Abstract
Polyplex formation (complexation) and gene release from the polyplexes (decomplexation) are major events in polymeric gene delivery, however the effect of the decomplexation rate on transfection has been rarely investigated. This study employed mixed polymers of poly(L-lysine) (PLL: MW ~7.4 kDa) and reducible PLL (RPLL) (MW ~6.7 kDa) to design decomplexation rate-controllable PLL100-xRPLLx/pDNA complexes (PRLx polyplexes). The transfection efficiency of a model gene (luciferase) in MCF7 and HEK293 cell lines increased with increasing x (RPLL content) in the PRLx polyplexes until peaking at x=2.5 and x=10, respectively, after which point transfection efficiency declined rapidly. In MCF7 cells, PRL2.5 polyplex produced 3- or 223-fold higher gene expression than PLL or RPLL polyplexes, respectively. Similarly, the transfection efficiency of PRL10 polyplex-transfected HEK293 cells was 3.8- or 67-fold higher than that of PLL or RPLL polyplexes, respectively. The transfection results were not apparently related to the particle size, surface charge, complexation/compactness, cellular uptake, or cytotoxicity of the tested polyplexes. However, the decomplexation rate varied by RPLL content in the polyplexes, which in turn influenced the gene transfection. The nuclear localization of pDNA delivered by PRLx polyplexes showed a similar trend to their transfection efficiencies. This study suggests that an optimum decomplexation rate may result in high nuclear localization of pDNA and transfection. Understanding in decomplexation and intracellular localization of pDNA may help develop more effective polyplexes.
Keywords: Decomplexation, Nuclear localization, Poly(L-lysine), Polymeric gene carrier, Reducible polymer
INTRODUCTION
Interest in polymeric gene carriers has been steadily increased due to their tailored structural characteristics (e.g., charges and architectures) and versatile design components endowing required functionalities (e.g., colloidal stabilization, cellular targetability, endosomal disruption, cytosolic transport, nuclear import, triggering drug release, and biocompatibility).1–4 Such functionalities have been thought to overcome various extracellular and intracellular barriers encountered in the gene delivery process, anticipating enhanced therapeutic effects of an exogenous gene delivered by polymeric carriers. In addition, it is critical for optimal transfection that the decomplexation to release genes from the carriers should occur at a right intracellular place and a right time.5 Our group has discovered that high transfection efficiency can be obtained by tuning the pH at which the polyplex is released from the endolysosome and that pH-tunable sulfonamide-based oligomers/polymers loaded in polyplexes affected gene expression but not gene silencing in gene delivery.6, 7 Despite the recognized significance of intracellular location and timing of gene, the related investigation is seldom conducted.
The release or decomplexation of plasmid DNA (pDNA) from electrostatically-driven polyplexes is mostly induced by competitive interactions with charged biological components which disrupt the ionic bond that binds the gene to the carrier and protects from degradation. For most cationic polyplexes, such as anionic polymers or molecules such as glycosaminoglycans in plasma,8–10 RNA in the cytoplasm,11 DNA in the nucleus,12 heparin,6, 13, 14 poly(acrylic acid) and sodium dodecyl sulfate in test tubes, it is difficult to control the unpacking process of polyplexes under a given extracellular and intracellular environment because their concentrations cannot be manipulated by extrinsic factors.
Longer polycations generally cause stronger attraction with negatively-charged genes than shorter polycations, resulting in tighter polyplexes.6, 14, 15 Thus, to facilitate polyanion-induced decomplexation of polyplexes especially in intracellular environments, degradable polymers responding to acidic pHs in the endolysosomes16 or glutathione in the cytoplasm and the nucleus14, 17, 18 have been developed. These polyplexes resulted in improved transfection efficiency. But the genes are susceptible to lytic enzymes in the endolysosomes. For pDNA, transfection efficiency could be dependent on the distance between its releasing site and the nucleus due to its poor mobility19 in intracellular environment. In addition, pDNA is deactivated or degraded by cytosolic nucleases, giving pDNA a 60–90 min half-life in HeLa and COS cells.20 Therefore, it is hypothesized that the intracellular release rate of genes affects transfection.
It has been generally recognized that the low transfection efficiency of poly(L-lysine) (PLL) polyplexes is caused by lack of endosomolytic activity of PLL. However, PLL polyplexes showed much slower decomplexation rate than poly(cyclooctene-g-oligolysine) (i.e., comb-shaped PLL) with nuclear localization signal (NLS), in spite of the fact that the former had a more nuclear uptake than the latter.21 Non-degradable and degradable PLL polymer-based polyplexes also showed molecular weight (MW)-dependent decomplexation and transfection efficiency.12, 15 For example, when forming linear polylysine/pDNA complexes, longer polylysine chains, (180 lysine units, MW 28 kDa) showed slower dissociation than shorter polylysine chains, (19 or 36 lysine units, 2.3 kDa or 4.4 kDa, respectively)3 due to the greater positive charge of the longer chains tightly binds the pDNA that interferes the release of the genes from the carrier, thus leading to increased inhibition of RNA synthesis and lower gene expression.12 However, reducible PLL (RPLL)-based polyplexes showed contradictory in vitro transfection results depending on their MWs compared to polyplexes prepared by PLL with comparable MWs. High MW RPLL (187 kDa) showed more effective transfection than PLL (205 kDa),22 whereas moderate MW RPLL (65 kDa) showed lower gene transfection efficiency than PLL (54 kDa).23 Based on these results, it appears that either “too fast” or “too slow” release of genes from polyplexes would bring low transfection efficiency.
This study investigated the effect of the decomplexation rate of pDNA on transfection efficiency. Commercial PLL with 7.4 kDa and synthesized reducible PLL (RPLL) with a comparable MW were selected. Using the polyplexes formed from a model gene and mixed PLL and RPLL, the influence of RPLL content on physicochemical characteristics (e.g., size, surface charge, complexation, and decomplexation) and biological characteristics (e.g., cellular uptake, cytotoxicity, transfection efficiency, and intracellular localization) was studied.
MATERIALS AND METHODS
Materials
Poly(L-lysine) hydrogen bromide (PLL·HBr; Mw 12 kDa), dimethyl sulfoxide (DMSO), 4-(2-hydroxy-ethyl)-1-piperazineethanesulfonic acid (HEPES), 3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyltetrazolium bromide (MTT), D-glucose, cystamine, sodium bicarbonate, recombinant human insulin, ethidium bromide (EtBr), paraformaldehyde (PFA), deuterium oxide (D 2O), dithiothreitol (DTT), heparin sodium salt (139 USP units/mg), Hoechst 33342, RPMI- 1640 medium, Dulbecco’s phosphate buffered saline (DPBS), Dulbecco’s modified eagle’s medium (DMEM), fetal bovine serum (FBS), trypsin-EDTA solution, and penicillin-streptomycin antibiotics were purchased from Sigma-Aldrich (St. Louis, MO). LysoTracker® Green dye and YOYO-1 were received from Invitrogen, Inc. (Carslbard, CA). A pDNA which encodes firefly luciferase (gWiz-Luc or pLuc) was bought from Aldevron, Inc. (Fargo, ND). A decapeptide (Cys-Lys8-Cys) was synthesized by American Peptide Company (Sunnyvale, CA). Luciferase assay kit and GSH-Glo™ glutathione assay kit were purchased from Promega Corporation (Madison, WI) and BCA protein assay kit was bought from Pierce Biotechnology, Inc. (Rockford, IL). Cy5-labeled pDNA was prepared by using Label IT® nucleic acid labeling kit of Cy5 dye (Mirus Bio LLC; Madison, WI).
Cells and Cell Culture
HEK293 cells (human embryonic kidney cell line) and MCF7 cells (human breast adenocarcinoma cell line) were used in this study. HEK293 cells were cultured in DMEM supplemented with 10% FBS and D-glucose (4.5 g/L). MCF7 cells were cultured in RPMI-1640 supplemented with 10% FBS, D-glucose (2 g/L), and insulin (4 mg/L). The cells were maintained and grown under a humidified air with 5% CO2 at 37 °C.
Synthesis and Characterization of Reducible Poly(Cys-Lys8-Cys) (RPLL)
As previously reported,22, 24 a decapeptide (Cys-Lys8-Cys) was oxidized to form reducible poly(Cys-Lys8-Cys) (RPLL) (Figure S1). Cys-Lys8-Cys (50 mg; 40 μmol) was dissolved in deionized water (DIW; 1 mL) and then cystamine (0.31 mg; 4 μmol) as a chain closer was added into the peptide solution. On adding DMSO (0.33 mL) into the solution, oxidative polymerization of the decapeptide was performed to form RPLL at room temperature (RT). After 1 day polymerization, the synthesized RPLL was dialyzed against DIW by using a dialysis membrane with a molecular weight cut-off (MWCO) 1 kDa for 2 days and then was lyophilized. As shown in Figure S2, the 1H-NMR spectra of RPLL in D2O were characterized and confirmed by characteristic peaks at δ 1.32 (–CH-CH2-CH2–), δ 1.57 (–CH-CH2-CH2–), δ 2.59 (–CH-CH2-NH2), δ 3.1 (–CH2-NH2CH-CO–), δ 3.25 (–S-CH2CH2-NH2), δ 3.8 (–S-CH2CH2-NH2), δ 4.17 (–NH-CHCH2-CO–), δ 4.3 (–CH2-NH2CH-CO–). Based on 1H-NMR analysis of RPLL, its estimated molecular weight (MW) was 6.7 kDa.
To determine polymer amount used for cell experiments, in vitro cytotoxicity of RPLL was evaluated by MTT-based cell viability as previously reported.7, 14 HEK293 cells and MCF7 cells were seeded at 2×103 cells and 5×103 cells per well of a 96-well plate, respectively and then were incubated for 24 hr in serum-containing culture medium. The cells were exposed to RPLL having different concentrations (0–100 μg/mL) for 24 hr. Then, MTT solution (10 μL; 5 mg/mL) was treated to the cells in the culture medium (0.1 mL) for additional 4 hr. After discarding the culture medium, DMSO was added to dissolve the formazan crystals which are produced by living cells. Their absorbance was measured at 570 nm to estimate the cell viability and was compared with that of PLL as a non-reducible counterpart.
Preparation and Physicochemical Characterization of Polyplexes
As previously reported,6, 14, 25 a cationic solution having polymers (i.e., PLL and RPLL) and an anionic solution containing pDNA were separately prepared by using HEPES buffer (20 mM, pH 7.4). Two solutions with an equivalent volume were mixed and then incubated for 30 min at RT. The formed polyplexes (20 μL per 1 μg of pDNA) were further evaluated. Complexation ratio of polyplexes was expressed based on the N/P ratio using amines (N) of polymers and phosphates (P) of pDNA.
Particle sizes and surface charges of polyplexes were evaluated as previously reported.6, 14, 25 After preparing polyplexes (100 μL; 5 μg pDNA), they were diluted with HEPES buffer (20 mM, pH 7.4) and pDNA in the polyplex solution was 2.5 μg/mL. The polyplexes were then monitored by using a Zetasizer 3000 (Malvern Instrument, UK) with a wavelength of 677 nm and a constant angle of 90° at RT.
The complexation ability of pDNA with PLL and RPLL was evaluated by a gel electrophoresis assay. After loading polyplexes (10 μL; 0.5 μg of pDNA) in 0.8% agarose gel having EtBr, the gel was run in 0.5x TBE buffer at a constant voltage (80 V) for 90 min. A UV illuminator was used to detect whether pDNA was exposed.
Compactness of pDNA was tested by using quenching fluorescent YOYO-1 dye intercalated with pDNA in polyplexes. After pDNA was intercalated with YOYO-1 dye (1 molecule per 10 bases of pDNA), YOYO-1-intercalated pDNA was used to form polyplexes. Fluorescent intensity of YOYO-1 in the polyplexes was measured at 495 nm (excitation) and 515 nm (emission). Compactness (%) of pDNA in polyplexes was calculated as a following equation.
where RFUPolyplex, RFUpDNA, and RFUBuffer are relative fluorescent units (RFU) of polyplexes, YOYO-1-intercalated pDNA, and HEPES buffer, respectively.
Decomplexation of pDNA from polyplexes was monitored by a gel electrophoresis assay in the presence of DTT and/or heparin. Polyplexes were exposed to 150 mM NaCl solution supplemented with heparin sodium salt (0–100 μg/mL) and/or DTT (20 mM) for 1 hr at 37 °C and a final concentration of pDNA was 25 μg/mL. Then the polyplexes were run and evaluated in 0.8% agarose gel as previously described in the complexation ability of polyplexes.
In addition, decomplexation kinetics of polyplexes was monitored by a dye-dequenching technique. The polyplexes with YOYO-1-intercalated pDNA (5 μg/mL) were treated with 150 mM NaCl solution having heparin (6.5 μg/mL) and DTT (10 mM) as a decomplexation solution (0.2 mL) at RT. Time-dependent decomplexation was evaluated by changing RFU of YOYO-1-intercalated pDNA exposed from the polyplexes using a plate reader. Free YOYO-1-intercalated pDNA and the decomplexation solution were set as 100% RFU and 0% RFU, respectively.
Biological Characterizations of Polyplexes
HEK293 cells and MCF7 cells were seeded at 1×105 cells/well and 5×105 cells/well (of a 6-well plate), respectively, for cellular uptake, in vitro transfection efficiency, and intracellular trafficking study, whereas their cell densities were 5×104 HEK293 cells/well and 2.5×105 MCF7 cells/well (of a 12-well plate) for in vitro cytotoxicity. The seeded cells were incubated for 24 hr in serum-containing culture medium and then were used for further studies.
For in vitro transfection study, cells were cultured in a 6-well-plate and the transfection experiments were performed as previously described.14, 25, 26 One hour before addition of polyplexes, serum-containing culture medium was replaced with serum-free and insulin-free transfection medium. After transfecting with polyplexes, the transfected cells were incubated for 4 hr. The transfection medium was then replaced with serum-containing culture medium and the cells were incubated for additional 44 hr. After the transfection experiments were completed, the cells transfected with pLuc were rinsed with DPBS and then lysed in a reporter lysis buffer. Relative luminescent unit (RLU) and protein content of transfected cells were evaluated by luciferase assay kit and BCA protein assay kit, respectively.
To evaluate whether transfection procedure of polyplex is toxic, in vitro cytotoxicity of polyplexes was evaluated by MTT-based cell viability and its experimental procedure mostly followed the aforementioned in vitro transfection study except for its cell density and pDNA dose (0.5 μg pDNA per well). After completing transfection procedures for 48 hr, MTT solution (0.1 mL; 5 mg/mL) was added to cells incubated in serum-containing culture medium (1 mL) for additional 4 hr. The in vitro cytotoxicity of the polymer, was evaluated using formazan crystals, which are metabolized by live cells. The formazan crystals were dissolved in DMSO and added to the cultured cells; their absorbance was subsequently monitored at 570 nm for converting cytotoxicity of polyplexes.
Cellular uptake of polyplexes was evaluated as previously reported.6, 14 Polyplexes prepared with YOYO-l-intercalated pDNA (1 μg pDNA per well) were treated to cells. After 4 hr incubation, the cells were detached and fixed with 4% PFA solution. The fluorescent polyplex-containing cells were monitored using flow cytometry (FACScan Analyzer, Becton–Dickinson; Franklin Lakes, NJ) with a primary argon laser (488 nm) and a fluorescence detector (530 ± 15 nm) to detect YOYO-1. Uptake levels of polyplexes in the cells were analyzed with a gated population of living 10,000 cells.
For intracellular trafficking of polyplexes, MCF7 cells were seeded on a coverglass in a six-well plate at 5×105 cells/well. Polyplexes prepared with Cy5-labeled pDNA (1 μg pDNA per well) were treated to the cells in serum-free transfection medium. After 3.5 hr transfection, Hoechst 33342 and LysoTracker®Green dye were added to stain the nuclei and the acidic vesicles (mostly relevant to late endosomes and lysosomes), respectively. At 4 hr incubation with polyplexes, the cells were rinsed with DPBS and fixed with 4% PFA solution for 5 min. The cells were evaluated using a laser scanning confocal microscope. Especially, nuclear localization of Cy5-labeled pDNA was quantified from fluorescence (FL) of two regions of interest (ROIs). The first region of fluorescence was obtained from the whole cell indicated as FLcell and the second region is from the nuclei, FLnuclei. Fluorescence intensity of Cy5-labeled pDNA was measured by using Image J software and the nuclear localization of Cy5-labeled pDNA was calculated from the following equation.27
Evaluation of intracellular glutathione concentration
HEK293 cells and MCF7 cells were seeded at 5×103 cells/well and 2.5×104 cells/well (of a 96-well plate), respectively and were cultured for 1 day. After discarding culture medium, the cells were rinsed and then were treated by following GSH Glo™ glutathione assay protocol.
Statistical analysis
The statistical significance of the data was evaluated by conducting an unpaired Student t-test at a confidence level of p<0.05.
RESULTS AND DISCUSSION
In order to control the decomplexation rates of pDNA in intracellular environments such as the cytoplasm and nucleus, this study designed pDNA complexes using lysine-based polycation mixtures comprising non-reducible PLL and RPLL as a decomplexation controller of polyplexes. PLL with MW 7.4 kDa (MW 12 kDa of PLL· HBr) and synthesized RPLL with a similar MW of 6.7 kDa were used to minimize MW effects of polycations on size and complexation of polyplexes. In (PLL100-xRPLLx)/pDNA complexes, (100-x)% and x% of primary amines for complexation with phosphate groups in pDNA were from PLL and RPLL, respectively and PRLx polyplexes as code names were used. Also, PRLx polyplexes were prepared at a fixed N/P ratio of 5 due to an optimum condition for transfection efficiency of PLL/pDNA complexes26 and polyplex formation based on electrophoresis (Figure S3).
Effects of RPLL content (X) in transfection efficiency of PRLx polyplexes
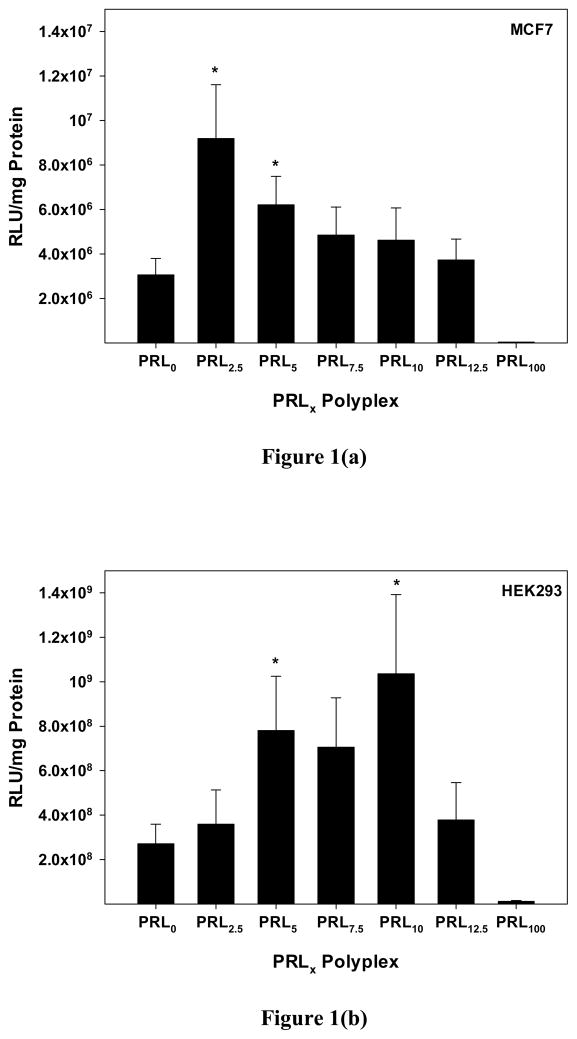
Prior to detailed analyses of various physicochemical and biological characteristics of PRLx polyplexes, we first examined whether the presence of RPLL in PRLx polyplexes affects their transfection efficiency in MCF7 and HEK293 cells. When we applied PRLx polyplexes to the cells, their transfection efficiencies increased, had a peak value at x=2.5 for MCF7 cells and x=10 for HEK293 cells and then decreased with increasing concentration of RPLL compared to that of PRL0 polyplexes (i.e., PLL/pDNA complexes) (Figure 1). Using MCF7 cells, PRL2.5 and PRL5 polyplexes showed 3-fold and 2-fold higher transfection efficiencies than PRL0 polyplexes, respectively and their differences were statistically significant (p=0.017 for PRL2.5 polyplexes and p=0.036 for PRL5 polyplexes compared with PRL0 polyplexes) by unpaired Student’s t-test. (Figure 1(a)). PRLx polyplex-transfected HEK293 cells produced 3.8-fold (p=0.042) higher transgene expression using PRL10 polyplexes and 2.9-fold (p=0.048) and 2.6-fold (p=0.066) higher transgene expression using PRL5 and PRL7.5 polyplexes, respectively, compared to PRL0 polyplexes (Figure 1(b)). When the transfection efficiencies of PRLx polyplexes in MCF7 cells were normalized with that of PRL100 polyplexes (i.e., RPLL/pDNA complexes), PRL0, PRL2.5, and PRL5 polyplexes, 74-fold, 223-fold, and 151-fold higher luciferase expression were observed, respectively (Figure S4). For HEK293 cells, PRL0, PRL5, PRL7.5, and PRL10 polyplexes resulted in 23-fold, 67-fold, 60-fold, and 88-fold better gene expression than RPL100 polyplexes (Figure S4). The transfection results suggest that RPLL content in PRLx polyplexes influences the transfection efficiencies. Neither the fast decomplexation of RPLL/pDNA complexes nor the slow decomplexation of PLL/pDNA complexes appear to be optimal for high ransfection efficiency. It seemed that non-degradable PLL helps to slow decomplexation of RPLL-rich PRLx polyplexes compared to RPLL polyplexes and that degradable RPLL induces faster decomplexation of PLL-rich PRLx polyplexes than PLL polyplexes. This led to efforts to determine how the change in RPLL content affects the physicochemical and biological characteristics of PRLx polyplexes.
Figure 1.
Transfection efficiencies of PLL100-xRPLLx/pDNA complexes (PRLx polyplexes) (N/P 5) in (a) MCF7 cells and (b) HEK293 cells. PLL/pDNA and RPLL/pDNA complexes were denominated as PRL0 and PRL100 polyplexes, respectively. * means p<0.05 compared with PRL0 polyplex by unpaired Student t-test. (Mean ± Standard Error; n≥4)
Effects of RPLL on particle size, surface charge, and complexation/compactness of PRLx polyplexes
The compactness and surface charge of pDNA polyplexes are affected by complexation conditions (e.g., dose of polymers, N/P ratios) and polymer characteristics (e.g., architectures, charge density, flexibility, hydrophobicity).5 Although the MW of RPLL used in this study is similar to that of PLL, the chain structure of RPLL is different from that of PLL (Figure S1). Thus, it is important to determine whether RPLL causes significant changes in particle size, surface charge, and complexation of PRLx polyplexes compared to those of PLL polyplexes.
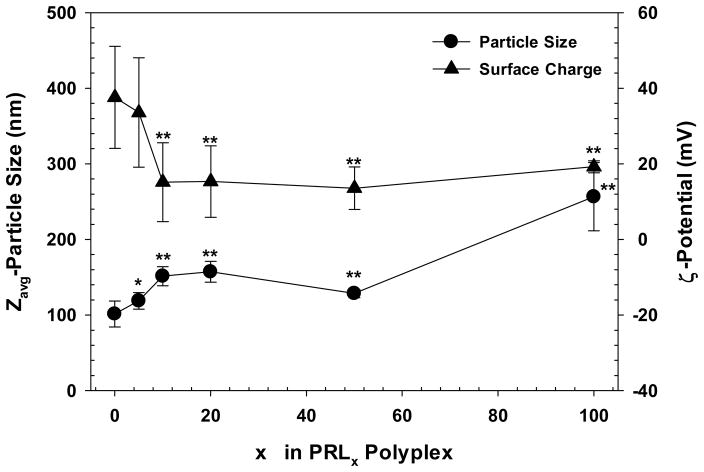
The particle size of RPL0 polyplex was about 100 nm in diameter. Particle sizes were slightly increased with increasing RPLL content in PRLx polyplexes, growing to about 120 nm for PRL5 polyplex (p=0.02) and then to around 150 nm (p<0.01) for PRLx polyplexes with x=10–50 (Figure 2). It might be due to the weaker pDNA complexation of RPLL compared with PLL resulting from the lower charge density of RPLL (157 Da per one primary amine) than PLL (129 Da per one primary amine). This conclusion is supported by the large particle size (250 nm diameter) of RPL100 polyplex.
Figure 2.
Particle size and surface charge of PRLx polyplexes (N/P 5) in HEPES buffer (25 mM, pH 7.4). * and ** mean p<0.05 and p<0.01, respectively, compared with PRL0 polyplex by unpaired Student t-test. (Mean ± Standard Deviation; n = 10)
PRL0 polyplex had a zeta-potential about 40 mV. Introducing RPLL to PRLx polyplexes, decreased the surface charge to about 35 mV at x=5 (p=0.53) and then reached a saturated value (~20 mV) at x=10–100 (p<0.01) (Figure 2). This observation is due to the presence of one carboxylic group associated with the cysteine on each decapeptide unit which negates the positive charge of the primary amine on the lysine. The presence of free carboxylic groups in RPLL provides a possible explanation for the increased particle size of PRLx polyplexes with higher RPLL content.
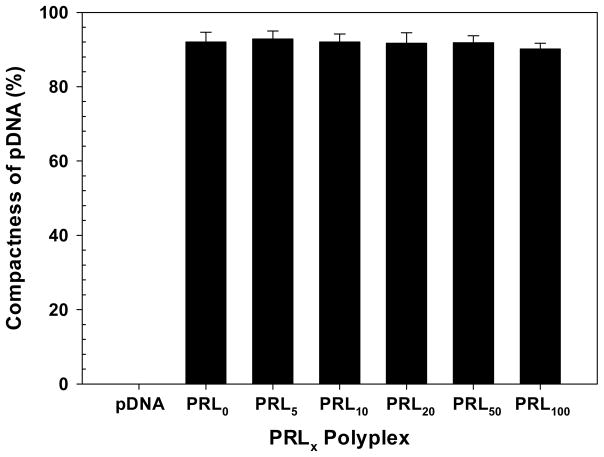
Condensation of pDNA in PLL and RPLL polyplexes (N/P 5) prepared in a HEPES buffer was confirmed by a gel electrophoresis method in 0.8% agarose gel (Figure S3). At N/P 5, both cationic polymers completely shielded pDNA on the polyplex surface. In addition, the compactness of pDNA in PRLx polyplexes was monitored by tracking quenching of YOYO-1- intercalated pDNA. As shown in Figure 3, the fluorescent intensity of unquenched YOYO-1- intercalated pDNA was set to 0% and complete quenching was given a value of 100%. All tested PRLx polyplexes (x=0–100) showed above 90% compactness of pDNA. The results indicate that different amounts of RPLL did not cause serious difference in the compactness of pDNA in PRLx polyplexes.
Figure 3.
Compactness (%) of pDNA in PRLx polyplexes (N/P 5) prepared in a HEPES buffer (25 mM, pH 7.4). (Mean ± Standard Deviation; n = 3)
Effects of RPLL in cellular viability and cell uptake of PRLx polyplexes
The surface charge and particle size of polyplexes influence cellular uptake which can further affect transfection efficiency. Also, excessively high positive surface charges of polyplexes could damage the cell membrane resulting in cellular toxicity and leading to reduced transfection efficiency. Thus, cellular uptake studies of PRLx polyplexes were performed by flow cytometry in MCF7 and HEK293 cells (Figure 4) and cytotoxicity of PRLx polyplexes was evaluated by MTT-based cell viability assay (Figure 5).
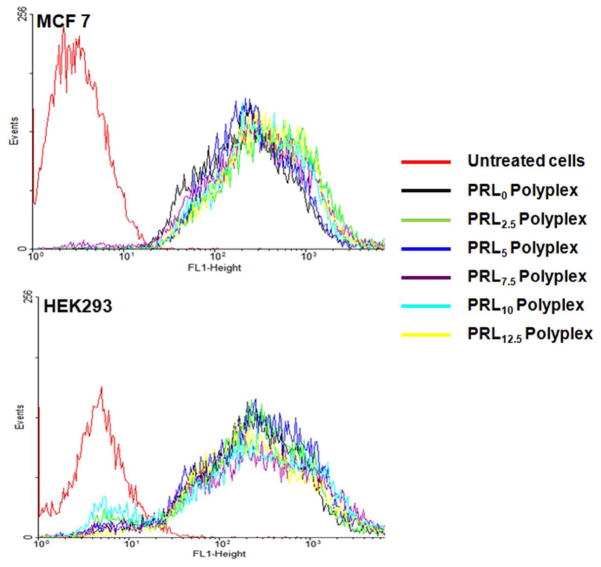
Figure 4.
Cellular uptake of PRLx polyplexes (N/P 5) after 4 hr transfection in (a) MCF7 and (b) HEK293 cells.
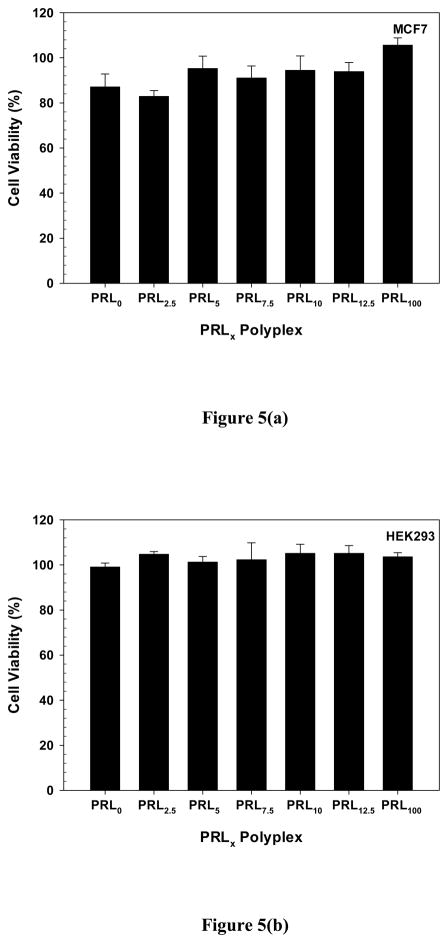
Figure 5.
Cytotoxicity of PRLx polyplexes (N/P 5) after 48 hr transfection in (a) MCF7 and (b) HEK293 cells. (Mean ± Standard Error; n = 6)
After forming PRLx polyplexes (x=0–12.5) with YOYO-1-intercalated pDNA, the cells were incubated with the polyplexes in serum-free and protein-free transfection medium for 4 hr. It was expected that cellular uptake of PRL0 polyplexes would be much higher than those of other PRLx polyplexes (x=2.5–12.5) because PRL0 polyplexes had much higher positive surface charges than other PRLx polyplexes (x=2.5–12.5). However, with both the cells, PRLx polyplexes showed similar uptake regardless of the amount of RPLL present (Figure 4). The results indicate that different transfection efficiencies of PRLx polyplexes (Figure 1) might be not caused by the cellular uptakes of PRLx polyplexes (Figure 4).
The cytotoxicity of polycations is influenced by their length, charge, degradability, hydrophobicity, and architecture28 and strongly affects the cytotoxicity, and thus transfection efficiency, of their polyplexes. PLL· HBr with MW 12 kDa showed more toxicity than RPLL 6.7 kDa in MCF7 and HEK293 cells (Figure S5). The IC50 of PLL· HBr was about 80 μg/mL for MCF7 cells and about 65 μg/mL for HEK293 cells. RPLL showed negligible cytotoxicity with above 80% cell viability in the tested range of polymer (0~100 μg/mL). Different cytotoxicities of PLL and RPLL might result from different degradation characteristics in intracellular environments because RPLL is degraded much more quickly than RPLL in the cytoplasm and the nucleus.
The cytotoxicity of PRLx polyplexes during a 48 hr transfection process was evaluated by an MTT-based cell viability assay using MCF7 and HEK293 cells. PRL0 and PRL100 polyplexes had about 88% and almost 100% cell viability against MCF7 cells, respectively. The difference might be caused by lower toxicity of RPLL than PLL. However, cell viability in the presence of PRLx polyplexes (x=2.5–12.5) differed only negligibly from PRL0 polyplexes. When applying the polyplexes into HEK293 cells, all tested PRLx polyplexes (x=0–100) were non-toxic compared to untreated control cells. Thus, the findings indicate that PRLx polyplex-dependent transfection efficiencies (Figure 1) were not influenced by the toxicity results of PRLx polyplexes.
Effects of RPLL in decomplexation of pDNA from PRLx polyplexes
pDNA must be decomplexed from the polyplex within the cell if the encoded protein is to be expressed. If pDNA release is too slow or absent gene expression will hardly occur due a lack of accessibility of transcriptional machinery to pDNA.5 Also, if polyplexes release pDNA too early, it may not reach the nucleus due to its limited mobility and vulnerability to enzymatic degradation.5, 19 Although PLL and RPLL had similar MW, their polyplexes caused very different transfection efficiencies. PLL/pDNA complexes (N/P 5) showed 74-fold and 23-fold higher luciferase expression than RPLL/pDNA complexes (N/P 5) in MCF7 and HEK293 cells, respectively (Figure 1 and Figure S4). With the hypothesis of the presence of relationship between the release rate of pDNA from a given system and transfection, decomplexation of pDNA from PRLx polyplexes were monitored in heparin- and DTT-containing NaCl solution.
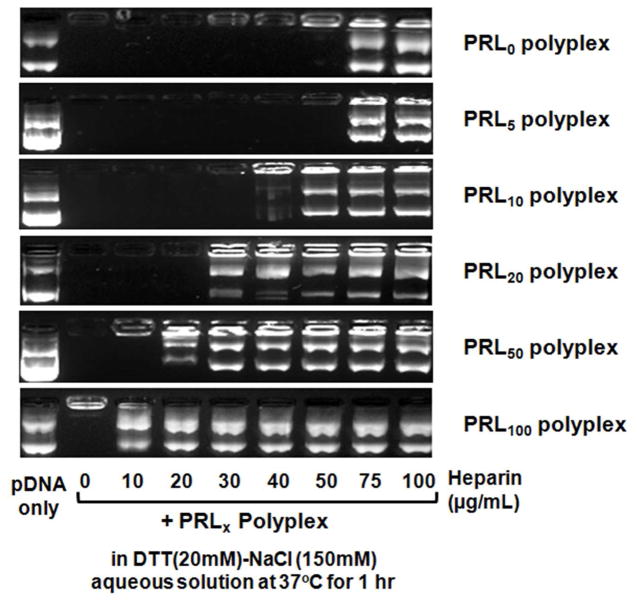
First, PRLx polyplexes (x=0, 5, 10, 20, 50, and 100) were incubated in 150 mM NaCl solution with DTT (20 mM) and different concentrations of heparin (0–100 μg/mL) at 37°C for 1 hr and then were electrophoresed in 0.8% agarose gel with EtBr. The stronger anionic character of heparin induced the release of weaker anionic pDNA when interacting with polyplexes.14 When reducible bonds in a polycation exist, DTT degrades disulfide bonds of reducible polymeric backbone. The static interaction strength with pDNA of the fragmented polycations become weaker, resulting in facilitated pDNA release.14 As shown in Figure 6, 50 μg/mL of heparin induced pDNA exposure of PRL0 polyplexes (i.e., PLL polyplexes) in the loading well and 75 μg/mL of heparin caused complete decomplexation of pDNA from PRL0 polyplexes. However, pDNA complexed in PRL100 polyplexes (i.e., RPLL polyplexes) was exposed at 0 μg/mL and was released at 10 μg/mL of heparin. With increasing the amount of RPLL in PRLx polyplexes, the required heparin concentration for complete decomplexation of pDNA was decreased. However, pDNA release from PRLx polyplexes with x=0–10 is difficult to discern under these experimental conditions due to the static analysis of the experiment.
Figure 6.
Decomplexation (or pDNA release) of PRLx polyplexes (N/P 5). After the polyplexes with pDNA (25 μg/mL) were exposed to DTT (20 mM) and heparin (0–100 μg/mL) in 150 mM NaCl aqueous solution at 37°C for 1 hr, the polyplex solution was loaded into 0.8% agarose gel and then was electrophoresed at 80 V for 90 min.
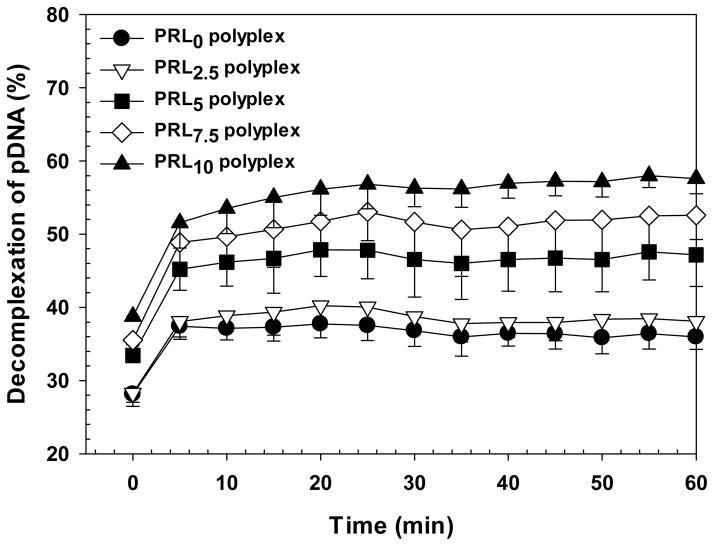
PRLx polyplexes (x=0–10) were exposed to a weakly reducing decomplexation solution (i.e., 150 mM NaCl solution with DTT (10 mM) and of heparin (6.5 μg/mL)) at RT and their pDNA release kinetics were monitored by de-quenching of YOYO-1-intercalated pDNA. Compared to the decomplexation kinetics of pDNA from PRL0 polyplexes, pDNA releases from PRLx polyplexes (x=2.5–10) were slightly accelerated with increasing RPLL content (Figure 7). These static and dynamic decomplexation results may support the transfection efficiency data of PRLx polyplexes and also indicate significance of decomplexation optimization regarding release of pDNA from polyplexes.
Figure 7.
Decomplexation (or pDNA release) kinetics of PRLx polyplexes (N/P 5) using YOYO- 1-labeled pDNA. The polyplexes with pDNA (5 μg/mL) were exposed to DTT (10 mM) and heparin (6.5 μg/mL) in 150 mM NaCl aqueous solution (0.2 mL) at RT. (n = 3; Mean ± Standard Deviation).
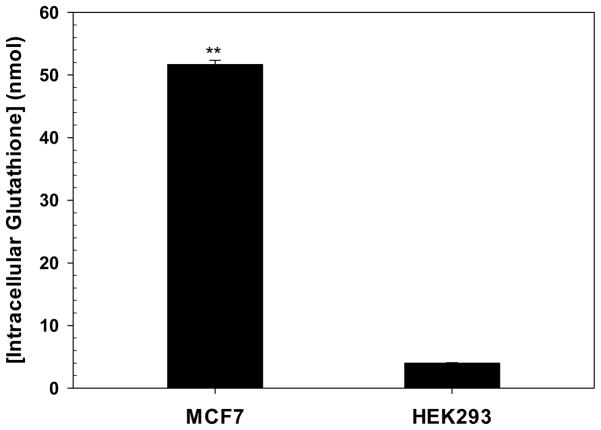
Transfection results of PRLx polyplexes (Figure 1) showed the highest transfection efficiency at a different amount of RPLL in PRLx polyplexes depending on cell type. That is, PRL2.5 polyplexes and PRL10 polyplexes showed maximum transfection efficiency in MCF7 and HEK293 cells, respectively. This result might be explained by different decomplexation rates of PRLx polyplexes which are further influenced by cell-dependent intracellular glutathione levels, which may accelerate RPLL degradation and thus pDNA release. In fact, tumor cells may have about 4-fold more intracellular glutathione levels than normal cells.29 And although tumor cell lines are originated from a identical organ, intracellular glutathione levels are different.30 Thus, to understand clearly why MCF7 cells required a smaller amount of RPLL in PRLx polyplexes to reach maximum transfection efficiency than HEK293 cells, intracellular glutathione levels of these two cells were monitored by a GSH-Glo glutathione assay protocol. As shown in Figure 8, MCF7 and HEK293 cells had 51.7 ± 0.7 nmol/25000 cells and 4.0 ± 0.1 nmol/5000 cells, respectively. As expected, MCF7 cells tumor cell line represented higher glutathione contents in the cells than the non-cancerous HEK293 cells. When applying same PRLx polyplexes, lower intracellular glutathione concentrations may cause slower decomplexation rate of PRLx polyplexes. Thus, to achieve similar decomplexation rate of PRLx polyplexes in MCF7 cells, PRLx polyplexes in HEK293 cells require higher RPLL contents because higher RPLL contents induce faster decomplexation rate of PRLx polyplexes. These factors help to explain why that PRL2.5 polyplexes and PRL10 polyplexes reached a maximum transfection efficiency in MCF7 and HEK293 cells, respectively.
Figure 8.
Intracellular glutathione levels of MCF7 and HEK293 cells. After seeding 25000 or 5000 cells into a well for MCF7 or HEK293 cells, the cells were culture for 24 hr and then were evaluated. ** means p<0.01 compared with HEK293 cells by unpaired Student t-test. (Mean ± Standard Error; n = 3)
In addition, when PLL with higher MWs than 7.4 kDa (used in this study) is applied for PRL polyplexes, RPLL content in the polyplexes should be increased to represent similar decomplexation rate of PLL 7.4 kDa-based PRL polyplexes because higher MW PLL makes stronger binding affinity with pDNA.
Effects of RPLL in intracellular localization of PRLx polyplexes
After pDNA is internalized into cells, nuclear localization of pDNA is a significant step required for transfection to take place. The polyplexes of polylysine having 28 kDa (about 180 repeating units) delivered pDNA in the nucleus and perinuclear area, whereas polylysines with 19 and 36 repeating units (about 2.3 kDa and 4.4 kDa, respectively) delivered pDNA in the nucleus.5, 12 Lower MW polylysine-mediated polyplexes expressed higher transfection efficiency than higher MW polylysine-mediated polyplexes.12 However, when using PLL with an intermediate MW (e.g., PLL with MW 7.4 kDa used in this study) to form polyplexes, it is unclear whether pDNA is localized only in the nucleus or not. It is also unclear how degradable polymers influence intracellular localization of pDNA.
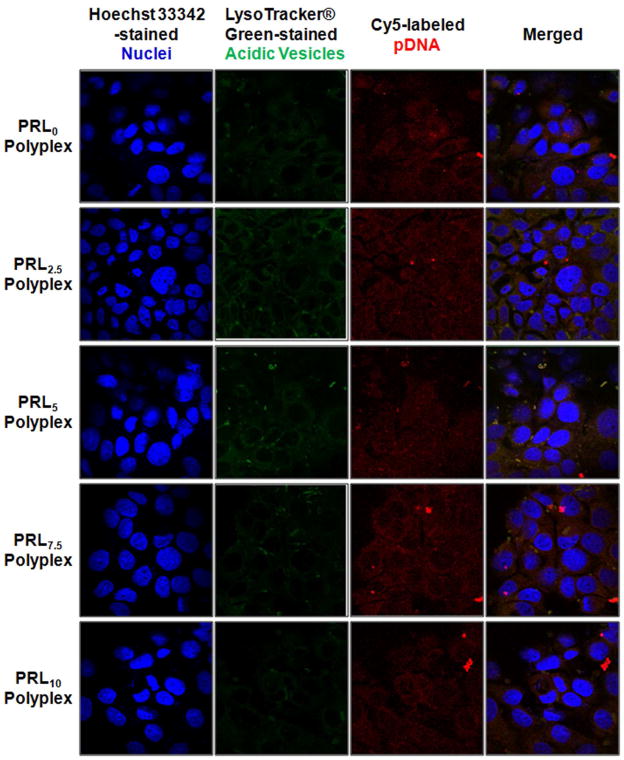
Thus, to understand these significant localization issues of pDNA delivered with degradable polycations, pDNA was labeled with Cy5 and the nucleus and acidic vesicles (i.e., late endosomes and lysosomes) were stained with Hoechst 33342 and LysoTracker™ Green, respectively. As shown in Figure 9, all tested PRLx polyplexes-transfected MCF7 cells showed similar distributions of acidic vesicles in the cytoplasm based on similar fluorescent intensity of LysoTracker™ Green. The presence of strong acidic vesicles was caused by weak endosomolytic function of PLL and RPLL. For intracellular distribution of pDNA, it was dependent on RPLL contents in PRLx polyplexes. In PRL0, PRL7.5, and PRL10 polyplexes, pDNA was mostly localized in the cytoplasm and little pDNA was found in the nucleus. However, it seemed that pDNA in PRL2.5 and PRL5 polyplexes distributed almost evenly throughout the cytoplasm and the nucleus.
Figure 9.
Intracellular distribution of Cy5-labeled pDNA delivered by PRLx polyplexes (N/P 5) at 4 hr post-transfection in MCF7 cells.
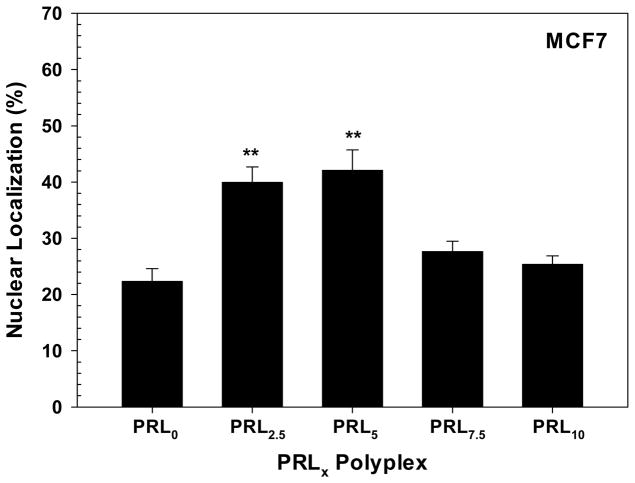
To clearly understand the nuclear localization of pDNA in PRLx polyplex-transfected MCF7 cells, the fluorescent intensity of pDNA was separately measured in the whole cell and in the nucleus (Figure 10). When transfecting MCF7 cells with PRL0 polyplexes, 22% of pDNA was found in the nucleus and the remaining 78% in the cytoplasm and acidic vesicles. Similarly, PRL7.5 and PRL10 polyplexes-transfected MCF7 cells had around 25% of pDNA in the nucleus. These PRLx polyplexes (x=0, 7.5, and 10) delivered 3–4-fold more pDNA in the cytoplasm and acidic vesicles than in the nucleus, but, as estimated in confocal microscope, PRL2.5 and PRL5 polyplexes delivered 40~42% of pDNA in the nucleus which is 1.8–1.9-fold more than that of PRL0 polyplexes. The different nuclear localization of pDNA may cause their different transfection efficiencies.
Figure 10.
Nuclear localization of Cy5-labeled pDNA delivered by PRLx polyplexes (N/P 5) at 4 hr post-transfection in MCF7 cells. Nuclear localization was calculated using relative fluorescence intensity ratio in cell and the nucleus (n = 10; Mean ± Standard Error).
Most researches for developing effective polymeric gene carriers have focused on new polymers with functionalities (e.g., colloidal stability, receptor-targetability, endosomolytic activity, nuclear translocation, quick pDNA release, and cytocompatibility) that can overcome various extracellular and intracellular hurdles during a journey of polyplexes from an administration site to the nucleus as a subcellular organelle of interest.1, 2 Although the significance of timely release of pDNA in the cytosol or the nucleus in polymeric transfection efficiency is recognized,5 the relationship between decomplexation rate, intracellular localization, and transfection efficiency of pDNA delivered with polymeric carriers has been widely ignored or not investigated. This study has shown that the pDNA contents in the nucleus is influenced by decomplexation rates of pDNA from polyplexes, and in turn influences the transfection efficiency of the polyplexes. These findings will give greater understanding in polymer-based gene delivery systems and will help to design effective functionalized polyplexes.
CONCLUSION
Polyplexes prepared with a mixture of PLL and RPLL were used to understand the relationship between decomplexation rate, intracellular localization, and transfection efficiency. Although the size and surface charge of PRLx polyplexes were influenced by the RPLL content, these characteristics did not influence cellular uptake of PRLx polyplexes. However, the highest transfection efficiency of PRLx polyplexes in this study may be caused by an optimal decomplexation rate (faster than that of PLL polyplexes and slower than that of RPLL polyplexes) and more nuclear localization of pDNA. These findings will help to design decomplexation-tunable polyplexes made of various combinations of non-degradable and degradable polymers.
Supplementary Material
Acknowledgments
This work was supported by NIH GM82866. We appreciate Hana Cho (The Catholic University of Korea) for her technical assistant and Joseph Nichols (The University of Utah) for thoughtful comments and helpful corrections.
Footnotes
The authors declare no competing financial interest.
Supporting Information Available. Supporting information includes: details about the synthetic scheme and 1H-NMR analysis of RPLL; Condensation of pDNA with either PLL or RPLL in agarose gel; Normalized transfection efficiencies of PLL100-xRPLLx/pDNA complexes (PRLx polyplexes) (N/P 5) in MCF7 cells and HEK293 cells; Dose-dependent cytotoxicity of PLL and RPLL in MCF7 and HEK293 cells. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kang HC, Lee M, Bae YH. Polymeric gene carriers. Crit Rev Eukaryot Gene Expr. 2005;15:317–342. doi: 10.1615/critreveukargeneexpr.v15.i4.30. [DOI] [PubMed] [Google Scholar]
- 2.Kang HC, Huh KM, Bae YH. Polymeric nucleic acid carrier: Current issues and novel design approaches. J Control Release. 2012;164:256–264. doi: 10.1016/j.jconrel.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner E. Polymers for siRNA delivery: inspired by viruses to be targeted, dynamic, and precise. Acc Chem Res. 2012;45:1005–1013. doi: 10.1021/ar2002232. [DOI] [PubMed] [Google Scholar]
- 4.Son S, Namgung R, Kim J, Singha K, Kim WJ. Bioreducible polymers for gene silencing and delivery. Acc Chem Res. 2012;45:1100–12. doi: 10.1021/ar200248u. [DOI] [PubMed] [Google Scholar]
- 5.Grigsby CL, Leong KW. Balancing protection and release of DNA: tools to address a bottleneck of non-viral gene delivery. J R Soc Interface. 2010;7:S67–82. doi: 10.1098/rsif.2009.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang HC, Bae YH. Co-delivery of small interfering RNA and plasmid DNA using a polymeric vector incorporating endosomolytic oligomeric sulfonamide. Biomaterials. 2011;32:4914–24. doi: 10.1016/j.biomaterials.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang HC, Bae YH. pH-Tunable endosomolytic oligomers for enhanced nucleic acid delivery. Adv Funct Mater. 2007;17:1263–1272. [Google Scholar]
- 8.Ruponen M, Yla-Herttuala S, Urtti A. Interactions of polymeric and liposomal gene delivery systems with extracellular glycosaminoglycans: physicochemical and transfection studies. Biochim Biophys Acta. 1999;1415:331–41. doi: 10.1016/s0005-2736(98)00199-0. [DOI] [PubMed] [Google Scholar]
- 9.Moret I, Esteban Peris J, Guillem VM, Benet M, Revert F, Dasi F, Crespo A, Alino SF. Stability of PEI-DNA and DOTAP-DNA complexes: effect of alkaline pH, heparin and serum. J Control Release. 2001;76:169–81. doi: 10.1016/s0168-3659(01)00415-1. [DOI] [PubMed] [Google Scholar]
- 10.Lu H, McDowell LM, Studelska DR, Zhang L. Glycosaminoglycans in Human and Bovine Serum: Detection of Twenty-Four Heparan Sulfate and Chondroitin Sulfate Motifs Including a Novel Sialic Acid-modified Chondroitin Sulfate Linkage Hexasaccharide. Glycobiol Insights. 2010:13–28. [PMC free article] [PubMed] [Google Scholar]
- 11.Huth S, Hoffmann F, von Gersdorff K, Laner A, Reinhardt D, Rosenecker J, Rudolph C. Interaction of polyamine gene vectors with RNA leads to the dissociation of plasmid DNA-carrier complexes. J Gene Med. 2006;8:1416–24. doi: 10.1002/jgm.975. [DOI] [PubMed] [Google Scholar]
- 12.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 13.Mishra D, Kang HC, Bae YH. Reconstitutable charged polymeric (PLGA)2-b-PEI micelles for gene therapeutics delivery. Biomaterials. 2011;32:3845–3854. doi: 10.1016/j.biomaterials.2011.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang HC, Kang HJ, Bae YH. A reducible polycationic gene vector derived from thiolated low molecular weight branched polyethyleneimine linked by 2-iminothiolane. Biomaterials. 2011;32:1193–1203. doi: 10.1016/j.biomaterials.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mann A, Thakur G, Shukla V, Singh AK, Khanduri R, Naik R, Jiang Y, Kalra N, Dwarakanath BS, Langel U, Ganguli M. Differences in DNA condensation and release by lysine and arginine homopeptides govern their DNA delivery efficiencies. Mol Pharmaceutics. 2011;8:1729–1741. doi: 10.1021/mp2000814. [DOI] [PubMed] [Google Scholar]
- 16.Kim YH, Park JH, Lee M, Kim YH, Park TG, Kim SW. Polyethylenimine with acid-labile linkages as a biodegradable gene carrier. J Control Release. 2005;103:209–19. doi: 10.1016/j.jconrel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Ou M, Xu R, Kim SH, Bull DA, Kim SW. A family of bioreducible poly (disulfide amine)s for gene delivery. Biomaterials. 2009;30:5804–14. doi: 10.1016/j.biomaterials.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Koo H, Jin GW, Mo H, Cho MY, Park JY, Choi JS, Park JS. Poly (ethylene oxide sulfide): new poly (ethylene glycol) derivatives degradable in reductive conditions. Biomacromolecules. 2005;6 (1):24–6. doi: 10.1021/bm049658l. [DOI] [PubMed] [Google Scholar]
- 19.Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. J Biol Chem. 2000;275:1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- 20.Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, Beatty B, Squire J, O’Brodovich H, Lukacs GL. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482–97. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- 21.Parelkar SS, Chan-Seng D, Emrick T. Reconfigurating polylysine architectures for controlling polyplex binding and non-viral transfection. Biomaterials. 2011;32:2432–2444. doi: 10.1016/j.biomaterials.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Oupicky D, Parker AL, Seymour LW. Laterally stabilized complexes of DNA with linear reducible polycations: strategy for triggered intracellular activation of DNA delivery vectors. J Am Chem Soc. 2002;124:8–9. doi: 10.1021/ja016440n. [DOI] [PubMed] [Google Scholar]
- 23.Read ML, Singh S, Ahmed Z, Stevenson M, Briggs SS, Oupicky D, Barrett LB, Spice R, Kendall M, Berry M, Preece JA, Logan A, Seymour LW. A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids. Nucleic Acids Res. 2005;33:e86. doi: 10.1093/nar/gni085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Read ML, Bremner KH, Oupicky D, Green NK, Searle PF, Seymour LW. Vectors based on reducible polycations facilitate intracellular release of nucleic acids. J Gene Med. 2003;5:232–45. doi: 10.1002/jgm.331. [DOI] [PubMed] [Google Scholar]
- 25.Kang HC, Samsonova O, Kang SW, Bae YH. The effect of environmental pH on polymeric transfection efficiency. Biomaterials. 2012;33:1651–62. doi: 10.1016/j.biomaterials.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang HC, Samsonova O, Bae YH. Trafficking microenvironmental pH of gene vector polycation in drug-sensitive and multidrug-resistant MCF7 breast cancer cell. Biomaterials. 2010;31:3071–3078. doi: 10.1016/j.biomaterials.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong JH, Kim SH, Christensen LV, Feijen J, Kim SW. Reducible poly (amido ethylenimine)-based gene delivery system for improved nucleus trafficking of plasmid DNA. Bioconjugate chemistry. 2010;21 (2):296–301. doi: 10.1021/bc9003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong JH, Kim SH, Christensen LV, Feijen J, Kim SW. Reducible poly (amido ethylenimine)-based gene delivery system for improved nucleus trafficking of plasmid DNA. Bioconjug Chem. 2010;21:296–301. doi: 10.1021/bc9003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuppusamy P, Li H, Ilangovan G, Cardounel AJ, Zweier JL, Yamada K, Krishna MC, Mitchell JB. Noninvasive imaging of tumor redox status and its modification by tissue glutathione levels. Cancer Res. 2002;62:307–12. [PubMed] [Google Scholar]
- 30.Manickam DS, Li J, Putt DA, Zhou QH, Wu C, Lash LH, Oupicky D. Effect of innate glutathione levels on activity of redox-responsive gene delivery vectors. J Control Release. 2010;141:77–84. doi: 10.1016/j.jconrel.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.