Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) latency associated-nuclear antigen 1 (LANA1) protein is constitutively expressed in all KSHV-infected cells, as well as in all forms of KSHV-associated malignancies. LANA1 is a multifunctional KSHV oncoprotein containing multiple repeat sequences that is important for viral episome maintenance and the regulation of cellular and viral gene expression. We characterize here multiple LANA1 isoforms and show that ∼50% of LANA1 is naturally generated as N-terminally truncated shoulder proteins that are detected on SDS-PAGE as faster-migrating shoulder bands designated LANA1S. Higher-molecular-weight LANA1S isoforms initiate downstream at noncanonical sites within the N-terminal region, whereas lower-molecular-weight LANA1S isoforms initiate downstream within the central repeat 1 domain. LANA1S proteins lack an N-terminal nuclear localization signal motif, and some isoforms differ from full-length, canonical LANA1 by localizing to perinuclear and cytoplasmic sites. Although LANA1 has until now been assumed to be solely active in the nucleus, this finding indicates that this major KSHV oncoprotein may have cytoplasmic activities as well. KSHV overcomes its limited genetic coding capacity by generating alternatively initiated protein isoforms that may have distinct biological functions.
INTRODUCTION
The Kaposi's sarcoma-associated herpesvirus (KSHV) (1) latency-associated nuclear antigen 1 (LANA1) is the primary viral genome maintenance protein (2, 3). In addition to its role in latent viral genome replication and segregation, it targets multiple cellular tumor suppressor genes, such as p53 and retinoblastoma (Rb) (4–6), and ribosomal subunit, chromatin remodeling, and transcriptional factor proteins (7–9). LANA1 was first discovered as a nuclear protein through adsorption studies using KSHV-negative cells and Kaposi's sarcoma patient sera (10). Patient antibodies to LANA1 were found to be specific for KSHV infection, and this was confirmed and extended in studies performed by Gao et al. (11) and Kedes et al. (12) to define the natural history of KSHV infection and KS.
LANA1 is a highly zwitterionic protein with basic N-terminal and C-terminal regions, separated by an acidic long central repeat (CR) region (13–16). The functions of LANA1 CR are unclear but appear to be critical for viral replication and genome maintenance (17). For KSHV strain BC1 (U75698) (18), the ORF73 gene product encoding LANA1 (Fig. 1A, LANA1 domains, antibody binding sites) is 1,162 amino acids (aa) long and can be divided into an N-terminal proline-, serine-rich domain (1 to 320 aa), an acidic central region (321 to 937 aa) subdivided into CR1, CR2, and CR3 subdomains, each having unique motifs of repeating glutamine, glutamic acid, asparagine amino acids (19), and a C-terminal domain (938 to 1,162 aa) that is rich in charged and hydrophobic amino acids (18, 20). Variations in LANA1 migration speeds on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) between KSHV strains are attributed to the loss or gain of repeat units from the highly repetitive central domain (21).
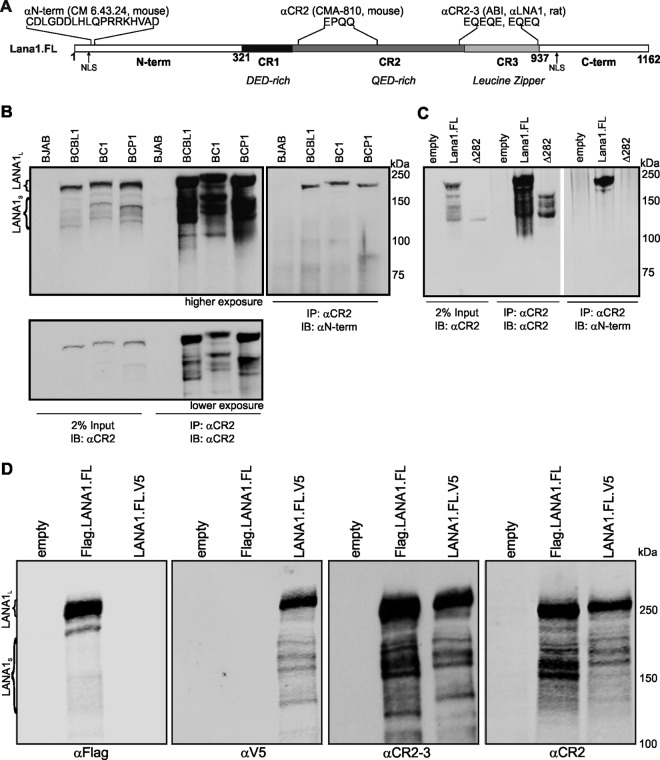
Fig 1.
KSHV LANA1 possesses N-terminally truncated protein isoforms. (A) Map of LANA1 domains and antibody binding sites. Full-length LANA1 (LANA1.FL) comprises N-terminal (N-term), C-terminal (C-term), and central repeat (CR1, CR2, and CR3) domains. αN-term antibody (CM 6.43.24) binding site (CDLGDDLHLQPKRRKHVAD) is next to the N-term NLS. αCR2 (CMA-810) recognizes the repetitive epitope EPQQ in CR2. αCR2-3 (ABI, αLNA1) recognizes repetitive EQEQ and EQEQE epitope found in CR2 and CR3 domains. (B) Coimmunoprecipitation/immunoblotting of LANA1 with antibodies to N-terminal and to CR2 epitopes reveals that LANA1 shoulders (LANA1S) are not detected with N-term antibody. Endogenously expressed LANA1 from BCBL1, BC1, and BCP1 cells was pulled down by CR2 antibody. Immunoprecipitated proteins and 2% of the total cell lysate served as an input control separated on an SDS–8% polyacrylamide gel and were immunoblotted either with αN-term or αCR2 antibody (given as higher and lower exposures). KSHV-negative BJAB cells were used as a control. Full-length LANA1 migrates as doublets at around 230 kDa (LANA1L) and the shoulder bands of LANA1 (LANA1S) at around 150 to 180 kDa. (C) Deletion of the N terminus eliminates LANA1 reactivity to N-term but not to CR2 and CR2-3 antibodies. Full-length LANA1 (LANA1.FL), an N-terminal truncation mutant lacking the first 281 aa (Δ282), and a control vector (empty) were overexpressed in HEK293 cells. The proteins were pulled down by CR2 antibody and analyzed by immunoblotting with αN-term or αCR2 antibody. (D) Epitope tagging confirms the absence of N-terminal peptide sequences for LANA1S isoforms. N-terminal FLAG-tagged (Flag.LANA1.FL) and C-terminal V5-tagged (LANA1.FL.V5) LANA1.FL expressing HEK293 cell lysates were immunoblotted with αFLAG, αV5, αCR2, and αCR2-3 antibodies.
The BC1 strain LANA1 is predicted to be an ∼135-kDa protein (theoretical pI/molecular weight, 3.83/135,213.27 [ExPASY]) (22), but it migrates aberrantly as a doublet at 226 to 234 kDa on SDS-PAGE (15, 23, 24). The doublet pattern results from a weak C-terminal noncanonical polyadenylation site that truncates approximately half of the LANA1 proteins, which are unable to bind the viral genome (25). Whole primary effusion lymphoma (PEL) cell immunoblots using patient sera (11) or monoclonal antibodies to LANA1 (26) revealed multiple shoulder bands at apparent molecular masses of ca. 150 to 180 kDa on PAGE. Several authors have assumed these bands represent degradation products of the major 226- to 234-kDa isoforms (27, 28), but this has not been demonstrated.
Nuclear localization is required for LANA1's functions in tethering the viral episome to chromatin during mitosis and for initiating latent plasmid replication. LANA1 nuclear localization signals (NLS) are encoded in both N-terminal (24 to 30 aa) and C-terminal regions (27, 29–31) The N-terminal NLS motif appears to be part of a bipartite NLS that can bind to multiple import receptors and thus might increase the ability of LANA1 to interact with different nuclear components (31). LANA1 residues 996 to 1139 in the C terminus bind to viral terminal repeat sequences and mediate episome replication (32) in concert with the chromatin-binding domain of LANA1 located at the N terminus at the initial LANA1 22 aa (28). Further, LANA1 N terminus binds to nucleosomes through histone H2A and H2B facilitating viral DNA replication (33).
Although LANA1 mRNA is expressed at low levels, LANA1 protein is abundantly detected in naturally infected human PEL-derived cells due to its long half-life (19). In the present study, we examine alternative noncanonical protein isoforms of KSHV LANA1. Translation initiation in eukaryotes usually follows the ribosomal scanning model, wherein the ribosomal subunit binds at the 5′ end of an mRNA and scans in the 3′ direction until it encounters an initiation AUG (methionine) codon in the context of the Kozak sequence, GCCRCCaugG (34). Despite the importance of the sequence surrounding the initiation codon, other factors may also determine the site of translation initiation. Exceptions occur in cases of leaky scanning (35, 36). In such cases, inefficient recognition of an AUG or recognition of non-AUG initiator codons (CUG, GUG, UUG, ACG, AUU, and AUA) results in continued scanning of a portion of the 43S preinitiation complexes and initiating at a downstream methionine, generally having an appropriate Kozak context (37).
Evidence is increasing that noncanonical translation initiation is more common in biological systems than previously thought. A recently developed global translation initiation sequencing method (GTI-seq) having single-base-pair resolution revealed an unexpected abundance of alternative translation initiation sites (TIS), including non-AUG sites, in human cells (38). According to that study, up to 27% of total TIS were located downstream of annotated TIS and 53% of these downstream TIS (dTIS) used non-AUG codons including CUG, AUG variants, and others. In addition to leaky scanning, reinitiation can occur on bicistronic or polycistronic mRNAs, where ribosomes resume scanning after the termination of a short upstream open reading frame (ORF) (39). Internal ribosome entry sites (IRES) bypass cap-dependent scanning at the 5′ end of the mRNA and recruit the initiation complex to internal start codons (40). In ribosome shunting (discontinuous scanning), cap-dependent ribosome scanning bypasses and initiates translation downstream of a region with inhibitory elements such as short ORFs or stem-loop structures (41, 42).
Alternative translation initiation was first defined for Sendai virus and Moloney murine leukemia virus (43, 44). KSHV K12 protein (kaposin) (45) has been shown to initiate translation from non-AUG sites. Translation of viral mRNAs, including the P gene of hepatitis B virus and the small Rep40-like protein encoded by adeno-associated virus type 5 using ribosomal scanning or internal initiation, has also been reported (46, 47).
We show here that LANA1 shoulder proteins are multiple noncanonical LANA1 translation products, called LANA1S. The highest-molecular-weight shoulder forms are generated by non-AUG-dTIS within the N terminus, while most of the lower-molecular-weight forms are initiated from TIS within the central repeat domain 1. These alternative translation products generate N-terminally truncated proteins that disrupt the N-terminal NLS. Some—but not all—shoulder bands are retained in the cytoplasm and partially colocalize with ribosomal and perinuclear mitochondrial structures. Viruses such as KSHV can in part overcome their limited genomic coding capacity during latency by generating alternatively translated protein isoforms. We demonstrate that LANA1, previously thought to be exclusively a nuclear protein, has the potential to interact with and dysregulate cellular cytoplasmic signaling proteins.
MATERIALS AND METHODS
Cell lines and antibodies.
HEK293 human embryonic kidney cells (American Type Culture Collection [ATCC]) and U2OS human osteosarcoma cells (kindly provided by Ole Gjoerup) were grown in Dulbecco modified Eagle medium (DMEM) with l-glutamine and 4.5 g of glucose/liter (Cellgro) supplemented with 10% fetal bovine serum (FBS; Sigma). BJAB, BCBL1, and BC1 cells (ATCC) were maintained in RPMI 1640 with l-glutamine (Cellgro) supplemented with 10% FBS. BCP1 cells were grown in RPMI 1640 medium with l-glutamine supplemented with 4.5 g of glucose/liter, 10 mM HEPES, 1 mM sodium pyruvate, and 20% FBS. We generated the mouse monoclonal antibodies anti-LANA1 (CMA-810, αCR2, EPQQ, 1:1,000) (48) and anti-LANA1 N-terminal (αN-term, CDLGDDLHLQPKRRKHVAD, 1:2,000). Rabbit monoclonal antibody for the ORF50 gene product was kindly provided by David Lukac (αORF50, 1:3,000). Rat anti-LNA1 antibody (αCR2-3) was purchased from Advanced Biotechnologies, Inc. The mouse V5 antibody (Invitrogen) and mouse Flag-M2 antibody (Sigma) were used for C-terminally and N-terminally tagged LANA1 detection, respectively. In subcellular fractionation experiments, α-tubulin (Sigma), LAMP1 (H5G11; Santa Cruz), Orc2 (BD Biosciences), lamin B1 (Invitrogen), and calreticulin (Abcam) antibodies were used to evaluate the subcellular fractions. Ribosomal protein S6 and the phospho-Rb antibodies pRb Ser780/795/807/811 were purchased from Cell Signaling. For confocal microscopy, we used mitochondrion-selective probes (MitoTracker; Invitrogen) and DRAQ5 (Cell Signaling) for nuclear staining. Antibodies were diluted according to the manufacturer's instructions.
Plasmids.
LANA1 constructs were generated by PCR with a BC1 strain based LANA1 plasmid DNA used as a template (18) and subcloned into pcDNA3.1/Zeo+ (Invitrogen) using the primers listed (Table 1). PCR products were digested with EcoRI/PflMI and ligated into LANA1 plasmid (LANA1.FL) used as a template. Δ161 comprises the amino acid sequence between aa 161 and 1162, and Δ282 comprises the amino acid sequence between aa 282 and 1162 starting with the third and fourth methionines, respectively. The 30.STOP and 159.STOP constructs have a STOP codon at amino acid positions 30 and 159, respectively, and were generated by overlapping PCR using primers (Table 1). The four in-frame methionines at amino acid positions 1, 6, 161, and 282 in the N-terminal domain were substituted with alanines by overlapping PCR using the indicated primer pairs (see Table 1). All of the plasmid sequences generated were confirmed by DNA sequencing. To generate pLANA1.d1EGFP, full-length LANA1 (from plasmid LANA1.FL) was digested with EcoRI/HindIII and subcloned into pd1EGFP-N1 vector (kindly provided by Don Ganem), which contains the PEST amino acid sequence (49) and thus targets fused proteins for degradation (19). The FLAG.LANA1.FL and LANA1.FL.V5 fusion constructs were generated by subcloning full-length LANA1 into pCMV.Tag2B (Flag; Stratagene) and pCDNA6.V5.HisB (V5; Invitrogen) via EcoRI/XhoI and NheI/HindIII restriction sites, respectively. Both constructs were sequenced after cloning. The construction of LANA1 enhanced green fluorescent protein (EGFP) and SIINFEKL fusion constructs was described previously (50). Central repeat deletion mutants of LANA1 ΔCR1, ΔCR2, and ΔCR3 were derived from EGFP-SIINFEKL fusion constructs (50). Briefly, the plasmid DNAs were linearized with HindIII, blunted with mung bean exonuclease (New England BioLabs), and subsequently digested with EcoRI restriction digestion enzyme. The released fragments were subcloned into parent vector pcDNA6.V5.HisB via EcoRI and EcoRV restriction sites. Enzymes were used as recommended by the manufacturer. All constructs were confirmed by DNA sequencing.
Table 1.
Primer sequences used for the generation of various LANA1 expression constructs
| Name | Orientationa | Sequence (5′–3′) |
|---|---|---|
| Δ161 | S | CAGAGAATTCAACAACCATGCGTCCGCCAC |
| AS | GCGGTATTCGCGAGATGG | |
| Δ282 | S | TGGGGGAATTCCACAACCATGTTGGTGCTC |
| AS | GCGGTATTCGCGAGATGG | |
| 30.STOP | S1 | CCAGAATTCCCGAGGATGGCGCCC |
| AS1 | GTCACATCTTTCTTAAGACCTGTTTCG | |
| S2 | CGAAACAGGTCTTAAGAAAGATGTGAC | |
| AS2 | GCGGTATTCGCGAGATGG | |
| 159.STOP | S1 | CCAGAATTCCCGAGGATGGCGCCC |
| AS1 | CGGACGCATAGGTTATGAAGAGTC | |
| S2 | GACTCTTCATAACCTATGCGTCCG | |
| AS2 | GCGGTATTCGCGAGATGG | |
| M1A | S1 | GGCAGTACATCTACGTATTAGTC |
| AS1 | TCCCGGGGGCGCCGCCCTCGGGAATTC | |
| S2 | GAATTCCCGAGGGCGGCGCCCCCGGGA | |
| AS2 | GCGGTATTCGCGAGATGG | |
| M6A | S1 | GGCAGTACATCTACGTATTAGTC |
| AS1 | CGACCTCAGGCGCGCTCCCGGGGGCGC | |
| S2 | GCGCCCCCGGGAGCGCGCCTGAGGTCG | |
| AS2 | GCGGTATTCGCGAGATGG | |
| M1A6A | S1 | GGCAGTACATCTACGTATTAGTC |
| AS1 | GCGCTCCCGGGGGCGCCGCCCTCGG | |
| S2 | CCGAGGGCGGCGCCCCCGGGAGCGCGC | |
| AS2 | GCGGTATTCGCGAGATGG | |
| M161A | S1 | CCAGAATTCCCGAGGATGGCGCCC |
| AS1 | GGGTGGCGGACGCGCAGGTGTTGAAGA | |
| S2 | TCTTCAACACCTGCGCGTCCGCCACCC | |
| AS2 | GCGGTATTCGCGAGATGG | |
| M282A | S1 | CCAGAATTCCCGAGGATGGCGCCC |
| AS1 | AAGGAGCACCAACGCGGCTGTGTCATC | |
| S2 | GATGACACAGCCGCGTTGGTGCTCCTT | |
| AS2 | GCGGTATTCGCGAGATGG |
S, sense; AS, antisense.
Western blotting and immunoprecipitation analysis.
The plasmid DNAs were prepared by using the Qiagen midi-prep procedure. HEK293 and U2OS cells were transfected with Lipofectamine 2000 (Invitrogen) and X-tremeGENE HP transfection reagent (Roche) according to the manufacturers' recommendations. Cells were harvested at 48 h posttransfection, rinsed once in phosphate-buffered saline (PBS), and lysed in radioimmunoprecipitation assay (RIPA) buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% NP-40 and also supplemented with protease inhibitors (Roche) for 30 min on ice. After centrifugation at 17,000 × g for 15 min, the protein concentrations were measured using DC protein assay kit reagents (Bio-Rad). Cell extracts were resolved on 4 to 20% Criterion TGX precast gels (Bio-Rad), NuPage 4 to 12% Bis-Tris gradient gels (Invitrogen), or self-cast 6 to 8% gels and then transferred to a nitrocellulose membrane (Hybond C Extra; Amersham). Membranes were incubated 1 h at room temperature in blocking buffer (5% nonfat dry milk, 1× Tris-buffered saline [TBS]) and immunoblotted with the respective antibodies in 5% bovine serum albumin–1× TBS–0.05% Tween 20 for 2 h at room temperature or overnight at 4°C. Goat anti-rat (Sigma), anti-mouse (Amersham), and anti-rabbit (Cell Signaling) horseradish peroxidase-conjugated secondary antibodies were diluted at 1:6,000 in blocking buffer. Proteins were detected by enhanced chemiluminescence (Perkin-Elmer). For infrared Western blot imaging, LI-COR 680 and LI-COR 800 fluorescently labeled secondary antibodies were diluted at 1:10,000, and the membranes were analyzed with an Odyssey imaging system (LI-COR Biosciences). For immunoprecipitations, the cells were lysed in 500 μl of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl 1% Triton X-100, 2 mM NaF, 1 mM NaVO3, protease inhibitor cocktail [Roche]) and precleared with 40 μl of 50% A/G Plus-agarose beads (Santa Cruz) with rotation for 1 h at 4°C. Precleared cell extracts were incubated at 4°C overnight with 3 μg of αCR2 (CMA-810) and coupled with 40 μl of 50% A/G Plus-agarose beads (for 2 h at 4°C). Immune complexes were washed in lysis buffer for 15 min, once with 0.5 M LiCl in 50 mM Tris-HCl for 10 min, and once again with lysis buffer. Subsequently, beads were collected after centrifugation for 1 min at 3,500 × g. Immunoprecipitated proteins were released from beads by boiling in 30 μl of 2× Laemmli buffer. Samples were resolved by SDS-PAGE and immunoblotted.
Protease inhibitor MG132, TPA, and lambda phosphatase treatment.
BJAB, BCBL1, BC1, and transfected HEK293 cells were either treated with proteasome inhibitor MG132 (20 μM, 12 h) or with dimethyl sulfoxide. In order to induce lytic replication in PEL cells, BJAB, BCBL1, and BC1 cells were incubated in medium containing 20 ng of 12-O-tetradecanoyl phorbol 13-acetate (TPA)/ml for 48 h. Cells were lysed in RIPA buffer, and equal amounts of protein were separated on a 4 to 12% gradient gel as described above. Transfected HEK293 cells and KSHV-negative BJAB cells and KSHV-positive BCBL1 cells were harvested, lysed, and briefly sonicated in NP-40 lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.6% SDS, 0.5% sodium deoxycholate, 1% NP-40, supplemented with protease inhibitors) for 30 min on ice. Equal amounts of protein lysate was then incubated with or without lambda phosphatase supplemented with 1× PMP buffer and 1× MnCl2 at 37°C for 1 h (New England BioLabs).
Nuclear and cytoplasmic fractionation.
The cells were harvested and washed in PBS and lysed in 250 μl of lysis buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM NaVO4, and 10 μg of aprotinin and leupeptin/ml) for 10 min on ice. The supernatant was removed (cytoplasmic fraction), and the pellet was resuspended in 150 μl of nuclei extraction buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 25% glycerol, 0.5 mM PMSF, 1 mM NaVO4, and 10 μg of aprotinin and leupeptin/ml). The nuclear fraction was sonicated briefly and centrifuged at full speed for 10 min at 4°C, and the supernatant was collected (nuclear fraction). The protein concentrations were estimated using DC protein assay reagents (Bio-Rad). Analysis of the LANA1 distribution in PEL-derived cells was performed using a subcellular protein fraction kit for cultured cells (Thermo Scientific) according to the manufacturer's instructions. Briefly, different lysis buffers were used to extract soluble cytoplasmic (CE), plasma, mitochondria, and endoplasmic reticulum (ER)/Golgi membranes (ME), the soluble nuclear extract (NE), and chromatin-bound nuclear proteins (CB). The protein concentrations were determined as described above, and adjusted lysates were run on gradient gels.
Immunofluorescence and confocal microscopy.
For immunofluorescence studies, U2OS cells were seeded at a confluence of 105 cells per well in a 24-well plate containing coverslips and were transfected with 0.1 to 0.3 μg of expression plasmids encoding LANA1 variants using X-tremeGENE HP transfection reagent (Roche). After 24 to 48 h, the cells were rinsed briefly with PBS and fixed with 2% paraformaldehyde for 20 min at room temperature and subsequently permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. After blocking (10% normal goat serum in PBS) for 1 h, the cells were incubated for 1 to 2 h in a humidified chamber at room temperature with primary antibody mouse αN-term and subsequently for 1 h with the secondary antibody (anti-mouse Alexa Fluor 568; Invitrogen), followed by rat monoclonal anti-LANA1 (1:1,000, αCR2-3) and anti-rat fluorescein isothiocyanate (FITC) (1:500; Sigma) staining. The cells were washed three times for 5 min after incubation with each antibody. Coverslips were mounted in Vectashield with DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories), and the cells were visualized with an Olympus microscope. For confocal microscopy, LANA1.FL transfected U2OS cells were washed briefly with PBS and incubated with 200 nM MitoTracker (Invitrogen) in serum-free DMEM for 30 min at 37°C. PEL cells were seeded on poly-l-lysine-coated coverslips incubated at 4°C for 15 min; unattached cells were rinsed with PBS and then fixed and permeabilized as described above, blocked with 10% normal goat serum, and incubated with rabbit αS6 (1:100; Cell Signaling) for 2 h at room temperature, followed by secondary antibody (anti-rabbit Alexa Fluor 568; Invitrogen). Stained cells were rinsed and incubated with mouse αCR2 (1:1,000) staining for 1 h at room temperature, followed by secondary antibody (anti-mouse Alexa Fluor 488; Invitrogen). The cells were rinsed and subsequently stained with DRAQ5 for 15 min at room temperature, rinsed in PBS, and mounted with Gelvatol mounting medium for fluorescence (Center for Biologic Imaging, University of Pittsburgh). Confocal images and three-dimensional (3D) visualization of BCBL1 cells were acquired using a Leica TCS SP confocal microscope.
RESULTS
LANA1 shoulder bands (LANA1S) lack N-terminal peptides.
To investigate LANA1 translation, we generated new mouse monoclonal antibodies directed against the LANA1 N terminus (CM6.43.24, αN-term), using an N-terminal peptide sequence between aa 33 and 50, and against the repetitive CR2 EPQQ epitope found throughout aa 446 to 596 (CMA-810, αCR2) (48). We used a commercially available rat monoclonal antibody recognizing the EQEQE in the CR3 domain (ABI, rat αLNA1) (26). In the present study, we found out that the same antibody reacts to the EQEQ epitope present in CR2 domain. Based on that, we define the rat monoclonal αLNA1 as an αCR2-3 antibody recognizing the EQEQE and EQEQ epitopes found throughout aa 698 to 872 (Fig. 1A).
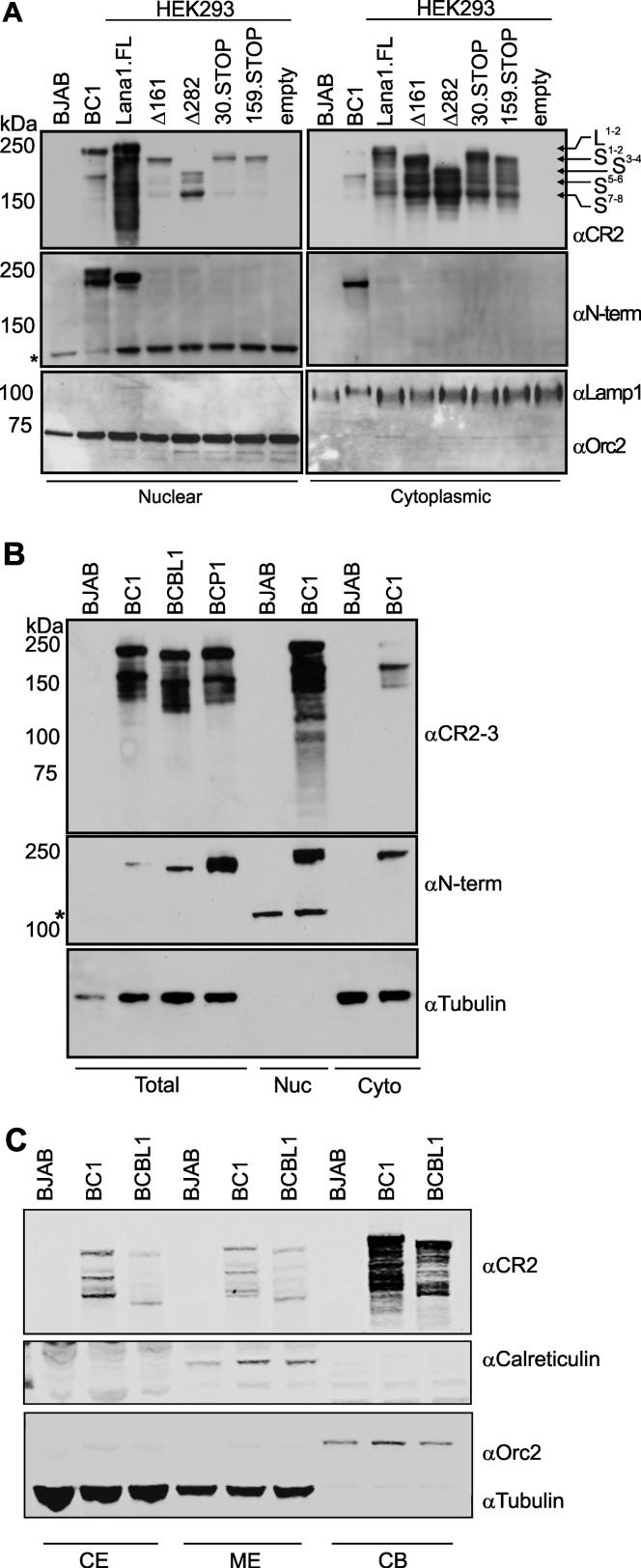
LANA1 proteins from KSHV-positive BCBL1, BC1, and BCP1 PEL cells were first immunoprecipitated with αCR2 antibody and then detected with either αCR2 or αN-term antibodies (Fig. 1B). PEL lysates showed a typical pattern of ∼230-kDa LANA1 doublets expected for the full-length protein (LANA1L) detected with both antibodies. LANA1 shoulder bands (LANA1S) that migrate at ca. 150 to 180 kDa (SDS–8% PAGE), however, are only detected by immunostaining with αCR2. Banding patterns on αCR2 immunoblotting are identical for the different cell lines except that the pattern proportionally shifts between cell lines, a finding consistent with the gain or loss of CR units as previously described (51). The αN-term antibody, however, only detects full-length LANA1L bands, suggesting that LANA1S lacks N terminus peptide sequences (Fig. 1B). As determined by quantitative infrared chromophore (LI-COR) immunoblotting, we estimate that LANA1S comprises ca. 50 to 60% of the total naturally expressed LANA1 in PEL cells (data not shown).
To confirm this, full-length LANA1 (LANA1.FL) was compared to a cloned construct with most of the N terminus deleted (Δ282 LANA1, see Fig. 3A). The Δ282 LANA1 was cloned to delete the first three potential start-methionines but to retain the fourth potential start-methionine. Despite differences in absolute expression abundance, most LANA1S isoforms that are expressed by LANA1.FL are also expressed by Δ282 LANA1. This is most readily seen after immunoprecipitation with αCR2 antibody (Fig. 1C). Δ282 LANA1 lacks the N-terminal epitope and cannot be detected by αN-term antibody (Fig. 1C).
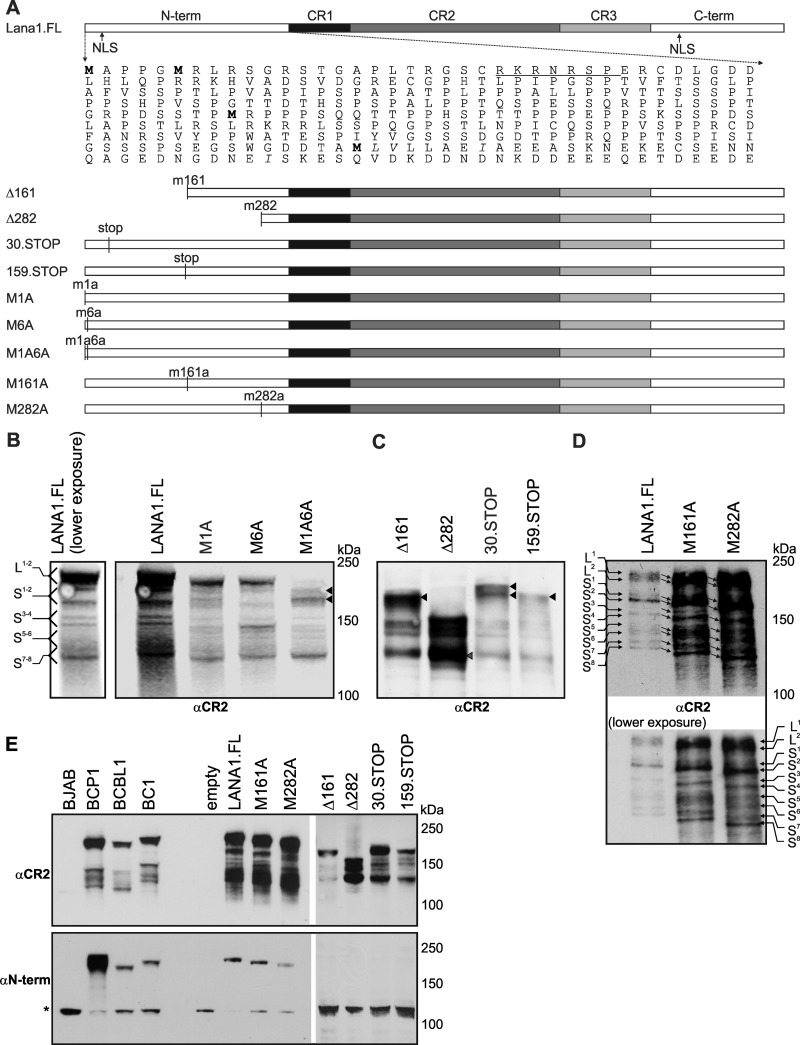
Fig 3.
Mutation analysis reveals that translation initiation sites for LANA1S occur at non-AUG codons in the N terminus and CR1 regions. (A) Map of the LANA1 N-terminal region, including four potential AUG (methionine) translation initiation site (TIS, boldface), four non-AUG TIS downstream of M282 (italics), and the nuclear localization signal (underlined). Deletion mutants starting at aa 161 (Δ161) and aa 282 (Δ282), stop codon insertions at aa 30 (30.STOP) and aa 159 (159.STOP), and methionine-to-alanine substitutions at aa 1 (M1A), aa 6 (M6A), aa 1 and 6 (M1A6A), aa 161 (M161A), and aa 282 (M282A) are shown. (B to D) Evidence for non-AUG-mediated alternative initiation in the N terminus. (B) Full-length LANA1 doublet is numbered as L1 and L2, and the shoulder bands are numbered from S1 to S8. Methionine-to-alanine substitution mutants M1A and M6A preserve all forms of LANA1, although intensities of individual bands can vary. Double-substitution mutant M1A6A lacks L1 and L2 but retains shoulder S1 to S8 bands. (C) Most of LANA1S isoforms are not generated from M161 or M82. LANA1s isoforms vary for Δ161, Δ282, 30.STOP, and 159.STOP mutants expressed in HEK293 cells. The LANA1S bands are similar for 30.STOP and M1A6A (see panel B), suggesting that S1 and S2 (arrowheads) initiate downstream from aa 30. The Δ161 and 159.STOP banding patterns lose S1, compared to M1A6A and 30.STOP, suggesting a TIS for S1 between residues 30 and 159. Δ282 lacks L1, L2, S1, and S2 but still preserves the low-molecular-weight shoulder bands (S3 to S8), suggesting a TIS for S2 between residues 159 and 282. (D) Alanine substitutions at M161 and M282 do not change the LANA1S banding pattern, indicating that these AUG codons are not TIS for LANA1S isoforms. Substitution mutants M161A and M282A do not alter the shoulder bands; however, the migration speed is increased for all isoforms (lower exposure, arrows). (E) Comparison of LANA1 construct expression to endogenous LANA1 expression in PEL cell lines. LANA1 constructs were transfected into HEK293 cells, and protein expression was analyzed by immunoblotting with αCR2 and αN-term antibodies and compared to LANA1 expression from BCP1, BCBL1, and BC1 cells (BJAB cells serve as a negative control). Total protein loading was variably adjusted between different samples to allow simultaneous detection of LANA1 isoforms on a single blot. *, A nonspecific band (present in BJAB cells) is detected with N-term antibody at ∼100 kDa.
N-terminal (FLAG.LANA1.FL) and C-terminal (LANA1.FL.V5) epitope-tagged full-length LANA1 constructs expressed in HEK293 cells have expression patterns similar to those seen in PEL cells (Fig. 1D). αCR2 and αCR2-3 antibodies detect both LANA1L and LANA1S protein isoforms generated by FLAG.LANA1.FL and LANA1.FL.V5. When these same lysates are immunoblotted with αFLAG antibody, only LANA1L is detected. LANA1.FL.V5 retain LANA1S bands detectable with αV5 antibody, indicating that LANA1S bands are generated from alternative initiation sites that bypass the N-terminal FLAG epitope.
Expression of LANA1S during MG132, lambda phosphatase, and TPA treatment.
LANA1S lacking N-terminal peptide sequences could be generated either by alternative translation initiation or by posttranslational modifications, such as proteolytic processing. To evaluate endoproteolytic cleavage and proteasomal degradation (52), PEL cells were treated with the proteasome inhibitor MG132 (20 μM) for 12 h and immunoblotted with αCR2 antibody, but no preferential increase in LANA1L compared to LANA1S was present (Fig. 2A). Similarly, MG132 treatment of a destabilized enhanced green fluorescent LANA1 protein (LANA1.d1EGFP) (50) expressed in HEK293 cells led to equal accumulation of all forms of LANA1. Treatment of LANA1-expressing cells with the pan-caspase inhibitor Z-VAD also did not alter the shoulder pattern (data not shown). Finally, no low-molecular-weight bands consistent with an N-terminal proteolytic cleavage product were detected with αN-term antibody by immunoblotting, indicating that no enzymatic or endoproteolytic cleavage has an effect on formation of LANA1 shoulder bands (data not shown).
Fig 2.
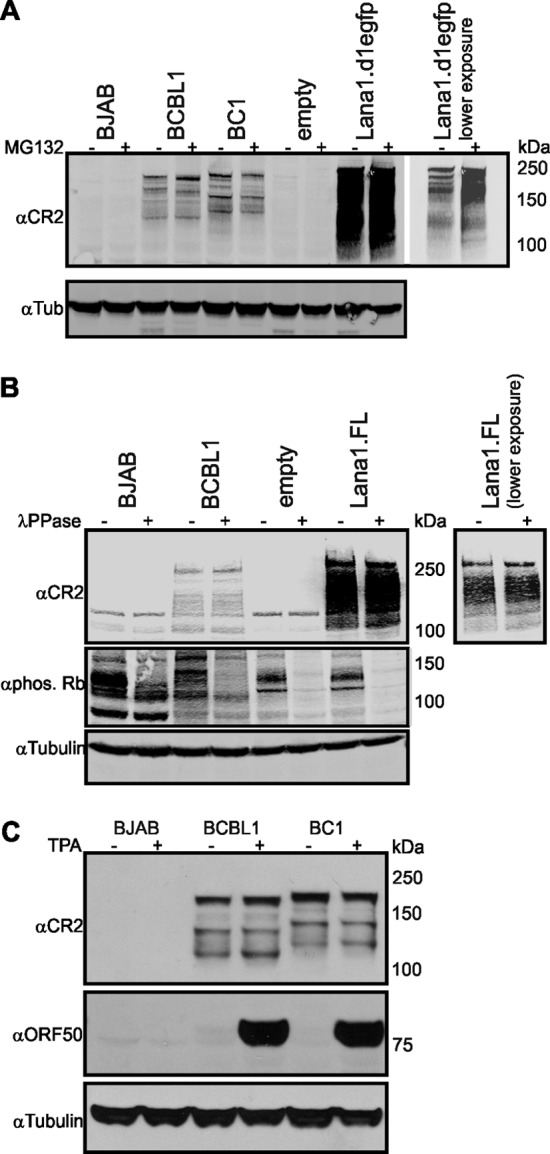
Expression of LANA1S during MG132, lambda phosphatase, and TPA treatment. (A) LANA1S isoforms are not diminished by MG132 treatment. HEK293 cells were transfected with empty vector or full-length LANA1-d1EGFP fusion construct harboring a PEST amino acid sequence. BJAB, BCBL1, BC1, and transfected HEK293 cells were either treated with proteasome inhibitor MG132 (20 μM, 12 h) or left untreated. Equal amounts of cell lysate were run on gradient gel and analyzed by immunoblotting with αCR2 and α-tubulin (αTub) antibody. (B) Aberrant LANA1 gel migration is not due phosphorylation. Lysates from BJAB, BCBL1, and HEK293 cells transfected with empty vector or LANA1.FL were treated with lambda phosphatase (λPPase) for 1 h at 37°C or left untreated. Samples were run on a gradient gel and immunoblotted for αCR2, α-phospho-Rb, and α-tubulin. α-Phospho-Rb shows the efficiency of the phosphatase treatment, and α-tubulin is used to depict equal loading. (C) LANA1S isoforms are constitutively expressed. To induce lytic replication, BJAB, BCBL1, and BC1 cells were incubated with medium containing 20 ng of TPA/ml for 48 h. Equal amounts of protein were separated on a gradient gel and immunoblotted αCR2, αORF50, and α-tubulin. KSHV ORF50 gene product is a lytic protein and was used as a positive control for the induction.
Lambda phosphatase treatment of LANA1 was used to measure shifts in electrophoretic migration for LANA1L and LANA1S caused by posttranslational phosphorylation. BJAB, BCBL1, and HEK293 cells expressing LANA1.FL, with or without lambda phosphatase treatment, were immunoblotted with αCR2. Phospho-Rb antibody detecting the endogenous levels of retinoblastoma protein (Rb) phosphorylated at serine residues was used as a control. As seen in Fig. 2B, phosphatase treatment reduced or eliminated Rb phosphorylation but did not markedly affect LANA1S banding patterns.
Like LANA1L, LANA1S bands are largely constitutively expressed in PEL cells. When the PEL cell lines BCLB1 and BC1 are treated with TPA (20 ng/ml for 48 h), robust expression of the lytic transactivator protein ORF50 is induced (Fig. 2C). In TPA-treated BCBL1 and BC1 cells, the lowest-molecular-weight LANA1S S7 and S8 levels are slightly increased. Aside from this, no changes in LANA1S band patterns are seen. Taken together, these experiments show that most LANA1S isoforms are constitutively expressed and are unlikely to be generated by common posttranslational modification processes.
LANA1S isoforms are generated through multiple, complex translation initiation sites.
The LANA1 N terminus region has three AUG-dTIS at amino acid positions 6, 161, and 282 that are potential start sites for leaky scanning and translation initiation downstream of the annotated-AUG (position 1) (Fig. 3A, indicated in boldface). Non-AUG-dTIS in the N terminus included four CUG, three UUG, five ACG, three AUU, three AUA, and seven GUG that are in-frame for LANA1 and could potentially serve as TIS to generate LANA1S isoforms recognized by αCR2 and αCR2-3 antibodies (Fig. 3A). We generated M1A, M6A, M1A6A, M161A, and M282A methionine-to-alanine substitution mutations at amino acid positions 1, 6, 160, and 282, N-terminal truncation mutations lacking the first 160 (Δ161) and 281 (Δ282) amino acids, and stop codon insertion mutants at amino acid positions 30 (30.STOP) and 159 (159.STOP) in BC1 strain LANA1.FL to search for alternative TIS (Fig. 3A). These mutants were expressed in HEK293 cells and analyzed by Western blotting. To facilitate the evaluation of the LANA1 banding pattern, the full-length LANA1 isoforms are numbered L1 and L2, and the shoulder bands are numbered S1 through S8 (Fig. 3B and D).
Both M1 and M6 conform the Kozak context and have either −3A/G or +4G, or both. Mutation of the start methionine to alanine (M1A) might enforce downstream LANA1 translation initiation (Fig. 3B). When this mutation was introduced, only a slight shift in LANA1L apparent molecular weight became apparent, without changes to the LANA1S banding pattern, which is most consistent with M6 becoming the new predominant initiation product. Likewise, the M6A substitution mutant has similar banding patterns to LANA1.FL (Fig. 3B). Although LANA1S S1 and S2 expression is reduced and S3 and S4 expression is enhanced for the M6A mutant, both bands are present at the same migration positions as for the wild-type protein. The M1A6A double mutant, however, eliminates L1 and L2 bands, while the lower-apparent-molecular-weight shoulder bands S1 to S8 are preserved (Fig. 3B). LANA1S S1 and S2 become prominent with the M1A6A mutations, suggesting that these bands arise from downstream initiation beyond methionine 6. The prominent doublet (Fig. 3B, M1A6A, black arrows) at ∼180 kDa seen with the M1A6A mutant may reflect a newly predominant isoform initiating at downstream methionines such as M161 or M282. Similar to the M1A6A mutant, the stop codon insertion mutant 30.STOP, generates S1 to S8 but not L1 and L2 (Fig. 3C, black arrows, Fig. 3E). Deletion of 1 to 161 aa (Δ161) and the 159.STOP insertion mutant have similar banding patterns but lack L1, L2, and S1 expression (Fig. 3C and E), suggesting that initiation for S1 occurs between aa 30 and 159. In the Δ282 deletion mutant, L1 to S2 are not present, but most of the low-molecular-weight shoulder bands remain detectable, which is consistent with S2 initiating between aa 161 and 282.
To determine whether M161 or M282 serve as TIS for LANA1S isoforms, both methionines were mutated to alanines (Fig. 3D and E). No changes in the patterns of any of the isoforms were seen, suggesting that M161 and M282 are not start sites for any of the LANA1S isoforms, although all M161 and M282 LANA1 proteins migrated slightly faster than predicted from changes in actual molecular weight using either αCR2 or αN-term antibodies (Fig. 3D, arrows indicate the shift in gel migration pattern, Fig. 3E).
Given the complexity of the LANA1S shoulder pattern, we next focused on the lowest-molecular-weight LANA1S S7-8 isoforms (Fig. 3C, Δ282, gray arrow), which migrate at ∼130 kDa and are unchanged in the substitution and deletion mutant proteins. These data suggest that S7 and S8 have a TIS downstream from methionine 282. The N terminus-CR1 transition region, between aa 282 and 321, encodes several non-AUG codons that could potentially play a role in translation initiation. We generated single or multiple alanine substitutions at amino acids L283, V284, I289, and I315 (Fig. 3A, shown in italics) and saw no changes in LANA1S S7 and S8 expression (data not shown), most likely reflecting dTIS in the central repeat region for the lowest-molecular-weight shoulder isoforms.
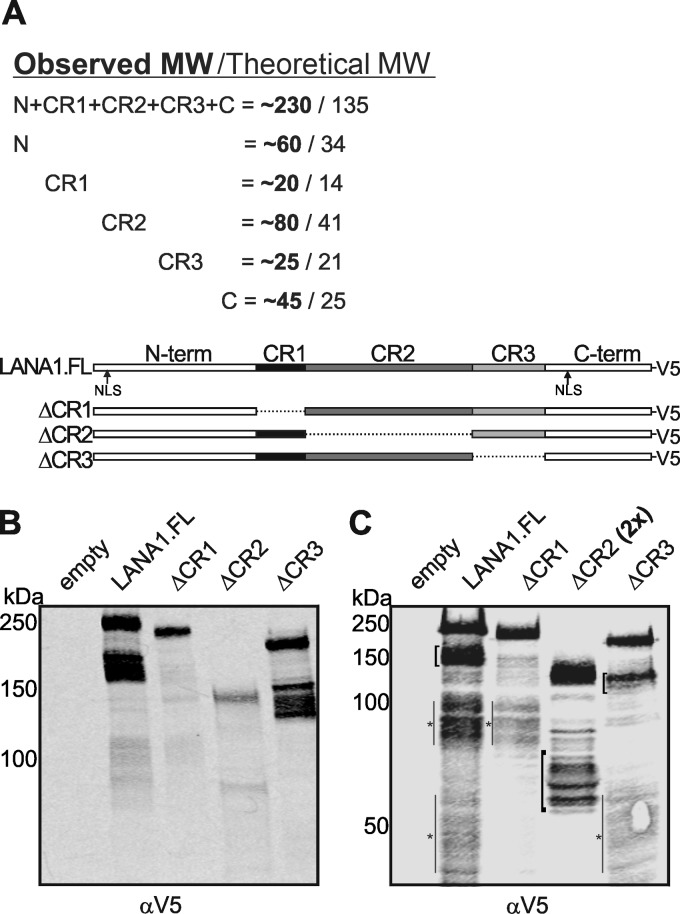
Central repeat domain 1 deletion eliminates most LANA low-molecular-weight shoulder bands.
To investigate the central repeat domains as potential initiation sites for LANA1S isoforms, we generated C-terminal V5 tagged central repeat (CR) deletion mutants of LANA1 (19). Because of the aberrantly retarded electrophoretic migration for LANA1, we first empirically determined (data not shown) the expected contributions for each LANA1 domain to the protein's overall gel mobility by deletion of individual domains (Fig. 4A). From this, the expected 150- to 180-kDa LANA1S isoforms (if present) would migrate at 130 to 160 kDa for ΔCR1, at 70 to 100 kDa for ΔCR2, and at 125 to 155 kDa for ΔCR3. The CR1 deletion reduced or eliminated the predominant LANA1S S3-S8 shoulder bands compared to full-length LANA1 (Fig. 4B). In contrast, shoulder bands were preserved with CR3 deletion but proportionally shifted in molecular weight. As previously reported, deletion of CR2 reduces stability of LANA1 (50). To detect ΔCR2 LANA1S, protein lysates were 2-fold overloaded compared to other LANA1 constructs, the gel was electrophoresed for a shorter time period, and the membrane was overexposed after incubation with the V5 antibody (Fig. 4C). LANAS1 to LANAS8 isoforms are denoted by brackets. Bands of the correct predicted size for ΔCR2 LANA1S (70 to 100 kDa) were detected in the overexposed gels.
Fig 4.
CR1 deletion eliminates low-molecular-weight LANA1S. (A) According to the migration pattern of different LANA1 expression constructs (data not shown), the apparent molecular weight of each domain is estimated. The observed molecular weights and theoretical molecular weights are expressed in thousands. LANA1.FL and different central repeat (CR) deletion mutants—ΔCR1, ΔCR2, and ΔCR3—were fused to a C-terminal V5 tag. (B) LANA1S isoforms are reduced or absent when the CR1 domain is deleted from LANA1 but not by the deletion of the CR2 or CR3 domains. Different LANA1 mutants were transfected in HEK293 cells, and lysates were analyzed by SDS-PAGE and Western blotting with αV5 antibody. LANA1.FL and ΔCR3 possesses shoulder bands at ca. 150 to 180 kDa and at ca. 125 to 155 kDa, respectively. (C) Deletion of CR2 is known to reduce the stability of LANA1. Lysates from panel B with the exception of 2-fold overloaded ΔCR2 were electrophoresed on a gradient gel for a shorter time period to visualize CR2-LANA1S, which is shifted ∼80 kDa compared to the LANA1.FL shoulders (black brackets). Additional low-abundance isoforms with <100-kDa bands containing the C-terminal V5 tag are marked with gray lines and asterisks.
When LANA1.FL was overexposed (Fig. 4C), however, additional low-abundance isoforms with <100-kDa bands containing the C-terminal V5 tag can also be detected (brackets with asterisks). No shift in these bands was present with CR1 deletion, suggesting that additional low-molecular-weight, low-abundance translation products may initiate from a site downstream of CR1. We cannot exclude that these low-abundance isoforms also contribute to the bands we identified as ΔCR2 LANA1S isoforms (Fig. 4C), illustrating the unexpected complexity of LANA1 translation patterns.
LANA1S localization to the cytoplasm.
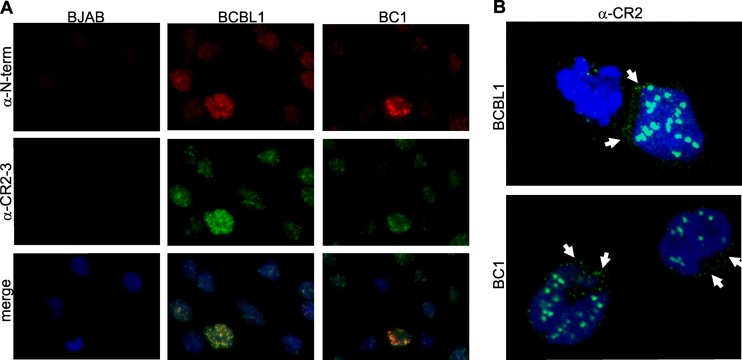
LANA1 isoforms lacking the first 22 to 30 aa do not encode the N-terminal LANA1 NLS (30) and cannot bind to chromatin (28). Therefore, we sought to determine whether these isoforms retained the ability to localize to the nucleus characteristic for canonical LANA1. Immunofluorescence microscopy of BCBL1 and BC1 PEL cells using N-term and CR2-3 antibodies reveals that most LANA1 protein is present in a typical punctate nuclear staining pattern (Fig. 5A). However, LANA1 can also be detected in the perinuclear cytoplasm of PEL cells by confocal microscopy (Fig. 5B, red arrows). This is most readily seen in 3D views of BCBL1 cells stained with αCR2 (see Movie S1 in the supplemental material) in which aggregates of LANA1 protein are present outside of the nuclear envelope.
Fig 5.
Perinuclear and cytoplasmic localization of endogenous LANA1. (A) Colocalization of mouse αN-term (red) and rat αCR2-3 (green) primary antibodies. BJAB, BCBL1, and BC1 cells were seeded on poly-l-lysine-coated coverslips. After fixation and permeabilization, th cells were costained with αN-term and αCR2-3 antibodies. Nuclei were stained with DAPI (blue). (B) Cytoplasmic and nuclear staining of endogenous LANA1 in BCBL1 and BC1 cells is seen with mouse αCR2 primary antibody. White arrows point to the perinuclear localization of LANA1. DRAQ5 was used for the nuclear staining. 3D visualization of LANA1 isoforms in BCBL1 by confocal microscopy is shown in Movie S1 in the supplemental material.
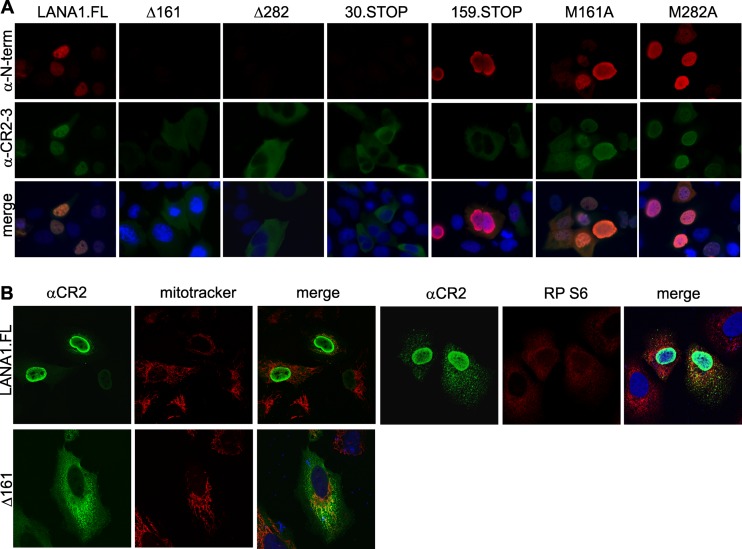
To determine whether LANA1 isoforms lacking the N-terminal NLS localize to cytoplasmic domains, LANA1.FL, Δ161, Δ282, 30.STOP, 159.STOP, M161A, and M282A constructs were expressed in U2OS cells and stained with αCR2-3 or αN-term antibodies. As seen in Fig. 6A, full-length LANA1L localizes mainly to the nucleus and to a lesser extent in the cytoplasm. Similarly, M161A and M282A, which express full-length LANA1L (Fig. 3D and E), retain nuclear localization and reside in the cytoplasm as well (Fig. 6A). The Δ161, Δ282, and 30.STOP proteins, however, primarily localize to the cytoplasm and can be detected with αCR2-3 or αCR2 antibodies, but not αN-term antibody since the epitope is absent in these constructs (Fig. 6A). The 159.STOP construct is predicted to generate an artificial N-terminal peptide (1 to 159 aa) retaining the N-terminal NLS and N-term epitope, as well as naturally translated C-terminal LANA1S isoforms (Fig. 3C). Consistent with this, αN-term antibody staining reveals nuclear localization of the N-terminal peptide, whereas αCR2-3 staining is cytoplasmic (Fig. 6A, 159.STOP).
Fig 6.
Subcellular distribution of LANA1 mutants. (A) LANA1.FL, N-terminal deletion constructs Δ161 and Δ282, stop codon insertion mutants 30.STOP and 159.STOP, and methionine-to-alanine substitution mutants M161A and M282A were overexpressed in U2OS cells. The cells were fixed with 2% paraformaldehyde and subsequently permeabilized with 0.2% Triton X-100. Primary mouse αN-term and rat αCR2-3 antibodies were used for the immunostaining. The nucleus was stained with DAPI (blue). CR2-3 antibody detects cytoplasmic isoforms for Δ161, Δ282, and 30.STOP constructs that are not detected with the N-term antibody. 159STOP generates a N-term antibody-positive peptide fragment that localizes to the nucleus and a CR2-3 antibody-positive cytoplasmic protein. (B) Partial confocal colocalization of LANA1 with perinuclear mitochondria and ribosomes in U2OS cells. Full-length LANA1 (LANA1.FL) or Δ161 was transfected in U2OS cells. One day after transfection, the cells were stained with mouse αCR2 and with either a mitochondrial marker (MitoTracker) or a ribosomal marker (RP S6, rabbit ribosomal protein S6). DRAQ5 (blue) was used for the nuclear staining.
LANA1S isoforms localize to various cytoplasmic structures (Fig. 6B). For example, LANA1S colocalizes with mitochondrion-containing structures in the perinuclear region of some, but not all cells, as measured by MitoTracker costaining (Fig. 6B, left panel). LANA1S can also partially colocalize with cytoplasmic ribosomal structures as detected by ribosomal S6 protein costaining (Fig. 6B, right panel) (9).
BJAB, BC1, BCBL1, and HEK293 cells expressing various LANA1 constructs were separated into nuclear and cytoplasmic fractions and immunoblotted with αCR2 and αN-Term antibodies (Fig. 7A and C). Fractionation quality was measured by immunoblotting with antibodies to lysosomal-associated membrane protein 1 (LAMP1; cytoplasmic) and origin recognition complex 2 (ORC2; nuclear). Fractionation reveals a more complex pattern than seen with immunofluorescence staining for LANA1. LANA1L naturally expressed in BC1 cells or from a full-length construct in HEK293 cells is primarily nuclear. Staining with αN-term antibody, however, reveals a portion of full-length LANA1L in the cytoplasm (Fig. 7A). In parallel with the immunofluorescence staining, for each mutation construct in which LANA1L is eliminated (Δ161, Δ282, 30.STOP, and 159.STOP), most LANA1S isoform proteins are expressed in the cytoplasm fraction and, to a lesser extent, in the nuclear compartment (Fig. 7A). Consistently, the lower-apparent-molecular-mass LANA1S isoforms of BC1, at ca. 130 to 150 kDa (LANA1 S3 to S8), are present in cytoplasmic fractions immunostained with αCR2 and αCR2-3 (Fig. 7B and C).
Fig 7.
Subcellular distribution of LANA1 isoforms. (A) Nuclear and cytoplasmic LANA1 localization for BJAB, BC1, and various LANA1 constructs transfected into HEK293 cells. Nuclear and cytoplasmic fractions were run on an SDS–8% polyacrylamide gel and immunoblotted with αCR2 and αN-term antibodies. LANA1 isoforms are numbered from L1 to S8. Orc2 and Lamp1 served as nuclear and cytoplasmic fraction markers, respectively. *, Nonspecific band in the nuclear fraction is detected with αN-term antibody at ∼100 kDa. (B) Total extracts of BJAB, BCBL1, and BC1 cells and nuclear (Nuc) and cytoplasmic (Cyto) fractions of BJAB and BC1 cells were separated on SDS–8% PAGE and immunoblotted with αCR2-3 and αN-term antibodies. Tubulin served as cytoplasmic fraction markers. *, Nonspecific band in the nuclear fraction is detected with αN-term antibody at ∼100 kDa. (C) Subcellular protein fractions (Thermo Scientific) were used to evaluate the endogenous LANA1 distribution in PEL cells. Equal amount of soluble cytoplasmic (CE), plasma, mitochondria and ER/Golgi membranes (ME), and chromatin-bound nuclear protein (CB) fractions were separated on a gradient gel and immunoblotted with αCR2, αOrc2 (nuclear fraction marker), α-tubulin (cytoplasmic fraction marker), and α-calreticulin (endoplasmic reticulum marker).
DISCUSSION
The size of a viral genome markedly constrains the genetic coding capacity needed by the virus to complete its life cycle. Viral genome size is limited both by nucleic acid encapsidation requirements and by the need to minimize viral antigen presentation. Viruses frequently encode highly multifunctional proteins that can perform multiple tasks from a single gene. Some viruses, such as polyomaviruses, use alternative splicing to generate functionally distinct proteins (53). Other viruses, including HIV, use frameshifting to amplify their genetic capacity from a single nucleic acid sequence (54).
We show here for the first time that endogenous and ectopically expressed LANA1 shoulder bands are N-terminally truncated isoforms that are present in both the nucleus and the cytoplasm using newly generated LANA1-specific antibodies recognizing epitopes in the N terminus and CR2 domain. Coimmunoprecipitation experiments with endogenous and ectopically expressed LANA1 reveal the absence of the LANA1 N-terminal peptides in the LANA1S shoulder bands. This was confirmed with protein detection using both N-terminal FLAG-tagged and C-terminal V5-tagged full-length LANA1 expressed in HEK293 cells.
LANA1 shoulders lacking N-terminal peptide sequences might be generated by alternative translation initiation, by partial N-terminal degradation, or by cleavage. If LANA1S bands are generated by proteasomal degradation, then we would expect that MG132 treatment should eliminate these bands in PEL cells. However, we did not find evidence that shoulder bands are generated by proteasomal processing, nor did we find evidence that posttranslational phosphorylation can account for the apparent shift in gel migration for LANA1S bands. LANA1 is a phosphoprotein, and a recent protein microarray study identified 63 human nuclear kinases that can phosphorylate LANA1 N terminus (55). Among these, 24 kinases phosphorylate the chromatin-binding domain (CBD) of LANA1 between aa 3 and 21. However, presumably due to the size and highly charged nature of LANA1, we did not identify changes in LANA1 phosphorylation by shifts in gel mobility and phosphorylation patterns cannot account for the LANA1 shoulder banding pattern. We also did not detect any cleavage product of the N-terminal domain recognized with the αN-term antibody, suggesting that the shoulders do not arise from proteolytic processing. Shoulder bands are also largely unchanged by lytic virus induction using TPA. A recent transcriptomic and proteomic study revealed that KSHV has tight latency in BC1 cells but not in BCBL1 cells (56, 57). We cannot exclude the possibility that lytic reactivation preferentially enhances expression of lowest-molecular-size LANA1S isoforms (ca. 150 kDa), but it is unlikely that these isoforms are solely generated during the virus lytic cycle. Although we also cannot exclude the possibility that other posttranslational modifications or proteasome-independent degradation contribute to formation of these bands, our data are most consistent with constitutive alternative translation products being generated from ORF73 mRNA in addition to the full-length LANA1L protein.
Several caveats to our analysis are appropriate. First, the protein expression pattern for LANA1 is exceedingly complex. LANA1 has a predicted size of ∼135 kDa but migrates as a doublet of 226 to 234 kDa on SDS-PAGE. The long-form bands of LANA1, L1 and L2, have been demonstrated to run as a doublet as a result of a noncanonical polyadenylation signal that generates a 76-aa deleted C-terminal form as described by Canham and Talbot (25). We reasoned that some of the shoulder forms might also arise from the C-terminal, noncanonical polyadenylation signal resulting in a multitude of similar doublet formations. The aberrant LANA1 migration (58), in large part due to the N-terminal proline-rich domain, causes shifts in migration for alternative or deleted forms that are much greater than predicted based on the theoretical molecular weights. In addition, the highly repetitive structure of the protein containing multiple CR2 and CR2-3 antibody repeat epitopes complicates the unique identification of each LANA1S isoform. This is particularly difficult in achieving clear resolution of individual high-molecular-weight bands. Thus, mapping studies to precisely identify the initiation sites for each and every individual shoulder band are beyond our current technical capacity.
Our analysis suggests that the slowest-migrating shoulder isoforms (LANA1 S1 and S2) result from alternative initiation at a non-AUG codon in the N-terminal sequence between aa 30 and 282. LANA1 N-terminal domain harbors four in-frame AUG codons. Using different methionine-to-alanine substitution, deletion, and stop codon insertion mutations, we failed to find evidence for translation initiation at M6, M161, or M282, suggesting a non-AUG downstream TIS (dTIS) for LANA1S S1-2 (38). Substitution mutations at amino acid positions 283, 284, 289, and 315 non-AUG codons in the region between M282 and CR1 did not affect LANA1S banding patterns, indicating that faster-migrating LANA1S isoforms (LANA1 S3 to S8) are likely to be generated from the central repeat region. Evidence points to CR1 as the major site for this initiation since CR1 deletion eliminates most of the low-molecular-weight isoforms.
The results presented here indicate that translation of LANA1 isoforms appears to occur by more than one mechanism. Leaky scanning is more likely to be involved in the generation of LANA1 shoulders rather than termination-reinitiation mechanism since an upstream ORF (uORF) is essential for the latter and the LANA1 expression constructs used in the present study lack uORFs. In a previous study, dTIS in human cells were found to be composed of 47% AUG and 53% non-AUG codons. The latter comprises CUG, AUG variants, and other codons as identified by GTI-seq (38). The LANA1 N terminus comprises four methionines at aa 1, 6, 161, and 282. Among these, M1, M6, and M282 have moderately strong Kozak context sequences with either −3A/G or +4G sites, or both. When M1 is deleted, the LANA1 banding pattern is largely unchanged, indicating that M6 can act as a TIS. Due to issues in protein resolution, we cannot determine whether M6 acts as a dTIS under natural conditions. Similar experiments with M161 and M282 do not reveal these methionines to have dTIS capacity, suggesting that non-AUG dTIS codons might be involved in shoulder formation. The reinitiation after 30.STOP and 159.STOP could be regulated by different mechanisms, including ribosomal shunting or IRES-like initiation at downstream non-AUG initiation codons.
As this protein's name implies (latency-associated nuclear antigen), the biological activities attributed to LANA1 have largely been ascribed to its localization in the nucleus. We show, however, that a substantial fraction of naturally generated LANA1 is actually constitutively present in the cytoplasm of KSHV-infected cells. The majority of LANA1 shoulders (LANA1S S3 to S8), as well as a minority population of full-length LANA1L forms are found in soluble cytoplasmic and membrane-bound fractions. Despite the lack of N-terminal NLS, an intact C-terminal NLS might be sufficient for the nuclear localization of some LANA1 shoulder forms, and we found both nuclear and cytoplasmic localization of LANA S3-8. Intriguingly, a portion of high-molecular-weight LANA1 having an intact N-terminal epitope can also be detected in cytoplasmic fractions. Consistent with the subcellular fraction analysis, confocal images suggest that LANA1S targets different cytoplasmic organelles (mitochondria, ribosomes). These findings help to explain incongruous published results, such as predicted LANA1 interactions with cytoplasmic proteins (7–9), opening up the possibility for studies of LANA1 interactions that may have previously been assumed to be artifactual. Ribosomal profiling studies reveal unexpected noncanonical cellular protein translation products are commonly made in cells (38). Our study indicates that a long-studied viral oncoprotein may make use similar alternative translation products to target multiple cellular processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mary Ann Accaviti for antibody production. We thank Phil Kline and David Lang for help with the manuscript. We thank João-Gonçalves for helpful comments.
L.F. received a doctoral fellowship from the Fundação para a Ciência e Tecnologia (Portugal). This study was supported by National Institutes of Health grants CA136363 and CA120726 to P.S.M. and Y.C., who are also funded as American Cancer Society Research Professors. This project used the UPCI Confocal Laser Microscopy and Li-COR Imaging Facility supported in part by award P30CA047904.
Footnotes
Published ahead of print 19 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03061-12.
REFERENCES
- 1. Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 265:1865–1869 [DOI] [PubMed] [Google Scholar]
- 2. Ballestas ME, Chatis PA, Kaye KM. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641–644 [DOI] [PubMed] [Google Scholar]
- 3. Ballestas ME, Kaye KM. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Radkov SA, Kellam P, Boshoff C. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121–1127 [DOI] [PubMed] [Google Scholar]
- 5. Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889–894 [DOI] [PubMed] [Google Scholar]
- 6. Sato-Matsumura KC, Matsumura T, Nabeshima M, Katano H, Sata T, Koizumi H. 2001. Serological and immunohistochemical detection of human herpesvirus 8 in Kaposi's sarcoma after immunosuppressive therapy for bullous pemphigoid. Br. J. Dermatol. 145:633–637 [DOI] [PubMed] [Google Scholar]
- 7. Kaul R, Verma SC, Robertson ES. 2007. Protein complexes associated with the Kaposi's sarcoma-associated herpesvirus-encoded LANA. Virology 364:317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shamay M, Liu J, Li R, Liao G, Shen L, Greenway M, Hu S, Zhu J, Xie Z, Ambinder RF, Qian J, Zhu H, Hayward SD. 2012. A protein array screen for Kaposi's sarcoma-associated herpesvirus LANA interactors links LANA to TIP60, PP2A activity, and telomere shortening. J. Virol. 86:5179–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen W, Dittmer DP. 2011. Ribosomal protein S6 interacts with the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 85:9495–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moore PS, Gao SJ, Dominguez G, Cesarman E, Lungu O, Knowles DM, Garber R, Pellett PE, McGeoch DJ, Chang Y. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J. Virol. 70:549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao S-J, Kingsley L, Hoover DR, Spira TJ, Rinaldo CR, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore PS. 1996. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N. Engl. J. Med. 335:233–241 [DOI] [PubMed] [Google Scholar]
- 12. Kedes DH, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918–924 [DOI] [PubMed] [Google Scholar]
- 13. Lieberman PM, Hu J, Renne R. 2007. Gammaherpesvirus maintenance and replication during latency, p 379–402 In Arvin A, Mocarski A, Moore PS, Roizman B, Whitley RJ. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 14. Moore PS, Chang Y. 2001. Molecular virology of Kaposi's sarcoma-associated herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:499–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rainbow L, Platt GM, Simpson GR, Sarid R, Gao SJ, Stoiber H, Herrington CS, Moore PS, Schulz TF. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kedes DH, Lagunoff M, Renne R, Ganem D. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Invest. 100:2606–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alkharsah KR, Schulz TF. 2012. A role for the internal repeat of the Kaposi's sarcoma-associated herpesvirus latent nuclear antigen in the persistence of an episomal viral genome. J. Virol. 86:1883–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kwun HJ, da Silva SR, Shah IM, Blake N, Moore PS, Chang Y. 2007. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics Epstein-Barr virus EBNA1 immune evasion through central repeat domain effects on protein processing. J. Virol. 81:8225–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu F, Day L, Gao SJ, Lieberman PM. 2006. Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi's sarcoma-associated herpesvirus lytic transcription. J. Virol. 80:5273–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao SJ, Zhang YJ, Deng JH, Rabkin CS, Flore O, Jenson HB. 1999. Molecular polymorphism of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent nuclear antigen: evidence for a large repertoire of viral genotypes and dual infection with different viral genotypes. J. Infect. Dis. 180:1466–1476 (Erratum, 180:1756.) [DOI] [PubMed] [Google Scholar]
- 22. Gasteiger EHC, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, 571–607 In Walker JM. (ed), The proteomics protocol handbook. Humana Press, Inc, Clifton, NJ [Google Scholar]
- 23. Gao S-J, Boshoff C, Jayachandra S, Weiss RA, Chang Y, Moore PS. 1997. KSHV ORF K9 (vIRF) is an oncogene that inhibits the interferon signaling pathway. Oncogene 15:1979–1986 [DOI] [PubMed] [Google Scholar]
- 24. Glenn M, Rainbow L, Aurad F, Davison A, Schulz TF. 1999. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J. Virol. 73:6953–6963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Canham M, Talbot SJ. 2004. A naturally occurring C-terminal truncated isoform of the latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus does not associate with viral episomal DNA. J. Gen. Virol. 85:1363–1369 [DOI] [PubMed] [Google Scholar]
- 26. Kellam P, Bourboulia D, Dupin N, Shotton C, Fisher C, Talbot S, Boshoff C, Weiss RA. 1999. Characterization of monoclonal antibodies raised against the latent nuclear antigen of human herpesvirus 8. J. Virol. 73:5149–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piolot T, Tramier M, Coppey M, Nicolas JC, Marechal V. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shinohara H, Fukushi M, Higuchi M, Oie M, Hoshi O, Ushiki T, Hayashi J, Fujii M. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917–12924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schwam DR, Luciano RL, Mahajan SS, Wong L, Wilson AC. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532–8540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garber AC, Shu MA, Hu J, Renne R. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cherezova L, Burnside KL, Rose TM. 2011. Conservation of complex nuclear localization signals utilizing classical and non-classical nuclear import pathways in LANA homologs of KSHV and RFHV. PLoS One 6:e18920 doi:10.1371/journal.pone.0018920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Komatsu T, Ballestas ME, Barbera AJ, Kelley-Clarke B, Kaye KM. 2004. KSHV LANA1 binds DNA as an oligomer and residues N-terminal to the oligomerization domain are essential for DNA binding, replication, and episome persistence. Virology 319:225–236 [DOI] [PubMed] [Google Scholar]
- 33. Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856–861 [DOI] [PubMed] [Google Scholar]
- 34. Kozak M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187–208 [DOI] [PubMed] [Google Scholar]
- 35. Kozak M. 1983. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol. Rev. 47:1–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gray NK, Wickens M. 1998. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 14:399–458 [DOI] [PubMed] [Google Scholar]
- 37. Kozak M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee S, Liu B, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U. S. A. 109:E2424–E2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozak M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jackson RJ, Kaminski A. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985–1000 [PMC free article] [PubMed] [Google Scholar]
- 41. Dominguez DI, Ryabova LA, Pooggin MM, Schmidt-Puchta W, Futterer J, Hohn T. 1998. Ribosome shunting in cauliflower mosaic virus. Identification of an essential and sufficient structural element. J. Biol. Chem. 273:3669–3678 [DOI] [PubMed] [Google Scholar]
- 42. Ryabova LA, Pooggin MM, Hohn T. 2006. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 119:52–62 [DOI] [PubMed] [Google Scholar]
- 43. Curran J, Kolakofsky D. 1988. Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J. 7:245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prats H, Kaghad M, Prats AC, Klagsbrun M, Lelias JM, Liauzun P, Chalon P, Tauber JP, Amalric F, Smith JA, et al. 1989. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc. Natl. Acad. Sci. U. S. A. 86:1836–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sadler R, Wu L, Forghani B, Renne R, Zhong W, Herndier B, Ganem D. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5722–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fouillot N, Tlouzeau S, Rossignol JM, Jean-Jean O. 1993. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J. Virol. 67:4886–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Farris KD, Pintel DJ. 2010. Adeno-associated virus type 5 utilizes alternative translation initiation to encode a small Rep40-like protein. J. Virol. 84:1193–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Corte-Real S, Collins C, Aires da Silva F, Simas JP, Barbas CF, III, Chang Y, Moore P, Goncalves J. 2005. Intrabodies targeting the Kaposi sarcoma-associated herpesvirus latency antigen inhibit viral persistence in lymphoma cells. Blood 106:3797–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802–805 [DOI] [PubMed] [Google Scholar]
- 50. Kwun HJ, da Silva SR, Qin H, Ferris RL, Tan R, Chang Y, Moore PS. 2011. The central repeat domain 1 of Kaposi's sarcoma-associated herpesvirus (KSHV) latency associated-nuclear antigen 1 (LANA1) prevents cis MHC class I peptide presentation. Virology 412:357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang YJ, Deng JH, Rabkin C, Gao SJ. 2000. Hot-spot variations of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen and application in genotyping by PCR-RFLP. J. Gen. Virol. 81:2049–2058 [DOI] [PubMed] [Google Scholar]
- 52. Chen ZJ. 2005. Ubiquitin signaling in the NF-κB pathway. Nat. Cell Biol. 7:758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gjoerup O, Chang Y. 2010. Update on human polyomaviruses and cancer. Adv. Cancer Res. 106:1–51 [DOI] [PubMed] [Google Scholar]
- 54. Brierley I, Dos Ramos FJ. 2006. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 119:29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Woodard C, Shamay M, Liao G, Zhu J, Ng AN, Li R, Newman R, Rho HS, Hu J, Wan J, Qian J, Zhu H, Hayward SD. 2012. Phosphorylation of the chromatin binding domain of KSHV LANA. PLoS Pathog. 8:e1002972 doi:10.1371/journal.ppat.1002972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dresang LR, Teuton JR, Feng H, Jacobs JM, Camp DG, 2nd, Purvine SO, Gritsenko MA, Li Z, Smith RD, Sugden B, Moore PS, Chang Y. 2011. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: insights on the detection and discovery of viral genes. BMC Genomics 12:625 doi:10.1186/1471-2164-12-625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gao SJ, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo CR, Saah A, Phair J, Detels R, Chang Y, Moore PS. 1996. KSHV antibodies among Americans, Italians, and Ugandans with or without Kaposi's sarcoma. Nat. Med. 2:925–928 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.