Abstract
In this study, the gentisate and protocatechuate pathways in Burkholderia xenovorans LB400 were analyzed by genomic and functional approaches, and their role in 3-hydroxybenzoate (3-HBA) and 4-hydroxybenzoate (4-HBA) degradation was proposed. The LB400 genome possesses two identical mhbRTDHI gene clusters encoding the gentisate pathway and one mhbM gene encoding a 3-HBA 6-hydroxylase that converts 3-HBA into gentisate. The pca genes encoding the protocatechuate pathway and the pobA gene encoding the 4-HBA 3-monooxygenase that oxidizes 4-HBA into protocatechuate are arranged in gene clusters and single genes mainly at the minor chromosome, but also at the major chromosome and the megaplasmid. Strain LB400 was able to grow on gentisate, protocatechuate, 3-HBA and 4-HBA. Transcriptional analyses showed that the mhbD gene encoding the gentisate 1,2-dioxygenase was expressed during growth on 3-HBA, 4-HBA and gentisate, whereas the pcaG gene encoding the protocatechuate 3,4-dioxygenase was expressed only during growth on 4-HBA and protocatechuate. The mhbM gene encoding the 3-HBA 6-hydroxylase was transcribed in strain LB400 during growth on HBAs, gentisate, protocatechuate and glucose. The pobA gene encoding the 4-HBA 3-monooxygenase was expressed during growth on HBAs and glucose. 3-HBA- and 4-HBA-grown LB400 cells showed gentisate 1,2-dioxygenase activity, whereas protocatechuate 3,4-dioxygenase activity was observed only in 4-HBA-grown cells. The mhbR gene encoding a MarR-type transcriptional regulator that probably regulates the expression of the MhbT transporter, and the pcaQ and pcaR genes encoding LysR-type transcriptional regulators that regulate pcaHG and pcaIJBDC genes, respectively, were transcribed during growth on both HBAs, gentisate, protocatechuate and glucose, suggesting a basal constitutive expression. The results indicate active gentisate, protocatechuate, 3-HBA and 4-HBA catabolic pathways in B. xenovorans LB400 and suggest that 3-HBA is channeled exclusively through the gentisate route, whereas 4-HBA is funneled into the protocatechuate central pathway and potentially into the gentisate pathway.
Introduction
Aerobic bacterial degradation of aromatic compound proceeds generally in two phases. In the first phase, the aromatic compound is prepared for ring cleavage by a variety of ring modification reactions. The second phase of aerobic degradation includes ring fission of the aromatic compound and subsequent reactions leading to the formation of Krebs cycle intermediates [1]–[4]. Betaproteobacteria of the Burkholderiales order possess an amazing metabolic versatility to degrade a large number of aromatic compounds [1], [3]. Burkholderia xenovorans LB400 is a bacterium able to degrade polychlorobiphenyls (PCBs) and diverse aromatic compounds [3]–[8]. LB400 genome has a size of 9.73 Mbp distributed in three replicons: the major chromosome (C1; 4.90 Mbp), the minor chromosome (C2; 3.36 Mbp) and the megaplasmid (MP; 1.47 Mbp) [3]. Genomic analysis of B. xenovorans strain LB400 revealed the presence of genes encoding an unusual high number of central and peripheral pathways for the degradation of aromatic compounds [3]. A recent study confirmed functionality of some predicted aromatic pathways [4]. However, the function of genes encoding diverse aromatic catabolic routes including the gentisate and protocatechuate pathways of B. xenovorans strain LB400 remains to be elucidated.
The gentisate pathway is a central route for the bacterial aerobic catabolism of benzoates such as 3-HBA and salicylate (2-hydroxybenzoate), phenolic compounds such as m-cresol and 2,5-xylenol, and polycyclic aromatic hydrocarbons including naphthalene and phenanthrene [9]–[14]. Gentisate is cleaved by gentisate 1,2-dioxygenase (MhbD) to form maleylpyruvate. Maleylpyruvate is then isomerized by a glutathione-dependent maleylpyruvate isomerase (MhbI) to yield fumarylpyruvate. A fumarylpyruvate hydrolase (MhbH) catalyzes fumarylpyruvate conversion into pyruvate and fumarate.
The gentisate catabolic pathway has been described as the central route for 3-HBA degradation in some bacteria. 3-HBA is degraded through the gentisate central pathway by the 3-HBA 6-hydroxylase in Burkholderia cepacia J2315 [15], Pseudomonas alcaligenes P25X1 [9], Klebsiella pneumoniae M5a1 [11], [16] and Salmonella typhimurium [12]. The 3-HBA 6-hydroxylase is encoded by the mhbM gene in K. pneumoniae M5a1 [11], [16]. Alternatively, 3-HBA could be degraded through the protocatechuate catabolic pathway by the 3-HBA 4-hydroxylase, which is encoded by the mobA gene in Comamonas testosteroni KH122 [17], [18].
The protocatechuate pathway is a central catabolic route for aromatic compounds, which is widely distributed among taxonomically diverse bacteria and fungi [1]–[3], [19], [20]. Protocatechuate is a key central intermediate in bacterial degradation of diverse aromatic compounds, including 4-HBA, 3-HBA, vanillate, ferulate, coniferyl alcohol, p-coumarate, phthalate, isophthalate and terephthalate. Protocatechuate oxygenolytic ring-cleavage is catalyzed by protocatechuate 3,4-dioxygenase (PcaGH) to generate 3-carboxy-cis,cis-muconate, which is converted into 4-carboxymuconolactone by 3-carboxy-cis,cis-muconate cycloisomerase (PcaB). 4-Carboxymuconolactone decarboxylase (PcaC) transforms the 4-carboxymuconolactone into β-ketoadipate enol-lactone, which is then hydrolyzed by β-ketoadipate enol-lactone hydrolase (PcaD) into β-ketoadipate. The enzyme β-ketoadipate succinyl-CoA tranferase (PcaIJ) converts β-ketoadipate into β-ketoadipyl-CoA, which is finally transformed into succinyl-CoA and acetyl-CoA by β-ketoadipyl-CoA thiolase (PcaF).
The protocatechuate central pathway is involved in 4-HBA degradation in some microorganisms. 4-HBA is hydroxylated by 4-HBA 3-monooxygenase encoded by the pobA gene to yield protocatechuate in Pseudomonas and Cupriavidus strains [2], [21], [22].
The aims of this study were to characterize the gentisate and protocatechuate central pathways in B. xenovorans LB400 using genomic and functional approaches. In addition, the role of these ring-cleavage pathways during 3-HBA and 4-HBA degradation in strain LB400 was studied. This report describes for the first time the metabolism of 3-HBA and 4-HBA in B. xenovorans LB400. This study demonstrates functional gentisate and protocatechuate catabolic pathways in B. xenovorans LB400 that are involved in the degradation of 3-HBA and 4-HBA.
Materials and Methods
Chemicals
3,4-dihydroxybenzoic acid (protocatechuate, >98% purity), 2,5-dihydroxybenzoic acid (gentisate, 98% purity), 3-hydroxybenzoic acid (>99% purity) and 4-hydroxybenzoic acid (≥99% purity) were obtained from Sigma-Aldrich (Saint Louis, MO, USA).
Bacterial Strain and Culture Conditions
B. xenovorans strain LB400 was cultivated in M9 mineral medium with trace solution and glucose (5 mM) [23], 3-HBA (5 and 10 mM), 4-HBA (5 mM), gentisate (5 mM) or protocatechuate (5 mM) as sole carbon and energy source at 30°C [3], [23]. Growth was determined by measuring turbidity at 525 nm and by counting colony-forming units (CFU). Aliquots taken from bacterial cultures were diluted and plated on Luria-Bertani (LB) medium. CFU mL−1 values were calculated as the mean ± SD of values from at least three independent experiments.
Isolation of RNA and RT-PCR
RNA was isolated from LB400 cells grown until mid-exponential growth phase (turbidity 525 nm 0.4–0.6) on 3-HBA, 4-HBA and glucose. RNA was isolated using RNeasy mini kit (Qiagen, Hilden, Germany). DNase I treatment was performed using the RNase-Free DNase Set (Qiagen, Hilden, Germany). The RNA was quantified using a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA). In this study, specific primers for mhbD (BxeA2627, BxeA4526), mhbT (BxeA2625, BxeA4528), mhbR (BxeA2624, BxeA4529), mhbM (BxeB2526), pcaG (BxeB2776), pcaQ (BxeB2772), pcaR (BxeB0642) and pobA (BxeA2040) genes were designed and used (Table 1). Reverse transcription-PCR (RT-PCR) was carried out with 40 µg of total RNA and the sequence-specific primers using SuperScript One-step RT-PCR with Platinum Taq (Invitrogen, Carlsbad, CA, USA). Amplification of the 16S rRNA gene was performed as control for DNA contamination using the primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′) as reported [4]. Negative and positive controls were included in each RT-PCR assay. The expression of the constitutively expressed 16S rRNA was used as a control to normalize across samples. At least three independent RNA samples were collected at each condition and two independent RT-PCR reactions for each sample were done to assess reproducibility.
Table 1. Primer sets designed and used in this study.
| Gene | Name | Sequence (5′- 3′) | Reference |
| mhbD1 mhbD2 | mhbDf | 5′-ATCCGGGGGACTTTATCATC-3′ | This study |
| mhbD1 mhbD2 | mhbDr | 5′-GCGATTCGAGTAGCTGAACA-3′ | This study |
| mhbR1 mhbR2 | mhbRf | 5′-GCAAGTTCGCCACAATTCCGTT-3′ | This study |
| mhbR1 mhbR2 | mhbRr | 5′-TTGCGTTACGGTAAAGGCGGAT-3′ | This study |
| pcaG | pcaGf | 5′-AAGACAGTTTCCTTCTCGCCCTG-3′ | This study |
| pcaG | pcaGr | 5′-CTCAAGCAAACGCCTTCGCAA-3′ | This study |
| pcaQ | pcaQf | 5′-AACTGTTCAGCGAGCCGTTGAT-3′ | This study |
| pcaQ | pcaQr | 5′-AAAAGGCAGCGGCAAACGTA-3′ | This study |
| pcaR | pcaRf | 5′-ATGCTCGACGTGCAGGAATTCA-3′ | This study |
| pcaR | pcaRr | 5′-TGAAACTTGCGATGCCAGTGCT-3′ | This study |
| mhbM | mhbMf | 5′-TGACCATTCTGGAACAGGCGAA-3′ | This study |
| mhbM | mhbMr | 5′-TATACGGCACATCAATCGCGCT-3′ | This study |
| pobA | pobAf | 5′-TTGATAGCGTGGTGCTGGAA-3′ | This study |
| pobA | pobAr | 5′-ACGCTATCTTTCGGATCGCA-3′ | This study |
| 16S rRNA | 27F | 5′-AGAGTTTGATCMTGGCTCAG-3′ | [4], [23] |
| 16S rRNA | 1492R | 5′-TACGGYTAC CTTGTTACGACTT-3′ | [4], [23] |
Preparation of Bacterial Cell Extracts
Cells grown until exponential phase with 3-HBA, 4-HBA or glucose were harvested by centrifugation (10,733 g for 10 min) at 4°C. Bacterial cells washed with sodium phosphate buffer (50 mM, pH 7.0) and concentrated were disrupted using ultrasonic cells disruptor Microson at 4°C. Cell lysates were clarified by centrifugation (19,300 g for 15 min) at 4°C. The protease inhibitor cocktail Tablet Complete mini (Roche, Indianapolis, USA) was added to cell extracts to reach a final concentration of 0.8 mg/ml. Protein concentration was measured using the Quantit-iT Protein Assay kit (Invitrogen, Carlsbad, CA, USA) that uses a fluorescent dye whose quantum yields are enhanced significantly when binding at the detergent-protein interface and measuring fluorescence at room temperature with the Qubit fluorometer (Invitrogen, Carlsbad, CA, USA) [24].
Dioxygenase Activity Assays
Gentisate 1,2-dioxygenase activity was determined spectrophotometrically by measuring the formation of maleylpyruvate at 330 nm [13]. The assay volume (10 ml) contained sodium phosphate buffer (50 mM, pH 7.0), crude extract (1000 µg of protein) and gentisate (0.6 mM). The reactions were carried out at 30°C and initiated by the addition of gentisate. Gentisate 1,2-dioxygenase activity was calculated using the molar extinction coefficient of maleylpyruvate 10,300 M−1 cm−1 [13], [25]. Protocatechuate 3,4-dioxygenase activity was determined spectrophotometrically by the disappearance of protocatechuate measuring the absorbance at 290 nm (A290 nm) [26]. The assay volume (10 ml) contained sodium phosphate buffer (50 mM, pH 7.0), crude extract (1000 µg of protein) and protocatechuate (0.6 mM). The reactions were carried out at 30°C and initiated by the addition of protocatechuate. Protocatechuate 3,4-dioxygenase activity was calculated using the difference of the molar extinction coefficient of protocatechuate and its product under these conditions that is 2,280 M−1 cm−1 [27]. One enzyme unit corresponds to the transformation under the conditions described above of 1 µmol of substrate to product per minute per milligram of protein at 30°C. The enzymatic activity assays were done in triplicate.
Results
Genomic Analysis of the Gentisate Pathway and Related Peripheral Routes
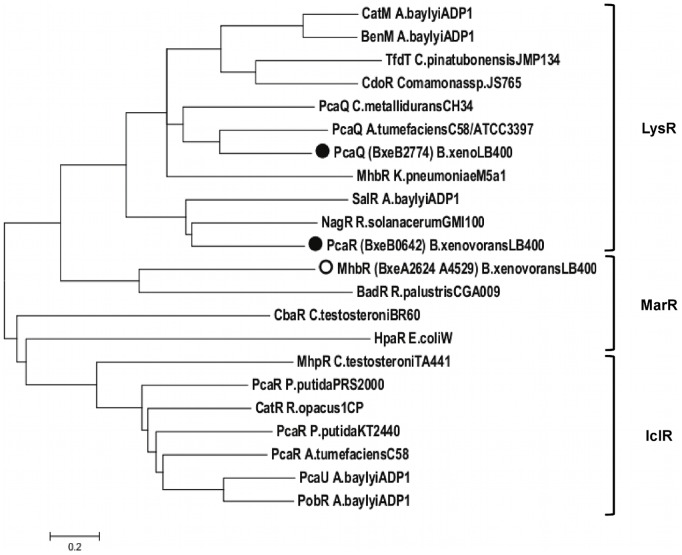
The mhb genes encoding the gentisate catabolic pathway were analyzed in the genome of B. xenovorans LB400. Table 2 describes the predicted genes encoding the gentisate central pathway and related peripheral routes in B. xenovorans LB400. Two identical mhbRTDHI gene clusters encoding the gentisate pathway were identified at C1 (Fig. 1). Both mhb gene clusters are located adjacent in two identical chromosomal regions comprising thirty genes. Sequence analyses suggest that mhbD, mhbH and mhbI genes of strain LB400 encode the gentisate 1,2-dioxygenase, maleylpyruvate isomerase and fumarylpyruvate hydratase, respectively. The mhbD, mhbH and mhbI gene products of strain LB400 possess >52% sequence identity with the respective MhbD, MhbH and MhbI proteins from K. pneumoniae M5a1 [13], [30]. Sequence analyses of both mhbDHI gene clusters neighborhoods in strain LB400, revealed that the BxeA2625 and BxeA4528 (hereafter, mhbT1 and mhbT2) genes encode major facilitator superfamily (MFS) transporters that belong to the aromatic acid:H+ symporter (AAHS) family. Genomic context suggests that the mhbT1 and mhbT2 genes encode a MFS transporters probably involved in gentisate transport in B. xenovorans LB400. The BxeA2624 and BxeA4529 genes (hereafter, mhbR1 and mhbR2) were located upstream and adjacent to the mhbT genes, and divergently transcribed. The mhbR gene product shares 30% sequence identity with MarR-type transcriptional regulator related to the mhb gene cluster from Polaromonas naphthalenivorans CJ2 (Table 2). Sequence alignment of transcriptional regulators associated to central aromatic pathways showed that the mhbR gene product from B. xenovorans LB400 is a MarR-type transcriptional regulator (Fig. 2), which probably regulates the expression of the mhbT gene. The MhbR regulator in K. pneumoniae M5a1 is a LysR-type activator of the mhbTDHI gene cluster [39]. Genome analyses indicated that two divergent σ70-type promoters were present between mhbR and mhbT. A regulatory binding site with a palindromic sequence for a MarR-type transcriptional regulator (5′-ATTGTTTCACAAACTAAT-3′) was identified upstream from the mhbT gene promoter, which is similar to the consensus MarR-type regulator binding site (5′-(T/A)(A/T)(T/A)CTT-N6-(A/G)A(G/A)(A/T)(T/AC)(A/T) -3′) [40].
Table 2. Predicted genes encoding the gentisate and protocatechuate pathways and peripheral reactions from B. xenovorans LB400.
| Gene* | ORF | aa | Related gene products | |||
| Protein | Function | Organism (Reference) | % Id (aa) | |||
| mhbR1 mhbR2 | BxeA2624 BxeA4529 | 166 | MhbR | Transcriptional regulator, MarR family | Polaromonas naphthalenivorans CJ2 [28] | 30 (161) |
| mhbT1 mhbT2 | BxeA2625BxeA4528 | 466 | PcaK | 4-Hydroxybenzoate transporter | Pseudomonas putida PRS2000 [29] | 33 (448) |
| mhbD1 mhbD2 | BxeA2627BxeA4526 | 348 | MhbD | Gentisate-1,2-dioxygenase | Klebsiella pneumoniae M5a1 [30] | 64 (345) |
| mhbH1 mhbH2 | BxeA2628BxeA4525 | 232 | MhbH | Fumarylpyruvate hydrolase | K. pneumoniae M5a1 [30] | 63 (232) |
| mhbI1mhbI2 | BxeA2629BxeA4524 | 216 | MhbI | Maleylpyruvate isomerase | K. pneumoniae M5a1 [30] | 53 (214) |
| mhbM | BxeB2526 | 396 | MhbM | 3-Hydroxybenzoate 6- hydroxylase | K. pneumoniae M5a1 [30] | 38(397) |
| pcaG | BxeB2776 | 195 | PcaG | Protocatechuate 3,4- dioxygenase, alpha subunit | Acinetobacter lwoffii K24 [31] | 90 (132) |
| pcaH | BxeB2775 | 234 | PcaH | Protocatechuate 3,4- dioxygenase, beta subunit | A. lwoffii K24 [31] | 96 (226) |
| pcaQ | BxeB2772 | 310 | PcaQ | Transcriptional regulator, LysR family | Agrobacterium tumefaciens A348 [32] | 48 (311) |
| pcaF1 | BxeB2167 | 400 | PcaF | β-Ketoadipyl-CoA thiolase | P. putida PRS2000 [33] | 71 (400) |
| pcaF2 | BxeA0469 | 400 | PcaF | β-Ketoadipyl- CoA thiolase | P. putida PRS2000 [33] | 69 (400) |
| pcaF3 | BxeA4255 | 402 | PcaF | β-Ketoadipyl-CoA thiolase | P. putida PRS2000 [33] | 42 (400) |
| pcaC | BxeB0647 | 130 | PcaC | 4-Carboxymucolactone decarboxylase | Acinetobacter baylyi ADP1 [34] | 54 (134) |
| pcaD1 | BxeB0646 | 263 | PcaD | β-Ketoadipate enol-lactone hydrolase | A. baylyi ADP1 [35] | 38 (266) |
| pcaD2 | BxeB0595 | 265 | PcaD | β-Ketoadipate enol-lactone hydrolase | A. baylyi ADP1 [35] | 31 (266) |
| pcaB1 | BxeB0645 | 459 | PcaB | 3-Carboxy-cis,cis-muconate cycloisomerase | P. putida PRS2000 [36] | 52 (422) |
| pcaB2 | BxeB1906 | 445 | PcaB | 3-Carboxy-cis,cis-muconate cycloisomerase | P. putida PRS2000 [36] | 34 (422) |
| pcaJ1 | BxeB0644 | 219 | PcaJ | β-Ketoadipate:succinyl-CoA transferase, beta subunit | P. putida PRS2000 [37] | 77 (213) |
| pcaJ2 | BxeC0574 | 265 | PcaJ | β-Ketoadipate:succinyl-CoA transferase, beta subunit | P. putida PRS2000 [37] | 53 (213) |
| pcaJ3 | BxeA1366 | 234 | PcaJ | β-Ketoadipate:succinyl-CoA transferase, beta subunit | P. putida PRS2000 [37] | 44 (213) |
| pcaI1 | BxeB0643 | 233 | PcaI | β-Ketoadipate:succinyl-CoA transferase, alpha subunit | P. putida PRS2000 [37] | 79 (231) |
| pcaI2 | BxeC0572 | 235 | PcaI | β-Ketoadipate:succinyl-CoA transferase, alpha subunit | P. putida PRS2000 [37] | 49 (231) |
| pcaR | BxeB0642 | 309 | PcaR | Transcriptional regulator, LysR family | Burkhoderia phytofirmans PsJN [38] | 95 (309) |
| pobA | BxeA2040 | 394 | PobA | 4-Hydroxybenzoate-3-monooxygenase | P. fluorescens Pf-5 [21] | 51 (394) |
Genes for those cases where biochemical or genetic evidence for function is available are underlined.
Figure 1. Predicted mhb genes encoding the gentisate central pathway and related genes in B. xenovorans LB400.
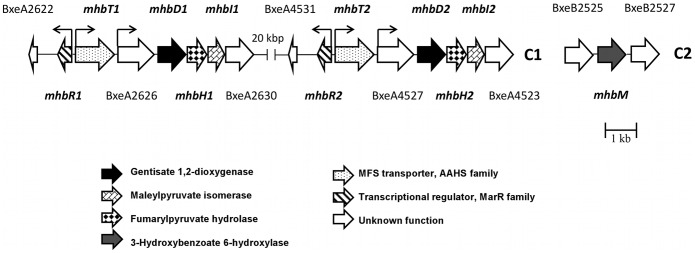
The mhb genes located at the major chromosome (C1) and the minor chromosome (C2). The genes encoding gentisate 1,2-dioxygenase are indicated with black arrows. The orientations of ORFs are represented by open arrows. The promoter regions for mhbR1, mhbR2, mhbT1, mhbT2, BxeA2626 and BxeA4527 are denoted with small black arrows, bent in the directions of transcription. The sizes of the genes and the intergenic regions are on scale.
Figure 2. Relationship between bacterial transcriptional regulators for gentisate and protocatechuate catabolic pathways.
LysR, MarR and IclR transcriptional regulators from B. xenovorans LB400 and other bacteria were depicted. The dendogram was obtained by the neighbor-joining method using MEGA 5.0 based on sequence alignment calculated by CLUSTAL W using the default options. Circles denote proteins encoded in the genome from B. xenovorans LB400. The sequence and their accession number are: CatM, Acinetobacter baylyi ADP1 (P07774); BenM, A. baylyi ADP1 (AAC46441); TfdT, Cupriavidus pinatubonensis JMP134 (AAC44724); CdoR, Comamonas sp. JS765 (AAC79916); MhbR, Klebsiella pneumoniae M5a1 (Q5EXK6); PcaQ, Cupriavidus metallidurans CH34 (ABF10579); PcaQ, Agrobacterium tumefaciens C58/ATCC3397 (P0A4T6); PcaQ, B. xenovorans LB400 (Q13RR8); SalR, A. baylyi ADP1 (AAF04311); NagR, Ralstonia sp. U2 (Q9EXL7); PcaR, B. xenovorans LB400 (ABE35311); MhbR, B. xenovorans LB400 (ABE30329); BadR, Rhodopseudomonas palustris CGA009 (AAC23923); CbaR, Comamonas testosteroni TA441 (AAG00065); HpaR, Escherichia coli W (Z37980); MhpR, C. testosteroni TA441 (P77569); PcaR, Pseudomonas putida PRS2000 (Q52154); CatR, Rhodococcus opacus (Q33539); PcaR, P. putida KT2440 (Q88N41); PcaR, A. tumefaciens C58 (Q7CV82); PcaU, A. baylyi ADP1 (AAC37157); PobR, A. baylyi ADP1 (A36893).
Genome analysis indicated that an independent σ70-type promoter is located upstream of the BxeA2626 (BxeA4527) and the mhbDHI genes. Upstream from the BxeA2626 (BxeA4527) gene promoter is located a LysR-type regulator binding sequence (5′-ATTCACTTCGAGAAT-3′) that has the conserved (5′-T-N11-A-3′) motif critical for binding of LysR-type transcriptional regulators [41]. The specific LysR-type transcriptional regulator gene of the mhbDHI gene cluster of strain LB400 has not been identified in this study.
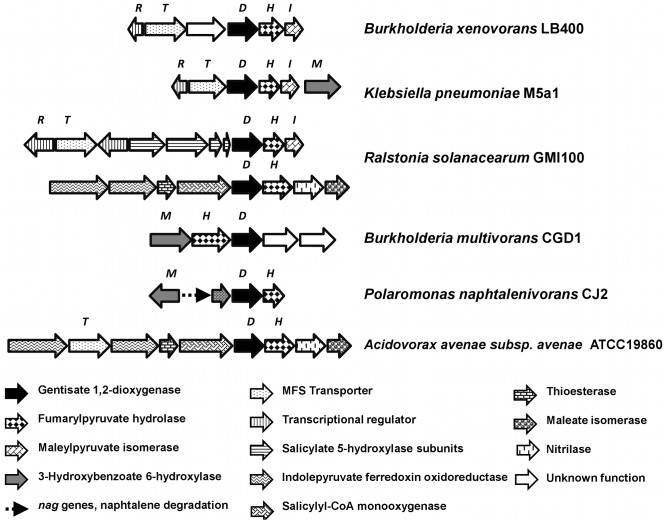
Gene organization of the mhb cluster in B. xenovorans LB400 is similar to the mhb cluster from K. pneumoniae M5a1, except for the presence of a coding sequence of unknown function between mhbT and mhbD genes in strain LB400, the presence of the mhbM gene encoding 3-HBA 6-hydroxylase located downstream to mhbI in strain M5a1 and that the mhbR genes encode different transcriptional regulators (Fig. 3). The mhb gene clusters from other Burkholderiales strains including Ralstonia solanacearum GMI100, B. multivorans CGD1, P. naphthalenivorans CJ2 and Acidovorax avenae subsp. avenae ATCC19860 showed different mhb gene organizations (Fig. 3).
Figure 3. Organization of the gene clusters involved in the catabolism of gentisate in B. xenovorans LB400 and other bacteria.
The genes encoding gentisate 1,2-dioxygenase are indicated with black arrows. The sizes of the genes and the intergenic regions are on scale.
A search of the gene encoding a 3-HBA 6-hydroxylase that funnel 3-HBA into the gentisate central pathway was performed in the LB400 genome. Sequence analysis indicates that the BxeB2526 gene (hereafter mhbM) located at C2 encodes a 3-HBA 6-hydroxylase (Fig. 1). The mhbM gene product from strain LB400 shared a 38% sequence identity with 3-HBA 6-hydroxylase (MhbM) from K. pneumoniae M5a1 [30]. A 4-HBA 1-hydroxylase activity that converts 4-HBA into gentisate has been described [42]. However, the gene sequence encoding 4-HBA 1-hydroxylase has not been reported yet, impeding a further genomic analysis.
Genomic Analysis of Protocatechuate Pathway and Related Peripheral Routes
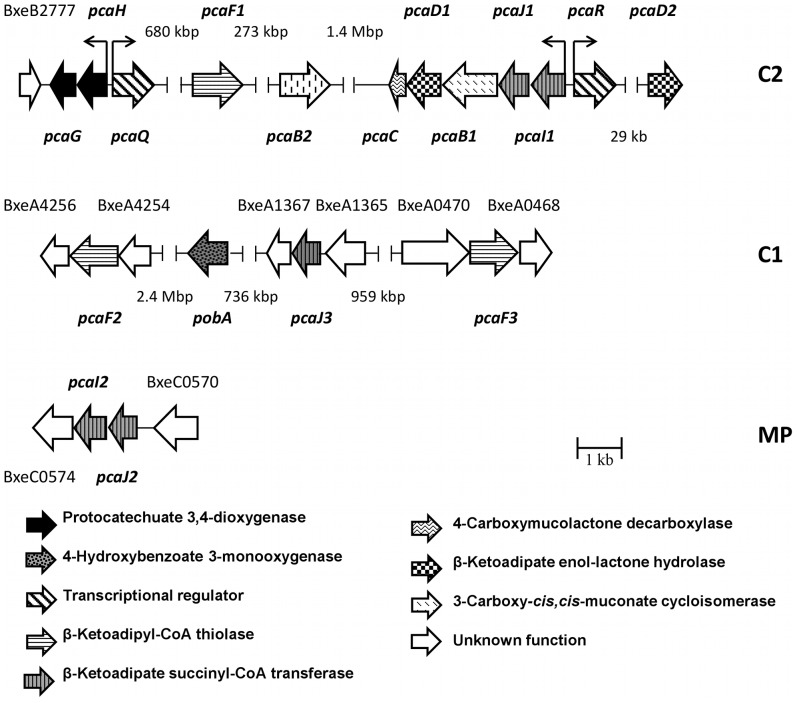
Predicted pca genes encoding the protocatechuate central pathway are distributed within the B. xenovorans LB400 genome (Fig. 4). Predicted pca genes encoding the protocatechuate pathway and related peripheral routes are listed in Table 2. Sequence analyses suggest that pcaG, pcaH, pcaQ, pcaF (BxeB2167; hereafter pcaF1), pcaC, pcaD (BxeB0646; hereafter pcaD1), pcaB (BxeB0645; hereafter pcaB1), pcaJ (BxeB0644; hereafter pcaJ1), pcaI (BxeB0643; hereafter pcaI1), pcaQ and pcaR genes encoding the protocatechuate catabolic pathway are located at C2. Copies of pcaB (BxeB1906; hereafter pcaB2) and pcaD genes (BxeB0595; hereafter pcaD2) were identified at C2. Two pcaF gene copies (hereafter pcaF2 and pcaF3, respectively), and a pcaJ gene copy (hereafter pcaJ3) were identified by sequence analyses at C1. In addition, pcaJ and pcaI genes copies (hereafter pcaJ2 and pcaI2, respectively) were identified by sequence analyses at MP. The pcaG and pcaH genes encoding the alpha and beta subunits from a protocatechuate 3,4-dioxygenase were located 2.3 Mbp distant region from pcaIJBDC gene cluster. The pcaG and pcaH gene products of strain LB400 showed ≥90% identity with the PcaG and PcaH proteins from Acinetobacter lwoffii K24 [31]. The spread in the organization of pca genes and genes encoding related peripheral pathways in B. xenovorans LB400 is also observed in Cupriadus metallidurans CH34 (Fig. 5). Predicted pcaQ gene located next to the pcaHG genes and divergently transcribed encodes a LysR-type transcriptional regulator (Fig. 2) that might regulate the expression of the pcaHG gene cluster. The PcaQ protein shared a 48% sequence identity with the LysR-type transcriptional regulator PcaQ from Agrobacterium tumefaciens A348 (Table 2) [43]. Two divergent σ70-type promoters are present between the pcaQ gene and the pcaGH gene cluster. A PcaQ LysR-type regulatory binding sequence (5′-ATAACGGCAGGTTAT-3′) was identified upstream of the pcaH gene promoter. This regulatory sequence has the consensus sequence for the PcaQ regulator recognition site sequence of Burkholderiales (5′-ATAACN5GTTAT-3′) [43]. Predicted pcaR gene (BxeB0642) located adjacent to the pcaIJBDC genes and divergently transcribed encodes a LysR-type transcriptional regulator (Fig. 2). The pcaR gene product shared a 95% sequence identity with a LysR-type transcriptional regulator adjacently located to the pcaIJBDC gene cluster from B. phytofirmans PsJN (Table 2) [38]. Genomic analysis of the pcaCD1B1J1I1R gene cluster showed that two divergent σ70-type promoters are present between the pcaR gene and the pcaCD1B1J1I1 gene cluster. A putative LysR-type regulatory binding sequence (5′-GTTCGCTAAACGAAT-3′) was identified upstream to the pcaR gene promoter. This sequence has the conserved (5′-T-N11-A-3′) motif critical for binding of LysR-type autoregulatory transcriptional regulators [41].
Figure 4. Genetic organization of the pca genes involved in protocatechuate catabolism in Burkholderia xenovorans LB400.
The pca genes are located in the minor chromosome (C2), major chromosome (C1) and megaplasmid (MP). The genes encoding the alpha and beta subunits of the protocatechuate 3,4-dioxygenase are indicated with black arrows. The orientations of ORFs are represented by open arrows. The promoter regions for pcaH, pcaQ, pcaI and pcaR are denoted with small black arrows, bent in the directions of transcription. The sizes of the genes and the intergenic regions are on scale.
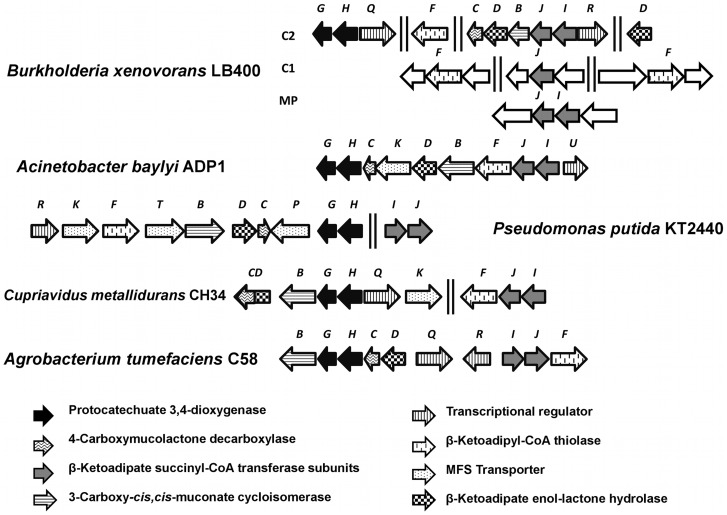
Figure 5. Organization of the pca genes involved in the catabolism of protocatechuate in B. xenovorans LB400 and other bacteria.
The genes encoding the alpha and beta subunits of the protocatechuate 3,4-dioxygenase are indicated with black arrows. The sizes of the genes and the intergenic regions are on scale.
The genes encoding the enzymes of the 3-HBA and 4-HBA peripheral reactions that funnel into the protocatechuate pathway were searched in the LB400 genome. The pobA gene (BxeA2040) encoding for a 4-HBA 3-monooxygenase that converts 4-HBA into protocatechuate was identified by sequence analyses at C1 (Fig. 4). The pobA gene protein from strain LB400 shares 51% sequence identity with the PobA enzyme from P. fluorescens strain Pf-5 [21]. In addition, protocatechuate has been described as a central route for 3-HBA degradation by a 3-HBA 4-hydroxylase encoded by the mobA gene in bacteria. The mobA gene was not found in the LB400 genome.
Growth of Strain LB400 on Gentisate, Protocatechuate and HBAs
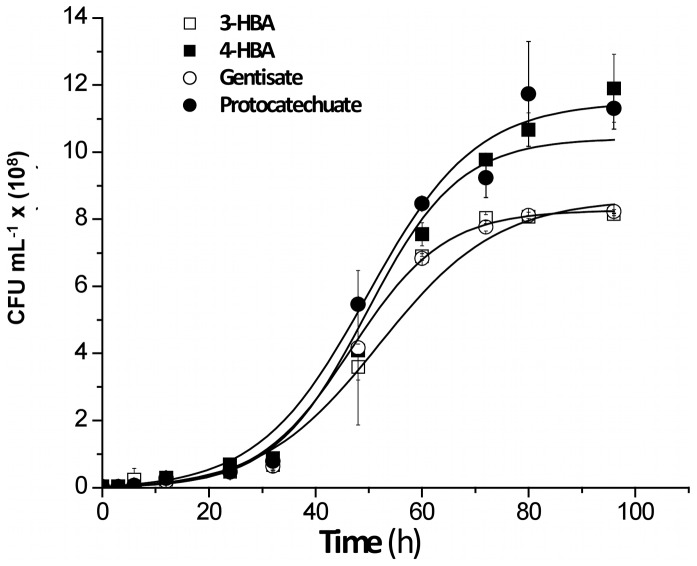
Sequence analyses of B. xenovorans LB400 genome revealed the presence of genes encoding the gentisate and the protocatechuate ring-cleavage pathways and 3-HBA and 4-HBA peripheral pathways. To investigate whether these catabolic pathways are functional, in a first approach growth assays were performed. The growth of B. xenovorans strain LB400 on the central metabolites gentisate and protocatechuate was studied. B. xenovorans LB400 was able to grow on gentisate (5 mM) and on protocatechuate (5 mM) as sole carbon and energy source (Fig. 6). In addition, the growth of B. xenovorans strain LB400 on 3-HBA or 4-HBA was studied. Strain LB400 was able to grow on 3-HBA (10 mM) and 4-HBA (5 mM) (Fig. 6). B. xenovorans LB400 attained higher cell concentration at stationary phase on protocatechuate (1.1 ×109 CFU mL−1) and 4-HBA (1.0×109 CFU mL−1). The growth of strain LB400 on gentisate or 3-HBA yielded 7.0×108 CFU mL−1 (Fig. 6). These results indicate functional 3-HBA and 4-HBA peripheral pathways and gentisate and protocatechuate central pathways in B. xenovorans LB400.
Figure 6. Growth of B. xenovorans LB400 on hydroxybenzoates, gentisate and protocatechuate.
LB400 cells were grown in M9 minimal medium using gentisate (5 mM), protocatechuate (5 mM), 3-HBA (10 mM) or 4-HBA (5 mM) as sole carbon and energy source. Each point is an average ± SDs of results from, at least, three independent assays.
Transcriptional Analyses of the mhb and pca Genes
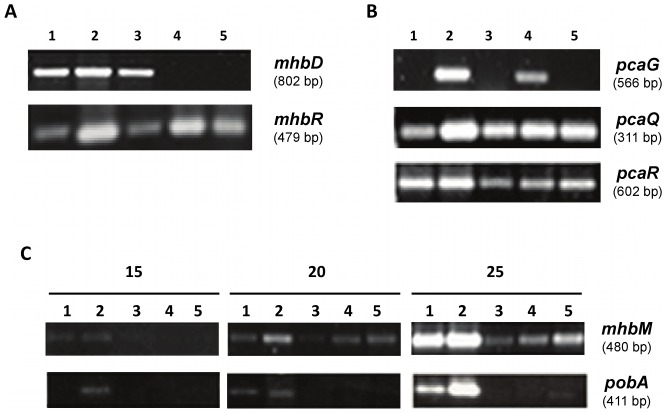
In order to determine the role of predicted genes encoding the gentisate and protocatechuate ring-cleavage routes and 3-HBA and 4-HBA peripheral pathways in strain LB400, transcriptional analyses of key catabolic genes were performed. In addition, the expression of genes encoding potential transcriptional regulators was analyzed. The expression of the mhbD gene encoding the enzyme gentisate 1,2-dioxygenase during exponential growth phase of LB400 cells grown on gentisate, protocatechuate, 3-HBA, 4-HBA, and glucose was analyzed. The mhbD gene was transcribed during growth of strain LB400 in gentisate, 3-HBA and 4-HBA (Fig. 7). In contrast, expression of the mhbD gene was not observed during growth on protocatechuate or glucose (Fig. 7). The mhbR gene encoding a MarR-type transcriptional regulator was transcribed during LB400 growth on 3-HBA, 4-HBA, gentisate, protocatechuate and glucose (Fig. 7). These results suggest that gentisate, 3-HBA and 4-HBA or its metabolites induce transcription of the mhbD gene in B. xenovorans LB400. On the other side, the mhbR gene has probably a basal constitutive expression. Transcription analysis of the mhbM gene encoding the 3-HBA 6-hydroxylase was performed. The mhbM gene of strain LB400 was expressed was during growth on 3-HBA, 4-HBA, gentisate, protocatechuate and glucose (Fig. 7). However, higher expression of the mhbM gene on HBAs than on glucose was observed.
Figure 7. Transcriptional analysis of the mhb and pca genes during LB400 growth on hydroxybenzoates, gentisate and protocatechuate.
LB400 cells were grown on 3-HBA (lane 1), 4-HBA (lane 2), gentisate (lane 3), protocatechuate (lane 4) and glucose (lane 5) as sole carbon source. (A) Transcription of mhbD and mhbR genes; (B) Transcription of pcaG, pcaQ and pcaR genes. (C) Transcription of mhbM and pobA genes. RT-PCR (15, 20 and 25 amplification cycles) assays were performed using RNA extracted from LB400 cells collected at mid exponential growth phase. Transcription of 16S rRNA gene was used as control (not shown).
Transcription of the pcaG gene encoding the key protocatechuate 3,4-dioxygenase alpha subunit during growth of strain LB400 with different aromatic substrates was studied. Predicted pcaG gene was expressed during growth of strain LB400 on protocatechuate and 4-HBA (Fig. 7). However, pcaG transcripts were not observed during growth of strain LB400 on gentisate, 3-HBA and glucose. These results suggest that by 4-HBA, protocatechuate or its metabolites induce the transcription of the pcaG gene. Both pcaQ and pcaR genes encoding the transcriptional regulators of the pcaGH genes and the pcaIJBDC gene cluster were transcribed during LB400 growth on HBAs, gentisate, protocatechuate and glucose (Fig. 7). Therefore, the pcaQ and pcaR genes have probably a basal constitutive expression.
Transcription of the pobA gene encoding the 4-HBA 3-monooxygenase of strain LB400 was analyzed. The pobA gene was expressed during LB400 growth only on 3-HBA, 4-HBA and glucose (Fig. 7). The pobA gene expression was higher during growth on HBAs than on glucose.
These results indicate that expression of the mhb and pca genes encoding catabolic enzymes is regulated in LB400 cells. These regulations are probably exerted by MarR-type and LysR-type transcriptional regulators. This study also suggests a regulated expression of the mhbM gene encoding the peripheral 3-HBA 6-hydroxylase and of the pobA gene encoding the peripheral 4-HBA 3-monooxygenase but with a basal constitutive expression of the mhbM gene. Overall, the transcriptional analyses suggest that 3-HBA is catabolized and funneled exclusively through the gentisate central pathway, whereas 4-HBA can be degraded through the protocatechuate and gentisate central pathways in strain LB400.
Gentisate and Protocatechuate Ring-cleavage Activities
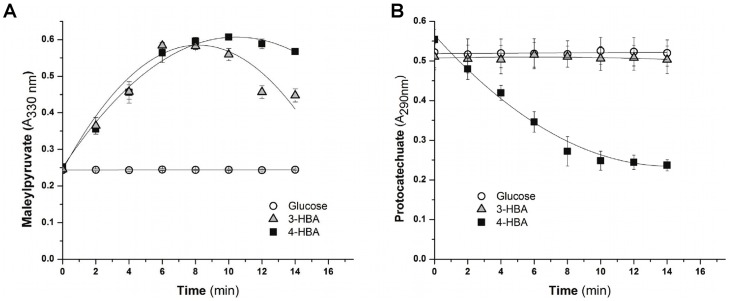
To further characterize the gentisate and protocatechuate central pathways in B. xenovorans strain LB400, the activities of the key enzymes gentisate 1,2-dioxygenase and protocatechuate 3,4-dioxygenase were measured. Both dioxygenase activities were determined in crude extract of cells cultured until mid exponential phase in 3-HBA, 4-HBA and glucose. Higher gentisate 1,2-dioxygenase activity was observed in cells grown on 3-HBA (0.053±0.001 U mg−1 protein) and 4-HBA (0.049±0.004 U mg−1 protein), whereas the gentisate 1,2-dioxygenase activity was not detected in glucose-growth cells (Fig. 8A). LB400 cells grown on 4-HBA showed high protocatechuate 3,4-dioxygenase activity (0.15±0.02 U mg−1 protein), whereas the protocatechuate dioxygenase activity was not observed in LB400 cells grown on 3-HBA and glucose (Fig. 8B). Enzyme activity assays suggest an active protocatechuate pathway in 4-HBA-grown cells, whereas the gentisate central pathway was found to be active in both 3-HBA- and 4-HBA-grown cells.
Figure 8. Gentisate and protocatechuate dioxygenase activities of B. xenovorans LB400.
Cells were grown on 3-HBA (black square), 4-HBA (empty square) and glucose (empty circles) as sole carbon source. (A) Gentisate 1,2-dioxygenase activity measured by maleylpyruvate formation. (B) Protocatechuate 3,4-dioxygenase activity measured by protocatechuate disappearance. Each point is an average ± SDs of values from three independent assays.
Discussion
This study has focused on a genomic analysis on the gentisate and protocatechuate catabolic pathways and its related peripheral pathways in B. xenovorans LB400. Experimental evidence was provided on the functionality of these ring-cleavage pathways, and their roles during 3-HBA and 4-HBA catabolism were proposed. Strain LB400 is able to grow on gentisate as sole carbon source, indicating a functional gentisate catabolic pathway. The gentisate catabolic pathway is present only in few Burkholderia strains [1]. Both mhbRTDHI gene clusters were found in two identical and adjacently located segments of 30 genes in LB400 major chromosome, indicating a gene cluster duplication process. Gene duplication is frequently observed in aromatic catabolic pathways and suggests functional redundancies, which could potentially increase the bacterial fitness and its adaptation to adverse environmental conditions. The mhbDHI gene cluster organization is similar in strains from Burkholderiales order including B. xenovorans LB400. However, the complete mhbRTDHI gene cluster with a coding sequence of unknown function between mhbT and mhbD was found in strain LB400, which has not been observed in other bacteria and its function remains to be elucidated.
Growth studies indicated that a 3-HBA peripheral pathway is present in B. xenovorans LB400. Gentisate has been described as the main ring-cleavage pathway for 3-HBA assimilation in bacteria, such as K. pneumoniae M5a1 [11], Corynebacterium glutamicum RES167 [44] and Rhodococcus sp. strain NCIMB 12038 [45]. In B. xenovorans strain LB400 the mhbM gene encoding the 3-HBA 6-monooxygenase is located at C2, whereas the mhbRTDHI gene cluster is located at C1 (Fig. 1). In contrast, in strains M5a1, RES167, NCIMB 12038 and in Comamonas and other Burkholderia strains, a 3-HBA 6 hydroxylase-encoding mhbM gene is usually found within the mhbDHI gene cluster. An increased transcription of mhbM gene in strain LB400 was observed during growth on 3-HBA. Therefore, we propose that the predicted mhbM gene product of strain LB400 is involved in the hydroxylation of 3-HBA to gentisate.
It is worth noting that B. xenovorans LB400 was able to grow using 10 mM 3-HBA, whereas no growth of LB400 cells was achieved on 5 mM 3-HBA. We propose that a 3-HBA (5 mM) did not support the growth of B. xenovorans strain LB400, probably due to an insufficient transport of 3-HBA across the membrane. These results suggest that strain LB400 lacks an active transport for 3-HBA. 4-HBA catabolism through the gentisate pathway has been described in bacterial strains from Bacillus genus [46] and the archaeal isolate Haloarcula sp. strain D1 [42]. B. xenovorans LB400 was able to grow using 4-HBA (5 mM) as sole carbon source. The LB400 growth on 4-HBA (5 mM) but not on 3-HBA (5 mM) suggests an efficient transport of 4-HBA rather than 3-HBA through the membrane by an active 4-HBA transport system. This is supported by our bioinformatics analyses that indicated that the mhbT gene product of strain LB400 has 33% sequence identity with the PcaK protein of P. putida PRS2000, which is an active transporter system for 4-HBA [29], [33]. The mhbT gene of B. xenovorans LB400 encodes a MFS transporter of the AAHS family that may be involved in the active transport of gentisate and 4-HBA.
Genomic analyses suggest that the mhbR gene, which is upstream and divergently transcribed to the mhbT gene, encodes for a transcriptional regulator of the mhbT gene in strain LB400. The mhbR gene of strain LB400 was transcribed during growth on 3-HBA, 4-HBA, gentisate, protocatechuate and glucose, suggesting a basal constitutive expression. The mhbR gene product from B. xenovorans LB400 shares 30% sequence identity with MarR-type transcriptional regulators from the mhb gene cluster from P. naphthalenivorans CJ2 [28] and C. testosteroni CNB-2 [47]. MarR regulators are winged helix-turn-helix DNA-binding proteins that respond to specific ligands and act as dimmers. In absence of the ligand, MarR regulators bind to palindromic sequences within the promoter, generally causing transcriptional repression [40]. We propose that the MarR-type regulator encoded by the mhbR gene from B. xenovorans LB400 is a transcriptional repressor that regulates the expression of the mhbT gene. Transcriptional analysis showed that the mhbD gene encoding a gentisate 1,2-dioxygenase was expressed during growth on gentisate, 3-HBA and 4-HBA, but not on glucose. Transcriptional analyses suggest that either 3-HBA, 4-HBA, gentisate or its metabolic intermediates induced the transcription of the mhb gene cluster. The expression of mhbD gene correlates with an increased gentisate 1,2-dioxygenase activity observed in LB400 cells grown on 3-HBA and 4-HBA. Genomics analysis showed the presence of a conserved sequence (5′-T-N11-A-3′) critical for LysR-type transcriptional regulator binding, located upstream of the -35 box of BxeA2626 and BxeA4527 genes. However, a LysR-type transcriptional regulator gene was not found in the neighborhoods of both mhb gene clusters. Although further studies are needed to survey the regulation mechanisms of 3-HBA and 4-HBA degradation pathways via gentisate in strain LB400, these genomic analyses suggest that mhbDHI cluster is regulated by a LysR-type transcriptional regulator in strain LB400. Overall, transcriptomic analyses and dioxygenase activity assays indicate that the gentisate pathway in strain LB400 is active during 3-HBA and 4-HBA degradation, suggesting that both substrates may be funneled into the gentisate central pathway (Fig. 9). We propose that a 4-HBA 1-hydroxylase could be involved in the conversion of 4-HBA into gentisate in B. xenovorans LB400. Experimental data with deuterated substrates in the archaeal strain Haloarcula sp. D1 supported the mechanism in which 4-HBA is converted into gentisate by hydroxylation of 4-HBA at C-1, with a concomitant 1,2-carboxyl group migration. The transformation is called NIH shift and is catalysed by a 4-HBA 1-hydroxylase [42]. Although no gene sequence information of the 4-HBA 1-hydroxylase enzyme is available to perform a genomic search, an active gentisate pathway during 4-HBA assimilation suggests a 4-HBA 1-hydroxylase activity in B. xenovorans strain LB400. Recently, we reported that B. xenovorans strain LB400 degrades 4-hydroxyphenylacetate via homogentisate, in which also a migration of the acetate group by a NIH shift reaction may also be involved [4]. Hydroxylation of monosubstituted aromatic rings at C-1, followed by a migration of the carboxymethyl side chain to C-2 has been described in P. acidovorans and Nitrosomonas europaea 19718 [48], [49]. Future studies are needed to characterize the genes encoding the 4-HBA 1-hydroxylase that converts 4-HBA into gentisate in bacteria, including B. xenovorans strain LB400.
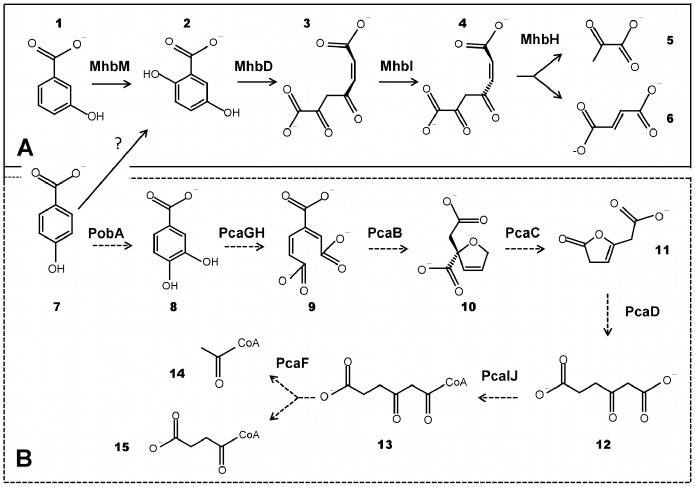
Figure 9. Models of 3-HBA and 4-HBA catabolic pathways in B. xenovorans LB400.
(A) 3-HBA and 4-HBA catabolism via the gentisate central pathway (continuous line). The substrates and products are: 3-hydroxybenzoate (1); gentisate (2); maleylpyruvate (3); fumarylpyruvate (4); pyruvate (5); fumarate (6) and 4-hydroxybenzoate (7). The enzymes are MhbM (3-HBA 6-hydroxylase), MhbD (gentisate 1,2-dioxygenase), MhbI (maleylpyruvate isomerase) and MhbH (fumarylpyruvate hydrolase). The gene encoding the 4-HBA 1-hydroxylase is unknown. (B) 4-HBA catabolism via protocatechuate (dotted line). The substrates and products are: 4-hydroxybenzoate (7); protocatechuate (8); 3-carboxy-cis,cis,-muconate (9); 4-carboxymucolactone (10); β-ketoadipate enol-lactone (11); β-ketoadipate (12); β-ketoadipyl-CoA (13); Acetyl-CoA (14); Succinyl-CoA (15).The enzymes are: PobA (4-hydroxybenzoate 3-monooxygenase), PcaGH (protocatechuate 3,4-dioxygenase), PcaB (3-carboxymuconate cycloisomerase), PcaC (4-carboxymuconolactone decarboxylase), PcaD (β-ketoadipate enol-lactone hydrolase), PcaJI (β-ketoadipate:succinyl-CoA tranferase) and PcaF (β-ketoadipyl-CoA thiolase).
Most bacteria metabolize 4-HBA by hydroxylation at C-3 to yield protocatechuate as central intermediate [1], [2]. The 4-HBA 3-monooxygenase encoded by the pobA gene catalyzes 4-HBA conversion into protocatechuate in Pseudomonas strains [21], [22]. Genomic analyses revealed the presence of the pca genes encoding the complete protocatechuate central pathway are located at C2 and pca gene copies are located at C1 and MP, while the pobA gene is separately located at C1 in strain LB400 genome. Spreading of pca genes and genes encoding its related peripheral reactions is also observed in Betaproteobacteria, such as Cupriavidus (Fig. 4), Burkholderia and Ralstonia strains [1]. Diverse pca gene organizations have been reported in bacteria [1], [2], [20]. The pcaGH genes are clustered with other pca genes of the protocatechuate route and related peripheral pathways in the Gammaproteobacteria A. baylyi ADP1 and P. putida KT2440, the Alphaproteobacteria strains Agrobacterium tumefaciens C58 and Sinorhizobium meliloti 1021, and the Betaproteobacteria from the Cupriavidus genus such as C. metallidurans CH34, C. pinatubonensis JMP143 and C. necator H16 [1]. In contrast, in B. xenovorans LB400 and other bacterial strains from the Burkholderia genus, the pcaGH genes are not clustered with other pca genes. B. xenovorans strain LB400 possesses the pcaQHG and the pcaRIJBDC gene clusters and separately pcaF gene copies. In addition, LB400 genome contains a pcaIJ gene cluster and pcaB, pcaD and pcaJ gene single-copies, indicating genetic redundancy.
B. xenovorans LB400 was able to grow on 4-HBA and protocatechuate, indicating functional 4-HBA peripheral and protocatechuate ring-cleavage pathways. Transcriptional analyses showed that pcaG gene encoding the protocatechuate 3,4-dioxygenase alpha subunit was expressed during protocatechuate and 4-HBA degradation by strain LB400. The expression of the pcaG gene during 4-HBA degradation correlates with an increased protocatechuate 3,4-dioxygenase activity observed in LB400 cells extracts grown on 4-HBA. In contrast, neither pcaG gene expression nor protocatechuate 3,4-dioxygenase activity was observed in 3-HBA and glucose-grown LB400 cells. These results suggest that 4-HBA is exclusively degraded via protocatechuate, in which a hydroxylation of 4-HBA at C-3 occurred for protocatechuate formation. The pobA gene of strain LB400 was transcribed during growth on 4-HBA, suggesting that the pobA gene product is involved in 4-HBA hydroxylation to produce protocatechuate in B. xenovorans strain LB400 (Fig. 9). B. xenovorans LB400 as well as most strains belonging to Burkholderia and Ralstonia genera that possess the pcaGH genes have a 4-HBA 3-hydroxylase-encoding gene, indicating that protocatechuate is the preferred catabolic pathway for 4-HBA degradation. In addition, these results suggest that either 4-HBA, protocatechuate or its metabolic intermediates induced the transcription of protocatechuate 3,4-dioxygenase-encoding genes. The pcaQ gene adjacently located to pcaGH genes encodes a LysR-type transcriptional regulator and was transcribed during growth on 4-HBA, protocatechuate, 3-HBA, gentisate and glucose, suggesting a constitutive expression of the pcaQ gene. Thus, the PcaQ LysR-type transcriptional regulator of strain LB400 probably controls the expression of the pcaGH genes during 4-HBA and protocatechuate degradation in strain LB400. LysR-type transcriptional regulators act as activators during aromatic compounds catabolism. In general, the inducers are metabolic intermediates of the catabolic pathway [2], [20]. The PcaQ regulator from strain LB400 probably acts as a transcriptional activator of the expression of the divergently transcribed pcaGH genes, like other PcaQ proteins that are transcriptional activators of the pca genes in Agrobacterium tumefaciens and Sinorhizobium meliloti [32], [43]. The pcaR gene adjacently located upstream of the pcaIJBDC gene cluster encodes a LysR-type transcriptional regulator and was transcribed during growth on 4-HBA, protocatechuate, 3-HBA, gentisate and glucose. In P. putida strains PRS2000 and KT2440, IclR-type transcriptional regulators regulate the pca gene cluster encoding enzymes for the degradation of 3-carboxy-cis,cis-muconate into Krebs cycle intermediates [2], [20]. We propose that the PcaR protein may regulate the expression of the pcaIJBDC gene cluster in strain LB400. The pcaRIJBDC gene organization with a LysR-type regulator with high sequence identity (95%) was observed in the closely relative strain B. phytofirmans PsJN. In accordance with this, it has been reported that all strains of the ‘B. cepacia complex’ as well as B. phymatum STM815 and B. glumae BGR1 possess a LysR-type transcriptional regulator gene clustered with the pcaIJBDC genes [1]. It is likely that the pcaR gene product of strain LB400 acts as a transcriptional activator of the expression of the divergently transcribed pcaIJBDC gene cluster. Further studies are required to confirm that PcaQ and PcaR proteins regulate the expression of the pcaHG and pcaIJBDC gene clusters in B. xenovorans LB400. The protocatechuate pathway is widespread among pathogenic and environmental bacteria, and only a small number of strains lack this pathway. Therefore, the protocatechuate catabolic pathway can be an important catabolic feature and play a key role to improve bacterial fitness in the environment and in pathogenesis.
In this report, we described functional gentisate and protocatechuate ring-cleavage pathways in B. xenovorans LB400, revealing inducible expression of key genes encoding gentisate and protocatechuate dioxygenases. Additionally, in this study 3-HBA and 4-HBA peripheral reactions in strain LB400 were reported. Based on genomic analyses, growth assays, gene expression analyses and key dioxygenase activities, we propose that 3-HBA degradation is channeled exclusively through the gentisate central pathway in B. xenovorans strain LB400, whereas 4-HBA can be degraded and funneled actively into the protocatechuate and gentisate central pathways (Fig. 9). This report reveals novel catabolic capabilities of B. xenovorans LB400, which can use two different ring-cleavage pathways for the metabolism of 3-HBA and 4-HBA. This study confirms that B. xenovorans LB400 possesses a high metabolic versatility, which is reflected on its extensive gene and functional redundancy for the degradation of aromatic compounds.
Acknowledgments
M.S. gratefully acknowledges Kenneth N. Timmis for his wise guidance and generous support.
Funding Statement
M.S. acknowledges financial support from FONDECYT (1110992 and 1070507) (http://www.fondecyt.cl), Center for Nanotechnology and Systems Biology (http://www.usm.cl) and USM (131109, 130948) (http://www.usm.cl) grants. The funders had no role in study design, data collection and analyses, decision to publish, or preparation of the manuscript.
References
- 1. Pérez-Pantoja D, Donoso R, Agulló L, Córdova M, Seeger M, et al. (2012) Genomic analysis of the potential for aromatic compounds biodegradation in Burkholderiales . Environ Microbiol 14: 1091–1117. [DOI] [PubMed] [Google Scholar]
- 2. Jiménez JI, Miñambres B, García JL, Díaz E (2002) Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol 4: 824–841. [DOI] [PubMed] [Google Scholar]
- 3. Chain P, Denef V, Konstantinidis K, Vergez L, Agulló L, et al. (2006) Burkholderia xenovorans LB400 harbors a multi-replicon, 9.7 Mbp genome shaped for versatility. Proc Natl Acad Sci USA 113: 15280–15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Méndez V, Agulló L, González M, Seeger M (2011) The homogentisate and homoprotocatechuate central pathways are involved in 3- and 4-hydroxyphenylacetate degradation by Burkholderia xenovorans LB400. PLoS ONE 6: e17583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seeger M, Timmis KN, Hofer B (1995) Conversion of chlorobiphenyls into phenylhexadienoates and benzoates by the enzymes of the upper pathway for PCB-degradation encoded by the bph locus of Pseudomonas sp. LB400. Appl Environ Microbiol 61: 2654–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seeger M, Zielinski M, Timmis K, Hofer B (1999) Regioespecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl Environ Microbiol 65: 3614–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seeger M, Cámara B, Hofer B (2001) Dehalogenation, denitration, dehydroxylation and angular attack on substituted biphenyls and related compounds by a biphenyl dioxygenase. J Bacteriol 183: 3548–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ponce B, Latorre VK, González M, Seeger M (2011) Antioxidant compounds improved PCB-degradation by Burkholderia xenovorans strain LB400. Enzyme Microb Technol 49: 509–516. [DOI] [PubMed] [Google Scholar]
- 9. Poh CL, Baily RC (1980) Evidence for isofunctional enzymes used in m-cresol and 2,5-xylenol degradation via the gentisate pathway in Pseudomonas alcaligenes . J Bacteriol 143: 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harpel MR, Lipscomb JD (1990) Gentisate 1,2-dioxygenase from Pseudomonas: substrate coordination to active site Fe+2 and mechanism of turnover. J Biol Mol 265: 22187–22196. [PubMed] [Google Scholar]
- 11. Jones DC, Cooper RA (1990) Catabolism of 3-hydroxybenzoate by the gentisate pathway in Klebsiella pneumoniae M5a1. Arch Microbiol 154: 489–495. [DOI] [PubMed] [Google Scholar]
- 12. Goetz FE, Harmuth LJ (1992) Gentisate pathway in Salmonella typhimurium: metabolism of m-hydroxybenzoate and gentisate. FEMS Microbiol Lett 97: 45–50. [DOI] [PubMed] [Google Scholar]
- 13. Suárez M, Ferrer E, Martín M (1996) Purification and biochemical characterization of gentisate 1,2-dioxygenase from Klebsiella pneumoniae M5a1. FEMS Microbiol Lett 143: 89–95. [DOI] [PubMed] [Google Scholar]
- 14. Ishiyama D, Vujaklija D, Davies J (2004) Novel pathway of salicylate degradation by Streptomyces sp. strain WA46. Appl Environ Microbiol 70: 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang LH, Hamzah RY, Yu YM, Tu SC (1987) Pseudomonas cepacia 3-hydroybenzoate 6-hydroxylase: induction, purification, and characterization. Biochem 26: 1099–1104. [DOI] [PubMed] [Google Scholar]
- 16. Suárez M, Martín M, Ferrer E, Garrido-Pertierra A (1995) Purification and characterization of 4-hydroxybenzoate 6-hydroxylase from a Klebsiella pneumoniae mutant strain. Arch Microbiol 164: 70–77. [DOI] [PubMed] [Google Scholar]
- 17. Michalover JL, Ribbons DW, Hughes H (1973) 3-Hydroxybenzoate 4-hydroxylase from Pseudomonas testosteroni . Biochem Biophys Res Commun 55: 888–896. [DOI] [PubMed] [Google Scholar]
- 18. Hiromoto T, Fujiwara S, Hosokawa K, Yamaguchi H (2006) Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site. J Mol Biol 364: 878–896. [DOI] [PubMed] [Google Scholar]
- 19. Pérez-Pantoja D, De la Iglesia R, Pieper D, González B (2008) Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol Rev 32: 736–794. [DOI] [PubMed] [Google Scholar]
- 20. Harwood C, Parales R (1996) The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50: 553–590. [DOI] [PubMed] [Google Scholar]
- 21. Van Berkel W, Westphal A, Eschrich K, Eppink M, de Kok A (1992) Substitution of Arg214 at the substrate-binding site of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens . Eur J Biochem 210: 411–419. [DOI] [PubMed] [Google Scholar]
- 22. Seibold B, Matthes M, Eppink MH, Lingens F, Van Berkel WJ, et al. (1996) 4-Hydroxybenzoate hydroxylase from Pseudomonas sp. CBS3. Purification, characterization, gene cloning, sequence analysis and assignment of structural features determining the coenzyme specificity. Eur J Biochem 239: 469–478. [DOI] [PubMed] [Google Scholar]
- 23. Agulló L, Cámara B, Martínez P, Latorre V, Seeger M (2007) Response to (chloro)biphenyls of the polychlorobiphenyl-degrader Burkholderia xenovorans LB400 involves stress proteins also induced by heat shock and oxidative stress. FEMS Microbiol Lett 267: 167–175. [DOI] [PubMed] [Google Scholar]
- 24. Noble JE, Bailey MJ (2009) Quantitation of protein. Methods Enzymol 463: 73–95. [DOI] [PubMed] [Google Scholar]
- 25. Wheelis ML, Palleroni NJ, Stanier RY (1967) The metabolism of aromatic acids by Pseudomonas testosteroni and P. acidovorans . Arch Microbiol 59: 302–314. [DOI] [PubMed] [Google Scholar]
- 26. Buchan A, Collier LS, Neidle EL, Moran MA (2000) Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacter lineage. Appl Environ Microbiol 66: 4662–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wright JD, Ratledge C (1991) Isolation of two Rhadotarula rubra strains showing differences in the degradation of aromatic compounds. Appl Microbiol Biotechnol 35: 94–99. [Google Scholar]
- 28. Yagi JM, Sims D, Brettin T, Bruce D, Madsen EL (2009) The genome of Polaromonas naphthalenivorans strain CJ2, isolated from coal tar-contaminated sediment, reveals physiological and metabolic versatility and evolution through extensive horizontal gene transfer. Environ Microbiol 11: 2253–2270. [DOI] [PubMed] [Google Scholar]
- 29. Nichols N, Harwood C (1997) PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. . J Bacteriol 179: 5056–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robson ND, Parrott S, Cooper RA (1996) In vitro formation of a catabolic plasmid carrying Klebsiella pneumoniae DNA that allows growth of Escherichia coli K-12 on 3-hydroxybenzoate. Microbiol 142: 2115–2120. [DOI] [PubMed] [Google Scholar]
- 31. Khang HY, Cho K, Song SY, Kim SJ, Lee SH, et al. (2002) Enhanced detection and characterization of protocatechuate 3,4-dioxygenase in Acinetobacter lwoffii K24 by proteomics using a column separation. Biochem Biophys Res Commun 295: 903–909. [DOI] [PubMed] [Google Scholar]
- 32. Parke D (1996) Characterization of PcaQ, a LysR-type transcriptional activator required for catabolism of phenolic compounds, from Agrobacterium tumefaciens . J Bacteriol 178: 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harwood C, Nichols N, Kim M, Ditty J, Parales R (1994) Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation and transport of 4-hydroxybenzoate. J Bacteriol 176: 6479–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hartnett C, Neidle EL, Ngai KL, Ornston LN (1990) DNA sequences of genes encoding Acinetobacter calcoaceticus protocatechuate 3,4-dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J Bacteriol 172: 956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hartnett GB, Ornston LN (1994) Acquisition of apparent DNA slippage structures during extensive evolutionary divergence of pcaD and catD genes encoding identical catalytic activities in Acinetobacter calcoaceticus . Gene 142: 23–29. [DOI] [PubMed] [Google Scholar]
- 36. Williams SE, Woolridge EM, Ransom SC, Landro JA, Babbit PC, et al. (1992) 3-Carboxy-cis,cis-muconate lactonizing enzyme from Pseudomonas putida is homologous to the class II fumarase family: a new reaction in the evolution of a mechanism motif. Biochem 31: 9768–9776. [DOI] [PubMed] [Google Scholar]
- 37. Parales RE, Harwood CS (1992) Characterization of the genes encoding β-ketoadipate:succinyl-coenzyme A transferase in Pseudomonas putida . J Bacteriol 174: 4657–4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weilharter A, Mitter B, Shin MV, Chain PS, Nowak J, et al. (2011) Complete genome sequence of the plant growth-promoting endophyte Burkholderia phytofirmans strain PsJN. J Bacteriol 193: 3383–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin LX, Liu H, Zhou NY (2010) MhbR, a LysR-type regulator involved in 3-hydroxybenzoate catabolism via gentisate in Klebsiella pneumoniae M5a1. Microbiol Res 165: 66–74. [DOI] [PubMed] [Google Scholar]
- 40. Perera IC, Groove A (2011) MarR homologs with urate-binding signature. Protein Sci 20: 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schell M (1993) Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol 47 597–626. [DOI] [PubMed] [Google Scholar]
- 42. Fairley DJ, Boyd DR, Sharma ND, Allen CC, Morgan P, et al. (2002) Aerobic metabolism of 4-hydroxybenzoic acid in Archaea via an unusual pathway involving an intramolecular migration (NIH shift). Appl Environ Microbiol 68: 6246–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. MacLean A, Haerty W, Golding B, Finan T (2011) The LysR-type PcaQ protein regulates expression of a protocatechuate-inducible ABC-type transport system in Sinorhizobium meliloti . Microbiol 157: 2522–2533. [DOI] [PubMed] [Google Scholar]
- 44. Shen XH, Jiang CY, Huang Y, Liu ZP, Liu SJ (2005) Functional identification of novel genes involved in the glutathione-independent gentisate pathway in Corynebacterium glutamicum . Appl Environ Microbiol 71: 3442–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu TT, Xu Y, Liu H, Luo S, Yin YJ, et al. (2011) Functional characterization of a gene cluster involved in gentisate catabolism in Rhodococcus sp. strain NCIMB 12038. Appl Microbiol Biotechnol 90: 671–678. [DOI] [PubMed] [Google Scholar]
- 46. Crawford RL (1976) Pathways of 4-hydroxybenzoate degradation among species of Bacillus . J Bacteriol 127: 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma YF, Zhang Y, Zhang JY, Chen DW, Zhu Y, et al. (2009) The complete genome of Comamonas testosteroni reveals its genetic adaptations to changing environments. Appl Environ Microbiol 75: 6812–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hareland WA, Crawford RL, Chapman PJ, Dagley S (1975) Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans . J Bacteriol 121: 272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vanelli T, Hooper AB (1995) NIH shift in the hydroxylation of aromatic compounds by the ammonia-oxidizing bacterium Nitrosomonas europaea . Biochem 34: 11743–11749. [DOI] [PubMed] [Google Scholar]