Abstract
The pathogenesis of incipient Alzheimer's disease (AD) has been resistant to analysis because of the complexity of AD and the overlap of its early-stage markers with normal aging. Gene microarrays provide new tools for addressing complexity because they allow overviews of the simultaneous activity of multiple cellular pathways. However, microarray data interpretation is often hindered by low statistical power, high false positives or false negatives, and by uncertain relevance to functional endpoints. Here, we analyzed hippocampal gene expression of nine control and 22 AD subjects of varying severity on 31 separate microarrays. We then tested the correlation of each gene's expression with MiniMental Status Examination (MMSE) and neurofibrillary tangle (NFT) scores across all 31 subjects regardless of diagnosis. These well powered tests revealed a major transcriptional response comprising thousands of genes significantly correlated with AD markers. Several hundred of these genes were also correlated with AD markers across only control and incipient AD subjects (MMSE > 20). Biological process categories associated with incipient AD-correlated genes were identified statistically (ease program) and revealed up-regulation of many transcription factor/signaling genes regulating proliferation and differentiation, including tumor suppressors, oligodendrocyte growth factors, and protein kinase A modulators. In addition, up-regulation of adhesion, apoptosis, lipid metabolism, and initial inflammation processes occurred, and down-regulation of protein folding/metabolism/transport and some energy metabolism and signaling pathways took place. These findings suggest a new model of AD pathogenesis in which a genomically orchestrated up-regulation of tumor suppressor-mediated differentiation and involution processes induces the spread of pathology along myelinated axons.
Alzheimer's disease (AD) has received intense study during past decades. Multiple processes have been implicated in AD, notably including abnormal β-amyloid (Aβ) production (1-7), tau hyperphosphorylation and neurofibrillary tangles (NFTs) (8, 9), synaptic pathology (10-12), oxidative stress (13-15), inflammation (5, 16-19), protein processing or misfolding (20, 21), calcium dyshomeostasis (15, 20-26), aberrant reentry of neurons into the cell cycle (27, 28), cholesterol synthesis (29, 30), and effects of hormones (23, 31) or growth factors (17, 32). Nevertheless, the pathogenic factors that initiate these processes remain elusive.
Several reasons account for the substantial resistance of AD pathogenesis to analysis. One is the vast extent and complexity of the disease, which affects numerous molecules, cells, and systems and impedes attempts to determine which alterations are specifically associated with early pathology. Another is that clinically normal subjects may exhibit considerable AD pathology, blurring criteria for distinguishing subjects with normal aging, mild cognitive impairment, or incipient AD (33-35).
We addressed the problems of high complexity and overlapping criteria by using a strategy combining powerful new gene microarray technology, which permits measurement of the expression of many thousands of genes simultaneously (36, 37), with statistical correlation analyses. This strategy allowed the linking of gene expression to cognitive and pathological markers of AD independently of AD diagnosis. We also focused on subjects with the earliest signs of AD. Several microarray studies of AD brain (38-42) and/or mouse models of AD (43) have been published. These studies have yielded important new insights, in particular, regarding changes in plasticity-related genes (e.g., ref. 43). However, few microarray studies use independent sample sizes sufficient to provide the statistical power needed to avoid high false positive (type I) and/or high false negative (type II) error (44, 45). In the present study, we ensured adequate power by using a separate array for each hippocampal sample of a large group of subjects (n = 31) and correlated the expression values of each of thousands of genes with pathological and cognitive indexes of incipient AD. Together, these approaches revealed a major and previously unrecognized transcriptional response with potentially important implications for the early pathogenesis of AD.
Methods
Human Brain Samples and Pathologic/Cognitive Assessment. Hippocampal specimens used in this study were obtained at autopsy from 35 subjects (16 female and 19 male; Table 1) through the Brain Bank of the Alzheimer's Disease Research Center at the University of Kentucky. At autopsy, coronal sections of the left hippocampus (3-5 mm) were immediately frozen in liquid nitrogen and stored at -80°C until analyzed. Adjacent sections were fixed in 10% formalin and used for neuropathologic evaluation. Except for borderline AD subjects (see below), all AD patients met Alzheimer's Disease and Related Disorders Association criteria for the clinical diagnosis of AD and Consortium to Establish a Registry for Alzheimer's Disease and National Institute of Aging-Reagan Institute neuropathology criteria for the diagnosis of AD. The frozen hippocampal tissues were warmed to -20°C to enable dissection of CA1 and CA3 under a Zeiss surgical microscope.
Table 1. Subjects in AD study.
| Control, n = 9 | Incipient, n = 7 | Moderate, n = 8 | Severe, n = 7 | |
|---|---|---|---|---|
| Age | 85.3 ± 2.7 | 90 ± 2.1 | 83.4 ± 1.1 | 84 ± 4.0 |
| NFT | 2.7 ± 1.0 | 9.4 ± 1.8 | 25.6 ± 3.5 | 32.7 ± 7.2 |
| Braak | 2.1 ± 0.4 | 5 ± 0.4 | 5.6 ± 0.2 | 5.9 ± 0.1 |
| MMSE | 27.7 ± 0.5 | 24.3 ± 1.1 | 16.5 ± 0.6 | 6 ± 1.4 |
| PMI | 2.6 ± 0.2 | 3.3 ± 0.6 | 3.2 ± 0.2 | 3 ± 0.1 |
Subjects were assigned to four groups reflecting different levels of AD severity or Control (see Methods). n, number of subjects in each group; Age, age at death; NFT, neurofibrillary tangle count; Braak, Braak stage; MMSE, adjusted Minimental Status Exam (see Methods); PMI, postmortem interval. Values are mean ± SEM.
The MiniMental State Examination (MMSE) is a reliable index of AD-related cognitive status at a given point in time (46). However, its rate of decline varies with severity, and mildly impaired patients show little MMSE decline even after several years (46). Recent MMSE data were available for most subjects but, in subjects for whom the interval between the most recent MMSE score and death was >1 year, the MMSE score was adjusted downward by one point per year. This approach likely underestimates MMSE decline for severely affected patients but seemed suitable for this study, given the slow MMSE decline in less impaired subjects (46) and our focus on such subjects. Postmortem scores on AD-related pathologic indices for Braak staging, hippocampal NFTs, and diffuse and neuritic senile plaques were determined as described (47). The MMSE and NFT values were selected as our primary markers for quantifying AD progression because of the Braak scale's limited range and because our NFT results correlated more closely with the MMSE (r = 0.45) than did our plaque values (r = 0.19), consistent with prior findings (33-35, 48). Further, the evidence that soluble rather than deposited Aβ may be more relevant to cognitive impairment is mounting (2, 5, 7).
Based primarily on MMSE criteria (35, 46), subjects were categorized initially into four groups, termed “Control” (MMSE >25), “Incipient AD” (MMSE 20-26), “Moderate AD” (MMSE 14-19), and “Severe AD” (MMSE <14) (Table 1). Several borderline cases (e.g., MMSE = 26) were assigned based on NFT, amyloid plaque, and Braak stage data. In addition, four subjects exhibited more cognitive deterioration (MMSE <20) than expected from their NFT or amyloid scores. Because these subjects were potentially affected by confounding conditions (49), they were excluded from the analyses, leaving n = 31 overall.
RNA Isolation and Affymetrix GeneChip Processing. Procedures for total RNA isolation, labeling, and microarray processing were similar to those described (44), except that human GeneChips (HG-U133A) and microarray suite 5 (mas5; 50) were used. Each subject's CA1 subfield RNA was processed and run on a separate chip. An average yield of 55 μg of biotin-labeled cRNA target was obtained from 8 μg of total RNA each per CA1 sample, of which 20 μg of cRNA was applied to one array. cRNA yield did not differ significantly among groups (P = 0.32), but the most severe AD group exhibited a trend toward lower cRNA levels, possibly reflecting greater cellular degeneration.
Microarray Data Analysis. Scaling and noise analyses were performed as described (44) and Affymetrix algorithms for signal intensity and presence P values (50), respectively, were used to determine expression (relative abundance) and detection reliability of transcripts. A gene probe set was rated “present” if it was detected on at least four chips in the study. Individual values were blanked and treated as missing values if they were >2 SD away from the group mean. Finally, probe sets were considered “genes” if they had been assigned a “gene symbol” annotation (Affymetrix database, www.affymetrix.com). Pearson's correlation tests and ANOVAs were performed in excel 9.0 on data copied from the mas5 pivot table, as described (44).
Biological Process Categorization by Gene Ontology. As noted, microarray studies face substantial false-positive concerns because of the large multiple comparison error (44, 45). Conversely, however, they can also strengthen statistical confidence by providing evidence of coregulation of multiple genes that are related by function or pathway (51-53). Here, we used a new software tool, the expression analysis systematic explorer (ease; http://david.niaid.nih.gov/david/ease.htm), to assign identified genes to “GO: Biological Process” categories of the Gene Ontology Consortium (www.geneontology.org) (51) and to test statistically (EASE Score, a modified Fisher's exact test) for significant coregulation (overrepresentation) of identified genes within each biological process category.
Results
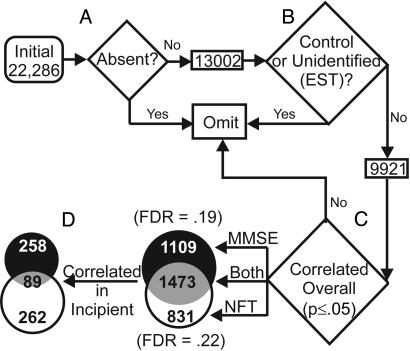
Gene Identification Algorithm (Fig. 1). To test thousands of genes for correlation with AD markers, while still managing multiple comparison error, we excluded all “absent” or undefined (expressed sequence tags) genes (Fig. 1 A and B), thereby reducing expected false positives (44). Pearson's test was then used to test each of the 9,921 remaining genes for its correlation with MMSE and NFT scores (Fig. 1C). A total of 3,413 genes were significantly associated (at P values of <0.05) with the MMSE, NFT, or both, across all 31 subjects (overall correlations). These correlated genes were termed “AD-related genes” (ADGs).
Fig. 1.
Gene identification algorithm. (A) Genes rated absent (see Methods) were excluded from analysis. (B) Only annotated probe sets (not expressed sequence tags) were included in the statistical analysis. (C) Pearson correlation was performed for every gene against both MMSE and NFT measures of each subject. Venn diagram shows the number of genes significantly correlated (P ≤ 0.05) with both MMSE and NFT or either index alone. For each index, the false discovery rate (FDR) was calculated. (D) For the genes found to correlate significantly across all subjects (overall, n = 31), another Pearson's correlation was performed post hoc among only the subjects rated either “Control” or “Incipient” (Incipient, n = 16).
For both the MMSE and NFT analyses, we calculated the false discovery rate, the number of false positives expected because of multiple comparisons divided by the total positives found. The false discovery rate provides a worst-case probability that any gene identified (e.g., at P < 0.05) by correlation is significant because of the error from multiple testing (44, 54). The observed false discovery rates (≈0.20; Fig. 1) are reasonably low for a microarray study, in particular, considering the relatively relaxed P value (P < 0.05), indicating good statistical power. [The false discovery rate generally decreases with more stringent P value criteria (44, 54). However, the confidence lost with a relaxed P value is substantially offset by the increased confidence gained from expanding the overall number of identified genes and strengthening the ease analysis of coregulation (44, 52, 53).]
Because NFT scores increase and MMSE scores decrease with AD severity, genes up-regulated with AD could only correlate positively with NFT scores or negatively with the MMSE, whereas genes down-regulated with AD could only correlate positively with the MMSE or negatively with NFT scores. Fig. 2 illustrates examples of the four patterns of correlation that were possible for ADGs. Overall, 1,977 ADGs were up-regulated and 1,436 were down-regulated. More were correlated with the MMSE than with NFT scores. The full set of all identified ADGs is included in Table 5, which is published as supporting information on the PNAS web site.
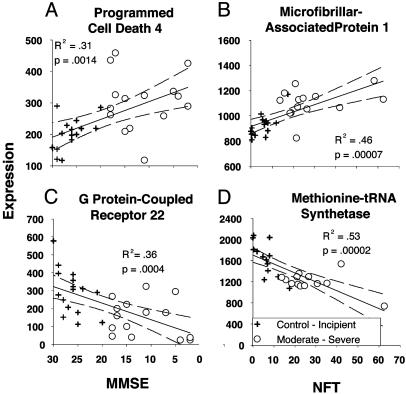
Fig. 2.
Examples of correlated genes illustrating the four directions of correlation through which genes were identified. For each gene, expression intensity is plotted on the y axis, and MMSE (A Left and C Left) or NFT (B Right and D Right) scores are plotted on the x axis; R2 value, P value (Pearson's test), linear fit (black line), and 95% confidence intervals (dashed lines) are also shown. The MMSE scale is reversed, so that more advanced AD increases to the right on both indexes. (A and B) Genes for which expression levels were up-regulated with AD, identified by negative or positive correlation with MMSE (A) or NFT (B) scores, respectively. (C and D) Genes for which expression levels were down-regulated with AD, identified by positive or negative correlation with MMSE (C) or NFT (D), respectively.
In a subsequent step (Fig. 1D), we identified post hoc those genes within this large set of ADGs that also correlated with AD markers across a smaller subgroup comprising incipient AD and control subjects (i.e., all subjects with MMSE ≥ 20 and NFT < 20) (n = 16). Within this subset, only genes correlated in the same direction as their overall correlations were considered. Of the 3,413 overall ADGs, 609 were found also to correlate significantly (at P values of ≤0.05) in the incipient subgroup, 258 with the MMSE, 262 with NFT scores, and 89 with both (termed “Incipient ADGs” or IADGs). More IADGs were up-regulated with AD (431 genes) than were down-regulated (178 genes) (see Table 6, which is published as supporting information on the PNAS web site, for alphabetical lists of all IADGs).
Biological Processes Associated with ADGs and IADGs. Using ease analysis (see Methods), we identified biological process categories that showed a disproportionately high number of coregulated genes (significant overrepresentation of ADGs or IADGs in those categories). The Gene Ontology Biological Process categories in which ADGs were overrepresented by ease score (in general, at P values of ≤0.05) are shown in Table 2. The overrepresented categories for IADGs are shown in Table 3. Because of the reduced number of genes and lower statistical power in this post hoc analysis, however, we set the significance level for identified categories of IADGs to P < 0.15.
Table 2. Biological process categories overrepresented by overall correlations (ADGs).
| Up-regulated (Total: 1,572/6,265; 25.1%) | ease | N/M/B | Down-regulated (Total: 1,126/6,265; 18.0%) | ease | N/M/B |
|---|---|---|---|---|---|
| Regulation of transcription (269/792; 34%) | 0.0000 | 21/38/41 | Energy pathways (57/151; 37.7%) | 0.0000 | 15/15/69 |
| Cell proliferation (210/666; 31.5%) | 0.0001 | 23/43/35 | ATP biosynthesis (16/23; 69.6%) | 0.0000 | 18/9/73 |
| Oncogenesis (24/47; 51.1%) | 0.0003 | 21/39/39 | Synaptic transmission (49/143; 34.3%) | 0.0000 | 9/30/61 |
| Protein amino acid phosphorylation (104/310; 33.5%) | 0.0006 | 23/30/47 | Coenzyme biosynthesis (20/40; 50%) | 0.0000 | 15/15/69 |
| Transition metal ion homeostasis (10/16; 62.5%) | 0.0076 | 18/45/36 | Cation transport (60/197; 30.5%) | 0.0000 | 13/18/69 |
| Positive regulation cell proliferation (25/62; 40.3%) | 0.0119 | 18/68/14 | Protein folding (30/86; 34.9%) | 0.0003 | 32/11/57 |
| Chromatin architecture (34/94; 36.2%) | 0.0186 | 25/43/33 | Tricarboxylic acid cycle (12/22; 54.5%) | 0.0006 | 27/27/47 |
| Nucleosome assembly (13/27; 48.1%) | 0.0219 | 11/56/33 | Glycolysis (14/29; 48.3%) | 0.0007 | 6/18/76 |
| Histogenesis and organogenesis (22/57; 38.6%) | 0.0319 | 22/17/61 | Neurogenesis (64/244; 26.2%) | 0.0011 | 19/27/53 |
| Cell adhesion (94/314; 29.9%) | 0.0346 | 19/46/35 | Amino acid catabolism (13/30; 43.3%) | 0.0038 | 33/0/67 |
| Development (235/850; 27.6%) | 0.0425 | 21/42/37 | Ubiquitin-dependent protein catabolism (27/87; 31%) | 0.0043 | 48/13/39 |
| Complement activation, classical (9/18; 50%) | 0.0576 | 10/40/50 | Secretion (14/37; 37.8%) | 0.0095 | 0/35/65 |
| Negative regulation cell proliferation (28/83; 33.7%) | 0.0762 | 09/50/41 | Protein transport (66/288; 22.9%) | 0.0245 | 26/25/49 |
| Isoprenoid metabolism (6/10; 60%) | 0.0789 | 00/83/17 | Neurotransmitter metabolism (6/11; 54.5%) | 0.0329 | 17/17/67 |
| Apoptosis (72/255; 29.5%) | 0.0818 | 13/32/55 | Axon guidance (8/19; 42.1%) | 0.0404 | 27/9/64 |
| Defense response (102/360; 28.3%) | 0.1010 | 15/57/28 | Calcium ion transport (11/32; 34.4%) | 0.0482 | 7/7/87 |
| Lipid metabolism (82/288; 28.5%) | 0.1250 | 15/47/38 | Microtubule-based process (20/73; 27.4%) | 0.0538 | 11/21/68 |
Biological process categories significantly overrepresented by ADGs (P ≤ 0.05; ease score) and a few other selected categories are shown. Numerous other similar significant categories are not included to reduce redundancy. Significant functional categories are those with a higher ratio of identified genes to all genes tested on the array for associations with that category, relative to the ratio of total identified genes in the study to all genes tested on the array for associations with all categories. Association numbers approximate but are not exactly equal to gene numbers in a category. After each category description (in parentheses) is the ratio of associations for that category and the percentage represented by that ratio. The analogous ratios for total identified up-regulated and down-regulated genes are shown in the headings (Total). ease, modified Fisher's exact test P value; N/M/B, percentage of genes included in category because they were significant by NFT correlation (N), MMSE correlation (M), or both (B). (The complete list of ADGs is given alphabetically in Table 5).
Table 3. Biological process categories overrepresented by incipient correlations (IADGs).
| Up-regulated (Total: 379/6,265; 6%) | ease | N/M/B | Down-regulated (Total: 154/6,265; 3%) | ease | N/M/B |
|---|---|---|---|---|---|
| Regulation of transcription, DNA... (64/781; 8%) | 0.008 | 30/49/21 | Protein folding (13/86; 15%) | 0.000 | 71/21/7 |
| Histogenesis and organogenesis (9/57; 16%) | 0.020 | 33/44/22 | Axon cargo transport (3/5; 60%) | 0.006 | 67/33/0 |
| Chromatin assembly/disassembly (8/52; 15%) | 0.035 | 22/78/0 | Synaptic transmission (10/143; 7%) | 0.008 | 33/67/11 |
| Cell proliferation (52/666; 8%) | 0.041 | 30/46/23 | Protein metabolism (46/1,415; 3%) | 0.028 | 64/28/9 |
| Cell adhesion (26/314; 8%) | 0.092 | 36/46/18 | Microtubule-based movement (4/33; 12%) | 0.046 | 50/50/0 |
| Development (61/850; 7%) | 0.103 | 38/43/19 | Electron transport (10/200; 5%) | 0.055 | 45/36/18 |
| Protein amino acid phosphorylation (25/310; 8%) | 0.122 | 38/46/15 | Cytokinesis (5/61; 8%) | 0.061 | 60/20/20 |
| Cell motility (16/182; 9%) | 0.134 | 35/41/24 | Intracellular transport (15/369; 4%) | 0.066 | 58/37/5 |
| Lipid metabolism (23/288; 8%) | 0.148 | 48/39/13 | GPCR signaling pathway (11/264; 4%) | 0.111 | 33/53/13 |
| Apoptosis (20/244; 8%) | 0.150 | 24/62/14 | Cell surface signal transduction (17/492; 4%) | 0.145 | 43/48/10 |
Biological process categories significantly overrepresented by IADGs (P ≤ 0.15; ease score) and a few other selected categories are shown. Numerous other similar significant categories are not included to reduce redundancy. Significant functional categories are those with a higher ratio of identified genes to all genes tested on the array for associations with that category, relative to the ratio of total identified genes in the study to all genes tested on the array for associations with all categories. The association numbers approximate but are not exactly equal to gene numbers in a category. After each category description (in parentheses) is the ratio of associations for that category and the percentage represented by that ratio. The analogous ratios for total identified up-regulated and down-regulated genes are shown in the headings (Total). ease, modified Fisher's exact test P value; N/M/B, percentage of genes included in category because they were significant by NFT correlation (N), MMSE correlation (M), or both (B). (The complete list of IADGs is given alphabetically in Table 6).
Although many overrepresented categories were similar between Tables 2 and 3, notable differences also occurred. The categories shown in Table 3 were of particular interest because they reflect groups of genes correlated with AD markers in the incipient subjects. Transcription factor, proliferation, and development processes were among the largest categories of up-regulated IADGs. In addition, extracellular matrix/cell adhesion/motility processes, comprising multiple laminins (A2,4), integrins (A1,6,7), tenascins, collagens, cadherins, proteoglycans, and amyloid precursor protein were up-regulated. Of note, several individual members of the semaphorin/plexin pathway, which inhibits axonal elongation, also were up-regulated IADGs (e.g., SEMA3B and plexin B2) (Table 6). Further, histogenesis, apoptosis, phosphorylation, and lipid metabolism, including prostaglandin synthesis, were overrepresented by up-regulated IADGs (Table 3). Although their categories were not overrepresented, several up-regulated IADGs reflected inflammatory and oxidative stress processes (e.g., IFN-γ, IL-18, interleukin receptors, and AOP2) (Table 6).
For down-regulated categories, a major difference was seen between ADGs and IADGs, in that multiple protein metabolism categories, including folding and transport (immunophilins, chaperones, and heat shock proteins), were overrepresented by IADGs (Table 3) but not ADGs (Table 2). One of the hallmarks of AD, reduced energy metabolism, which dominated the down-regulated categories of ADGs (Table 2), was only reflected in one category, electron transport, of down-regulated IADGs (Table 3).
Calcium Signaling Regulation. Altered Ca2+ signaling is suspected of a role in AD and brain aging and also was identified in a recent microarray study of aging (44). Although signaling pathways in general, including Ca2+ pathways and transport systems, were down-regulated in AD (Tables 2 and 3), some individual up-regulated Ca2+-dependent IADGs included the cAMP response element-binding protein (CREB) cofactor (EP300), a calpain inhibitor (calpastatin), S100A4, and the Ca2+-dependent death-associated protein kinase (DAPK2) (Table 6).
Transcription Factors (TFs). The TF category was the most significantly overrepresented by up-regulated IADGs and ADGs. Table 4 shows the TF-category IADGs correlated with NFT, MMSE scores, or both (only those correlated at P values of ≤0.025 are shown). Review of the functions of the identified TFs revealed that a disproportionately high number are tumor suppressors (TSs) or TS cofactors (boldface), including several of the retinoblastoma (RB) family (also see Table 6 for additional RB members). Many other identified TFs are related to lipid/cholesterol biosynthesis and adipocyte differentiation (underlined). Numerous zinc finger TFs favoring transcriptional repression also were identified. Paradoxically, however, a considerable number of the remaining TFs are associated with growth or proliferation. In general, more up-regulated TFs for TS and lipogenesis were correlated with NFT scores than with MMSE, whereas more growth-related TFs were correlated with MMSE (Table 4; see Table 6 for gene descriptions).
Table 4. Up-regulated IADGs categorized as TFs.
| + NFT | ||||
| ZNF253 | CEBPA | RBAK | THG-1 | KLF2 |
| SREBF1 | NF1-C | PML | ZNF268 | RBL 1(p107)* |
| C20orf104 | RBBP1 | GL 12 | ZBRK1 | PPARBP* |
| RXRB | CERD4 | ASCL 1 | GTF21 | |
| − MMSE | ||||
| SMARCC2 | RUNX2 | ZNF198 | SP1B | SP3 |
| BRD1 | TIX1 | CHD2 | HMGB3 | ENSR1 |
| ZNF32 | LOC51580 | HOXB5 | HOXC4 | Rpo1-2 |
| ZNF7 | C22orf | NCOA3 | TCF3 | PRKR |
| ZNF43 | ID4 | EP300 | PB1 | ZNF136* |
| ZNF254 | ZNF237 | ZNF83 | ZNF84 | |
Gene symbols for TF IADGs positively correlated with NFT, negatively correlated with MMSE, or both (*) are shown separately (only those with P ≤ 0.025). IADGs for TS (boldface) or lipogenic (underlined) functions are highlighted. (Full descriptions of all IADGs, alphabetically listed, are available in Table 6).
TF category IADGs correlated with both NFT and MMSE scores.
TS. The high proportion of TS-related TFs prompted us to inspect other biological process categories for genes with TS functions. Many IADGs with TS or cellular differentiation functions were found in the phosphorylation, apoptotic, cell cycle, and other categories (e.g., TGF-β, GSK3B, PDCD4, FZR1, SFRP1, AIM1, DAPK2, and CDK2AP1). Conversely, inspection of the down-regulated TF categories (not shown) revealed many TFs important for growth and proliferation, including several of the MYC family (MGA and IRLB) and DP1(TFDP1), a member of the growth-promoting E2F family targeted by the RB family of TSs (Table 6).
PKA Pathways. The cAMP-dependent protein kinase (PKA) pathway stimulates growth in some cell types and differentiation and inhibition of growth in others (55). Several PKA-related genes were up-regulated IADGs, including A kinase-anchoring molecules (AKAP9, AKAP13, and CAP350), adenylate cyclase 7, and the PKA RIIα subunit (Table 6).
Discussion
Overview of Gene Changes in Incipient AD. These studies reveal that widespread changes in genomic regulation of multiple cellular pathways are major correlates of incipient AD. As noted, it has been recognized previously that inflammation, synaptic dysfunction, energy failure, glial reactivity, protein misfolding, lipogenesis and cell cycle disturbances accompany AD. However, the major transcriptional orchestration seen here in incipient AD may provide a new perspective on the possible origins of these deleterious processes. In addition, the widespread activation of growth, differentiation, and TS pathways, and the apparent collapse of protein transport machinery so early in the disease, suggest clues on the early pathogenesis of AD. These conclusions are supported by reasonably high levels of statistical confidence for individual genes and by statistical evidence of coregulation of genes within related pathways and categories.
Activation of TS Pathways. Multiple TSs, some of which regulate the cell cycle, were identified here within the TF (Table 4) and other categories. Previous studies have found evidence of cell cycle reentry in neurons of the AD brain (27, 28), and a handful of studies have also examined TSs in relation to AD, largely in terms of their roles in apoptotic pathways (e.g., p53) (28, 56-58). However, TSs have other actions unrelated to apoptosis and can, in fact, be antiapoptotic (57, 59). Notably, TSs play critical roles in cellular differentiation related to development and tumor suppression. For example, overexpression of some TSs (e.g., RB proteins) induces cell cycle arrest, differentiation, and process extension in astrocytomas (58, 60). TS expression also is necessary for neurite extension and synaptogenesis in neuronal development (57, 61). Moreover, in some cell types, TSs operate by inducing cellular senescence and inhibiting protein biosynthesis (62).
Many of the other identified processes reflect differentiation or senescence pathways and could therefore mediate TS actions. For example, the extensive extracellular matrix (ECM) remodeling and cell adhesion changes observed, presumably mediated largely by astrocytes, are similar to processes seen during astrocyte differentiation or cell-type-specific responses to injury (63-65). Analogous ECM/adhesion pathways are also used by peripheral cells to suppress cancer cell invasion and metastasis during oncogenesis (66). Similarly, inflammatory responses, mediated by glial cells in the brain (17, 18), also can be tumor-suppressive (67-69). Further, the generalized shutdown of protein metabolism seen here in early AD (Table 3) is a common manifestation of TS-mediated cellular senescence in some cell types (62). Also, the up-regulation of lipid metabolism pathways (Tables 3 and 4) may reflect activation of oligodendrocyte (OG) myelination programs (44, 70), which are expressed primarily in terminally differentiated OGs and can be directly activated by TSs (71). In turn, altered ECM, impaired microtubular transport of proteins, and disturbed myelination could be unfavorable for axonal elongation and maintenance. Interestingly, GSK3B, an up-regulated IADG (Table 6) that exhibits TS activity in the Wnt pathway, also plays a role in the hyperphosphorylation of tau (8, 9), in which it acts cooperatively with PKA (72). Thus, multiple processes identified in incipient AD could reflect TS-mediated differentiation or senescence responses of specific brain cell types and result in early pathology of myelinated axons.
This possibility, in turn, raises the question of what might trigger TS activation. TSs can be activated by developmental factors, DNA/cellular damage, or dysregulation of the cell cycle. Therefore, oxidative stress, inflammation, or abnormal Ca2+ signaling are clearly candidate activators of TSs. In addition, TSs act as negative feedback regulators of growth and are often elevated in response to excess growth factor (GF) production in tumors (73). Many up-regulated GFs also were identified here (Table 4), perhaps originating in OGs and their progenitors, which retain substantial growth potential in adult brain. Consistent with this possibility, several of the up-regulated IADGs, including PDGFB, FYN, and FGFR3, play major roles in OG proliferation, differentiation, and myelinogenesis (74, 75). However, if correct, this interpretation would still leave unanswered the question of what stimulates excess GF release from OGs (but see ref. 44).
Implications of NFT- and MMSE-Specific Correlated Processes. Although more IADGs were correlated with MMSE than NFT scores, several major processes were notable exceptions (Table 3), in particular, up-regulated lipid metabolism and down-regulated protein folding/metabolism. Among up-regulated TFs (Table 4), IADGs for TSs, lipid biosynthesis, and transcriptional repression were more often correlated with NFT scores. Further, up-regulation of semaphorins (SEMA3B) and axonal semaphorin receptors (plexinB2 and ESDN), which mediate growth cone collapse and axonal retraction and guide OG migration (76-78), were correlated with NFT scores (Table 6). In contrast, TFs for growth (Table 4) and genes related to adhesion and inflammation were more often correlated with MMSE.
A Model of AD Progression Along Myelinated Axons. The transcriptional responses identified here and their marker-specific correlations suggest a previously unreported model of incipient AD pathology (Fig. 3). Alterations in axons or myelin sheaths initially stimulate growth/remyelination responses in localized OGs, which in turn secrete GFs that activate adjacent neurons and glial cells. This triggers compensatory TS responses specific to cell type that induce protein aggregation, affect axonal-myelin interactions, and result in NFTs. As NFT density increases, wider ECM, amyloid precursor protein, and inflammatory changes may be triggered that impact cognition. This model could help to explain why AD pathogenesis appears to march along myelinated axons from the entorhinal cortex to hippocampus and neocortex (48, 79), leaving NFTs and plaques in its wake.
Fig. 3.
Schematic model. In this model, OGs are activated either by damage to myelin (asterisks in red) or by endogenous deregulation, resulting in GF production and remyelination (lipogenic) growth responses. GFs from OGs trigger oligodendrocyte progenitor (OGP) cells to divide, but they also reach other cell types through extracellular space and perhaps through the myelin sheath into axons and adjacent OGs. Excess GFs from OGs trigger TS pathways specific to various brain cell types, which, in turn, induce unfavorable ECM changes by astroglia (A), proinflammatory cytokines (INF) from microglia (MG) and astroglia, and repression of protein synthesis (PS) in neurons (N). These TS responses impair axonal protein transport, induce axonal retraction, activate additional remyelination programs, and culminate in NFTs and, perhaps, altered amyloid precursor protein processing. This process begins in the entorhinal cortex and spreads sequentially through adjacent OGs (1-4) along myelinated axons to the hippocampus and neocortex.
In summary, the present studies revealed widespread and apparently orchestrated transcriptional responses associated with early signs of AD pathology. Dissecting the bases for these early responses should yield important insights into pathogenic mechanisms and suggest therapeutic approaches to AD.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Snyder for drawing our attention to possible changes in semaphorins and their potential relevance to axonal pathology and Drs. David Wekstein and Daron Davis for important contributions to subject recruitment and assessment. This research was supported by National Institute on Aging Grants AG10836 and AG05144.
Abbreviations: MMSE, MiniMental Status Examination; NFT, neurofibrillary tangle; AD, Alzheimer's disease; ADG, AD-related gene; IADG, incipient ADG; TS, tumor suppressor; TF, transcription factor; RB, retinoblastoma; ECM, extracellular matrix; GF, growth factor; OG, oligodendrocyte.
References
- 1.Tanzi, R. E. & Bertram, L. (2001) Neuron 32, 181-184. [DOI] [PubMed] [Google Scholar]
- 2.Klein, W. L., Krafft, G. A. & Finch, C. E. (2001) Trends Neurosci. 24, 219-224. [DOI] [PubMed] [Google Scholar]
- 3.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353-356. [DOI] [PubMed] [Google Scholar]
- 4.Price, D. L. & Sisodia, S. S. (1998) Annu. Rev. Neurosci. 21, 479-505. [DOI] [PubMed] [Google Scholar]
- 5.Mucke, L., Masliah, E., Yu, G. Q., Mallory, M., Rockenstein, E. M., Tatsuno, G., Hu, K., Kholodenko, D., Johnson-Wood, K. & McConlogue, L. (2000) J. Neurosci. 20, 4050-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullan, M. & Crawford, F. (1994) Mol. Neurobiol. 9, 15-22. [DOI] [PubMed] [Google Scholar]
- 7.Morgan, D. (2003) Neurochem. Res. 28, 1029-1034. [DOI] [PubMed] [Google Scholar]
- 8.Noble, W., Olm, V., Takata, K., Casey, E., Mary, O., Meyerson, J., Gaynor, K., LaFrancois, J., Wang, L., Kondo, T., et al. (2003) Neuron 38, 555-565. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, G. V. & Bailey, C. D. (2002) J. Alzheimer's Dis. 4, 375-398. [DOI] [PubMed] [Google Scholar]
- 10.Sze, C. I., Troncoso, J. C., Kawas, C., Mouton, P., Price, D. L. & Martin, L. J. (1997) J. Neuropathol. Exp. Neurol. 56, 933-944. [DOI] [PubMed] [Google Scholar]
- 11.Scheff, S. W. & Price, D. A. (2001) J. Alzheimer's Dis. 3, 495-505. [DOI] [PubMed] [Google Scholar]
- 12.Masliah, E., Mallory, M., Hansen, L., DeTeresa, R., Alford, M. & Terry, R. (1994) Neurosci. Lett. 174, 67-72. [DOI] [PubMed] [Google Scholar]
- 13.Aksenov, M. Y., Aksenova, M. V., Butterfield, D. A., Geddes, J. W. & Markesbery, W. R. (2001) Neuroscience 103, 373-383. [DOI] [PubMed] [Google Scholar]
- 14.Bickford, P. C., Gould, T., Briederick, L., Chadman, K., Pollock, A., Young, D., Shukitt-Hale, B. & Joseph, J. (2000) Brain Res. 866, 211-217. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, G. E. (2002) Free Radical Biol. Med. 32, 1061-1070. [DOI] [PubMed] [Google Scholar]
- 16.Gemma, C., Mesches, M. H., Sepesi, B., Choo, K., Holmes, D. B. & Bickford, P. C. (2002) J. Neurosci. 22, 6114-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mrak, R. E. & Griffin, W. S. (2001) Neurobiol. Aging 22, 903-908. [DOI] [PubMed] [Google Scholar]
- 18.Finch, C. E., Morgan, T., Rozovsky, I., Xie, Z., Weindruch, R. & Prolla, T. (2002) in Microglia in the Degenerating and Regenerating CNS, ed. Streit, W. J. (Springer, New York).
- 19.Rogers, J., Webster, S., Lue, L. F., Brachova, L., Civin, W. H., Emmerling, M., Shivers, B., Walker, D. & McGeer, P. (1996) Neurobiol. Aging 17, 681-686. [DOI] [PubMed] [Google Scholar]
- 20.Nixon, R. A., Mathews, P. M. & Cataldo, A. M. (2001) J. Alzheimer's Dis. 3, 97-107. [DOI] [PubMed] [Google Scholar]
- 21.Forman, M. S., Lee, V. M. & Trojanowski, J. Q. (2003) Trends Neurosci. 26, 407-410. [DOI] [PubMed] [Google Scholar]
- 22.Disterhoft, J. F., Moyer, J. R., Jr., & Thompson, L. T. (1994) Ann. N. Y. Acad. Sci. 747, 382-406. [DOI] [PubMed] [Google Scholar]
- 23.Landfield, P. W., Thibault, O., Mazzanti, M. L., Porter, N. M. & Kerr, D. S. (1992) J. Neurobiol. 23, 1247-1260. [DOI] [PubMed] [Google Scholar]
- 24.Nixon, R. A., Saito, K. I., Grynspan, F., Griffin, W. R., Katayama, S., Honda, T., Mohan, P. S., Shea, T. B. & Beermann, M. (1994) Ann. N. Y. Acad. Sci. 747, 77-91. [DOI] [PubMed] [Google Scholar]
- 25.Mattson, M. P., LaFerla, F. M., Chan, S. L., Leissring, M. A., Shepel, P. N. & Geiger, J. D. (2000) Trends Neurosci. 23, 222-229. [DOI] [PubMed] [Google Scholar]
- 26.Thibault, O., Porter, N. M., Chen, K. C., Blalock, E. M., Kaminker, P. G., Clodfelter, G. V., Brewer, L. D. & Landfield, P. W. (1998) Cell Calcium 24, 417-433. [DOI] [PubMed] [Google Scholar]
- 27.Arendt, T., Holzer, M., Stobe, A., Gartner, U., Luth, H. J., Bruckner, M. K. & Ueberham, U. (2000) Ann. N. Y. Acad. Sci. 920, 249-255. [DOI] [PubMed] [Google Scholar]
- 28.Bowser, R. & Smith, M. A. (2002) J. Alzheimer's Dis. 4, 249-254. [DOI] [PubMed] [Google Scholar]
- 29.Petanceska, S. S., DeRosa, S., Olm, V., Diaz, N., Sharma, A., Thomas-Bryant, T., Duff, K., Pappolla, M. & Refolo, L. M. (2002) J. Mol. Neurosci. 19, 155-161. [DOI] [PubMed] [Google Scholar]
- 30.Puglielli, L., Konopka, G., Pack-Chung, E., Ingano, L. A., Berezovska, O., Hyman, B. T., Chang, T. Y., Tanzi, R. E. & Kovacs, D. M. (2001) Nat. Cell Biol. 3, 905-912. [DOI] [PubMed] [Google Scholar]
- 31.Green, P. S. & Simpkins, J. W. (2000) Ann. N.Y. Acad. Sci. 924, 93-98. [DOI] [PubMed] [Google Scholar]
- 32.Hoffer, B. & Olson, L. (1997) J. Neural Transm. 49, Suppl., 1-10. [DOI] [PubMed] [Google Scholar]
- 33.Terry, R. (1999) in Alzheimer's Disease, eds. Terry, R., Katzman, R., Bick, K. & Sisodia, S. S. (Lippincott Williams &Wilkins, Philadelphia), pp. 187-206.
- 34.Schmitt, F. A., Davis, D. G., Wekstein, D. R., Smith, C. D., Ashford, J. W. & Markesbery, W. R. (2000) Neurology 55, 370-376. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell, T. W., Mufson, E. J., Schneider, J. A., Cochran, E. J., Nissanov, J., Han, L. Y., Bienias, J. L., Lee, V. M., Trojanowski, J. Q., Bennett, D. A., et al. (2002) Ann. Neurol. 51, 182-189. [DOI] [PubMed] [Google Scholar]
- 36.Schena, M., Heller, R. A., Theriault, T. P., Konrad, K., Lachenmeier, E. & Davis, R. W. (1998) Trends Biotechnol 16, 301-306. [DOI] [PubMed] [Google Scholar]
- 37.Barlow, C. & Lockhart, D. J. (2002) Curr. Opin. Neurobiol. 12, 554-561. [DOI] [PubMed] [Google Scholar]
- 38.Colangelo, V., Schurr, J., Ball, M. J., Pelaez, R. P., Bazan, N. G. & Lukiw, W. J. (2002) J. Neurosci. Res. 70, 462-473. [DOI] [PubMed] [Google Scholar]
- 39.Ginsberg, S. D., Hemby, S. E., Lee, V. M., Eberwine, J. H. & Trojanowski, J. Q. (2000) Ann. Neurol. 48, 77-87. [PubMed] [Google Scholar]
- 40.Loring, J. F., Wen, X., Lee, J. M., Seilhamer, J. & Somogyi, R. (2001) DNA Cell Biol. 20, 683-695. [DOI] [PubMed] [Google Scholar]
- 41.Pasinetti, G. M. (2001) J. Neurosci. Res. 65, 471-476. [DOI] [PubMed] [Google Scholar]
- 42.Yao, P. J., Zhu, M., Pyun, E. I., Brooks, A. I., Therianos, S., Meyers, V. E. & Coleman, P. D. (2003) Neurobiol. Dis. 12, 97-109. [DOI] [PubMed] [Google Scholar]
- 43.Dickey, C. A., Loring, J. F., Montgomery, J., Gordon, M. N., Eastman, P. S. & Morgan, D. (2003) J. Neurosci. 23, 5219-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blalock, E. M., Chen, K. C., Sharrow, K., Herman, J. P., Porter, N. M., Foster, T. C. & Landfield, P. W. (2003) J. Neurosci. 23, 3807-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, R. A., Galecki, A. & Shmookler-Reis, R. J. (2001) J. Gerontol. A Biol. Sci. Med. Sci. 56, B52-B57. [DOI] [PubMed] [Google Scholar]
- 46.Clark, C. M., Sheppard, L., Fillenbaum, G. G., Galasko, D., Morris, J. C., Koss, E., Mohs, R. & Heyman, A. (1999) Arch. Neurol. 56, 857-862. [DOI] [PubMed] [Google Scholar]
- 47.Geddes, J. W., Tekirian, T. L., Soultanian, N. S., Ashford, J. W., Davis, D. G. & Markesbery, W. R. (1997) Neurobiol. Aging 18, S99-S105. [DOI] [PubMed] [Google Scholar]
- 48.Hyman, B. T. (1997) Neurobiol. Aging 18, S27-S32. [DOI] [PubMed] [Google Scholar]
- 49.Galasko, D., Hansen, L. A., Katzman, R., Wiederholt, W., Masliah, E., Terry, R., Hill, L. R., Lessin, P. & Thal, L. J. (1994) Arch. Neurol. 51, 888-895. [DOI] [PubMed] [Google Scholar]
- 50.Affymetrix (2001) Affymetrix Microarray Suite User's Guide (Affymetrix, Santa Clara, CA), Version 5.
- 51.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirnics, K., Middleton, F. A., Marquez, A., Lewis, D. A. & Levitt, P. (2000) Neuron 28, 53-67. [DOI] [PubMed] [Google Scholar]
- 53.Prolla, T. A., Allison, D. B. & Weindruch, R. (2001) J. Gerontol. A Biol. Sci. Med. Sci. 56, B327-B330. [DOI] [PubMed] [Google Scholar]
- 54.Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N. & Golani, I. (2001) Behav. Brain Res. 125, 279-284. [DOI] [PubMed] [Google Scholar]
- 55.Stork, P. J. & Schmitt, J. M. (2002) Trends Cell Biol. 12, 258-266. [DOI] [PubMed] [Google Scholar]
- 56.de la Monte, S. M., Sohn, Y. K. & Wands, J. R. (1997) J. Neurol. Sci. 152, 73-83. [DOI] [PubMed] [Google Scholar]
- 57.Slack, R. S., Skerjanc, I. S., Lach, B., Craig, J., Jardine, K. & McBurney, M. W. (1995) J. Cell Biol. 129, 779-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galderisi, U., Melone, M. A., Jori, F. P., Piegari, E., Di Bernardo, G., Cipollaro, M., Cascino, A., Peluso, G., Claudio, P. P. & Giordano, A. (2001) Mol. Cell Neurosci. 17, 415-425. [DOI] [PubMed] [Google Scholar]
- 59.Lipinski, M. M., Macleod, K. F., Williams, B. O., Mullaney, T. L., Crowley, D. & Jacks, T. (2001) EMBO J. 20, 3402-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyajima, M., Nornes, H. O., Sato, K. & Neuman, T. (1996) J. Neurosci. Res. 46, 108-113. [DOI] [PubMed] [Google Scholar]
- 61.van Kesteren, R. E., Syed, N. I., Munno, D. W., Bouwman, J., Feng, Z. P., Geraerts, W. P. & Smit, A. B. (2001) J. Neurosci. 21, RC161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campisi, J. (2001) Trends Cell Biol. 11, S27-S31. [DOI] [PubMed] [Google Scholar]
- 63.Powell, E. M., Meiners, S., DiProspero, N. A. & Geller, H. M. (1997) Cell Tissue Res. 290, 385-393. [DOI] [PubMed] [Google Scholar]
- 64.Fawcett, J. W. & Asher, R. A. (1999) Brain Res. Bull. 49, 377-391. [DOI] [PubMed] [Google Scholar]
- 65.McKeon, R. J., Jurynec, M. J. & Buck, C. R. (1999) J. Neurosci. 19, 10778-10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minamitani, T., Ariga, H. & Matsumoto, K. (2002) Biol. Pharm. Bull. 25, 1472-1475. [DOI] [PubMed] [Google Scholar]
- 67.Sasaki, H. & Fukushima, M. (1994) Anticancer Drugs 5, 131-138. [DOI] [PubMed] [Google Scholar]
- 68.Xie, R. L., Gupta, S., Miele, A., Shiffman, D., Stein, J. L., Stein, G. S. & van Wijnen, A. J. (2003) J. Biol. Chem. 278, 26589-26596. [DOI] [PubMed] [Google Scholar]
- 69.Bordin, S. & Tan, X. (2001) Cell Signalling 13, 119-123. [DOI] [PubMed] [Google Scholar]
- 70.Nagarajan, R., Svaren, J., Le, N., Araki, T., Watson, M. & Milbrandt, J. (2001) Neuron 30, 355-368. [DOI] [PubMed] [Google Scholar]
- 71.Wei, Q., Miskimins, W. K. & Miskimins, R. (2003) Mol. Cell. Biol. 23, 4035-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, J. Z., Wu, Q., Smith, A., Grundke-Iqbal, I. & Iqbal, K. (1998) FEBS Lett. 436, 28-34. [DOI] [PubMed] [Google Scholar]
- 73.Howard, C. M., Claudio, P. P., De Luca, A., Stiegler, P., Jori, F. P., Safdar, N. M., Caputi, M., Khalili, K. & Giordano, A. (2000) Cancer Res. 60, 2737-2744. [PubMed] [Google Scholar]
- 74.Durand, B. & Raff, M. (2000) BioEssays 22, 64-71. [DOI] [PubMed] [Google Scholar]
- 75.Oh, L. Y., Denninger, A., Colvin, J. S., Vyas, A., Tole, S., Ornitz, D. M. & Bansal, R. (2003) J. Neurosci. 23, 883-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bagri, A., Cheng, H. J., Yaron, A., Pleasure, S. J. & Tessier-Lavigne, M. (2003) Cell 113, 285-299. [DOI] [PubMed] [Google Scholar]
- 77.Kuhn, T. B., Brown, M. D., Wilcox, C. L., Raper, J. A. & Bamburg, J. R. (1999) J. Neurosci. 19, 1965-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spassky, N., de Castro, F., Le Bras, B., Heydon, K., Queraud-LeSaux, F., Bloch-Gallego, E., Chedotal, A., Zalc, B. & Thomas, J. L. (2002) J. Neurosci. 22, 5992-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Braak, H. & Braak, E. (1998) J. Neural Transm. Suppl. 53, 127-140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.