Abstract
MicroRNAs (miRNAs) have been identified as key molecules in regulatory networks. The fine-tuning role of miRNAs in addition to the regulatory role of transcription factors has shown that molecular events during development are tightly regulated. In addition, several miRNAs play crucial roles in the response to abiotic stress induced by drought, salinity, low temperatures, and metals such as aluminium. Interestingly, several miRNAs have overlapping roles with regard to development, stress responses, and nutrient homeostasis. Moreover, in response to the same abiotic stresses, different expression patterns for some conserved miRNA families among different plant species revealed different metabolic adjustments. The use of deep sequencing technologies for the characterisation of miRNA frequency and the identification of new miRNAs adds complexity to regulatory networks in plants. In this review, we consider the regulatory role of miRNAs in plant development and abiotic stresses, as well as the impact of deep sequencing technologies on the generation of miRNA data.
Keywords: miRNAs, development, abiotic stress, nutrients, deep sequencing
MicroRNAs, Their Synthesis and Processing
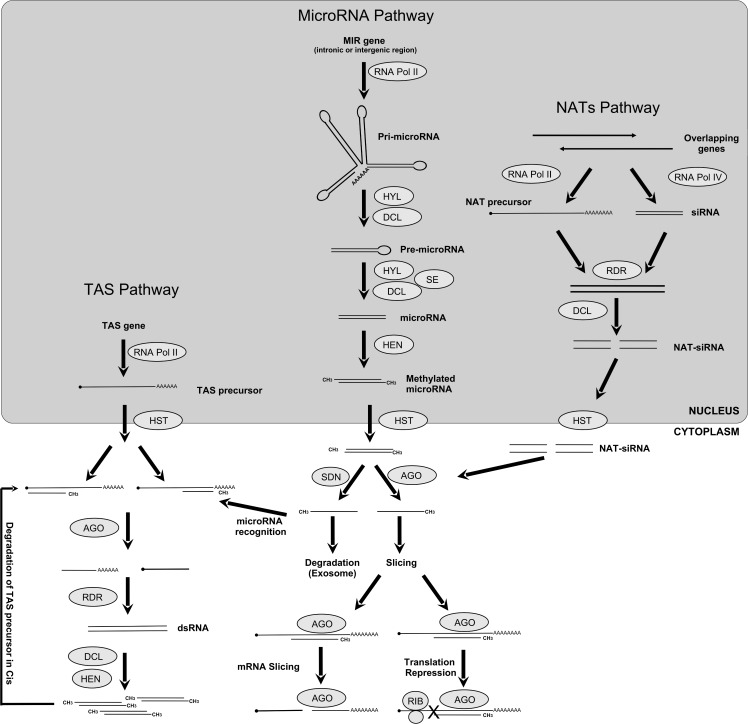
Gene transcription is a key mechanism regulated by transcription factors and also by distinct small RNAs of 21 to 24 nucleotide of length that can act at the transcriptional and post-transcriptional level (Jamalkandi and Masoudi-Nejad, 2009; Voinnet, 2009). In plants, the regulation of gene expression mediated by small RNAs initiates after the generation of double stranded RNAs and/or single strand RNAs that are folded into stem-loop/hairpin structures in the cells. These are recognized by RNase III-like enzymes called Dicer-Like (DCL), processed into small interfering RNAs, and loaded into protein complexes (RISC) to effectuate gene silencing after the recognition of different complementary target RNAs and or DNA. Distinct biochemical pathways generate different classes of small RNAs: short interfering RNAs (siRNAs), piwi-interacting RNAs occurring exclusively in animals (piRNAs), trans-acting siRNAs (TAS), naturally anti-sense siRNAs (NAT) and microRNAs (miRNAs) (Ramachandran and Chen, 2008; Chen, 2009; Jamalkandi and Masoudi-Nejad, 2009; Liu and Paroo, 2010). TAS pathway - RNA Pol II transcribes TAS genes into a TAS precursor, which is recognized by a complementary siRNA and sliced by Argonaute (AGO) proteins into small RNA which serves as a template for RNA Dependent RNA Polymerases (RDR) to make dsRNAs. This siRNA duplex originated by Dicer-Like directs cleavage of the TAS precursor in cis or another target mRNAs in trans. MicroRNA pathway: a MIR gene is transcribed by RNA Pol II into a precursor pri-microRNA which is stabilized and cleaved by a protein complex composed of DCL and Hyponastic Leaves (HYL) into a premicroRNA, which is further processed into a mature micro-RNA. The HUA Enhancer (HEN) methylates the resulting mature microRNA form in the 2’-hydroxy termini of both strands. This methylated mature form is exported to cytoplasm through HASTY protein (HST).
Once in the cytoplasm, AGO proteins recognize the mature microRNA and direct it to the target gene. Later, the AGO can induce the slicing of mRNA target or repress the translation complex. The other microRNA strand is directed to the exosome and degraded by Small RNA Degrading Nuclease (SDN). Natural Acting Small RNAs (NAT) Pathway: overlapping genes can be transcribed by RNA Pol II, resulting in a NAT precursor complementary to an siRNA, which serves as a template to the RDR proteins. The DCL protein cleaves this double-stranded precursor into dsRNAs, which are exported to the cytosol by HST protein. The NAT-siRNAs loaded into AGO complexes induce mRNA degradation in the same way as for the microRNA pathway (Figure 1) (Voinnet, 2009; Krol et al., 2010).
Figure 1.
General view of the small RNA pathways in plants. Cis/Trans-Acting small interfering RNAs (TAS) Pathway: A TAS gene is transcribed by RNA Pol II into a TAS precursor, later this precursor is recognized by a complementary siRNA and sliced by Argonaute (AGO) proteins into small RNA which serves as a template for RNA Dependent RNA Polymerases (RDR) to make dsRNAs. This siRNA duplex originated by Dicer-Like directs cleavage of the TAS precursor in cis, or another target mRNAs in trans. MicroRNA Pathway: A MIR gene, usually located in intergenic or intronic region, is transcribed by RNA Pol II into a precursor RNA named pri-microRNA, which is stabilized and cleaved by a protein complex composed by Dicer-like proteins (DCL) and Hyponastic Leaves (HYL) into a pre-microRNA which is further processed by the same complex plus Serrate (SE) into a mature microRNA. The HUA Enhancer (HEN) methylates the resulting mature microRNA form in the 2’-hydroxy termini of both strands. This methylated mature form is exported to cytoplasm through HASTY protein (HST). Once in the cytoplasm, AGO proteins recognize one strand of the mature microRNA and direct it to the target gene. Later, the AGO can induce the slicing of mRNA target or repress the translation complex. The other microRNA strand is directed to the Exosome and degraded by Small RNA Degrading Nuclease (SDN). Natural Acting Small RNAs (NAT) Pathway: Overlapping genes can be transcribed by RNA Pol II, resulting in a NAT precursor complementary to an siRNA, which serves as a template to the RDR proteins. The DCL protein cleaves this double-stranded precursor into dsRNAs, which are exported to the cytosol by HST protein. The NAT-siRNAs loaded into AGO complexes induce mRNA degradation in the same way as for microRNA pathway.
Although there are three major classes of small RNAs, miRNAs have been widely characterised in numerous biological conditions in plants. MiRNA genes originated from inverted duplications and random sequences in the genome (Felippes et al., 2008; Voinnet, 2009). They are transcribed by RNA Pol II into long primary polyadenylated RNA molecules and processed into mature miRNAs by Dicer-Like proteins (Parizotto et al., 2004). In plants, several biological experiments indicate that miRNAs play key roles during development and in response to environmental stresses (Figure 2) (Sunkar, 2010). The growing number of miRNAs has revealed the high complexity of genomes and biochemical and metabolic pathways in plants. Different miRNAs can act as regulators, from very early developmental phases to the reproductive phase (Chen, 2009). Although the study of the regulatory roles of miRNAs uncovered a new field in plant biology, the roles of several miRNAs remain to be discovered.
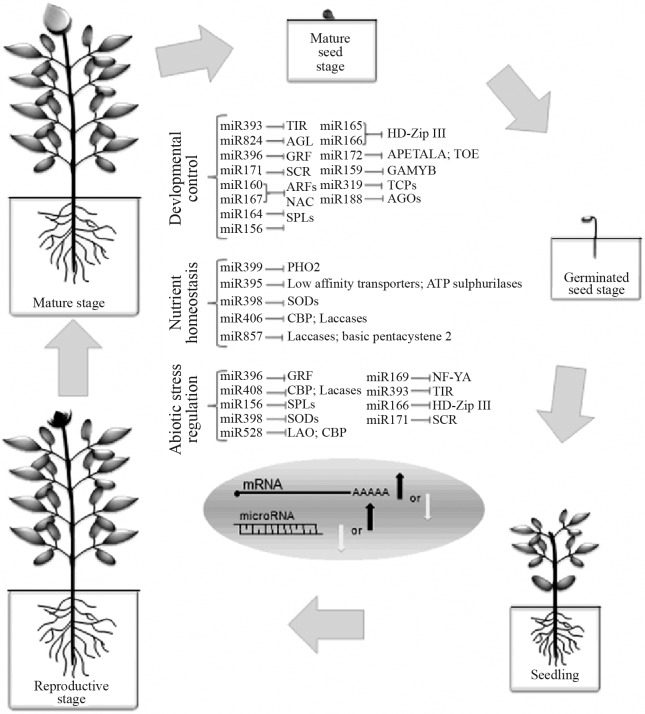
Figure 2.
Mature miRNAs act in plant development, in response to abiotic stresses, and also in the control of nutrient homeostasis. Depending on the plant species, the miRNA/target genes are modulated differently in the same biological conditions. Only miRNA targets already confirmed by expression analysis and/or degradome sequencing were included in this diagram. TIR: F-box protein; AGL: agamous like; GRF: growth regulating factor; SCR: scarecrow like; ARF: auxin response factor; NAC (NAM): no apical meristem; SPL; squamosa promter binding like; HDZip III; homeodomain transcription factor; APETALA; GAMyb: Gibberelic Acid Myb transcription factor; TCPs: Teosinte Branched/cycloidea transcription factor; AGOs: Argonaute; PHO2: phosphate 2 - E2 Conjugase protein; Low affinity transporter; ATP sulphurilases; SODs: superoxide dismutase; CBP: Copper Ion Binding Protein; Laccases; Basic pentacysteine 2; L-AO: L- ascorbate oxidase; NF-YA: CCAAT-box binding transcription factor.
Here, in a concise review, the regulatory action of miRNAs in development and response to abiotic stress will be discussed. Briefly, the new sequencing technologies will also be addressed, as far as they apply to the characterisation and identification of new miRNAs.
miRNAs and Development in Plants
In plants, mutations in the genes involved in biogenesis and the regulatory roles of miRNAs produce strong effects on development. These effects demonstrate the crucial role of miRNAs in development (Ramachandran and Chen, 2008; Chen, 2009; Xie et al., 2010). The Argonaute genes (AGO), especially the miR168a and miR168b-regulated AGO1, have a fundamental role in the stabilisation and regulatory action of other miRNAs (Vaucheret et al., 2004). In Arabidopsis thaliana, due to overlapping functions among different members of the MIR168 family, mutations in the MIR168a gene did not affect plant development under normal growth conditions. (Vaucheret, 2009). Although some miRNA families are numerous, there are few examples in the literature that uncover functions for individual members in plants (Chen, 2009). One good example is the TF (transcription factors) coding genes No Apical Meristem (NAM-NAC) and cup-shaped cotyledon (CUC) that are regulated by the miR164 family in A. thaliana, which are important in root and shoot development (Baker et al., 2005; Guo et al., 2005; Nikovics et al., 2006; Sieber et al., 2007; Raman et al., 2008). Triple mutants of miR164abc revealed that the genes athMIR164a and athMIR164b partially overlap athMIR164c function during floral development, as the phenotype became more severe in the triple mutant (Sieber et al., 2007). Individual mutants for athMIR164a and athMIR164b result in plants with more roots, which diversify the functional role of the miR164 family (Guo et al., 2005). To allow for proper root development, the short root (SHR) and scarecrow like (SCR) proteins activate the MIRNA165a and MIR166b genes, which in turn negatively regulate the TF HD-ZipIII (Carlsbecker et al., 2010). The osaxr mutant rice plant insensitive for auxin revealed numerous miRNAs and complex regulatory signals involved in root development (Meng et al., 2009).
It is already known that miR156 regulates the squamosa promoter binding- like (SPL) genes and that plants overexpressing miR156 are semi-dwarf, have altered numbers of leaves, and have longer vegetative phase (Xie et al., 2006; Wang et al., 2008; Zhang et al., 2011b). The down-regulated expression of miR156 from the juvenile to the adult phase is in contrast to the up-regulation of miR172, which is an important regulator of the floral patterning genes such as APETALA2, TOE1 and TOE2 (Aukerman and Sakai, 2003; Wu et al., 2009; Zhu and Helliwell, 2010). Interestingly, the dominant corngrass (Cg1) mutant, which contains two tandem MIR156 genes, showed an over-expression of miR156 in the meristem and lateral organs, and reduced levels of miR172, suggesting that the regulatory roles for these two miRNAs in the transition from the juvenile to the reproductive phase in maize and other plants is conserved (Chuck et al., 2007). Recently, ablation of leaf primordia delayed the transition to the reproductive phase, revealing that transcriptional signals that modulate miR156 regulatory action were found to be crucial for phase change in plants (Yang et al., 2011). In maize, it was determined that the spatial expression of miR156, which regulates the TF ZmTSH4, is crucial for the establishment of the lateral meristems (Chuck et al., 2010). Strikingly, the posttranscriptional regulation of OsSPL14 by miR156 defines the rice plant architecture, with mutants displaying a reduced number of tillers and more branches in the panicles (Jiao et al., 2010; Miura et al., 2010). Throughout the different developmental stages of soybean seeds, 26 new miRNAs and their target genes were identified using deep sequencing and degradome approaches (Song et al., 2011).
Hormone signalling and gene expression under miRNA control have deterministic roles in plant development (Liu and Chen, 2009; Liu et al., 2009). In A. thaliana, miR159-targeting members of the gibberelic acid MYB (GAMYB) family regulates seed germination and anther formation (Reyes and Chua, 2007). The overexpression of miR159 and the inhibition of the MYB gene expression delayed flowering and caused male sterility (Millar and Gubler, 2005). In Arabidopsis plants, miR159ab deregulation of the GAMYB-like genes resulted in reduction of the cell proliferation and programmed cell death (Alonso-Peral et al., 2010). Jasmonic acid biosynthesis is regulated by the teosinte branched/cycloidea proteins (TCP), which are TFs. These TFs have functional roles in development and leaf senescence and are regulated by miR319 (Schommer et al., 2008). Functional genes in the auxin signalling pathway (ARFs – auxin response factors) are miRNA targets. Plants with miR160-resistant forms of the ARF10, ARF16 and ARF17 genes showed pleiotropic effects in shoots and roots (Mallory et al., 2005; Liu et al., 2007). The overexpression of miR160 resulted in plants with less sensitivity to giberelic acid during germination (Liu et al., 2007). In flowering, during stamens and gynoecium development, miR167 has an important role when targeting ARF6 and ARF8 genes (Wu et al., 2006). The SCR family is targeted by miR171c to promote the proper development of auxiliary meristems during branching (Wang et al., 2010). Cell proliferation in A. thaliana is attenuated by the upregulation of miR396, which downregulates the growth regulating factor genes (GRF) that are crucial regulators in the cell cycle (Rodriguez et al., 2010). The occurrence of stomata, crucial for plant transpiration, depends partially on miR824 targeting the agamous like 16 gene (AGL16) (Kutter et al., 2007).
miRNAs in Response to Abiotic Stresses
In addition to the role of miRNAs in plant development, they are dramatically affected under abiotic stresses, where they regulate several coding genes in plants (Reyes et al., 2010; Sunkar, 2010). An understanding of how miRNAs act when they regulate gene expression and which coding genes are miRNA targets during stress responses, such as drought, salinity, metals, temperature and nutrient homeostasis, will help the generation of more tolerant plants (Sunkar, 2010).
The Regulatory Role of miRNAs in Plants Under Drought, Salinity, Aluminum, and Low Temperatures
It has been suggested that miR393 is one of the key miRNAs during stress responses because of its altered expression in A. thaliana, Oryza sativa, Medicago truncatula, Phaseolus vulgaris and other plants under drought, salinity, low temperature, and aluminium stress conditions (Sunkar and Zhu, 2004; Zhao et al., 2007; Liu et al., 2008; Arenas-Huertero et al., 2009; Trindade et al., 2010). However, the molecular evidence that miR393 regulates its targets in several environmental conditions remains to be considered. Recently, Arabidopsis plants overexpressing osaMIR393 became more tolerant to salt excess, suggesting a regulatory role in salinity tolerance (Gao et al., 2011). It is known that miRNAs from the miR169 family respond differently to drought, salinity, low temperatures and aluminium in plants (Zhao et al., 2007; Liu et al., 2008; Zhou et al., 2008; Zhao et al., 2009). In response to salinity and drought stress in rice, the expression of the nuclear transcription factor YA (NF-YA) genes is modulated by members of the miR169 family (Zhao et al., 2009). In A. thaliana, nf-ya plants and plants overexpressing miR169 are more sensitive to drought (Li et al., 2008). On the contrary in tomato, plants overexpressing miR169c, which targets a gene involved in the opening and closing of stomata, are more tolerant to drought (Zhang et al., 2011a). A reduction in the expression of miR530a, miR1445, miR1446a-e and miR1447 in Populus trichocarpa was detected in plants under drought and salinity, which is different from the miR1450 pattern of expression, downregulation under drought conditions and upregulation under high salinity (Lu et al., 2008). In Triticum dicoccoides, the ancestor of cultivated wheat, the upregulation of miR1450 revealed an inverse response when compared with Populus trichocarpa under drought conditions (Kantar et al., 2011). Although the gene MIR1450 is present in both monocot and dicots, the expression patterns suggest regulatory differences under drought (Lu et al., 2008; Kantar et al., 2011). The formation of the superoxide anion O2− in response to stresses is converted into less toxic molecules by superoxide dismutases SOD1 and SOD2 proteins, whose mRNAs are targeted by miR398 (Sunkar et al., 2006; Jagadeeswaran et al., 2009; Trindade et al., 2010; Kantar et al., 2011). The inverse correlation between miRNAs miR156, miR166, miR171, miR408 and their targets were detected in barley plants under drought (Kantar et al., 2010). In different tissues from different developmental stages in rice plants under drought conditions in soil, miRNAs miR156, miR171 and miR408 were also detected (Zhou et al., 2010). In Medicago truncatula, miR408 acts to regulate plantacyanin genes in response to drought (Trindade et al., 2010). In acidic soils, the availability of aluminium in low pH conditions inhibits root growth, which affects plant development dramatically (Ryan et al., 2011). Comparing japonica and indica subspecies, which differ in aluminium tolerance, we have characterised the expression of miRNAs in rice plants treated with aluminium. Using RT-qPCR, it was possible to detect sixteen differentially expressed miRNAs in rice roots, which reveals a complex miRNA response in rice under aluminium stress. The inverse regulation of miR528 and its targets L-ascorbate oxidase (L-AO) and copper ion binding protein genes was also observed. This finding corresponds to the first report on the characterisation of the miRNA response in plants under aluminium stress (Lima et al., 2011).
miRNAs and Their Regulatory Role in the Response to UV-B Radiation, Hypoxia, and Oxidative Stress
The redox state of the cellular environment and the generation of ROS as a consequence of UV-B radiation and hypoxia reprograms plant responses due to eminent irreversible damage (Blokhina and Fagerstedt, 2010; De Gara et al., 2010). The induction of different microRNAs in maize plants under low oxygen points to a diverse role of miRNAs in morphological and physiological adaptations in root cells and in sulphur and oxidative metabolism (Zhang et al., 2008). The repression of miR398 and the upregulation of SOD proteins has a crucial role in Arabidopsis plants under oxidative stress (Sunkar et al., 2006). The downregulation of miR395 and the induction of miR398, as well as the respective inversion of expression of their targets in response to UV-B in Populus tremula, suggests there are important differences in the stress-induced metabolic adjustments compared with Arabidopsis (Jia et al., 2009). By deep sequencing, the identification of miRNAs in rice plants under hydrogen peroxide treatment has broadened the roles for miRNAs in plants under oxidative stress. Targets of these hydrogen peroxide-responsive miRNAs are involved in different cellular responses and metabolic processes including transcriptional regulation, nutrient transport, auxin homeostasis, cell proliferation and programmed cell death, which indicates the diversity of miRNAs function in plants’ responses under oxidative stress (Li et al., 2010a).
The Importance of miRNAs in Nutrient Homeostasis
The uptake of nutrients is a compulsory requirement of plants, and the homeostasis of nutrients is critical for the maintenance of growth and development (Giehl et al., 2009; Yang and Finnegan, 2010). Sulphur is transported into the cell as sulphate and has a structural role in protein folding (Rausch and Wachter, 2005). Under sulphate deficiency, miR395 down-regulates low affinity transporters and ATP sulphurilases (Jones-Rhoades and Bartel, 2004). Interestingly, in Arabidopsis roots, both the sulphate transporter AST68 and miR395 were induced. The spatial expression patterns suggested that miR395 limits the expression of its targets in the phloem cells (Kawashima et al., 2009). Phosphate (Pi) homeostasis is under miR399 regulation. Under low cellular phosphate levels, Pi responsive gene (PHR1) activates miR399, which negatively regulates the phosphate 2 gene (PHO2), which has a role in protein degradation pathways (Bari et al., 2006). An alternative regulation in phosphate signalling is the expression of a non-coding RNA called IPS (Induced by Phosphate Starvation), which has a miR399 binding site with some mismatches that impair IPS cleavage by miR399 under ideal phosphate conditions. The sequestration of miRNAs by IPS (target mimicry) blocks the down-regulation of PHO2 by miR399 (Franco-Zorrilla et al., 2007). Although target mimicry needs further investigation in other plants, it is functional and widespread in Arabidopsis (Todesco et al., 2010). In addition, by deep sequencing, the detection of several miRNAs revealed a much more complex regulatory network in phosphate signalling (Hsieh et al., 2009; Gu et al., 2010).
Metals like copper and iron are also essential micro-nutrients to plants. MiRNAs miR398, miR408 and miR857 are part of a signalling network that functions in the regulation of copper levels in plant cells. Under copper deficiency, these miRNAs were induced and negatively regulated their targets (Yamasaki et al., 2007; Burkhead et al., 2009). Several miRNAs were up-regulated in response to low iron levels in Arabidopsis (Kong and Yang, 2010). Interestingly, a member of the miR854 family that is induced in plants under iron deficiency conditions is also conserved in animals, and its targets can be regulated via translation inhibition (Arteaga-Vazquez et al., 2006; Kong and Yang, 2010). In nutrient metabolism, miR169 regulates its target NF-YA genes, which have an important role in the balance of nitrogen in plants (Zhao et al., 2011).
Next Generation Sequencing Technologies: How to Deal with an Increasing Amount of Data?
Deep sequencing technologies are revolutionising molecular biology (Brautigam and Gowik, 2010), lowering the costs of sequencing and increasing throughput by several orders of magnitude (Paszkiewicz and Studholme, 2010). A deep sequencing approach was successfully applied for de novo sequencing of plant genomes (Imelfort and Edwards, 2009), metagenomics studies in grapevines (Coetzee et al., 2010), sequencing of natural strains in Arabidopsis (Ossowski et al., 2008), RNA sequencing of different tissues from soybean (Severin et al., 2010) and miRNA identification in many organisms. In addition, deep sequencing approaches applied to the characterisation of miRNA frequency and the identification of new miRNAs that regulate development and abiotic stress responses brought more complexity to regulatory networks in plants (Li et al., 2010a; Song et al., 2011). The growing number of miRNAs identified mainly by deep sequencing is increasing with sequence data in databases as miRBase (Kozomara and Griffiths-Jones, 2010) and the Plant MicroRNA Database (PMRD) (Zhang et al., 2010), a specific databank for plant miRNAs. This database allows for the retrieval of a target gene, promoter sequence, and expression profile for some miRNA genes. However, the rapid increase in molecular data in databases represents only the tip of the iceberg, and these data demand more laborious analyses for the identification of miRNA function.
Concluding Remarks and Future Perspectives
The regulatory role of miRNAs in plants is definitely a subject that will require much more investigation in plant biology. As presented in this review, several miRNAs have been determined to be involved in plant development and abiotic stress responses. They are positioned for the fine-tuning of distinct regulatory networks. In addition, the identification of new miRNAs and their targets adds more complexity to gene expression regulatory networks. The increasing number of miRNAs identified by deep sequencing, in different and multiple experimental conditions, points to a need for further biological investigation. To address the question raised in the previous section we consider that the latest strategies for the understanding of individual miRNA function as miRNA target mimicry (Franco-Zorrilla et al., 2007), identification of miRNA target genes by degradome approach (German et al., 2008; Li et al., 2010b), and silencing of miRNAs (Eamens and Wang, 2011) are suitable molecular tools to unravel the functional role of the increasing number of miRNAs (Figure 3). Finally, since miRNAs regulate numerous transcription factors during development and in response to different stresses, high throughput analysis as RNA-seq (deep sequencing of mRNAs), proteomics and metabolomics should be always considered as complementary approaches to investigate the global effects of the conservation and diversity of miRNA responses in different biological conditions and in different plant species.
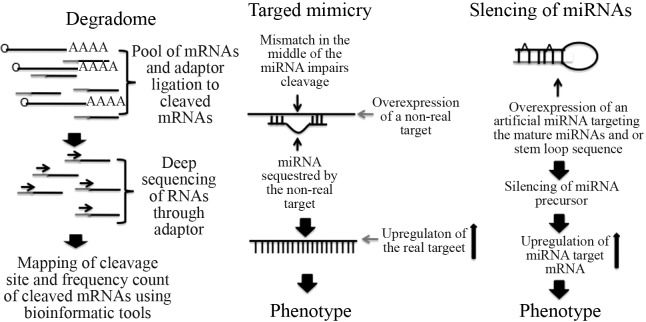
Figure 3.
General representation of the degradome sequencing approach, miRNA target mimicry, and silencing of miRNAs. The degradome sequencing approach has been used to detect miRNA target genes through deep sequencing of cleaved mRNAs and mapping of cleaved sites. Target mimicry is a suitable molecular tool that is based on the expression of a transgene carrying a non-real target of a miRNA that is partially complementary to the miRNA. This RNA contains three to four nucleotides that are not complementary to a core region in the middle of the miRNA, which causes the sequestration of miRNAs, blocks cleavage and up-regulates the real target mRNAs. Silencing of miRNAs triggered by artificial miRNAs targeting the mature and or precursor miRNA, directs cleavage and RNA silencing of the precursor miRNAs and upregulation of miRNA target RNAs.
Acknowledgments
This work was supported by CNPq grant number 476005/2008-6 and 559636/2009-1. JCL received a PhD fellowship from CNPq, and RM a CNPq research grant number 307868/2011-7. GLM was awarded a PhD fellowship from CAPES.
References
- Alonso-Peral MM, Li J, Li Y, Allen RS, Schnippenkoetter W, Ohms S, White RG, Millar AA. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 2010;154:757–771. doi: 10.1104/pp.110.160630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero C, Perez B, Rabanal F, Blanco-Melo D, De la Rosa C, Estrada-Navarrete G, Sanchez F, Covarrubias AA, Reyes JL. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol Biol. 2009;70:385–401. doi: 10.1007/s11103-009-9480-3. [DOI] [PubMed] [Google Scholar]
- Arteaga-Vazquez M, Caballero-Perez J, Vielle-Calzada JP. A family of microRNAs present in plants and animals. Plant Cell. 2006;18:3355–3369. doi: 10.1105/tpc.106.044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol. 2005;15:303–315. doi: 10.1016/j.cub.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhina O, Fagerstedt KV. Oxidative metabolism, ROS and NO under oxygen deprivation. Plant Physiol Biochem. 2010;48:359–373. doi: 10.1016/j.plaphy.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Brautigam A, Gowik U. What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biol. 2010;12:831–841. doi: 10.1111/j.1438-8677.2010.00373.x. [DOI] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KA, Abdel-Ghany SE, Cohu CM, Pilon M. Copper homeostasis. New Phytol. 2009;182:799–816. doi: 10.1111/j.1469-8137.2009.02846.x. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vaten A, Thitamadee S, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S. The hetero-chronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- Chuck G, Whipple C, Jackson D, Hake S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development. 2010;137:1243–1250. doi: 10.1242/dev.048348. [DOI] [PubMed] [Google Scholar]
- Coetzee B, Freeborough MJ, Maree HJ, Celton JM, Rees DJ, Burger JT. Deep sequencing analysis of viruses infecting grapevines: Virome of a vineyard. Virology. 2010;400:157–163. doi: 10.1016/j.virol.2010.01.023. [DOI] [PubMed] [Google Scholar]
- De Gara L, Locato V, Dipierro S, de Pinto MC. Redox homeostasis in plants. The challenge of living with endogenous oxygen production. Respir Physiol Neurobiol. 2010;173(Suppl):S13–19. doi: 10.1016/j.resp.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Eamens AL, Wang MB. Alternate approaches to repress endogenous microRNA activity in Arabidopsis thaliana. Plant Sign Behav. 2011;6:349–359. doi: 10.4161/psb.6.3.14340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FF, Schneeberger K, Dezulian T, Huson DH, Weigel D. Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA. 2008;14:2455–2459. doi: 10.1261/rna.1149408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Gao P, Bai X, Yang L, Lv D, Pan X, Li Y, Cai H, Ji W, Chen Q, Zhu Y. osa-MIR393: A salinity- and alkaline stress-related microRNA gene. Mol Biol Rep. 2011;38:237–242. doi: 10.1007/s11033-010-0100-8. [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- Giehl RF, Meda AR, von Wiren N. Moving up, down, and everywhere: Signaling of micronutrients in plants. Curr Opin Plant Biol. 2009;12:320–327. doi: 10.1016/j.pbi.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Gu M, Xu K, Chen A, Zhu Y, Tang G, Xu G. Expression analysis suggests potential roles of microRNAs for phosphate and arbuscular mycorrhizal signaling in Solanum lycopersicum. Physiol Plant. 2010;138:226–237. doi: 10.1111/j.1399-3054.2009.01320.x. [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to down-regulate auxin signals for arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imelfort M, Edwards D. De novo sequencing of plant genomes using second-generation technologies. Brief Bioinform. 2009;10:609–618. doi: 10.1093/bib/bbp039. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G, Saini A, Sunkar R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta. 2009;229:1009–1014. doi: 10.1007/s00425-009-0889-3. [DOI] [PubMed] [Google Scholar]
- Jamalkandi SA, Masoudi-Nejad A. Reconstruction of Arabidopsis thaliana fully integrated small RNA pathway. Funct Integr Genomics. 2009;9:419–432. doi: 10.1007/s10142-009-0141-z. [DOI] [PubMed] [Google Scholar]
- Jia X, Ren L, Chen QJ, Li R, Tang G. UV-B-responsive microRNAs in Populus tremula. J Plant Physiol. 2009;166:2046–2057. doi: 10.1016/j.jplph.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Kantar M, Lucas SJ, Budak H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta. 2011;233:471–484. doi: 10.1007/s00425-010-1309-4. [DOI] [PubMed] [Google Scholar]
- Kantar M, Unver T, Budak H. Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct Integr Genomics. 2010;10:493–507. doi: 10.1007/s10142-010-0181-4. [DOI] [PubMed] [Google Scholar]
- Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 2009;57:313–321. doi: 10.1111/j.1365-313X.2008.03690.x. [DOI] [PubMed] [Google Scholar]
- Kong WW, Yang ZM. Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiol Biochem. 2010;48:153–159. doi: 10.1016/j.plaphy.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2010;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kutter C, Schob H, Stadler M, Meins F, Jr, Si-Ammour A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell. 2007;19:2417–2429. doi: 10.1105/tpc.107.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Li H, Zhang YX, Liu JY. Identification and analysis of seven H2O2-responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica) Nucleic Acids Res. 2010a;39:2821–2833. doi: 10.1093/nar/gkq1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010b;62:742–759. doi: 10.1111/j.1365-313X.2010.04187.x. [DOI] [PubMed] [Google Scholar]
- Lima JC, Arenhart RA, Margis-Pinheiro M, Margis R. Aluminum triggers broad changes in microRNA expression in rice roots. Genet Mol Res. 2011;10:2817–2832. doi: 10.4238/2011.November.10.4. [DOI] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Micro-array-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14:836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Chen YQ. Insights into the mechanism of plant development: Interactions of miRNAs pathway with phyto-hormone response. Biochem Biophys Res Commun. 2009;384:1–5. doi: 10.1016/j.bbrc.2009.04.028. [DOI] [PubMed] [Google Scholar]
- Liu Q, Paroo Z. Biochemical principles of small RNA pathways. Annu Rev Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhang YC, Wang CY, Luo YC, Huang QJ, Chen SY, Zhou H, Qu LH, Chen YQ. Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett. 2009;583:723–728. doi: 10.1016/j.febslet.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Lu S, Sun YH, Chiang VL. Stress-responsive microRNAs in Populus. Plant J. 2008;55:131–151. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Huang F, Shi Q, Cao J, Chen D, Zhang J, Ni J, Wu P, Chen M. Genome-wide survey of rice microRNAs and microRNA-target pairs in the root of a novel auxin-resistant mutant. Planta. 2009;230:883–898. doi: 10.1007/s00425-009-0994-3. [DOI] [PubMed] [Google Scholar]
- Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18:2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkiewicz K, Studholme DJ. De novo assembly of short sequence reads. Brief Bioinform. 2010;11:457–472. doi: 10.1093/bib/bbq020. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 2008;13:368–374. doi: 10.1016/j.tplants.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008;55:65–76. doi: 10.1111/j.1365-313X.2008.03483.x. [DOI] [PubMed] [Google Scholar]
- Rausch T, Wachter A. Sulfur metabolism: A versatile platform for launching defence operations. Trends Plant Sci. 2005;10:503–509. doi: 10.1016/j.tplants.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Reyes JL, Arenas-Huertero C, Sunkar R. Cloning of stress-responsive microRNAs and other small RNAs from plants. Methods Mol Biol. 2010;639:239–251. doi: 10.1007/978-1-60761-702-0_14. [DOI] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137:103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PR, Tyerman SD, Sasaki T, Furuichi T, Yamamoto Y, Zhang WH, Delhaize E. The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J Exp Bot. 2011;62:9–20. doi: 10.1093/jxb/erq272. [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chetelat A, Cubas P, Farmer EE, Nath U, Weigel D. Control of jasmo-nate biosynthesis and senescence by miR319 targets. PLoS Biology. 2008;6:e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010;10:e160. doi: 10.1186/1471-2229-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: Role of the MIR164 family in developmental robustness. Development. 2007;134:1051–1060. doi: 10.1242/dev.02817. [DOI] [PubMed] [Google Scholar]
- Song QX, Liu YF, Hu XY, Zhang WK, Ma B, Chen SY, Zhang JS. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011;11:e5. doi: 10.1186/1471-2229-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R. MicroRNAs with macro-effects on plant stress responses. Semin Cell Dev Biol. 2010;21:805–811. doi: 10.1016/j.semcdb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genetics. 2010;6:e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade I, Capitao C, Dalmay T, Fevereiro MP, Santos DM. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta. 2010;231:705–716. doi: 10.1007/s00425-009-1078-0. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS One. 2009;4:e6442. doi: 10.1371/journal.pone.0006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 2008;20:1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Mai YX, Zhang YC, Luo Q, Yang HQ. MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant. 2010;3:794–806. doi: 10.1093/mp/ssq042. [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006;142:280–293. doi: 10.1104/pp.106.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Khanna K, Ruan S. Expression of microRNAs and its regulation in plants. Semin Cell Dev Biol. 2010;21:790–797. doi: 10.1016/j.semcdb.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M. Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem. 2007;282:16369–16378. doi: 10.1074/jbc.M700138200. [DOI] [PubMed] [Google Scholar]
- Yang L, Conway SR, Poethig RS. Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development. 2011;138:245–249. doi: 10.1242/dev.058578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Finnegan PM. Regulation of phosphate starvation responses in higher plants. Ann Bot. 2010;105:513–526. doi: 10.1093/aob/mcq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zou Z, Gong P, Zhang J, Ziaf K, Li H, Xiao F, Ye Z. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol Lett. 2011a;33:403–409. doi: 10.1007/s10529-010-0436-0. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zou Z, Zhang J, Zhang Y, Han Q, Hu T, Xu X, Liu H, Li H, Ye Z. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 2011b;585:435–439. doi: 10.1016/j.febslet.2010.12.036. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wei L, Zou X, Tao Y, Liu Z, Zheng Y. Submergence-responsive MicroRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Ann Bot. 2008;102:509–519. doi: 10.1093/aob/mcn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZH, Yu JY, Li DF, Zhang ZY, Liu FX, Zhou X, Wang T, Ling Y, Su Z. PMRD: Plant microRNA database. Nucleic Acids Res. 2010;38:D806–D813. doi: 10.1093/nar/gkp818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ge L, Liang R, Li W, Ruan K, Lin H, Jin Y. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol. 2009;10:e29. doi: 10.1186/1471-2199-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BT, Liang RQ, Ge LF, Li W, Xiao HS, Lin HX, Ruan KC, Jin YX. Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun. 2007;354:585–590. doi: 10.1016/j.bbrc.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Zhao M, Ding H, Zhu J, Zhang F, Li W. Involvement of miR169 in the nitrogen-startvation responses in Arabidopsis. New Phytol. 2011;190:906–915. doi: 10.1111/j.1469-8137.2011.03647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot. 2010;61:4157–4168. doi: 10.1093/jxb/erq237. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang G, Sutoh K, Zhu JK, Zhang W. Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim Biophys Acta. 2008;1779:780–788. doi: 10.1016/j.bbagrm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Zhu QH, Helliwell CA. Regulation of flowering time and floral patterning by miR172. J Exp Bot. 2010;62:487–495. doi: 10.1093/jxb/erq295. [DOI] [PubMed] [Google Scholar]