Abstract
The outer domain of the HIV-1 gp120 envelope glycoprotein contains the epitope for broadly neutralizing antibodies directed to the CD4-binding site, many of which are able to neutralize over 90% of circulating HIV-1 isolates. While the outer domain is conformationally more stable than other portions of the HIV-1 envelope, efforts to express the outer domain as an immunogen for eliciting broadly neutralizing antibodies have not been successful, potentially because natural outer domain variants do not bind strongly to antibodies such as VRC01. In this study, we optimized the antigenic properties of the HIV-1 Env outer domain to generate OD4.2.2, from the KER2018 strain of clade A HIV-1, enabling it to bind antibodies such as VRC01 with nanomolar affinity. The crystal structure of OD4.2.2 in complex with VRC-PG04 was solved at 3.0-Å resolution and compared to known crystal structures including (i) the structure of core gp120 bound by VRC-PG04 and (ii) a circularly permutated version of the outer domain in complex with antibody PGT128. Much of the VRC-PG04 epitope was preserved in the OD4.2.2 structure, though with altered N and C termini conformations. Overall, roughly one-third of the outer domain structure appeared to be fixed in conformation, independent of alterations in termini, clade, or ligand, while other portions of the outer domain displayed substantial structural malleability. The crystal structure of OD4.2.2 with VRC-PG04 provides atomic-level details for an HIV-1 domain recognized by broadly neutralizing antibodies and insights relevant to the rational design of an immunogen that could elicit such antibodies by vaccination.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) viral spike is comprised of three gp120 envelope glycoproteins and three gp41 transmembrane molecules and utilizes multiple mechanisms, including extraordinary sequence diversity, an evolving glycan shield, and conformational masking, to evade the humoral immune response (1–3). Despite these mechanisms of evasion, antibodies have been identified that recognize a few conserved regions on the viral spike. These vulnerable regions include the CD4 receptor-binding site on gp120, two sites of N-linked glycan at residues N160 and N332 on gp120, and the membrane-proximal external region of gp41. Antibodies that target these sites are capable of neutralizing over 90% of circulating HIV-1 isolates, while those against the CD4-binding site including VRC01 (with over 90% breadth) (4) and VRC-PG04 (76% breadth) are broad and extremely potent (5). The b12 antibody also targets the CD4-binding site, but its breadth of neutralization is a more modest ∼35% (6). To date, attempts to induce broadly neutralizing CD4-binding site antibodies through immunization with monomeric gp120 immunogens (7, 8) or trimeric gp140 immunogens in multiple vaccine trials have been unsuccessful (9, 10). Studies aimed at understanding the development of CD4-binding-site-specific antibodies in infected individuals (11, 12) showed that even though antibodies targeting the CD4-binding site can develop within a few weeks of infection, broadly neutralizing antibodies targeting this site appear to take years to develop and occur in a relatively small proportion of infected individuals (6, 11, 13).
To counteract mechanisms of HIV-1 immune evasion and to facilitate the development of neutralizing antibodies focused against the CD4-binding site, an independent outer domain molecule has been proposed as a minimal immunogen (14). The outer domain is recognized by broadly neutralizing antibodies, such as b12 (6), VRC01 (4), and HJ16 (15), and recapitulation of these antibodies through vaccination is highly desired.
The initial design of an outer domain construct, named OD1 by Yang, Sodroski, and colleagues (16), revealed the potential as well as the difficulties inherent in this strategy. While capable of binding to antibody 2G12 and antibodies specific for the third variable loop (V3) region of gp120 (16), the dissociation rate with b12 was substantially increased for OD1 relative to full-length gp120 (6), and the OD1 molecule failed to elicit broadly neutralizing antibodies in rabbit immunizations. In subsequent work the outer domain from the clade B virus TA1 R3A was expressed in a cell surface-transmembrane context (17). The cell surface presentation of the outer domain led to increased stability, and the lipid bilayer was proposed to mimic the interaction of the outer domain with the remainder of gp120. This variant was capable of binding b12 with affinity comparable to that of the native Env spike and was also able to absorb neutralizing antibodies from serum (such as b12), indicating that it preserved essential features of the CD4-binding site. Here, we describe the design and antigenic optimization of a soluble outer domain variant, termed OD4.2.2, from the clade A KER2018 strain of HIV-1. We present the crystal structure of OD4.2.2 in complex with the broadly neutralizing antibody VRC-PG04, determined in monoclinic and trigonal lattices, and compare this structure to previous liganded and unliganded HIV-1 gp120 structures to provide insight into this critical vaccine immunogen.
MATERIALS AND METHODS
Cell lines, media, and antibodies.
293F human embryonic kidney cell lines were purchased from Invitrogen (Carlsbad, CA) and were maintained in Freestyle 293 expression medium (Invitrogen). Monoclonal antibodies (Abs) used in binding studies (VRC01, VRC-PG04, and b12) were expressed by transient cotransfection of both the heavy- and light-chain gene-containing constructs into 293F cells in suspension culture at 37°C, and supernatant was harvested after 6 days with antibody purified from the supernatant using a protein A affinity column (Qiagen). Antigen-binding fragment (Fab) was produced by endoproteinase Lys-C digestion of purified antibody in 25 mM Tris-Cl–1 mM EDTA (pH 8) for 6 h at 37°C. The cleavage reaction was stopped with 1 mM TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone) and 0.4 mM leupeptin and then passed over a protein A column (Qiagen) to remove the Fc fragment. Flowthrough fractions containing the Fab fragment were dialyzed into 50 mM Na acetate, pH 5.0, buffer, applied to a 10-ml MonoS (Qiagen) column, and eluted with a 0 to 250 mM NaCl gradient. Peak fractions were pooled, concentrated, and further purified by size exclusion chromatography (10/60 Superdex-200; GE) using gel filtration buffer (350 mM NaCl, 2.5 mM Tris, pH 7.1, 0.02% NaN3).
Outer domain construct preparation and design.
The outer domain construct OD1 was made by PCR of amino acids 252 to 482 from a clade B virus strain named R2 (18) (this is a CCR5-coreceptor utilizing virus, not to be confused with CCR2-coreceptor utilizing viruses), which was isolated from a long-term nonprogressor patient (19). The outer domain gene was fused to the human CD5 signal peptide sequence (MPMGSLQPLATLYLLGMLVASVLA) at the 5′ end and to a 3′ sequence encoding a human rhinovirus (HRV) 3C protease site and a six-His purification tag. OD1.1 was identical to OD1 with the exception of an N363Q mutation. The OD2 molecule incorporated the removal of the β20-β21 (β20–21) strands, and residues 424 to 432 were joined with a simple G-G dipeptide connector. This was designed to retain the outer domain hydrogen bonding network while removing areas of structural differences between the CD4- and b12-bound structures of core gp120 (Protein Data Bank [PDB] accession numbers 2NXY and 2NY7, respectively) (6). Subsequent OD2.x versions incorporated truncations of the V3 loop designed to retain hydrogen bonding at the V3 base while providing variation in the connecting loop. The OD2.1 molecule is truncated at the V3 loop, leaving 9 amino acids on each side of the stem connected by NTRKGAGII [named V3 (9, 9)]. OD2.2 with a V3-GSL loop also contains 9 amino acids on each side of the stem and is connected by six residues, NTRGRR, giving the loop an overall basic charge (17). Mutagenesis of outer domain molecules was carried out using a QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The R2 strain outer domain molecule was optimized up to version OD2.2.
Following production and testing of OD2.2, outer domain molecules from multiple clade A and clade C viruses were produced with the same design features and further assessed for stability and binding to VRC01. The outer domain construct from KER2018 bound tightly to VRC01 and was further optimized for antigenicity and ultimately for crystallization studies. All outer domain constructs were expressed under the cytomegalovirus (CMV) promoter in the CMV/R vector (20).
Outer domain production.
The clade B virus R2 outer domain proteins were expressed in 293F cells in the presence of 10 mg/liter swainsonine and 2.5 mg/liter kifunensine for 5 days with an average yield of 60 mg/liter of cells (initial expression and binding studies with protein expressed in 293F cells in the absence of kifunensine and swainsonine showed no binding of VRC01 to the OD proteins [data not shown]). Supernatant was applied to a Ni2+-nitrilotriacetic acid resin (Qiagen) and purified using an elution buffer consisting of 20 mM Tris-Cl, pH 7.5, 200 mM NaCl, and 250 mM imidazole. The eluted fractions containing OD protein were further purified on a Superdex 200 gel filtration column (GE Healthcare) using gel filtration buffer (350 mM NaCl, 2.5 mM Tris, pH 7.1, 0.02% NaN3), and the purified fractions were concentrated to ∼10 mg/ml. The clade A KER2018 outer domain constructs (OD2.2 to -4.2.2) were expressed in 293F cells without glycan-processing modifiers while the OD4.2.2 protein used for crystallization was expressed in N-acetylglucosaminyltransferase I-negative (GnTI−) 293S cells with yields typically of >20 mg/liter of cells for each of these proteins.
Binding studies using biolayer interferometry.
A fortéBio Octet Red384 instrument was used to measure binding kinetics of outer domain molecules to VRC01, VRC-PG04, and b12 antibodies and also to a group of poorly neutralizing antibodies, b6, b13 (21) F105, F91 (22), and 15e (23), binding to OD4.2.2 and core KER2018 and YU2 full-length gp120 protein. All the assays were performed with agitation set to 1,000 rpm in phosphate-buffered saline (PBS) buffer supplemented with 1% bovine serum albumin (BSA) in order to minimize nonspecific interactions. The final volume for all the solutions was 100 μl/well. Assays were performed at 30°C in solid black 96-well plates (Geiger Bio-One). Antibody (25 μg/ml) in PBS buffer was used to load anti-IgG Fc probes for 300 s. Typical capture levels were between 0.5 and 1 nm, and variability within a row of eight tips did not exceed 0.1 nm. Biosensor tips were then equilibrated for 300 s in PBS buffer supplemented with 1% BSA prior to binding measurements of the analyte in solution (0.15 to 10 μM) for 300 s; binding was then allowed to dissociate for 300 s. Dissociation wells were used only once to prevent contamination. Parallel correction to subtract systematic baseline drift was carried out by subtracting the measurements recorded for a sensor loaded with CD4-binding-site antibodies but incubated with PBS buffer supplemented with 1% BSA. To remove nonspecific binding responses, a membrane-proximal external region-specific antibody, 2F5, which does not bind to the outer domain or core molecules, was loaded onto the sensor and incubated with analytes, and the nonspecific response was subtracted from CD4-binding site antibody response data. Data analysis and curve fitting were done using Octet software, version 7.0. Experimental data were fitted with the binding equations describing a 1:1 interaction. Global analyses of the complete data sets assuming binding was reversible (full dissociation) were carried out using nonlinear least-squares fitting allowing a single set of binding parameters to be obtained simultaneously for all concentrations used in each experiment.
Binding studies using surface plasmon resonance.
Binding affinities of OD4.2.2 to different antibodies were also assessed by surface plasmon resonance on a Biacore T-200 (GE Healthcare) at 20°C with buffer HBS-EP+ (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% surfactant P-20). Monoclonal mouse anti-human IgG (Fc) antibody, which binds an epitope in the CH2 domain of IgG Fc, was immobilized onto a CM5 chip at 8,000 to 10,000 response units (RU) according to protocols provided in the Human Antibody Capture Kit (GE Healthcare). Antibodies VRC01, VRC-PG04, and NIH45-46 were captured with the immobilized antihuman Fc antibody up to a level of ∼250 to 500 RU. OD4.2.2 at 2-fold increasing concentrations (50 nM to 200 nM) was injected over the captured antibody channel and a blank channel at a flow rate of 30 μl/min for 3 min and allowed to dissociate for 5 min. Fab fragments from antibodies b3 (21), m14 (24), and m18 (25) were directly immobilized onto a CM5 chip to a level of ∼600 RU; OD4.2.2, KER2018, core, and YU2 gp120 full-length protein at 2-fold increasing concentrations from 7.8 nM were injected over the immobilized Fab molecules at a flow rate of 30 μl/min for 5 min and allowed to dissociate for 5 min. The flow channels were regenerated with two 35-μl injections of 3.0 M MgCl2 at a flow rate of 70 μl/min. All sensorgrams were corrected with appropriate blank references and fitted globally with Biacore Evaluation software using a 1:1 Langmuir model of binding.
Isothermal titration calorimetry.
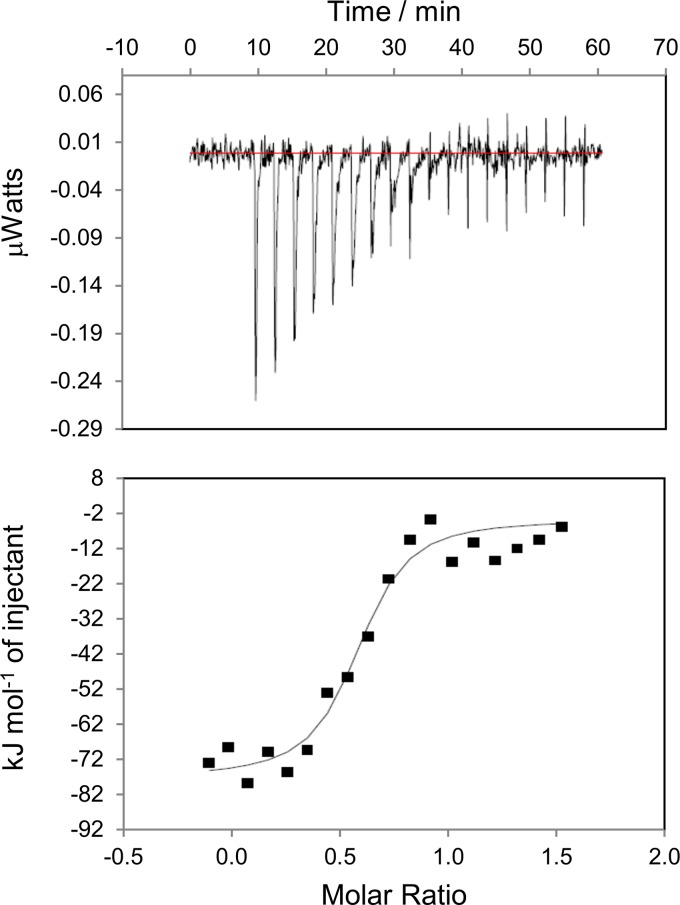
Isothermal titration calorimetry experiments were performed using an iTC200 Microcalorimeter from MicroCal, Inc. (Northampton, MA). OD4.2.2 and VRC-PG04 Fab were dialyzed in the same PBS buffer (Gibco) overnight at room temperature. Concentrations of all ligands were determined using A280 absorbance values and known extinction coefficients for both proteins. OD4.2.2 was titrated with VRC-PG04 Fab using 20 injections of 2 μl per injection at 25°C. The concentration of the OD4.2.2 molecule was 3 to 5 μM, and the concentration of the VRC-PG04 Fab used was between 50 and 75 μM, with experiments carried out in triplicate. The calorimetric data-fitting results showed that the binding stoichiometry was ∼0.6, indicating that only 60% of the OD4.2.2 molecule in solution was in a fully active state and could be bound by VRC-PG04 Fab. Data were analyzed with Microcal ORIGIN software using a single-site binding model; even though OD4.2.2 was only partially active, the ORIGIN-derived values for entropy and enthalpy should be unaffected by this substoichiometric binding.
OD4.2.2 preparation for crystallography.
OD4.2.2 as a fusion protein with an HRV 3C protease cleavage site, followed by an engineered ferritin molecule used as a chaperon to increase the homogeneity of the glycosylation, was produced in GnTI− cells as described for 293 expression. Recombinant protein was purified by mannose-specific lectin (Galanthus nivalis) resin (EY Laboratories) specific for α1,3- and α1,6-linked high-mannose structures, followed by cleavage of ferritin using HRV 3C protease. OD4.2.2 was purified away from ferritin by gel filtration using gel filtration buffer. OD4.2.2 was then deglycosylated at pH 6.0 using EndoHf endoglycosidase (New England BioLabs) for ∼30 h at 37°C. Digestion progress was assessed by SDS-PAGE visualization, and upon observation of a single protein band with minimal smearing, the digestion reaction was returned to pH 7.4, and fully deglycosylated molecules were separated from partially deglycosylated molecules by purification over concanavalin A (ConA) resin. This sample was then purified by gel filtration to remove the EndoHf enzyme. Pure deglycosylated protein was allowed to form complexes with VRC01 and VRC-PG04. The VRC01 and VRC-PG04 Fab proteins were incubated with a molar excess of OD4.2.2 protein in separate complex formation experiments. Fractions containing antibody-OD complex were concentrated to 16 mg/ml prior to crystal condition screening using a Honeybee crystallization robot, utilizing the vapor diffusion method in hanging drops at 20°C by mixing 0.2 μl of protein complex with 0.2 μl of reservoir solution.
Protein crystallization and data collection.
Initial robotic screening yielded crystal hits for the VRC-PG04-OD4.2 complex under two conditions: condition 1 using 10% polyethylene glycol (PEG) 400, 20% PEG 8000, 100 mM Na acetate, pH 5.5, and 500 mM NaCl; condition 2 using 52.5% PEG 400, 100 mM HEPES, pH 7.5, and 200 mM CaCl2. These crystals were manually reproduced in hanging drops by mixing 1 μl of protein complex with 1 μl of the initial reservoir solution and modification of both PEG concentrations for condition 1 and various PEG 400 concentrations for crystal condition 2. Crystallization condition optimization for both conditions allowed growth of larger single crystals which were harvested and flash-cooled in liquid nitrogen using 25% 2R,3R-butanediol as a cryoprotectant. Crystals from condition 1 gave diffraction to ∼2.85 Å in a trigonal format while crystals from condition 2 gave data to 3.0-Å resolution in a monoclinic format. Data were collected at a wavelength of 1.00 Å at the Southeast Regional Collaborative Access Team (SER-CAT) beamline ID-22 (Advanced Photon Source, Argonne National Laboratory).
Structure determination, model building, and refinement.
Diffraction data sets were scaled and processed using the XDS program, and crystal unit cell analysis indicated one antibody-OD4.2 complex present in each crystallographic asymmetric unit for both space groups. Molecular replacement searches were carried out using PHASER (26) with molecule component models extracted from the published VRC-PG04-gp120 complex structure (PDB accession number 3SE9) (5). Briefly, sequential searches with the constant domain of Fab VRC-PG04, the loop-deleted outer domain of HIV-1 clade E strain 93TH057, and the variable region of Fab VRC-PG04 were performed to obtain solutions for a full antibody-OD complex. To reduce model bias, the complementarity-determining regions (CDR) of the variable domain of Fab VRC-PG04 were truncated before molecular replacement searches. Model building was carried out using Coot (27), and refinement was performed using Refmac5 (14), PHENIX (28), and Buster-TNT (29). Iterative cycles of model building utilizing translation-libration-screw motion (TLS) refinement with 13 groups resulted in 190 residues of the OD4.2.2 molecule and the full Fab being modeled in the trigonal crystal form, while in the monoclinic form, 184 residues of the OD4.2.2 molecule and the entire Fab molecule were observed. Data collection and refinement statistics are presented in Table 1.
Table 1.
Crystallographic data and refinement statistics
| Parameter | Value for the indicated form of OD4.2.2-VRC-PG04a |
|
|---|---|---|
| 3-fold symmetry crystalb | 2-fold symmetry crystalc | |
| Data collection statistics | ||
| Space group | P3221 | P21 |
| Unit cell dimensions | ||
| a, b, c (Å) | 158.9, 158.9, 80.3 | 47.6, 73.47, 109.3 |
| β (°) | 100.9 | |
| Resolution range (Å) | 43.6–3.0 (3.11–3.0) | 50.0–2.85 (3.05–2.85) |
| Rmerge (%) | 14.6 | 22.5 |
| I/σI | 21.82 (2.63) | 6.21(2.01) |
| Completeness (%) | 97.2 (88.3) | 85.4 (76.3) |
| No. of reflections | 86,674 | 356,422 |
| No. of unique reflections | 22,975 | 14,947 |
| Refinement | ||
| Resolution (Å) | 43.6–3.0 | 50.0–2.85 |
| Rwork/Rfree (%) | 18.6/24.5 | 24.8/32.0 |
| B factors (Å) | ||
| VRC-PG04 Hd | 73.6 | 34.3 |
| VRC-PG04 Ld | 71.7 | 39.7 |
| KER2018 OD4.2.2 | 85.3 | 51.9 |
| Water molecules | 31.4 | N/A |
| RMSDs | ||
| Bond length (Å) | 0.009 | 0.009 |
| Bond angle (°) | 1.299 | 1.190 |
| Ramachandran plot (%) | ||
| Outliers | 0.49 | 1.17 |
| Favored | 94.49 | 93.49 |
Values in parentheses are for the highest-resolution shells.
PDB code 4I3R. Crystallization conditions: 10% PEG 400, 20% PEG 8000, 100 mM Na acetate, pH 5.5, 500 mM NaCl.
PDB code 4I3S. Crystallization conditions: 52.5 % PEG 400, 100 mM HEPES, pH 7.5, 200 mM CaCl2.
H and L, heavy and light chains of VRC-PG04 Fab, respectively.
Structure analysis and figure preparation.
The Ramachandran plot, as determined by MolProbity (30), shows 94.5% of all residues in favored regions and 99.5% of all residues in allowed regions for the trigonal crystal form and 93.5% of all residues in favored regions and 98.8% of all residues in allowed regions for the monoclinic crystal form. Structural comparison and alignments were carried out using LSQKAB (31, 32). Buried surface area analyses and contact residue analysis were carried out using Pdbsum (33) and PISA (34), respectively. All structural images were created using PyMOL (PyMOL Molecular Graphics System, version 1.1; Schrödinger, LLC).
Protein structure accession numbers.
Atomic coordinate and structure factors of the trigonal and monoclinic X-ray crystal structures of the OD4.2.2 with VRC-PG04 were deposited in the Protein Data Bank under accession codes 4I3R and 4I3S, respectively.
RESULTS
Design and antigenic optimization of an independent outer domain molecule.
The HIV-1 gp120 envelope glycoprotein is comprised of an inner domain, an outer domain, and a bridging sheet minidomain induced upon CD4-receptor binding (35). Although CD4 binds to both the outer domain and bridging sheet, the critical initial site of contact appears to reside primarily on the outer domain (36). To produce a functionally independent outer domain molecule with the same antigenic properties as those observed in the outer domain within the context of a full-length gp120 molecule, a number of features were varied. These variants included changes in HIV-1 strain origin, glycosylation pattern, length and sequence of loops, and presence of a stabilizing disulfide bond, and, in addition, recognition by antibodies b12, VRC01, and VRC-PG04 was assessed.
Initially, a gp120 outer domain—originating from a clade B virus strain R2 identified from a long-term-nonprogressor patient (18)—was used based on the premise that this molecule might be better able to elicit neutralizing antibodies that target the CD4-binding site. The initial construct, named R2 OD1 was made by creating a construct spanning residues 252 to 482 of the clade B strain R2 gp120 (Fig. 1A and 2) (gp120 residues are referred to by standard HXBc2 numbering system). Recognition of OD1 by antibodies b12, VRC01, and VRC-PG04 was in the submicromolar range, with b12 displaying fast on-/off-rates, while VRC01 and VRC-PG04 displayed reduced association and dissociation rates (Fig. 3 and Table 2; see also Fig. S1 to S3 in the supplemental material).
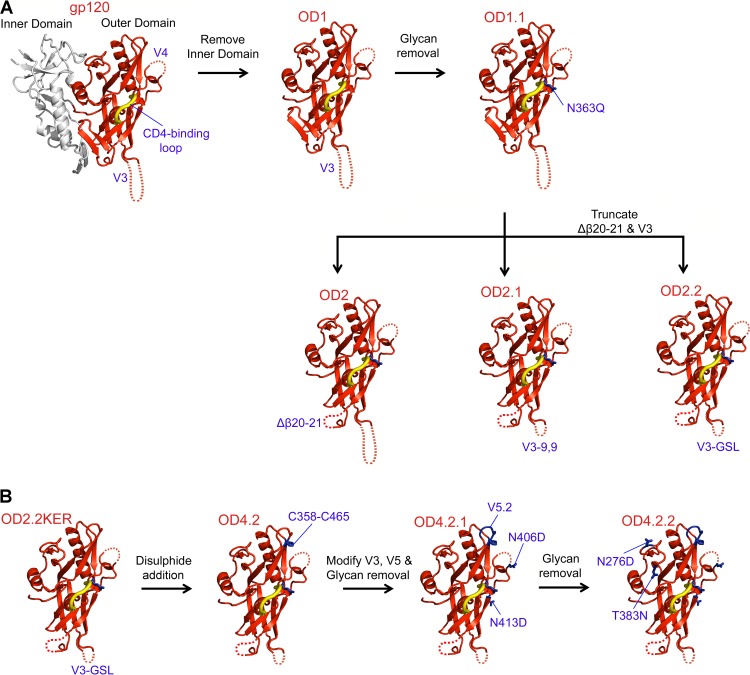
Fig 1.
Design of HIV-1 gp120 KER2018 clade A OD4.2.2. Sequential rational design based on gp120 structure coupled with binding studies allowed the production of a molecule with high affinity to VRC01-like antibodies. (A) Models of the sequential optimization of the outer domain of HIV-1 gp120 clade B R2 strain shown in red cartoon representation, with the gp120 inner domain shown in gray. Mutations/deletions are indicated by labels and dotted lines. (B) Sequential optimization of HIV-1 clade A KER2018 OD4.2.2 design. Specific mutations are labeled, and the residues are shown in stick representation in blue.
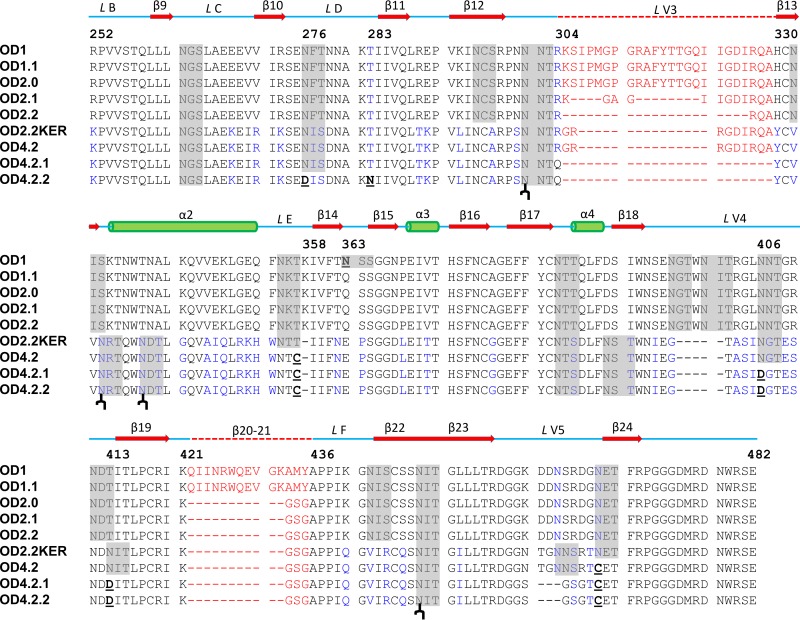
Fig 2.
Sequence alignment of outer domain designs. Residues which differ between versions are shown in blue. Residues underlined and in bold were mutated in the outer domain designs. V3-loop and the β20–21 strand residues are in red. Secondary structure elements and names are shown above the sequences with residue numbers also indicated. Predicted glycosylation sites are highlighted in gray, and the four glycosylation sites observed in OD4.2.2 are indicated ( ) below the OD4.2.2 sequence.
) below the OD4.2.2 sequence.
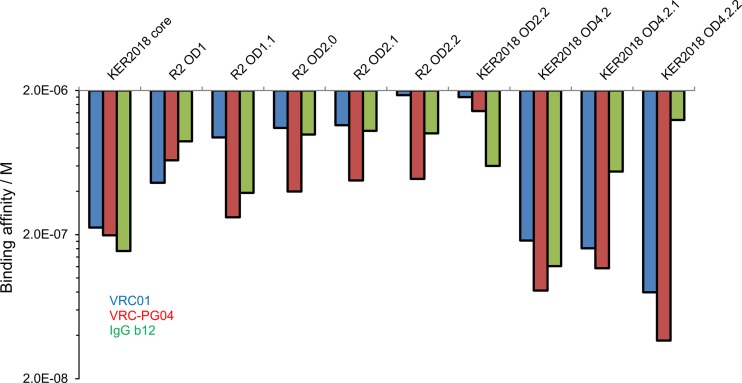
Fig 3.
Binding affinity constants of VRC01, VRC-PG04, and IgG b12 to core and outer domain constructs as assessed using Octet Biolayer Interferometry. Sequential truncations of the outer domain molecule resulted in lower binding affinity while structural stabilization, mutagenesis, and glycan removal led to increased affinity for VRC01 and VRC-PG04 antibodies.
Table 2.
Binding affinities of HIV-1 gp120 molecules to VRC01, VRC-PG04, and IgG b12 as measured by biolayer interferometrya
| Protein | VRC01 |
VRC-PG04 |
IgG b12 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Kon(M−1s−1) | Koff(s−1) | KD (M) | Kon(M−1s−1) | Koff(s−1) | KD (M) | Kon(M−1s−1) | Koff(s−1) | KD (M) | |
| KER2018 core | 1.79 × 103 | 4.00 × 10−4 | 2.23 × 10−7 | 2.77 × 103 | 5.50 × 10−4 | 1.99 × 10−7 | 4.90 × 104 | 7.60 × 10−3 | 1.54 × 10−7 |
| R2 OD1 | 2.21 × 103 | 1.06 × 10−5 | 4.79 × 10−7 | 2.37 × 103 | 1.56 × 10−3 | 6.58 × 10−7 | 7.44 × 104 | 6.59 × 10−2 | 8.86 × 10−7 |
| R2 OD1.1 | 1.46 × 103 | 1.39 × 10−5 | 9.52 × 10−7 | 3.40 × 103 | 9.00 × 10−4 | 2.65 × 10−7 | 1.52 × 105 | 5.95 × 10−2 | 3.91 × 10−7 |
| R2 OD2.0 | 1.71 × 103 | 1.89 × 10−3 | 1.11 × 10−6 | 3.17 × 103 | 1.26 × 10−3 | 3.97 × 10−7 | 1.49 × 105 | 5.95 × 10−2 | 3.91 × 10−7 |
| R2 OD2.1 | 1.82 × 103 | 2.09 × 10−3 | 1.15 × 10−6 | 3.34 × 103 | 1.59 × 10−3 | 4.76 × 10−7 | 1.87 × 105 | 2.01 × 10−1 | 1.07 × 10−6 |
| R2 OD2.2 | 1.06 × 103 | 1.96 × 10−3 | 1.85 × 10−6 | 2.75 × 103 | 1.34 × 10−3 | 4.87 × 10−7 | 1.74 × 105 | 1.76 × 10−1 | 1.01 × 10−6 |
| KER2018 OD2.2 | 3.31 × 103 | 5.98 × 10−3 | 1.81 × 10−6 | 3.27 × 103 | 4.72 × 10−3 | 1.44 × 10−6 | 3.12 × 104 | 1.86 × 10−2 | 5.96 × 10−7 |
| KER2018 OD4.2 | 1.06 × 104 | 1.94 × 10−3 | 1.83 × 10−7 | 2.55 × 104 | 2.12 × 10−3 | 8.31 × 10−8 | 6.10 × 105 | 6.90 × 10−2 | 1.21 × 10−7 |
| KER2018 OD4.2.1 | 1.84 × 104 | 2.95 × 10−3 | 1.60 × 10−7 | 1.80 × 104 | 2.09 × 10−3 | 1.16 × 10−7 | 1.40 × 105 | 7.68 × 10−2 | 5.48 × 10−7 |
| KER2018 OD4.2.2 | 1.39 × 104 | 1.11 × 10−3 | 7.99 × 10−8 | 1.86 × 104 | 6.84 × 10−4 | 3.68 × 10−8 | 1.09 × 105 | 1.33 × 10−1 | 1.22 × 10−6 |
Kon, on-rate constant; Koff, off-rate constant; KD, equilibrium dissociation constant. Estimate of error was less than 10% of measured values.
Analysis of the OD1 sequence identified a potential site of N-linked glycosylation at residue 363, which was adjacent to the functionally critical CD4-binding loop. To test the effects of removal of this potential glycan, an R2 OD1.1 molecule was designed that incorporated an N363Q mutation. R2 OD1.1 bound b12 with affinities of 390 nM, VRC01 at 947 nM, and VRC-PG04 at 264 nM (Fig. 3 and Table 2; see also Fig. S1 to S3). The observed change in affinity indicates that N363 is likely glycosylated, and its proximity to the CD4-binding site can influence recognition by neutralizing antibodies.
Next, β20–21 strands (OD2) and the V3 loop (OD2.1 and OD2.2) were truncated. In the context of viral Env, these truncations had been observed to improve the binding of b12 in strains derived from HIV-1 clade B virus IIIB and simian-human immunodeficiency virus (SHIV) 89.6 (37). In the context of the soluble outer domain, these truncations however, led to reduced binding (Fig. 3 and Table 2; see also Fig. S1 to S3 in the supplemental material). Although these mutations did not increase binding, the V3 loop and other flexible loops on gp120 appear to be immunodominant (38), and removing/truncating these regions reduced protein determinants that might compete for immune recognition of the critical VRC01 epitope on the outer domain. We thus decided that the small reduction in affinity was a reasonable trade-off for removal of potential immunodominant regions.
To enhance affinity further, we next tested combinations of strain variation and the presence of stabilizing disulfides. One variant, OD4.2, derived from the clade A strain KER2018 and containing a disulfide bond between residues 358 and 465, showed high-affinity binding to b12, VRC01, and VRC-PG04, with affinity constants of 121 nM, 183 nM, and 83 nM, respectively.
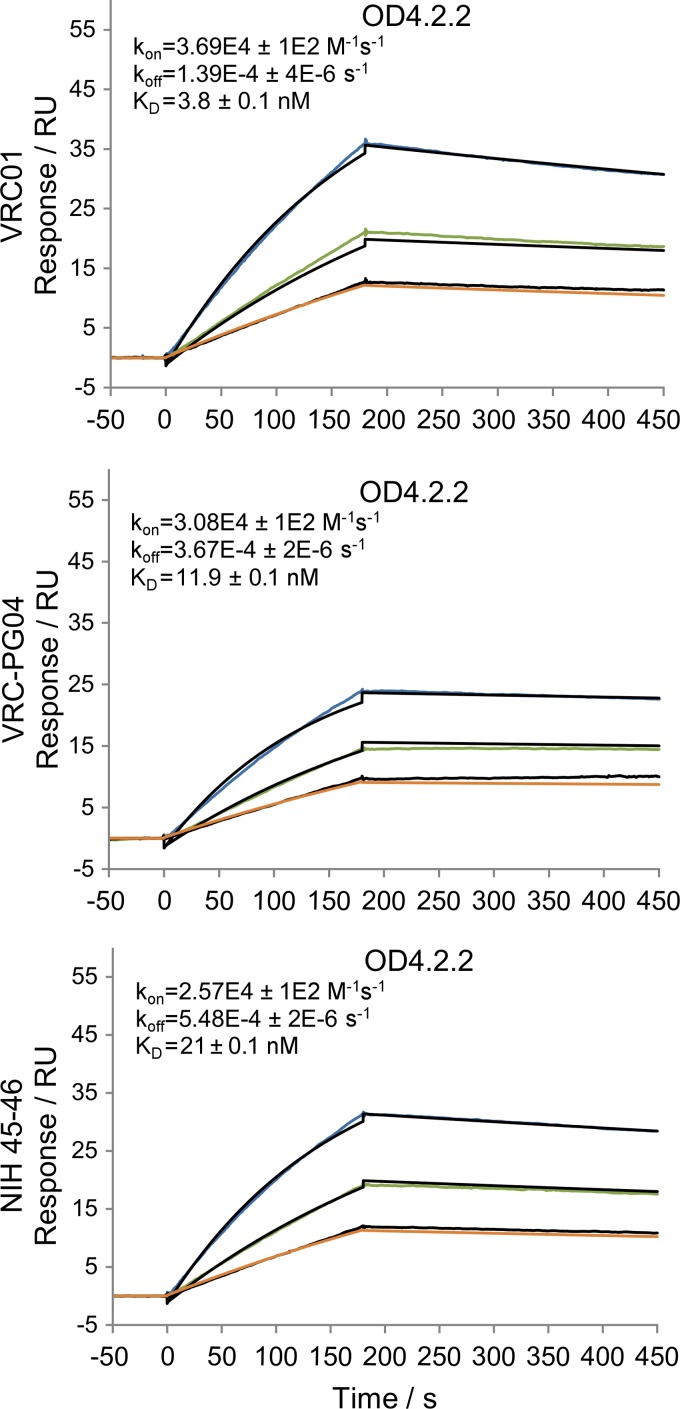
Alteration of residues 358 and 465 led not only to formation of the designed disulfide bond but also to the loss of a glycan at residue 356. We tested removal of additional N-linked glycans at residues 406 and 413 as well as shortening of the fifth variable loop (V5) from a nine-residue GGNTGNNSR sequence to a six-residue GGSGSG sequence (Fig. 1, V5.2) to produce OD4.2.1. These alterations reduced b12 recognition by ∼2-fold but had little effect on VRC01 or VRC-PG04 recognition (Fig. 3 and Table 2). Then, we tested the removal of an N-linked glycan at residue 276 and a potential site of O-linked glycosylation at residue 283 (OD4.2.2) (Fig. 1 and 2). Following these mutations, the affinity of OD4.2.2 for VRC01 and VRC-PG04 was greater than that of KER2018 core gp120 affinity for these antibodies although b12 affinity was weaker (Table 2). When assessed by surface plasmon resonance, OD4.2.2 bound VRC01 at an affinity of 3.8 nM, VRC-PG04 at 11.9 nM, and NIH45-46 at 21.3 nM (Fig. 4).
Fig 4.
Surface plasmon resonance sensorgrams of VRC01, VRC-PG04, and NIH45-46 antibodies immobilized using an anti-Fc antibody and OD4.2.2 flowed over the respective antibodies. The black lines represent the best fit of the kinetic data to a 1:1 binding model. Experiments were carried out at 25°C in PBS buffer (pH 7.4). koff, on-rate constant (M−1s−1); kon, off-rate constant (s−1); KD, equilibrium dissociation constant.
Finally, a broad group of poorly or nonneutralizing antibodies which target the CD4-binding site of HIV-1 gp120 were assessed for binding to OD4.2.2, core KER2018, and YU2 full-length protein (Table 3; see also Fig. S4 in the supplemental material). These antibodies can neutralize tier 1 isolates but cannot neutralize tier 2 isolates. None of these antibodies bound to the OD4.2.2 molecule, while all of these antibodies bind to the KER2018 core gp120 and YU2 full-length gp120 molecules with affinities ranging from micromolar to subnanomolar amounts.
Table 3.
Binding affinities of HIV-1 gp120 molecules KER2018 core and YU2 full-length gp120 to a group of poorly neutralizing CD4-binding site-directed antibodies as measured by surface plasmon resonance and biolayer interferometry
| CD4-binding site-directed antibody | KER2018 corea |
Full-length YU2a |
||||
|---|---|---|---|---|---|---|
| Kon(M−1s−1) | Koff (s−1) | KD (M) | Kon(M−1s−1) | Koff (s−1) | KD (M) | |
| b3 | 3.23 × 103 | 5.76 × 10−4 | 1.78 × 10−7 | 1.40 × 105 | 1.18 × 10−5 | 8.42 × 10−11 |
| m14 | 6.37 × 103 | 5.99 × 10−3 | 9.41 × 10−7 | 1.54 × 105 | 3.13 × 10−4 | 2.03 × 10−9 |
| m18 | 1.10 × 104 | 9.61 × 10−3 | 8.77 × 10−7 | 2.63 × 105 | 1.01 × 10−4 | 3.82 × 10−10 |
| b6 | 8.86 × 103 | 3.81 × 10−5 | 4.31 × 10−9 | 7.87 × 105 | 4.63 × 10−4 | 5.89 × 10−10 |
| b13 | 1.63 × 104 | 3.97 × 10−2 | 2.44 × 10−6 | 1.22 × 106 | 1.23 × 10−3 | 1.01 × 10−9 |
| F91 | 1.43 × 104 | 2.30 × 10−2 | 1.61 × 10−6 | 1.12 × 106 | 2.41 × 10−3 | 2.15 × 10−9 |
| F105 | 1.46 × 104 | 2.98 × 10−2 | 2.04 × 10−6 | 9.14 × 105 | 3.46 × 10−3 | 3.78 × 10−9 |
| 15e | 1.36 × 104 | 9.64 × 10−3 | 7.09 × 10−7 | 1.81 × 105 | 1.77 × 10−3 | 9.77 × 10−9 |
Kon, on-rate constant; Koff, off-rate constant; KD, equilibrium dissociation constant. Estimate of error was less than 10% of measured values.
In summary, sequential modifications from OD1 to OD4.2.2 led to a stable outer domain molecule made up of only 190 residues which had substantially increased affinity for VRC01 and VRC-PG04 compared to previous outer domain molecules. Although these changes reduced b12 affinity by ∼10-fold, antibody b12 is of substantially lower breadth than VRC01 and VRC-PG04, and we felt it critical to focus on the most effective antibodies—in this case, creating an outer domain immunogen specific for VRC01-like antibodies and incapable of binding to a large spectrum of poorly neutralizing antibodies.
Characterization of the thermodynamics of OD4.2.2 recognition by VRC-PG04.
To assess the thermodynamics of outer domain recognition by VRC-PG04, we carried out isothermal calorimetric measurements to assess the heat of interaction released by titrating OD4.2.2 with VRC-PG04 Fab in a microcalorimeter (iTC200; Microcal). The observed change in enthalpy, ΔH, for titrations was −20.4 ± 0.9 kcal/mol with a ΔG of −9.8 ± 0.9 kcal/mol, which corresponded to an affinity of 75 nM (Fig. 5). The calculated entropy (−TΔS, where T is the absolute temperature and ΔS is the change in entropy) from these measurements was 10.6 ± 1.3 kcal/mol, indicative of an entropically driven interaction, in which each of the interacting molecules does not have a substantial decrease in entropy relative to the bound state, but the interaction nonetheless results in substantial release of solvent from the binding surface. These results differ dramatically from the result with the full-length version of gp120 binding to CD4 or VRC01, in which −TΔS is 40 to 50 kcal/mol (39, 40), and the interaction is driven by the heat of folding related to gp120 conformational rearrangements, with substantial opposing reduction in entropy. Overall, the thermodynamics of OD4.2.2 binding to VRC-PG04 suggest an interaction between two well-folded molecules that is substantially more “lock-and-key” than that observed with full-length gp120 interactions with either CD4 or VRC01.
Fig 5.
Calorimetry data for the titration of OD4.2.2 with VRC-PG04 antibody in PBS buffer (pH 7.4). Measurements were carried out in triplicate with a representative titration result shown. The top panel shows raw data with the area under each spike proportional to the heat produced at each injection of VRC-PG04 Fab. The lower panel shows integrated areas normalized to the number of moles of VRC-PG04 Fab.
Crystal structures of OD4.2.2 in complex with VRC-PG04.
Crystallization was attempted with OD4.2, OD4.2.1, and OD4.2.2, either alone or in complexes with the Fab of VRC01, VRC03, and VRC-PG04. Small-volume robotic screening of 576 crystal growth conditions for each sample produced crystals only for the complex of OD4.2.2 with the Fab of VRC-PG04. Two OD4.2.2-VRC-PG04 crystal forms were observed, one with growth morphology that was 3-fold symmetric and the other with morphology that was 2-fold symmetric. Crystals of each of these crystal forms were reproduced manually and cryo-protected with 25% 2R,3R-butanediol to enable data collection. The 3-fold symmetry crystals diffracted to 3.0 Å while the 2-fold symmetry crystals diffracted to 2.85-Å resolution; the latter diffraction displayed greater anisotropy with visible splitting of reflections at higher resolution. Molecular replacement revealed the space group for the 3-fold symmetry crystals to be P3221 with a single complex of VRC-PG04 Fab and outer domain in the asymmetric unit, and final refinement yielded Rcryst/Rfree values of 18.6%/24.5% (Table 1). Meanwhile, molecular replacement revealed the space group for the 2-fold symmetry crystals to be P21 with a single complex of VRC-PG04 Fab and outer domain in the asymmetric unit, and refinement yielded Rcryst/Rfree of 24.8%/32.0% (Table 1). The refined models obtained in the trigonal and monoclinic lattices were highly similar to one another, with root mean square deviation (RMSD) of 0.7 Å over all atoms (see Fig. S5 in the supplemental material).
Electron density appeared superior with the trigonal crystals versus the monoclinic ones, with greater continuity in the fourth variable loop, allowing a complete model trace for this loop, and electron density for the N and C termini that allowed one and five additional residues, respectively, to be traced relative to the model obtained from the monoclinic crystals. In addition, the monoclinic lattice displayed contacts which might influence the conformations of the N and C termini of the OD4.2.2 molecule. For these reasons, the following structural description is derived from the trigonal cocrystals of OD4.2.2 with VRC-PG04.
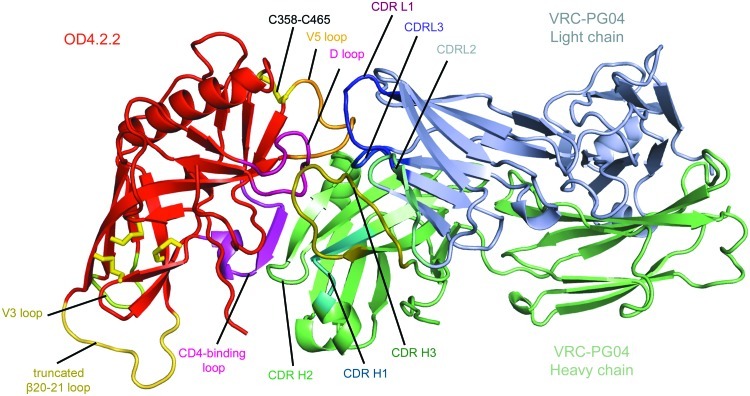
The crystal structure of OD4.2.2 in complex with Fab VRC-PG04 is shown (Fig. 6). The OD4.2.2 interface with VRC-PG04 had 2,175 Å2 of buried surface: 938 Å2, 189 Å2, and 1,048 Å2 from VRC-PG04 heavy chain, light chain, and OD4.2.2, respectively. The heavy chain of VRC-PG04 formed the majority of contacts including 12 hydrogen bonds, two salt bridges, and 133 nonbonded contacts. These involved 19 residues of VRC-PG04 interacting with 23 residues of OD4.2.2. The light chain of VRC-PG04 formed an additional 10 nonbonded contacts utilizing two residues of VRC-PG04 to contact five outer domain residues. All six CDR loops were well ordered with an average B factor of 65 Å2 compared with 90 Å2 for the overall B factor of the complex. Two of the loops, CDR L1 and CDR L2, made no contacts. Of the remaining four loops, CDR H2 and CDR H3 formed the most extensive contacts, interacting with the CD4-binding loop and loop D of OD4.2.2, respectively.
Fig 6.
Structure of the outer domain from clade A KER2018, variant OD4.2.2, in complex with the antigen binding domain of antibody VRC-PG04 shown in cartoon representation. Both the light- and heavy-chain regions of VRC-PG04 interact with the outer domain of gp120 in a manner typical of VRC01-like antibodies with the CDR H2 forming strong interactions with the CD4-binding loop. The OD4.2.2 molecule is shown in red except for the V5 loop (orange), D loop (pink), V3 loop (yellow), and truncated β20–21 loop (yellow-orange); disulfide bonds found in the outer domain molecule are shown in yellow stick format. VRC-PG04 is in green and light blue for the heavy and light chains, respectively, except for CDR H1 (teal), CDR H2 (green), CDR H3 (dark green), and CDR L1 (purple), CDR L2 (cyan), and CDR L3 (blue).
Comparison of OD4.2.2-VRC-PG04 complex and coree-VRC-PG04 complex.
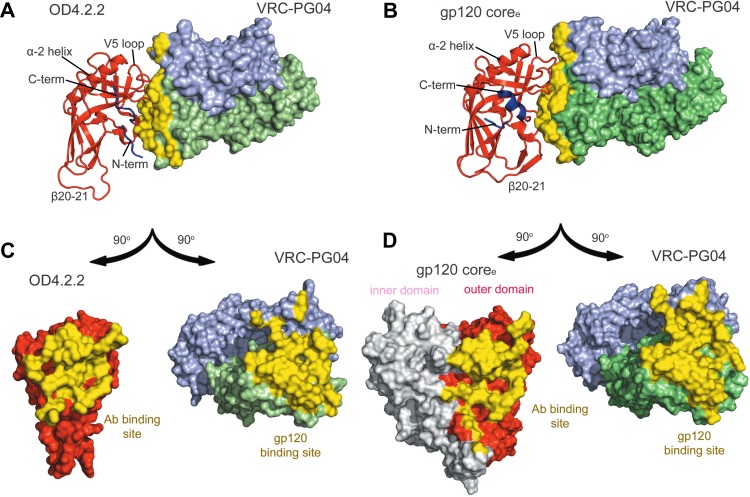
The structure of VRC-PG04 was determined previously in complex with an HIV-1 gp120 extended core (coree) (PDB code 3SE9) (5), which is about twice the size of OD4.2.2 and comprises a gp120 with truncations in V1/V2 and V3 as well as the N and C termini. Comparison of the VRC-PG04-bound structures of OD4.2.2 and clade A/E gp120 coree revealed 83 residues with less than 1 Å RMSD (Fig. 7). These include the critical CD4-binding loop and the loop D region of OD4.2.2, which are virtually indistinguishable from those observed in the core structure, and the antibody interactions with these regions are also preserved.
Fig 7.
Comparison of the OD4.2.2 molecule and clade E coree HIV-1 gp120 molecule in complex with VRC-PG04. Both the gp120 epitopes and antibody paratopes have significant regions of similarity, but additional contacts are made between the heavy chain of VRC-PG04 and OD4.2.2 compared to its binding site observed in the HIV-1 coree complex structure, while VRC-PG04 light-chain contacts are reduced. (A) OD4.2.2 (cartoon format with the N and C termini regions in blue) in complex with VRC-PG04 (surface representation). The buried surface area for VRC-PG04 is shown in yellow. (B) The outer domain of HIV-1 gp120 coree (cartoon format; the inner domain is omitted for clarity) in complex with VRC-PG04 (surface representation) is shown with the VRC-PG04 buried surface area in yellow. (C) OD4.2.2 and VRC-PG04 are rotated 90° in opposite directions, respectively, to allow visualization of the buried surface area between the epitope of the gp120 outer domain and the paratope of the VRC-PG04 antibody with the buried interactive surfaces shown in yellow. (D) HIV-1 gp120 coree molecule and the VRC-PG04 antibody are rotated to allow visualization of their respective epitopes and paratopes which are shown in yellow surface representation.
Despite substantial similarity, differences were also observed, especially at the N and C termini and V5 regions of OD4.2.2 (Fig. 7). The N terminus (residues 252 to 256),which typically interacts with the α-1 helix found in the inner domain of core gp120 structures, is displaced ∼20 Å in the OD4.2.2 complex toward the VRC-PG04 interface, where it interacts directly with VRC-PG04, burying a total of 186 Å2 of surface area between the OD4.2.2 N terminus and the heavy chain of VRC-PG04. Meanwhile, the C terminus (residues 474 to 482), which typically forms the α-5 helix, unfolds into a strand that interacts with outer domain strands β9 and β10.
The V5 loop (residues 457 to 465) also adopts a different orientation. In the coree complex structure, the V5 loop contacts the light chain with a total buried surface area of 305 Å2. In contrast, in the OD4.2.2 complex structure, the V5 loop does not contact the light chain. Instead, it contacts the CDR H3 region of VRC-PG04 with a total buried surface area of 189 Å2. This alteration in the V5 region may be related to the removal of an N-linked glycan at residue 465 in OD4.2.2 (in the coree complex, this glycan contributes 124 Å2 of buried surface area to the light chain of VRC-PG04). Alternatively, the introduction of the disulfide bond between residue 358 and the C terminus of V5 (at residue 465) may also alter the orientation of V5.
Overall, the OD4.2.2 and coree interfaces with VRC-PG04 display both similarities and differences. In the absence of structures corresponding to OD design intermediates, it is unclear whether the antigenic optimization enhances the mimicry between OD4.2.2 and coree recognition of VRC-PG04. The large potential structural diversity of the outer domain thus complicated correlations between ligand affinity and structural mimicry.
Comparison of OD4.2.2 with other HIV-1 gp120 core structures.
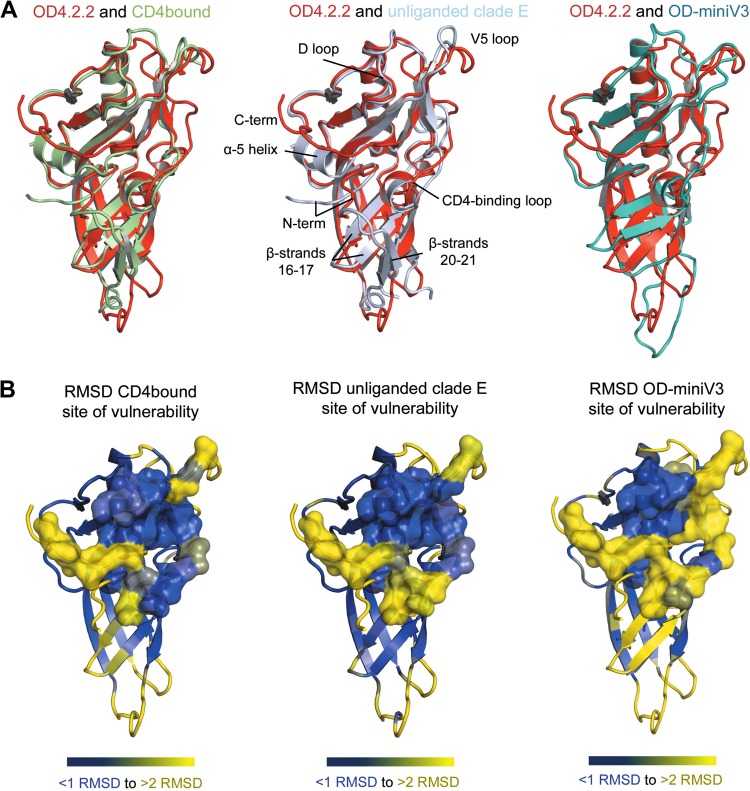
In addition to the VRC-PG04-bound coree molecule, the OD4.2.2 structure was compared to HIV-1 gp120 core structures in the CD4-bound and unliganded contexts (Fig. 8). As with the VRC-PG04-bound coree comparison, the overall structure of OD4.2.2 was similar to that seen in the core structures, with the major differences occurring in the N and C termini and the V5 regions (Fig. 8).
Fig 8.
Structural comparison of OD4.2.2 to HIV-1 gp120 in the CD4-bound form as observed in the CD4-liganded complex and unliganded clade E coree and the HXBc2 circularly permutated OD-miniV3. Comparisons indicate that there are significant differences in the HIV-1 site of vulnerability, the N- and C-terminal region of the outer domain, and regions adjacent to β-strands 16 and 17. (A) Structural overlay of OD4.2.2 (red) with the gp120 outer domain of CD4-bound gp120 (green) and unliganded clade E gp120 (light blue) and the circularly permutated HXBc2 OD-miniV3 outer domain (teal). (B) OD4.2.2 is shown in cartoon representation colored according to RMSD comparisons with other outer domains of gp120 (blue, <1 Å RMSD; yellow, >2 Å RMSD); the previously defined site of vulnerability is shown in semitransparent surface representation.
In terms of the site of vulnerability associated with the CD4 contact region of the outer domain, comparison of OD4.2.2 with both the CD4-liganded and unliganded core structures revealed that most of this critical region is preserved. The primary differences result from structural changes in the OD4.2.2 N terminus and V5 regions. In the unliganded structure, the CD4-binding loop is altered relative to that of the CD4-bound structure and also the VRC-PG04-bound OD4.2.2 structure, indicating that there is plasticity in this region even in core gp120 structures. Overall, the majority of the sites of vulnerability displayed RMSDs of less than 1 Å between equivalent atoms, indicating substantial mimicry in this critical region of the outer domain.
Comparison of OD4.2.2 with OD-miniV3.
Recently, the structure of a circularly permutated outer domain construct from the clade B HXBc2 strain, which contained a minimized V3 loop (OD-miniV3), was determined in complex with the N332-directed PGT128 antibody (PDB code 3TYG) (Fig. 8) (41). This circularly permutated outer domain molecule was connected at the native outer domain N and C termini (residues 254 and 477) using a nine-amino-acid linker region (IARCQIAGT). The artificial N and C termini were placed at the termini of the V4 loop at residues 413 and 395A, respectively, thus removing the V4 loop.
Overall structural comparison of OD4.2.2 with OD-miniV3 (Fig. 8) indicated that although there are large sequence differences (Fig. 9) (56% sequence identity), a significant proportion of the molecule had a similar structure with an RMSD of <1 Å between equivalent atoms. In total, a defined set of 73 residues showed less than 1 Å RMSD between the two outer domain structures (Fig. 8 and 10), and 57 of the 73 residues shared structural similarity with the gp120 core structures.
Fig 9.
Sequence alignment of outer domain molecules (OD4.2.2 and OD-miniV3) and the outer domains of the CD4-bound HXBc2 molecule and unbound clade E molecule. Residues highlighted in red form the site of vulnerability of the CD4-receptor binding site.
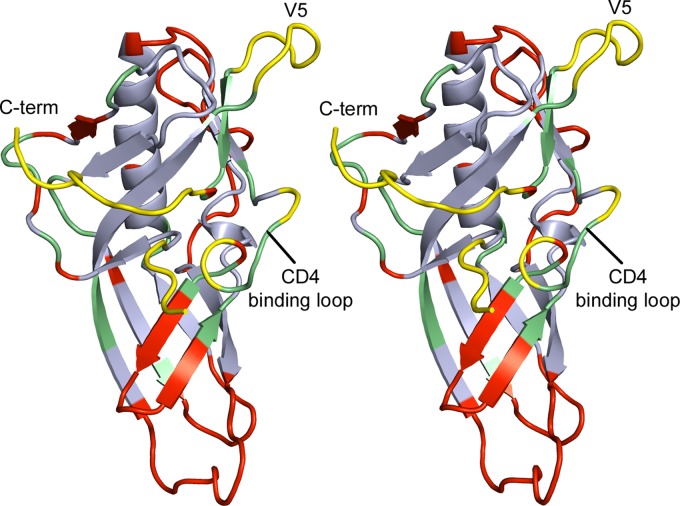
Fig 10.
Stereo view of the OD4.2.2 molecule. Analysis and comparisons of this structure indicate regions which are conserved between outer domain molecules regardless of sequence or design (light blue), regions which are in the CD4-bound conformation (green), areas which are dissimilar across the two outer domain molecules (red), and regions which should be targeted for further optimization/stabilization (yellow) in order to produce an optimal outer domain immunogen.
We next examined the site of vulnerability associated with the outer domain contact for CD4. In terms of sequence, 22 of 24 residues that comprise the site of vulnerability on the outer domains are identical. Despite this similarity, substantial structural differences were observed in the site of vulnerability between OD4.2.2 and OD-miniV3. Of the ∼5 sequence segments that make up this site (Fig. 10), only the loop D region (residues 274 to 283) display less than 1 Å RMSD. Regions with substantial alterations include the V5, CD4-binding loop, and N terminus.
The previously noted removal of a single V5 glycan as well as the introduction of a disulfide bond may alter the conformation of the V5 region in OD4.2.2 compared to that of the OD-miniV3. Meanwhile, strands β16 and β17, which lie under the CD4-binding loop, adopt a different conformation in OD-miniV3. Although it is difficult to pinpoint the basis of this structural difference, we note that the clade B R2 outer domain variants displayed reductions in affinity upon truncation of β20–21 strands and the V3 loop, and OD-miniV3 utilizes substantially different V3 and β20–21 amino acid sequences. These observed binding and structural differences indicate that modifications in these regions can have greater overall effects. Finally, the region of OD-miniV3 that corresponds to the N and C termini in OD4.2.2 is disordered in the OD-miniV3 structure. This disorder, coupled to the variation that we observe in the N and C termini of OD4.2.2, leads us to believe that this region is highly mobile, potentially existing in multiple conformations. Together, these differences suggest that OD-miniV3 would have to alter its binding surface substantially to be recognized with high affinity by VRC-PG04.
Overall, the structural variation between OD4.2.2 and OD-miniV3 highlights the substantial structural malleability of the gp120 outer domain. To preserve the outer domain contact for CD4, it seems essential to stabilize a number of the segments that comprise this site, including the outer domain N terminus, regions in proximity to the CD4-binding loop, V5 loop, and also the outer domain C terminus. In particular, the novel orientations of the N and C termini of OD4.2.2 and the relatively high RMSD for these areas in comparisons between multiple gp120 molecules indicate that these residues are a particularly malleable region of the outer domain. These residues may constitute an intermediate layer between the outer domain and the inner domain vital for viral entry; indeed, mutagenesis of these residues in the context of the functional viral spike dramatically affects infectivity of HIV-1 (42). Finally, we note that our OD4.2.2 is in complex with VRC-PG04, so while there are substantially higher similarities in the sites of vulnerability between OD4.2.2 and other core gp120 molecules than with the OD-miniV3, the role of VRC-PG04 in stabilizing the observed OD4.2.2 conformation is unclear in the absence of a ligand-free OD4.2.2 structure.
DISCUSSION
Elicitation of neutralizing antibodies that target the CD4-binding site is a major goal of HIV-1-vaccine development. Since the majority of the CD4-binding site is located on the outer domain of the gp120 molecule, the outer domain may represent a minimal HIV-1 immunogen. Through an iterative design process coupled with assessment of antibody recognition, we were able to produce a soluble outer domain with antigenic properties which appeared to be similar to or better than those of full-length Env molecules. Thermodynamic measurements further suggested that substantial structural rearrangements were not required for interaction with broadly neutralizing antibodies such as VRC-PG04.
The iterative procedure to improve the antigenicity of a soluble outer domain molecule is the first reported case of an outer domain-only construct with CD4-binding-site antigenicity for broadly neutralizing antibodies comparable to that of larger HIV-1 Env constructs. The antigenic optimization of this construct, OD4.2.2, was accomplished while reducing the size of gp120 from its full length of ∼500 residues to a more compact 190 residues, removing the immunodominant V3 and β20-21 regions, and eliminating the affinity to poorly or nonneutralizing antibodies. Crystallization of OD4.2.2 in complex with VRC-PG04 permitted an atomic-level examination of both OD4.2.2 structure and VRC-PG04 epitope. Comparison of the CD4-binding site of vulnerability on OD4.2.2 with the same site in the context of other gp120 molecules indicated that despite antigenic optimization, aspects of OD4.2.2 differed structurally. To produce an outer domain immunogen capable of eliciting VRC01-like antibodies, further optimization of the outer domain, especially in regions corresponding to the V5 and N and C termini, likely needs to be carried out to enhance OD4.2.2 structural mimicry of the site of CD4 contact (Fig. 10).
Overall, our results suggest that although antigenic optimization can lead to high-affinity interactions between outer domain molecules and VRC01-like antibodies, the potential of HIV-1 Env for conformational mobility and molecular variation may be underappreciated. A combination of atomic-level structures alongside antigenic optimization and immune-response characterization may enable the development of immunogens capable of eliciting broadly neutralizing antibodies. Further assessment of the affinity of outer domain-only molecules to germ line and intermediate antibodies will gauge the capability of outer domain molecules to elicit and guide the development of a broadly neutralizing immune response (43). It will be interesting to see whether structural mimicry, antigenic similarity, or another factor such as germ line affinity plays the dominant role in eliciting antibodies like VRC01.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Structural Biology Section, Structural Bioinformatics Section, Laboratory of Virology, and the Vector Core Section at the Vaccine Research Center for insightful comments and discussions and J. Stuckey for assistance with figures.
Support for this work was provided by the Intramural Research Program of the National Institutes of Health (National Institute of Allergy and Infectious Diseases). Use of sector 22 (SER-CAT) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
Footnotes
Published ahead of print 12 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02717-12.
REFERENCES
- 1. Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19–42 [DOI] [PubMed] [Google Scholar]
- 2. Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 3. Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 4. Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, Keefer MC, McElrath MJ, Walker MC, Wagner KF, McNeil JG, McCutchan FE, Burke DS. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J. Infect. Dis. 173:340–348 [DOI] [PubMed] [Google Scholar]
- 8. Moore JP, Cao Y, Qing L, Sattentau QJ, Pyati J, Koduri R, Robinson J, Barbas CF, III, Burton DR, Ho DD. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grundner C, Li Y, Louder M, Mascola J, Yang X, Sodroski J, Wyatt R. 2005. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology 331:33–46 [DOI] [PubMed] [Google Scholar]
- 10. Kang YK, Andjelic S, Binley JM, Crooks ET, Franti M, Iyer SP, Donovan GP, Dey AK, Zhu P, Roux KH, Durso RJ, Parsons TF, Maddon PJ, Moore JP, Olson WC. 2009. Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine 27:5120–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, Mascola JR. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lynch RM, Tran L, Louder MK, Schmidt SD, Cohen M, Dersimonian R, Euler Z, Gray ES, Abdool Karim S, Kirchherr J, Montefiori DC, Sibeko S, Soderberg K, Tomaras G, Yang ZY, Nabel GJ, Schuitemaker H, Morris L, Haynes BF, Mascola JR. 2012. The development of CD4 binding site antibodies during HIV-1 infection. J. Virol. 86:7588–7595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu X, Zhou T, O'Dell S, Wyatt RT, Kwong PD, Mascola JR. 2009. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. J. Virol. 83:10892–10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 15. Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, Willems B, Zekveld MJ, Dreja H, O'Sullivan E, Pade C, Orkin C, Jeffs SA, Montefiori DC, Davis D, Weissenhorn W, McKnight A, Heeney JL, Sallusto F, Sattentau QJ, Weiss RA, Lanzavecchia A. 2010. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5:e8805 doi:10.1371/journal.pone.0008805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang X, Tomov V, Kurteva S, Wang L, Ren X, Gorny MK, Zolla-Pazner S, Sodroski J. 2004. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J. Virol. 78:12975–12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu L, Zhou T, Yang ZY, Svehla K, O'Dell S, Louder MK, Xu L, Mascola JR, Burton DR, Hoxie JA, Doms RW, Kwong PD, Nabel GJ. 2009. Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J. Virol. 83:5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quinnan GV, Jr, Zhang PF, Fu DW, Dong M, Alter HJ. 1999. Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res. Hum. Retroviruses 15:561–570 [DOI] [PubMed] [Google Scholar]
- 19. Vujcic LK, Quinnan GV., Jr 1995. Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res. Hum. Retroviruses 11:783–787 [DOI] [PubMed] [Google Scholar]
- 20. Wu L, Yang ZY, Xu L, Welcher B, Winfrey S, Shao Y, Mascola JR, Nabel GJ. 2006. Cross-clade recognition and neutralization by the V3 region from clade C human immunodeficiency virus-1 envelope. Vaccine 24:4995–5002 [DOI] [PubMed] [Google Scholar]
- 21. Barbas CF, III, Collet TA, Amberg W, Roben P, Binley JM, Hoekstra D, Cababa D, Jones TM, Williamson RA, Pilkington GR, Haigwood NL, Cabezas E, Satterthwait AC, Sanz I, Burton DR. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812–823 [DOI] [PubMed] [Google Scholar]
- 22. Posner MR, Hideshima T, Cannon T, Mukherjee M, Mayer KH, Byrn RA. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J. Immunol. 146:4325–4332 [PubMed] [Google Scholar]
- 23. Ho DD, McKeating JA, Li XL, Moudgil T, Daar ES, Sun NC, Robinson JE. 1991. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J. Virol. 65:489–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang MY, Xiao X, Sidorov IA, Choudhry V, Cham F, Zhang PF, Bouma P, Zwick M, Choudhary A, Montefiori DC, Broder CC, Burton DR, Quinnan GV, Jr, Dimitrov DS. 2004. Identification and characterization of a new cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody. J. Virol. 78:9233–9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang MY, Shu Y, Phogat S, Xiao X, Cham F, Bouma P, Choudhary A, Feng YR, Sanz I, Rybak S, Broder CC, Quinnan GV, Evans T, Dimitrov DS. 2003. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J. Immunol. Methods 283:17–25 [DOI] [PubMed] [Google Scholar]
- 26. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart O, Vonrhein C, Womack T. 2011. BUSTER, version 2.10.0. Global Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 30. Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 32. Kabsch W. 1976. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32:922–923 [Google Scholar]
- 33. Laskowski RA. 2009. PDBsum new things. Nucleic Acids Res. 37:D355–D359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 35. Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li MQ, Lee SS, Gan ZG, Tan Y, Meng JH, He ML. 2007. Achieving a high coverage—the challenge of controlling HIV spread in heroin users. Harm Reduct. J. 4:8 doi:10.1186/1477-7517-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berkower I, Patel C, Ni Y, Virnik K, Xiang Z, Spadaccini A. 2008. Targeted deletion in the β20-β21 loop of HIV envelope glycoprotein gp120 exposes the CD4 binding site for antibody binding. Virology 377:330–338 [DOI] [PubMed] [Google Scholar]
- 38. Zwart G, Langedijk H, van der Hoek L, de Jong JJ, Wolfs TF, Ramautarsing C, Bakker M, de Ronde A, Goudsmit J. 1991. Immunodominance and antigenic variation of the principal neutralization domain of HIV-1. Virology 181:481–489 [DOI] [PubMed] [Google Scholar]
- 39. Li Y, O'Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. 2011. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J. Virol. 85:8954–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. U. S. A. 97:9026–9031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pietzsch J, Scheid JF, Mouquet H, Klein F, Seaman MS, Jankovic M, Corti D, Lanzavecchia A, Nussenzweig MC. 2010. Human anti-HIV-neutralizing antibodies frequently target a conserved epitope essential for viral fitness. J. Exp. Med. 207:1995–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haynes BF, Kelsoe G, Harrison SC, Kepler TB. 2012. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 30:423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.