Abstract
Small noncoding HIV-1 leader exon 3 is defined by its splice sites A2 and D3. While 3′ splice site (3′ss) A2 needs to be activated for vpr mRNA formation, the location of the vpr start codon within downstream intron 3 requires silencing of splicing at 5′ss D3. Here we show that the inclusion of both HIV-1 exon 3 and vpr mRNA processing is promoted by an exonic splicing enhancer (ESEvpr) localized between exonic splicing silencer ESSV and 5′ss D3. The ESEvpr sequence was found to be bound by members of the Transformer 2 (Tra2) protein family. Coexpression of these proteins in provirus-transfected cells led to an increase in the levels of exon 3 inclusion, confirming that they act through ESEvpr. Further analyses revealed that ESEvpr supports the binding of U1 snRNA at 5′ss D3, allowing bridging interactions across the upstream exon with 3′ss A2. In line with this, an increase or decrease in the complementarity of 5′ss D3 to the 5′ end of U1 snRNA was accompanied by a higher or lower vpr expression level. Activation of 3′ss A2 through the proposed bridging interactions, however, was not dependent on the splicing competence of 5′ss D3 because rendering it splicing defective but still competent for efficient U1 snRNA binding maintained the enhancing function of D3. Therefore, we propose that splicing at 3′ss A2 occurs temporally between the binding of U1 snRNA and splicing at D3.
INTRODUCTION
During human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR)-driven transcription, RNA polymerase II generates a pre-mRNA that encodes at least 15 viral proteins (1). The preformed 43S ribosomal subunit recognizes the CAP structure moving along its template until it encounters a translational start codon, defined by its surrounding sequence (2). Thus, the position of the Gag and Gag/Pol open reading frames (ORFs) proximal to the 5′ end of the unspliced viral mRNA ensured their efficient recognition. However, proper HIV-1 replication is intimately connected to the expression of seven other ORFs located distal to CAP that encode the viral proteins Vif, Vpr, Tat, Rev, Nef, Vpu, and Env. Alternative splicing removes inhibitory upstream AUGs, thereby placing downstream ORFs near the CAP structure and allowing their efficient translation by the scanning ribosome. The particular HIV-1 protein encoded by a spliced mRNA is almost always specified by the ORF that is immediately downstream of the 3′ splice site (3′ss) used to create the mRNA. The sole exception is the env ORF within the bicistronic vpu-env mRNAs, whose translation is dependent on a minimal upstream ORF within the HIV-1 vpu leader (3, 4).
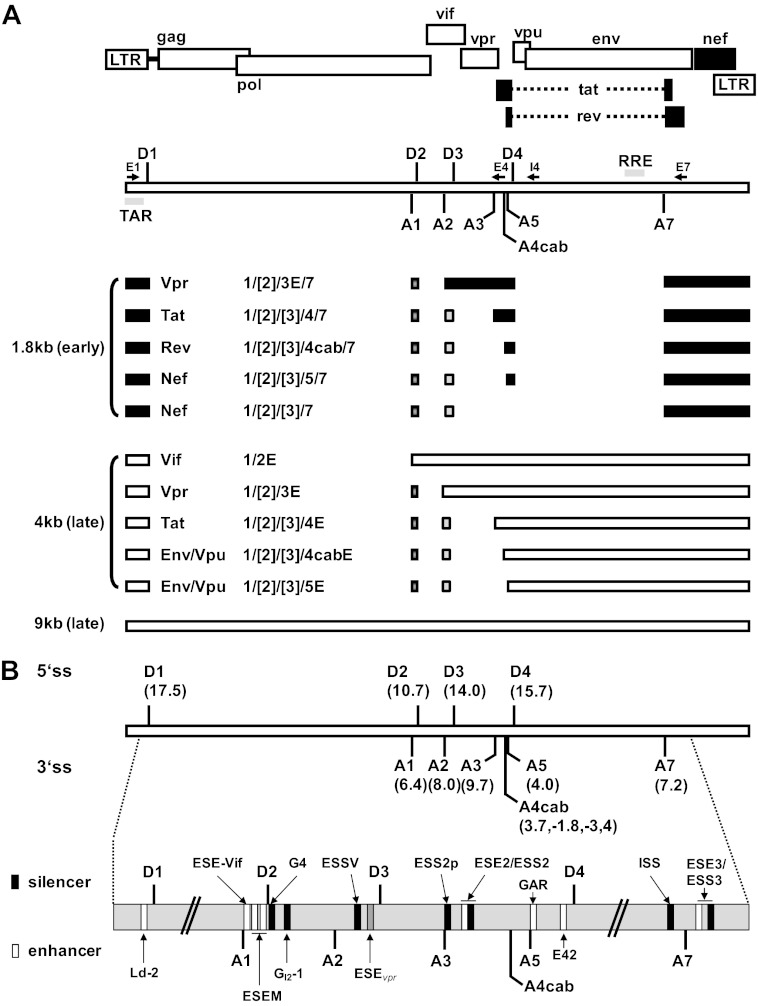
On the basis of their intron contents, three different-sized viral mRNA classes can be defined: the unspliced (9-kb), intron-containing (4-kb), and intronless (1.8-kb) viral RNAs (Fig. 1A) (5; for a recent review, see reference 6). The accumulation of these viral mRNA classes occurs in a temporal order (7, 8). In the early phase of viral gene expression, the HIV-1 pre-mRNA is extensively spliced, leading to intronless 1.8-kb mRNA species such as the tat, rev, and nef mRNAs. Rev is necessary for the onset of the late phase of viral gene expression that is characterized by a shift within the cytoplasmic mRNA pool toward isoforms with increased intron content and in which the normal nuclear retention mechanisms are bypassed (9). Rev recognizes an RNA secondary structure, called the Rev-responsive element (RRE), within the env coding sequence. Rev-RRE interactions target the intron-containing (4-kb) and unspliced (9-kb) viral mRNAs for CRM1 export receptor pathway-mediated transport into the cytoplasm, which essentially relies on the multimerization capacity of Rev (10). During the late phase of viral gene expression, the accessory and structural proteins Vif, Vpr, Vpu, and Env are translated from the respective intron-containing viral mRNAs (4 kb). In addition, the unspliced viral mRNA (9 kb) is used for translation of the structural and enzymatic components or enclosed as genomic RNA in progeny virions. Viral mRNA diversity is further increased by the alternative inclusion of either one or both of the two noncoding leader exons, 2 and 3. Noncoding leader exon 3 is flanked by 3′ss A2 and 5′ss D3. The formation of intron-containing vpr mRNA, however, requires the activation of 3′ss A2 but silencing of 5′ss D3, since the ORF of Vpr starts within the downstream intron of exon 3. Thus, vpr mRNA processing and exon 3 inclusion are mutually exclusive. Nevertheless, both splicing patterns are negatively regulated by an exonic splicing silencer (ESSV) within exon 3 (Fig. 1B) (11–13). ESSV contains three (pyrimidine)UAG motifs which promote the binding of members of the hnRNP A/B protein family to the viral mRNA, inhibiting splicing at the upstream 3′ss A2. In the absence of functional ESSV, the levels of exon 3 and vpr mRNA splicing are excessively increased, This leads to a severe perturbation of the balance between spliced and unspliced viral mRNAs that is detrimental to virus particle production (11, 13).
Fig 1.
Alternative splicing of the HIV-1 pre-mRNA. (A) Schematic of the HIV-1 genome. The ORFs are indicated by open boxes. The LTRs are located at both ends of the provirus. All HIV-1 proteins are encoded in a single primary RNA. Alternative splicing allows all viral proteins to be efficiently translated within the host cell. The 5′ (SD) and 3′ (SA) splice sites are depicted. Alternatively spliced noncoding exons 2 and 3 within Rev-independent (1.8-kb size class) and Rev-dependent (4-kb size class) spliced mRNAs are shown as boxes (exon 2, dark gray; exon 3, light gray). The positions of the primers used in RT-PCRs for the analyses of viral mRNA splicing are indicated by arrows (E1 [fwd], exon1; E4 [rev], exon 4; I4 [rev], intron 4; E7 [rev], exon 7). (B) Intrinsic strength of the 5′ss (D1 to D4) and 3′ss (A1 to A7) distributed along the HIV-1 pre-mRNA. Each value in parentheses reflects the predicted intrinsic strength (5′ss, HBond score [www.uni-duesseldorf.de/rna]; 3′ss, MaxEnt score [http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html]). The nomenclature of the viral splice sites is from reference 9. Positions of known enhancer (white) and silencer (black) sequences within the HIV-1 pre-mRNA are shown. Exon 3 is flanked by 3′ss A2 and 5′ss D3. The positions of the Ld-2 (35), ESE-Vif (36), ESEM (14), G4 (36), GI2-1 (M. Widera, and H. Schaal, submitted for publication), ESSV (11–13), ESS2p (37), ESE2 (38, 39), ESS2 (40–42), GAR, guanosine-adenosine rich (GAR) ESE (16, 17, 27), E42 (27), ISS (15), ESE3 (43), and ESS3 (43–45; adapted from references 27 and 46) sequences are shown.
In this work, we identified an exonic splicing enhancer (termed ESEvpr) between the repressing ESSV and the 5′ss D3 that acts positively on its recognition by the U1 snRNP. We show that the defect in virus particle production seen in the context of ESSV-negative provirus was efficiently rescued by additional inactivation of this ESE but that vpr mRNA processing critically depended on the presence of intact ESEvpr. Furthermore, we identified Tra2-alpha and Tra2-beta as the splicing regulatory proteins by mass spectrometry and overexpression analyses. Finally, replacement of 5′ss D3 with a splicing-incompetent but U1 binding-competent 5′ss-like sequence revealed that the ESEvpr-mediated U1 snRNP stabilization to HIV-1 exon 3 is essential for vpr mRNA expression. This argues for a function for U1 snRNP binding to 5′ss D3, unrelated to splicing, that resulted in the activation of 3′ss A2, potentially via the formation of an exon definition complex.
MATERIALS AND METHODS
Oligonucleotides.
The oligonucleotides used in this study were obtained from Metabion GmbH (Martinsried, Germany).
Primers for site-directed mutagenesis.
The oligonucleotide primers used for site-directed mutagenesis are described in Table 1.
Table 1.
DNA oligonucleotides used in this work
| Plasmid | Primer | Primer sequence |
|---|---|---|
| LTR ex2 ex3 | 1814 | 5′ GCG CGC ACG GCA AGA 3′ |
| 1817 | 5′ CTT TAC GAT GCC ATT GGG A 3′ | |
| 2346 | 5′ GAA GCG CGC ACG GCA AGA GGC GAG 3′ | |
| 2347 | 5′ CGC GAA TTC AGG CCT CTC TC 3′ | |
| 2381 | 5′ GGG CTC GAG ACT AGT GGC TGA CTT CCT GGA TG 3′ | |
| 2384 | 5′ GGG ACT AGT CAA GAA ATG GAG AAA AAA A 3′ | |
| 2385 | 5′ GTA CCC GGG CAC CAA TAA CTG CCT TA 3′ | |
| 2386 | 5′ GGG CAT ATG TAT GTT TCA GGG AAA GCT AGG GGA 3′ | |
| LTR ex2 ex3 D3down | 3817 | 5′ TTT TCA GAA TCT GCT ATA AGA AAT ACC ATA TTA GGA CGT ATA GTT AGT CCT AGG TGT GAA TAT CAA GCA GGA CAT AAC AAG GTT GGT TCT CTA CAG TAC TTG GCA CTA G 3′ |
| LTR ex2 ex3 ESE −25T>C, −16A>G D3down | 3818 | 5′ TTT TTC AGA ATC TGC TAT AAG AAA TAC CAT ATT AGG ACG TAT AGT TAG TCC TAG GTG CGA ATA TCA GGC AGG ACA TAA CAA GGT TGG TTC TCT ACA GTA CTT GGC ACT AG 3′ |
| LTR ex2 ex3 ESSV− D3down | 3819 | 5′ TTT TTC AGA ATC TGC TAT AAG AAA TAC CAT ATT AGG ACG TAT AGT ATC GCC ACG TTG TGA ATA TCA AGC AGG ACA TAA CAA GGT TGG TTC TCT ACA GTA CTT GGC ACT AG 3′ |
| LTR ex2 ex3 ESSV− ESE −25T>C, −16A>G D3down | 3820 | 5′ TTT TTC AGA ATC TGC TAT AAG AAA TAC CAT ATT AGG ACG TAT AGT ATC GCC ACG TTG CGA ATA TCA GGC AGG ACA TAA CAA GGT TGG TTC TCT ACA GTA CTT GGC ACT AG 3′ |
| LTR ex2 ex3 D3up | 3813 | 5′ TTT TCA GAA TCT GCT ATA AGA AAT ACC ATA TTA GGA CGT ATA GTT AGT CCT AGG TGT GAA TAT CAA GCA GGA CAT AAC AAG GTA GGT AGT CTA CAG TAC TTG GCA CTA GCA G 3′ |
| LTR ex2 ex3 ESE −25T>C, −16A>G D3up | 3814 | 5′ TTT TTC AGA ATC TGC TAT AAG AAA TAC CAT ATT AGG ACG TAT AGT TAG TCC TAG GTG CGA ATA TCA GGC AGG ACA TAA CAA GGT AGG TAG TCT ACA GTA CTT GGC ACT AGC AG 3′ |
| LTR ex2 ex3 ESSV− D3up | 3815 | 5′ TTT TTC AGA ATC TGC TAT AAG AAA TAC CAT ATT AGG ACG TAT AGT ATC GCC ACG TTG TGA ATA TCA AGC AGG ACA TAA CAA GGT AGG TAG TCT ACA GTA CTT GGC ACT AGC AG 3′ |
| LTR ex2 ex3 ESSV− ESE −25T>C, −16A>G D3up | 3816 | 5′ TTT TTC AGA ATC TGC TAT AAG AAA TAC CAT ATT AGG ACG TAT AGT ATC GCC ACG TTG CGA ATA TCA GGC AGG ACA TAA CAA GGT AGG TAG TCT ACA GTA CTT GGC ACT AGC AG 3′ |
| LTR ex2 ex3 ESSV− D3+1G>C | 3889 | 5′ TTT TTC AGA ATC TGC TAT AAG AAA TAC CAT ATT AGG ACG TAT AGT ATC GCC ACG TTG TGA ATA TCA AGC AGG ACA TAA CAA GCT TGG TTC TCT ACA GTA CTT GGC ACT AG 3′ |
| LTR ex2 ex3 ESSV− ESE −25T>C, −16A>G D3 +1G>C | 3890 | 5′ TTT TTC AGA ATC TGC TAT AAG AAA TAC CAT ATT AGG ACG TAT AGT ATC GCC ACG TTG CGA ATA TCA GGC AGG ACA TAA CAA GCT TGG TTC TCT ACA GTA CTT GGC ACT AG 3′ |
| LTR ex2 ex3 ESSV− GTV | 3885 | 5′ TTT TTC AGA ATC TGC TAT AAG AAA TAC CAT ATT AGG ACG TAT AGT ATC GCC ACG TTG TGA ATA TCA AGC AGG ACA TAA CCA GCT AAG TAT TCT ACA GTA CTT GGC ACT AGC AG 3′ |
| LTR ex2 ex3 ESSV− ESE −25T>C, −16A>G GTV | 3886 | 5′ TTT TTC AGA ATC TGC TAT AAG AAA TAC CAT ATT AGG ACG TAT AGT ATC GCC ACG TTG CGA ATA TCA GGC AGG ACA TAA CCA GCT AAG TAT TCT ACA GTA CTT GGC ACT AGC AG 3′ |
| 2588 | 5′ CTT TAC GAT GCC ATT GGG A 3′ | |
| pUCB U1 αD3 | 3924 | 5′ GCC CGA AGA TCT CGA TCC TAG CTT GCA GGG GAG ATA CCA TGA TC 3′ |
| 3926 | 5′ TTT TCA CTC GAG CCT CCA CTG TAG 3′ | |
| pUCB U1 αD3 (+1G>C) | 3925 | 5′ GCC CGA AGA TCT CGA TCC TAC CTT GCA GGG GAG ATA CCA TGA TC 3′ |
| 3926 | 5′ TTT TCA CTC GAG CCT CCA CTG TAG 3′ |
Primers used for semiquantitative and quantitative real-time RT-PCR.
The oligonucleotide primers used for semiquantitative and quantitative real-time reverse transcription (RT)-PCR are described in Table 2.
Table 2.
Primers used for semiquantitative and quantitative real-time RT-PCR
| Viral mRNA type | Primer | Primer sequence |
|---|---|---|
| Classes: | ||
| E1 | 1544 (E1) | 5′ CTT GAA AGC GAA AGT AAA GC 3′ |
| E7 | 3392 (E7) | 5′ CGT CCC AGA TAA GTG CTA AGG 3′ |
| I4 | 640 (I4) | 5′ CAA TAC TAC TTC TTG TGG GTT GG 3′ |
| E4 | 3632 (E4) | 5′ TGG ATG CTT CCA GGG CTC 3′ |
| All classes | 3387 | 5′ TTG CTC AAT GCC ACA GCC AT 3′ |
| 3388 | 5′ TTT GAC CAC TTG CCA CCC AT 3′ | |
| Unspliced | 3389 | 5′ TTC TTC AGA GCA GAC CAG AGC 3′ |
| 3390 | 5′ GCT GCC AAA GAG TGA TCT GA 3′ | |
| Multiply spliced | 3391 | 5′ TCT ATC AAA GCA ACC CAC CTC 3′ |
| 3392 | 5′ CGT CCC AGA TAA GTG CTA AGG 3′ | |
| vif | 3395 | 5′ GGC GAC TGG GAC AGC A 3′ |
| 3396 | 5′ CCT GTC TAC TTG CCA CAC 3′ | |
| vpr | 3397 | 5′ CGG CGA CTG AAT CTG CTA T 3′ |
| 3398 | 5′ CCT AAC ACT AGG CAA AGG TG 3′ | |
| Exon 3 inclusion | 3397 | 5′ CGG CGA CTG AAT CTG CTA T 3′ |
| 3636 | 5′ CCG CTT CTT CCT TGT TAT GTC 3′ | |
| Exon 3 exclusion | 3629 | 5′ GGC GGC GAC TGG AAG AAG C 3′ |
| 3637 | 5′ GAG AAG CTT GAT GAG TCT GAC 3′ | |
| GH1a | 1224 | 5′ TCT TCC AGC CTC CCA TCA GCG TTT GG 3′ |
| 1225 | 5′ CAA CAG AAA TCC AAC CTA GAG CTG CT 3′ |
Transfection control.
HIV-1-based subgenomic splicing reporter.
The HIV-1 NL4-3 (GenBank accession no. M19921)-derived parental plasmid LTR ex2 ex3 contains the two small noncoding leader exons 2 and 3 and the 5′ part of tat exon 1 interspersed with their authentic intronic sequences.
LTR ex2 ex3 was constructed as follows. First, the EcoRI/PstI fragment of the previously described LTR SD SA tatCAT minigene (14) was replaced with a PCR product obtained with primer pair 1814/1817 and pNLA1 (15)—a cDNA derivate of pNL4-3—as the template, leading to LTR SD ex2 ex3 SA. Subsequently, viral splice site D1 was inserted via BssHII/EcoRI restriction sites by using an amplicon obtained by PCR with primer pair 2346/2347 and SV-1-env (16) as the template, generating LTR D1 ex2 ex3 SA. In the next step, the NdeI/SalI fragment of LTR D1 ex2 ex3 SA was replaced with an NdeI/XhoI-digested PCR product amplified with primer pair 2386/2381 by using pNLA-1 as the template. This fragment contained viral splice site A3, an SpeI restriction site inserted via reverse primer 2381 and duplicated sequences downstream of 3′ss A3, generating LTR D1 ex2 ex3 A3 dupl. To remove the duplicated sequences obtained during this cloning step, the SpeI/XmaI fragment of LTR D1 ex2 ex3 A3 dubl was replaced with a PCR product obtained with primer pair 2384/2385 and LTR SD SAtatCAT as the template, leading to LTR ex2 ex3/pNLA1. Finally, the LTR ex2 ex3 splicing reporter, whose viral nucleotide sequences are identical to those of pNL4-3, was cloned by insertion of the NdeI/EcoRI fragment from pNL4-3 into LTR ex2 ex3/pNLA1.
LTR ex2 ex3 ESSV and ESEvpr mutants were constructed by PCR mutagenesis. For construction, the AlwNI/SpeI fragment of LTR ex2 ex3 was replaced with the respective PCR products using an appropriate forward mutagenesis primer and 2588 as the reverse PCR primer containing AlwNI and SpeI restriction sites. All plasmid sequences can be obtained on request.
Proviral HIV-1 plasmids.
pNL4-3 mutants were constructed by replacing the region between PflMI and EcoRI of proviral clone pNL4-3 with the respective mutated LTR ex2 ex3 minigene fragments, which were generated as described above.
U1 snRNA expression plasmids.
pUCBU1αD3 and pUCBU1αD3(+1G>C) were constructed by the insertion of a PCR product amplified with primer pairs 3924/3926 and 3925/3926, respectively, containing BglII and XhoI restriction sites into the template pUCBU1 (kindly provided by Alan M. Weiner) into pUCBΔU1 (17).
Cell culture and RT-PCR analysis.
HeLa and HEK 293T cells were maintained in Dulbecco's high-glucose modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum and 50 μg/ml each penicillin and streptomycin (Invitrogen). Transfections were done in six-well plates with 2.5 × 105 cells per plate using FuGENE6 reagent (Roche) according to the manufacturer's instructions. Total-RNA samples were collected 30 h after transfection from either HeLa or HEK 293T cells transfected with subgenomic or proviral constructs and pXGH1 as a control. For RT, 4 μg of RNA was subjected to DNA digestion with 10 U of DNase I (Roche). DNase I was heat inactivated at 70°C for 5 min, and cDNA synthesis was allowed to occur for 1 h at 50°C and 15 min at 72°C by using 200 U Superscript III RNase H− reverse transcriptase (Invitrogen), 7.5 pmol oligo(dT)12-18 (Invitrogen) as the primer, 20 U of RNasin (Promega), and 10 mM each deoxynucleoside triphosphate (Qiagen). For semiquantitative analysis of minigene mRNAs, cDNA was used as the template for a PCR with forward primer 1544 and reverse primer 3632. For a transfection control, a PCR was performed with primers 1224 and 1225 to specifically detect GH1 mRNA. For analysis of exon 3 inclusion in viral tat mRNAs and vpr mRNA splicing, a PCR was performed using primers 1544 (E1) and 3632 (E4). For analysis of 1.8-kb HIV-1 mRNAs, a PCR was carried out with forward primer 1544 (E1) and reverse primer 3392 (E7). Partially spliced 4.0-kb HIV-1 mRNAs were detected with primers 1544 (E1) and 640 (I4). PCR products were separated on 8% nondenaturing polyacrylamide gels, stained with ethidium bromide, and visualized with a Lumi-Imager (Roche).
Quantitative real-time PCR assays for the detection of single viral mRNA species were done with primer pairs 3389/3390 for unspliced mRNA, 3391/3392 for multiple spliced mRNA, 3395/3396 for vif mRNA, 3397/3398 for vpr mRNA, 3397/3636 for exon 3 inclusion, and 3629/3637 for exon exclusion. For normalization, primers 3387 and 3388 were used and the level of overall viral mRNAs present in each sample was monitored. Fluorescence emission was read by a LightCycler 1.5 (Roche). Data are presented as the average of three independent RT-PCR experiments.
Antibodies.
The following primary antibodies were used for immunoblot analysis. A mouse antibody against α-actin (A2228) was obtained from Sigma-Aldrich. A mouse antibody against hnRNP A1 (9H10) was purchased from Santa Cruz Biotechnology. A rabbit antibody against Tra2-beta (ab50846) was obtained from Abcam. A sheep antibody against HIV-1 p24 was purchased from Biochrom AG. Rabbit antiserum against Vif and rabbit antiserum against Vpr were obtained through the NIH AIDS Research and Reference Reagent Program from Dana Gabuzda (18) and Jeffrey Kopp, respectively. For detection, we used a horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (A6154) from Sigma-Aldrich, an HRP-conjugated anti-mouse antibody (NA931) from GE Healthcare (Munich, Germany), and an HRP-conjugated anti-sheep antibody from Jackson ImmunoResearch Laboratories Inc. (West Grove, PA).
Protein analysis.
Transfected cells were lysed in radioimmunoprecipitation assay buffer (25 mM Tris HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail [Roche]). Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, and subjected to an immunoblotting procedure. The membranes were probed with the respective primary and secondary antibodies and developed with ECL chemiluminescence reagents (GE Healthcare).
Protein isolation by RNA affinity chromatography.
Short RNA oligonucleotides were obtained from Metabion. The RNA oligonucleotides were covalently coupled to agarose beads (Sigma). Immobilized RNAs were incubated in HeLa nuclear extract (Cilbiotech) diluted to a concentration of 40% with buffer D (20 mM HEPES-KOH [pH 7.9], 5% [vol/vol] glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol). To remove unspecific bound proteins, samples were washed five times with 1 ml buffer D containing 4 mM magnesium chloride (800 rpm, 2 to 3 min, Eppendorf microcentrifuge). Precipitated proteins were eluted from the RNA by heating to 95°C for 10 min in protein sample buffer. Protein samples were subjected to mass spectrometry or loaded onto an SDS-polyacrylamide gel for Western blot analysis.
Mass spectrometry and mass spectrometric data analysis.
Protein samples from RNA affinity purification experiments were loaded onto an SDS-polyacrylamide gel, concentrated in the stacking gel, stained with silver, reduced, alkylated, and digested with trypsin. Peptides were extracted from the gel and subjected to liquid chromatography in 0.1% trifluoroacetic acid. For peptide separation over a 140-min gradient, an Ultimate 3000 Rapid Separation liquid chromatography system (Dionex/Thermo Scientific, Idstein, Germany) equipped with an Acclaim PepMap 100 C18 column (75-μm inside diameter, 50-cm length, 2-μm particle size; Dionex/Thermo Scientific, Idstein, Germany) was used. Mass spectrometry was carried out with an Orbitrap Elite high-resolution instrument (Thermo Scientific, Bremen, Germany) operated in positive mode and equipped with a nanoelectrospray ionization source. The capillary temperature was set to 275°C, and the source voltage was set to 1.5 kV. Survey scans were carried out with the Orbitrap analyzer over a mass range of 350 to 1,700 m/z at a resolution of 60,000 (at 400 m/z). The target value for the automatic gain control was 1,000,000, and the maximum fill time was 200 ms. The 20 most intense doubly and triply charged peptide ions (minimal signal intensity, 500) were isolated, transferred to the linear ion trap (LTQ) part of the instrument, and fragmented by collision-induced dissociation. Peptide fragments were analyzed by using a maximal fill time of 200 ms and an automatic gain control target value of 100,000. The available mass range was 200 to 2,000 m/z at a resolution of 5,400 (at 400 m/z). Two fragment spectra were summed, and already fragmented ions were excluded from fragmentation for 45 s.
Raw files were further processed for protein and peptide identification and quantification using MaxQuant software suite version 1.3.0.5 (Max Planck Institute of Biochemistry, Planegg, Germany). Within the software suite, database searches were carried out by using 86,875 human sequences from the UniProtKB/SwissProt database, including the Trembl part (release 06.2012), with the following parameters: mass tolerance Fourier-transformed mass spectra (Orbitrap) first/second search, 20 ppm/6 ppm; mass tolerance fragment spectra (linear ion trap), 0.4 Da; fixed modification, carbamidomethyl; variable modification, methionine oxidation and acetylation at protein N termini. Label-free quantification was done by using the “match between runs” option with a 2-min time window. Peptides and proteins were accepted at a false-discovery rate (FDR) of 1%, and proteins identified with a minimum of two peptides and quantitative information available for all 10 measured samples were subjected to subsequent statistical analysis. To discriminate selective from nonselective binding protein besides calculating conventional Student t tests on log-transformed data, the significance analysis of microarrays (SAM) algorithm (19) implemented in Perseus version 1.2.7.4 (Max Planck Institute of Biochemistry, Planegg, Germany) was used (FDR threshold, 0.05; constant S0, 1.2). The algorithm accounts both for the change in protein abundance and standard deviation of measurements.
RESULTS
ESEvpr is necessary for vpr mRNA processing.
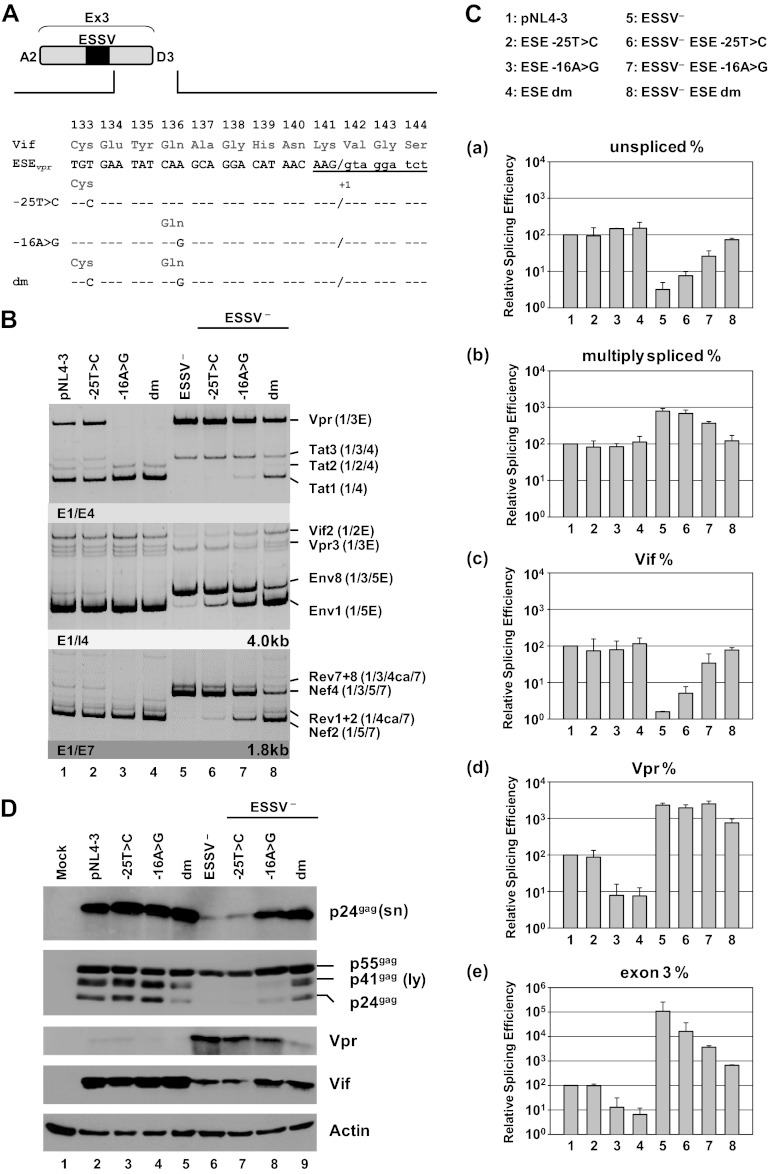
Using an enhancer-dependent splicing reporter (16), we systematically screened exon 3 for splicing regulatory elements and found the 25-nucleotide-long fragment between ESSV and D3 to contain an enhancer sequence (20). On the basis of hexamer score changes, we singled out two point mutations suspected to impair ESEvpr enhancer function (Fig. 2A) (S. Theiss, S. Erkelenz, and H. Schaal, unpublished data) (21).
Fig 2.
ESEvpr is necessary for exon 3 inclusion and vpr mRNA processing. (A) The wild-type ESEvpr sequence and the amino acid sequence encoded by the overlapping vif ORF are shown below exon 3. Mutated ESEvpr nucleotide residues are denoted by their positions relative to the GT dinucleotide of viral 5′ss D3. The black box represents the upstream ESSV. Uppercase letters represent exonic positions, and lowercase letters represent intronic positions. (B) HEK 293T cells (2.5 × 105) were transiently transfected with 1 μg of each of the proviral plasmids. At 30 h after transfection, total-RNA samples were collected and used for RT-PCR analyses with different sets of primer pairs. HIV-1 mRNA species are indicated to the right of the gels in accordance with the nomenclature published previously (5). (C) cDNA samples were prepared as described for panel B and used in real-time PCR assays to specifically quantitate the relative abundances of unspliced (a), multiply spliced (b), Vif (c), and Vpr (d) mRNA species and exon 3 inclusion ratios (e). For normalization, primers 3387 and 3388 were used to detect the total viral mRNA content of each sample. Data represent expression ratios relative to that of wild-type pNL4-3 (bar 1), which was set to 100%. Values and error bars show the average ± standard deviation of three independent transfection experiments. Bars correspond to lanes in panel B. (D) HEK 293T cells (2.5 × 105) were transiently transfected with 1 μg of each of the proviral plasmids. At 48 h posttransfection, viral supernatants were collected, layered onto 20% sucrose solution, and centrifuged at 28,000 rpm for 90 min at 4°C to pellet the released virions. In addition, cells were harvested and resuspended in lysis buffer. Supernatants and cellular lysates were resolved by 12% SDS-PAGE and electroblotted onto nitrocellulose membranes. To determine virus particle production and the expression of viral proteins, samples were probed with primary antibodies specifically detecting structural p24gag (CA) and the viral infectivity factors Vif and Vpr. Equal amounts of cell lysates were controlled for by the detection of α-actin. E, extended exon; dm, double mutation; sn, supernatant; ly, lysate.
In order to confirm their relevance for exon 3 splice site activation and vpr mRNA formation, we used proviral clone pNL4-3 (GenBank accession no. M19921) and mutant forms thereof to transfect HEK 293T cells. Semiquantitative RT-PCRs were set up with different primer pairs to detect exon 3 inclusion and vpr mRNA processing within intron-containing and intronless viral RNAs (Fig. 2B). In the presence of the repressing ESSV, the ESEvpr single mutation −16A>G, as well as the −25T>C −16A>G double mutation, led to nearly undetectable levels of exon 3 inclusion in the tat, nef, and env mRNAs (Tat3, Env8, Nef4, Rev7+8) (Fig. 2B, lanes 1 to 4). Additionally, it was impossible to detect vpr mRNAs (Fig. 2B, lanes 1 to 4), indicating that ESEvpr is also required for the activation of 3′ss A2. In line with previous observations (13), inactivation of ESSV resulted in a shift from exon 3-less to exon 3-containing nef, rev, tat, and env mRNAs. Additionally, we observed a considerable increase in the expression of vpr mRNAs (Fig. 2B, cf. lanes 1 and 5). However, the inclusion of exon 3 in the nef, rev, tat, and env mRNA species could be gradually reduced to near-wild-type levels, starting from −25T>C, followed by −16A>G and then the double mutation (Fig. 2B, lanes 6 to 8). Taken together, these results demonstrated that ESEvpr contributed to the regulation of exon 3 inclusion in each of the viral mRNA species. To thoroughly examine ESEvpr for its impact on the regulation of HIV-1 exon 3 splicing, quantitative RT-PCR analyses were performed. Different primer pairs were used to specifically quantitate the relative levels of viral unspliced, spliced, vpr, vif, and exon 3-containing mRNAs. Quantitative RT-PCR assays showed that ESEvpr mutations did not significantly alter the levels of unspliced, spliced, and vif mRNAs in the context of the ESSV-positive virus (Fig. 2C, parts a to c, bars 1 to 4). However, the single point mutation −16A>G alone was able to downmodulate the relative amount of vpr mRNA, indicating that ESEvpr was necessary for the activation of 3′ss A2 even in the presence of ESSV (Fig. 2C, part d, bars 1 to 4). Furthermore, the levels of exon 3-containing mRNA species were greatly reduced (Fig. 2C, part e, bars 3 and 4). Consistent with previous work (13), disruption of ESSV resulted in a large reduction (∼10- to 20-fold) in the level of unspliced mRNA (Fig. 2C, part a, cf. bars 1 and 5). Moreover, the relative amount of multiply spliced mRNAs was upregulated approximately 10-fold (Fig. 2C, part b, cf. bars 1 and 5). In addition, loss of ESSV function because of mutagenesis induced a strong decrease in vif mRNA levels of up to 20-fold (Fig. 2C, part c, cf. bars 1 and 5). In contrast, expression of vpr and exon 3-containing viral mRNAs was detected at highly elevated levels (Fig. 2C, parts d and e, cf. bars 1 and 5). These results were consistent with recent studies showing that disruption of ESSV causes a dramatic deregulation of viral splicing. However, second-site mutations within the ESEvpr element could compensate for the lack of ESSV activity (Fig. 2C, parts a to e, bars 6 to 8). ESEvpr double mutations restored at least normal levels of unspliced, spliced, and vif mRNAs (Fig. 2C, parts a to c, cf. bars 1 and 8). vpr mRNA levels were also decreased in the case of the ESEvpr double mutation both with and without ESSV (Fig. 2C, part d, cf. bars 1 and 4, and 5 and 8), albeit they did not completely return to wild-type levels (Fig. 2C, part d, cf. bars 1 and 8), possibly because of residual enhancer activity. This notion was supported by the finding that the expression of exon 3-including mRNAs also did not entirely return to normal levels (Fig. 2C, part e, cf. bars 1 and 8).
Furthermore, we performed Western blot analyses to evaluate the levels of both intracellular viral proteins and virus particles released into the supernatant (Fig. 2D). In agreement with the data obtained from real-time PCR assays, the levels of Gag and Vif proteins were mostly unaffected by the ESEvpr mutants in the context of the intact ESSV (Fig. 2D, lanes 2 to 5). Moreover, similar viral capsid (CA, p24gag) levels within the supernatant samples indicated that virus particle production was not significantly changed (Fig. 2D, lanes 2 to 5). However, in the absence of ESSV, the levels of the Gag and Vif proteins were strongly reduced (Fig. 2D, lane 6). Moreover, we observed a striking defect in Gag processing, characterized by loss of the Gag precursor p55 cleavage products p41 and p24 (Fig. 2D, lane 6). As expected from the RT-PCR results, Vpr protein expression was drastically increased in the ESSV mutant (Fig. 2D, lane 6). As anticipated on the basis of earlier studies (13), mutation of ESSV led to a defect in the liberation of virus particles into the cell supernatant, as indicated by the detection of only small amounts of p24gag (Fig. 2D, lane 6). This has been hypothesized to result from insufficient amounts of intracellular Gag, which is needed to drive virus assembly at the cellular plasma membrane. The expression and normal processing of structural proteins and Vif within the cells were reinstated following the double mutation of ESEvpr (Fig. 2D, lane 9). Furthermore, the failure to efficiently produce virus particles of ESSV-negative clones could be rescued by the −16A>G mutation and the double mutation (Fig. 2D, lanes 8 and 9). Finally, Vpr protein amounts were strongly reduced following the insertion of the −25T>C −16A>G double mutation in the absence of ESSV, demonstrating again that ESEvpr is required for 3′ss A2 activation.
In summary, ESEvpr appears to promote the use of splice sites A2 and D3. Additionally, these observations demonstrate that a functional enhancer is critical for the expression of vpr mRNA, noteworthy even when ESSV is active, indicating a delicate interplay between ESSV and ESEvpr in the regulation of viral HIV-1 exon 3 splicing.
The splicing factors Tra2-alpha and Tra2-beta bind to the ESEvpr sequence.
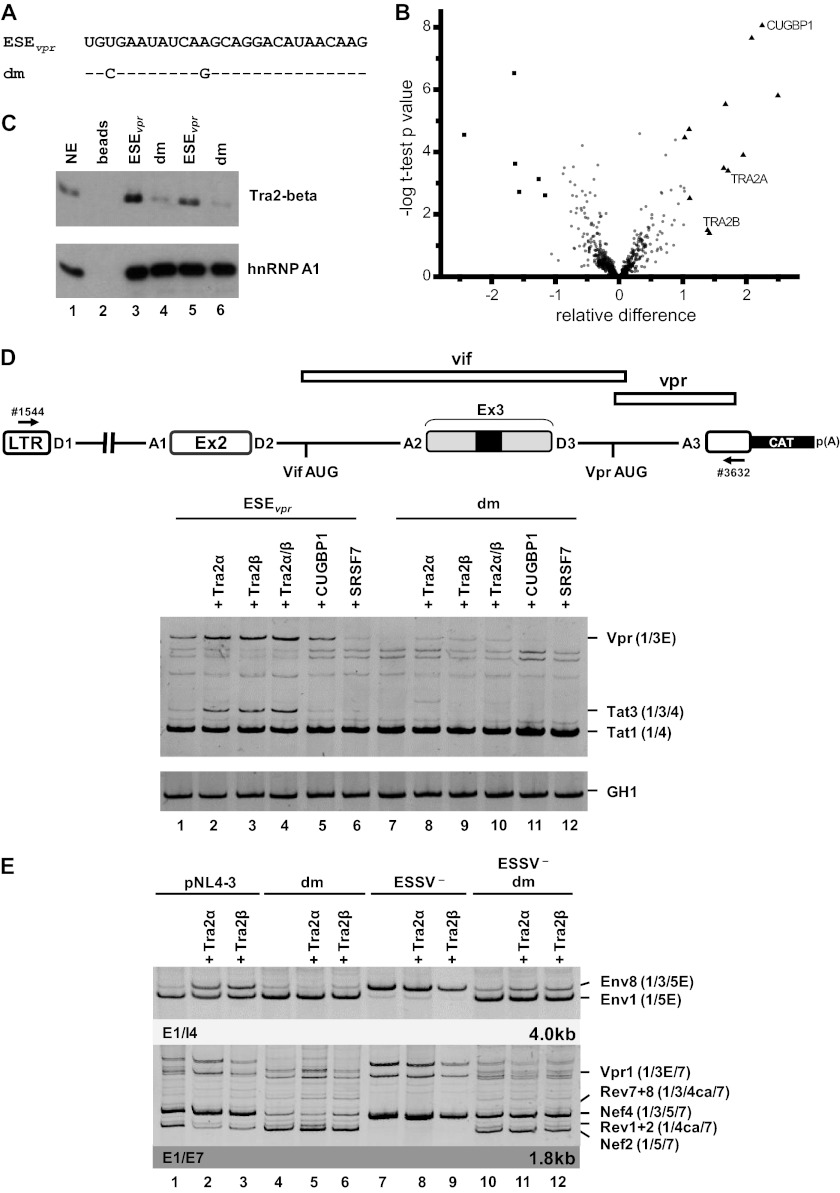
To identify cellular factors that bind to the ESEvpr sequence, RNA affinity purification experiments were performed. Therefore, we incubated short, in vitro-synthesized RNA substrates of either the wild-type or the double-mutated (−25T>C −16A>G) ESEvpr sequence (each n = 5) in HeLa cell nuclear extracts (Fig. 3A). After SDS-PAGE purification, proteins were in-gel digested with trypsin. The peptides obtained were separated by liquid chromatography and mass spectrometry for label-free quantitative analysis. This allowed us to quantify 602 RNA affinity-purified proteins in each of the 10 samples analyzed. To discriminate unspecific binding proteins from proteins bound to the ESEvpr sequence specifically affected by the double mutation, FDR-controlled statistical analysis based on the SAM method (19) was used. This algorithm assigns a score based on the change in protein abundance relative to the standard deviation of repeated measurements and estimates the FDR by using permutations. This approach revealed 6 proteins in the double-mutated ESEvpr group, as well as 12 proteins in the wild-type group, to be significantly enriched (Table 3; Fig. 3B). Eight of those 12 proteins could be assigned to the gene ontology biological process term mRNA processing, representing a significant enrichment of this biological process (FDR-adjusted P value, 0.01). Besides binding to several members of the cleavage stimulation factor complexes, as well as the cleavage polyadenylation-stimulating factor complexes, we found a significant increase in the proteins Tra2-alpha, Tra2-beta, and CUGBP1 in the wild-type ESEvpr sequence. Because of their known role in regulating alternative splicing, we chose them for further validation (22–25).
Fig 3.
ESEvpr is bound by the splicing factors Tra2-alpha and Tra2-beta. (A) In vitro-transcribed RNA substrates used for RNA pulldown experiments (dm, double mutation). (B) Volcano plot of RNA binding proteins purified by RNA pulldown with a nonmutated or a mutated ESEvpr sequence with HeLa cell nuclear extract. The precipitated proteins were digested with trypsin and subjected to quantitative mass spectrometry analysis. The x axis of the volcano plot shows the relative difference in protein abundance as calculated by the SAM method, whereas the y axis shows the −log t-test P value of the groupwise comparison of protein abundances. Besides the majority of probably unspecifically binding proteins (circles), some proteins preferably bound to the wild-type ESEvpr sequence (triangles) or the mutated ESEvpr variant (squares). The proteinsTra2-alpha and Tra2-beta were selected for validation experiments. (C) Immunoblot analysis with an antibody specific for Tra2-beta and hnRNPA1 confirmed significantly smaller amounts of Tra2-beta for the double mutant. (D) HeLa cells (2.5 × 105) were transiently cotransfected with 1 μg of each of the HIV-1-based LTR ex2 ex3 splicing reporters, 0.2 μg of SVctat (47); 1 μg of pXGH5 (GH1) as a transfection control, and 1 μg of pcDNA3.1(+), an expression plasmid for Tra2-alpha, Tra2-beta, CUGBP1, and SRSF7. At 30 h posttransfection, total RNA was isolated and subjected to semiquantitative RT-PCR analyses with primers 1544 and 3632. For measurement of equal transfection efficiencies, a separate PCR was carried out with a primer pair (1224/1225) specific for human growth hormone 1 (GH1). (E) RT-PCR analyses of intronless (2-kb) and intron-containing (4-kb) viral mRNA species following the transient transfection of HEK 293T cells with 1 μg of the respective proviral construct and 1 μg of pcDNA3.1(+), an expression plasmid for either Tra2-alpha or Tra2-beta. E, extended exon; dm, double mutation.
Table 3.
Proteins identified by mass spectrometry
| Protein | IDs | Gene(s) | t-test P value | MS intensity ratio ESE/dm | No. of unique peptides |
|---|---|---|---|---|---|
| RNA-binding protein 4B | Q9BQ04, E9PM61, E9PLB0, Q9BWF3-2, E7EQS3, D6R9K7, Q9BWF3-3 | RBM4B, RBM4 | 2.83E-05 | 0,18 | 3 |
| Flap endonuclease 1 | P39748 | FEN1 | 2.96E-07 | 0,32 | 16 |
| Zinc finger protein 207 | H0Y3M2, E1P660, O43670, O43670-2, A8MTG3 | ZNF207 | 0.000239198 | 0,33 | 5 |
| Serine hydroxymethyltransferase | P34897, P34897-3, P34897-2, B4DLV4, H0YIZ0, G3V4W5, G3V5L0 | SHMT2 | 0.00190648 | 0,31 | 11 |
| RNA-binding protein 14 | Q96PK6 | RBM14 | 0.000738779 | 0,43 | 3 |
| Mitotic checkpoint protein BUB3 | O43684, O43684-2, B4DDM6 | BUB3 | 0.00246569 | 0,44 | 7 |
| Cleavage stimulation factor subunit 2 | E7EWR4, P33240, P33240-2, B4DUD5, E9PID8 | CSTF2 | 3.50E-05 | 2,02 | 5 |
| Cleavage stimulation factor subunit 3 | Q12996, F5H0G6 | CSTF3 | 1.89E-05 | 2,15 | 14 |
| Putative DNase TATDN3 | G3V151, Q17R31, Q17R31-2, E9PJE5, E9PNH3, E9PP81, E9PRA1 | TATDN3 | 0.00305696 | 2,20 | 11 |
| Transformer-2 protein homolog beta | P62995, E7EQD1, P62995-3, H7BXF3 | TRA2B | 0.0323067 | 2,49 | 8 |
| Single-stranded DNA-binding protein, mitochondrial | Q04837, E7EUY5, C9K0U8 | SSBP1 | 0.0400618 | 2,53 | 4 |
| Cleavage and polyadenylation specificity factor subunit 7 | Q8N684-3, Q8N684, Q8N684-2, F5H669, F5H047, F5H6M0 | CPSF7 | 0.000334073 | 3,00 | 11 |
| Cleavage and polyadenylation specificity factor subunit 5 | O43809, H3BND3 | NUDT21 | 2.97E-06 | 3,18 | 14 |
| Transformer-2 protein homolog alpha | Q13595, B4DUA9, B4DQI6, Q13595-2 | TRA2A | 0.000404988 | 3,27 | 3 |
| Cleavage and polyadenylation specificity factor subunit 6 | Q16630-2, Q16630, C9JGC2, F8WJN3, Q16630-3 | CPSF6 | 0.000126486 | 3,68 | 9 |
| Squamous cell carcinoma antigen recognized by T cells 3 | Q15020, B7ZKM0 | SART3 | 2.24E-08 | 4,27 | 4 |
| CUGBP Elav-like family member 1 | G5EA30, Q92879-4, Q92879, F8W940, Q92879-3, Q92879-2, E9PKU1, F5H0D8 | CELF1 | 8.77E-09 | 4,74 | 9 |
| U6 snRNA-associated Sm-like protein LSm2 | Q9Y333 | LSM2 | 1.56E-06 | 5,51 | 5 |
Western blot analyses confirmed that while the levels of hnRNP A1 were not changed by the double mutation, Tra2-beta was precipitated with significantly reduced efficiency by the mutated ESEvpr sequence (Fig. 3C, cf. lanes 3 and 4 and lanes 5 and 6). To unravel whether the splicing factors identified are functionally involved in exon 3 splice site activation, we performed coexpression experiments and analyzed their effects on HIV-1 exon 3 and vpr mRNA splicing. In the context of an HIV-1-based minigene (Fig. 3D), coexpression of Tra2-alpha, -beta, and both increased exon 3 splice site activation in the presence of the wild-type ESEvpr sequence (Fig. 3D, cf. lanes 1 to 4), while it failed to promote exon 3 inclusion following inactivation of the enhancer (Fig. 3D, cf. lanes 7 to 10). Coexpression of CUGBP1 or SRSF7, however, did not increase exon 3 splice site activation in either in the context of wild-type ESEvpr (Fig. 3D, cf. lanes 1, 5, and 6) or that of double mutant ESEvpr (Fig. 3D, cf. lanes 7, 11, and 12). Tra2 proteins were also coexpressed together with pNL4-3 and the derived mutants (Fig. 3E), reiterating their role in ESEvpr-controlled exon 3 splice site activation. Once again, it was found that Tra2-alpha and -beta overexpression increased the inclusion of exon 3 in the case of wild-type ESEvpr but not that of double mutant ESEvpr (Fig. 3E, cf. lanes 1 to 6). Therefore, we concluded that Tra2 proteins bind to newly found ESEvpr and are thereby involved in the activation of exon 3 inclusion and vpr mRNA splicing.
A modified U1 snRNA fully complementary to 5′ss D3 strongly activates exon 3 inclusion and vpr mRNA expression.
To determine the importance of ESEvpr for exon 3 splice site use under conditions of optimal 5′ss D3 recognition, we generated a 5′-end-mutated U1 snRNA matching all 11 nucleotides of 5′ss D3 (Fig. 4A). HEK 293T cells were transiently cotransfected with this modified U1 snRNA expression vector and proviral DNA containing either wild-type or mutant exon 3 sequences. In general, RT-PCR analysis of RNA isolated from the transfected cells revealed a dramatic shift toward vpr and/or exon 3 spliced mRNAs upon the coexpression of the mutated U1 snRNA, indicated by larger amounts of vpr mRNA species (Fig. 4B, e.g., Vpr3, cf. lanes 1 to 8 and 9 to 16) and the increased levels of exon 3-containing mRNA species (Fig. 4B, e.g., Tat3 [cf. lanes 1 to 8 and 9 to 16] or Nef4 [cf. lanes 1 to 4 and 9 to 12]). On the basis of these results, we concluded that the coexpressed U1 snRNA seemed to assemble correctly into mature snRNPs and that U1—as expected and anticipated by a recent publication (26)—increased 5′ss D3 recognition. Interestingly, the strong increase in exon 3 splice site activation appeared to suppress the inclusion of exon 2 in the viral mRNA species (Fig. 4B, Tat2, cf. lanes 1 and 9), indicating that a balanced exon 3 splicing activity is also necessary to permit the use of exon 2 splice sites A1 and D2. However, in the absence of functional ESEvpr, U1 snRNA coexpression predominantly activated vpr mRNA splicing, while only a minor influence on exon 3 inclusion was observed (Fig. 4B, Vpr and to a lesser extent Tat3, cf. lanes 3 to 4 and 11 to 12), indicating not only that ESEvpr may enhance the early recognition of 5′ss D3 but also supports its use later in the splicing reaction. It is worth noting that inactivation of both ESSV and ESEvpr allowed efficient vpr mRNA splicing and exon 3 inclusion upon the coexpression of modified U1 snRNA (Fig. 4B, Vpr3 and Tat3, lanes 8 and 16), rather arguing for a repressive activity of ESSV that addresses splicing after initial 5′ss recognition and that is counteracted by active ESEvpr. Western blot analysis of the p24 levels within the supernatant suggested that the coexpression of the U1 snRNA could induce excessive exon 3 splicing even in the presence of ESSV, thereby dramatically reducing virus particle production (Fig. 4B, cf. lanes 1 and 2 and lanes 9 and 10). However, binding of the coexpressed U1 snRNA relied on the presence of ESEvpr to elicit excessive activation of the exon 3 splice sites, causing severely reduced viral particle production (Fig. 4B, cf. lanes 9 and 12). When ESSV was disrupted, U1 coexpression efficiently inhibited viral particle production independent of ESEvpr activity (Fig. 4B, cf. lanes 5 to 8 and 13 to 16). These findings emphasize that the complementarity between the 5′ss D3 and the U1 snRNA is not the sole determinant of exon 3 inclusion. However, the absence of functional ESEvpr could be (at least partially) bypassed by increasing base pairing between U1 snRNA and 5′ss D3.
Fig 4.
Coexpression of a modified U1 snRNP with full complementarity to 5′ss D3 induces HIV-1 exon 3 splicing and vpr mRNA expression. (A) Schematic drawing of a 5′-end-modified U1 snRNA (right) perfectly matching the 5′ss D3 sequence. Mutated nucleotides are indicated by gray capital letters. Additional base pairing interactions between 5′ss D3 and the optimized 5′ end of the U1 snRNA are indicated by vertical gray lines. (B) HEK 293T cells (2.5 × 105) were transiently cotransfected with 1 μg of both a proviral plasmid and a U1 snRNA expression plasmid. Total RNA was isolated and subjected to RT-PCR analyses. PCR products were resolved by PAGE and stained with ethidium bromide. RT-PCR samples are shown at the top. The main viral mRNA species are indicated on the right. Viral supernatants were collected as well and analyzed for viral p24gag concentrations by immunoblotting (bottom). E, extended exon; dm, double mutation; sn, supernatant.
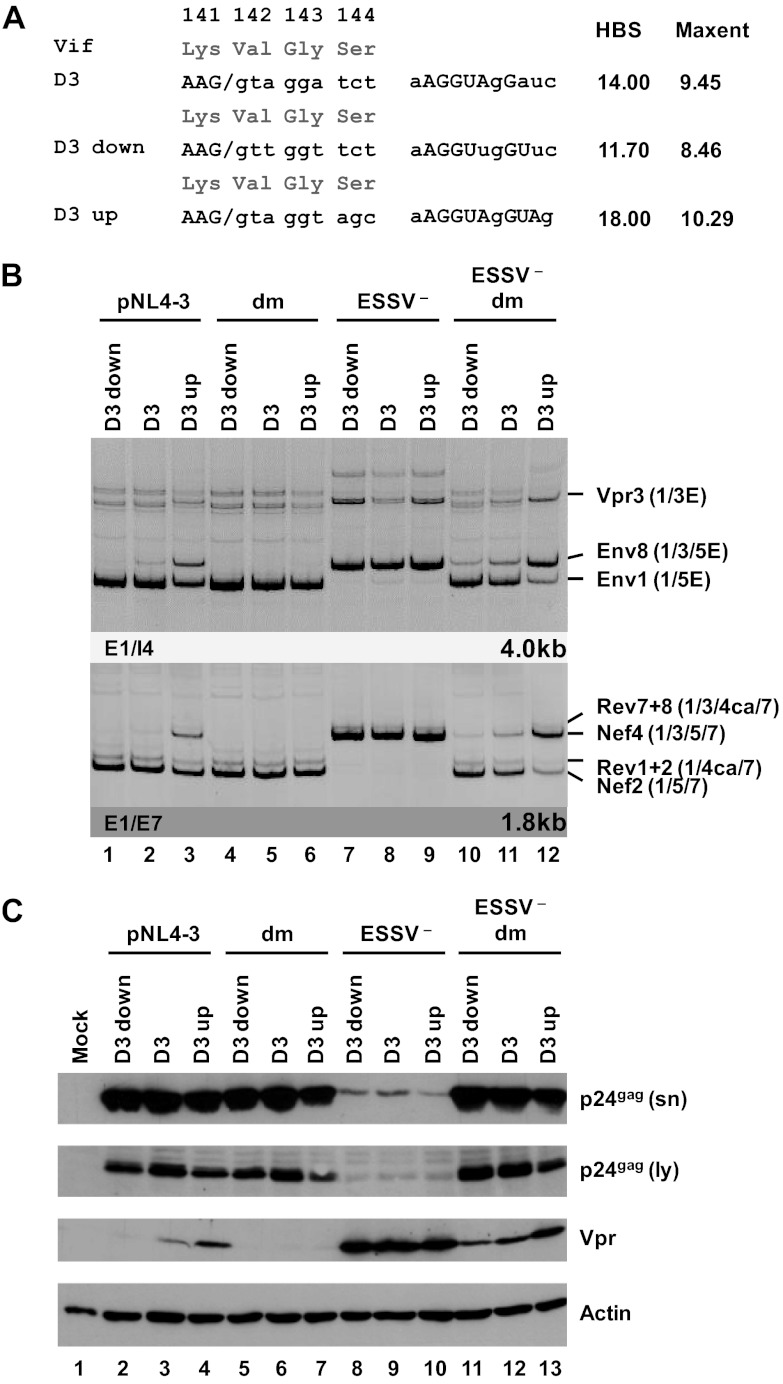
vpr mRNA expression can be modulated by up and down mutations of 5′ss D3.
The use of 3′ss A2 results in the formation of vpr-mRNA but only when splicing at the downstream 5′ss D3 is suppressed because this would remove the Vpr translational initiation codon within intron 3 from the mature transcripts. Remarkably, ESEvpr was shown to be critical for vpr mRNA expression, although it is located close to 5′ss D3 and separated from 3′ss A2 by the repressor ESSV. It was found previously that efficient recognition of a 5′ss by the U1 snRNP exerts positive feedback on the assembly of splicing factors at the upstream 3′ss—most likely via interactions across the exon (27–29). To analyze the interdependence of ESEvpr, 5′ss D3, and vpr mRNA expression, mutations predicted to either decrease (D3 down) or increase (D3 up) the intrinsic strength of the viral 5′ss D3 were tested in the context of a replication-competent provirus (Fig. 5A). Mutations were chosen so that the overlapping Vif ORF was not changed. Following transient transfection of HEK 293T cells with proviral DNA, the exon 3 abundance within the viral mRNA species was determined for each of the mutant proviruses by RT-PCR analysis (Fig. 5B). As expected, the extent of the complementarity between U1 snRNA and the 5′ss basically correlated with the amounts of exon 3 present in the viral transcripts. In the presence of ESEvpr activity, weakening of 5′ss D3 caused a decrease in the levels of exon 3-containing isoforms within both major viral mRNA classes, whereas an increase in the complementarity of 5′ss D3 partially overcame the general repression of exon 3 splicing by dominant negative ESSV (Fig. 5B, lanes 1 to 3). This was in line with the hypothesis that the stability of U1 snRNP binding to a 5′ss plays a pivotal role in the recognition of the entire exon. However, mutant forms of ESEvpr showed no detectable exon 3 inclusion, regardless of their intrinsic 5′ss strength (Fig. 5B, lanes 4 to 6), indicating a strict requirement for functional ESEvpr to enable stable binding of the U1 snRNP to 5′ss D3. Mutant ESSV was also associated with a poor response of the vpr3 mRNA to the distinct 5′ss variants in the experiment shown (Fig. 5B, lanes 7 to 9) and lacked any response in parallel experiments (data not shown). Efficient exon 3 inclusion was detected in each case, irrespective of up or down mutations within D3 (Fig. 5B, e.g., Nef4, lanes 7 to 9), suggesting that in the absence of ESSV activity, recognition of a weaker 5′ss can be compensated for by stronger activation of 3′ss A2. Finally, when both ESSV and ESEvpr were mutated, the intrinsic 5′ss strength again up- or downmodulated the frequency of exon 3 inclusion in the viral mRNAs (Fig. 5B, e.g., Nef4, lanes 10 to 12). This reinforces the notion that in a less favorable environment with regard to enhancer strength, the efficiency of exon inclusion exhibits a higher dependency on the ability of a splice site to bind the U1 snRNP on its own. Taken together, the data show that the overall efficiency of HIV-1 exon 3 splicing is adjusted by the individual strength of the preceding exonic splicing regulatory elements and the intrinsic strength of 5′ss D3. Western blot analyses were consistent with these results and revealed that Vpr expression was under the combined control of ESSV, ESEvpr, and 5′ss D3 (Fig. 5C). Correspondingly, increasing base pairing between the 5′ end of U1 snRNA and 5′ss D3 was accompanied by a higher abundance of Vpr protein within the transfected cells, whereas a reduction of the intrinsic strength showed the opposite effect on Vpr expression (Fig. 5C, lanes 2 to 4 and 11 to 13). However, in the presence of only one intact splicing regulatory element, either ESEvpr or ESSV, exon 3 splicing efficiency was either too low or too high to allow tuning by alterations of 5′ss strength (Fig. 5C, Vpr, lanes 5 to 7 and 8 to 10). These results substantiate the observations that U1 snRNP binding to 5′ss D3 enhances the use of upstream 3′ss A2 and that splicing of exon 3 can be considered the integrated outcome of exonic elements (ESEvpr and ESSV) and the intrinsic strength of 5′ss D3.
Fig 5.
5′ss D3 up and down mutations modulate HIV-1 exon 3 splicing and vpr mRNA formation. (A) Silent mutations predicted to decrease or increase the complementarity to the 5′ end of the endogenous U1 snRNA were introduced into viral 5′ss D3. Exonic nucleotides are denoted in uppercase letters, and intronic nucleotides are denoted in lowercase letters. Complementarity and predicted intrinsic strength by HBond score (HBS) and MaxEnt score algorithms are both shown next to the 5′ss sequence. Nucleotides complementary to the U1 snRNA are in capital letters, while mismatches to the U1 snRNA are in lowercase letters. (B) HEK 293T cells (2.5 × 105) were transiently transfected with 1 μg of each of the different infectious clones. RNA was isolated from the cells, DNase I digested, and reverse transcribed. The resultant cDNA served as the DNA template in semiquantitative PCRs using primer pairs E1/I4 and E1/E7 to specifically detect viral 4.0- and 1.8-kb viral mRNAs, respectively. Proviral mutants are shown above the panels. The main HIV-1 mRNA species are indicated at the right. (C) Protein lysates and viral supernatants were collected from HEK 293T cells transfected with 1 μg of pNL4-3 or mutant derivatives. Samples were loaded on 12% SDS-polyacrylamide gels and, after separation, transferred to nitrocellulose membranes. Viral proteins and α-actin (as a loading control) were determined by probing with specific primary antibodies. For detection, appropriate HRP-conjugated antibodies and ECL detection reagent were applied. HBS, HBond score; MaxEnt, MaxEnt score; dm, double mutation; E, extended exon; sn, supernatant; ly, lysate.
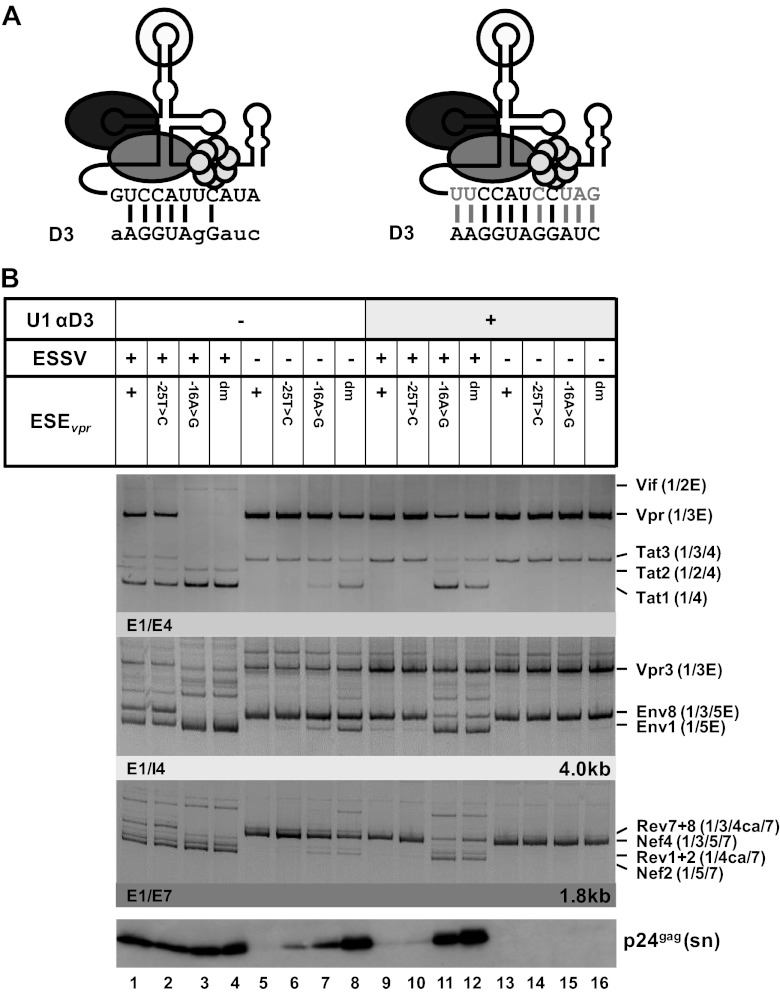
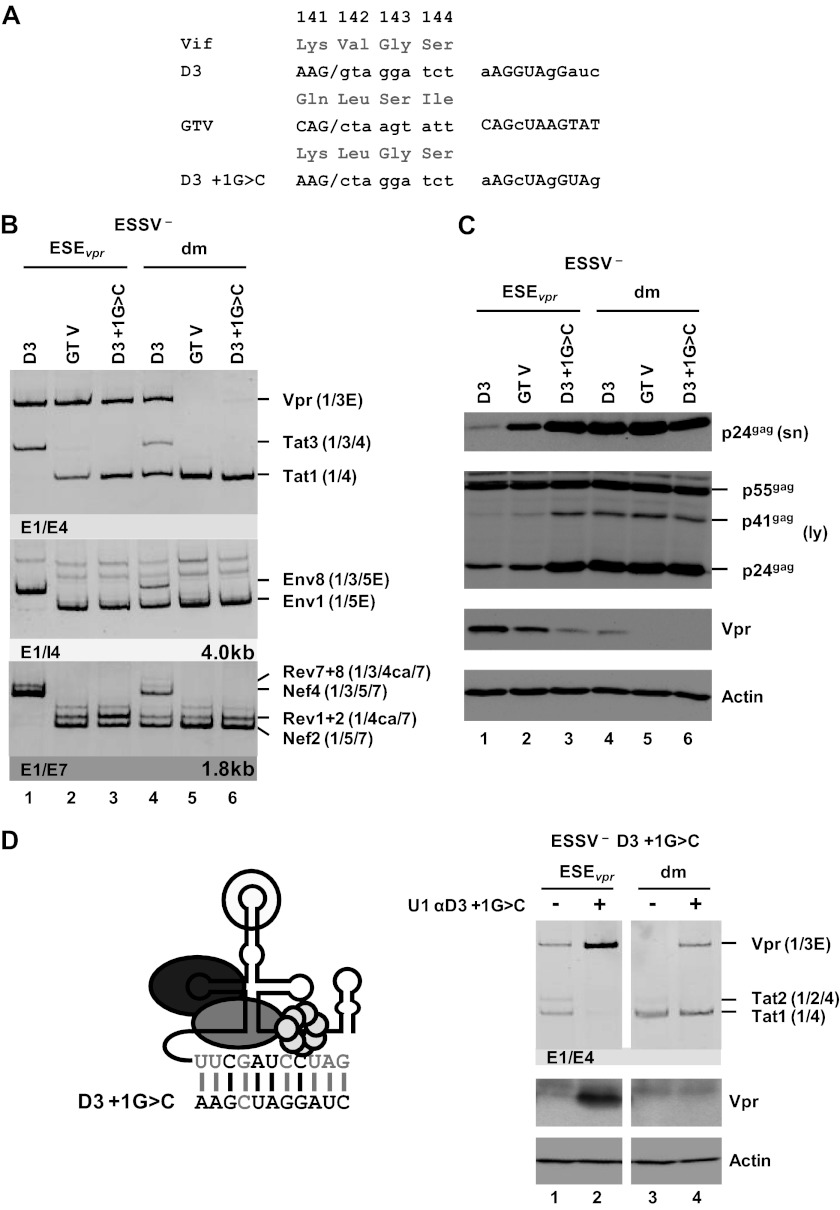
Binding of the U1 snRNP to a nonfunctional 5′ss is sufficient to augment splicing at upstream 3′ss A2.
The presented results suggest that U1 snRNP binding to the 5′ss fulfills two functions during pre-mRNA splicing; i.e., (i) it enhances the formation of exon definition complexes and therefore promotes recognition of the upstream 3′ss (27–29), and (ii) it commits the bound 5′ss to splice site pairing with a 3′ss across the downstream intron into the prespliceosome (30). It is hypothesized here that vpr mRNA splicing requires exon definition, so that 5′ss D3 is recognized by U1 snRNA but splicing at D3 must occur with lower efficiency. To gain a broader understanding of how vpr mRNA expression is regulated by U1 snRNP binding, 5′ss D3 was replaced with GTV (17), a U1 binding-competent but splicing-incompetent sequence with nonetheless nearly full complementarity to U1 snRNA (Fig. 6A). As a control, 5′ss D3 was also inactivated by a G-to-C mutation at position +1. The resulting set of variants ranged from a functional 5′ss, which supported efficient binding of the U1 snRNP, as well as splicing (D3), to sequences allowing either efficient binding (GTV) or neither efficient binding nor splicing (D3+1G>C). These variants were tested in the context of ESSV-negative or ESSV/ESEvpr double-negative proviruses. Splicing efficiency at 3′ss A2 was determined by semiquantitative RT-PCR analyses.
Fig 6.
U1 snRNP binding to a splicing-incompetent 5′ss enhances vpr mRNA expression. (A) 5′ss D3 was replaced with a splicing-incompetent sequence that perfectly matches the free 5′ end of the cellular U1 snRNA except for position +1 (GTV). As a control, 5′ss D3 was disabled for splicing by a G-to-C mutation at position +1, decreasing its complementarity to the U1 snRNA (D3+1G>C). Complementarity patterns are shown next to the 5′ss sequences. Matches to the U1 snRNA are indicated by uppercase letters, and residues not complementary are in lowercase letters. (B) HEK 293T cells (2.5 × 105) were transiently transfected with 1 μg of each of the proviral constructs and analyzed by semiquantitative RT-PCR. RT-PCR products were resolved by PAGE, followed by ethidium bromide staining. Mutants are depicted at the top. Main viral mRNAs are indicated on the right. (C) Cellular lysates and viral supernatants were obtained from transfected HEK 293T cells and loaded onto 12% SDS-polyacrylamide gels. After transfer to nitrocellulose membranes, viral proteins were determined with specific antibodies for p24gag and Vpr. To ensure the loading of equal protein amounts, the membrane was also probed with an antibody to cellular α-actin. (D) Schematic drawing of a 5′-end-modified U1 snRNA perfectly matching the 5′ss D3+1G>C sequence (left). Mutated nucleotides are indicated by gray capital letters. Additional base pairing interactions between 5′ss D3 and the optimized 5′ end of the U1 snRNA are indicated by vertical gray lines. HEK 293T cells (2.5 × 105) were transiently transfected with 1 μg of both proviral pNL4-3 DNA and U1 snRNA expression plasmid. Total RNA and cellular lysates were isolated and subjected to RT-PCR or Western blot analysis (right). E, extended exon; dm, double mutation; sn, supernatant; ly, lysate.
While 5′ss D3 was efficiently used in HEK 293T cells transfected with an ESSV mutant, neither GTV nor D3+1G>C was spliced and thus did not allow the accumulation of exon 3-containing mRNAs (Fig. 6B, Tat3, Env8, and Nef4, lanes 1 to 3). In the presence of ESEvpr, replacement of D3 with GTV or D3+1G>C monotonically increased the tat1-to-vpr mRNA expression ratio (Fig. 6B, Vpr and Tat1, lanes 1 to 3), which was consistent with greater complementarity between GTV and U1 snRNA than between GTV and D3+1G>C. Even in the absence of ESEvpr, wild-type D3 still retained some vpr mRNA expression and exon 3 inclusion, although at a higher level of tat1 mRNA expression (Fig. 6B, Vpr and Tat1, lane 4), while the disruption of both ESEvpr and wild-type D3 abolished all vpr mRNA expression (lanes 5 and 6). These results were in agreement with the hypothesis that U1 snRNP binding to splicing-incompetent U1 snRNA binding sites alone suffices to augment cross-exon interactions and splicing at 3′ss A2.
The change in mRNA expression ratios induced by these mutations was entirely consistent with the decreasing amount of Vpr protein detected by Western blot analysis (Fig. 6C, Vpr). Moreover, it was observed that decreasing vpr mRNA splicing by both reducing U1 snRNA complementarity at D3 and mutating ESEvpr could rescue virus particle production of ESSV-negative provirus (Fig. 6C, p24gag [sn/ly]).
The finding that ESEvpr-dependent binding of U1 snRNA promotes the use of 3′ss A2 was further confirmed by the coexpression of a U1 snRNA fully complementary to splicing-inactive 5′ss D3 (+1G>C) (Fig. 6D), which largely activated vpr mRNA splicing (Fig. 6D, top), as well as Vpr protein expression (Fig. 6D, bottom).
In summary, these findings recapitulate earlier studies showing that the two functions of U1 snRNP binding to the 5′ss can be dissected, supporting (i) the formation of exon definition complexes and (ii) the assembly of a prespliceosome across a downstream intron (27–29).
DISCUSSION
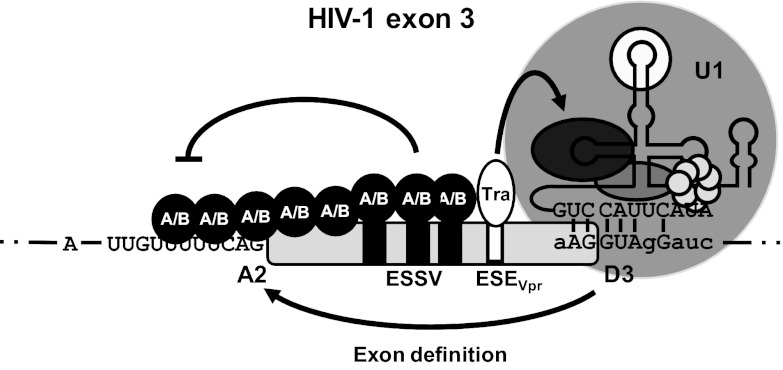
Splice site recognition is commonly found to be under the combined control of multiple nearby splicing regulatory elements that can either compete or cooperate to regulate splicing activation. It was previously shown that the HIV-1 noncoding leader exon 3 harbors a negative splicing regulatory element—termed ESSV—within its central portion that selectively represses upstream 3′ss A2 (12) and concomitantly inhibits exon 3 inclusion in the different viral mRNA species (13). ESSV disruption results in the strong accumulation of vpr mRNA and exon 3-containing isoforms, which leads to viral replication incompetence. However, ESSV does not act alone; using an in silico-based mutagenesis strategy, we identified an enhancer sequence—termed ESEvpr—upstream of 5′ss D3 that was essential for exon 3 splicing in the context of wild-type ESSV and that provided excess exon 3 splice site activation when ESSV was inactive. This notion was strengthened by the finding that ESSV/ESEvpr double-negative provirus retrieved the capability to efficiently replicate, probably because of reduced exon 3 splicing and a recovery of the ratio of unspliced to spliced viral mRNAs. Therefore, it was concluded that balanced exon 3 splicing is under the combined control of ESSV and ESEvpr (Fig. 7). In addition, we identified the splicing factors Tra2-alpha and Tra2-beta as proteins that bind to the ESEvpr sequence, thereby promoting the inclusion of exon 3 in the viral mRNAs.
Fig 7.
HIV-1 exon 3 splicing is under the combined control of ESSV and ESEvpr. Splice site recognition at HIV-1 exon 3 is regulated by ESSV and a novel exonic splicing enhancer (ESEvpr) embedded in the region upstream of 5′ss D3. ESSV is associated with hnRNP A/B proteins, which may multimerize along the 5′ end of exon 3, occluding 3′ss A2. Tra2-alpha and Tra2-beta bind to the ESEvpr sequence, potentially enhancing recruitment of the U1 snRNP to 5′ss D3, which in turn may promote interactions across the upstream exon and activation of 3′ss A2. U1, U1 snRNP.
Interestingly, we further demonstrated that ESEvpr is crucial for vpr mRNA expression. While 3′ss A2 needs to be activated, splicing at 5′ss D3 removes the vpr-coding intron located downstream and therefore needs to be repressed. However, whereas the use of 5′ss D3 in the splicing reaction excludes vpr mRNA expression, we showed that its early recognition by the U1 snRNP is required. This is in line with the exon definition hypothesis in which binding of the U1 snRNP to the 5′ss promotes interactions across the exon and thereby activation of the upstream 3′ss (27–29). By use of 5′ss D3 variations with either decreased or increased complementarity to the U1 snRNA, we demonstrated that the splicing enhancer promotes U1 snRNP recruitment and thereby exon definition. Accordingly, up and down mutations of 5′ss D3 increased or decreased vpr mRNA expression levels, underlining the importance of 5′ss D3 recognition for regulated levels of Vpr. These results recapitulated previous findings demonstrating that reduced recognition of 5′ss D2 leads to lower levels of Vif expression (29). Notably, vif mRNA formation requires the activation of upstream 3′ss A1, while the downstream intron needs to be retained, which is comparable to the mechanism providing vpr mRNA, as shown in the accompanying paper by Widera et al. (31). The use of splice sites D2 and D3 in the splicing reaction counteracts the expression of vif and vpr mRNAs, respectively. However, the aforementioned observations indicate that their initial recognition by the U1 snRNA is essential for obtaining Vif and Vpr expression. This idea was highlighted by the finding that ESEvpr-dependent recognition of splicing-incompetent U1 snRNP binding sites sufficed to enhance exon definition and activation of 3′ss A2 for vpr mRNA formation. Therefore, our results—in agreement with earlier findings (29)—indicate that recognition of 5′ss D2 and D3 must occur with higher efficiency than their use in the splicing reaction in order to permit the expression of Vif and Vpr. It was shown previously that exon definition complexes can readily progress into intron definition complexes, finally performing the splicing reaction across an upstream intron (32). However, it was also found that splicing regulatory elements can negatively control the conversion of exon-to-intron definition complexes (33). Therefore, a model seems plausible in which ESEvpr promotes the formation of an exon definition complex that then can be efficiently changed into an upstream intron definition complex but in which the same process may occur less efficiently across the downstream intron, ultimately allowing the formation of vpr mRNAs. Herein, splicing factors binding to the vpr intron might play a particular role through interference with the assembly of the spliceosome, as described for the polypyrimidine tract binding protein (33).
Alternatively, it has been suggested that high-mobility group A protein 1a renders the U1 snRNP inactive at 5′ss D3 for progression with the splicing reaction, thereby mediating vpr mRNA expression (34). Although disabled from carrying out the splicing reaction, this “dead-end” U1 snRNP should maintain its capability to enhance the assembly of exon definition complexes and thus activation of 3′ss A2. However, since we showed here that ESEvpr-mediated early recognition of 5′ss D3 is critical for Vpr expression, interference with the removal of the downstream vpr intron appears to occur at a later step during spliceosome assembly. Whether this is achieved by either one of the alternative suggested models needs to be further clarified.
ACKNOWLEDGMENTS
We thank Joshua Madsen and Martin C. Stoltzfus for critical readings of the manuscript and helpful comments. We are grateful to Alain Cochrane and Kinji Ohno for providing expression plasmids for Tra2-alpha, -beta, and CUGBP1. We thank Björn Wefers for excellent technical assistance.
The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1HXB2 Vif antiserum from Dana Gabuzda and HIV-1NL4-3 Vpr antiserum (1-46) from Jeffrey Kopp. This work was supported by the DFG (SCHA 909/3-1); the Heinz Ansmann Foundation for AIDS Research, Düsseldorf (H.S.); and the Jürgen Manchot Stiftung (H.S).
Footnotes
Published ahead of print 19 December 2012
REFERENCES
- 1. Frankel AD, Young JA. 1998. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 67:1–25 [DOI] [PubMed] [Google Scholar]
- 2. Kozak M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson JL, Johnson AT, Howard JL, Purcell DF. 2007. Both linear and discontinuous ribosome scanning are used for translation initiation from bicistronic human immunodeficiency virus type 1 env mRNAs. J. Virol. 81:4664–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krummheuer J, Johnson AT, Hauber I, Kammler S, Anderson JL, Hauber J, Purcell DF, Schaal H. 2007. A minimal uORF within the HIV-1 vpu leader allows efficient translation initiation at the downstream env AUG. Virology 363:261–271 [DOI] [PubMed] [Google Scholar]
- 5. Purcell DF, Martin MA. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stoltzfus CM. 2009. Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication. Adv. Virus Res. 74:1–40 [DOI] [PubMed] [Google Scholar]
- 7. Kim SY, Byrn R, Groopman J, Baltimore D. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klotman ME, Kim S, Buchbinder A, DeRossi A, Baltimore D, Wong-Staal F. 1991. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc. Natl. Acad. Sci. U. S. A. 88:5011–5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cullen BR. 1991. Regulation of human immunodeficiency virus replication. Annu. Rev. Microbiol. 45:219–250 [DOI] [PubMed] [Google Scholar]
- 10. Hoffmann D, Schwarck D, Banning C, Brenner M, Mariyanna L, Krepstakies M, Schindler M, Millar DP, Hauber J. 2012. Formation of trans-activation competent HIV-1 Rev:RRE complexes requires the recruitment of multiple protein activation domains. PLoS One 7:e38305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bilodeau PS, Domsic JK, Mayeda A, Krainer AR, Stoltzfus CM. 2001. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol. 75:8487–8497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Domsic JK, Wang Y, Mayeda A, Krainer AR, Stoltzfus CM. 2003. Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol. Cell. Biol. 23:8762–8772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Madsen JM, Stoltzfus CM. 2005. An exonic splicing silencer downstream of the 3′ splice site A2 is required for efficient human immunodeficiency virus type 1 replication. J. Virol. 79:10478–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kammler S, Otte M, Hauber I, Kjems J, Hauber J, Schaal H. 2006. The strength of the HIV-1 3′ splice sites affects Rev function. Retrovirology 3:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tange TO, Damgaard CK, Guth S, Valcarcel J, Kjems J. 2001. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 20:5748–5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Caputi M, Freund M, Kammler S, Asang C, Schaal H. 2004. A bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu, and nef gene expression. J. Virol. 78:6517–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kammler S, Leurs C, Freund M, Krummheuer J, Seidel K, Tange TO, Lund MK, Kjems J, Scheid A, Schaal H. 2001. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA 7:421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goncalves J, Jallepalli P, Gabuzda DH. 1994. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J. Virol. 68:704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Otte M. 2006. Identifizierung von cis-wirkenden Sequenzen in den alternativen HIV-1 Leaderexons und ihre funktionelle Bedeutung für die Spleißregulation. Ph.D. dissertation Heinrich-Heine-Universität, Düsseldorf, Germany [Google Scholar]
- 21. Fairbrother WG, Yeh RF, Sharp PA, Burge CB. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297:1007–1013 [DOI] [PubMed] [Google Scholar]
- 22. Daoud R, Mies G, Smialowska A, Olah L, Hossmann KA, Stamm S. 2002. Ischemia induces a translocation of the splicing factor tra2-beta 1 and changes alternative splicing patterns in the brain. J. Neurosci. 22:5889–5899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F, Rosenberg C, Hulette C, Jellinger K, Hampel H, Riederer P, Moller HJ, Andreadis A, Henkel K, Stamm S. 2006. The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and Tra2-beta1 changes in sporadic Alzheimer's disease. J. Neurochem. 96:635–644 [DOI] [PubMed] [Google Scholar]
- 24. Sciabica KS, Hertel KJ. 2006. The splicing regulators Tra and Tra2 are unusually potent activators of pre-mRNA splicing. Nucleic Acids Res. 34:6612–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Watermann DO, Tang Y, Zur Hausen A, Jager M, Stamm S, Stickeler E. 2006. Splicing factor Tra2-beta1 is specifically induced in breast cancer and regulates alternative splicing of the CD44 gene. Cancer Res. 66:4774–4780 [DOI] [PubMed] [Google Scholar]
- 26. Mandal D, Feng Z, Stoltzfus CM. 2010. Excessive RNA splicing and inhibition of HIV-1 replication induced by modified U1 small nuclear RNAs. J. Virol. 84:12790–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Asang C, Hauber I, Schaal H. 2008. Insights into the selective activation of alternatively used splice acceptors by the human immunodeficiency virus type-1 bidirectional splicing enhancer. Nucleic Acids Res. 36:1450–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoffman BE, Grabowski PJ. 1992. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 6:2554–2568 [DOI] [PubMed] [Google Scholar]
- 29. Mandal D, Exline CM, Feng Z, Stoltzfus CM. 2009. Regulation of Vif mRNA splicing by human immunodeficiency virus type 1 requires 5′ splice site D2 and an exonic splicing enhancer to counteract cellular restriction factor APOBEC3G. J. Virol. 83:6067–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lim SR, Hertel KJ. 2004. Commitment to splice site pairing coincides with A complex formation. Mol. Cell 15:477–483 [DOI] [PubMed] [Google Scholar]
- 31. Widera M, Erkelenz S, Hillebrand F, Krikoni A, Widera D, Kaisers W, Deenen R, Gombert M, Dellen R, Pfeiffer T, Kaltschmidt B, Münk C, Bosch V, Köhrer K, Schaal H. 2013. An intronic G run within HIV-1 intron 2 is critical for splicing regulation of vif mRNA. J. Virol. 87:2707–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schneider M, Will CL, Anokhina M, Tazi J, Urlaub H, Luhrmann R. 2010. Exon definition complexes contain the tri-snRNP and can be directly converted into B-like precatalytic splicing complexes. Mol. Cell 38:223–235 [DOI] [PubMed] [Google Scholar]
- 33. Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. 2008. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 15:183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsuruno C, Ohe K, Kuramitsu M, Kohma T, Takahama Y, Hamaguchi Y, Hamaguchi I, Okuma K. 2011. HMGA1a is involved in specific splice site regulation of human immunodeficiency virus type 1. Biochem. Biophys. Res. Commun. 406:512–517 [DOI] [PubMed] [Google Scholar]
- 35. Asang C, Erkelenz S, Schaal H. 2012. The HIV-1 major splice donor D1 is activated by splicing enhancer elements within the leader region and the p17-inhibitory sequence. Virology 432:133–145 [DOI] [PubMed] [Google Scholar]
- 36. Exline CM, Feng Z, Stoltzfus CM. 2008. Negative and positive mRNA splicing elements act competitively to regulate human immunodeficiency virus type 1 vif gene expression. J. Virol. 82:3921–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jacquenet S, Mereau A, Bilodeau PS, Damier L, Stoltzfus CM, Branlant C. 2001. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J. Biol. Chem. 276:40464–40475 [DOI] [PubMed] [Google Scholar]
- 38. Hallay H, Locker N, Ayadi L, Ropers D, Guittet E, Branlant C. 2006. Biochemical and NMR study on the competition between proteins SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 at the HIV-1 Tat exon 2 splicing site. J. Biol. Chem. 281:37159–37174 [DOI] [PubMed] [Google Scholar]
- 39. Zahler AM, Damgaard CK, Kjems J, Caputi M. 2004. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 279:10077–10084 [DOI] [PubMed] [Google Scholar]
- 40. Amendt BA, Hesslein D, Chang LJ, Stoltzfus CM. 1994. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 14:3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caputi M, Mayeda A, Krainer AR, Zahler AM. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Si Z, Amendt BA, Stoltzfus CM. 1997. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 25:861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Staffa A, Cochrane A. 1995. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 15:4597–4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Amendt BA, Si ZH, Stoltzfus CM. 1995. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 15:4606–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Si ZH, Rauch D, Stoltzfus CM. 1998. The exon splicing silencer in human immunodeficiency virus type 1 Tat exon 3 is bipartite and acts early in spliceosome assembly. Mol. Cell. Biol. 18:5404–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stoltzfus CM, Madsen JM. 2006. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr. HIV Res. 4:43–55 [DOI] [PubMed] [Google Scholar]
- 47. Schaal H, Pfeiffer P, Klein M, Gehrmann P, Scheid A. 1993. Use of DNA end joining activity of a Xenopus laevis egg extract for construction of deletions and expression vectors for HIV-1 Tat and Rev proteins. Gene 124:275–280 [DOI] [PubMed] [Google Scholar]