Abstract
N-myc downstream regulated gene 1 (NDRG1/Cap43/Drg-1) has previously been shown to be dysregulated in esophageal squamous cell carcinoma (ESCC). In this study, we investigated the role of NDRG1 in the neoplastic progression of ESCC using ectopic gain-of-function and loss-of-function approaches. Stable transfectants of the KYSE30 ESCC cell line with altered NDRG1 levels were generated by lentiviral transduction. Although no measurable effects on in vitro cell proliferation were observed with altered NDRG1 expression, the ectopic overexpression of NDRG1 was positively linked to recognized markers of metastasis, angiogenesis and apoptotic evasion. Accordingly, in the nude mouse xenograft model system, NDRG1 overexpression promoted the in vivo growth of KYSE30 derived xenografts, which could be attributed to the reduced apoptotic and enhanced angiogenic activities associated with this gene. These processes were mediated in part by increased NFκB activity in NDRG1 overexpressing cells. Nevertheless, no significant phenotypic changes were observed in response to NDRG1 knock-down, suggesting that this gene might not be essential for the neoplastic progression of ESCC. Taken together, our results suggest that NDRG1 may play positive but dispensable roles in the progression of esophageal squamous cell carcinoma.
Keywords: NDRG1, esophageal carcinoma, proliferation, angiogenesis, apoptosis
Introduction
Esophageal squamous cell carcinoma (ESCC) ranks as one of the deadliest tumors with a high incidence in developing countries including parts of Southern Africa, the Middle East and the Far East.1 Indeed, its unfavorable prognosis is further complicated by the lack of knowledge about the molecular biology of this disease. Recently, an immunohistochemical (IHC) study of Japanese ESCC patients showed that overexpression of N-myc downstream regulated gene 1 (NDRG1) was associated with advanced tumor stages as an independent marker of unfavorable prognosis,2 suggesting a role for NDRG1 in esophageal tumorigenesis.
Despite evidence linking NDRG1 to cellular differentiation and tissue maturation during fetal and postnatal development,3 the functions of NDRG1 in cancer remain poorly understood. The expression of NDRG1 is strongly induced by various stress signals and stimuli related to carcinogenesis, including DNA damage, hypoxia, DNA methylation and histone deacetylation targeting drugs, carcinogens, as well as differentiation inducers.3 Various oncogenes and tumor suppressor genes participate in the regulation of NDRG1 expression including, p53,4 HIF-1α,5,6 AP-1,7 Egr-18 and PTEN,9 N-myc, c-myc10,11 and VHL.12 These observations are consistent with reports that NDRG1 expression is frequently altered during neoplastic processes. However, observations regarding the functional roles of NDRG1 in tumor progression are highly contradictory, from both immunohistochemical analysis of clinical samples to functional studies using ectopic NDRG1 overexpression or knock-down.
Previous studies suggested NDRG1 to be a tumor suppressor, as ectopic NDRG1 overexpression showed inhibitory effects on proliferation, metastasis and angiogenesis in colon,4,13 breast,9,14 prostate9,15 and pancreatic cancers.16 It was also reported in colon and lung cancer cells that NDRG1 was necessary to sensitize cells to doxorubicin induced apoptosis.4 In addition, immunohistochemistry observations in breast, colon, prostate and pancreatic cancers reported reduced NDRG1 expression in tumor tissue, especially when the tumor became metastatic.9,13-16
On the other hand, many other investigations support the role of NDRG1 as an oncogene. Besides ESCC2 it was shown that NDRG1 is upregulated during human and mouse skin carcinogenesis.17,18 Elevated NDRG1 levels in cancerous tissue was also observed in human oral squamous cell carcinoma,19 cervical adenocarcinoma,20 renal,21 colon,22 and liver cancers.23,24 In these cases, increased NDRG1 was associated with at least one of the following parameters: advanced tumor grade, metastasis, vascular invasion and poor prognosis. Moreover, ubiquitous upregulation of NDRG1 in tumor tissue compared with normal tissue was observed in multiple human cancer types including brain, breast, lung, colon, kidney, liver and prostate cancers.5 In functional studies, NDRG1 knock-down by RNA interference was shown to reduce cell proliferation and invasion, and induce apoptosis in hepatocellular carcinoma cell lines.25 NDRG1 could also mediate resistance to genotoxicity-mediated apoptosis in cultured colon cancer cells, in colon xenografts and even in colon cancer patients (clinical trial of irinotecan).26,27 Consistently, the anti-apoptotic role of NDRG1 in the context of hypoxia was also observed in human trophoblasts, with a normal genetic background.28 Considering these contradictory observations one could hypothesize that the function of NDRG1 in cancer may be tissue or even cell type specific.3
Therefore, further exploration through ectopic gain- and loss-of-function of NDRG1 was performed in ESCC cell lines, since the conflicting data in the literature do not clarify the role of NDRG1 in esophageal cancer.
Results
Altering NDRG1 expression has no substantial effect on in vitro cell growth in KYSE30 cells
To determine the potential role of NDRG1 in esophageal squamous cell cancer (ESCC), stable transfectants with altered NDRG1 levels were established using KYSE30 cells. A lentiviral vector system containing a GFP reporter gene to monitor transduction efficiency was used, and altered NDRG1 expression level was verified using western blot analysis (Fig. 1A). Flow cytometry analysis of the GFP protein in the transfectant “pool” showed an up to 85% GFP positive rate even 6 weeks (> 20 passages) after lentiviral transduction, indicating the cells were effectively and stably transduced (Fig. S1A).
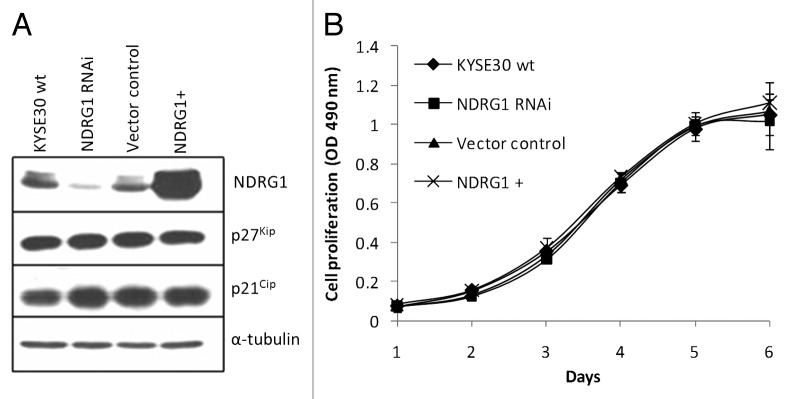
Figure 1. Ectopic alteration of NDRG1 expression level in KYSE30 cells and its effect on cell proliferation. “NDRG1+” refers to transfectants ectopically overexpressing NDRG1, here and elsewhere. (A) Western blot analysis for NDRG1, p27Kip and p21Cip. (B) Anchorage dependent proliferation of wild type KYSE30 cells and transfectants with altered NDRG1. The MTS proliferation assay was measured at 490 nm. Each point represents the mean of four readings in one experiment. Bars, ± SD.
We next compared the growth rates of wild type KYSE30 and derivative transfectants in culture by MTS assay. No significant difference in growth rates was observed in both adherent and non-adherent model systems (Fig. 1B; Fig. S2). This was confirmed by the similar DNA content profiles revealed by FACS analysis of propidium iodide stained cells (Fig. S1B) as well as the similar expression levels of several cell cycle regulators such as p21Cip1 and p27Kip1 (Fig. 1A)In summary, our results suggest that NDRG1 protein levels might not directly impact on cell proliferation in cultured KYSE30 cells.
NDRG1 is positively related to surrogate markers of metastasis
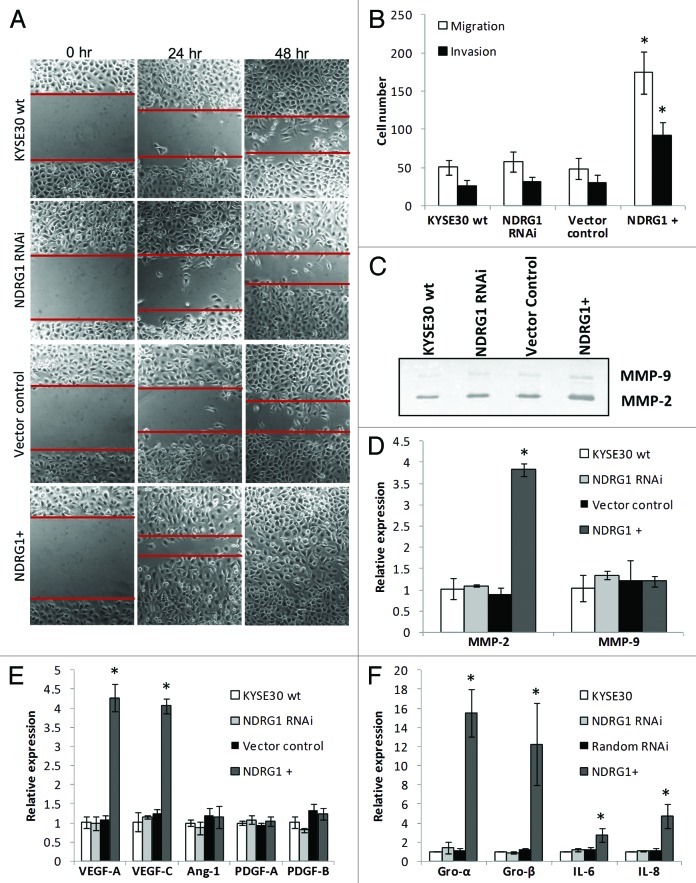
Next, the effects of NDRG1 on surrogate markers of metastasis in KYSE30 cells were investigated using several approaches. First, significantly enhanced motility of NDRG1-overexpressing KYSE30 cells was observed in scratch/wound-healing assays performed on Matrigel coated surfaces (Fig. 2A). Consistent results were also obtained on measuring cell migration/invasion under chemotaxis in transwell filters. Compared with wild type KYSE30 and vector control transfectants, markedly increased migration and invasion was observed in ectopic NDRG1-overexpressing transfectants (Fig. 2B). Besides motility, the invasiveness of cells was also determined by the ability of cells to degrade extracellular matrix components. Consistent with the evidence observed in the Matrigel invasion assay (Fig. 2B), our zymography results indicated elevated MMP-2 activity in response to NDRG1 overexpression, with the MMP-9 activity remaining relatively similar (Fig. 2C). Comparable observations were also shown in quantitative real-time RT-PCR assays, showing that elevated MMP-2 but not MMP-9 mRNA levels may be a downstream effector of NDRG1 in modulating cellular invasiveness (Fig. 2D).
Figure 2. The effect of NDRG1 on cell migration, invasion and expression of angiogenic cytokines. (A) Representative images (100 × magnification) of the scratch/wound-healing assay. The scratched “gaps” are highlighted with black lines. (B) Transwell migration and invasion assay. Cells that migrated through plain (migration) or Matrigel coated (invasion) porous membranes were quantified 48 h after seeding. (C) Gelatin zymography to detect gelatinases MMP-2 and MMP-9 activity. An aliquot of concentrated culture media representative of the same number of KYSE30 wild type cells and transfectants was subjected to the zymography assay. One of the three independent experiments is shown. (D) Quantitative real-time RT-PCR to determine the effect of NDRG1 on the mRNA levels of MMP-2 and MMP-9. The expression has been normalized to β-actin mRNA. (E and F) The mRNA levels of VEGF-A, VEGF-C, Angiopiotein-1 (Ang-1), PDGF-A, PDGF-B, GRO-α/CXCL1, GRO-β/CXCL-2, IL-6/INF-β and IL-8/CXCL-8 were determined by quantitative real-time RT-PCR. Expression has been normalized to β-actin mRNA. Columns, mean of the normalized data from three independent experiments; Bars, ± SD; * p < 0.05 vs. wild type control.
Angiogenesis and lymphangiogenesis play important roles in solid tumor growth and metastasis. Besides facilitating access to oxygen and nutrients for the maintenance of tumor growth, angiogenesis and lymphangiogenesis also provide access routes for the metastasis of cancer cells. In previous studies, angiogenic cytokines such as VEGF-A, VEGF-C, PDGF-B and angiopoietin-1 were all positively related to ESCC progression and malignancy.33,38,39 Using quantitative real-time RT-PCR, elevated mRNA levels of VEGF-A and VEGF-C but not PDGF-A, PDGF-B and angiopoietin-1 were detected in NDRG1 overexpressing cells (Fig. 2E). Furthermore, an increase in VEGF-A protein secreted into the media, as determined by ELISA, was also observed with 300.7 ± 48.1, 311.6 ± 51.4, 293.3 ± 82.1 and 567.9 ± 49.1 pg VEGF-A/106 cells detected in wild type, NDRG1 RNAi (p = 0.660), vector control (p = 0.771) and NDRG1 overexpressing KYSE30 cells (p = 0.021), respectively.
Recently, pro-inflammatory chemokines have been implicated as effective angiogenic promoters in several cancers, including ESCC.40,41 Besides directly mediating angiogenesis in endothelial cells, chemokines may also enhance the infiltration of macrophages/monocytes and neutrophils as additional potent manufacturers of angiogenic cytokines and chemokines. Interestingly, the mRNA levels of several pro-inflammatory chemokines such as GRO-α/CXCL1, GRO-β/CXCL-2 and IL-8/CXCL-8 were found significantly elevated in response to ectopic NDRG1 overexpression, suggesting the pleiotropic supportive role of NDRG1 on angiogenesis in KYSE30 cells (Fig. 2F). In addition, we also observed NDRG1 may enhance the mRNA level of IL-6/INF-β, an apoptotic suppressor and metastatic promoter previously shown to be related to ESCC progression.42
In contrast, no significant phenotypic response of NDRG1 knock-down was observed in the above assays. Taken together, in KYSE30 cells, we propose NDRG1 as a positive modulator of metastasis but this gene may not be necessary for the maintenance of the metastatic phenotype.
Ectopic NDRG1 overexpression has protective effect against apoptosis
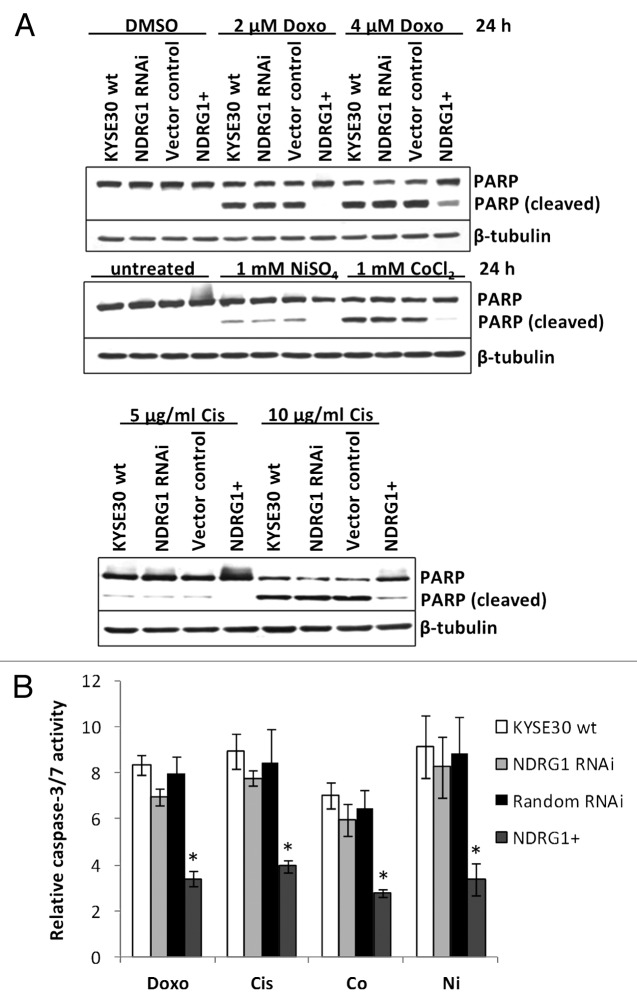
Apoptosis is another crucial regulator of carcinogenesis, especially in vivo, where cancer cells are continuously challenged by various stresses (e.g., DNA damage, hypoxic response). Considering that the expression of NDRG1 can be modulated by various stress conditions,3 we next checked whether NDRG1 was able to modulate the cellular response to apoptotic stimuli. After 24h treatment with DNA damaging agents (2 μM doxorubicin or 5 μg/mL cisplatin) or hypoxia mimics (1 mM Ni2+ or Co2+), KYSE30 cells underwent apoptosis, which was observed by cleavage of PARP protein (Fig. 3A) and elevated caspase 3/7 activity (Fig. 3B). Compared with wild type KYSE30 cells, reduced apoptosis (measured by both PARP cleavage and caspase 3/7 activity) was observed in transfectants with ectopic NDRG1 overexpression, while similar levels of apoptosis were found in both NDRG1 knock-down and vector control transfectants. Our results suggest a protective role for NDRG1 against apoptosis initiated by cellular stresses such as genotoxicity and hypoxia.
Figure 3. The effect of NDRG1 on apoptosis. (A) Subconfluent cells were challenged with indicated reagents for 24 h. Total cell lysate (including the floaters) was subjected to western blot analysis to detect the intact (p116) and cleaved (p85) forms of PARP. (B) Caspase3/7 activities were determined after 24 h treatment of indicated reagents, and were plotted as fold induction against untreated cells. Abbreviations for treatments: Doxo: 2 μM doxorubicin; Cis: 5 μg/mL cisplatin; Ni: 1 mM NiSO4; Co: 1 mM CoCl2. Columns, mean of the normalized data from three independent experiments; Bars, ± SD; * p < 0.05 vs. wild type control.
NDRG1-mediated chemokine expression is NFκB-dependent
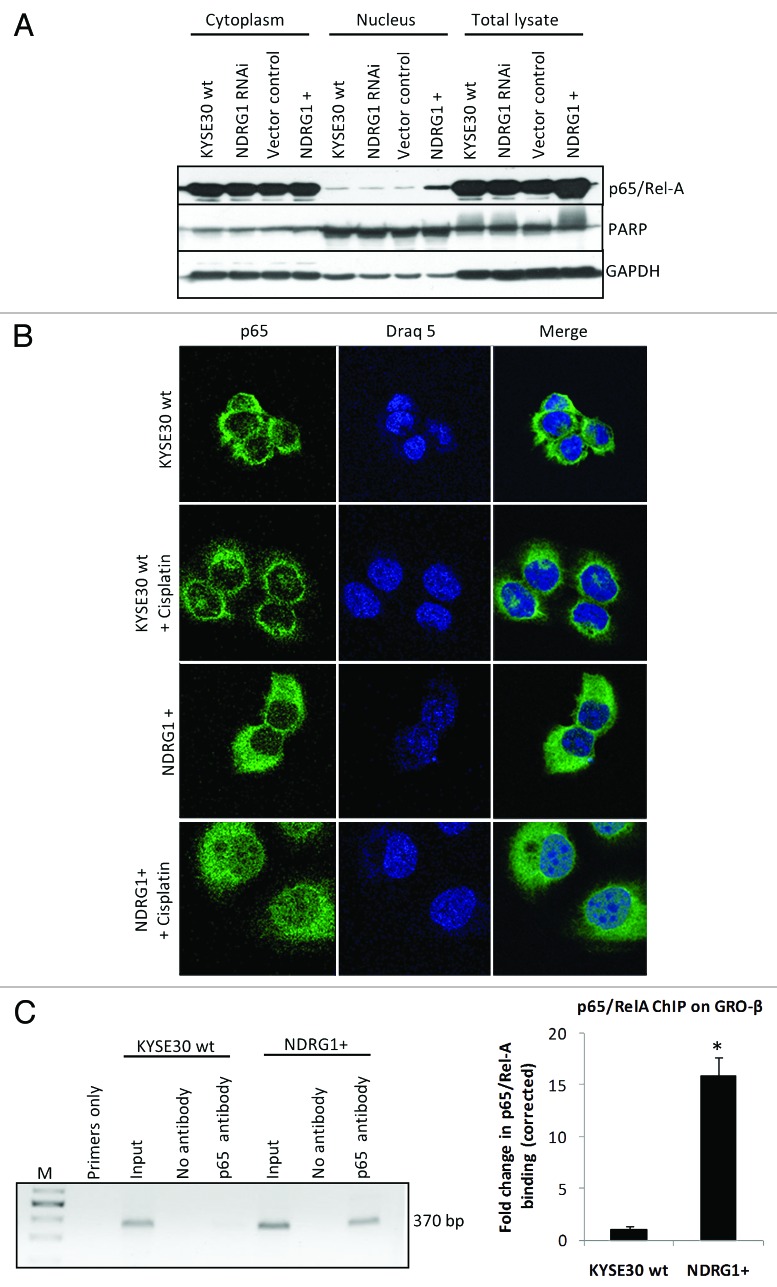
Considering the role of NFκB in regulating chemokine expression, we next examined whether this transcription factor participates in NDRG1-mediated chemokine upregulation. In KYSE30 cells overexpressing NDRG1, a significantly higher level of nuclear p65/RelA (a subunit of the NFκB complex) was demonstrated by western blot analysis on subcellular fractions (Fig. 4A), suggesting that NDRG1 overexpression was associated with elevated levels of nuclear p65 (NFκB). Likewise, immunofluorescence confocal microscopy images (taken through the plane of the nucleus) showed elevated levels (2–3 fold) of p65/RelA in the nucleus of NDRG1 overexpressing KYSE30 cells (KYSE30 NDRG1+), relative to control cells (Fig. 4B). Furthermore, treatment of KYSE30 NDRG1+ cells with cisplatin led to a substantial accumulation of p65/RelA in the nucleus of these cells, 2- to 3-fold higher than that observed in wild type cells treated with cisplatin or untreated KYSE30 NDRG1+ cells (Fig. 4B). In addition, a chromatin immunoprecipitation (ChIP) assay detected elevated binding of p65/RelA on the promoter of GRO-β/CXCL-2 (Fig. 4C) in KYSE30 NDRG1+ cells, indicating that increased levels of p65/RelA in the nucleus of cells was associated with increased binding of NFκB to its target genes.
Figure 4. NDRG1 overexpression enhances p65 nuclear localization and binding to the NFκB motif. (A) Whole cell lysates, along with cytoplasmic and nuclear protein fractions from KYSE30 cells and transfectants were subjected to western blot analysis to detect levels of p65 in the nucleus. Antibodies to PARP and GAPDH were used to determine the degree of cytoplasmic and nuclear cross-contamination, respectively. (B) Confocal microscope images, taken through the plane of the nucleus, showing p65 localization in KYSE30 cells and KYSE30 NDRG1 + cells untreated, or treated with Cisplatin (C), ChIP assays to demonstrate the p65/Rel-A dependent upregulation of GRO-β/CXCL-2 in NDRG1 overexpressing KYSE30 cells. ChIP assay was performed with p65/Rel-A antibody and null antibody as control in parental and NDRG1 overexpressing KYSE30 cells, respectively. Quantitative PCR was performed with specific primers flanking the p65/Rel-A consensus site of the GRO-β/CXCL-2 promoter. Both the PCR product (370 bp) and the qPCR result of the fold change corrected with non-specific interaction, detected in the no-antibody control ChIP, are shown; * p < 0.05 vs. wild type control.
Ectopic overexpression of NDRG1 promotes the progression of KYSE30 derived xenografts in nude mice
Xenograft studies were performed to check the validity of our in vitro observations in vivo. Six nude mice were randomly divided to each group and xenografts were generated by dorsal flank injection of 5 × 106 wild type KYSE30 cells as well as transfectants. Primary tumors developed in all nude mice and became palpable within one week after inoculation. However, due to the unexpected widespread, massive necrosis which liquefied the xenograft tissues and prevented accurate measurement, all the mice in the NDRG1 RNAi and vector control groups were euthanized in week 3 for ethical reasons and were excluded from this study. Consequently, the wild type KYSE30 cells were used as control. As shown in Figure 5A, significant bigger and more vascularized xenografts were generated from NDRG1 overexpressing cells.
Figure 5. Nude mice xenograft model to explore the in situ growth of KYSE30 wild type cells (KYSE30 wt) and transfectants with constitutive NDRG1 overexpression (NDRG1+) (n = 6 in each group). (A) 5 × 106 cells were inoculated subcutaneously into the dorsal flank. Photographs of the dissected xenografts with the same magnification scale are shown on the right. Dots, mean of the xenograft volume of all the six mice in the group; Bars, ± SD; significant difference in tumor volume was observed from week 2, * p < 0.05. (B) Analysis of proliferative activity, apoptosis and vascularization in histological sections of the xenografts derived from KYSE30 wt and NDRG1+ cells, with the quantitative results shown on the left and representative images (× 100) shown on the right. Columns, mean data of the sections from the six mice in the group; Bars, ± SD; (*) p < 0.05. (a) Proliferative activity was assessed by Ki-67 staining; (b) vascularization was determined by staining for CD31 and (c) apoptosis was measured by terminal TUNEL assay, as described in the methods section.
Immunohistochemical analysis was next performed on the xenograft sections (Fig. 5B). As expected, significantly higher NDRG1 levels were observed in xenografts derived from NDRG1 overexpressing cells, indicating that overexpression occurred and was maintained throughout the study period (data not shown). Although the Ki-67 proliferation index was higher in NDRG1 overexpressing xenografts (36.2 ± 5.1%) compared with wild type xenografts (29.8 ± 7.5%) (p = 0.021), the small difference in absolute value lies within the same range and might not fully explain the large increase in xenograft tumor volume (Fig. 5A). This is consistent with our in vitro data, where NDRG1 had no measurable effect on cell proliferation. Besides, NDRG1 overexpressing xenografts demonstrated enhanced angiogenic activity, which was visualized by CD31 immunohistochemical staining of murine endothelial cells. In xenografts with NDRG1 overexpression, the vascularized area filled with typical clusters of endothelia (an indication of well-developed microvessel network) occupied an average of 26.2% of the total area of the section (Fig. 5B). In contrast, a microvessel network could seldom be seen in xenografts derived from wild type cells (vascularization < 2% of the total area). Consistent with the in vitro observation shown in Figure 2, NDRG1 significantly correlates with increased angiogenesis in xenografts (p = 0.006), which provides the oxygen and nutrients to ensure rapid tumor growth. Furthermore, markedly decreased apoptotic activity was observed in the NDRG1 overexpressing xenografts through in situ TUNEL assay (p < 0.001) (Fig. 5B), which accordingly reflects the anti-apoptotic effect of NDRG1 observed in the cell culture system, although the decreased apoptosis could also be attributed to the elevated angiogenesis as a crucial stress alleviator of tumor cell growth in vivo. These observations suggest the participation of NDRG1 in the cellular adaptive responses against the in vivo growing environment. Angiogenesis may lead to a more favorable environment for cell growth, while apoptotic evasion protects cells from existing insults. Considering that NDRG1 has no effect on cell proliferation in culture, the significant effect of NDRG1 overexpression on the Ki-67 index may also be a reflection of enhanced adaptation driven by NDRG1.
Taken together, data from the nude mice studies are completely consistent with our results obtained from the in vitro experiments. For KYSE30 cells, we propose an oncogenic role for NDRG1, functioning as a stress alleviator in the development of ESCC.
Discussion
Although the biological function of NDRG1 has been explored in several types of cancer, very little is known about its role in esophageal squamous cell carcinoma (ESCC). Here, we provide the first evidence, through both in vitro and in vivo gain-of-function (ectopic overexpression) studies, that NDRG1 may have an oncogenic action in the progression of ESCC by promoting metastasis, angiogenesis and apoptotic evasion. In addition, although NDRG1 has been shown to be irrelevant to the proliferation of KYSE30 cells in culture, its positive effects on angiogenesis and apoptotic evasion may promote tumor growth in vivo as demonstrated in the xenograft study. Considering the mutual relationship between these neoplastic processes, it is conceivable that NDRG1 may mediate a series of programmatic changes, either directly or indirectly, and eventually facilitates progression of the tumor cell to a more malignant phenotype.
Nevertheless, no significant phenotypic alteration was observed through loss-of-function studies, although an effective knock-down efficiency (> 80%) was elicited in the transfectants investigated. It is possible that the remaining NDRG1 activity was sufficient to promote tumorigenesis. Despite the lack of direct evidence in the field of ESCC, we noticed that the role of NDRG1 may also be dispensable in normal physiological processes. NDRG1 has been implicated in the development and maintenance of murine brain and kidney due to its active regulation and abundant expression in these organs. However, no apparent abnormalities were observed in the brain and kidney of NDRG1 double knockout mice, morphologically or even functionally.43,44 Although functional studies have demonstrated that NDRG1 participates in trophoblast differentiation, a prerequisite event for the implantation of the embryo into the endometrium,28 NDRG1 deficient mice are fertile,43 suggesting NDRG1 may not be necessary for functional trophoblasts. Hence, we propose the existence of compensatory processes for NDRG1 function in both physiological and neoplastic backgrounds, clearly requiring further exploration.
Interestingly, the function of NDRG1 revealed in this study is consistent with the expression profile of this protein, as observed by other authors, suggesting that the findings in the ectopic functional studies were not an artifact. In a previous large-scale analysis of ESCC tissue samples from 124 patients, NDRG1 upregulation was shown strongly correlated with cancer progression (by TNM clinical classification), while the Kaplan-Meier analysis indicated NDRG1 expression was independently associated with an increased risk and poor prognosis.2 Moreover, multivariate analysis in the same study showed significant positive correlation between NDRG1 expression and several clinico-pathologic characteristics reflecting invasion, metastasis and angiogenesis. In line with other studies,5 NDRG1 was upregulated by hypoxia mimicking agents nickel and cobalt in KYSE30 and several other ESCC cell lines (data not shown), indicating NDRG1 as a potential hypoxia responsive gene in ESCC. Hypoxia, is a common phenomenon in solid tumors, including ESCC.45 As shown in our study, NDRG1 facilitates the adaptive responses to hypoxia such as angiogenesis and apoptosis evasion, which eventually contribute to the malignant phenotype.46 Furthermore, the pro-metastatic property of NDRG1 can also be viewed as a reflection of the outcome from hypoxia adaptation.
Until recently, the molecular mechanism of how NDRG1 exerts its function in ESCC still remains largely unclear. In our study, NDRG1 was observed to be capable of modulating the mRNA levels of many other genes, suggesting the ability of NDRG1 to influence the cellular transcriptome. Our results certainly implicate NFκB in mediating at least part of the phenotype in NDRG1 overexpressing cells, given our compartment fractionation and ChIP data. In fact, the candidacy of NFκB as an effector mediating NDRG1’s biological function is further supported by the solid evidence that NFκB signaling promotes tumor growth, metastasis, angiogenesis and apoptosis evasion in ESCC47. In addition, as shown in Figure 4B, NFκB is activated in response to cisplatin in KYSE30 cells, while NDRG1 overexpression further enhances this process. Interestingly, a study in patients with esophageal adenocarcinoma revealed similar NFκB activation after platinum-based therapy and linked the acquired NFκB activation with poor overall patient survival.48 Therefore, it may be hypothesized that NFκB may confer NDRG1-induced chemo-resistance (Fig. 3) and evasion of apoptosis (Fig. 5B).
In conclusion, our study suggests a pro-oncogenic but dispensable role of NDRG1 in ESCC as a modulator of metastasis, angiogenesis and apoptotic evasion.
Material and Methods
Cell line and reagents
Human ESCC cell line KYSE30, gifted from Dr. Shimada,29 was grown as described using RPMI1640 plus 10% fetal bovine serum (FBS). Materials for cell culture were all from Gibco (Invitrogen, Carlsbad, CA).
Antibodies were purchased as follows: NDRG1 from Kinasource; NF-κB p65, PARP, p21Cip1, p27Kip1, α- and β-tubulin from Santa Cruz Biotechnology; Ki-67 from Zymed; CD31 from Research Diagnostics Inc.. All the chemicals, unless specified, were purchased from Sigma-Aldrich.
Establishment of transfectants with altered NDRG1 expression
The bicistronic lentiviral vectors, pWPI and pLVTHM,30 packaging plasmid psPAX2 and envelope plasmid pMD2.G, were purchased from Addgene (http://www.addgene.org). pWPI, with a human EF1 promoter and a green fluorescent protein (GFP) reporter, was used to express NDRG1. Within the pLVTHM vector, an additional H1 RNA polymerase III promoter permits the expression of a short hairpin RNA (shRNA) for RNA interference (RNAi). The sense sequences for NDRG1 RNAi and scrambled control were 5′-GGAGTCCTTCAACGATTTG-3′ and 5′-GGGTCTTAGAACTAGTTCC-3′ respectively.28 Due to the similarity of pWPI and pLVTHM vectors, the scrambled RNAi control vector was used as vector control for both NDRG1 overexpression and knock-down experiments. All constructs were confirmed by sequencing.
Lentiviral particles were produced using 293FT cells (Invitrogen) cultured in Dulbecco's modified Eagle medium (DMEM) plus 10% FBS. Subconfluent cultures, in 75 cm2 flasks, were co-transfected with 8.5 μg of lentiviral vector, 3.9 μg of psPAX2, 1.3 μg of pMD2G and 33.5 μL FuGENE HD transfection reagent (Roche Diagnostics). The medium was changed 12h later for virus containing medium, which had been collected 36h post-transfection, filtered through a 0.45 μm PVDF filter (Millipore) and stored at -80°C.
Before transduction, virus containing medium was titrated using a serial dilution method.31 At subconfluence, KYSE30 cells were incubated with lentiviral particles, at a multiplicity of infection of 2–3 and 8 μg/mL hexadimethrine bromide for 16h, before the medium was changed. The transduction efficiency and stability of the transfectants were monitored by GFP FACS analysis.
Quantitative Real-time RT-PCR
Total RNA was extracted from cells using Trizol LS Reagent (Invitrogen). For first-strand cDNA synthesis, 1 μg of total RNA was reverse transcribed using ImProm-II™ Reverse Transcriptase system (Promega). Quantitative real-time RT-PCR was subsequently performed using the KAPA SYBR qPCR Master mix (KAPABiosystems) containing one of the primer pairs listed in Table 1. The comparative threshold cycle (CT) method32 was used for the calculation of expression fold change between samples, standardized using the β-actin housekeeping gene. Primers for VEGF-A, VEGF-C and PDGF-B were designed to detect all the transcript variants.
Table 1. Primer sequence, product length and product annealing temperature for quantitative Real-Time PCR study.
| Transcript | Primer sequence | Length (bp) | Annealing temperature °C |
|---|---|---|---|
| VEGF-A |
For 5′-CCTCCGAAACCATGAACTTT-3′ |
236 |
60°C |
| |
Rev 5′-TTCTTTGGTCTGCATTCACATT-3′ |
|
|
| VEGF-C |
For 5′-GCCAACCTCAACTCAAGGAC-3′ |
200 |
60°C |
| |
Rev 5′-CCCACATCTGTAGACGGACA-3′ |
|
|
| Angiopoietin-1 |
For 5′-TTCCTTTCCTTTGCTTTCCTC-3′ |
184 |
53°C |
| |
Rev 5′-CTGCAGAGCGTTTGTGTTGT-3′ |
|
|
| PDGF-B |
For 5′-TTATGAGATGCTGAGTGACCAC-3′ |
156 |
55°C |
| |
Rev 5′-CCTTCTTCCACGAGCCAAG-3′ |
|
|
| MMP-2 |
For 5′-TGGCGATGGATACCCCTTT-3′ |
117 |
57.5°C |
| |
Rev 5′-TTCTCCCAAGGTCCATAGCTCAT-3′ |
|
|
| MMP-9 |
For 5′-CCTGGGCAGATTCCAAACCT-3′ |
88 |
53°C |
| |
Rev 5′-GCAAGTCTTCCGAGTAGTTTTGGAT-3′ |
|
|
| GRO-α |
For 5′-CCCCAAGAACATCCAAAGTG-3′ |
122 |
60°C |
| |
Rev 5′-TAACTATGGGGGATGCAGGA-3′ |
|
|
| GRO-β |
For 5′-CTCAAGAATGGGCAGAAAGC-3′ |
132 |
60°C |
| |
Rev 5′-TCAGGAACAGCCACCAATAA-3′ |
|
|
| IL-6 |
For 5′-TCTCCACAAGCGCCTTCG-3′ |
193 |
60°C |
| |
Rev 5′-CTCAGGGCTGAGATGCCG-3′ |
|
|
| IL-8 |
For 5′-TGCCAAGGAGTGCTAAAG-3′ |
197 |
60°C |
| |
Rev 5′-CTCCACAACCCTCTGCAC-3′ |
|
|
| β-actin |
For 5′-ATCGTGCGTGACATTAAGGA-3′ |
178 |
53–60°C |
| Rev 5′-AGGAAGGAAGGCTGGAAGAG-3′ |
Western immunoblotting
Total or compartmental (cytoplasmic and nuclear) protein lysate from subconfluent cells were prepared and subjected to western blot analysis as described elsewhere.33 Antibody detection was performed using the horseradish peroxidase-conjugated secondary antibodies and chemiluminescence reagents from SuperSignal West Dura kit (Pierce).
In vitro cell growth assay
Cell growth was measured by MTS assay (Promega). Cells were seeded in 96-well plates at a density 1,500 or 15,000 per well to determine anchorage-dependent or -independent cell growth, respectively. For anchorage-independent cell growth, cells were seeded into poly-2-hydroxyethyl methacrylate coated plates and cultured in medium containing 1.5% methylcellulose.34
Scratch/wound-healing assay
Cell mobility was studied using scratch assays as described previously.35 Briefly, cells were plated in Matrigel (100 μg/mL) coated dishes to reach ~90–100% confluence. A sterilized P200 tip was used to scratch lines in the cell monolayer. One hour before scratching, the medium was replaced with medium containing 0.1% FBS to minimize the cell proliferation. Photos of the same region were taken with the same magnification at 0, 8 and 24 h post-wounding.
Migration and invasion assay
For invasion assays transwell filters (Costar), with an 8 μm pore size, coated with Matrigel (BD Biosciences) were used. Cells (4.0 × 104) in RPMI medium with 0.1% FBS were plated in the top chambers over chemo-attractant (20% FBS and 200 ng/mL EGF) in the bottom chambers. After 48 h incubation, non-migrated cells as well as Matrigel in the upper surface of the filters were removed with a cotton swab. Migrated cells, on the lower side of the membrane, were fixed in methanol, stained with crystal violet (0.2% w/v in 2% methanol) and counted (averaged from 6 randomly chosen fields of view for each trans-well). Migration assays were performed in the same way, omitting Matrigel coating from filters. Experiments were normalized using a parallel MTS assay and performed in triplicate to confirm reproducibility.
Gelatin zymography
To prepare conditioned medium, subconfluent cultures were incubated in RPMI-1640 medium with 0.1% FBS for 24 h. Immediately after collecting conditioned media for lyophilization, cells in dishes were trypsinized and counted for normalization. Sample equivalent to 1 × 105 cells was loaded directly on a 10% polyacrylamide gel containing 0.1% SDS and 0.4 mg/ml gelatin. After non-reducing electrophoresis, the gel was washed with 50 mM TRIS-HCl (pH 7.5) containing 2.5% Triton X-100 and zymography buffer (50 mM TRIS-HCl (pH 7.5), 0.002% Brij-35, 10 mM CaCl2 and 5 μM ZnCl2) for 20 min, and then incubated in zymography buffer at room temperature for 16 h. Proteolytic regions indicating gelatinolytic activity were displayed by Coomassie brilliant blue R-250 staining.
Determination of VEGF-A by ELISA
The concentration of VEGF-A secreted into the medium was measured by DuoSet ELISA Development kit (R&D Systems). Conditioned media (with 1% FBS) was prepared as for gelatin zymography and directly assayed without lyophilization. Results were normalized with the number of cells at the time of harvest and reported as picograms per 106 cells.
Immunocytochemistry and confocal microscopy
Cells were grown on gelatin-coated glass coverslips, and where indicated, treated with 5 μg/ml cisplatin for 24 h. Cells were fixed in 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 10 min and then saturated with 5% BSA for 30 min. Cells were incubated with mouse anti-NFκB p65 [sc-8008] (Santa Cruz Biotech) for 1h followed by an incubation with goat anti-mouse Alexa 488 (Molecular Probes, Invitrogen) for 1h; both diluted in 1% BSA-PBS. The cells were then incubated with Draq 5 diluted 1/1000 in 1% BSA-PBS for 1 h. Cells were mounted with mowiol polymerizing solution, and observed under a confocal microscope (Leica TCS-SP1). Data for different cell types and/or conditions was acquired at the same PMT to facilitate quantification of the expression and localization of NFκB. The z-stacks were acquired at 0.25 μm over 7–10 μm.
Chromatin immunoprecipitation assay
Cells were seeded at 1 × 107 cells per 150 mm tissue culture dish and left overnight. Chromatin immunoprecipitation (ChIP) was performed as described elsewhere.36 Chromatin was immunoprecipitated using 3 μg of antibody against p65/Rel-A (NFκB) [sc-8008] (Santa Cruz) or no antibody as a control. DNA was purified with the WizardR SV Gel and PCR Clean-Up System (Promega) as per the manufacturer’s instructions. Purified DNA was amplified by real-time PCR using primers (Forward: 5′ATCTGACCCACGACGCACTG3′, Reverse: 5′ GGAGGAGAGCTGGCAAGGAG3′) designed to span the NFκB binding site within the GRO-β promoter. Real-time PCR cycling conditions were 95°C for 3 min, followed by 40 cycles of 95°C for 1 sec and 56°C for 20 sec.
Caspase-3/7 assay
Caspase-3/7 activity was determined using the Caspase-Glo 3/7 activity assay (Promega), according to the manufacturer's instructions. Briefly, 8,000 cells/well were seeded into a 96-well plate in triplicate overnight before apoptosis induction for 24 h. Caspase 3/7 activities were determined by relative luminescence units quantified using a Luminoskan Ascent Luminometer (Thermo LabSystems). The fold induction of caspase 3/7 normalized against that of untreated cells.
Nude mice xenograft model
Six athymic nude mice (BALB/c, aged 4–5 weeks) were randomly divided into two groups and 5 × 106 cells in PBS were injected s.c. into the dorsal flank. Tumor dimensions were measured by caliper every week and the tumor volume was calculated using the following formula: V = 0.5 × L × W2 (L = larger diameter and W = smaller diameter). Growth curves were plotted using average tumor volume. Mice were sacrificed 5 weeks after tumor cell inoculation. All xenografts were fixed in 4% paraformaldehyde, embedded in paraffin and cut into 4 μm sections for further pathological examination and immunohistochemical analysis.
Immunohistochemical analysis on nude mice xenograft sections
TUNEL assays were performed to assess the in situ apoptotic activity, using the DeadEnd™ Colorimetric TUNEL System (Promega), following the manufacturer’s instructions. Immunohistochemical staining for NDRG1 and Ki-67 was performed by the S-P (peroxidase labeled streptavidin) method using the UltraSensitive™ SP Staining kit from Maixin_Bio and processed as described before.37 Antigen retrieval for CD31 (an endothelial surface marker) was performed with proteinase K (Dako) solution. Sections were then incubated with a 1:50 dilution of rat anti-mouse CD31 primary antibody in 1% BSA in PBS. The CD31 primary antibody was detected with a Biotin-Streptavidin-horse radish peroxidase system (Histomark) using 3,3′,5,5′-tetramethylbenzidine as chromogen. Sections were counterstained with a 0.25% neutral red solution before mounting. For each stain, appropriate negative controls were performed. No interfering background staining was observed (data not shown).
Images were obtained using a Nikon 90i light microscope. Ki-67 labeling index (proliferation marker) was determined as the percentage of immunoreactive nuclei to the total number of nuclei in three random 200 × fields. An average of 4000 nuclei was counted in each section. Apoptosis activity was calculated as the average number of TUNEL positive nuclei in three independent microscopic fields with most intensive staining at 200 × magnification (~4,000 cells). The angiogenic activity was determined by the percentage of vascularized area showing endothelial cell clusters vs. the total area of the section. Images of a complete cross-section were acquired at 32 × magnification. All of the ensuing analysis was performed with Visiopharm Integrated Systems (Visiopharm A/S). All counts and scoring were performed by a pathologist blind to the study. Average number and SD were calculated from all the sections for each group.
Statistical analysis
For imaging data, a representative image from one of the three independent experiments is shown. Quantitative data was obtained from three independent experiments; statistical significance was determined by Student’s t-test in comparison with samples from the wild type control. For in vivo studies the non-parametric Mann Witney test was used. SPSS 17.0 statistical package (SPSS Inc.) was used for statistical analysis and a p value of less than 0.05 was considered statistically significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to acknowledge the following organizations for their funding contribution toward this study: NIH 5RO1CA112020, Joint China/South Africa Science and Technology Agreement, France/South Africa Scientific Cooperation Agreement UID 68670, International Science and Technology Cooperation and Exchanges Programs (No. 2008DFA31130) of China (to X. Z.), the National Research Foundation of South Africa, the Ernst and Ethel Eriksen Trust, the Harry Oppenheimer Memorial Trust, the Deutscher Akademischer Austausch Dienst and the University of Cape Town.
Supplemental Material
Supplemental material may be found here:
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/22956
Reference List
- 1.Lam AK. Molecular biology of esophageal squamous cell carcinoma. Crit Rev Oncol Hematol. 2000;33:71–90. doi: 10.1016/S1040-8428(99)00054-2. [DOI] [PubMed] [Google Scholar]
- 2.Sohda M, Mochida Y, Kato H, Miyazaki T, Nakajima M, Fukuchi M, et al. Overexpression of Cap43 is associated with malignant status of esophageal cancer. Anticancer Res. 2009;29:965–70. [PubMed] [Google Scholar]
- 3.Kovacevic Z, Richardson DR. The metastasis suppressor, Ndrg-1: a new ally in the fight against cancer. Carcinogenesis. 2006;27:2355–66. doi: 10.1093/carcin/bgl146. [DOI] [PubMed] [Google Scholar]
- 4.Stein S, Thomas EK, Herzog B, Westfall MD, Rocheleau JV, Jackson RS, 2nd, et al. NDRG1 is necessary for p53-dependent apoptosis. J Biol Chem. 2004;279:48930–40. doi: 10.1074/jbc.M400386200. [DOI] [PubMed] [Google Scholar]
- 5.Cangul H. Hypoxia upregulates the expression of the NDRG1 gene leading to its overexpression in various human cancers. BMC Genet. 2004;5:27. doi: 10.1186/1471-2156-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le NT, Richardson DR. Iron chelators with high antiproliferative activity up-regulate the expression of a growth inhibitory and metastasis suppressor gene: a link between iron metabolism and proliferation. Blood. 2004;104:2967–75. doi: 10.1182/blood-2004-05-1866. [DOI] [PubMed] [Google Scholar]
- 7.Salnikow K, Kluz T, Costa M, Piquemal D, Demidenko ZN, Xie K, et al. The regulation of hypoxic genes by calcium involves c-Jun/AP-1, which cooperates with hypoxia-inducible factor 1 in response to hypoxia. Mol Cell Biol. 2002;22:1734–41. doi: 10.1128/MCB.22.6.1734-1741.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang P, Tchou-Wong KM, Costa M. Egr-1 mediates hypoxia-inducible transcription of the NDRG1 gene through an overlapping Egr-1/Sp1 binding site in the promoter. Cancer Res. 2007;67:9125–33. doi: 10.1158/0008-5472.CAN-07-1525. [DOI] [PubMed] [Google Scholar]
- 9.Bandyopadhyay S, Pai SK, Hirota S, Hosobe S, Tsukada T, Miura K, et al. PTEN up-regulates the tumor metastasis suppressor gene Drg-1 in prostate and breast cancer. Cancer Res. 2004;64:7655–60. doi: 10.1158/0008-5472.CAN-04-1623. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Kretzner L. The growth-inhibitory Ndrg1 gene is a Myc negative target in human neuroblastomas and other cell types with overexpressed N- or c-myc. Mol Cell Biochem. 2003;250:91–105. doi: 10.1023/A:1024918328162. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Chen S, Zhang W, Zhang J, Liu X, Shi H, et al. Human differentiation-related gene NDRG1 is a Myc downstream-regulated gene that is repressed by Myc on the core promoter region. Gene. 2008;417:5–12. doi: 10.1016/j.gene.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Masuda K, Ono M, Okamoto M, Morikawa W, Otsubo M, Migita T, et al. Downregulation of Cap43 gene by von Hippel-Lindau tumor suppressor protein in human renal cancer cells. Int J Cancer. 2003;105:803–10. doi: 10.1002/ijc.11152. [DOI] [PubMed] [Google Scholar]
- 13.Guan RJ, Ford HL, Fu Y, Li Y, Shaw LM, Pardee AB. Drg-1 as a differentiation-related, putative metastatic suppressor gene in human colon cancer. Cancer Res. 2000;60:749–55. [PubMed] [Google Scholar]
- 14.Bandyopadhyay S, Pai SK, Hirota S, Hosobe S, Takano Y, Saito K, et al. Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene. 2004;23:5675–81. doi: 10.1038/sj.onc.1207734. [DOI] [PubMed] [Google Scholar]
- 15.Bandyopadhyay S, Pai SK, Gross SC, Hirota S, Hosobe S, Miura K, et al. The Drg-1 gene suppresses tumor metastasis in prostate cancer. Cancer Res. 2003;63:1731–6. [PubMed] [Google Scholar]
- 16.Maruyama Y, Ono M, Kawahara A, Yokoyama T, Basaki Y, Kage M, et al. Tumor growth suppression in pancreatic cancer by a putative metastasis suppressor gene Cap43/NDRG1/Drg-1 through modulation of angiogenesis. Cancer Res. 2006;66:6233–42. doi: 10.1158/0008-5472.CAN-06-0183. [DOI] [PubMed] [Google Scholar]
- 17.Dang C, Gottschling M, Manning K, O’Currain E, Schneider S, Sterry W, et al. Identification of dysregulated genes in cutaneous squamous cell carcinoma. Oncol Rep. 2006;16:513–9. [PubMed] [Google Scholar]
- 18.Gómez-Casero E, Navarro M, Rodríguez-Puebla ML, Larcher F, Paramio JM, Conti CJ, et al. Regulation of the differentiation-related gene Drg-1 during mouse skin carcinogenesis. Mol Carcinog. 2001;32:100–9. doi: 10.1002/mc.1069. [DOI] [PubMed] [Google Scholar]
- 19.Chang JT, Wang HM, Chang KW, Chen WH, Wen MC, Hsu YM, et al. Identification of differentially expressed genes in oral squamous cell carcinoma (OSCC): overexpression of NPM, CDK1 and NDRG1 and underexpression of CHES1. Int J Cancer. 2005;114:942–9. doi: 10.1002/ijc.20663. [DOI] [PubMed] [Google Scholar]
- 20.Nishio S, Ushijima K, Tsuda N, Takemoto S, Kawano K, Yamaguchi T, et al. Cap43/NDRG1/Drg-1 is a molecular target for angiogenesis and a prognostic indicator in cervical adenocarcinoma. Cancer Lett. 2008;264:36–43. doi: 10.1016/j.canlet.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 21.Nishie A, Masuda K, Otsubo M, Migita T, Tsuneyoshi M, Kohno K, et al. High expression of the Cap43 gene in infiltrating macrophages of human renal cell carcinomas. Clin Cancer Res. 2001;7:2145–51. [PubMed] [Google Scholar]
- 22.Wang Z, Wang F, Wang WQ, Gao Q, Wei WL, Yang Y, et al. Correlation of N-myc downstream-regulated gene 1 overexpression with progressive growth of colorectal neoplasm. World J Gastroenterol. 2004;10:550–4. doi: 10.3748/wjg.v10.i4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sibold S, Roh V, Keogh A, Studer P, Tiffon C, Angst E, et al. Hypoxia increases cytoplasmic expression of NDRG1, but is insufficient for its membrane localization in human hepatocellular carcinoma. FEBS Lett. 2007;581:989–94. doi: 10.1016/j.febslet.2007.01.080. [DOI] [PubMed] [Google Scholar]
- 24.Chua MS, Sun H, Cheung ST, Mason V, Higgins J, Ross DT, et al. Overexpression of NDRG1 is an indicator of poor prognosis in hepatocellular carcinoma. Mod Pathol. 2007;20:76–83. doi: 10.1038/modpathol.3800711. [DOI] [PubMed] [Google Scholar]
- 25.Yan X, Chua MS, Sun H, So S. N-Myc down-regulated gene 1 mediates proliferation, invasion, and apoptosis of hepatocellular carcinoma cells. Cancer Lett. 2008;262:133–42. doi: 10.1016/j.canlet.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 26.Motwani M, Sirotnak FM, She Y, Commes T, Schwartz GK. Drg1, a novel target for modulating sensitivity to CPT-11 in colon cancer cells. Cancer Res. 2002;62:3950–5. [PubMed] [Google Scholar]
- 27.Shah MA, Kemeny N, Hummer A, Drobnjak M, Motwani M, Cordon-Cardo C, et al. Drg1 expression in 131 colorectal liver metastases: correlation with clinical variables and patient outcomes. Clin Cancer Res. 2005;11:3296–302. doi: 10.1158/1078-0432.CCR-04-2417. [DOI] [PubMed] [Google Scholar]
- 28.Chen B, Nelson DM, Sadovsky Y. N-myc down-regulated gene 1 modulates the response of term human trophoblasts to hypoxic injury. J Biol Chem. 2006;281:2764–72. doi: 10.1074/jbc.M507330200. [DOI] [PubMed] [Google Scholar]
- 29.Shimada Y, Imamura M, Wagata T, Yamaguchi N, Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer. 1992;69:277–84. doi: 10.1002/1097-0142(19920115)69:2<277::AID-CNCR2820690202>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 30.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–61. doi: 10.1128/JVI.77.16.8957-8951.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–5. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Nagata J, Kijima H, Hatanaka H, Tokunaga T, Kamochi J, Abe Y, et al. Angiopoietin-1 and vascular endothelial growth factor expression in human esophageal cancer. Int J Mol Med. 2002;10:423–6. [PubMed] [Google Scholar]
- 34.Kantak SS, Kramer RH. E-cadherin regulates anchorage-independent growth and survival in oral squamous cell carcinoma cells. J Biol Chem. 1998;273:16953–61. doi: 10.1074/jbc.273.27.16953. [DOI] [PubMed] [Google Scholar]
- 35.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–33. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 36.Leaner VD, Donninger H, Ellis CA, Clark GJ, Birrer MJ. p75-Ras-GRF1 is a c-Jun/AP-1 target protein: its up regulation results in increased Ras activity and is necessary for c-Jun-induced nonadherent growth of Rat1a cells. Mol Cell Biol. 2005;25:3324–37. doi: 10.1128/MCB.25.8.3324-3337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang B, Khachigian LM, Esau L, Birrer MJ, Zhao X, Parker MI, et al. A key role for early growth response-1 and nuclear factor-kappaB in mediating and maintaining GRO/CXCR2 proliferative signaling in esophageal cancer. Mol Cancer Res. 2009;7:755–64. doi: 10.1158/1541-7786.MCR-08-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loges S, Clausen H, Reichelt U, Bubenheim M, Erbersdobler A, Schurr P, et al. Determination of microvessel density by quantitative real-time PCR in esophageal cancer: correlation with histologic methods, angiogenic growth factor expression, and lymph node metastasis. Clin Cancer Res. 2007;13:76–80. doi: 10.1158/1078-0432.CCR-06-1324. [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto S, Yamada Y, Narikiyo M, Ueno M, Tamaki H, Miki K, et al. Prognostic significance of platelet-derived growth factor-BB expression in human esophageal squamous cell carcinomas. Anticancer Res. 2007;27(4B):2409–14. [PubMed] [Google Scholar]
- 40.Krzystek-Korpacka M, Matusiewicz M, Diakowska D, Grabowski K, Blachut K, Konieczny D, et al. Elevation of circulating interleukin-8 is related to lymph node and distant metastases in esophageal squamous cell carcinomas--implication for clinical evaluation of cancer patient. Cytokine. 2008;41:232–9. doi: 10.1016/j.cyto.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Strieter RM, Belperio JA, Phillips RJ, Keane MP. CXC chemokines in angiogenesis of cancer. Semin Cancer Biol. 2004;14:195–200. doi: 10.1016/j.semcancer.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Oka M, Yamamoto K, Takahashi M, Hakozaki M, Abe T, Iizuka N, et al. Relationship between serum levels of interleukin 6, various disease parameters and malnutrition in patients with esophageal squamous cell carcinoma. Cancer Res. 1996;56:2776–80. [PubMed] [Google Scholar]
- 43.Okuda T, Higashi Y, Kokame K, Tanaka C, Kondoh H, Miyata T. Ndrg1-deficient mice exhibit a progressive demyelinating disorder of peripheral nerves. Mol Cell Biol. 2004;24:3949–56. doi: 10.1128/MCB.24.9.3949-3956.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuda T, Kokame K, Miyata T. Differential expression patterns of NDRG family proteins in the central nervous system. J Histochem Cytochem. 2008;56:175–82. doi: 10.1369/jhc.7A7323.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haustermans K, Hofland I, Van de Pavert L, Geboes K, Varia M, Raleigh J, et al. Diffusion limited hypoxia estimated by vascular image analysis: comparison with pimonidazole staining in human tumors. Radiother Oncol. 2000;55:325–33. doi: 10.1016/S0167-8140(00)00206-1. [DOI] [PubMed] [Google Scholar]
- 46.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 47.Li B, Li YY, Tsao SW, Cheung AL. Targeting NF-kappaB signaling pathway suppresses tumor growth, angiogenesis, and metastasis of human esophageal cancer. Mol Cancer Ther. 2009;8:2635–44. doi: 10.1158/1535-7163.MCT-09-0162. [DOI] [PubMed] [Google Scholar]
- 48.Izzo JG, Wu X, Wu TT, Huang P, Lee JS, Liao Z, et al. Therapy-induced expression of NF-kappaB portends poor prognosis in patients with localized esophageal cancer undergoing preoperative chemoradiation. Dis Esophagus. 2009;22:127–32. doi: 10.1111/j.1442-2050.2008.00884.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.