Abstract
Viruses in the Ebolavirus and Marburgvirus genera (family Filoviridae) have been associated with large outbreaks of hemorrhagic fever in human and nonhuman primates. The first documented cases occurred in primates over 45 years ago, but the amount of virus genetic diversity detected within bat populations, which have recently been identified as potential reservoir hosts, suggests that the filoviruses are much older. Here, detailed Bayesian coalescent phylogenetic analyses are performed on 97 whole-genome sequences, 55 of which are newly reported, to comprehensively examine molecular evolutionary rates and estimate dates of common ancestry for viruses within the family Filoviridae. Molecular evolutionary rates for viruses belonging to different species range from 0.46 × 10−4 nucleotide substitutions/site/year for Sudan ebolavirus to 8.21 × 10−4 nucleotide substitutions/site/year for Reston ebolavirus. Most recent common ancestry can be traced back only within the last 50 years for Reston ebolavirus and Zaire ebolavirus species and suggests that viruses within these species may have undergone recent genetic bottlenecks. Viruses within Marburg marburgvirus and Sudan ebolavirus species can be traced back further and share most recent common ancestors approximately 700 and 850 years before the present, respectively. Examination of the whole family suggests that members of the Filoviridae, including the recently described Lloviu virus, shared a most recent common ancestor approximately 10,000 years ago. These data will be valuable for understanding the evolution of filoviruses in the context of natural history as new reservoir hosts are identified and, further, for determining mechanisms of emergence, pathogenicity, and the ongoing threat to public health.
INTRODUCTION
Viruses of the Ebolavirus and Marburgvirus genera (family Filoviridae) are nonsegmented, negative-strand RNA viruses that are, with few exceptions, responsible for severe hemorrhagic fever outbreaks in Africa with high case fatality (1). Filovirus transmission among humans occurs through direct human-to-human contact or contact with their infectious bodily fluids, particularly in the late stages of infection, when viral loads are highest. Historically, the largest outbreaks have been health care facility based, with virus spread increased by poor barrier nursing techniques and reuse of needles or other medical devices in the absence of disinfection. Virus dissemination within communities is further exacerbated by common African burial practices that include communal washing of bodies, particularly by women, who are also the primary caregivers within families.
Within the genus Ebolavirus, five viruses are recognized (Ebola virus, Sudan virus, Reston virus, Taï Forest virus, and Bundibugyo virus), each representing a distinct virus species (Zaire ebolavirus, Sudan ebolavirus, Reston ebolavirus, Taï Forest ebolavirus, and Bundibugyo ebolavirus). In contrast to the genus Ebolavirus, the genus Marburgvirus contains a single virus species (Marburg marburgvirus), albeit consisting of two distinct viruses, Marburg virus and Ravn virus (2), which are approximately 20% divergent from one another (3). The filoviruses are primarily African in origin, with the exception of Reston virus, which so far has been found only in the Philippines or in nonhuman primates originating in the Philippines. In addition, an Ebola-like filovirus, Lloviu virus, was recently identified in bats in Spain (4) and likely represents another distinct genus.
While Ebola virus infections can result in a case fatality approaching 90% (1), the case fatalities associated with other viruses of the genus Ebolavirus appear to be considerably lower. The case fatality associated with Sudan virus infections ranges from 53 to 66% (5, 6), and that of Bundibugyo virus is estimated near 40% based on epidemiologic findings from the 2007 Uganda outbreak (7–9). Reston virus, while highly pathogenic for nonhuman primates, does not appear to cause disease in humans (10, 11). In the Philippines, several abattoir workers on infected pig farms were found to be seropositive for Reston virus yet reported no clinical symptoms (12). Finally, Taï Forest virus has been described only in a single nonfatal human case (13). Marburg and Ravn virus infections can result in case fatalities ranging from approximately 20% to 90% (3, 14, 15).
Genetic investigations of filovirus outbreaks have found them to be of two general types, those that are the result of a single introduction into the human population followed by person-to-person spread and those outbreaks that result from multiple introductions followed by short chains of human transmission. During person-to-person transmission, there appears to be little molecular evolution of the virus (3, 16). Initial introduction into the human population is often thought to result from contact with infected carcasses of nonhuman primates or other mammals or direct contact with an infected reservoir host (17–21). Despite numerous attempts to identify the natural reservoir(s) of the filoviruses over the past ≥30 years, only recently have bats been implicated as possible reservoirs for the ebolaviruses and marburgviruses (20–24). Over the past 10 years, filovirus RNA and antibodies have been detected in several bat species, but it was not until 2007 that Marburg and Ravn virus isolates were recovered from Egyptian fruit bats (Rousettus aegyptiacus) associated with a small outbreak of Marburg hemorrhagic fever in southwestern Uganda (21). In contrast to the low virus genetic diversity observed with person-to-person virus transmission during human outbreaks, potential bat reservoir populations appear to harbor genetically diverse virus populations at single geographical locations (20, 21).
Although the filoviruses are thought to have originated quite some time ago, age estimates have ranged from a few thousand (25, 26) to hundreds of thousands (4) to a few million (27, 28) years ago. Further confusing the issue, some authors have presented data indicating that viruses of at least one filovirus species (Zaire ebolavirus) share a most recent common ancestor (MRCA) in the very recent past (29–32). Previous studies have attempted to estimate rates of nonsynonymous substitutions and divergence times among viruses within the Filoviridae; however, most have been limited in terms of sample size (25) or have examined only a single species, in particular, Zaire ebolavirus (29, 31, 32). Here, we perform detailed Bayesian coalescent phylogenetic analyses on 97 virus whole-genome sequences (55 of which are newly reported here) to comprehensively examine the evolutionary rates of the filoviruses and estimate dates of common ancestry within this family. Such information is valuable for understanding the evolution of viruses in this family in the context of natural history, pathogenicity, and ongoing threat.
MATERIALS AND METHODS
Viral RNA extraction, reverse transcription-PCR (RT-PCR), and whole-genome sequencing by primer walking were performed as previously described (3, 33) or as follows, with primers specific for each virus species. The Ebola virus genome was amplified in seven overlapping RT-PCR fragments (see Table S1 in the supplemental material). The Sudan virus genome was amplified in five overlapping fragments, with a few exceptions (see Table S1 in the supplemental material). In some cases, fragment A was amplified in 2 smaller fragments (A1 and A2). In other cases, nested reactions were performed (fragments C and D) using the original fragment primers found in Table S1 in the supplemental material for the first round and a second set of primers (also shown in Table S1 in the supplemental material) for the nested reaction.
Individual data sets were constructed for each virus species and for the family as a whole (see Table S2 in the supplemental material). Bundibugyo and Taï Forest viruses and the recently described Lloviu virus were excluded from intraspecific analyses since only a single genomic sequence has been generated for each virus. These viruses were, however, included in analyses examining the family Filoviridae as a whole. Multiple sequence alignments were generated by Clustal X (34) or the MAFFT function (35) in SeaView (36) with subsequent proofing by eye.
Bayesian coalescent phylogenetic analysis, implemented in the BEAST software package, was used to determine the molecular evolutionary rate and an estimate of the time to the most recent common ancestor for each of the data sets (37). The HKY+G nucleotide substitution model was chosen for its simplicity, to avoid overparameterizing the analyses, and for direct comparison among data sets. Preliminary analyses consisting of 10,000,000 generations were performed to identify the most appropriate clock (strict versus relaxed uncorrelated lognormal versus relaxed uncorrelated exponential) and demographic (constant versus Bayesian skyline population size) models (38) for each species-specific data set. A Yule process, rather than a coalescent prior, was chosen to more accurately account for the speciation process when analyzing the family Filoviridae as a whole. Model selection was based on an analysis of marginal likelihoods (39), calculated in Tracer version 1.5. Once the nucleotide substitution, molecular clock, and demographic models were chosen (Table 1), final analyses were run for a total of 10,000,000 to 390,000,000 generations to ensure effective sample sizes (ESSs) of at least 200. Maximum clade credibility trees were constructed using the software programs TreeAnnotator and FigTree (37). Additionally, the Path-O-Gen program (available from http://tree.bio.ed.ac.uk/software/pathogen/) was used to run root-to-tip regressions in order to independently test the assumption of a molecular clock (i.e., temporal signal) for each of the individual species data sets.
Table 1.
Parameters used in Bayesian coalescent phylogenetic analyses for each of the filovirus data setsa
| Species | Substitution model | Clock model | Population model |
|---|---|---|---|
| Zaire ebolavirus | HKY+G | Relaxed uncorrelated lognormal | Constant |
| Sudan ebolavirus | HKY+G | Strict | Constant |
| Reston ebolavirus | HKY+G | Strict | Constant |
| Taï Forest ebolavirus | NA | NA | NA |
| Bundibugyo ebolavirus | NA | NA | NA |
| Marburg marburgvirus | HKY+G | Relaxed uncorrelated exponential | Constant |
| Family Filoviridae | HKY+G | Strict | Yule |
NA, not applicable.
Nucleotide sequence accession numbers.
Sequences generated in this study were deposited in GenBank under accession numbers KC242783 to KC242801, JX477165, JX477166, and JX458825 to JX458858.
RESULTS AND DISCUSSION.
Zaire ebolavirus.
The Zaire ebolavirus data set consisted of 22 Ebola virus whole-genome sequences. These included samples from outbreaks in the Democratic Republic of Congo (DRC) (formerly Zaire) that were collected in the mid- to late 1970s near Yambuku (3 samples), in the mid-1990s around Kikwit (3 samples), and most recently between 2007 and 2008 near Luebo, Democratic Republic of Congo (9 samples). Seven additional sequences were generated from cases collected in Gabon during the mid-1990s and in 2002. The outbreaks in Yambuku and Kikwit were the largest known Ebola virus outbreaks on record (>300 cases each). Both outbreaks were hospital based and likely the result of single virus introductions into the human population followed by rapid spread between patients, health care workers, and their families. Sequencing of portions of the highly variable glycoprotein (GP) gene from fatal and nonfatal patients at the beginning and end of the Kikwit outbreak found no sequence variation whatsoever (16). In contrast, the recurrent but smaller outbreaks in Gabon were characterized by shorter transmission chains of multiple virus lineages and were epidemiologically and genetically linked to repeated contact with infected nonhuman primates or other mammals scavenged (or hunted) in the nearby forests (18).
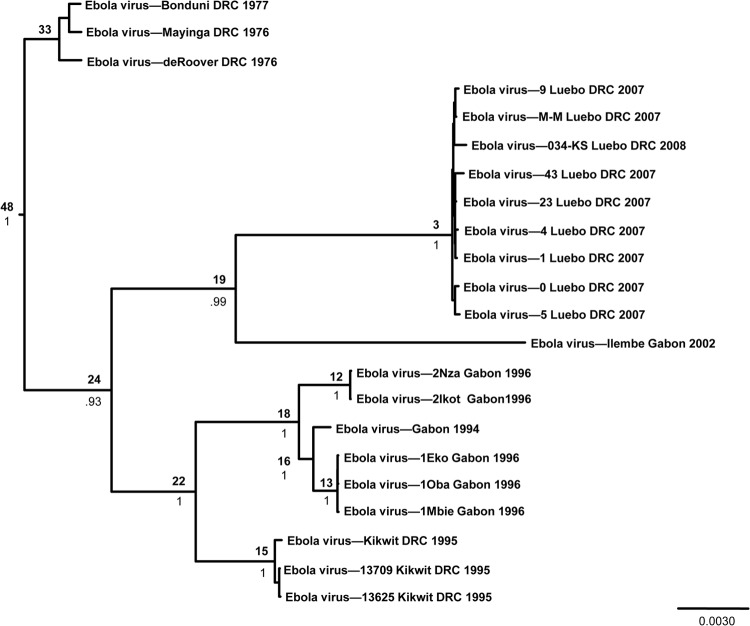
The overall genetic diversity within the Zaire ebolavirus species is low, with a maximum 2.7% nucleotide difference between sequences. Within a single outbreak, genetic diversity was even lower. For example, sequences from patients in Luebo, Democratic Republic of Congo, collected in 2007 and 2008 were less than 0.07% divergent. While a degree of spatial clustering was apparent due to the inherent nature of human outbreaks, viruses clustered temporally (Fig. 1). Interestingly, viruses from the Democratic Republic of Congo fell into three separate clades: those from the 1970s occupied the basal position of the tree, whereas viruses in the remaining two clades were sister to viruses collected from Gabon during similar time spans (i.e., 1994 to 1996 and 2002 to 2008). A similar pattern was observed among viruses collected in the Democratic Republic of Congo between 1976 and 2008 when examining the glycoprotein (GP) and nucleoprotein (NP) genes (30); however, those results suggested that viruses from Luebo shared greater similarity with a virus collected in 1976 in Yambuku than with a 1995 virus from Kikwit (30). Our results, based on whole-genome sequences, suggested that these viruses were equidistant (approximately 2% divergent).
Fig 1.
Bayesian coalescent analysis of Ebola virus. The maximum clade credibility tree is shown with the MRCA, in years before 2008, at each node. Posterior probability values are shown beneath MRCA estimates. Scale is in substitutions/site.
Bayesian coalescent analyses estimated that the most recent common ancestor (MRCA) of these extant viruses occurred around 48 years prior to 2008, or around 1960 (Table 2 and Fig. 1), 16 years before Ebola virus was first recognized (40, 41). In addition, our analyses suggested that viruses similar to those found during the Luebo outbreak may have been circulating since 2005, 2 years before the outbreak was first recognized. The molecular evolutionary rate was relatively rapid and was estimated at 7.06 × 10−4 nucleotide substitutions/site/year (Table 2). An independent root-to-tip regression analysis using the Path-O-Gen program resulted in an estimate contained within the 95% highest posterior density (HPD) interval from the Bayesian coalescent analysis (2.2 × 10−4 nucleotide substitutions/site/year; correlation coefficient = 0.89). While it is possible that Ebola virus emerged only recently, the low level of genetic diversity despite a moderately fast molecular evolutionary rate, coupled with the previously hypothesized ancestral age of the filoviruses overall, suggests that Ebola virus has more likely undergone a recent genetic bottleneck, a hypothesis that others have proposed as well (29, 30, 32). Given the dependence of a zoonotic virus on its host population, it is easy to envision a scenario in which the number of susceptible hosts declines, for a variety of possible reasons, and results in a small effective viral population size and a subsequent loss of genetic diversity that would limit our ability to trace the virus back through time. Alternatively, the virus may have jumped into a new host or into a new geographic area in the recent past, resulting in a founder effect (29).
Table 2.
Molecular evolutionary rate and TMRCA for viruses belonging to each of the filovirus speciesa
| Species | Yr of first documented outbreak | TMRCA estimate (yr) | No. of yrs before the present for TMRCA estimate | Substitution rate (× 10−4 nucleotide substitutions/site/yr) |
|---|---|---|---|---|
| Zaire ebolavirus | 1976 | 1960 | 48 (32–65) | 7.06 (1.97–10.7) |
| Sudan ebolavirus | 1976 | 1173 | 838 (290–1,452) | 0.46 (0.14–0.78) |
| Reston ebolavirus | 1989 | 1979 | 30 (27–33) | 8.21 (7.30–9.18) |
| Taï Forest ebolavirus | 1994 | NA | NA | NA |
| Bundibugyo ebolavirus | 2007 | NA | NA | NA |
| Marburg marburgvirus | 1967 | 1302 | 707 (113–1,423) | 5.67 (0.49–8.97) |
TMRCA, time to most recent common ancestor. Values in parentheses are 95% highest posterior density (HPD) intervals.
Sudan ebolavirus.
Sudan virus first emerged in 1976 in Nzara, southern Sudan, where it appeared again in 1979. Both outbreaks were hospital based, with the 1976 outbreak causing 284 cases, 53% of which were fatal. Sudan virus did not reappear until 2000, when it was found to be the agent responsible for the largest ebolavirus outbreak on record, 425 cases, in Gulu, Uganda. Genetic studies of the Gulu outbreak found no evidence of virus evolution during human-to-human transmission or between viruses from fatal or nonfatal cases (6). Since 2000, Sudan virus has emerged two more times, causing a limited outbreak in 2004 in Yambio, Sudan (only 25 km from Nzara), and a single case in 2011 in Luwero district outside Kampala, Uganda. The full-length genome sequences of these viruses are examined here. Compared to viruses within the species Zaire ebolavirus, those within the species Sudan ebolavirus appear to be much more geographically constrained, with all five outbreaks occurring within a 400-mile range, presumably a reflection of the limited distribution of the natural reservoir or other ecological constraints.
The Sudan ebolavirus data set consisted of 5 Sudan virus whole-genome sequences, one from each outbreak. Interestingly, viruses clustered spatially rather than temporally, with the Ugandan isolates forming a well-supported clade distinct from those collected in Sudan (Fig. 2). Genetic diversity of the isolates collected within each country was low (less than 0.4% for Sudan and approximately 0.6% for Uganda) and, in the case of viruses collected in Sudan, highlights the ability of this species to maintain genetic stability over a nearly 30-year span. The overall diversity of viruses within the species was also low, with a maximum of 5.2% nucleotide divergence between samples collected in Sudan in 2004 and Uganda in 2011.
Fig 2.
Bayesian coalescent analysis of Sudan virus. The maximum clade credibility tree is shown with the MRCA, in years before 2011, at each node. Posterior probability values are shown beneath MRCA estimates. Scale is in substitutions/site.
The molecular evolutionary rate was estimated at 0.46 × 10−4 nucleotide substitutions/site/year, and viruses within this species appear to have shared a MRCA more than 800 years before the present (Table 2 and Fig. 2). A similar rate, 0.39 × 10−4 nucleotide substitutions/site/year, was obtained from the root-to-tip regression (correlation coefficient = 0.89). This evolutionary rate estimate is much slower than that calculated for the more pathogenic Ebola virus and suggests that Sudan virus may be significantly older. Alternatively, given the hypothesis that Ebola virus has undergone a recent genetic bottleneck, it may just be that we are able to trace the MRCA further back for Sudan virus. Analyses suggested that the virus isolates from Sudan shared a MRCA approximately 79 years before 2011 while the Ugandan viruses shared a MRCA around 99 years ago (Fig. 2).
Given the close geographic proximity of northern Uganda and southern Sudan, it is interesting to see such a genetic division between samples from the two neighboring countries. According to these analyses, the virus has been circulating for hundreds of years, with the extant lineages arising around 100 years before the present. This would appear to be ample time for the viruses to cocirculate and may suggest that some unknown ecological constraint has kept these two lineages from becoming panmictic or genetically homogeneous over time. It is possible that the two lineages are being maintained in different hosts—either in distinct species or subspecies or in populations that have been geographically isolated. The Albertine Rift, which runs along the border of the Democratic Republic of Congo and Uganda, Rwanda, etc., and the mountains that flank this region (the Virungas in eastern Democratic Republic of Congo, the Ruwenzoris in western Uganda, etc.) have acted as biogeographic barriers for a number of small mammals and birds historically (42–47), so it is not unreasonable to think that this region may serve as a biogeographic barrier for Sudan virus and/or its reservoir host as well. Additional samples of Sudan virus will be needed to explore this issue further given the small sample size currently available.
Reston ebolavirus.
To date, the origins of all known Reston virus outbreaks have been epidemiologically linked to the Philippines. First appearing in 1989 in infected monkeys consigned from a breeding/export facility in the Philippines to laboratories in Reston, VA, the virus was highly lethal in the imported animals but caused asymptomatic infections in four animal care workers that worked with the monkeys (48, 49). The same Philippine export facility was the source of two more outbreaks following the export of infected monkeys to Sienna, Italy, in 1992 and Alice, TX, in 1996. More recently, in 2008 and 2009, Reston virus appeared repeatedly in Philippine swine in association with high-density pig farming operations. Interestingly, follow-up studies found that pigs experimentally infected with Reston virus (50) can shed virus from the mucosa in the nasopharynx in the absence of clinical disease, consistent with the possibility that the virus could circulate undetected in pigs. The Reston ebolavirus data set analyzed here included 7 Reston virus whole-genome sequences, each from outbreaks with epidemiologic linkage to the Philippines. Four of these viruses were collected from pigs during the 2008/2009 outbreak in the Philippines, with the other 3 sequences representing isolates associated with animals that were imported from the Philippines and subsequently involved in nonhuman primate outbreaks in Pennsylvania in 1989/1990 and Texas in 1996.
In the recent Reston virus outbreaks in the Philippines, there appears to be no temporal or spatial clustering patterns. The virus genetic diversity found on the same farm (farm A) from 2008 to 2009 (a 1-year period) was 0.079%, while simultaneous sampling in 2008 of a single farm (farm CE) found the divergence to be approximately 4.5%. Further interpretations of these data are difficult in the absence of detailed records of porcine movement between farms before and during these outbreak periods. Similar to Ebola virus, it appears as though Reston virus is evolving at a rate (8.21 × 10−4 nucleotide substitutions/site/year) significantly faster than that of Sudan virus (Table 2). The root-to-tip regression suggested a highly similar rate estimate of 8.5 × 10−4 nucleotide substitutions/site/year (correlation coefficient = 0.99). Likewise, extant Reston viruses share an ancestor in the recent past, approximately 30 years before 2009 (Table 2 and Fig. 3), or in 1979, 10 years prior to the initial identification of Reston virus in a shipment of cynomolgus monkeys from the Philippines to the United States (11). Again, based on the assumption that the filoviruses are ancient viruses, it appears likely that Reston virus (like Ebola virus) has also undergone a recent genetic bottleneck. During the late 1970s, there was an increase in human population in the Philippines along with an increase in logging and deforestation (see the report at www.fao.org/docrep/003/X6967E/x6967e07.htm). It is possible that the reservoir of Reston virus was negatively affected by these changes, potentially resulting in a genetic bottleneck. Alternatively, perhaps the virus jumped into pigs around that time, and based on the demonstrated ability of experimentally infected pigs to shed virus in the absence of disease, the virus has circulated among pigs since that time, with nonhuman primates serving as accidental or spillover hosts. Another alternative is that the bottleneck indicates a founder effect from a recent introduction into the Philippines, possibly from mainland Asia, given that the Philippines is an island nation, or even from shipments of monkeys sent from Africa as part of the exotic pet trade.
Fig 3.
Bayesian coalescent analysis of Reston virus. The maximum clade credibility tree is shown with the MRCA, in years before 2009, at each node. Posterior probability values are shown beneath MRCA estimates. Scale is in substitutions/site.
Marburg marburgvirus.
The Marburg marburgvirus data set included 60 sequences consisting of virus genomes from 48 human samples and 12 bat isolates. Of these full-length sequences, 34 (27 human and 7 bat) are newly reported here. Marburg virus was first identified in 1967 in monkeys consigned from Uganda to Europe (19), while Ravn virus was identified from a fatal human case in Kenya 20 years later. More recently, the cave-dwelling Egyptian fruit bat, Rousettus aegyptiacus, has been identified as a potential natural reservoir for Marburg and Ravn viruses (Marburg marburgvirus), consistent with historical linkage of Marburg hemorrhagic fever (MHF) outbreaks directly to subterranean environments, particularly African gold mines or caves, where these bats can live in high-density colonies ranging in size up to several hundred thousand. Outbreaks among gold miners in Durba, DRC, from 1998 to 2000 and Ibanda, Uganda, in 2007 consisted of at least 11 combined spillover events from the natural reservoir, followed by short chains of human-to-human transmission.
The largest Marburg virus outbreak on record, consisting of 252 cases and 227 deaths, occurred in Angola in 2005. The origin of the Angola outbreak was never determined, but epidemiologic investigations found health clinics to be implicated as primary conduits for virus dissemination. Genetic studies, like those for the Ebola virus outbreak in Kikwit and the Sudan virus outbreak in Gulu, found very little virus evolution during the outbreak, with some whole-genome sequences (19,114 nucleotides [nt]) in the Angola outbreak showing 100% identity despite coming from patients infected 6 weeks apart.
As others have demonstrated previously, genetic diversity between Ravn virus and Marburg virus was high, at approximately 20% (3, 20, 21). Despite significant divergence at the nucleotide level, Marburg and Ravn viruses still exhibit a high proportion of amino acid conservation, with the exception of the glycoprotein gene (3), a finding which adds to the body of evidence supporting these viruses as members of a single species. Ecological investigations (20, 21) have found the entire known genetic spectrum of this species to simultaneously circulate among R. aegyptiacus bats at any one location, including both Ravn and Marburg viruses.
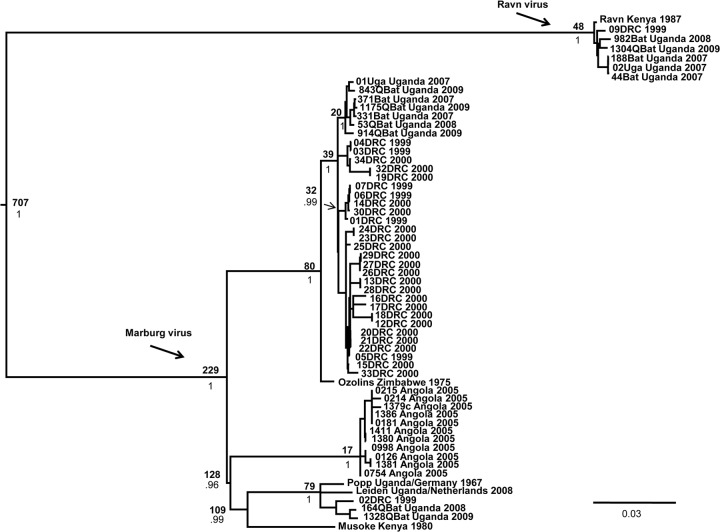
Aside from the human outbreak in Angola in 2005, temporal and spatial structure was nonexistent. As mentioned above, multiple distinct lineages of the Marburg marburgvirus species, including both Marburg and Ravn viruses, were identified in single caves associated with large bat populations in both Uganda and the Democratic Republic of Congo (Fig. 4, note Bat sequences). Others have shown the same relationship based on partial genetic sequences (20, 21), indicating the potential for a long-term association between Marburg and Ravn viruses and their hypothesized reservoir hosts. The lack of spatial and temporal structure also suggests that the viruses may have a long evolutionary history within the region and have had sufficient time to geographically mix (panmixia). Indeed, this fits with the hypothesis of bats as the reservoir host. High contact rates due to sheer density among these cave-dwelling bats could easily account for the levels of transmission necessary to maintain the virus in the environment over time. Furthermore, given the vagility or potential movement of bats in general, it is not surprising to expect significant gene flow among viruses associated with the bats, even over a potentially large region. What remains a mystery, however, is the maintenance of two distinct viruses in the absence of spatial structure.
Fig 4.
Bayesian coalescent analysis of viruses of the species Marburg marburgvirus. The maximum clade credibility tree is shown with the MRCA, in years before 2009, at each node. Posterior probability values are shown beneath MRCA estimates. Scale is in substitutions/site.
The molecular evolutionary rate for Marburg and Ravn viruses was estimated at 5.67 × 10−4 nucleotide substitutions/site/year (Table 2), which is moderate in relation to Ebola and Reston viruses but still much quicker than the molecular evolutionary rate of Sudan virus. The estimate obtained from the root-to-tip regression (2.0 × 10−4 nucleotide substitutions/site/year; correlation coefficient = 0.76) was contained within the 95% HPD interval generated in the Bayesian coalescent analysis. The estimated time to the MRCA for the Marburg and Ravn viruses overall was approximately 700 years before 2009, with the Ravn viruses sharing a MRCA much more recently, approximately 48 years ago (Fig. 4). Marburg viruses may be older, as they share a MRCA over 200 years ago (Fig. 4).
For all virus species, not just Marburg marburgvirus, it is possible that phylogenetic dating methodologies might underestimate the age of older viral divergence events due to saturation of synonymous substitutions (26), resulting in a loss of the true extent of evolutionary change over time. This is especially true for more-variable genes, like the glycoprotein gene, that undergo strong purifying selection (26, 51).
Filoviridae.
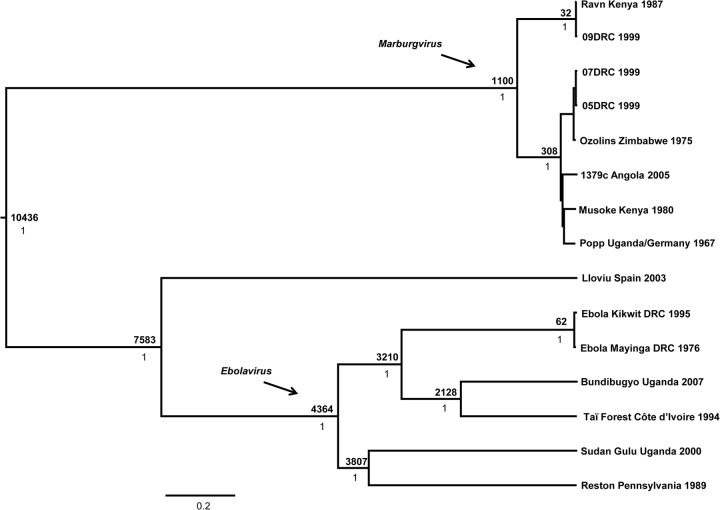
Finally, in order to examine overall evolutionary history of the family Filoviridae, we constructed a data set consisting of representatives from each species within the Ebolavirus and Marburgvirus genera as well as the recently described Lloviu virus. This method assumes that each sequence represents a distinct species; therefore, one representative per species was selected for inclusion in the analysis, with a couple of exceptions. In the case of the Marburg marburgvirus species, we included several Marburg and Ravn virus representatives to account for the vast amount of genetic diversity present within the species. In addition, we included 2 representatives of Ebola virus to test whether the overall family tree would capture a time to MRCA similar to the estimate generated by the single-species data set. The molecular evolutionary rate was estimated at 8.95 × 10−4 (95% HPD interval, 0.94 × 10−4 to 2.33 × 10−4) nucleotide substitutions/site/year, slightly faster than but similar to the rates estimated for Ebola virus and Reston virus. According to our analyses, which were designed to take into account the patterns associated with speciation processes, particularly emergence and extinction events, the family Filoviridae shares a MRCA approximately 10,400 (95% HPD interval, 6,535 to 16,244) years ago (Fig. 5), which is about the time the Earth emerged from the last ice age. Our estimate is similar to a previous calculation (7,100 to 7,900 years) based on synonymous and nonsynonymous substitution ratios (25) and far younger than other recent assessments using non-clock-based methods that rely on the assumption of filovirus-like elements that appear to have been integrated into the genome of a variety of mammals and subjected to different evolutionary pressures (27, 28). Additional approximations of MRCA that resulted in older estimates than those generated in this study differed in terms of more-limited numbers of whole-genome taxa as well as methodology (4, 26). One such study (4) employed a coalescent prior, which is most suitable for analyzing relationships among members of the same population or species, to generate a hypothesized date of MRCA for the family. Our study utilized a Yule prior, which is most suitable for examining relationships among members of different species.
Fig 5.
Bayesian coalescent analysis of viruses of the family Filoviridae. The maximum clade credibility tree is shown with the MRCA, in years before 2007, at each node. Posterior probability values are shown beneath MRCA estimates. Scale is in substitutions/site.
Another study (26) suggested that high levels of sequence divergence and potential saturation issues may bias Bayesian estimates toward younger age estimations, and this may be the case with our data set as well; however, that particular study examined only viruses of the genus Ebolavirus. At the very least, our analysis provides another hypothesis and a minimum estimate of most recent common ancestry for extant viruses within the family Filoviridae. Our results correspond well with at least one previous estimate (25) for the group, and our family-wide analysis was able to recover an estimate of MRCA for Ebola virus very similar to that of the analysis examining Ebola virus alone, indicating a good degree of robustness in the data. Regardless, filoviruses are clearly much older than what their initial detection in the 1960s might suggest. The species appear to be evolving continuously, with viruses of some species (Zaire ebolavirus and Reston ebolavirus, for example) likely undergoing recent genetic bottlenecks. The processes contributing to the genetic bottlenecks of these viruses, yet not those of others, remain unknown and highlight the importance of examining the biology of the filoviruses as well as their distinct reservoir hosts. As we learn more about the reservoirs of the other filoviruses, we will be able to use characteristics of the hosts to make predictions regarding the viruses. For example, the range of the virus will depend on the range of the host to some extent, along with other external abiotic factors that influence the host directly (and, thus, the pathogen indirectly). Furthermore, virus evolution clearly depends on the host—for the number of susceptible hosts available for the virus to infect, the number of potential opportunities that a virus has for transmission due to the social or sexual interactions of the host species, and more. Much work remains to decipher exactly how these viruses have evolved over time and in relation to their host species.
Supplementary Material
ACKNOWLEDGMENTS
We thank Patricia Leman for laboratory assistance and analysis.
The findings and conclusions reported here do not necessarily represent the views of the Centers for Disease Control and Prevention or the Department of Health and Human Services.
Footnotes
Published ahead of print 19 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03118-12.
REFERENCES
- 1. Sanchez A, Geisbert TW, Feldmann H. 2007. Filoviridae: Marburg and Ebola Viruses, p 1409–1448 In Knipe DM, Howley PM. (ed), Fields virology. Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 2. Kuhn JH, Becker S, Ebihara H, Geisbert TW, Johnson KM, Kawaoka Y, Lipkin WI, Negredo AI, Netesov SV, Nichol ST, Palacios G, Peters CJ, Tenorio A, Volchkov VE, Jahrling PB. 2010. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 155:2083–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Towner JS, Khristova ML, Sealy TK, Vincent MJ, Erickson BR, Bawiec DA, Hartman AL, Comer JA, Zaki SR, Stroher U, Gomes DA Silva F, del Castillo F, Rollin PE, Ksiazek TG, Nichol ST. 2006. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J. Virol. 80:6497–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Negredo A, Palacios G, Vazquez-Moron S, Gonzalez F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, de la Cruz Martinez M, Herrera JE, Pizarro M, Hutchison SK, Echevarria JE, Lipkin WI, Tenorio A. 2011. Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog. 7:e1002304 doi:10.1371/journal.ppat.1002304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baron RC, McCormick JB, Zubeir OA. 1983. Ebola virus disease in southern Sudan: hospital dissemination and intrafamilial spread. Bull. World Health Organ. 61:997–1003 [PMC free article] [PubMed] [Google Scholar]
- 6. Towner JS, Rollin PE, Bausch DG, Sanchez A, Crary SM, Vincent M, Lee WF, Spiropoulou CF, Ksiazek TG, Lukwiya M, Kaducu F, Downing R, Nichol ST. 2004. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J. Virol. 78:4330–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. MacNeil A, Farnon EC, Wamala J, Okware S, Cannon DL, Reed Z, Towner JS, Tappero JW, Lutwama J, Downing R, Nichol ST, Ksiazek TG, Rollin PE. 2010. Proportion of deaths and clinical features in Bundibugyo Ebola virus infection, Uganda. Emerg. Infect. Dis. 16(12):1969–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Towner JS, Sealy TK, Khristova ML, Albarino CG, Conlan S, Reeder SA, Quan P-L, Lipkin WI, Downing R, Tappero JW, Okware S, Lutwama J, Bakamutumaho B, Kayiwa J, Comer JA, Rollin PE, Ksiazek TG, Nichol ST. 2008. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 4:e1000212 doi:10.1371/journal.ppat.1000212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wamala JF, Lukwago L, Malimbo M, Nguku P, Yoti Z. 2010. Ebola hemorrhagic fever associated with novel virus strain, Uganda 2007-2008. Emerg. Infect. Dis. 16:1087–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barrette RW, Xu L, Rowland JM, McIntosh MT. 2011. Current perspectives on the phylogeny of Filoviridae. Infect. Genet. Evol. 11(7):1514–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jahrling PB, Geisbert TW, Dalgard DW, Johnson ED, Ksiazek TG, Hall WC, Peters CJ. 1990. Preliminary report: isolation of Ebola virus from monkeys imported to the USA. Lancet 335:502–505 [DOI] [PubMed] [Google Scholar]
- 12. Barrette RW, Metwally SA, Rowland JM, Xu L, Zaki SR, Nichol ST, Rollin PE, Towner JS, Shieh W-J, Batten B, Sealy TK, Carrillo C, Moran KE, Bracht AJ, Mayr GA, Sirios-Cruz M, Catbagan DP, Lautner EA, Ksiazek TG, White WR, McIntosh MT. 2009. Discovery of swine as a host for the Reston ebolavirus. Science 325:204–206 [DOI] [PubMed] [Google Scholar]
- 13. Le Guenno B, Formenty P, Wyers M, Gounon P, Walker F, Boesch C. 1995. Isolation and partial characterization of a new strain of Ebola virus. Lancet 345:1271–1274 [DOI] [PubMed] [Google Scholar]
- 14. Bausch DG, Borchert M, Grein T, Roth C, Swanepoel R, Libande ML, Talarmin A, Bertherat E, Muyembe-Tamfum JJ, Tugume B, Colebunders R, Konde KM, Pirard P, Olinda LL, Rodier GR, Campbell P, Tomori O, Ksiazek TG, Rollin PE. 2003. Risk factors for Marburg hemorrhagic fever, Democratic Republic of the Congo. Emerg. Infect. Dis. 9:1531–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martini GA, Siegert R. 1971. Preface, p v In Martini GA, Siegert R. (ed), Marburg virus disease. Springer-Verlag, New York, NY [Google Scholar]
- 16. Rodriguez LL, De Roo A, Guimard Y, Trappier SG, Sanchez A, Bressler D, Williams AJ, Rowe AK, Bertolli J, Khan AS, Ksiazek TG, Peters CJ, Nichol ST. 1999. Persistence and genetic stability of Ebola virus during the outbreak in Kikwit, Democratic Republic of the Congo, 1995. J. Infect. Dis. 179(Suppl 1):S170–S176 [DOI] [PubMed] [Google Scholar]
- 17. Leroy EM, Epelboin A, Mondonge V, Pourrut X, Gonzalez JP, Muyembe-Tamfum JJ, Formenty P. 2009. Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, Democratic Republic of Congo, 2007. Vector Borne Zoonotic Dis. 9:723–728 [DOI] [PubMed] [Google Scholar]
- 18. Leroy EM, Rouquet P, Formenty P, Souquiere S, Kilbourne A, Froment JM, Bermejo M, Smit S, Karesh W, Swanepoel R, Zaki SR, Rollin PE. 2004. Multiple Ebola virus transmission events and rapid decline of Central African wildlife. Science 303:387–390 [DOI] [PubMed] [Google Scholar]
- 19. Smith MW. 1982. Field aspects of the Marburg virus outbreak: 1967. Primate Supply 7:11–15 [Google Scholar]
- 20. Swanepoel R, Smit SB, Rollin PE, Formenty P, Leman PA, Kemp A, Burt FJ, Grobbelaar AA, Croft J, Bausch DG, Zeller H, Leirs H, Braack LEO, Libande ML, Zaki S, Nichol ST, Ksiazek TG, Paweska JT, International Scientific and Technical Committee for Marburg Hemorrhagic Fever Control in the Democratic Republic of the Congo 2007. Studies of reservoir hosts for Marburg virus. Emerg. Infect. Dis. 13:1847–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Towner JS, Amman BR, Sealy TK, Carroll SAR, Comer JA, Kemp A, Swanepoel R, Paddock CD, Balinandi S, Khristova ML, Formenty PBH, Albarino CG, Miller DM, Reed ZD, Kayiwa JT, Mills JN, Cannon DL, Greer PW, Byaruhanga E, Farnon EC, Atimnedi P, Okware S, Katongole-Mbidde E, Downing R, Tappero JW, Zaki SR, Ksiazek TG, Nichol ST, Rollin PE. 2009. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 5:e1000536 doi:10.1371/journal.ppat.1000536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Delicat A, Paweska JT, Gonzalez JP, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576 [DOI] [PubMed] [Google Scholar]
- 23. Pourrut X, Souris M, Towner JS, Rollin PE, Nichol ST, Gonzalez JP, Leroy E. 2009. Large serological survey showing cocirculation of Ebola and Marburg viruses in Gabonese bat populations, and a high seroprevalence of both viruses in Rousettus aegyptiacus. BMC Infect. Dis. 9:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Towner JS, Pourrut X, Albarino CG, Nkogue CN, Bird BH, Grard G, Ksiazek TG, Gonzalez JP, Nichol ST, Leroy EM. 2007. Marburg virus infection detected in a common African bat. PLoS One 2:e764 doi:10.1371/journal.pone.0000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suzuki Y, Gojobori T. 1997. The origin and evolution of Ebola and Marburg viruses. Mol. Biol. Evol. 14:800–806 [DOI] [PubMed] [Google Scholar]
- 26. Wertheim JO, Kosakovsky Pond SL. 2011. Purifying selection can obscure the ancient age of viral lineages. Mol. Biol. Evol. 28:3355–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taylor DJ, Dittmar K, Ballinger MJ, Bruenn JA. 2011. Evolutionary maintenance of filovirus-like genes in bat genomes. BMC Evol. Biol. 11:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taylor DJ, Leach RW, Bruenn J. 2010. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol. Biol. 10:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Biek R, Walsh PD, Leroy EM, Real LA. 2006. Recent common ancestry of Ebola Zaire virus found in a bat reservoir. PLoS Pathog. 2:e90 doi:10.1371/journal.ppat.0020090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grard G, Biek R, Muyembe-Tamfum JJ, Fair J, Wolfe N, Formenty P, Paweska J, Leroy E. 2011. Emergence of divergent Zaire Ebola virus strains in Democratic Republic of the Congo in 2007 and 2008. J. Infect. Dis. 204(Suppl 3):S776–S784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walsh PD, Biek R, Real LA. 2005. Wave-like spread of Ebola Zaire. PLoS Biol. 3:e371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wittmann TJ, Biek R, Hassanin A, Rouquet P, Reed P, Yaba P, Pourrut X, Real LA, Gonzalez JP, Leroy EM. 2007. Isolates of Zaire ebolavirus from wild apes reveal genetic lineage and recombinants. Proc. Natl. Acad. Sci. U. S. A. 104:17123–17127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Towner JS, Sealy TK, Ksiazek TG, Nichol ST. 2007. High-throughput molecular detection of hemorrhagic fever virus threats with applications for outbreak settings. J. Infect. Dis. 196(Suppl 2):S205–S212 [DOI] [PubMed] [Google Scholar]
- 34. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galtier N, Gouy M, Gautier C. 1996. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543–548 [DOI] [PubMed] [Google Scholar]
- 37. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88 doi:10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suchard MA, Weiss RE, Sinsheimer JS. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18:1001–1013 [DOI] [PubMed] [Google Scholar]
- 40. Emond RT, Evans B, Bowen BT, Lloyd G. 1977. A case of Ebola virus infection. Br. Med. J. 2:541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson KM. 1978. Ebola haemorrhagic fever in Zaire, 1976. Bull. World Health Organ. 56:271–293 [PMC free article] [PubMed] [Google Scholar]
- 42. Bowie CK, Fjeldsa J, Hackett SJ, Bates JM, Crowe TM. 2006. Coalescent models reveal the relative roles of ancestral polymorphism, vicariance, and dispersal in shaping phylogeographical structure of an African montane forest robin. Mol. Phylogenet. Evol. 38:171–188 [DOI] [PubMed] [Google Scholar]
- 43. Carleton M, Kerbis Peterhans JC, Stanley WT. 2006. Review of the Hylomyscus denniae group (Rodentia: Muridae) in eastern Africa, with comments on the generic allocation of Epimys endorobae Heller. Proc. Biol. Soc. Washington 119:293–325 [Google Scholar]
- 44. Carleton MD, Stanley WT. 2005. Review of the Hylomyscus denniae complex (Rodentia: Muridae) in Tanzania, with description of a new species. Proc. Biol. Soc. Washington 118:619–646 [Google Scholar]
- 45. Fahr J, Vierhaus H, Hutterer R, Kock D. 2002. A revision of the Rhinolophus maclaudi species group with the description of a new species from West Africa (Chiroptera: Rhinolophidae). Myotis 40:95–126 [Google Scholar]
- 46. Huhndorf MH, Kerbis Peterhans JC, Loew SS. 2007. Comparative phylogeography of three endemic rodents from the Albertine Rift, east central Africa. Mol. Ecol. 16:663–674 [DOI] [PubMed] [Google Scholar]
- 47. Taylor PJ, Maree S, Van Sandwyk J, Kerbis Peterhans JC, Stanley WT, Verheyen E, Kaliba P, Verheyen W, Kaleme P, Bennett NC. 2009. Speciation mirrors geomorphology and palaeoclimatic history in African laminate-toothed rats (Muridae: Otomyini) of the Otomys denti and Otomys lacustris species-complexes in the ‘Montane Circle’ of East Africa. Biol. J. Linn. Soc. Lond. 96:913–941 [Google Scholar]
- 48. Centers for Disease Control and Prevention 1990. Update: filovirus infections among persons with occupational exposure to nonhuman primates. MMWR Morb. Mortal. Wkly. Rep. 39:266–267, 273 [PubMed] [Google Scholar]
- 49. World Health Organization 2009. WHO experts consultation on Ebola Reston pathogenicity in humans. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/resources/publications/HSE_EPR_2009_2.pdf [Google Scholar]
- 50. Marsh GA, Haining J, Robinson R, Foord A, Yamada M, Barr JA, Payne J, White J, Yu M, Bingham J, Rollin PE, Nichol ST, Wang LF, Middleton D. 2011. Ebola Reston virus infection of pigs: clinical significance and transmission potential. J. Infect. Dis. 204(Suppl 3):S804–S809 [DOI] [PubMed] [Google Scholar]
- 51. Suchard MA, Rambaut A. 2009. Many-core algorithms for statistical phylogenetics. Bioinformatics 25:1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.