Abstract
Since HIV-1 Tat has been associated with neurocognitive dysfunction, we investigated 60 HIV-1 subtype B infected individuals who were characterized for neurocognitive functioning and had paired CSF and blood plasma samples available. To avoid issues with repeated sampling, we generated population-based HIV-1 tat sequences from each compartment and evaluated these data using a battery of phylogenetic, statistical and machine learning tools. These analyses identified position HXB2 5905 within the cysteine-rich domain of tat as a signature of CSF-derived HIV-1, and a higher number of mixed bases in CSF, measure of diversity, was associated with HIV-associated neurocognitive disorder. Since identified mutations were synonymous, we evaluated the predicted secondary RNA structures, which showed that this mutation altered secondary structure. As a measure of divergence, the genetic distance between the blood and CSF derived tat was inversely correlated with current and nadir CD4+ T cell counts. These data suggest that specific HIV-1 features of tat influence neurotropism and neurocognitive impairment.
Keywords: HIV, central nervous system, tat, compartmentalization
Introduction
HIV-1 crosses the blood-brain barrier during primary infection, eventually resulting in neurological complications in up to 50% of untreated individuals (Grant et al. 1987; Ho et al. 1985; Price. 1996). Anatomic compartments play an important role in the dynamics and evolution of HIV, as distinct tissues provide different cellular targets, immunologic pressures and variations in drug concentration, thus driving viral evolution differently in various compartments (Nickle et al. 2003). Previous reports have shown that, although each host’s viral population is genetically distinct from populations in other individuals, signature polymorphisms can be identified across hosts that distinguish cerebrospinal fluid (CSF)-derived virus from virus in the blood during clade B HIV infection (Pillai et al. 2006; Ritola et al. 2005; Strain et al. 2005). Additionally, distinct reverse transcriptase and V3 genotypes are often discordant between CSF and blood plasma virus (Pillai et al. 2006; Wong et al. 1997), and analysis of positions in and near the V3 loop of env have been associated with impairment of neurocognitive performance (Pillai et al. 2006). Many of these previous studies were small (range 4–27 subjects) and relied on the analysis of clonal sequences to evaluate the viral populations in the anatomic compartments, and these clonal sequences represent a form of repeated measures, which can confound interpretation of results (Choi et al. 2011). In this study, we evaluated a larger sample and used population-based tat sequences to investigate the viral genetic motifs associated with neurocognitive impairment and anatomic compartment of origin.
Several viral and cellular factors contribute to the neuronal damage after HIV-1 invasion into the brain (Kaul et al. 2001). Among the viral factors, HIV proteins such as Tat and Env are known to be neurotoxic in vitro (Kaul et al. 2001). In the brain, Tat may activate uninfected macrophages/microglia or astrocytes to release cytokines, chemokines, or other toxins, which may adversely affect neuronal function or apoptosis (Kruman et al. 1998; Li et al. 2009). The Tat protein also has chemotactic properties for monocytes (Albini et al. 1998), and the increased migration of the activated monocytes to the brain is correlated with HIV associated neurocognitive disorder (HAND) (Kaul et al. 2001). Also, several studies have compared HIV-1 subtypes B and C tat for their neuropathogeneic properties (Mishra et al, 2008; Ranga et al, 2004), and a genetic distinction between HIV-1 subtypes B and C in tat at residue 31 where subtype C has a Cys and subtype B has a serine has been implicated (Mishra et al, 2008; Ranga et al, 2004). Specifically, Ranga et al. reported this difference could result in the attenuation of chemotactic properties of Tat (Ranga et al, 2004), and Mishra et al. suggested that this mutation could attenuate the neurotoxic properties of Tat (Mishra et al, 2008). In this, the largest study of HIV-1 tat to date, we investigated various viral characteristics of HIV-1 subtype B tat that are associated with viral populations in the blood and CSF and neurocognitive dysfunction.
Results
Study cohort and rate of HAND
Of our 61 subjects, 60 had subtype B HIV-1 and one had subtype D. Only those with subtype B infection contributed to the overall analysis. Most were men (82%) with a mean age of 41.9 years. Their mean current CD4+ T cell count was 342 cells/µL, with a mean HIV RNA levels in CSF and blood plasma of 3.6 and 4.7 log10copies/mL, respectively. Thirty-six (60%) subjects were classified as having only incidental comorbidities, and 17 (28.3%) had contributing conditions; another 7 (11.7%) had confounding comorbidities that could preclude a HAND diagnosis. At evaluation, 35% of total participants (21/60) were neuropsychologically impaired, and impairment rates in the comorbidity groups were: 25% (9/36) incidental, 44% (8/18) contributing, and 57% (4/7) confounded. In univariate analysis, there were no significant differences in demographic and clinical characteristics such as age, sex, duration of education, current and nadir CD4+ T-cell counts, current plasma and CSF HIV RNA levels, and CSF WBC counts between those with and without neurocognitive impairments (Table 1). To evaluate differences between subjects with HAND and without HAND, we excluded the 7 subjects with confounding comorbidities, and in univariate analysis there was no difference in the demographic and clinical characteristics between those with and without HAND when those with confounding comorbidities were excluded (Table 2).
Table 1.
Demographic, clinical and viral characteristics of subjects
| Characteristics |
Overall (n=60) |
NC impairment (n=21) |
NC normal (n=39) |
P value |
|---|---|---|---|---|
| Age, mean years±SD | 41.9±8.7 | 41.0±8.5 | 42.4±8.8 | 0.55 |
| Sex, no. of male (%) | 49 (82%) | 17 (81%) | 32 (82%) | 0.92 |
| Education, mean years±SD | 12.3±2.6 | 12.0±2.5 | 12.4±2.7 | 0.59 |
| Comorbidity subgroups, no. of subjects (%) | 0.12 | |||
| Incidental | 36 (60%) | 9 (43%) | 27 (69%) | |
| Contributing | 17 (28%) | 8 (38%) | 9 (23%) | |
| Confounding | 7 (12%) | 4 (19%) | 3 (8%) | |
| CSF HIV RNA, mean log10 copies/mL±SD | 3.6±0.8 | 3.7±0.9 | 3.6±0.8 | 0.87 |
| Plasma HIV RNA, mean log10copies/mL±SD | 4.6±0.7 | 4.7±0.6 | 4.6±0.7 | 0.62 |
| Current CD4+ T cell, mean cells/µL (range)±SD | 342±216 | 348±242 | 339±204 | 0.88 |
| Nadir CD4+ T cell counts, median cells/µL (range) | 250±191 | 263±199 | 243±189 | 0.69 |
| AIDS diagnosis, no. of subjects (%) | 28 (47%) | 9 (43%) | 19 (49%) | 0.79 |
| Past ARV use, no. of subjects (%) | 14 (23%) | 7 (33%) | 7 (18%) | 0.21 |
| CSF WBC counts, median cells/µL (IQR) | 6.5 (3–14.25) | 6 (3–20) | 7 (3–14) | 0.97 |
| Mean genetic distance between CSF and blood derived tat (%) | 0.3% | 0.4% | 0.3%, | 0.83 |
| Nucleotide at position 5905 of CSF derived tat, no. of thymidine or Y*(%) | 13 (22%) | 6 (29%) | 7 (18%) | 0.35 |
| Median no. of mixed bases in CSF-derived tat (IQR) | 2** (0–3) | 3** (1–5.5) | 1** (0–3) | 0.05 |
| Median no. of mixed bases in blood-derived tat (IQR) | 2** (1–3.75) | 2** (1–3.5) | 2** (1–4) | 0.77 |
ARV= history of antiretroviral use; NC= Neurocognitive
mixed base of cytosine and thymine,
the median number of mixed bases, Bold: P<0.05
Table 2.
Demographic, clinical and viral characteristics of subjects without confounding comorbidities
| Characteristics |
Overall (n=53) |
HAND (+) (n=17) |
HAND (−) (n=36) |
P value |
|---|---|---|---|---|
| Age, mean years±SD | 42.3±8.8 | 42.1±8.5 | 42.4±9.1 | 0.90 |
| Sex, no. of male (%) | 43 (81%) | 13 (76%) | 30 (83%) | 0.71 |
| Education, mean years±SD | 12.6±2.5 | 12.4±2.7 | 12.8±2.4 | 0.59 |
| Comorbidity subgroups, no. of subjects (%) | 0.13 | |||
| Incidental | 36 (68%) | 9 (53%) | 27 (75%) | |
| Contributing | 17 (32%) | 8 (47%) | 9 (25%) | |
| CSF HIV RNA, mean log10 copies/mL±SD | 3.7±0.8 | 3.8±0.9 | 3.6±0.8 | 0.48 |
| Plasma HIV RNA, mean log10copies/mL±SD | 4.7±0.7 | 4.8±0.6 | 4.6±0.7 | 0.41 |
| Current CD4+ T cell, mean cells/µL (range)±SD | 332±210 | 300±215 | 347±209 | 0.46 |
| Nadir CD4+ T cell counts, median cells/µL (range) | 240±186 | 220±180 | 249±190 | 0.61 |
| AIDS diagnosis, no. of subjects (%) | 25 (47%) | 8 (47%) | 17 (47%) | 0.99 |
| Past ARV use, no. of subjects (%) | 14 (26%) | 7 (41%) | 7 (19%) | 0.11 |
| CSF WBC counts, median cells/µL (IQR) | 7 (3–14) | 5 (2–11) | 7 (3–15) | 0.67 |
| Mean genetic distance between CSF and blood derived tat (%) | 0.3% | 0.5% | 0.3%, | 0.61 |
| Nucleotide at position 5905 of CSF derived tat, no. of thymidine or Y*(%) | 11 (21%) | 5 (29%) | 6 (17%) | 0.30 |
| Median no. of mixed bases in CSF-derived tat (IQR) | 2** (0–3) | 3** (1–7.5) | 1** (0–3) | 0.005 |
| Median no. of mixed bases in blood-derived tat (IQR) | 2** (1–3) | 3** (1–4.5) | 2** (0.25–3) | 0.36 |
ARV= history of antiretroviral use; HAND= HIV associated neurocognitive disorder; += present; −= absent.
mixed base of cytosine and thymine,
the median number of mixed bases in sequences from each group, Bold: P<0.05
Characteristics of CSF and blood derived HIV-1 tat
For quality control, we performed phylogenetic analysis of all sequences evaluated together, and as expected, CSF and blood derived HIV-1 tat sequences from the same subject shared a most recent common ancestor (Figure S1). Among 4320 amino acid positions (72 amino acid positions × 60 sequences) of paired CSF and blood tat sequences, 64 (1.5%) positions showed diversity. As a potential measure of compartmentalization and divergence between blood and CSF viral populations, we evaluated the genetic distances between blood and CSF derived tat sequences and found that the mean genetic distance between CSF and blood derived sequences was overall relatively close at 0.3±1.2% (0–6.3%). Along these lines, we also performed selection analysis using FEL (Kosakovsky Pond and Frost. 2005) and identified 5 sites (codons T40, N61, S62, Q66, and S68) that were inferred to be under positive selection across all tat sequences but not specific to viral populations derived from each compartment. As a measure of viral population diversity (Poon et al. 2010) in each compartment, we then investigated the number of total mixed bases in each population-based sequence, and this measure was also not different between CSF and blood derived tat sequences (2.52±2.12 and 2.52±2.76, P=0.39, Mann-Whitney U test).
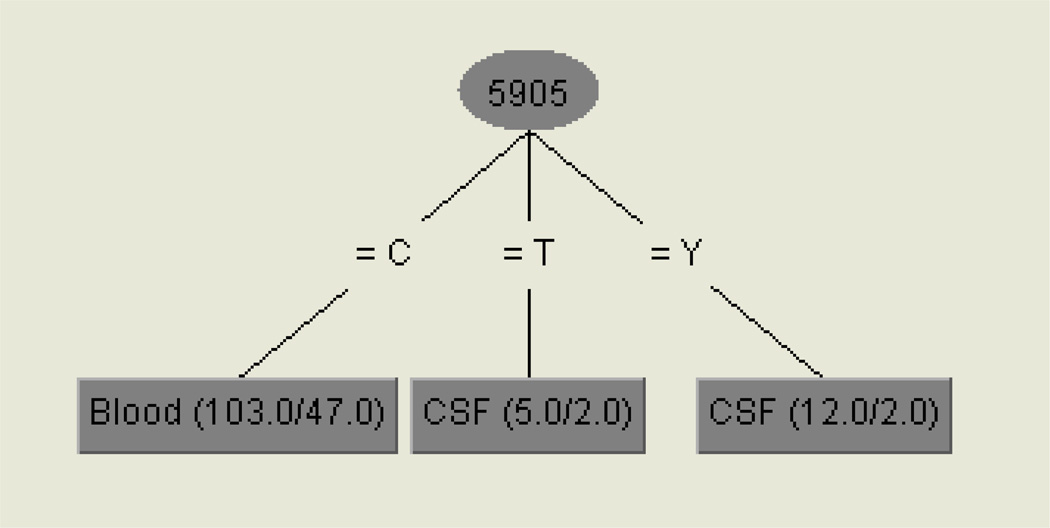
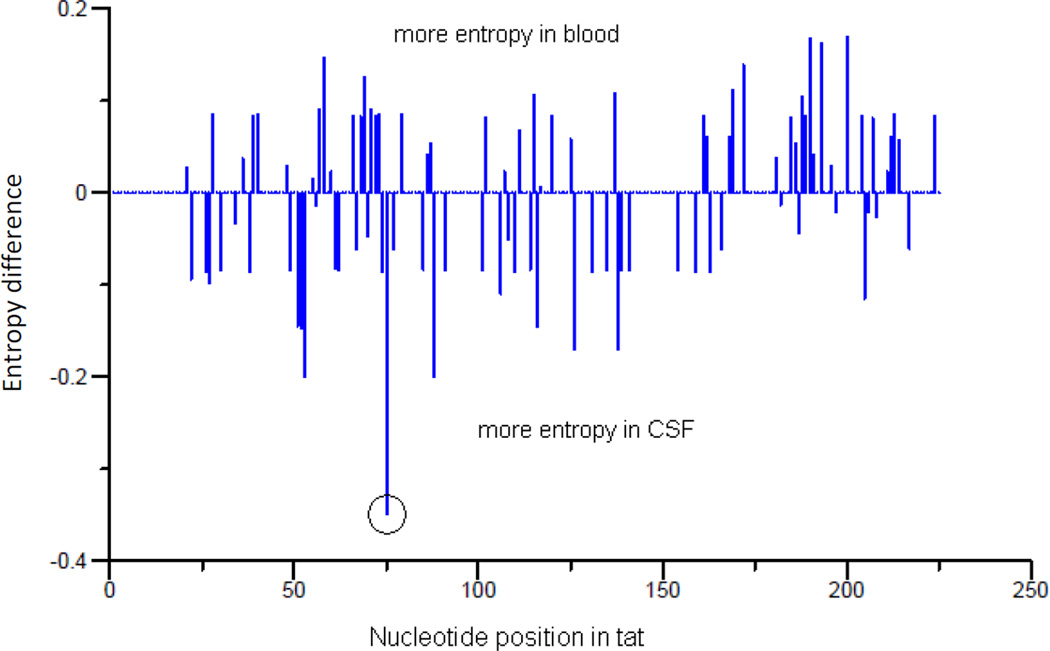
To look for evidence of a genetic signature shared by CSF-derived sequences across individuals (i.e. potential neurotropism), we employed a machine learning approach, as previously described for env (Pillai et al. 2006). Different from previous investigations, only one population-based tat sequence per compartment of each subject was evaluated to avoid the issue of repeated measures. This classification model identified nucleotide position 75 (HXB2 position 5905) of tat as a genetic ‘signature’ for viral populations derived from the CSF (Figure 1). Specifically, a thymine or ‘Y’ (mixed base of cytosine and thymine) at tat nucleotide HXB2 position 5905 was significantly correlated with virus derived from the CSF (P=0.03, Fisher’s exact test). The net difference in site-specific variability between CSF and blood (as measured by Shannon entropy) was also highest at this position (Figure 2). Since this mutation was a synonymous or ‘silent’ mutation, there was no phenotypic signature associated with this mutation (Figure S2).
Fig. 1.
Genetic signature associated with CSF sequences.
Legend. The values in parentheses are the number of instances/number of incorrect classifications. Nucleotide position (5905) is based on HXB2 position and one-letter nucleotide codes are used. Position 5905 C was associated with blood and 5905T or Y CSF.
Fig. 2.
Difference in Shannon entropy between CSF- and blood-derived tat sequences
Legend: Position 5905 (circle) of HIV-1 tat had the greatest difference in Shannon entropy between CSF and blood. Y axis (Entropy difference) represents the entropy difference between CSF and blood.
Clinical correlates associated with tat
Similar to the methods described above, we then investigated tat characteristics that were associated with HAND. We found no signature residue or nucleotide position that was associated with neurocognitive impairment or HAND, and the genetic distance between CSF and blood derived sequences did not differ between subjects with and without neurocognitive impairment or HAND (Tables 1 and 2). However, the number of mixed bases in CSF (i.e. increased viral population diversity) was significantly higher in subjects with HAND than those without HAND (Table 2)(P=0.005 by Mann Whitney U). Although not associated with viral loads or neurocognitive impairment, the greater the genetic distance between blood and CSF derived viruses, the lower the current and nadir CD4+ T cell counts of participants (correlation coefficient=−0.281 and −0.284, respectively, all P=0.03, Spearman’s rank correlation) (Tables 1 and 2).
Secondary RNA structures for HIV-1 B subtype tat with and without a signature synonymous mutation
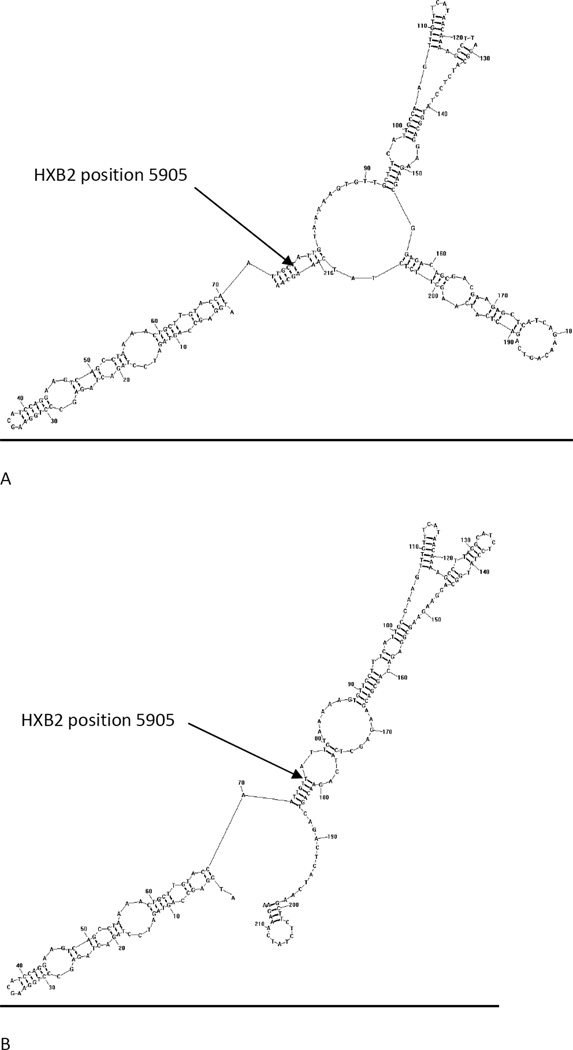
To better evaluate the possible changes associated with a synonymous mutation being a signature for CSF derived virus, we next estimated changes in secondary RNA structure associated with the C5905T motif. Secondary RNA structures with lowest free energy for HIV-1 B subtype tat (HXB2 position 5831–6046) and mutant tat with the signature synonymous mutation (C5905T) were predicted with the algorithm implemented in RNAstructure v5.2 (Reuter and Mathews. 2010). Figure 3 shows that the mutation altered the predicted secondary structure, but the calculated free energy of two structures was not significantly different (−53.3 kcal/mol for wild type and −51.9 kcal/mol for mutant), which suggests that each structure is equally valid. The substitution changed the shape and length of two stems by pairing bases in the estimated stem structure (Figure 3).
Fig. 3.
Secondary RNA structures for wild-type tat of HIV-1 subtype B(HXB2) (A) and mutant tat with the CSF-signature synonymous mutation (C5905T) (B)
Discussion
The HIV-1 tat sequence can be subdivided into distinct regions on the basis of its amino acid composition: a N-terminal activation region, a cysteine-rich role domain, a core region, a basic region and a glutamine rich region (Bayer et al. 1995). Each of these regions is essential for Tat function. Among those, amino acid residues 22–38 comprise a highly conserved cysteine-rich domain of Tat. The cysteine-rich domain seems to contribute to the mechanism of Tat toxicity (Aksenov et al. 2009; Egele et al. 2008; Misumi et al. 2004), including neurotoxicity in vitro (Li et al. 2009; Nath et al. 1996). A substitution of a specific cysteine within this domain significantly attenuated its neurotoxicity (Aksenov et al. 2009), and the lower neurotoxic potential of clade C HIV-1 has implicated a mutation of cysteine 31 within this domain (Mishra et al. 2008; Ranga et al. 2004). In this study, we identified a nucleotide position (HXB2 position 5905) within the cysteine-rich domain as a synonymous signature of CSF-derived subtype B HIV-1. However, the biological significance of the synonymous mutation remains unclear.
The cysteine-rich domain of Tat is highly conserved, and might serve as a binding domain for zinc-mediated linkage of Tat monomers (Frankel et al. 1988; Huang and Wang. 1996). While a silent mutation within the region might not affect zinc binding, synonymous changes can alter RNA secondary structure and influence RNA stability (Watts et al. 2009). RNA secondary structure plays an important role in the HIV life cycle (Berkhout. 1992; Galetto et al. 2004; Parkin et al. 1992; Wang et al. 2008), and we showed that the HXB2 position 5905 mutations altered the predicted secondary RNA structure including the shape and length of stems. At present, we do not have informative data with respect to whether the RNA secondary structure affects tat function and neurotoxicity. In vitro mutagenesis studies might identify the functional significance of this mutation.
Compartmentalization of HIV-1 env (Power et al. 1994; Ritola et al. 2005; Strain et al. 2005) and diversity of env and pol (Hightower et al. 2011) have been associated with HAND. Standard evaluation of both anatomic compartmentalization and diversity has required single genome sequencing, clonal amplification and ultra-deep sequencing, but these resource intensive methods are complicated by issues of repeated sampling (Choi et al. 2011). To address these methodological limitations, we used population-based sequences derived from both blood and CSF HIV RNA populations and then measured: (i) the genetic distance between CSF and blood derived sequences as a measure of divergence and surrogate for compartmentalization, and (ii) the total mixed base index (a count of total mixed bases divided by the sequence length) as measure of diversity. In this study, the median number of mixed bases of CSF-derived tat was significantly higher among subjects with HAND, which suggests HIV population diversity of tat most likely correlates with HAND. The genetic distance between blood and CSF derived sequences, as a measure of potential compartmentalization, however, was not significantly associated with HAND. Interestingly, genetic distance between blood and CSF derived viruses was inversely correlated with current and nadir CD4+ T cell counts, and may provide evidence of correlation between duration of infection, disease progression and viral evolution in anatomic compartments (Keys et al. 1993). Further, a previous report that lower CD4 counts are associated with greater autonomous CNS production of HIV is also consistent with this observation (Robertson et al. 2007). As a hypothesis, viral adaptation that occurs separately in blood and CSF compartments demonstrates a phenotype that is most identifiable as a drop in CD4 counts (i.e. blood adaptation) than a decline in neurocognitive functioning (i.e. CNS adaptation), and low CD4 counts might increase the risk for neuroadaptation but neuroadaptation may not be sufficient to cause neurovirulence.
To our knowledge, this is the largest study of its kind investigating HIV-1 tat. The reported trends are consistent with previous studies investigating other coding regions, but similar to previous studies, this study is also most likely limited by the relatively small number of subjects. In summary, nucleotide position (HXB2 position 5905) within the cysteine-rich domain appears to be a signature of CSF-derived HIV-1, and HIV population diversity of tat seems to be higher in subjects with HAND. Although the genetic distance between the blood and CSF derived tat was not associated with neurocognitive impairment, it was inversely correlated with current and nadir CD4+ T cell counts. The biological significance of the 5905 mutation is unclear, but this study provides evidence that it is associated with neurotropism, and should be evaluated in vitro mutagenesis studies.
Materials and Methods
Subjects and specimens
Sixty-one individuals enrolled in the CNS HIV Antiretroviral Therapy Effects Research (CHARTER) cohort between 2002 and 2009 were examined (Heaton et al. 2010). No subjects took antiretroviral therapy for at least 2 months before specimen collection, had blood plasma HIV RNA of >500 copies/mL and no evidence of systemic or CNS opportunistic infections or malignancy based on clinical, laboratory and/or neuroimaging studies. Subjects were selected from the cohort for having matched plasma and CSF available and having detectable plasma and CSF viral loads. Blood was collected in acid citrate dextrose tubes and CSF was collected without additive within 1 h from venipuncture. Blood plasma and cell-free CSF were aliquoted, frozen and stored at −80°C until processing.
Neurobehavioral assessment
Subjects completed a detailed neuropsychological assessment measuring their functioning in 7 cognitive domains known to be commonly affected by HIV: verbal fluency, executive functioning, speed of information processing, learning, recall, working memory, and motor skill, as previously described (Moore et al. 2010; Woods et al. 2004). The best available normative standards were used, which correct for effects of age, education, sex, and ethnicity, as appropriate. Test scores were converted to demographically corrected standard scores (t scores). To classify presence and severity of neurocognitive impairment, we applied an objective algorithm shown to yield excellent interrater reliability (Woods et al. 2004). This algorithm conforms to the Frascati criteria for diagnosing HAND (Antinori et al. 2007), which requires presence of at least mild impairment in at least 2 of the 7 ability domains. All sub-classification of HAND require a determination that the neurocognitive impairment and functional disability are believed to be due to effects of HIV on the brain, and are not readily attributable to a comorbid condition. To facilitate interrater reliability of these determinations, we utilized the online supplement to the previous report (Antinori et al. 2007), which provided detailed guidelines for classifying the most commonly encountered comorbid conditions with respect to whether they should be considered incidental, contributing, or confounding (Heaton et al. 2010).
Nucleotide sequencing
HIV-1 RNA was extracted and quantified from 500 µL of blood plasma and from 500 µL of CSF using the QIAmp Viral RNA Mini Kit (QIAGEN, Valencia, CA) according to the manufacturer’s instructions. Reverse transcription of HIV-1 RNA extracted from blood and CSF to make cDNA was accomplished using RETROscript kit per the manufacturer’s instructions (Applied Biosystems). Then 5µl of blood HIV cDNA underwent PCR with primers and conditions specific for HIV-1 tat for sequencing (Yukl et al. 2009). All PCR products were sequenced using Prism Dye terminator kits (ABI) on an ABI 3100 Genetic Analyzer. The tat sequences of subjects have been submitted to GenBank (GenBank ID: JF338918-JF339037).
Sequence analysis
Sequences were initially edited and aligned by Clustral W (Thompson et al. 1994). The alignment was then manually edited in Bioedit, version 7.05 to preserve frame insertions and deletions if present (Hall et al. 1999). Phylogenetic trees were generated from tat sequences that were obtained from a matrix of synonymous nucleotide distances using the Hasegawa-Kishino-Yang model of evolution with a 4:1 ratio of transversion to transitions in the PhyML program on Los Alamos National Laboratory HIV Sequence Database website (http://www.hiv.lanl.gov/content/sequence/PHYML/interface.html, accessed January 15, 2011) (Guindon et al. 2005). Pairwise distance was calculated by Tamura-Nei (93) estimator using HyPhy package (Pond et al. 2005). Sequences were examined for inter-subtype recombination, using RIP 3.0 (http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html, accessed December 15, 2010), and conserved residues using VESPA (http://www.hiv.lanl.gov/content/sequence/VESPA/vespa.html, accessed December 15, 2010)(Korber and Myers. 1992). All classification experiments in this analysis were conducted using WEKA (Waikato Environment for Knowledge Analysis), an open-source collection of data processing and machine learning algorithms (Witten and Frank. 2000). The J48 decision tree inducer, based on the C4.5 algorithm, was implemented with the parameter ‘MinNumObj’ set at a value of 11 to limit the complexity of theories and minimize the risk of over-fitting (Baliga et al. 1992). Classifiers were evaluated using 100 iterations of stratified 10-fold cross-validations, a procedure designed to conservatively reflect the performance of classification models on novel data sets (Witten and Frank. 2000). Shannon entropy was calculated to identify signatures of CSF-derived HIV-1 tat, using Entropy (http://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy.html, accessed December 15, 2010). The residue specific entropy was computed from the frequency f(Ai) of amino acid A at position i according to -∑A f(Ai) ln[f(Ai)] (Reza. 1994). A batch file implemented in HyPhy was used to provide a measure of viral population diversity by counting total mixed bases, synonymous mixed bases and nonsynonymous mixed bases (Pond et al. 2005; Hightower et al. 2011). Fixed effects likelihood (FEL) methods identified purifying and diversifying selection across the phylogeny (FEL) and along internal branches (iFEL). Model selection was used to select a DNA substitution model, evidence for recombination was evaluated with GARD (Kosakovsky Pond et al. 2006), and analyses were implemented in web-based Datamonkey (accessed December 10, 2010)(Pond and Frost. 2005). Secondary RNA structures with lowest free energy for the 1st exon of wild type tat (HXB2 position 5831–6046) and mutant tat with signature non-synonymous mutations were predicted with the algorithm implemented in RNAstructure v5.2 (Reuter and Mathews. 2010).
Statistical analysis
Fisher’s exact test was used to compare categorical or binary measures, and independent t-tests or Mann-Whitney U tests were used to compare continuous variables. Spearman’s rank correlation was used to evaluate correlations between continuous variables. All P values were 2-tailed and P<0.05 was considered statistically significant. All analyses were performed using SPSS for Windows 12.0 (SPSS, Chicago, Illinois, USA).
Supplementary Material
Acknowledgment
This work was supported by grants from the National Institutes of Health: MH22005, AI69432, AI043638, MH62512, MH083552, AI077304, AI36214, AI047745, AI74621, AI080353, and the James B. Pendleton Charitable Trust. This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2011-220-E00015)
The CNS HIV Anti-Retroviral Therapy Effects Research (CHARTER) group is affiliated with the Johns Hopkins University, Mount Sinai School of Medicine, University of California, San Diego, University of Texas, Galveston, University of Washington, Seattle, Washington University, St. Louis and is headquartered at the University of California, San Diego and includes: Director: Igor Grant, M.D.; Co-Directors: J. Allen McCutchan, M.D., Ronald J. Ellis, M.D., Ph.D., Thomas D. Marcotte, Ph.D.; Center Manager: Donald Franklin, Jr.; Neuromedical Component: Ronald J. Ellis, M.D., Ph.D. (P.I.), J. Allen McCutchan, M.D., Terry Alexander, R.N.; Laboratory, Pharmacology and Immunology Component: Scott Letendre, M.D. (P.I.), Edmund Capparelli, Pharm.D.; Neurobehavioral Component: Robert K. Heaton, Ph.D. (P.I.), J. Hampton Atkinson, M.D., Steven Paul Woods, Psy.D., Matthew Dawson; Virology Component: Davey M. Smith, M.D. (P.I.), Joseph K. Wong, M.D.; Imaging Component: Christine Fennema-Notestine, Ph.D. (Co-P.I.), Michael J. Taylor, Ph.D. (Co-P.I.), Rebecca Theilmann, Ph.D.; Data Management Component: Anthony C. Gamst, Ph.D. (P.I.), Clint Cushman,; Statistics Component: Ian Abramson, Ph.D. (P.I.), Florin Vaida, Ph.D.; Protocol Coordinating Component: Thomas D. Marcotte, Ph.D. (P.I.), Rodney von Jaeger, M.P.H.; Johns Hopkins University Site: Justin McArthur (P.I.), Mary Smith; Mount Sinai School of Medicine Site: Susan Morgello, M.D. (Co-P.I.) and David Simpson, M.D. (Co-P.I.), Letty Mintz, N.P.; University of California, San Diego Site: J. Allen McCutchan, M.D. (P.I.), Will Toperoff, N.P..; University of Washington, Seattle Site: Ann Collier, M.D. (Co-P.I.) and Christina Marra, M.D. (Co-P.I.), Trudy Jones, M.N., A.R.N.P.; University of Texas, Galveston Site: Benjamin Gelman, M.D., Ph.D. (P.I.), Eleanor Head, R.N., B.S.N.; and Washington University, St. Louis Site: David Clifford, M.D. (P.I.), Muhammad Al-Lozi, M.D., Mengesha Teshome, M.D.
Contributor Information
Jun Yong Choi, Department of Internal Medicine and AIDS Research Institute, Yonsei University College of Medicine, 250 Seongsanno, Seodaemun-gu, Seoul 120–752, Korea.
George K. Hightower, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0679, USA
Joseph K. Wong, University of California San Francisco, 4150 Clement St, 111W3, San Francisco, CA 94121, USA
Robert Heaton, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0679, USA.
Steven Woods, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0679, USA.
Igor Grant, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0679, USA.
Thomas D. Marcotte, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0679, USA
Ronald J. Ellis, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0679, USA
Scott L. Letendre, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0679, USA
Ann C. Collier, University of Washington, Harborview Medical Center, 325 9th Avenue, Seattle, WA 98104, USA
Christina M. Marra, University of Washington, Harborview Medical Center, 325 9th Avenue, Seattle, WA 98104, USA
David B. Clifford, Washington University in St Louis, 660 South Euclid Avenue, Saint Louis, MO 63110, USA
Benjamin B. Gelman, University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77555, USA
Justin C. McArthur, Johns Hopkins University, 600 N Wolfe St, Baltimore, MD 21231, USA
Susan Morgello, Mountain Sinai School of Medicine, 1 Gustave L. Levy Place, New York, NY 10029, USA.
David M. Simpson, Mountain Sinai School of Medicine, 1 Gustave L. Levy Place, New York, NY 10029, USA
J. Allen McCutchan, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0679, USA.
Douglas D. Richman, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0679, USA Veterans Affairs San Diego Healthcare System, San Diego, CA 92161, USA.
Davey M. Smith, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0679, USA Veterans Affairs San Diego Healthcare System, San Diego, CA 92161, USA.
References
- Aksenov MY, Aksenova MV, Mactutus CF, Booze RM. Attenuated neurotoxicity of the transactivation-defective HIV-1 Tat protein in hippocampal cell cultures. Exp Neurol. 2009;219:586–590. doi: 10.1016/j.expneurol.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini A, Ferrini S, Benelli R, Sforzini S, Giunciuglio D, Aluigi MG, Proudfoot AE, Alouani S, Wells TN, Mariani G, Rabin RL, Farber JM, Noonan DM. HIV-1 Tat protein mimicry of chemokines. Proc Natl Acad Sci U S A. 1998;95:13153–13158. doi: 10.1073/pnas.95.22.13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, Gisslen M, Grant I, Heaton RK, Joseph J, Marder K, Marra CM, McArthur JC, Nunn M, Price RW, Pulliam L, Robertson KR, Sacktor N, Valcour V, Wojna VE. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69:1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga G, Case J, Jain S, Suraj M. Machine Learning of Higher-Order Programs. Lecture Notes in Computer Science. 1992;620:9–20. [Google Scholar]
- Bayer P, Kraft M, Ejchart A, Westendorp M, Frank R, Rosch P. Structural studies of HIV-1 Tat protein. J Mol Biol. 1995;247:529–535. doi: 10.1006/jmbi.1995.0158. [DOI] [PubMed] [Google Scholar]
- Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Korsakovsky Pond SL, Vazquez H, Williams SL, McCammon JA, Richman DD, Smith DM. Molecular features of the V1-V4 coding region of sexually transmitted HIV-1; 18th Conference on Human Retroviruses and Opportunistic Infections; 2011. [Google Scholar]
- Egele C, Barbier P, Didier P, Piemont E, Allegro D, Chaloin O, Muller S, Peyrot V, Mely Y. Modulation of microtubule assembly by the HIV-1 Tat protein is strongly dependent on zinc binding to Tat. Retrovirology. 2008;5:62. doi: 10.1186/1742-4690-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel AD, Bredt DS, Pabo CO. Tat protein from human immunodeficiency virus forms a metal-linked dimer. Science. 1988;240:70–73. doi: 10.1126/science.2832944. [DOI] [PubMed] [Google Scholar]
- Galetto R, Moumen A, Giacomoni V, Veron M, Charneau P, Negroni M. The structure of HIV-1 genomic RNA in the gp120 gene determines a recombination hot spot in vivo. J Biol Chem. 2004;279:36625–36632. doi: 10.1074/jbc.M405476200. [DOI] [PubMed] [Google Scholar]
- Grant I, Atkinson JH, Hesselink JR, Kennedy CJ, Richman DD, Spector SA, McCutchan JA. Evidence for early central nervous system involvement in the acquired immunodeficiency syndrome (AIDS) and other human immunodeficiency virus (HIV) infections. Studies with neuropsychologic testing and magnetic resonance imaging. Ann Intern Med. 1987;107:828–836. doi: 10.7326/0003-4819-107-6-828. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Heaton RK, Clifford DB, Franklin DR, Jr, Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75:2087–2096. doi: 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower GK, Wong JK, Letendre SL, Umlauf A, Ellis RJ, Ignacio CC, Heaton RK, Grant I, Richman DD, Smith DM. Higher HIV-1 genetic diversity is associated with AIDS and neuropsychological impairment; 18th Conference on Human Retroviruses and Opportunistic Infections; 2011. [Google Scholar]
- Ho DD, Rota TR, Schooley RT, Kaplan JC, Allan JD, Groopman JE, Resnick L, Felsenstein D, Andrews CA, Hirsch MS. Isolation of HTLV-III from cerebrospinal fluid and neural tissues of patients with neurologic syndromes related to the acquired immunodeficiency syndrome. N Engl J Med. 1985;313:1493–1497. doi: 10.1056/NEJM198512123132401. [DOI] [PubMed] [Google Scholar]
- Huang HW, Wang KT. Structural characterization of the metal binding site in the cysteine-rich region of HIV-1 Tat protein. Biochem Biophys Res Commun. 1996;227:615–621. doi: 10.1006/bbrc.1996.1554. [DOI] [PubMed] [Google Scholar]
- Kaul M, Garden GA, Lipton SA. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature. 2001;410:988–994. doi: 10.1038/35073667. [DOI] [PubMed] [Google Scholar]
- Keys B, Karis J, Fadeel B, Valentin A, Norkrans G, Hagberg L, Chiodi F. V3 sequences of paired HIV-1 isolates from blood and cerebrospinal fluid cluster according to host and show variation related to the clinical stage of disease. Virology. 1993;196:475–483. doi: 10.1006/viro.1993.1503. [DOI] [PubMed] [Google Scholar]
- Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Kruman II, Nath A, Mattson MP. HIV-1 protein Tat induces apoptosis of hippocampal neurons by a mechanism involving caspase activation, calcium overload, and oxidative stress. Exp Neurol. 1998;154:276–288. doi: 10.1006/exnr.1998.6958. [DOI] [PubMed] [Google Scholar]
- Li W, Li G, Steiner J, Nath A. Role of Tat protein in HIV neuropathogenesis. Neurotox Res. 2009;16:205–220. doi: 10.1007/s12640-009-9047-8. [DOI] [PubMed] [Google Scholar]
- Mishra M, Vetrivel S, Siddappa NB, Ranga U, Seth P. Clade-specific differences in neurotoxicity of human immunodeficiency virus-1 B and C Tat of human neurons: significance of dicysteine C30C31 motif. Ann Neurol. 2008;63:366–376. doi: 10.1002/ana.21292. [DOI] [PubMed] [Google Scholar]
- Misumi S, Takamune N, Ohtsubo Y, Waniguchi K, Shoji S. Zn2+ binding to cysteine-rich domain of extracellular human immunodeficiency virus type 1 Tat protein is associated with Tat protein-induced apoptosis. AIDS Res Hum Retroviruses. 2004;20:297–304. doi: 10.1089/088922204322996536. [DOI] [PubMed] [Google Scholar]
- Moore DJ, Letendre SL, Morris S, Umlauf A, Deutsch R, Smith DM, Little S, Rooney A, Franklin DR, Gouaux B, Leblanc S, Rosario D, Fennema-Notestine C, Heaton RK, Ellis RJ, Atkinson JH, Grant I. Neurocognitive functioning in acute or early HIV infection. J Neurovirol. 2010 doi: 10.1007/s13365-010-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A, Psooy K, Martin C, Knudsen B, Magnuson DS, Haughey N, Geiger JD. Identification of a human immunodeficiency virus type 1 Tat epitope that is neuroexcitatory and neurotoxic. J Virol. 1996;70:1475–1480. doi: 10.1128/jvi.70.3.1475-1480.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickle DC, Jensen MA, Shriner D, Brodie SJ, Frenkel LM, Mittler JE, Mullins JI. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol. 2003;77:5540–5546. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin NT, Chamorro M, Varmus HE. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai SK, Pond SL, Liu Y, Good BM, Strain MC, Ellis RJ, Letendre S, Smith DM, Gunthard HF, Grant I, Marcotte TD, McCutchan JA, Richman DD, Wong JK. Genetic attributes of cerebrospinal fluid-derived HIV-1 env. Brain. 2006;129:1872–1883. doi: 10.1093/brain/awl136. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Poon AF, Swenson LC, Dong WW, Deng W, Kosakovsky Pond SL, Brumme ZL, Mullins JI, Richman DD, Harrigan PR, Frost SD. Phylogenetic analysis of population-based and deep sequencing data to identify coevolving sites in the nef gene of HIV-1. Mol Biol Evol. 2010;27:819–832. doi: 10.1093/molbev/msp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C, McArthur JC, Johnson RT, Griffin DE, Glass JD, Perryman S, Chesebro B. Demented and nondemented patients with AIDS differ in brain-derived human immunodeficiency virus type 1 envelope sequences. J Virol. 1994;68:4643–4649. doi: 10.1128/jvi.68.7.4643-4649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RW. Neurological complications of HIV infection. Lancet. 1996;348:445–452. doi: 10.1016/S0140-6736(95)11035-6. [DOI] [PubMed] [Google Scholar]
- Ranga U, Shankarappa R, Siddappa NB, Ramakrishna L, Nagendran R, Mahalingam M, Mahadevan A, Jayasuryan N, Satishchandra P, Shankar SK, Prasad VR. Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J Virol. 2004;78:2586–2590. doi: 10.1128/JVI.78.5.2586-2590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza FM. An introduction to information theory. New York: Dover; 1994. [Google Scholar]
- Ritola K, Robertson K, Fiscus SA, Hall C, Swanstrom R. Increased human immunodeficiency virus type 1 (HIV-1) env compartmentalization in the presence of HIV-1-associated dementia. J Virol. 2005;79:10830–10834. doi: 10.1128/JVI.79.16.10830-10834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KR, Smurzynski M, Parsons TD, Wu K, Bosch RJ, Wu J, McArthur JC, Collier AC, Evans SR, Ellis RJ. The prevalence and incidence of neurocognitive impairment in the HAART era. AIDS. 2007;21:1915–1921. doi: 10.1097/QAD.0b013e32828e4e27. [DOI] [PubMed] [Google Scholar]
- Strain MC, Letendre S, Pillai SK, Russell T, Ignacio CC, Gunthard HF, Good B, Smith DM, Wolinsky SM, Furtado M, Marquie-Beck J, Durelle J, Grant I, Richman DD, Marcotte T, McCutchan JA, Ellis RJ, Wong JK. Genetic composition of human immunodeficiency virus type 1 in cerebrospinal fluid and blood without treatment and during failing antiretroviral therapy. J Virol. 2005;79:1772–1788. doi: 10.1128/JVI.79.3.1772-1788.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Barr I, Guo F, Lee C. Evidence of a novel RNA secondary structure in the coding region of HIV-1 pol gene. RNA. 2008;14:2478–2488. doi: 10.1261/rna.1252608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten IH, Frank E. Data mining : practical machine learning tools and techniques with Java implementations. San Francisco, Calif: Morgan Kaufmann; 2000. [Google Scholar]
- Wong JK, Ignacio CC, Torriani F, Havlir D, Fitch NJ, Richman DD. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods SP, Rippeth JD, Frol AB, Levy JK, Ryan E, Soukup VM, Hinkin CH, Lazzaretto D, Cherner M, Marcotte TD, Gelman BB, Morgello S, Singer EJ, Grant I, Heaton RK. Interrater reliability of clinical ratings and neurocognitive diagnoses in HIV. J Clin Exp Neuropsychol. 2004;26:759–778. doi: 10.1080/13803390490509565. [DOI] [PubMed] [Google Scholar]
- Yukl S, Pillai S, Li P, Chang K, Pasutti W, Ahlgren C, Havlir D, Strain M, Gunthard H, Richman D, Rice AP, Daar E, Little S, Wong JK. Latently-infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity. Virology. 2009;387:98–108. doi: 10.1016/j.virol.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.