lower abundances). In addition to the w and a-B ion types, the ETcaD spectra also contained some a/z ions, but with a lower abundance of internal ions than CID. Employing multiple MS/MS methods offers the most flexible hierarchical strategy for structural characterization of the more challenging nucleic acid modifications and adducts.
The tandem mass spectra for crosslinked ODNs are complicated, even for ones containing only 14 nucleotides. It should be feasible to employ the UVPD, IRMPD, and ET-PD hybrid methods to evaluate the sequence-selective reactivities of cisplatin with medium-sized ODNs (6-25 mer) and characterize the resulting adducts. Application of these methods to biologically relevant DNA structure will be far more challenging due to the larger sizes of the products. However, by combining enzymatic (endonuclease/exonuclease) digestion with the LC-MS/MS methods described here, structure elucidation of DNA crosslinks should be feasible. We anticipate that the MS/MS methods should also be useful for elucidating the structures and locating the sites of crosslinks formed between ODNs and other DNA-interactive agents. Addressing these grander problems would be significantly facilitated by utilization of new software tools, such as the recently developed oligonucleotide mass assembler (OMA) and oligonucleotide peak analyzer (OPA) package [60], to assist the interpretation of the MS/MS spectra. The ability to investigate even more complicate macromolecular complexes, such as those containing DNA, ligands, and proteins, offers another frontier that may benefit from a large array of MS/MS options.
Supplementary Material
Figure 1.
Relative distributions of all ion types upon CID, IRMPD, UVPD, ETD, ETcaD, ETIRMPD for (a) the 3- (or 3-•) charge state and (b) 3+ (or 3+•) charge state of unmodified ss1. The relative portion of each category of ion is obtained by dividing the summed abundance for all fragments in a particular category by the total abundance of all fragment ions.
Acknowledgment
Funding from NIH (RO1 GM65956) and the Welch Foundation (F1155) is gratefully acknowledged.
Abbreviations
- CID
Collision-induced dissociation
- IRMPD
Infrared multiphoton dissociation
- UVPD
Ultraviolet photodissociation
- ECD
electron capture dissociation
- EDD
electron detachment dissociation
- ETD
electron transfer dissociation
- NETD
electron transfer dissociation in negative polarity mode
- ETcaD
electron transfer (ET) followed by CID
- ET-IRMPD
electron transfer (ET) followed by IRMPD
- ET-UVPD
electron transfer (ET) followed by UVPD
References
- 1.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2(3):188. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 2.Ou TM, Lu YJ, Tan JH, Huang ZS, Wong KY, Gu LQ. G-quadruplexes: Targets in anticancer drug design. ChemMedChem. 2008;3(5):690. doi: 10.1002/cmdc.200700300. [DOI] [PubMed] [Google Scholar]
- 3.Miller KM, Rodriguez R. G-quadruplexes: selective DNA targeting for cancer therapeutics? Expert Rev. Clin. Pharmacol. 2011;4(2):139. doi: 10.1586/ecp.11.4. [DOI] [PubMed] [Google Scholar]
- 4.Rajski SR, Williams RM. DNA cross-linking agents as antitumor drugs. Chem. Rev. 1998;98(8):2723. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 5.Brunton LL, Lazo JS, Parker KL. Goodman & Gilman's the pharmacological basis of therapeutics. McGraw-Hill; New York: 2006. [Google Scholar]
- 6.Wheate NJ, Brodie CR, Collins JG, Kemp S, Aldrich-Wright JR. DNA Intercalators in Cancer Therapy: Organic and Inorganic Drugs and Their Spectroscopic Tools of Analysis. Mini-Rev. Med. Chem. 2007;7(6):627. doi: 10.2174/138955707780859413. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff G, Hoffmann S. DNA-Binding of Drugs Used in Medicinal Therapies. Curr. Med. Chem. 2002;9(3):321. doi: 10.2174/0929867023371085. [DOI] [PubMed] [Google Scholar]
- 8.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta-Gene Struct. Expression. 1998;1400(1-3):83. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 9.Prestayko AW, Daoust JC, Issell BF, Crooke ST. Cisplatin (cis-diamminedichloroplatinum-II). Cancer Treat. Rev. 1979;6(1):17. doi: 10.1016/s0305-7372(79)80057-2. [DOI] [PubMed] [Google Scholar]
- 10.Lippert B. Cisplatin: Chemistry and biochemistry of a leading anticancer drug. VHCA & Wiley-VCH; Zurich, Switzerland: 1999. [Google Scholar]
- 11.Rosenberg B, Van Camp L, Krigas T. Inhibition of Cell Division in Escherichia coli by Electrolysis Products from a Platinum Electrode. Nature. 1965;205(4972):698. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- 12.Boulikas T, Vougiouka M. Recent clinical trials using cisplatin, carboplatin and their combination chemotherapy drugs (review). Oncol. Rep. 2004;11(3):559. [PubMed] [Google Scholar]
- 13.Loehrer PJ, Einhorn LH. Cisplatin. Annals of internal medicine. 1984;100(5):704. doi: 10.7326/0003-4819-100-5-704. [DOI] [PubMed] [Google Scholar]
- 14.Muggia F. Platinum compounds 30 years after the introduction of cisplatin: Implications for the treatment of ovarian cancer. Gynecol. Oncol. 2009;112(1):275. doi: 10.1016/j.ygyno.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 15.Wong E, Giandomenico CM. Current Status of Platinum-Based Antitumor Drugs. Chem. Rev. 1999;99(9):2451. doi: 10.1021/cr980420v. [DOI] [PubMed] [Google Scholar]
- 16.Robins AB. The reaction of 14C-labelled platinum ethylenediamine dichloride with adenine compounds and DNA. Chem.-Biol. Interact. 1973;7(1):11. doi: 10.1016/0009-2797(73)90011-2. [DOI] [PubMed] [Google Scholar]
- 17.Robins AB. The reaction of 14C-labelled platinum ethylenediamine dichloride with nucleic acid constituents. Chem.-Biol. Interact. 1973;6(1):35. doi: 10.1016/0009-2797(73)90084-7. [DOI] [PubMed] [Google Scholar]
- 18.Mansy S, Rosenberg B, Thomson AJ. Binding of cis- and trans-dichlorodiammineplatinum(II) to nucleosides.i. Location of the binding sites. J. Am. Chem. Soc. 1973;95(5):1633. doi: 10.1021/ja00786a045. [DOI] [PubMed] [Google Scholar]
- 19.Fichtinger-Schepman AMJ, Van der Veer JL, Den Hartog JHJ, Lohman PHM, Reedijk J. Adducts of the antitumor drug cis-diamminedichloroplatinum(II) with DNA: formation, identification, and quantitation. Biochemistry. 1985;24(3):707. doi: 10.1021/bi00324a025. [DOI] [PubMed] [Google Scholar]
- 20.Fichtinger-Schepman AMJ, van Oosterom AT, Lohman PHM, Berends F. cis-Diamminedichloroplatinum(II)-induced DNA Adducts in Peripheral Leukocytes from Seven Cancer Patients: Quantitative Immunochemical Detection of the Adduct Induction and Removal after a Single Dose of cis-Diamminedichloroplatinum(II). Cancer Res. 1987;47(11):3000. [PubMed] [Google Scholar]
- 21.Mangrum JB, Farrell NP. Excursions in polynuclear platinum DNA binding. Chem. Commun. 2010;46(36):6640. doi: 10.1039/c0cc01254h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jamieson ER, Lippard SJ. Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev. 1999;99(9):2467. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 23.Poklar N, Pilch DS, Lippard SJ, Redding EA, Dunham SU, Breslauer KJ. Influence of cisplatin intrastrand crosslinking on the conformation, thermal stability, and energetics of a 20-mer DNA duplex. Proc. Natl. Acad. Sci. U. S. A. 1996;93(15):7606. doi: 10.1073/pnas.93.15.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, McLuckey SA. Gas-phase fragmentation of oligonucleotide ions. Int. J. Mass Spectrom. 2004;237(2-3):197. [Google Scholar]
- 25.Le Pla RC, Ritchie KJ, Henderson CJ, Wolf CR, Harrington CF, Farmer PB. Development of a Liquid Chromatography–Electrospray Ionization Tandem Mass Spectrometry Method for Detecting Oxaliplatin–DNA Intrastrand Cross-Links in Biological Samples. Chem. Res. Toxicol. 2007;20(8):1177. doi: 10.1021/tx700088j. [DOI] [PubMed] [Google Scholar]
- 26.Beck JL, Colgrave ML, Ralph SF, Sheil MM. Electrospray ionization mass spectrometry of oligonucleotide complexes with drugs, metals, and proteins. Mass Spectrom. Rev. 2001;20(2):61. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- 27.Anichina J, Zhao Y, Hrudey SE, Schreiber A, Li X-F. Electrospray Ionization Tandem Mass Spectrometry Analysis of the Reactivity of Structurally Related Bromo-methyl-benzoquinones toward Oligonucleotides. Anal. Chem. 2011;83(21):8145. doi: 10.1021/ac201646z. [DOI] [PubMed] [Google Scholar]
- 28.Barry JP, Vouros P, Vanschepdael A, Law SJ. Mass AND Sequence Verification of modified oligonucleotides using electrospray tandem mass spectrometry. J. Mass Spectrom. 1995;30(7):993. [Google Scholar]
- 29.Iannitti-Tito P, Weimann A, Wickham G, Sheil MM. Structural analysis of drug-DNA adducts by tandem mass spectrometry. Analyst. 2000;125(4):627. doi: 10.1039/a908920i. [DOI] [PubMed] [Google Scholar]
- 30.Zhang QR, Yu ET, Kellersberger KA, Crosland E, Fabris D. Toward building a database of bifunctional probes for the MS3D investigation of nucleic acids structures. J. Am. Soc. Mass Spectrom. 2006;17(11):1570. doi: 10.1016/j.jasms.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Martin LB, Schreiner AF, Vanbreemen RB. Characterization of cisplatin adducts of oligonucleotides by fast-atom-bombardment mass-spectrometry. Anal. Biochem. 1991;193(1):6. doi: 10.1016/0003-2697(91)90035-r. [DOI] [PubMed] [Google Scholar]
- 32.Costello CE, Comess KM, Plaziak AS, Bancroft DP, Lippard SJ. Fast-atom-bombardment and highperformance tandem mass-spectrometry of platinum(II) oligoribonucleotide fragments. Int. J. Mass Spectrom. Ion Processes. 1992;122:255. [Google Scholar]
- 33.Egger AE, Hartinger CG, Ben Hamidane H, Tsybin YO, Keppler BK, Dyson PJ. High Resolution Mass Spectrometry for Studying the Interactions of Cisplatin with Oligonucleotides. Inorg. Chem. 2008;47(22):10626. doi: 10.1021/ic801371r. [DOI] [PubMed] [Google Scholar]
- 34.Nyakas A, Eymann M, Schuerch S. The Influence of Cisplatin on the Gas-Phase Dissociation of Oligonucleotides Studied by Electrospray Ionization Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2009;20(5):792. doi: 10.1016/j.jasms.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 35.Nyakas A, Stucki SR, Schurch S. Tandem Mass Spectrometry of Modified and Platinated Oligoribonucleotides. J. Am. Soc. Mass Spectrom. 2011;22(5):875. doi: 10.1007/s13361-011-0106-z. [DOI] [PubMed] [Google Scholar]
- 36.McLuckey S, Van Berkel G, Glish G. Tandem Mass Spectrometry of Small, Multiply Charged Oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3(1):60. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 37.McLuckey SA, Habibi-Goudarzi S. Decompositions of multiply charged oligonucleotide anions. J. Am. Chem. Soc. 1993;115(25):12085. [Google Scholar]
- 38.Wan KX, Gross ML, Shibue T. Gas-phase stability of double-stranded oligodeoxynucleotides and their noncovalent complexes with DNA-binding drugs as revealed by collisional activation in an ion trap. J. Am. Soc. Mass Spectrom. 2000;11(5):450. doi: 10.1016/S1044-0305(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 39.Rosu F, Pirotte S, Pauw ED, Gabelica V. Positive and negative ion mode ESI-MS and MS/MS for studying drug–DNA complexes. Int. J. Mass Spectrom. 2006;253(3):156. [Google Scholar]
- 40.Smith SI, Brodbelt JS. Rapid characterization of cross-links, mono-adducts, and non-covalent binding of psoralens to deoxyoligonucleotides by LC-UV/ESI-MS and IRMPD mass spectrometry. Analyst. 2010;135(5):943. doi: 10.1039/b924023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce SE, Guziec LJ, Guziec FS, Brodbelt JS. Characterization of Aziridinylbenzoquinone DNA Cross-Links by Liquid Chromatography-Infrared Multiphoton Dissociation-Mass Spectrometry. Chem. Res. Toxicol. 2010;23(6):1097. doi: 10.1021/tx1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazzitelli CL, Brodbelt JS. Probing Ligand Binding to Duplex DNA Using KMnO4 Reactions and Electrospray Ionization Tandem Mass Spectrometry. Anal. Chem. 2007;79(12):4636. doi: 10.1021/ac070145p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groessl M, Tsybin YO, Hartinger CG, Keppler BK, Dyson PJ. Ruthenium versus platinum: interactions of anticancer metallodrugs with duplex oligonucleotides characterised by electrospray ionisation mass spectrometry. J. Biol. Inorg. Chem. 2010;15(5):677. doi: 10.1007/s00775-010-0635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan K, Gross M, Shibue T. Gas-phase stability of double-stranded oligodeoxynucleotides and their noncovalent complexes with DNA-binding drugs as revealed by collisional activation in an ion trap. J. Am. Soc. Mass Spectrom. 2000;11(5):450. doi: 10.1016/S1044-0305(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 45.Reyzer ML, Brodbelt JS, Kerwin SM, Kumar D. Evaluation of complexation of metal-mediated DNA-binding drugs to oligonucleotides via electrospray ionization mass spectrometry. Nucleic Acids Res. 2001;29(21):e103. doi: 10.1093/nar/29.21.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith SI, Guziec LJ, Guziec FS, Hasinoff BB, Brodbelt JS. Evaluation of relative DNA binding affinities of anthrapyrazoles by electrospray ionization mass spectrometry. J. Mass Spectrom. 2007;42(5):681. doi: 10.1002/jms.1205. [DOI] [PubMed] [Google Scholar]
- 47.Guan ZQ, Kelleher NL, Oconnor PB, Aaserud DJ, Little DP, McLafferty FW. 193 nm photodissociation of larger multiply-charged biomolecules. Int. J. Mass Spectrom. Ion Processes. 1996;157:357. [Google Scholar]
- 48.Wilson JJ, Brodbelt JS. Infrared Multiphoton Dissociation of Duplex DNA/Drug Complexes in a Quadrupole Ion Trap. Anal. Chem. 2007;79(5):2067. doi: 10.1021/ac061946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith SI, Brodbelt JS. Characterization of Oligodeoxynucleotides and Modifications by 193 nm Photodissociation and Electron Photodetachment Dissociation. Anal. Chem. 2010;82(17):7218. doi: 10.1021/ac100989q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zubarev RA, Kelleher NL, McLafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J. Am. Chem. Soc. 1998;120(13):3265. [Google Scholar]
- 51.Budnik BA, Haselmann KF, Zubarev RA. Electron detachment dissociation of peptide di-anions: an electron–hole recombination phenomenon. Chem. Phys. Lett. 2001;342(3-4):299. [Google Scholar]
- 52.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2004;101(26):9528. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper HJ, Hakansson K, Marshall AG. The role of electron capture dissociation in biomolecular analysis. Mass Spectrom. Rev. 2005;24(2):201. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- 54.Schultz KN, Hakansson K. Rapid electron canture dissociation of mass-selectively accumulated oligodeoxynucleotide dications. Int. J. Mass Spectrom. 2004;234(1-3):123. [Google Scholar]
- 55.Yang J, Mo JJ, Adamson JT, Hakansson K. Characterization of oligodeoxynucleotides by electron detachment dissociation Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2005;77(6):1876. doi: 10.1021/ac048415g. [DOI] [PubMed] [Google Scholar]
- 56.Smith SI, Brodbelt JS. Electron transfer dissociation of oligonucleotide cations. Int. J. Mass Spectrom. 2009;283:85. doi: 10.1016/j.ijms.2009.02.012. (Copyright (C) 2011 American Chemical Society (ACS). All Rights Reserved.)
- 57.Smith SI, Brodbelt JS. Hybrid Activation Methods for Elucidating Nucleic Acid Modifications. Anal. Chem. 2011;83(1):303. doi: 10.1021/ac102411a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brodbelt JS, Wilson JJ. Infrared multiphoton dissociation in quadrupole ion traps. Mass Spectrom. Rev. 2009;28(3):390. doi: 10.1002/mas.20216. [DOI] [PubMed] [Google Scholar]
- 59.Little DP, Aaserud DJ, Valaskovic GA, McLafferty FW. Sequence information from 42-108-mer DNAs (complete for a 50-mer) by tandem mass spectrometry. J. Am. Chem. Soc. 1996;118(39):9352. [Google Scholar]
- 60.Nyakas A, Blum LC, Stucki SR, Reymond J-L, Schurch S. OMA and OPA-Software-supported mass spectra analysis of native and modified nucleic acids. J. Am. Soc. Mass Spectrom. 2012 doi: 10.1007/s13361-012-0529-1. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.