Abstract
Psychophysics generally relies on estimating a subject’s ability to perform a specific task as a function of an observed stimulus. For threshold studies, the fitted functions are called psychometric functions. While fitting psychometric functions to data acquired using adaptive sampling procedures (e.g., “staircase” procedures), investigators have encountered a bias in the spread (“slope” or “threshold”) parameter that has been attributed to the serial dependency of the adaptive data. Using simulations, we confirm this bias for cumulative Gaussian parametric maximum likelihood fits on data collected via adaptive sampling procedures, and then present a bias-reduced maximum likelihood fit that substantially reduces the bias without reducing the precision of the spread parameter estimate and without reducing the accuracy or precision of the other fit parameters. As a separate topic, we explain how to implement this bias-reduction technique using generalized linear model fits as well as other numeric maximum likelihood techniques such as the Nelder-Mead simplex. We then provide a comparison of the iterative bootstrap and observed information matrix techniques for estimating parameter fit variance from adaptive sampling procedure data sets. The iterative bootstrap technique is shown to be slightly more accurate; however, the observed information technique executes in a small fraction (0.005%) of the time required by the iterative bootstrap technique, which is an advantage when a real-time estimate of parameter fit variance is required.
Keywords: Maximum likelihood estimation, generalized linear models, GLM, psychophysics, threshold, vestibular
Introduction
In earlier papers in this series, we presented how signal detection theory relates to the measurement of vestibular thresholds (Merfeld 2011) and presented an investigation of fitting vestibular threshold data across frequencies (Lim and Merfeld 2012). While we remain focused on vestibular applications, this paper focuses on a more general problem – how to perform psychometric function fits that yield less biased parameter estimates for adaptive sampling procedure data than existing approaches (e.g., Wichmann and Hill 2001a; Treutwein and Strasburger 1999).
In the statistical literature, bias reduction techniques have long been described (Quenouille 1956; Cox and Hinkley 1974; McCullagh and Nelder 1989; Firth 1993; Kosmidis and Firth 2010). Firth (1993) divided these bias reduction techniques into three categories. 1) Jackknife techniques that are computationally intensive (e.g., Quenouille, 1949, 1956), 2) a direct calculation approach that calculates an estimated bias and directly subtracts it (e.g., Cox and Hinkley 1974), and 3) an approach that shifts the score function so as to minimize the bias (Firth, 1993). Despite this long and fruitful history, to our knowledge, psychometric functions have never before been fit using bias-reduction techniques despite knowledge that the spread parameter demonstrated bias when data were acquired using adaptive test paradigms (Leek, Hanna and Marshall 1992; Treutwein and Strasburger 1999; Kaernbach 2001; Leek 2001).
To resolve this long-standing problem of biased parameter estimates, this paper applies bias-reduced fits to forced choice, binary, psychometric data collected using adaptive sampling procedures. In the main body of the paper, we present results after subtracting the bias at each iteration of a reweighted least squares procedure followed by a nonlinear transformation of the estimates. This technique removes the bias caused by adaptive sampling. We also present results obtained by numerically optimizing the modified scores (Online Resource 1) and obtain indistinguishable answers when the problem statement is identical. While generally applicable, we avoid jackknife approaches herein simply because we are interested in obtaining a bias-reduced parameter estimate in near real-time.
The estimates obtained from the bias-reduced algorithms are statistically more accurate (i.e., less biased) and equally precise (i.e., same variance) when compared to existing methods given a fixed number of trials. Fewer trials for a given level of accuracy and precision suggest that these methods utilize available data more efficiently. This is especially important for our vestibular application because motion stimuli take time to present. For example, if one wants to obtain data at a motion stimulus frequency of 0.05 Hz, then each trial will last at least 20 seconds. In this context, efficient use of available data is important to keep test length manageable, especially for the clinical setting where several different threshold assays (e.g., yaw rotation, y-translation, z-translation, etc.) may be required at multiple frequencies. If tests last too long, subjects can be less attentive, lapses can occur, and subjects can even fall asleep.
In the first section of the results (“Accuracy of parameter estimates”), we confirm that for adaptive sampling procedures the maximum likelihood estimate (MLE) on a psychometric function’s spread parameter (σ) provided by both generalized linear model (GLM) fits as well as other numerical maximum likelihood methods are downwardly biased (Kaernbach 2001; Leek 2001). This inaccuracy reduces as the number of trials (n) increases, but its impact is substantial for fewer than 200 trials. For example, when n = 50, we measure a parameter estimate bias on the spread parameter (σ) that underestimates its value on average by 30% of its standard deviation. We demonstrate how to correct for this parameter bias by eliminating the first order asymptotic parameter bias term that generally diminishes with 1/n. Correcting for this parameter bias is important as it enables the direct comparison of data sets obtained using different sampling procedures and different termination criteria (e.g. different numbers of trials).
Experimenters may also want to know the variance associated with the estimated parameters for each individual data set. In the past, observed information and iterative bootstrap techniques have been employed to measure the variance on parameter estimates (McKee et al. 1985; Wichmann and Hill 2001b) for data acquired using non-adaptive sampling test paradigms. In the second section of the results (“Estimating the precision of parameter estimates”), we show how to apply these methods to data collected via adaptive sampling procedures. We also demonstrate that the observed information technique – while slightly less accurate than the iterative bootstrap technique – can be executed in a fraction (0.005%) of the time.
Methods
The psychometric function
The psychometric function assumptions and notation used in this paper are the same as those used in earlier papers in this series (Merfeld 2011; Lim and Merfeld 2012). Specifically, the psychometric function is assumed to be a cumulative Gaussian distribution. Subject responses are binary and are 0 for negative responses (e.g., I perceive I moved to the right) and 1 for positive responses (e.g., I perceive that I moved to the left). The linear translation of the psychometric function along the abscissa is referred to as the “psychometric bias” or “vestibular bias” and is represented mathematically as μ. This value corresponds to the point of subjective equality (PSE) where the subject’s likelihood of responding with a 0 or 1 is equal and also serves as the decision boundary for a one-interval direction-recognition binary response task (Merfeld 2011). This “vestibular bias”, μ, should not be confused with the parameter bias, which describes the inaccuracy of psychometric function parameter estimates. To help distinguish between these two terms, we refer to parameter bias as “bias” and vestibular bias using μ from this point onward. The spread of the psychometric function, which can be directly related to the function’s slope, will be characterized by σ. For vestibular applications, this parameter corresponds to the standard deviation of the equivalent physiological noise (Merfeld 2011), which is often referred to as the threshold. Our estimates of μ and σ will be represented by μ̂ and σ̂ respectively.
Mathematically, the following equation defines our psychometric function (ψ):
where Φ is the cumulative standard Gaussian distribution and z is a “dummy” integration variable. Here, we also introduce the probit model parameterization of ψ that will be used to fit the psychometric function via a generalized linear model (GLM) fit.
where b = [b1 b2]T are the GLM fit parameters that characterize the psychometric function. Through some simple algebra it can be shown that μ = − b1/b2 and σ = 1/b2 (Dobson and Barnett 2008). For simplicity, we choose not to include a lapse rate (Wichmann and Hill 2001a) or nonlinear asymmetry (Roditi and Crane 2012).
Psychometric function fits
For all of our analyses herein, we implement maximum likelihood estimation by fitting the psychometric function, ψ, to binary data using a generalized linear model (GLM) (Dobson and Barnett 2008; McCullagh and Nelder 1990). We choose to work with GLM fits as they provide a unifying framework for many commonly used statistical techniques (Dobson and Barnett 2008), and such a framework provides insights on problems that are not provided by Nelder-Mead or other numerical fit algorithms. In fact, such an insight provided by GLM fits specifically led to the bias reduction methods reported herein. Furthermore, GLM fits are often used to fit psychometric functions (e.g. Knoblauch and Maloney 2008; Lim and Merfeld 2012, Yssaad-Fesselier & Knoblauch 2006; Zupan and Merfeld 2008), and are a natural choice for our vestibular application where subject responses are binary (e.g., Did I move left or right?) and the psychometric function ranges from 0% to 100%. We use the function glmfit.m from the statistics toolbox in MATLAB R2011b with a binomial response distribution and the probit link to implement the GLM fits.
We have also implemented this bias reduction approach using another numerical maximum likelihood technique (Nelder-Mead simplex), which yielded answers identical to the GLM approach. We show the calculations underlying this method in Online Resource 1.
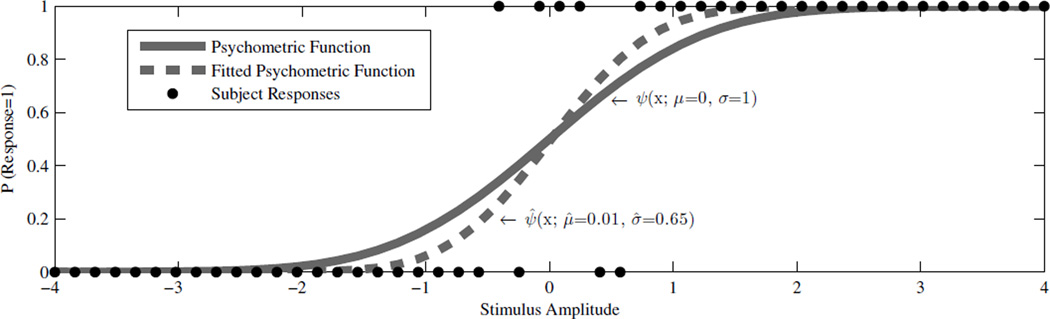
The following example demonstrates how a psychometric function can be fit to binary response data and is included to provide visual intuition for such fits. Consider a subject with a psychometric function that has a vestibular bias, μ, of 0 and spread, σ, of 1. We simulated an experiment on this subject with 50 stimuli spaced uniformly from −4σ to +4σ (Fig. The binary responses, actual psychometric function and fitted psychometric function are provided in Fig. 1. As will be discussed in detail later, this is an example where the estimated fit underestimates the spread (σ) and thus the slope is too steep at the midpoint.
Fig. 1.
Simulation of an experiment on a subject with μ = 0 and σ = 1 with 50 stimuli spaced uniformly from −4σ to +4σ. The black dots show the subject’s binary response (0 or 1) to each stimulus. In this experiment the subject responds 0 if they perceive a negative stimulus and 1 if they perceive a positive stimulus. The solid line is the actual underlying psychometric function and the broken line is the maximum likelihood Gaussian fit to the data
Spread parameter bias
Adaptive sampling procedures are procedures in which the physical stimulus of each trial is determined by the responses and stimuli of the previous trial or sequence of trials. These procedures were originally developed to increase the efficiency and robustness of psychophysical measurements (for review, see Leek 2001). Initially, they focused on measuring a single point on the psychometric function, called the threshold, but in the past few decades, researchers have started to estimate other psychometric function characteristics such as the slope, lapse rate and asymmetry (Treutwein and Strasburger 1999; Leek 2001; Wichmann and Hill 2001a; Roditi and Crane 2012). When using these methods to measure the slope of the psychometric function, researchers found that, for an experiment with a small number of trials, their estimates taken near the midpoint of the psychometric function were, on average, too steep (Leek et al. 1992; Treutwein and Strasburger 1999). While a complete explanation of this parameter estimation bias is complex and not precisely understood, it has been shown that a major component of the bias is caused by the serial dependency of the adaptive data (Kaernbach 2001; Klein 2001). Simply stated, adaptive sampling procedures only allow for certain, correlated experimental configurations (i.e. stimulus and response vector pairings) and, for data sets with a small number of trials, these configurations result in biased slope estimates.
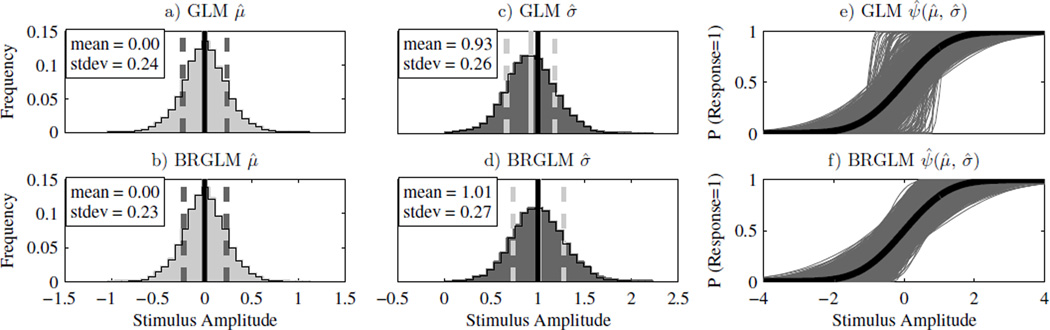
The slope at the midpoint of a psychometric function can be related to the spread parameter. For our cumulative normal psychometric function the slope at the midpoint is . Thus, when adaptive data causes the slope at the midpoint to be upwardly biased (too large), the spread parameter, σ, will be downwardly biased (too small). For example, we find a 7.5% downward bias on σ̂ when estimates are obtained using maximum likelihood fits on 10,000 simulated 100 trial long, 3-Down/1-Up adaptive staircase data sets (Fig. 2c).
Fig. 2.
GLM and BRGLM μ̂ and σ̂ distributions for a 3-down/1-up staircase with n = 50 trials, μ = 0 and σ = 1. Panels a) and c) show the histograms for μ̂ and σ̂ using GLM fits. Panels b) and d) show the histograms for μ̂ and σ̂ using BRGLM fits. In panels a–d), the solid black line is the actual parameter value, the solid gray line is the mean of the parameter estimates, and the dashed gray lines indicate one standard deviation either side of the mean. Panels e) and f) show the GLM and BRGLM psychometric function fits for all 10,000 data sets in gray, and the actual psychometric function in black
In this paper the severity of the parameter estimate bias will be divided into three categories. This categorization is used to provide a graphical view of simulation results when presented as a table. A highly biased estimator (Category III) will be defined as an estimator that is biased by more than 25% of its standard deviation. This categorization comes from a common rule of thumb in statistics that defines a poor estimator as one that is biased by more than 25% of its standard deviation (Efron and Tibshirani 1993). An estimator with an inconsequential bias (“unbiased”, Category I) will be defined as an estimator that is biased by less than 10% of its standard deviation, and a moderately biased estimator (Category II) will be defined as an estimator that is biased between 10% and 25% of its standard deviation. The estimator, σ̂, in Fig. 2c has a downward bias that is 7.5% of σ and a standard deviation that is 25.8% of σ. Therefore, in this example, σ̂ is biased by 29% of its standard deviation and is classified as highly biased (Category III). To avoid confusion we emphasize that in this paper σ refers to the spread parameter of the psychometric function and not the standard deviation of a parameter estimate.
A number of solutions have been offered to remove or reduce this bias. Hall suggested that one could calculate the expected percent size of the bias in advance and remove it by multiplying the biased estimate by a scale factor (1981). One problem with this method is that, when the estimate is downwardly biased, as for the spread parameter (σ), the multiplier is greater than one, which causes the variance on the estimated spread parameter to increase. We will demonstrate that, by using bias-reduced maximum likelihood estimation (McCullagh and Nelder 1989; Firth 1993; Kosmidis 2007, Kosmidis and Firth 2010), we can correct for the downward bias on σ̂ without significantly increasing the variance on μ̂ or σ̂, and without decreasing the accuracy on μ̂. Furthermore, standard maximum likelihood estimation can lead to asymmetric distributions when the number of trials is small and/or the vestibular bias (μ) is large. This characteristic also improves with biased-reduced maximum likelihood estimation.
In contrast, when an estimate is upwardly biased, we note that the scale factor method will require a multiplier less than one, which will actually reduce both the bias and variance of the estimate. We will use this fact, in combination with bias-reduced maximum likelihood estimation, to further improve both the accuracy and precision of spread estimates from very small data sets (circa 25 trials).
Bias-reduced maximum likelihood estimation
In experiments with a large number of trials the bias on σ̂ is negligible compared to the variance. In experiments with fewer trials the bias becomes significant as illustrated in the previous section. We can improve our estimation by removing the order n−1 term of the asymptotic bias expansion from the maximum likelihood estimate (Firth 1993). While it is not clear how this asymptotic bias is related to the bias caused by the serial dependency of the adaptive data, we have repeatedly found (reported in results herein) that when using any of several adaptive test protocols, bias reduction on b̂1 and b̂2 followed by a nonlinear transformation to μ̂ and σ̂ leads to unbiased μ̂ and σ̂ estimates. Bias reduction directly on μ̂ and σ̂ did not produce unbiased estimates (see Online Resource 1) and therefore the nonlinear transformation, surprisingly, appears to be a beneficial step for this application.
We utilize two methods for implementing bias-reduced estimation. The first method is applicable to GLM routines and the second can be applied to constrained numeric maximum likelihood fits like the ones described by Wichmann and Hill (2001a) and Treutwein and Strasburger (1999).
Bias-reduced generalized linear model
To implement the bias-reduced generalized linear model (BRGLM) routine, we modified the MATLAB function glmfit.m to create a new MATLAB function brglmfit.m. MATLAB code implementing the bias-reduced GLM fit is available upon request. In this modified function, during each iteration of the GLM reweighted least squares algorithm, the order n−1 asymptotic bias term is calculated and subtracted from our coefficient parameter estimate, b̂. The order n−1 asymptotic bias term is calculated using the following formula (McCullagh and Nelder 1989):
where X is the stimulus vector (with a first column of ones if the constant b1 term is to be included in the model) and W is the quadratic weights vector (diagonalized into a matrix) which is inversely related to the variance of subject’s binary responses. For non-canonical models such as the probit link, the components of ξ are given by:
where are the derivatives of the GLM link function (ui),mi = b̂1 + b̂2xi and Qii are the diagonal elements of Q = X(XTWX)−1XT. For the probit model, the link function is the cumulative standard Gaussian distribution [ui = ψi = Φ(mi) = Φ(b̂1 + b̂2xi)], and therefore ξi = Qiimi/2 (McCullagh and Nelder 1989).
Bias-reduction in numeric maximum likelihood fits
The above approach to bias reduction applies to GLM fits only. In Online Resource 1, we show how to find bias-reduced psychometric function parameter estimates for other numeric methods that calculate maximum likelihood fits (e.g. Wichman and Hill 2001a). When implemented, these calculations yielded fits identical to those presented for GLM fits.
Parameter estimate variance: the iterative bootstrap technique
The iterative bootstrap technique has been popularized in the last decade by Wichmann and Hill in the context of estimating confidence intervals on fitted psychometric function parameters. In their paper, Wichmann and Hill describe a bootstrap method that relies on a large number of simulated repetitions of the original experiment (2001b). These Monte Carlo simulations bootstrap to the original data set’s psychometric function parameter estimates and use these estimates to re-simulate the subject’s responses. This technique can be applied to adaptive sampling procedures as we show herein. However, for adaptive sampling procedures, it is crucial to re-simulate only the subject’s response vector, y, to the experimentally observed stimulus vector, x, and not to re-simulate the entire experiment – i.e. both the stimulus vector and the subject’s response vector. This is a meaningful distinction only for adaptive sampling procedures as non-adaptive protocols would, by definition, never alter the stimuli. Since the stimulus vector is fixed, we fit the simulated bootstrap data sets using a standard (i.e. non-bias-reduced) maximum likelihood technique because the bias derived from the serial dependency of adaptive data is not present for non-adaptive (fixed) data sets. Indeed, preliminary simulations confirmed that fitting the bootstrap data with a standard maximum likelihood technique yielded, on average, more accurate variance estimates than when fitting the bootstrap data using bias-reduced maximum likelihood estimation.
Furthermore, the actual variance to which our variance estimates will be compared can be calculated by conducting an iterative bootstrap simulation evaluated at the actual psychometric function parameters.
Parameter estimate variance: the observed information matrix technique
As will be shown, the observed information technique (Casella and Berger 2001) of measuring parameter estimate variance is slightly less accurate on average than the bootstrap method for experiments with a small number of trials. However, it executes much faster making it preferable when trying to get a measure of variance in real-time (e.g. after every trial). The purpose of including the observed information technique in this paper is to provide a quantitative comparison of its accuracy and execution time to the iterative bootstrap technique.
The observed information technique estimates the parameter estimate covariance matrix by taking the inverse of the observed information matrix—the negative Hessian matrix of the log-likelihood function—evaluated at the estimated parameter values. It should be noted that this method is related to, but not the same as, the asymptotic technique described by Foster and Bischof (1991) which only considers the inverse of the diagonal terms in the observed information matrix, thereby ignoring the off-diagonal terms, as a measure of the variance. Preliminary simulations showed that the full observed information matrix approach is more accurate and robust when n is small compared to this alternate asymptotic technique, and so we proceed using the complete observed information matrix. The observed information matrix was calculated with respect to the (μ, σ) parameterization of the log likelihood function (see Online Resource 1).
Simulations
Accuracy of parameter estimates
To test our bias reduction, we conducted Monte Carlo simulations. Three different adaptive sampling procedures and one non-adaptive sampling procedure were used to generate data sets. For our adaptive sampling procedures we used a 3-Down/1-Up (3D/1U) PEST (parameter estimation by sequential testing) staircase, a 4-Down/1-Up (4D/1U) PEST staircase, and a novel maximum likelihood (MLE) procedure.
An N-Down/M-Up staircase decreases in stimulus magnitude after N correct responses at one level and increases in magnitude after M incorrect responses at one level. The size of the change in stimulus magnitude is determined using parameter estimation by sequential testing (PEST) rules developed by Taylor and Creelman (1967):
After each reversal, halve the step size
A step in the same direction as the last uses the same step size, with the following exception.
A third step in the same direction uses a doubled step size. Each additional step in the same direction is also doubled with the following exception.
If a reversal immediately follows a step doubling, then one extra same size step is taken before doubling.
Minimum and maximum step sizes are specified. The magnitudes of the minimum and maximum step sizes were chosen to be 0.38dB (1.25log10(2)) and 6.02dB (20log10(2)) respectively.
A 3-Down/1-Up PEST staircase targets a correct response rate of 79.4% while a 4-Down/1-Up PEST staircase targets a correct response rate of 84.1% (Leek 2001).
Our novel maximum likelihood procedure begins with a 2-Down/1-Up PEST staircase until the data can be fit using the function glmfit.m without triggering a statistical warning from MATLAB (circa 16±4 trials). Then, for each subsequent trial the previous data are fit and the next stimulus is chosen based on a targeted 90% correct response rate. A 90% correct response rate was chosen because it is a level where a substantial amount of information can be obtained on both μ and σ; however, for the purposes of this paper, any percent correct response rate could have been targeted. All adaptive sampling procedures started at a stimulus level of 8σ.
Our non-adaptive sampling procedure uses a fixed number of trials such that 12% of the trials were at ±1.5σ, 40% were at ±1σ, 36% were at ±0.75σ, and 12% were at ±0.5σ. For example, when the total number of trials was 50, there were 6 trials at ±1.5σ, 20 trials at ±1σ, 18 trials at ±0.75σ, and 6 trials at ±0.5σ, with an equal number of positive and negative trials at each level.
The simulations were performed with n=50, 100 and 200 trials for both μ = 0 and μ = 0.5σ. Each procedure was simulated to create 10,000 data sets for each setting of n and μ. In total, we have 2 (μ = 0 and μ = 0.5σ) × 4 (3D/1U, 4D/1U, MLE, Non-Adaptive) × 3 (n = 50, 100 and 200) × 10,000 = 240,000 data sets. Each data set was fit using a GLM fit and a BRGLM fit.
Estimating the precision of parameter estimates
Both the iterative bootstrap technique and the observed information technique were tested using a 3-Down/1-Up PEST staircase procedure. The staircase procedure was simulated with n=50, 100 and 200 trials for both μ = 0 and μ = 0.5σ. 10,000 data sets were simulated for each seting of n and μ. The parameter estimates for each simulated data set were calculated using a BRGLM fit. From these parameter estimates, the estimated and actual variance for each data set was calculated using the approaches described in the previous section. For the iterative bootstrap technique each data set was re-simulated 2000 times to match the bootstrap technique used by Wichmann and Hill (2001b).
Simulation hardware and software
Simulations were implemented in MATLAB R2011b (The Mathworks, Inc, Massachusetts) on the Harvard Orchestra computation cluster. Simulations were run on an IBM BladeCenter HS21 XM with a 3.16 GHz Xeon processor and 8 GB of RAM.
Results
Accuracy of parameter estimates
We primarily report the means and standard deviations of the μ̂ and σ̂ parameter estimates obtained via generalized linear model (GLM) and bias-reduced GLM (BRGLM) fits. It is important to clearly state that in this first section of the results the standard deviations of the parameter estimates represent the overall parameter estimate precision associated with the fit in combination with the testing procedure (e.g., staircase, maximum likelihood, or non-adaptive) used to obtain the data set.
Fig. 2 compares the GLM fit and BRGLM fit parameter estimate distributions of μ̂ and σ̂ when n = 50 trials, μ = 0, σ = 1 and the 3-Down/1-Up adaptive staircase procedure was used to generate data sets. The GLM fit σ̂ distribution (Fig. 2c) is highly biased (category III) while the BRGLM fit (Fig. 2d) is unbiased (category I), causing the GLM fit to produce more psychometric function estimates with slopes that are too steep at the midpoint (Fig. 2e and Fig. 2f). We also notice that both μ̂ distributions (Fig. 2a and Fig. 2b) are unbiased (category I) and nearly identical, and that the standard deviations of the two σ̂ distributions are almost the same. Note that the parameter estimates in this and subsequent figures are in “units” of stimulus amplitude.
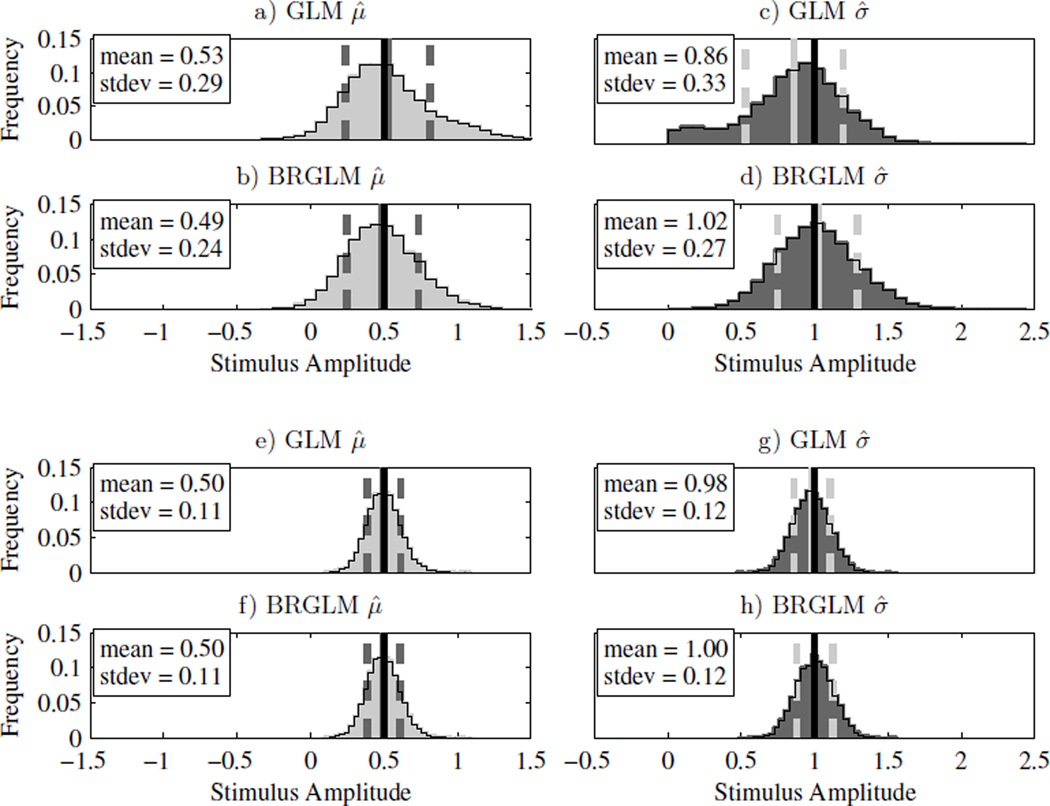
Fig. 3a–d) show the fitted parameter distributions with the same simulation parameter values as above but with a vestibular bias (μ) of 0.5σ instead of zero. Note that all results shown scale with μ and σ. For example simulation results with μ = 1 and σ = 2 are scaled versions of simulations with μ = 0.5 and σ = 1. We see that both the μ̂ (Fig. 3a) and σ̂ (Fig. 3c) GLM estimates have asymmetric distributions while the BRGLM estimates (Fig. 3b and Fig. 3d) are relatively symmetric and bell-shaped. In this case, the magnitude of the skewness of the distributions, a normalized statistical measure of this asymmetry, is reduced by about a factor of 2 – from 0.59 to 0.27 for μ̂ and from 0.41 to 0.22 for σ̂ – when bias reduction is used. Furthermore, the GLM fit μ̂ distribution (Fig. 3c) is highly biased (category III) and has a standard deviation that is 33% the size of σ, while the BRGLM fit σ̂ distribution (Fig. 3d) is unbiased (category I) and has a smaller standard deviation that is 27% of σ. Additionally, both μ̂ distributions (Fig. 3a and Fig. 3b) are unbiased (category I) but the BRGLM μ̂ distribution (Fig. 3b) has a smaller standard deviation.
Fig. 3.
GLM and BRGLM μ̂ and σ̂ distributions for a 3-down/1-up staircase with μ = 0.5σ and σ = 1 for n = 50 trials (panels a–d) and n = 200 trials (panels e – h). Panels a, c, e and g) show the histograms for μ̂ and σ̂ using GLM fits. Panels b, d, f and h) show the histograms for μ̂ and σ̂ using BRGLM fits. The solid black line is the actual parameter value, the solid gray line is the mean of the parameter estimates, and the dashed gray lines indicate one standard deviation either side of the mean
Fig. 3e–h) show the fitted parameter distributions with the same simulation settings as in Fig. 3a–d) except now the number of trials has increased from 50 to 200. The GLM σ̂ distribution (Fig. 3g) is moderately biased (category II) while the BRGLM fit distribution (Fig. 3h) is unbiased (category I). Furthermore, both have approximately the same standard deviation. Both the μ̂ distributions (Fig. 3e and Fig. 3f) are also unbiased (category I) and almost identical.
Up to this point, for all the adaptive sampling procedure simulations, the BRGLM estimates were clearly better – both less biased and less skewed with equal variance – than the corresponding GLM estimates. Fig. 4 presents the case where we have used a non-adaptive sampling procedure with 100 trials and μ = 0. Both σ̂ distributions are asymmetric (the skewness is 0.74 for GLM and 0.75 for BRGLM), and the BRGLM σ̂ distribution (Fig. 4d) is moderately biased (category II), while the GLM σ̂ distribution (Fig. 4c) is unbiased (category I). Both μ̂ distributions (Fig. 4a and Fig. 4b) are unbiased and almost identical. Note, however, that the peak of the GLM σ̂ distribution (Fig. 4c) occurs at a value less than 1, while the peak of the BRGLM σ̂ distribution (Fig. 4d) occurs nearer to 1.
Fig. 4.
GLM and BRGLM μ̂ and σ̂ distributions for a non-adaptive sampling procedure with n = 100 trials, μ = 0 and σ = 1. Panels a) and c) show the histograms for μ̂ and σ̂ using GLM fits. Panels b) and d) show the histograms for μ̂ and σ̂ using BRGLM fits. The solid black line is the actual parameter value, the solid gray line is the mean of the parameter estimates, and the dashed gray lines indicate one standard deviation either side of the mean
Table 1 lists the means and standard deviations for all simulations, including the illustrative examples shown in Fig. 2, 3 and 4. The key result is that in all cases that use an adaptive sampling procedure, the GLM σ̂ estimates are more biased than the BRGLM estimates. We also notice that both GLM and BRGLM μ̂ estimates are always unbiased. Furthermore, for adaptive sampling procedures with a small number of trials (n = 50) and vestibular bias (μ = 0.5σ), the standard deviations of BRGLM estimates are significantly smaller than those of the GLM estimates. Otherwise the standard deviations of the two techniques are more or less equivalent.
Table 1.
GLM and BRGLM μ̂ and σ̂ means±(standard deviations) for all simulations
| μ | # of Trials | Procedure | GLM: μ̂ | BRGLM: μ̂ | GLM: σ̂ | BRGLM: σ̂ |
|---|---|---|---|---|---|---|
| μ = 0 |
n = 50 |
3D/1U | 0.00±(0.24) | 0.00±(0.23) | 0.93±(0.26) | 1.01±(0.27) |
| 4D/1U | 0.01±(0.30) | 0.01±(0.25) | 0.91±(0.26) | 1.02±(0.25) | ||
| MLE | 0.00±(0.24) | 0.00±(0.24) | 0.93±(0.24) | 1.00±(0.26) | ||
| Non-Adaptive | 0.00±(0.22) | 0.00±(0.21) | 1.00±(0.27) | 1.06±(0.27) | ||
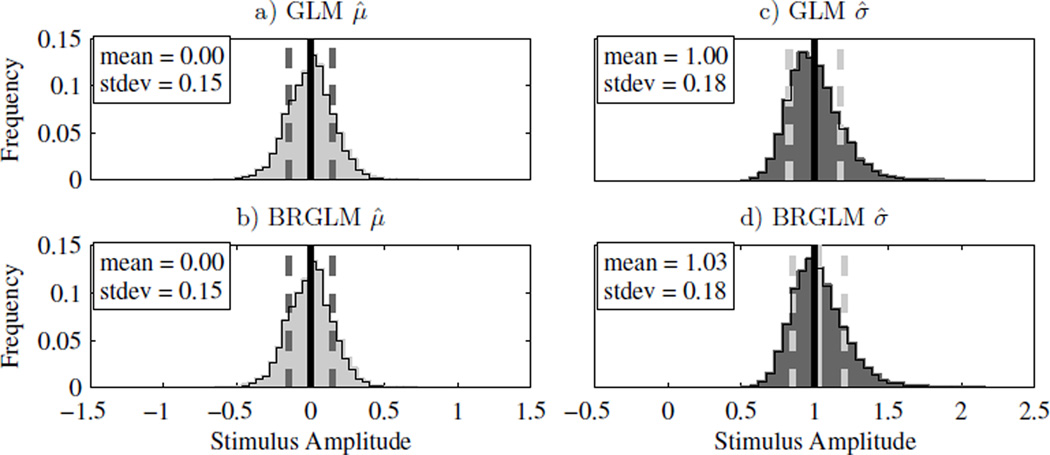
| n = 100 | 3D/1U | 0.00±(0.15) | 0.00±(0.15) | 0.97±(0.17) | 1.00±(0.18) | |
| 4D/1U | 0.00±(0.17) | 0.00±(0.17) | 0.97±(0.16) | 1.00±(0.16) | ||
| MLE | 0.00±(0.17) | 0.00±(0.17) | 0.97±(0.15) | 1.00±(0.16) | ||
| Non-Adaptive | 0.00±(0.15) | 0.00±(0.15) | 1.00±(0.18) | 1.03±(0.18) | ||
| n = 200 | 3D/1U | 0.00±(0.10) | 0.00±(0.10) | 0.98±(0.12) | 1.00±(0.12) | |
| 4D/1U | 0.00±(0.11) | 0.00±(0.11) | 0.99±(0.11) | 1.00±(0.11) | ||
| MLE | 0.00±(0.12) | 0.00±(0.12) | 0.99±(0.10) | 1.00±(0.10) | ||
| Non-Adaptive | 0.00±(0.10) | 0.00±(0.10) | 1.00±(0.12) | 1.01±(0.12) | ||
| μ = 0.5σ |
n = 50 |
3D/1U | 0.53±(0.29) | 0.49±(0.24) | 0.86±(0.33) | 1.02±(0.27) |
| 4D/1U | 0.59±(0.37) | 0.48±(0.26) | 0.80±(0.37) | 1.05±(0.26) | ||
| MLE | 0.49±(0.25) | 0.49±(0.24) | 0.91±(0.27) | 0.99±(0.28) | ||
| Non-Adaptive | 0.52±(0.24) | 0.51±(0.24) | 0.99±(0.30) | 1.07±(0.29) | ||
| n = 100 | 3D/1U | 0.50±(0.17) | 0.49±(0.16) | 0.95±(0.19) | 1.00±(0.18) | |
| 4D/1U | 0.52±(0.21) | 0.50±(0.18) | 0.94±(0.21) | 1.01±(0.17) | ||
| MLE | 0.50±(0.17) | 0.50±(0.17) | 0.97±(0.16) | 1.00±(0.16) | ||
| Non-Adaptive | 0.51±(0.16) | 0.51±(0.16) | 1.00±(0.19) | 1.03±(0.19) | ||
| n = 200 | 3D/1U | 0.50±(0.11) | 0.50±(0.11) | 0.98±(0.12) | 1.00±(0.12) | |
| 4D/1U | 0.50±(0.12) | 0.50±(0.12) | 0.98±(0.12) | 1.00±(0.12) | ||
| MLE | 0.50±(0.12) | 0.50±(0.12) | 0.99±(0.10) | 1.00±(0.10) | ||
| Non-Adaptive | 0.50±(0.11) | 0.50±(0.11) | 1.00±(0.13) | 1.02±(0.13) | ||
Category I Bias (<10% of stdev) ![]()
![]()
Bias correction for fewer than 50 trials
Simulations were also run for a 3D/1U staircase procedure with n = 25 trials (Table 2). We see that when BRGLM fits were used and μ was set to 0, the bias to standard deviation ratio on σ̂ was reduced from 61% to 19%, the standard deviation of σ̂ was reduced from 0.46 to 0.42, and the standard deviation on μ̂ was reduced from 0.51 to 0.37. Similarly, when BRGLM fits were used and μ was set to 0.5σ, the bias to standard deviation ratio on σ̂ was reduced from 80% to 34%, the bias to standard deviation ratio on μ̂ was reduced from 22% to 8%, the standard deviation of σ̂ was reduced from 0.50 to 0.41, and the standard deviation on μ̂ was reduced from 0.51 to 0.38.
Table 2.
GLM and BRGLM μ̂ and σ̂ means±(standard deviations) for a 3D/1U staircase procedure, n =25 and σ = 1
| μ | GLM: μ̂ | BRGLM: μ̂ | GLM: σ̂ | BRGLM: σ̂ |
|---|---|---|---|---|
| μ = 0 | 0.00±(0.51) | 0.00±(0.37) | 0.72±(0.46) | 1.08±(0.42) |
| μ = 0.5σ | 0.61±(0.51) | 0.47±(0.38) | 0.60±(0.50) | 1.14±(0.41) |
Category I Bias (<10% of stdev) ![]()
![]()
Since the biased-reduced σ̂ estimates in Table 2 are positively biased, we can use a scale factor (α) to improve both the accuracy (reduced bias) and precision (lower variance) of our estimates. To determine the value of α, we utilized Monte Carlo simulations to calculate the expected bias on the BRGLM σ̂ estimates (E[σ̂] – σ) for different values of σ (σ = 0.1 to 4) with μ set to 0. For this range, the expected bias scales linearly with σ (E[σ̂] – σ = 0.081σ) and is independent of the starting stimulus relative to σ.
Similarly, if we let μ be non-zero by setting it equal to kσ, where k is a constant, the expected bias still scales linearly with σ, but the size of the scaling increases non-linearly with increasing values of |k| (Online Resource 2). For the vestibular system |μ| is typically small and much less than σ (Crane 2012). For this range, the scale factor is almost constant as a function of |k|, and consequently, we can use the scale factor that we calculated for k = 0 as a good approximation. However, if the range of k were larger, we could take this nonlinearity into account to improve our scale factor estimates (Online Resource 2). Therefore, for |k| ≤ 0.5, we can use ασ̂ = (1.081)−1 σ̂ = 0.925σ̂ to remove the bias on σ̂. In a similar manner we calculated a scale factor of α = (0.724)−1 = 1.382 for the GLM fit σ̂ estimates. Table 3 shows the results of scaling the BRGLM σ̂ estimates by α = 0.925 and the GLM σ̂ estimates by α = 1.382 for σ = 1 and μ = 0, 0.2σ and 0.5σ. Note that the standard deviation for σ̂ is 40–45% lower when the bias-correction scaling is performed after a BRGLM fit (0.39, 0.39, 0.38) than when bias-correction scaling is performed after a GLM fit (0.63, 0.65, 0.69).
Table 3.
GLM σ̂, GLM ασ̂, BRGLM σ̂ and BRGLM ασ̂ means±(standard deviations) for a 3D/1U staircase procedure and n =25
| μ | GLM: σ̂ | GLM: ασ̂|α = 1.382 | BRGLM: σ̂ | BRGLM: α σ̂|α = 0.925 |
|---|---|---|---|---|
| μ = 0 | 0.72±(0.46) | 1.00± (0.63) | 1.08±(0.42) | 1.00±(0.39) |
| μ = 0.2σ | 0.70±(0.47) | 0.97±(0.65) | 1.09±(0.42) | 1.01±(0.39) |
| μ = 0.5σ | 0.59±(0.50) | 0.82±(0.69) | 1.14±(0.41) | 1.05±(0.38) |
Category I Bias (<10% of stdev) ![]()
![]()
The key results in Table 3 are: 1) the scaled GLM estimates (ασ̂) are less biased but have larger standard deviations than the GLM σ̂ estimates, and 2) the scaled BRGLM estimates (ασ̂) are less biased and have smaller standard deviations than all other estimates. We also note that a similar technique could be used to improve the non-adaptive BRGLM σ̂ estimates in Table 1 which demonstrate a similar overestimation to that demonstrated in Tables 2 and 3.
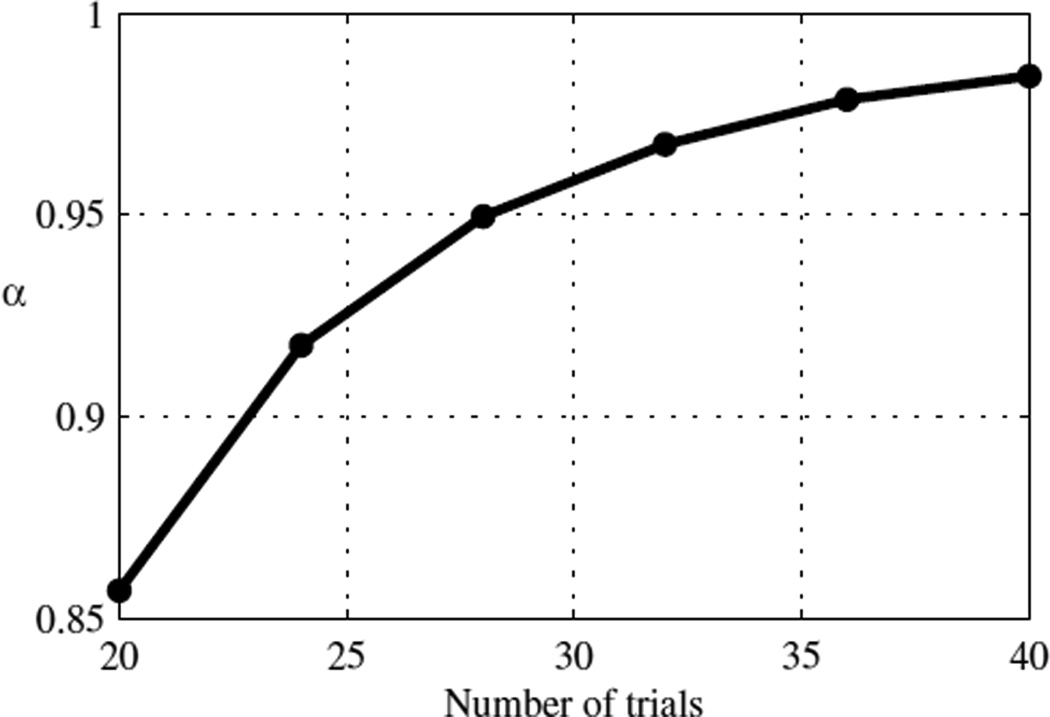
We emphasize that a different α has to be calculated if the number of trials or the adaptive sampling procedure used is changed. Fig. 5 shows the value of α for n = 20 to 40 for the 3D/1U staircase.
Fig. 5.
The scale factor, α, calculated with μ = 0 for BRGLM σ̂ estimates collected via a 3D/1U staircase for n = 20 to n = 40
Additional simulations to verify generality of results
We performed simulations with several different procedures and assumptions to confirm the generality of our results. Specifically, we re-simulated bias-reduction for each of the cases reported in Table 1 with 1) an initial stimulus of 1σ instead of 8σ, 2) a psychometric function and fitting procedure based on a logistic distribution instead of a Gaussian distribution, 3) an alternate ending criteria that terminated the staircase procedure after 5 minima (circa 52±9 trials), and 4) another ending criteria that terminated an adaptive sampling procedure when the coefficient of variation (CV) on the σ parameter estimate reached 0.25 (circa 57±10 trials). For this alternate ending criteria, the observed information technique was used to calculate the coefficient of variation (i.e. the ratio of the estimated standard deviation of σ̂i to σ̂i, where σ̂i was the bias-reduced estimate of σ for the ith data set).
In Table 4, we report our results for the above 4 cases using a 3D/1U staircase procedure with an underlying psychometric function with no vestibular bias (μ) and a physiological noise standard deviation (σ) of 1. We see that bias-reduction works with each of the four changes. The results in table 4 are representative of all the results we obtained for each of the different simulation scenarios reported in Table 1.
Table 4.
Simulation results for a 3D/1U staircase procedure with no vestibular bias (μ) and a physiological noise standard deviation (σ) of 1 for four alternative assumptions: 1) an initial stimulus of 1σ instead of 8 σ, 2) a psychometric function and fitting procedure based on a logistic distribution instead of a Gaussian distribution, 3) an alternate ending criteria that terminated the staircase procedure after 5 minima, and 4) another ending criteria that terminated the adaptive sampling procedure when the CV (see text) on the σ parameter estimate reached 0.25.
| GLM: μ̂ | BRGLM: μ | GLM: σ̂ | BRGLM: σ̂ | |
|---|---|---|---|---|
| 1) Initial stimulus at 1σ (n = 50) | 0.00±(0.21) | 0.00±(0.21) | 0.94±(0.24) | 1.00±(0.25) |
| 2) Logistic distribution (n = 50) | 0.00±(0.23) | 0.00±(0.22) | 0.93±(0.27) | 1.02±(0.29) |
| 3) End criteria: 5th minima (n = 52±9) | 0.00±(0.25) | 0.00±(0.23) | 0.93±(0.26) | 1.01±(0.27) |
| 4) End criteria: CV (n = 57±10) | 0.00±(0.19) | 0.00±(0.19) | 0.94±(0.23) | 0.99±(0.23) |
Category I Bias (<10% of stdev) ![]()
![]()
Estimating the precision of parameter estimates
We are interested in calculating and characterizing the error of the iterative bootstrap and observed information techniques. The error was calculated for each data set by taking the square root of the variance estimate using either the observed information or iterative bootstrap technique and then subtracting that value from the square root of the actual variance for that data set. Thus if the error were positive, the variance estimate was too large and if the error were negative, the variance estimate was too small.
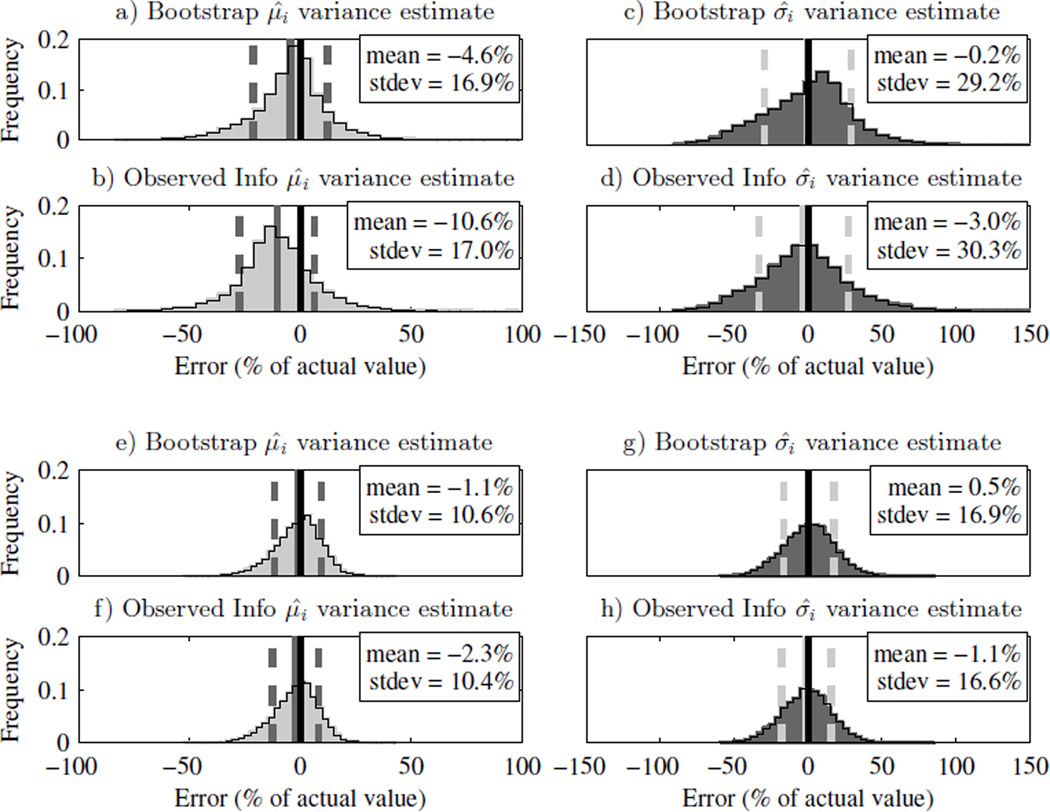
Fig. 6a–d) compare the error of the square root of the variance estimates of μ̂ and σ̂ when n = 50 trials, μ = 0, σ = 1, and the 3-Down/1-Up staircase adaptive procedure was used to generate data sets. We see that, on average, both techniques underestimate the variance on μ̂ and σ̂, and note that the observed information technique (Fig. 6b and Fig. 6d) is somewhat less accurate than the iterative bootstrap (Fig. 6a and Fig. 6c).
Fig. 6.
Iterative bootstrap and observed information variance estimate error on μ̂i and σ̂i (the estimates for the ith data set) for a 3-down/1-up staircase 0.5 and 1for n = 50 trials (panels a–d) and n = 200 trials (panels e–h). Panels a, c, e and g) show the histograms for the error on the variance estimates of μ̂i and σ̂i using the iterative bootstrap technique. Panels b, d, f and h) show the histograms for the error on the variance estimates of μ̂i and σ̂i using the observed information technique. The solid black line shows 0% error, the solid gray line is the mean error, and the dashed gray lines indicate one standard deviation either side of the mean error
Fig. 6e–h) show the variance estimate error with the same simulation settings as in Fig. 6a–d) except now the number of trials has increased from 50 to 200 and a vestibular bias of μ = 0.5σ has been included. In this case, the number of trials is large and the observed information and iterative bootstrap techniques provide similar results.
Table 5 lists the variance estimate error means and standard deviations for all 3-Down/1-Up simulations. The iterative bootstrap technique is generally more accurate than the observed information technique. Furthermore, the bootstrap method is also, in most cases, more precise. As the number of trials increases, the discrepancy between the accuracy of the iterative bootstrap and observed information techniques decreases and by n = 200, they yield nearly the same answer.
Table 5.
Iterative bootstrap and observed information variance estimate error (% of actual value) means±(standard deviations) for all 3-Down/1-Up simulations
| μ | # of Trials | Bootstrap: μ̂i | Observed: μ̂i | Bootstrap: σ̂i | Observed: σ̂i |
|---|---|---|---|---|---|
| μ = 0 | n = 50 | −4.6±(16.9) | −10.6±(17.0) | −0.2±(29.2) | −3.0±(30.3) |
| n = 100 | −1.0±(13.2) | −3.2±(12.9) | 0.9±(27.4) | −0.8±(26.8) | |
| n = 200 | −0.4±(9.5) | −1.4±(9.2) | 0.4±(20.0) | −0.8±(19.7) | |
| μ = 0.5σ | n = 50 | −5.7±(20.0) | −8.6±(20.1) | −2.2±(24.9) | −1.8±(29.6) |
| n = 100 | −3.1±(14.5) | −5.5±(14.0) | −0.1±(20.8) | −4.1±(21.0) | |
| n = 200 | −1.1±(10.6) | −2.3±(10.4) | 0.5±(16.9) | −1.1±(16.6) | |
Table 6 shows the single data set iterative bootstrap and observed information approach execution time means and standard deviations for all 3-Down/1-Up simulations. We see that the observed information technique executes on the order of 0.0005 seconds while the iterative bootstrap technique (with 2000 iterations) executes on the order of 10 seconds. The bootstrap takes longest for n = 50 as the GLM fit requires more time for small n.
Table 6.
Iterative bootstrap and observed information matrix average execution times in seconds for all 3-Down/1-Up simulations
| μ | # of Trials | Bootstrap (seconds) | Observed (seconds) |
|---|---|---|---|
| μ = 0 | n=50 | 12.9 | 3.2×10−4 |
| n=100 | 12.2 | 3.6×10−4 | |
| n=200 | 12.3 | 5.3×10−4 | |
| μ = 0.5σ | n=50 | 14.8 | 3.5×10−4 |
| n=100 | 13.8 | 3.9×10−4 | |
| n=200 | 13.8 | 5.4×10−4 | |
Discussion
Accuracy of parameter estimates
This paper focused on fitting data obtained using adaptive sampling procedures. Our simulations confirm that, for adaptive one-interval forced-choice paradigms, maximum likelihood estimates on the psychometric function’s spread parameter, σ, are downwardly biased (Lim and Merfeld 2012; Kaernbach 2001; Leek 2001) for 200 trials or less. Using bias-reduced maximum likelihood estimation we were able to substantially correct for this bias. These corrections worked on all of the adaptive sampling procedures that we tested without increasing the variance on μ̂ and σ̂ or decreasing the accuracy of μ̂. In the case of a small number of trials (n = 50) and the presence of a vestibular bias (μ = 0.5σ) bias-reduced maximum likelihood estimation not only corrected the bias, but also reduced the variance and skewness of the estimators. Furthermore, bias-reduction substantially improved σ̂ estimates for a staircase procedure with very small n (i.e. 25 trials), and these bias-reduced estimates were further improved by utilizing a pre-determined scale factor.
Despite our focus on adaptive sampling procedures, we also showed that maximum likelihood estimates on data collected from non-adaptive sampling procedures had a mode (i.e., distribution peak) that underestimated the actual value of σ – like the fits of adaptive sampling procedures described in detail herein. However, the distributions had a positive skew that counter-acted this underestimation to yield nearly unbiased mean estimates of σ. Using bias-reduced maximum likelihood estimation on these data sets yielded distribution modes that demonstrated less underestimation, but the positive skewness remained – leading to a mean that slightly overestimated the mean. Such skewed distributions make unbiased fits more complicated for non-adaptive sampling procedures and are beyond the scope of this paper.
One aspect that remains unexplained is the fact that while bias-reduced maximum likelihood estimation removes the order n−1 asymptotic bias term from the maximum likelihood estimate, it is not clear how this is directly related to the bias that arises from the serial dependency of the adaptive sampling procedure (Kaernbach 2001; Klein 2001). This serial dependency exists because adaptive data are not collected independently, as the responses from previous trials determine the location of subsequent trials. On the other hand, non-adaptive sampling procedures are independent as the levels to be tested were chosen by the experimenter in advance. For adaptive data, these multiple trial dependencies and their effects are complex and cause the bias we see in the σ estimates (Kaernbach 2001). Our observations could simply be coincidental. However, the consistency of our numerical findings makes it seem much more likely that some relationship links these two biases through the b1, b2) parameterization of the psychometric function and the nonlinear transformation to the (μ, σ) domain. Whatever the reason may be, we have demonstrated that our bias-reduced maximum likelihood technique worked well with all adaptive sampling procedures tested.
We emphasize that the bias reduction techniques did not uniformly work this well. When data were acquired using non-adaptive procedures, the mode for the estimate of σ shifted to be less biased, but the average σ parameter estimate yielded an overestimate (Table 1). The behavior difference for the parameter mean and parameter mode was due to a skewed distribution that was present both with and without bias correction for non-adaptive stimuli (but not adaptive stimuli). The overestimation for the average bias corrected estimate of σ was not surprising because the average estimate of σ was unbiased when estimated using a standard GLM fit. Parameter estimation for data acquired using non-adaptive procedures is not the focus of this paper; this topic deserves further investigation in a separate study.
Though we show that it is possible to obtain unbiased estimates for adaptive sampling procedures, one might reasonably ask: why is bias reduction necessary? As one answer, bias reduction allows the direct comparison of data sets having different numbers of trials. Recall that bias decreases as the number of trials increases. Thus, for an adaptive staircase procedure, one cannot directly compare data for different subjects unless the number of trials is the same. Bias reduction also allows us to compare data obtained using different staircase procedures – for example comparing data obtained using 3D/1U and 4D/1U staircases. Finally, bias reduction allows us to compare thresholds obtained using staircase procedures to those obtained via any other procedure (e.g., non-adaptive methods). For example, in the clinic, bias reduction provides an unbiased estimate that can be compared to an unbiased normative data set obtained using slightly different methodology.
To illustrate these points, imagine that a normative data set (with μ = 0) was obtained using an MLE procedure with n = 200, while a patient’s data set (also with μ = 0) was obtained using a staircase procedure with n = 50. If the patient had a normal threshold, and GLM fits were used to fit both the patient and normative data, then the patient’s threshold would be, on average, 6% lower than the normative average (Table 1). On the other hand, if BRGLM fits were used to fit both the patient and normative data, then the patient’s threshold would only be, on average, 1% higher than the normative average (Table 1). The same holds true for scientific literature as unbiased threshold estimates allow the direct comparison of data obtained using different methodologies.
Estimating the precision of parameter estimates
Our comparison of the iterative bootstrap and the observed information techniques for estimating parameter variance showed that the iterative bootstrap technique was more accurate. However, the results were surprisingly similar even for a small number of trials, and as the number of trials increased to 200, the variance estimates of the two techniques almost became identical. Thus, for real-time variance estimations (e.g. after every trial), the observed information approach, which executes about 20,000 times faster than the iterative bootstrap technique, can be used to save time and still provide accurate parameter variance estimates.
When applying the bootstrap technique to adaptive sampling procedures, variance estimates were obtained by using the bias-reduced parameter estimates from the original data set to re-simulate the subject’s response vector, y, to the experimentally observed stimulus vector, x. Since the stimulus vector was fixed when running the bootstrap simulations, the simulated bootstrap data sets were fit using a standard (i.e. non-bias-reduced) maximum likelihood technique because the bias derived from the serial dependency of adaptive data is not present for non-adaptive (fixed) data sets.
Percent correct detection
In this paper we chose to define the vertical axis of our psychometric function as the probability that the subject’s response is positive – with a fit ranging between 0 and 1 – rather than the probability that the subject’s response is correct – with a fit ranging between 0.5 and 1. Preliminary 3D/1U adaptive staircase simulations not included herein showed that analyzing the percent correct (ranging between 50 and 100%) during our direction recognition task yielded a 57% greater standard deviation in the threshold estimate than when analyzing the data using a fit between 0 and 100%. This was true even when optimal conditions were established for the percent correct detection analysis (e.g., no vestibular bias), and the fit model exactly matched the subject model. While we have not explored this in detail, we think that the primary explanation of the relatively poor performance of percent correct detection model fits is that the expected response variance p(1 − p) is high in the neighborhood of the 50% plateau. This plateau needs to be established for a good fit of a model that varies between 50% and 100% and is an issue that was previously discussed by Merfeld (2011) and Jakel and Wichmann (2006).
Furthermore, we need to make a number of questionable assumptions to apply percent correct analysis. First, this analysis requires an assumption of zero vestibular bias. Second, the standard lognormal analysis assumes that either the underlying neural noise is asymmetric (i.e., lognormal) or that the signal underlying the response scales in a log normal manner for very small stimuli near threshold. None of these assumptions seem reasonable nor consistent with experimental data. Therefore, while not a focus of this study, we did not find any reason to encourage us to further investigate percent correct fits of data acquired during a recognition task.
Relation to earlier methods
In this short section we briefly describe our understanding of how our bias-reduction method relates to others described in earlier statistics literature. We first learned of bias correction from the second edition of “Generalized Linear Models” (McCullagh and Nelder, 1989). We used the equations in this book to calculate the bias vector and iteratively subtract it from the parameter estimates in MATLAB’s glmfit m-file. This method yielded most of the bias-reduced results (e.g., Table 1) presented herein. Later, we learned of the modified score approach (Firth, 1993), which we decided to implement to evaluate relative performance. As expected, for the identical problem definition, these yielded identical results as shown in online resource 1.
The methods we developed seem similar to some found in the general statistical literature, so we briefly summarize some relevant literature here. Bias correction, in the context of GLM fits, is analyzed by McCullagh and Nelder (1989) and Cordeiro and McCullagh (1991). More recently, Kosmidis and Firth (2009) applied an iterative scheme to generalized linear model fits via an adjustment of the “working observations”, and, shortly thereafter, Kosmidis and Firth (2010) described a general iterative method that calculates and subtracts bias from the score function.
We note two aspects that make this contribution unique. First, with one exception discussed earlier (Hall 1981), we are unaware of any earlier work that has specifically applied these or any other bias correction techniques to the problem of estimating parameter bias when fitting psychometric functions, which is a focus of this paper. Second, a major component of the bias that we are trying to reduce has long been known (Leek, Hanna and Marshall 1992; Treutwein and Strasburger 1999; Kaernbach 2001; Leek 2001) and is ascribed in part to the serial dependency of adaptive data (Kaernbach 2001; Klein 2001). To our knowledge, none of the pre-existing literature investigates parameter bias reduction for this parameter bias component that is present in data acquired using adaptive methods (e.g., staircase procedures). In fact, our finding that bias reduction on the (b1, b2) parameterization yields better results for estimating (μ̂, σ̂) from adaptive data having serial dependency than a (μ, σ) parameterization was a surprise.
Supplementary Material
Acknowledgments
We thank Faisal Karmali, Wei Wang, Torin Clark and Luzia Grabherr for reviewing a draft of this manuscript. This research was supported by the National Institutes of Health/National Institute of Deafness and Other Communication Disorders grant DC04158. The Orchestra computational cluster is supported by the National Institutes of Health shared equipment grant 1S10RR028832.
References
- Casella G, Berger RL. Statistical Inference, Second Edition. Duxbury: Pacific Grove; 2001. [Google Scholar]
- Cox DR, Hinkley DV. Theoretical Statistics. London: Chapman and Hall; 1974. [Google Scholar]
- Cordeiro GM, McCullagh P. Bias correction in generalized linear models. J Roy Stat Soc B Met. 1991:629–643. [Google Scholar]
- Crane BT. Fore-aft translation aftereffects. Exp Brain Res. 2012;219:477–487. doi: 10.1007/s00221-012-3105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A, Barnett A. An Introduction to Generalized Linear Models, Third Edition. Boca Raton: Chapman and Hall/CRC; 2008. [Google Scholar]
- Efron B, Hinkley DV. Assessing the accuracy of the maximum likelihood estimator: Observed versus expected Fisher information. Biometrika. 1978;65:457–487. [Google Scholar]
- Efron B, Tibshirani RJ. An introduction to the bootstrap. New York: Chapman and Hall; 1993. [Google Scholar]
- Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- Foster DH, Bischof WF. Thresholds from psychometric functions: superiority of bootstrap to incremental and probit variance estimators. Psychol Bull. 1991;109:152–159. [Google Scholar]
- Hall JL. Hybrid adaptive procedure for estimation of psychometric functions. J Acoust Soc Am. 1981;69:1763–1769. doi: 10.1121/1.385912. [DOI] [PubMed] [Google Scholar]
- Jakel F, Wichmann FA. Spatial four-alternative forced-choice method is the preferred psychophysical method for naïve observers. J Vis. 2006;6:1307–1322. doi: 10.1167/6.11.13. [DOI] [PubMed] [Google Scholar]
- Kaernbach C. Slope bias of psychometric functions derived from adaptive data. Percept Psychophys. 2001;63:1389–1398. doi: 10.3758/bf03194550. [DOI] [PubMed] [Google Scholar]
- Klein SA. Measuring, estimating, and understanding the psychometric function: a commentary. Percept Psychophys. 2001;63:1421–1455. doi: 10.3758/bf03194552. [DOI] [PubMed] [Google Scholar]
- Knoblauch K, Maloney LT. Estimating classification images with generalized linear and additive models. J Vis. 2008;8:1–19. doi: 10.1167/8.16.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmidis I. Ph.D. thesis. Warwick, England: Dept. of Statistics, Univ.; 2007. Bias reduction in exponential family nonlinear models. [Google Scholar]
- Kosmidis I, Firth D. Bias reduction in exponential family nonlinear models. Biometrika. 2009;96:793–804. [Google Scholar]
- Kosmidis I, Firth D. A generic algorithm for reducing bias in parametric estimation. Electron J Statist. 2010;4:1097–1112. [Google Scholar]
- Leek MR. Adaptive procedures is psychophysical research. Percept Psychophys. 2001;63:1279–1292. doi: 10.3758/bf03194543. [DOI] [PubMed] [Google Scholar]
- Leek MR, Hanna TE, Marshall L. Estimation of psychometric functions from adaptive procedures. Percept Psychophys. 1992;51:247–256. doi: 10.3758/bf03212251. [DOI] [PubMed] [Google Scholar]
- Lim K, Merfeld DM. Signal Detection Theory and Vestibular Perception: II. Fitting Perceptual Thresholds as a Function of Frequency. Exp Brain Res. 2012 doi: 10.1007/s00221-012-3217-2. <submitted>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P, Nelder J. Generalized Linear Models, 2nd Ed. Boca Raton: Chapman and Hall; 1989. [Google Scholar]
- McKee SP, Klein SA, Teller DY. Statistical properties of forced-choice psychometric functions: Implications of probit analysis. Atten Percept Psychophys. 1985;37:286–298. doi: 10.3758/bf03211350. [DOI] [PubMed] [Google Scholar]
- Merfeld DM. Signal detection theory and vestibular thresholds: I. Basic theory and practical considerations. Exp Brain Res. 2011;210:389–405. doi: 10.1007/s00221-011-2557-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenouille MH. Notes on bias in estimation. Biometrika. 1956;43:353–360. [Google Scholar]
- Roditi RE, Crane BT. Directional asymmetries and age effects in human self-motion perception. J Assoc Res Otolaryngol. 2012;13:381–401. doi: 10.1007/s10162-012-0318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MM, Creelman CD. PEST: Efficient estimates on probability functions. J Acoust Soc Am. 1967;41:782–787. [Google Scholar]
- Treutwein B, Strasburger H. Fitting the psychometric function. Percept Psychophys. 1999;61:87–106. doi: 10.3758/bf03211951. [DOI] [PubMed] [Google Scholar]
- Wichmann FA, Hill NJ. The psychometric function: I. Fitting, sampling, and goodness of fit. Percept Psychophys. 2001a;63:1293–1313. doi: 10.3758/bf03194544. [DOI] [PubMed] [Google Scholar]
- Wichmann FA, Hill NJ. The psychometric function: II. Bootstrap-based confidence intervals and sampling. Percept Psychophys. 2001b;63:1314–1329. doi: 10.3758/bf03194545. [DOI] [PubMed] [Google Scholar]
- Yssaad-Fesselier R, Knoblauch K. Modeling psychometric functions in R. Behav Res Methods. 2006;38:28–41. doi: 10.3758/bf03192747. [DOI] [PubMed] [Google Scholar]
- Zupan LH, Merfeld DM. Interaural self-motion linear velocity thresholds are shifted by roll vection. Exp Brain Res. 2008;191:505–511. doi: 10.1007/s00221-008-1540-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.