Abstract
Streptococcus suis serotype 2 is a highly invasive, extracellular pathogen in pigs with the capacity to cause severe infections in humans. This study was initiated by the finding that IgM degradation products are released after opsonization of S. suis. The objective of this work was to identify the bacterial factor responsible for IgM degradation. The results of this study showed that a member of the IdeS family, designated IdeSsuis (Immunoglobulin M-degrading enzyme of S. suis), is responsible and sufficient for IgM cleavage. Recombinant IdeSsuis was found to degrade only IgM but neither IgG nor IgA. Interestingly, Western blot analysis revealed that IdeSsuis is host specific, as it exclusively cleaves porcine IgM but not IgM from six other species, including a closely related member of the Suidae family. As demonstrated by flow cytometry and immunofluorescence microscopy, IdeSsuis modulates binding of IgM to the bacterial surface. IdeSsuis is the first prokaryotic IgM-specific protease described, indicating that this enzyme is involved in a so-far-unknown mechanism of host-pathogen interaction at an early stage of the host immune response. Furthermore, cleavage of porcine IgM by IdeSsuis is the first identified phenotype reflecting functional adaptation of S. suis to pigs as the main host.
INTRODUCTION
The respiratory, alimentary, and genital tracts of healthy pigs are commonly colonized by Streptococcus suis (1, 2). However, S. suis is also one of the most important porcine pathogens, causing different pathologies such as meningitis, septicemia, arthritis, and endocarditis (3). Furthermore, S. suis serotype 2 is an important zoonotic agent in Asia, causing meningitis in adults (4, 5), but transmission of S. suis between humans has not yet been confirmed.
Various virulence or virulence-associated factors of S. suis serotype 2 have been identified during recent years, among which the capsule is so far the only known essential virulence factor protecting the pathogen against phagocytosis (6, 7). A number of surface-associated and secreted proteins of S. suis serotype 2 exhibit the same (or very similar) functions as homologous factors of other pathogenic streptococci. Important examples are peptidoglycan polysaccharide deacetylase, the opacity factor of S. suis, the factor H-binding protein, the fibronectin- and fibrinogen-binding protein of S. suis, enolase, and suilysin (8–13). On the other hand, a surface-associated or secreted factor with a function unique for S. suis has not yet been described. Furthermore, although many assays were carried out with cells of porcine origin, clear evidence for functional adaptation to pigs as the main host is still lacking for S. suis.
Infections generally elicit an early antigen-specific IgM response followed by affinity maturation and isotype switching. Furthermore, “natural” IgM antibodies, which are present prior to the infection, play an important role in protection against different pathogens and link innate to adaptive immunity (14, 15). Secreted IgM (sIgM) in humans is present mainly as a pentamer. Cysteine residues forming intradomain disulfide bonds are crucial for the pentamer structure and are conserved among human and porcine IgMs (16). Binding of the IgM pentamer to surfaces of pathogens leads to activation of the classical complement cascade, as IgM, including porcine IgM, contains a C1q binding motif (17, 18). Humoral immune responses involving IgM and immune cells might also be mediated by a recently identified Fc receptor of B and T cells specifically binding only IgM (FcμR) (19). Furthermore, monomeric membrane IgM (mIgM) is the major B-cell receptor (20). In pigs, IgM is especially important, as (i) mIgM is the only B-cell receptor, since pigs lack IgD; (ii) IgM synthesis in newborn piglets starts much earlier than IgG and IgA synthesis; and (iii) the porcine placenta does not allow transfer of any Ig, and IgM in colostrum is crucial for protection against pathogens, which is carried out by complement-mediated killing (16, 17, 21).
The human pathogen Streptococcus pyogenes secretes a very specific IgG endopeptidase, named IdeS, which efficiently cleaves human IgG in the hinge region. IdeS activity results in the formation of F(ab)2 and Fc fragments (22). As the latter are known to prime neutrophils, diffusion and circulation of IdeS might cause activation of neutrophils in noninfected tissue parts (23). Furthermore, IdeS, also known as Mac-1, has been shown to directly interfere with neutrophil effector functions by inhibiting Fc receptor recognition of IgG (24). IgG endopeptidases homologous to IdeS are expressed by Streptococcus equi subsp. equi and S. equi subsp. zooepidemicus, i.e., IdeE and IdeZ, respectively (25, 26). Here we report that the homologue of IdeS in S. suis, designated IdeSsuis (Immunoglobulin M-degrading enzyme of S. suis), is distinct from the other members of this family in that it is not an IgG endopeptidase but a highly specific and efficient IgM protease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. suis strain 10 is a virulent serotype 2 strain that has been used by different groups for mutagenesis and experimental infections of pigs (7–9, 27). The capsule-deficient isogenic mutant strain 10cpsΔEF was kindly provided by Hilde Smith (AWG, Lelystad, Netherlands) (7). Streptococci were grown on Columbia agar plates with 6% sheep blood or in Bacto Todd-Hewitt broth (THB). Lactococcus lactis subsp. cremoris MG1363 (here L. lactis) was grown at 30°C in M17 medium (Oxoid) supplemented with 0.5% glucose. Escherichia coli strains were cultured in Luria-Bertani (LB) medium. In appropriate cases, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml for E. coli; chloramphenicol, 3.5 μg/ml for S. suis and 8 μg/ml for E. coli; erythromycin, 2 μg/ml for S. suis, 5 μg/ml for L. lactis, and 400 μg/ml for E. coli; and spectinomycin, 100 μg/ml for S. suis and 50 μg/ml for E. coli.
Animal experiments.
In this study, different samples from experimentally infected piglets were investigated, all of which were part of a previous study (24). The protocol for this animal experiment was approved by the Committee on Animal Experiments of the Lower Saxonian State Office for Consumer Protection and Food Safety (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit [LAVES]; permit no. 33.9-42502-04-07/1243). This study was performed in strict accordance with the principles and recommendations outlined in the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (European Treaty Series, no. 123; http://www.conventions.coe.int/Treaty/en/Treaties/Html/123.htm) and the German Animal Protection Law (Tierschutzgesetz).
All application techniques and sampling of blood conducted in this study were in line with the recommendations of the German Society for Laboratory Animal Science (Gesellschaft für Versuchstierkunde) and the German Veterinary Association for the Protection of Animals (Tierärztliche Vereinigung für Tierschutz e. V.) (http://www.gv-solas.de/auss/tie/index.html). Generation of hyperimmune sera in rabbits and swine in our institute is registered at the LAVES under no. 87A044 and 12A226, respectively.
Sequence analysis.
Multiple-sequence analyses were conducted with T-coffee version 8.93 (including ClustalW 1.83) using the SIB Bioinformatics Resource Portal Expasy (www.expasy.ch). SignalP 4.0 was run to identify a putative signal peptide and the respective cleavage sites (www.cbs.dtu.dk/services/SignalP/).
DNA techniques and primer sequences.
Routine DNA manipulations were performed as described previously (28). Primers were designed based on the sequence of SSU0496 in the genome of S. suis P1/7 (www.sanger.ac.uk). Chromosomal DNA of strain 10 served as the template in all PCRs conducted for generation of inserts. DNA fragments were amplified with Phusion polymerase (Promega, Mannheim, Germany). All primer sequences are provided in Table S1 in the supplemental material. Comprehensive restriction and PCR analysis was conducted with all newly constructed plasmids. The inserts of a number of different plasmids were also sequenced in this study. They encompassed different sequences of ideSsuis from strain 10 ranging from nucleotides −496 to 3552 (ideSsuis nucleotides 1 to 3426). Sequencing of different plasmids confirmed the entire published P1/7 sequence of this region for strain 10.
Cloning of ideSsuis.
For expression of recombinant His-tagged IdeSsuis (rIdeSsuis), lacking only the signal sequence of IdeSsuis, the 3,353-bp (encoding amino acids [aa] 34 to 1141 of IdeSsuis) amplification product generated with the primer pair ideSpostSSBamHI plus ideSterPstI (see Table S1 in the supplemental material) was cloned into pET45b after BamHI and PstI digestion to give pETideSsuis. Sequencing of the entire insert was conducted to verify this cloning and the sequence of ideSsuis. The plasmids pETideSsuis_homologue, pETideSsuis_L_domain, and pETideSsuis_C_domain encoding the truncated His-tagged proteins rIdeSsuis_homologue, rIdeSsuis_L_domain, and rIdeSsuis_C_domain, respectively, were constructed as follows. The 1,216-bp, 871-bp, and 2,171-bp amplification products of primer pairs ideSsuispostSSBamHI plus ideSsuis_hom_rev, ideSsuispostSSBamHI plus ideSsuis_L_domain_rev, and ideSsuisCfor_BamHI plus ideSsuisterPstI, respectively, were cloned into pET45b after BamHI and PstI digestion (see Table S1 in the supplemental material).
For heterologous expression of IdeSsuis in L. lactis under the control of the constitutive promoter P23, the plasmid pOriideSsuis was constructed. For this, the primer pair ideSsuisBamHIforL plus ideSsuisterPstI (see Table S1 in the supplemental material) was used to amplify the 3,462-bp product including the complete open reading frame of ideSsuis. After BamHI and PstI digestion, the insert was cloned in the shuttle vector pOri23 (29).
Targeted mutagenesis of ideSsuis and its complementation.
In-frame deletion mutagenesis of ideSsuis was conducted in S. suis strain 10 with the thermosensitive plasmid pSET5ΔideSsuis. To construct this vector, a 614-bp 5′-ideSsuis amplification product generated with the primer pair preProIdeSuisPstI plus postSSideSuisBamHI and a 612-bp 3′-ideSsuis amplification product amplified with the primer pair preEndideSsuisBamHI plus postEndideSsuisEcoRI were cut with the restriction enzymes indicated in the names of the primers (see Table S1 in the supplemental material). Both amplicons were inserted between the PstI and EcoRI site of pSET5s (30) to generate pSET5ΔideSsuis. Restriction analysis and sequencing were carried out to verify this plasmid and in particular the in-frame deletion. The temperature-sensitive replication of this vector enabled allelic exchange of ideSsuis as described previously (9). The mutant strain 10ΔideSsuis was confirmed by comprehensive Southern blot analysis using 4 different probes (the 464-bp amplicon of deleted ideSsuis DNA using the primer pair ideSsuis_mid_for plus ideSsuismid_rev2, the two inserts of pSET5ΔideSsuis, and the 500-bp amplicon of pSET5 backbone DNA generated with pSET5_for and pSET5_rev) and two different digestions of DNA (HincII and BamHI).
For complementation of 10ΔideSsuis, the vector pGA14ideSsuis was used. This vector was constructed by cloning the 3,800-bp amplification product generated with the primer pair ideSsuis_SacI_for plus ideSsuis_SacI_rev after SacI digestion into pGA14 (31). The cloning of pSET5ΔideSsuis and pGA14ideSsuis was verified by restriction analysis and sequencing.
Expression and purification of recombinant IdeSsuis proteins.
E. coli BL21(DE3) isolates carrying plasmid pETideSsuis, pETideSsuis_homologue, pETideSsuis_L_domain, or pETideSsuis_C_domain were grown in LB broth plus ampicillin at 37°C to an optical density at 600 nm (OD600) of 0.5. Protein expression was induced by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the bacteria were incubated for an additional 4 h. Purification of rIdeSsuis, rIdeSsuis_homologue, rIdeSsuis_L_domain, and rIdeSsuis_C_domain by Ni2+-nitrilotriacetic acid affinity chromatography under native conditions was carried out as recommended by the manufacturer (Macherey-Nagel).
SDS-PAGE and Western blot analysis.
If not stated otherwise, samples for SDS-PAGE and Western blot analysis were prepared with nonreducing sample buffer. For anti-IgM, anti-IgG, and anti-IgA Western blots, proteins were separated in 6%, 12%, and 10% separating gels, respectively, and 4% stacking gels, if not stated otherwise. For Western blot analysis, proteins were transferred to nitrocellulose membrane (Roth) by standard procedures (28). After blocking of membranes (5% dry milk powder in Tris-buffered saline-Tween 20 [TBST]), incubation with antibodies was conducted in TBST with 1% dry milk powder. All antibodies used in Western blot analyses are specified together with the final dilution in Table S2 in the supplemental material. Between incubations, blots were washed thoroughly 4 times. Blots incubated with horseradish peroxidase-conjugated antibodies were developed using Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific) as recommended by the manufacturer.
Immunization of rabbits and piglets.
Polyclonal antisera against purified rIdeSsuis, rIdeSsuis_homologue, and rIdeSsuis_C_domain were raised in New Zealand White rabbits (Charles River Laboratories) through three consecutive immunizations (50% Freund's incomplete adjuvant; Sigma-Aldrich). An immune serum against L. lactis pOri23 was drawn from a pig 14 days after a single application of a bacterin containing 20% Emulsigen (MVP Laboratories Inc., Omaha, NE) as adjuvant and approximately 109 formaldehyde-killed bacteria. Blood for the bactericidal assay was drawn from a piglet immunized once with an S. suis strain 10 bacterin containing 20% Emulsigen and approximately 109 formaldehyde-killed bacteria.
Analysis of immunoglobulin degradation.
Bacterial supernatants were concentrated with Amicon Ultra 15-ml centrifugal filters with a 30-kDa cutoff (Merck Millipore) either 6-fold (L. lactis) or 24-fold (S. suis). Incubation of concentrated bacterial supernatants or 5 μg/ml recombinant IdeSsuis constructs with diluted lithium heparin plasma (16 IU heparin/ml) and serum was conducted for 2.5 h on a rotator at 37°C if not stated otherwise. The final dilution of plasma or serum was 1:100 in all samples. For determination of the inhibitor profile, concentrated culture supernatants were incubated 30 min with different inhibitors prior to adding diluted porcine plasma for 2.5 h at 37°C (all samples were covered with aluminum foil). The following inhibitor conditions were tested: 6.1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) (Calbiochem); 26.7 mM EDTA (Roth), pH 7.5; 533 μM bestatin hydrochloride (Sigma-Aldrich); 80 μM E-64 (Sigma-Aldrich); 20 mM iodoacetamide (Roth); and 6 μM Z-LVG-CHN2 (Bachem). Colostrum was obtained from a sow within 4 h after parturition and incubated in a final dilution of 1:400 with the concentrated culture supernatants.
Purification of porcine IgG and IgM.
Porcine IgG was purified from 2 ml serum using 250 mg protein A Sepharose (Sigma-Aldrich). Briefly, after application of the serum to the columns, they were washed with 20 mM sodium phosphate, pH 7.0, and IgGs were eluted with 100 mM sodium citrate, pH 4. Eluted fractions were immediately neutralized with 1 M Tris-HCl (pH 9.5). All fractions were investigated in Coomassie blue-stained SDS-PAGE gels and anti-IgG Western blots.
Prior IgM purification ammonium sulfate was added to porcine plasma, diluted with phosphate-buffered saline (PBS) (pH 7.5) 1:2 to a final concentration of 0.8 M. Subsequently, IgM was bound onto HiTrap IgM purification HP (GE Healthcare), prepared according to the manufacturers' instructions. After extensive washing (20 mM sodium phosphate, 0.8 M ammonium sulfate, pH 7.5), IgMs were eluted in 20 mM sodium phosphate buffer, pH 7.5. Eluted fractions included substantial amounts of IgG. Thus, eluted fractions were pooled and applied to a matrix of protein G Sepharose (HiTrap G HP; GE Healthcare) for binding of IgGs. The matrix was washed with 20 mM sodium phosphate buffer, pH 7.5. SDS-PAGE, Coomassie blue staining, and Western blot analysis were conducted to identify fractions including IgM but not IgG. Flowthrough fractions containing IgM but not IgG were used for IgM cleavage analysis. Protein G Sepharose-bound IgGs were eluted in 0.1 M glycine-HCl, pH 2.7. The pH was immediately adjusted by addition of 1 M Tris, pH 8.0.
Analysis of the specificity of IdeSsuis by incubation of body fluids with rIdeSsuis.
Lithium heparin plasma (1:100 diluted; 16 IU heparin/ml), 1:100 diluted serum, 1:20 diluted joint fluids, and undiluted cerebrospinal fluids were incubated with 5 μg/ml rIdeSsuis for 3.5 h, separated by reducing SDS-PAGE with stacking and separation gels of 4% and 10% acrylamide, respectively, and stained with silver to evaluate protein degradation. Silver staining was conducted as described previously (32). Cerebrospinal and joint fluids were either from piglets with fibrino-suppurative meningitis and synovialitis caused by S. suis infection or from piglets with no lesions. For comparison, body fluids were also incubated with 1 μg/ml proteinase K. Comparative Western blot analysis was conducted as described above but under reducing conditions.
Generation of culture supernatants for IgM cleavage analysis after opsonization of bacteria.
Opsonization of S. suis and L. lactis was conducted with convalescent-phase sera and immune sera, respectively. Convalescent-phase sera were drawn 9 days after challenge of live-vaccinated-piglets with S. suis strain 10 (33) and were known to mediate opsonophagocytic killing. Bacteria used in this assay were grown to late exponential phase (OD600 = 0.8). Heat inactivation of S. suis was conducted for 30 min at 70°C. Killing of bacteria was also performed by treatment with 0.2% formaldehyde for 12 h. One milliliter of viable or dead culture was centrifuged, washed with PBS, and resuspended in 300 μl serum. After 30 min of rotation at 8°C, bacteria were centrifuged, washed three times with PBS, and resuspended in 1 ml THB. Prior to and after incubation of bacteria for different time periods (2.5 h, 3.5 h, 4.5 h, or 5.5 h) at 37°C (S. suis) or 30°C (L. lactis), bacteria were centrifuged and frozen in liquid nitrogen for further analysis (immunofluorescence microscopy and flow cytometry). The culture supernatant was concentrated with Amicon Ultra 0.5-ml centrifugal filters with a 10-kDa cut off (Merck Millipore) to 20 μl for Western blot analysis.
Immunofluorescence microscopy.
Fifty microliters of opsonized bacteria was allowed to dry on a microscope slide. After fixation in 3.7% paraformaldehyde and blocking with 5% fetal calf serum, opsonized bacteria were incubated with a polyclonal fluorescein isothiocyanate (FITC)-labeled goat anti-pig IgM serum (1:500; abcam, Cambridge, United Kingdom). Repeated washing with PBS was conducted between all steps. Samples were examined using the inverted immunofluorescence microscope Nikon Eclipse Ti-S equipped with a 60× 0.5- to 1.25-numerical-aperture (NA) oil Plan Fluor objective (Nikon) driven by NIS Elements software BR 3.2.
Flow cytometry analysis of bacteria.
For detection of surface-associated IdeSsuis, bacteria were grown to an OD600 of 0.8. One milliliter of the bacterial culture was washed five times with PBS and resuspended in 200 μl PBS. Forty microliters of the suspension was incubated with rabbit anti-rIdeSsuis antiserum for 1 h at 8°C. For comparison, bacteria were also incubated with rabbit antisera against rMRP (a surface-associated protein) and rSLY (a secreted cytolysin) and with preimmune sera (33, 34). Bacteria were washed twice with PBS and subsequently incubated in PBS with 1:1,000-labeled Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) on a rotator at 8°C for 1 h.
For analysis of IgM antigen bound to the bacterial surface, frozen bacteria collected after opsonization and prior to as well as after subsequent incubation at 37°C (see above) were resuspended in 400 μl PBS. Fifty microliters of this suspension was added to 450 μl PBS with 5% goat serum and rotated for 1 h at 8°C for blocking. Bacteria were centrifuged and incubated with the monoclonal antibody against porcine IgM used also for Western blot analysis in a 1:225 dilution in PBS for 1 h at 8°C. Bacteria were washed three times with PBS and subsequently incubated in PBS with 1:500 phycoerythrin-labeled goat anti-mouse IgG (Bio Legend) on a rotator at 8°C for 1 h.
After incubation with fluorescent antibodies, bacteria were washed three times and resuspended in PBS with 0.375% formaldehyde for further flow cytometry analysis. Fluorescent bacteria were measured using a FACScan instrument (488-nm Argon laser; Becton, Dickinson). Further analysis was performed with the software WinMDI (version 2.9.). For each determination 10,000 events were acquired, and initial analysis of bacterial cells was carried out by dot plot analysis (forward scatter [fsc] versus sideward scatter [ssc]) to define the cell population of interest. Subsequently, fluorescent bacteria were detected at channels FL-1 and FL-2, and results were illustrated as a histogram.
Bactericidal assay.
Killing of S. suis in porcine blood was investigated in freshly drawn heparinized blood from a piglet immunized once with a bacterin. Sampling of blood was conducted between the 15th and 18th days postvaccination. Stocks of frozen bacterial suspensions including 15% glycerol were thawed prior to the bactericidal assay, and 1.5 × 105 CFU were mixed with 500 μl of heparinized blood containing approximately 3.5 × 106 neutrophil granulocytes, giving a multiplicity of infection (MOI) of approximately 0.04. White blood cells were counted using a hemocytometer chamber, and leukocytes were differentiated by Wright-stained blood smears (kindly conducted by Martin Ganter, Tierärzliche Hochschule Hannover). CFU were determined at t of 0 min and 120 min of incubation on a rotator at 37°C.
RT-PCR from RNA of S. suis grown in blood ex vivo.
Ten milliliters of fresh porcine blood was inoculated with 1 × 107 or 1 × 108 CFU of S. suis strain 10 for 4-h or 30-min incubation, respectively, at 37°C on a rotator. Bacterial RNA was extracted immediately afterwards essentially as described previously (35). For comparison, RNA was extracted by the same procedure from bacteria grown in 10 ml THB to early exponential and stationary phases. Reverse transcriptase (RT)-PCR was conducted as described previously (36), except that the oligonucleotide primers ideSsuis_qPCR_for (GCGGTGGTTTTGTAAAGGAA) and ideSsuis_qPCR_rev (TTCCAACCACCAATGTACCA) were used. The real-time PCR program included initial denaturation at 95°C for 20 min and 25 cycles of denaturation at 95°C for 20 s, annealing at 55°C for 30 s, and amplification at 72°C for 20 s.
Statistical analysis.
The Mann-Whitney test was performed to analyze differences between the wild-type (wt) S. suis, the isogenic deletion mutant strain 10ΔideSsuis, and the complemented deletion mutant 10ΔideSsuis(pGA14ideSsuis). Data collected from flow cytometry were analyzed with t tests. Probabilities of <0.05 were considered significant.
RESULTS
S. suis degrades opsonizing IgM.
We initiated this work by investigating the fate of S. suis-specific porcine IgM binding to the surface of this pathogen. In previously conducted experimental infections of piglets with S. suis (33), specific IgMs that bound to the surface of the isogenic encapsulated and unencapsulated S. suis serotype 2 strain were elicited (Fig. 1). Subsequent incubation of S. suis in THB resulted in the release of specific IgM degradation products into the supernatant (Fig. 1B to D). Importantly, IgM degradation products were not recorded when heat- or formaldehyde-inactivated S. suis cells were used (Fig. 1C and D), but they could be observed after incubation of concentrated culture supernatants with porcine plasma (Fig. 2, compare first and last lanes). Based on these results, we hypothesized that S. suis secreted a protease cleaving IgM. To prove this hypothesis, we investigated whether protease inhibitors might abolish IgM cleavage activity upon coincubation of plasma with culture supernatants of S. suis. The addition of iodoacetamide but not AEBSF, bestatin, E-64, Z-LVG-CHN2, or EDTA inhibited IgM cleavage under these experimental conditions (Fig. 2). Inhibition by iodoacetamide suggested that a cysteine protease is involved in the observed cleavage of porcine IgM. Remarkably, however, cysteine protease-specific inhibitors E-64 and Z-LVG-CHN2 did not affect IgM cleavage. As the cysteine IgG protease IdeS of S. pyogenes is also not inhibited by E-64 (22), we investigated whether an IdeS homologue encoded by the genomic sequence of S. suis might be responsible for cleavage of porcine IgM.
Fig 1.
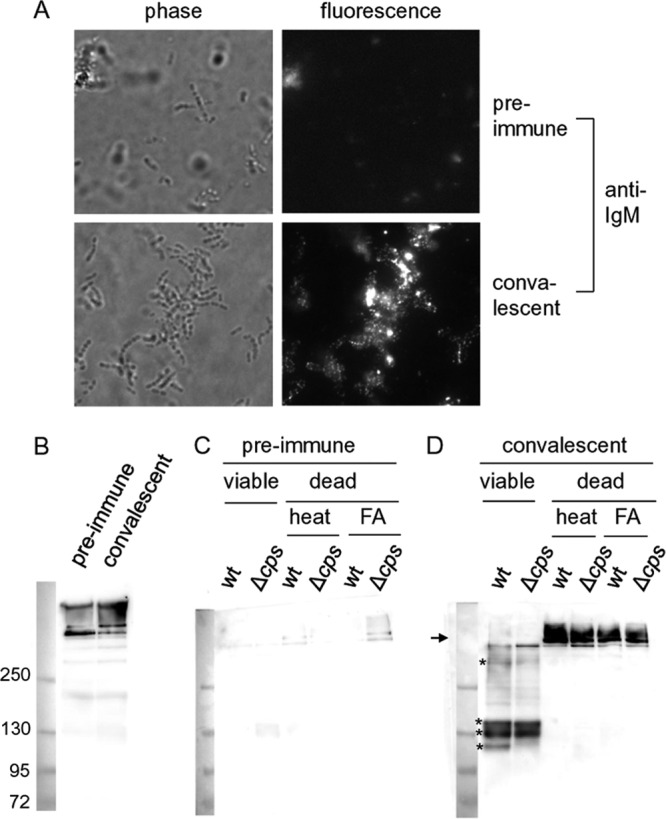
Degradation of IgM binding to the bacterial surface by viable but not by dead S. suis. (A) Binding of specific IgM antibodies to the surface of S. suis wild-type strain 10 (wt) (27). Wild-type S. suis was incubated with either preimmune or convalescent-phase serum. Immunofluorescence microscopy was conducted with an FITC-tagged polyclonal anti-porcine IgM antibody. (B) Anti-IgM Western blot analysis with a polyclonal antibody (PAB) against porcine IgM of the preimmune and convalescent-phase sera used in experiments shown in panel A. (C, D) Anti-IgM Western blot analysis (PAB) of supernatants of viable and dead S. suis cells incubated for 2.5 h after opsonization with the preimmune (C) and convalescent-phase (D) sera whose blots are shown in panel B. Wild-type S. suis and its unencapsulated mutant strain 10cpsΔEF (Δcps) were killed by either heat inactivation or formaldehyde (FA) treatment. The position of the porcine IgM multimer is marked with an arrow, and the degradation products of porcine IgM are indicated by asterisks. The molecular mass marker is shown on the left of each of the blots.
Fig 2.

Inhibitor profile of IgM degradation activity detected in supernatants of an S. suis culture. Anti-IgM Western blot analysis (PAB) of diluted porcine plasma incubated with concentrated supernatants of S. suis strain 10 without and with the indicated protease inhibitors as well as with dimethyl sulfoxide (DMSO) as solvent control for Z-LVG-CHN2. PBS incubated with diluted porcine plasma served as negative control (final dilution of porcine plasma, 1:100). The position of the porcine IgM multimer is marked with an arrow, and the cleavage products of porcine IgM are indicated by asterisks on the left side for the first positive lane. The marker bands are shown on the left side (sizes in kDa).
Different S. suis strains express a large protein, IdeSsuis, with similarity to IdeS.
The genomic sequences of S. suis serotype 2 strains (www.sanger.ac.uk) contain an open reading frame (SSU0496 in P1/7, here designated ideSsuis) encoding a putative 124-kDa protein with similarity to the 38-kDa IdeS protein of S. pyogenes (also described as Mac-1), the 39-kDa IdeE protein of S. equi subsp. equi, the 44-kDa IdeE2 protein, and the 42-kDa IdeZ protein of S. equi subsp. zooepidemicus, respectively. In silico analysis revealed that the similarity of IdeSsuis to these IgG endopeptidases was restricted to aa 141 to 426 of the total 1,141 aa of IdeSsuis (see Fig. S1 in the supplemental material). In this region, IdeSsuis showed 30% identity and 50% similarity to IdeS of S. pyogenes. In the multiple T-coffee sequence alignment analysis of IdeS, IdeE, IdeE2, IdeZ, and IdeSsuis, all obtained a score of 78 or 79, with the exception of IdeSsuis, which received a score of 73 (see Fig. S1 in the supplemental material).
Analysis by SignalP 4.0 trained on Gram-positive bacteria predicted a signal sequence at the N terminus and a signal sequence cleavage site between positions 32 and 33 (D = 0.66). Expression of IdeSsuis was investigated in different S. suis strains with an antiserum raised against full-length recombinant IdeSsuis (rIdeSsuis). IdeSsuis was detectable in culture supernatants of all S. suis strains investigated, including strains of the important clonal complexes 1, 27, and 16 (previously called 87) (Fig. 3). The respective band was located between 130 and 250 kDa as referred to by the molecular mass standard. However, variations in sizes were recorded for IdeSsuis proteins expressed by different S. suis strains. The IdeSsuis band was also recognized by antisera raised specifically against the N-terminal and C-terminal parts of IdeSsuis, demonstrating that under these conditions mature IdeSsuis includes the large C-terminal part showing no similarity to IdeS (see Fig. S2 in the supplemental material). As IdeSsuis has a putative C-terminal transmembrane domain (37), localization of IdeSsuis on the bacterial surface was also investigated via flow cytometry. IdeSsuis was detectable on the bacterial surface of the serotype 2 strain 10, in contrast to suilysin, a secreted cytolysin (see Fig. S3 in the supplemental material). However, the mean fluorescence intensity (MFI) for bacteria stained with α-IdeSsuis was much lower than the MFI of bacteria incubated with α-MRP (means ± standard deviations for 3 experiments were as follows: anti-IdeSsuis-stained wt strain, 10.13 ± 1.6 MFI, and 10ΔideSsuis strain, 5.3 ± 0.4 MFI, with P of 0.013; anti-MRP-stained wt strain, 390 ± 88.2 MFI, and 10ΔideSsuis strain, 396 ± 77.2, with P of 0.95; see Fig. S3 in the supplemental material). In conclusion, the results demonstrate that IdeSsuis is released by different S. suis strains into the culture medium. Flow cytometry profiles of S. suis serotype 2 indicate that IdeSsuis, in accordance with in silico analysis, at least transiently remains attached to the bacterial surface.
Fig 3.
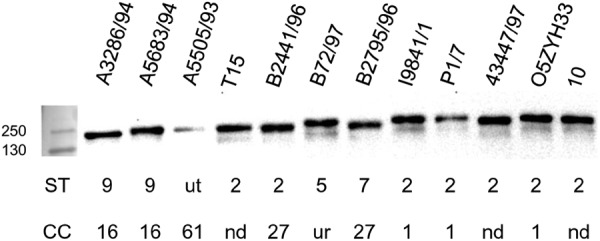
Detection of IdeSsuis in supernatants of S. suis wild-type strains of different serotypes (ST) and clonal complexes (CC). Anti-IdeSsuis Western blot analysis of 24-fold-concentrated supernatants of the indicated S. suis strains after separation of proteins in 10% SDS-PAGE under reducing conditions. One S. suis strain (A5505/93) is untypeable (ut) in serotyping, and one strain (B72/97) belongs to the unrelated (ur) sequence type 95 [49]. The sequence types of three additional strains investigated have not yet been determined (nd). The marker bands are shown on the left side (sizes in kDa).
IdeSsuis is an efficient and specific IgM protease.
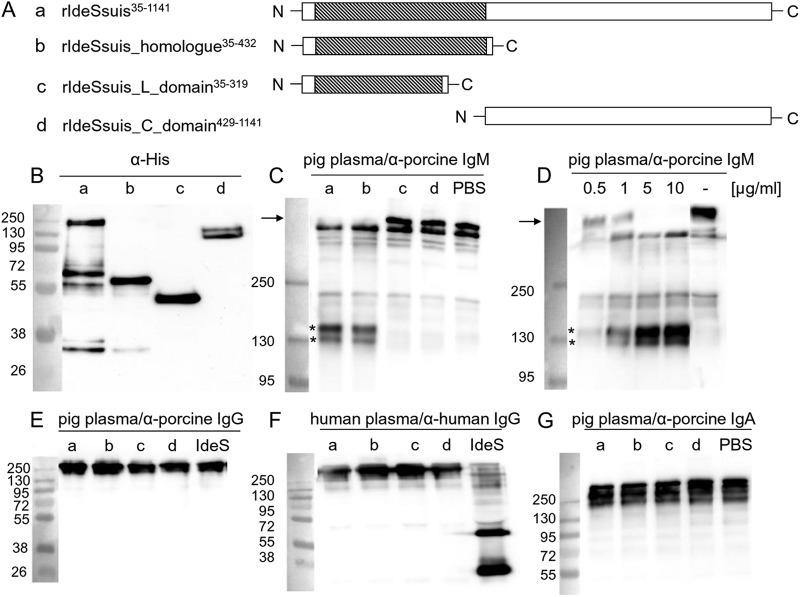
Different recombinant His-tagged derivatives of IdeSsuis were generated to investigate the putative cleavage of IgM (Fig. 4A and B). Western blot analysis conducted with two different anti-porcine IgM antibodies (poly- and monoclonal) demonstrated that porcine IgM is cleaved by full-length rIdeSsuis and the rIdeSsuis_homologue but not by the rIdeSsuis_L_domain or the rIdeSsuis_C_domain constructs (Fig. 4C; see Fig. S4A in the supplemental material). Thus, amino acids 35 to 432 of IdeSsuis with similarity to IdeS from S. pyogenes are sufficient for IgM cleavage. Cleavage of IgM was detectable already after 10 min using 0.5 μg/ml rIdeSsuis (Fig. 4D).
Fig 4.
Cleavage of IgM but not IgG and IgA by recombinant IdeSsuis constructs. (A) Illustration of rIdeSsuis and its truncated derivatives. Regions similar to IdeS of S. pyogenes are shaded. (B) Anti-His Western blot analysis of purified rIdeSsuis (a) and its truncated derivatives (b to d) illustrated in panel A. (C) Anti-IgM Western blot (PAB) of diluted porcine plasma incubated with 5 μg/ml of rIdeSsuis (a) or its truncated derivatives (b to d) illustrated in panel A. (D) Anti-IgM Western blot analysis (PAB) of diluted porcine plasma incubated with increasing concentrations of rIdeSsuis as indicated or with PBS for 10 min at 37°C. (E, F) Anti-porcine (E) and anti-human (F) IgG Western blot analysis of diluted porcine and human plasma, respectively, incubated with 5 μg/ml of rIdeSsuis (a) or its truncated derivatives (b to d) shown in panel A. Recombinant IdeS of S. pyogenes was included as positive control for cleavage of human IgG. (G) Anti-porcine IgA Western blot analysis of diluted porcine plasma incubated with 5 μg/ml rIdeSsuis (a) or its truncated derivatives (b to d) illustrated in panel A. The final dilution of porcine plasma was 1:100 in all samples. The marker bands are shown on the left (sizes in kDa).
As IdeSsuis was found to be similar to the IgG endopeptidase IdeS, we thoroughly investigated the putative cleavage of porcine IgG by rIdeSsuis using one polyclonal antiserum and two monoclonal antibodies with different specificities against porcine IgG (subclass-specific antibodies against porcine IgG are not yet available). Though sera and purified porcine IgGs from different pigs (neonatal colostrum-deprived piglets, weaning SPF piglets, conventional weaning, and fattening pigs as well as sows) were used as IgG sources, cleavage of porcine IgG by rIdeSsuis or any of the truncated IdeSsuis constructs was not detectable (Fig. 4E; see Fig. S5 in the supplemental material). Furthermore, rIdeSsuis, in contrast to rIdeS, did not cleave human IgG (Fig. 4F). Finally, porcine IgA in serum was also not cleaved after incubation with rIdeSsuis (Fig. 4G). These results show that IdeSsuis cleaves neither porcine or human IgG nor porcine IgA.
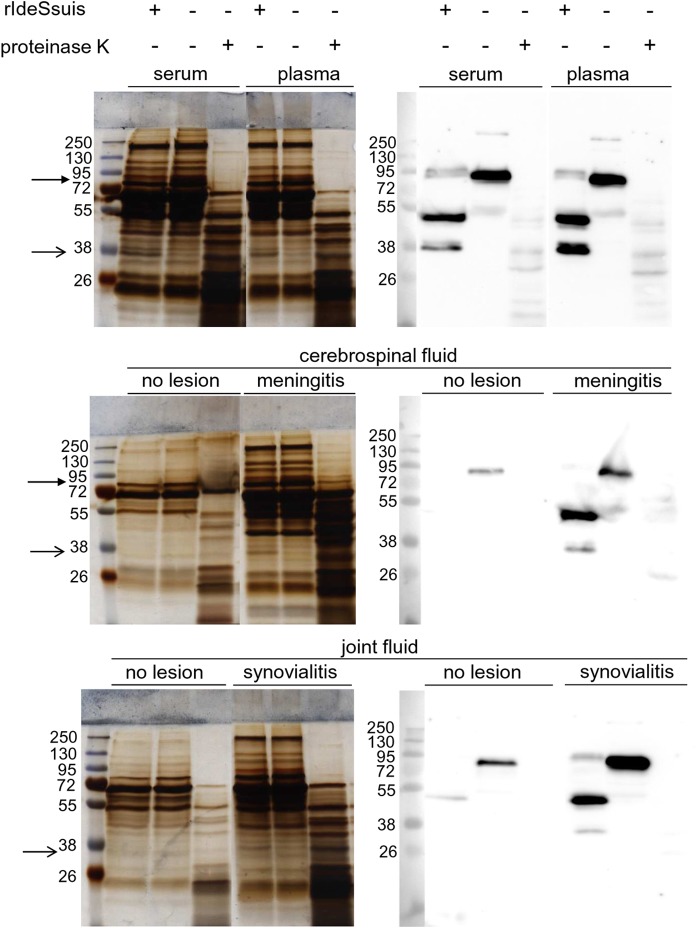
To further assess the specificity of IdeSsuis, different body fluids of piglets were incubated with 5 μg/ml rIdeSsuis and investigated by SDS-PAGE analysis for a change in the protein band pattern. Silver staining allowed detection of a single additional band in the pattern of serum and plasma after incubation with rIdeSsuis (Fig. 5, simple arrows). This band was running close to the 38-kDa band of the marker. Anti-IgM Western blot analysis under reducing conditions revealed an IdeSsuis-dependent IgM cleavage product at the same position. Furthermore, one band of serum and plasma running between 95 and 72 kDa disappeared after incubation with rIdeSsuis (Fig. 5, arrow with filled head). Anti-IgM Western blot analysis suggested that this is the band of the heavy chain of porcine IgM including the cleavage site of IdeSsuis (Fig. 5). In contrast, incubation of porcine plasma and serum under the same conditions with 1 μg/ml proteinase K resulted in a completely different band pattern (Fig. 5). As meningitis and arthritis are important manifestations of S. suis infections in piglets, cerebrospinal and joint fluids from piglets with and without respective lesions (meningitis and synovialitis) were also incubated with rIdeSsuis. As shown in Fig. 5, cerebrospinal and joint fluids from affected but not from unaffected piglets showed a different band pattern after incubation with rIdeSsuis. As in the case of serum and plasma, only a single additional 38-kDa band was observed, and an IdeSsuis-dependent IgM cleavage product of the same size was detectable in the respective Western blot analysis. Furthermore, the only protein band of cerebrospinal fluid that disappeared after rIdeSsuis incubation was again of the same size as the heavy chain of porcine IgM (Fig. 5, arrow with filled head). In conclusion, the results of SDS-PAGE analysis and Western blot analysis indicate that IdeSsuis is a highly specific IgM protease.
Fig 5.
IdeSsuis is an IgM-specific protease. Plasma, serum, and cerebrospinal and joint fluids of piglets were incubated with rIdeSsuis (5 μg/ml) or, for comparison, with proteinase K (1 μg/ml) as indicated on the top, separated in SDS-PAGE under reducing conditions, silver stained (left side), and analyzed by anti-IgM Western blotting (right side). Cerebrospinal and joint fluids were as indicated from piglets with meningitis and synovialitis, respectively, or from piglets without these lesions. The position of the heavy chain of porcine IgM is marked with an arrow with a filled head, and the position of the additional band (approximately 38 kDa) present after rIdeSsuis incubation is indicated with a simple arrow (in accordance with detection of an IgM cleavage product of the same size in the anti-IgM Western blot).
Expression of IdeSsuis by S. suis is required and sufficient for IgM cleavage.
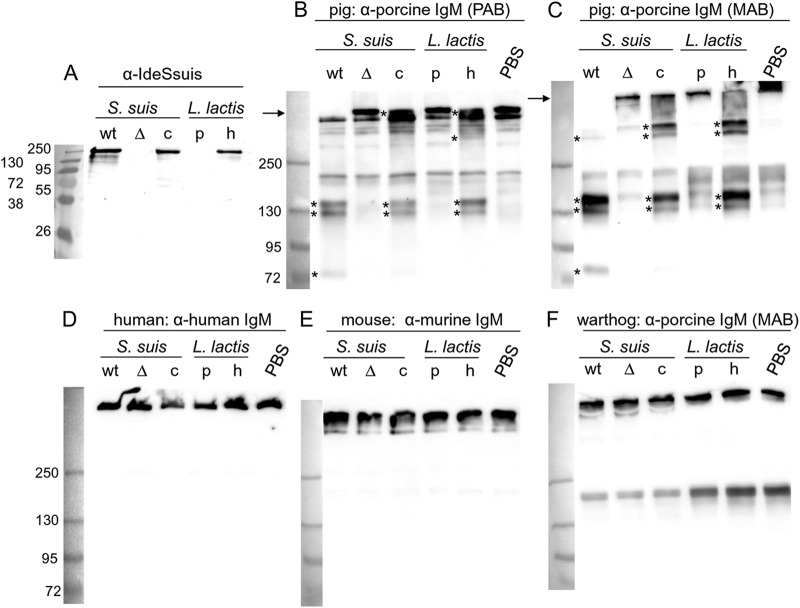
As rIdeSsuis was found to be a specific IgM protease, we generated an ideSsuis deletion mutant by precise in-frame deletion to prove that the IgM cleavage activity of S. suis supernatants was attributable solely to IdeSsuis. As expected, the wild-type (wt) and the complemented mutant strains, but not the isogenic ideSsuis mutant strain 10ΔideSsuis, secreted IdeSsuis (Fig. 6A). Importantly, only S. suis supernatants of the wt and the complemented isogenic mutant contained IgM-cleaving activity (Fig. 6B and C). The cleaving activity of the S. suis wt supernatant elicited three or four bands after separation by nonreducing SDS-PAGE and subsequent Western blot analysis (asterisks in Fig. 6B and C): one major band running between the 130- and 250-kDa marker bands, one band close to the 130-kDa marker, and one running just above the 72-kDa band. Based on our results shown in Fig. 6C, cleavage of the IgM multimer (arrow in Fig. 6C) was complete after incubation with the supernatant of the wt but not with the supernatant of the complemented isogenic mutant 10ΔideSsuis(pGA14ideSsuis) (Fig. 6C, lane c). Intermediate IgM cleavage by the supernatant of the complemented mutant under these conditions was associated with one prominent and one or two faint high-molecular-mass bands running between the 250-kDa molecular mass marker and the remaining band of the high-molecular-mass IgM (Fig. 6C). These results indicate that the porcine IgM multimer contains numerous IdeSsuis cleavage sites and that IgM cleavage occurs in subsequent steps.
Fig 6.
IdeSsuis cleaves IgM from pigs but not IgM from humans, mice, and warthogs. (A) Anti-IdeSsuis Western blot analysis of 24-fold-concentrated supernatants of S. suis strains 10 (wt), 10ΔideSsuis (Δ), and 10ΔideSsuis(pGA14ideSsuis) (c) as well as the 6-fold-concentrated supernatants of L. lactis pOri23 (p) and L. lactis(pOriideSsuis) (h). (B to F) Anti-IgM Western blot analysis of diluted plasma or serum from pig (B and C), human (D), mouse (E), and warthog (F) incubated with the concentrated supernatants investigated as shown in panel A or with PBS. Detection of IgM was conducted with polyclonal antibodies (PAB) against porcine (B), human (D), or mouse (E) IgM or with a monoclonal antibody (MAB) against porcine IgM (C and F). The positions of the IgM multimers are marked with arrows on the left side of the blots, and the cleavage products of porcine IgM are indicated by asterisks on their left side. The final dilution of porcine plasma or serum was 1:100 in all samples. The marker bands are shown on the left (sizes in kDa).
To further verify cleavage of IgM by IdeSsuis, porcine IgM was purified and subsequently used as a substrate for rIdeSsuis and IdeSsuis present in the supernatant of wt S. suis. Three high-molecular-mass bands were detectable in the Coomassie blue-stained gel under nonreducing conditions after two-step IgM purification (see Fig. S6 in the supplemental material). It is noteworthy that the most prominent band was recognized by the polyclonal anti-IgM serum but not the monoclonal anti-IgM antibody, suggesting that the IgM preparation included α-2-macroglobulin and that the polyclonal anti-IgM serum used in this study was also recognizing α-2-macroglobulin (38). Importantly, Western blot analysis clearly demonstrated that rIdeSsuis and the supernatant of S. suis cleaved IgM in this preparation (see Fig. S6B and C in the supplemental material). Though the pattern of cleavage products showed differences in the two Western blots, the major IgM cleavage product running between the 130-kDa and 250-kDa bands of the molecular mass marker was confirmed in both Western blots with the poly- and monoclonal anti-IgM antibodies (see Fig. S6B and C in the supplemental material).
As S. suis lost IgM cleaving activity through deletion of ideSsuis, we further asked whether expression of IdeSsuis by a different nonpathogenic Gram-positive bacterium might be sufficient to achieve IgM cleavage activity in the bacterial supernatant. For this, full-length IdeSsuis was expressed in Lactococcus lactis. Release of IdeSsuis by L. lactis in the culture supernatant was confirmed by Western blotting (Fig. 6A, lane h) and was found to be sufficient to cleave porcine IgM (Fig. 6B and C, lanes h).
As Igs in colostrum are crucial for the protection of newborn piglets, IgM cleavage by the supernatants of the different S. suis and L. lactis strains was investigated not only in plasma and serum but also in colostrum (see Fig. S7 in the supplemental material). Incubation of colostrum with the supernatant of the wt S. suis strain, but not of the isogenic and complemented mutants, resulted in complete cleavage of porcine IgM in colostrum. IgM cleavage in colostrum, albeit incomplete, was also recorded with the supernatant of the L. lactis strain expressing IdeSsuis (see Fig. S7 in the supplemental material).
IdeSsuis is a porcine-specific IgM protease.
To investigate the host specificity of IdeSsuis, putative cleavage of IgM from other species was analyzed using plasma or sera from (i) species that occasionally are infected by S. suis, i.e., humans and cattle; (ii) mice, which are currently most frequently used in experimental models for S. suis infection; (iii) the pig-related species warthog (Phacochoerus africanus africanus, family Suidae); and (iv) two species rarely associated with S. suis, i.e., cats and dogs. Incubation of serum or plasma from any of these species with the supernatants of S. suis strain 10 and L. lactis secreting IdeSsuis (Fig. 6D to F; see Fig. S8B to D in the supplemental material) or the different recombinant IdeSsuis constructs (see Fig. S4B to D and S8B to D in the supplemental material) did not result in any cleavage of IgM. Noteworthy, warthog IgM was detectable with different anti-porcine IgM antibodies, including the monoclonal antibody. Thus, sensitivity to IdeSsuis, but not immunorecognition by different anti-IgM antibodies, allowed differentiation of IgM from pig and warthog. In addition to IgM of pigs (Sus scrofa domestica, family Suidae), cleavage of IgM from wild boars (Sus scrofa, family Suidae) was also observed (see Fig. S8A in the supplemental material). In conclusion, IgM of pigs and wild boars but not of any other tested species is a substrate of IdeSsuis.
IdeSsuis cleaves IgM bound to the bacterial surface.
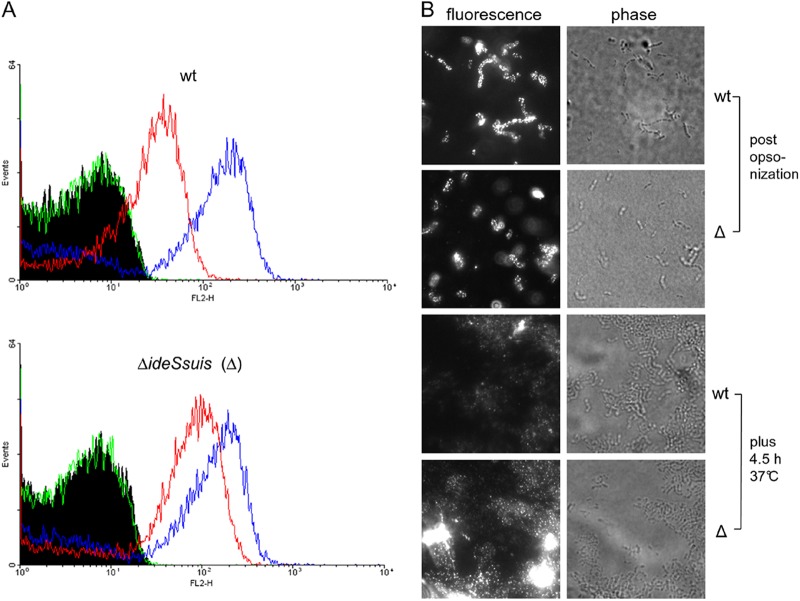
As this work was originally initiated to study the fate of specific IgM antibodies binding to the surface of S. suis, we asked whether antigen-bound IgM might be a target for IdeSsuis. Opsonization was conducted with the same sera and under the same conditions as in our previous comparison of viable and dead bacteria (Fig. 1). IgM cleavage products were recorded after incubation of the wt strain for 2.5 h after opsonization, whereas in the culture supernatants of the isogenic 10ΔideSsuis mutant no cleavage was observed during any of the tested incubation times ranging from 2.5 to 5.5 h (see Fig. S9A and B in the supplemental material). Importantly, flow cytometry analysis revealed significant differences in the intensity of surface-bound IgM antigen between the wt and the 10ΔideSsuis mutant at t of 4.5 h but not at t of zero hour after opsonization (means ± standard deviations for 3 experiments are as follows: wt, 44 ± 8 MFI, and 10ΔideSsuis, 101 ± 15 MFI; P = 0.01 for t of 4.5 h; Fig. 7A). These differences were confirmed by immunofluorescence microscopy using a different α-porcine IgM antibody (Fig. 7B). Together these results show that expression of IdeSsuis by S. suis leads to the reduction of surface-bound IgM antigen. We further investigated whether expression of IdeSsuis is sufficient to elicit cleavage of IgM antibodies binding to a different Gram-positive bacterium. For this purpose, L. lactis secreting IdeSsuis was opsonized with a porcine hyperimmune serum against L. lactis. In accordance with previous results, IgM cleavage products could also be detected in the culture supernatant of L. lactis secreting IdeSsuis (see Fig. S9C in the supplemental material). Thus, IdeSsuis expression modulates IgM opsonization of S. suis and results in the release of IgM cleavage products.
Fig 7.
S. suis modulates IgM binding by IdeSsuis expression. S. suis strain 10 (wt) and its isogenic mutant 10ΔideSsuis (Δ) were opsonized with a convalescent-phase serum at 8°C and subsequently incubated in THB for 4.5 hours at 37°C. (A) Flow cytometry analysis of bacteria after opsonization with convalescent-phase serum (blue lines) and subsequent incubation for 4.5 h at 37°C (red lines). Surface-bound antigen was detected with the monoclonal antibody against porcine IgM as primary antibody and a phycoerythrin-tagged anti-mouse antibody as secondary antibody. The green line shows the result for the control lacking the primary antibody. (B) Immunofluorescence microscopy analysis conducted with a polyclonal FITC-labeled anti-pig IgM antibody. Representative results for three independent experiments.
IdeSsuis promotes survival in porcine blood.
Bacteremia is considered to be a crucial step in the pathogenesis of S. suis diseases. As IgM antibodies might be an important part of host defense at this stage, we investigated transcription of ideSsuis in porcine blood ex vivo. RT-PCR demonstrated that ideSsuis is transcribed by S. suis strain 10 after 30 min as well as after 4 h of incubation of bacteria in freshly drawn porcine blood (Fig. 8A). Furthermore, we investigated the survival of S. suis in the blood of a piglet immunized once with a homologous bacterin. The isogenic mutant 10ΔideSsuis is significantly attenuated in survival under these conditions in comparison to the wt and the complemented strain (Fig. 8B). Altogether, these results suggest that cleavage of IgM by IdeSsuis occurs during S. suis bacteremia and that this novel function is crucial for survival of S. suis in the presence of specific IgM.
Fig 8.
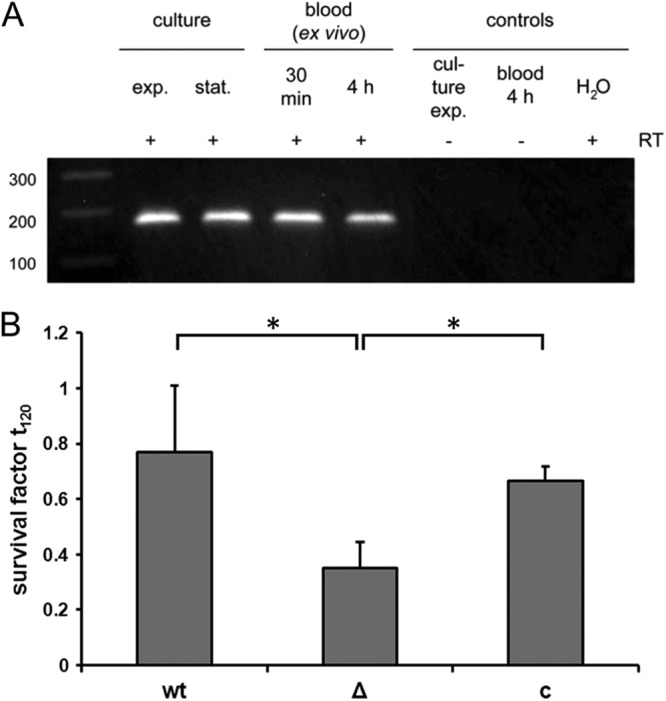
The ideSsuis gene is transcribed ex vivo in porcine blood (A), and IdeSsuis promotes increased survival in blood of a piglet vaccinated once with a bacterin (B). (A) RT-PCR detection of ideSsuis mRNA of S. suis strain 10 in vitro grown to early exponential (exp.) or stationary (stat.) phase or incubated ex vivo with porcine blood for the indicated times. The marker bands are shown on the left side (sizes in bp). RT, reverse transcriptase. (B) Survival of S. suis strain 10 (wt), strain 10ΔideSsuis (Δ), and strain 10ΔideSsuis(pGA14ideSsuis) (c) in porcine blood, which was drawn from a piglet between 15 and 18 days after immunization with a bacterin (single application).
DISCUSSION
Recently, it has been suggested that the open reading frame SSU0496 (designated ideSsuis in this work) encodes an enzyme sharing peptidase activity with IdeS (37). As the amino acids of the catalytic center of IdeS are conserved in IdeSsuis, we investigated a putative IgG protease function of IdeSsuis. As a reliable purification method of any porcine IgG subclass has not been described and IgG subclass-specific antibodies are not yet available for pigs (17), it is currently not possible to test specifically each of the six porcine IgG subclasses for their possible cleavage by IdeSsuis. Nevertheless, we used very different sources of porcine IgG (including serum and purified porcine IgGs from very different pigs) and three different antibodies (poly- and monoclonal) with known differences in IgG recognition to investigate the putative IgG cleavage. As we did not detect cleavage of IgG by IdeSsuis in any of our experiments, we conclude that IdeSsuis does not share the IgG peptidase activity with the other members of the IdeS family.
However, this work demonstrates that IdeSsuis is an isotype and host-specific bacterial IgM protease expressed by S. suis. Although SpeB expressed by S. pyogenes and SspA from Staphylococcus aureus have previously been described to degrade IgM (39, 40), there are substantial differences between IdeSsuis on the one side and SpeB and SspA on the other side with regard to specificity and efficiency of IgM proteolysis. First, recent data indicate that SpeB cannot cleave intact IgG or IgM under physiological conditions (U. von Pawel-Rammingen, unpublished data), while our results show that IdeSsuis cleaves the intact IgM multimer in serum or plasma. Second, SspA and SpeB degrade the heavy chains of different immunoglobulin classes (IgM, IgA, and IgG) (40), whereas IdeSsuis was found to cleave only porcine IgM. Thus, to the best of our knowledge IdeSsuis is the first specific prokaryotic IgM protease described. IgM cleavage of IdeSsuis was found to be very efficient, as substantial cleavage of IgM by rIdeSsuis already occurred within 10 min and subsequent to 8°C opsonization. IgM cleavage products were detectable in culture supernatants as early as after 2.5 h of incubation. Based on the results of this work, we found that the specificity and efficiency of the IgM cleavage by IdeSsuis might be comparable to those of the IgG cleavage activity of IdeS (22).
IdeSsuis is unique in function and size within the IdeS family. The large size of IdeSsuis in comparison to that of IdeS is, however, not crucial for IgM cleavage, as a truncated recombinant IdeSsuis protein retained IgM cleavage activity. Whether or not IdeSsuis might have further function(s) related to the additional 715 C-terminal amino acids remains so far elusive. The inhibitor profile of IdeSsuis is similar to that of IdeS, except that Z-LVG-CHN2 inhibits IgG cleavage by IdeS (22) but does not inhibit cleavage of IgM by IdeSsuis (Fig. 2). We suggest nevertheless that IdeSsuis most likely belongs to the clan of cysteine proteases, but final classification has to await a detailed enzymatic characterization, currently being performed in our laboratories.
As porcine IgM contains the C1q activating motif (17), porcine IgM, like IgM of other species, is considered an important activator of the classical complement cascade. Therefore, IdeSsuis cleavage of IgM might well affect activation of the classical complement cascade, especially in piglets with low titers of specific IgG3, the only porcine IgG subclass containing a prominent C1q binding motif (17). Experiments are currently in progress in our laboratory to investigate this hypothesis.
Cleavage of human IgG in the hinge region by IdeS has major implications on the interactions between S. pyogenes and phagocytes expressing the Fcγ receptor. Priming of neutrophils through released Fc fragments as a consequence of IdeS activity is thought to cause activation of phagocytes at a distance from the site of infection (23). As IdeSsuis was shown to cleave porcine IgM but not IgG in this study, priming of neutrophils by the immunoglobulin fragments released as a consequence of IdeSsuis activity is not expected.
The fascinating finding that a host-specific IgM protease is closely related to a host-specific IgG protease (IdeS) raises the question whether this phenomenon is related to functional differences among the immunoglobulins of the different species. Pigs express six different IgG subclasses, five of which have at least two allotypes (41). Based on amino acid sequence data, it was predicted that all porcine IgG subclasses should be able to bind to the FcγRs and that the very distinct porcine IgG3 should have the highest affinity (42). In contrast, only two human IgG subclasses, IgG1 and IgG3, which are the main substrates of IdeS, bind to FcγRs. Therefore, the diversity of porcine IgG might have made it very difficult, if not impossible, for S. suis to evolve an IgG endopeptidase cleaving all porcine IgGs with the potential to activate FcyR-positive effector cells. On the other hand, at birth all antibody-expressing B cells are IgM+ in pigs, in contrast to other species (17, 43, 44). As S. suis generally colonizes piglets immediately after birth, one might speculate that expression of IdeSsuis substantially influences antibody-containing cells in the neonatal period. Thus, we plan to investigate cleavage of membrane-bound IgM by IdeSsuis and the consequence for IgM+ host cells in the future.
It is currently not understood what determines pigs and wild boars to be the natural host of S. suis. Indeed, IdeSsuis is the first described factor of S. suis that clearly reflects adaptation to interaction with the porcine immune system, as IdeSsuis was found to cleave porcine IgM but not IgM from six other species. The high specificity of IdeSsuis is pinpointed by the observation that IgM from warthogs was not cleaved, even though warthogs belong to the same family as pigs (Suidae). Cleavage of porcine but not human IgM by IdeSsuis suggests that this protein contributes to immune escape during infections of piglets but not during zoonotic infections. Experimental infections of piglets are planned to elucidate the impact of IgM cleavage by IdeSsuis on the pathogenesis of S. suis diseases.
Identification of the protease cleaving IgM bound to the bacterial surface should help to improve vaccines against S. suis. Induction of opsonizing antibodies is crucial for the protective efficacy of an S. suis bacterin (27, 45). As complement is important for the immunoglobulin-mediated killing elicited by an S. suis bacterin (unpublished results) and IdeSsuis promoted survival in blood of a vaccinated piglet, we hypothesize that neutralization of the IdeSsuis IgM protease activity might substantially improve the protective efficacy of bacterins or other future vaccines inducing opsonizing antibodies.
In summary, this work describes the specific and efficient cleavage of IgM by the IdeSsuis protein. Further elucidation of the structure and function of IdeSsuis should provide a better understanding of this fascinating coevolution of host immunoglobulins and specific bacterial immunoglobulin endoproteases.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Smith (DLO-Lelystad, Netherlands) for S. suis strains 10 and 10cpsΔEF, H. Wisselink (DLO-Lelystad, Netherlands) for S. suis T15, I. Sobottka (Universitätsklinikum Hamburg-Eppendorf, Germany) for S. suis 43447/97, J. Tang (Research Institute for Medicine of Nanjing Command, China) for S. suis O5ZYH33, and Dieter Reinscheid (Fachhochschule Bonn-Rhein-Sieg, Germany) for L. lactis pOri23. Daisuke Takamatsu (National Institute of Animal Health, Japan) kindly provided the plasmid pSET5s. We thank Tina Risch (Allwetterzoo Münster, Germany) for the serum of a warthog. Antibodies against IgM from nonporcine species were kindly provided by Hans-Joachim Schuberth (Tierärztliche Hochschule Hannover) and Marcus Fulde (Institute for Medical Microbiology and Hospital Immunology, Hannover Medical School). Maren Seitz, Andreas Nehrlich, and Jörg Willenborg are acknowledged for support in flow cytometry, immunofluorescence microscopy, and real-time RT-PCR, respectively. The Clinic for Pigs, Small Ruminants, Forensic Medicine, and Ambulatory Service (Tierärztliche Hochschule Hannover) kindly conducted vaccination of a pig with L. lactis pOri23.
This study was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG), Bonn, Germany (SFB587), and by the Swedish research council (project 2009-4997).
Footnotes
Published ahead of print 14 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01875-12.
REFERENCES
- 1. Baele M, Chiers K, Devriese LA, Smith HE, Wisselink HJ, Vaneechoutte M, Haesebrouck F. 2001. The gram-positive tonsillar and nasal flora of piglets before and after weaning. J. Appl. Microbiol. 91:997–1003 [DOI] [PubMed] [Google Scholar]
- 2. Su Y, Yao W, Perez-Gutierrez ON, Smidt H, Zhu WY. 2008. Changes in abundance of Lactobacillus spp. and Streptococcus suis in the stomach, jejunum and ileum of piglets after weaning. FEMS Microbiol. Ecol. 66:546–555 [DOI] [PubMed] [Google Scholar]
- 3. Gottschalk M. 2011. Streptococcosis, p 841–855 In Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW. (ed), Diseases of swine, 10th e., Wiley-Blackwell, Oxford, United Kingdom [Google Scholar]
- 4. Mai NT, Hoa NT, Nga TV, Linh l Chau DTT, Sinh DX, Phu NH, Chuong LV, Diep TS, Campbell J, Nghia HD, Minh TN, Chau NV, de Jong MD, Chinh NT, Hien TT, Farrar J, Schultsz C. 2008. Streptococcus suis meningitis in adults in Vietnam. Clin. Infect. Dis. 46:659–667 [DOI] [PubMed] [Google Scholar]
- 5. Wertheim HF, Nghia HD, Taylor W, Schultsz C. 2009. Streptococcus suis: an emerging human pathogen. Clin. Infect. Dis. 48:617–625 [DOI] [PubMed] [Google Scholar]
- 6. Charland N, Harel J, Kobisch M, Lacasse S, Gottschalk M. 1998. Streptococcus suis serotype 2 mutants deficient in capsular expression. Microbiology 144:325–332 [DOI] [PubMed] [Google Scholar]
- 7. Smith HE, Damman M, van der Velde J, Wagenaar F, Wisselink HJ, Stockhofe-Zurwieden N, Smits MA. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67:1750–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Greeff A, Buys H, Verhaar R, Dijkstra J, van Alphen L, Smith HE. 2002. Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect. Immun. 70:1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baums CG, Kaim U, Fulde M, Ramachandran G, Goethe R, Valentin-Weigand P. 2006. Identification of a novel virulence determinant with serum opacification activity in Streptococcus suis. Infect. Immun. 74:6154–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fittipaldi N, Sekizaki T, Takamatsu D, Dominguez-Punaro ML, Harel J, Bui NK, Vollmer W, Gottschalk M. 2008. Significant contribution of the pgdA gene to the virulence of Streptococcus suis. Mol. Microbiol. 70:1120–1135 [DOI] [PubMed] [Google Scholar]
- 11. Jacobs AAC, Loeffen PLW, van den Berg AJG, Storm PK. 1994. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect. Immun. 62:1742–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Esgleas M, Li Y, Hancock MA, Harel J, Dubreuil JD, Gottschalk M. 2008. Isolation and characterization of alpha-enolase, a novel fibronectin-binding protein from Streptococcus suis. Microbiology 154:2668–2679 [DOI] [PubMed] [Google Scholar]
- 13. Pian Y, Gan S, Wang S, Guo J, Wang P, Zheng Y, Cai X, Jiang Y, Yuan Y. 2012. Fhb, a novel factor H-binding surface protein, contributes to the antiphagocytic ability and virulence of Streptococcus suis. Infect. Immun. 80:2402–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ochsenbein AF, Zinkernagel RM. 2000. Natural antibodies and complement link innate and acquired immunity. Immunol. Today 12:624–630 [DOI] [PubMed] [Google Scholar]
- 15. Ochsenbein AF, Fehr T, Lutz C, Suter M, Brombacher F, Hengartner H, Zinkernagel RM. 1999. Control of early viral and bacterial distribution and disease by natural antibodies. Science 286:2156–2159 [DOI] [PubMed] [Google Scholar]
- 16. Sun J, Butler JE. 1997. Sequence analysis of pig switch μ, Cμ, and Cμm. Immunogenetics 46:452–460 [DOI] [PubMed] [Google Scholar]
- 17. Butler JE, Zhao Y, Sinkora M, Wertz N, Kacskovics I. 2009. Immunoglobulins, antibody repertoire and B cell development. Dev. Comp. Immunol. 33:321–333 [DOI] [PubMed] [Google Scholar]
- 18. Czajkowsky DM, Shao Z. 2009. The human IgM pentamer is a mushroom-shaped molecule with a flexural bias. Proc. Natl. Acad. Sci. U. S. A. 106:14960–14965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kubagawa H, Oka S, Kubagawa Y, Torii I, Takayama E, Kang DW, Gartland GL, Bertoli LF, Mori Takatsu HH, Kitamura T, Ohno H, Wang JY. 2009. Identity of the elusive IgM Fc receptor (FcmuR) in humans. J. Exp. Med. 206:2779–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coffman RL, Cohn M. 1977. The class of surface immunoglobulin on virgin and memory B lymphocytes. J. Immunol. 118:1806–1815 [PubMed] [Google Scholar]
- 21. Butler JE, Sun J, Navarro P. 1996. The swine Ig heavy chain locus has a single JH and no identifiable IgD. Int. Immunol. 8:1897–1904 [DOI] [PubMed] [Google Scholar]
- 22. von Pawel-Rammingen U, Johansson BP, Björck L. 2002. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J. 21:1607–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Söderberg Johansson J, Pawel-Rammingen U. 2008. The streptococcal protease IdeS modulates bacterial IgGFc binding and generates 1/2Fc fragments with the ability to prime polymorphonuclear leucocytes. Mol. Immunol. 45:3347–3353 [DOI] [PubMed] [Google Scholar]
- 24. Lei B, Deleo FR, Hoe NP, Graham MR, Mackie SM, Cole RL, Liu M, Hill HR, Low DE, Federle MJ, Scott JR, Musser JM. 2001. Evasion of human innate and acquired immunity by a bacterial homolog of CD11b that inhibits opsonophagocytosis. Nat. Med. 7:1298–1305 [DOI] [PubMed] [Google Scholar]
- 25. Lannergard J, Guss B. 2006. IdeE, an IgG-endopeptidase of Streptococcus equi ssp. equi. FEMS Microbiol. Lett. 262:230–235 [DOI] [PubMed] [Google Scholar]
- 26. Hulting G, Flock M, Frykberg L, Lannergard J, Flock JI, Guss B. 2009. Two novel IgG endopeptidases of Streptococcus equi. FEMS Microbiol. Lett. 298:44–50 [DOI] [PubMed] [Google Scholar]
- 27. Baums CG, Kock C, Beineke A, Bennecke K, Goethe R, Schröder C, Waldmann KH, Valentin-Weigand P. 2009. Streptococcus suis bacterin and subunit vaccine immunogenicities and protective efficacies against serotypes 2 and 9. Clin. Vaccine Immunol. 16:200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Que YA, Haefliger JA, Francioli P, Moreillon P. 2000. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect. Immun. 68:3516–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takamatsu D, Osaki M, Sekizaki T. 2001. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46:140–148 [DOI] [PubMed] [Google Scholar]
- 31. Perez-Martinez G, Kok J, Venema G, van Dijl JM, Smith H, Bron S. 1992. Protein export elements from Lactococcus lactis. Mol. Gen. Genet. 234:401–411 [DOI] [PubMed] [Google Scholar]
- 32. Rabilloud T. 1999. Silverstaining of 2-D electrophoresis gels. Methods Mol. Biol. 112:297–305 [DOI] [PubMed] [Google Scholar]
- 33. Kock C, Beineke A, Seitz M, Ganter M, Waldmann KH, Valentin-Weigand P, Baums CG. 2009. Intranasal immunization with a live Streptococcus suis isogenic ofs mutant elicited suilysin-neutralization titers but failed to induce opsonizing antibodies and protection. Vet. Immunol. Immunopathol. 132:135–145 [DOI] [PubMed] [Google Scholar]
- 34. Beineke A, Bennecke K, Neis C, Schröder C, Waldmann KH, Baumgärtner W, Valentin-Weigand P, Baums CG. 2008. Comparative evaluation of virulence and pathology of Streptococcus suis serotypes 2 and 9 in experimentally infected growers. Vet. Microbiol. 128:423–430 [DOI] [PubMed] [Google Scholar]
- 35. Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956 [DOI] [PubMed] [Google Scholar]
- 36. Willenborg J, Fulde M, de Greeff A, Rohde M, Smith HE, Valentin-Weigand P, Goethe R. 2011. Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. Microbiology 157:1823–1833 [DOI] [PubMed] [Google Scholar]
- 37. Holden MT, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, Cronin A, Goodhead I, Mungall K, Quail MA, Price C, Rabbinowitsch E, Sharp S, Croucher NJ, Chieu TB, Mai NT, Diep TS, Chinh NT, Kehoe M, Leigh JA, Ward PN, Dowson CG, Whatmore AM, Chanter N, Iversen P, Gottschalk M, Slater JD, Smith HE, Spratt BG, Xu J, Ye C, Bentley S, Barrell BG, Schultsz C, Maskell DJ. 2009. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS ONE 4(7):e6072 doi:10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Buysscher EV, Berman DT. 1975. Preparation of monospecific antiserums against porcine immunoglobulins, using agarose-linked immunosorbents. Am. J. Vet. Res. 36:1323–1326 [PubMed] [Google Scholar]
- 39. Collin M, Olsėn A. 2001. Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect. Immun. 69:7187–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prokesova L, Potuznikova B, Potempa J, Zikan J, Radl J, Hachova L, Baran K, Porwit-Bobr Z, John C. 1992. Cleavage of human immunoglobulins by serine proteinase from Staphylococcus aureus. Immunol. Lett. 31:259–265 [DOI] [PubMed] [Google Scholar]
- 41. Butler JE, Wertz N. 2006. Antibody repertoire development in fetal and neonatal piglets. XVII. IgG subclass transcription revisited with emphasis on new IgG3. J. Immunol. 177:5480–5489 [DOI] [PubMed] [Google Scholar]
- 42. Butler JE, Wertz N, Deschacht N, Kacskovics I. 2009. Porcine IgG: structure, genetics, and evolution. Immunogenetics 61:209–230 [DOI] [PubMed] [Google Scholar]
- 43. Brown PJ, Bourne FJ. 1976. Development of immunoglobulin-containing cell populations in intestine, spleen, and mesenteric lymph node of the young pig, as demonstrated by peroxidase-conjugated antiserums. Am. J. Vet. Res. 37:1309–1314 [PubMed] [Google Scholar]
- 44. Allen WD, Porter P. 1973. The relative distribution of IgM and IgA cells in intestinal mucosa and lymphoid tissues of the young unweaned pig and their significance in ontogenesis of secretory immunity. Immunology 24:493–501 [PMC free article] [PubMed] [Google Scholar]
- 45. Büttner N, Beineke A, de Buhr N, Lilienthal S, Merkel J, Waldmann KH, Valentin-Weigand P, Baums CG. 2012. Streptococcus suis serotype 9 bacterin immunogenicity and protective efficacy. Vet. Immunol. Immunopathol. 146:191–200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.