Abstract
Gene targeting in embryonic stem cells has become the principal technology for manipulation of the mouse genome, offering unrivalled accuracy in allele design and access to conditional mutagenesis. To bring these advantages to the wider research community, large-scale mouse knockout programmes are producing a permanent resource of targeted mutations in all protein-coding genes. Here we report the establishment of a high-throughput gene-targeting pipeline for the generation of reporter-tagged, conditional alleles. Computational allele design, 96-well modular vector construction and high-efficiency gene-targeting strategies have been combined to mutate genes on an unprecedented scale. So far, more than 12,000 vectors and 9,000 conditional targeted alleles have been produced in highly germline-competent C57BL/6N embryonic stem cells. High-throughput genome engineering highlighted by this study is broadly applicable to rat and human stem cells and provides a foundation for future genome-wide efforts aimed at deciphering the function of all genes encoded by the mammalian genome.
Following the complete sequencing of the human and mouse genomes, the functional analysis of each of the twenty thousand or so protein-coding genes remains an important goal and a major technical challenge. Several genome-wide mutagenesis strategies have been applied in the mouse, including ethyl-nitrosourea (ENU) mutagenesis, transposon mutagenesis, gene trapping and gene targeting. Gene trapping in mouse embryonic stem (ES) cells1,2 has been the most productive so far, providing hundreds of thousands of random insertional mutations in more than half of the protein-coding genes in the mouse3–5. Notably, these ES cell resources can be archived indefinitely and are easily distributed to the scientific community for the purpose of generating knockout mice. However, gene-trap alleles cannot be precisely engineered and the strategy favours genes expressed in mouse ES cells.
Given the limitations of gene trapping, it is clear that the generation of a complete set of gene knockouts in the mouse will require the application of gene-targeting technology in ES cells6–8. Gene targeting can be used to engineer virtually any alteration in the mammalian genome by homologous recombination in mouse ES cells, from point mutations to large chromosomal rearrangements9,10. Over the past 20 years, gene targeting has been used to elucidate the function of more than 5,000 mammalian genes. Scaling this technology to the remainder of the genome presents numerous technical challenges and requires the production of targeted ES cells on an unprecedented scale, beyond the scope of conventional methodologies.
The first targeting pipeline for ES cells was reported several years ago before the completion of the mouse genome sequence (Velocigene)11. Bacterial artificial chromosome (BAC)-based targeting vectors were constructed to replace the coding sequence of the target gene with a lacZ reporter and promoter-driven selection cassette. Oligonucleotides required for the construction of targeting vectors by recombineering were based on cDNA sequences surrounding the translation initiation and termination signals of each target gene, thus requiring no previous knowledge of the underlying genomic structure of the gene. In a single recombineering step, modified BAC clones were generated with high efficiency and used to target genes in ES cells. Correctly targeted events, which involved the deletion of up to 70-kilobases (kb) of genomic sequence, were identified using a novel high-throughput allele-counting assay. The deletion of large regions of genomic sequence, although effective for eliminating the function of the target gene, can have unintended consequences on the regulation of adjacent and distant transcriptional units12,13.
To support and accelerate progress towards the genetic analysis of all mammalian genes, large-scale knockout consortia were established in 2006 with the goal of generating a complete resource of reporter-tagged null mutations in C57BL/6 mouse ES cells14. C57BL/6 is one of the best characterized inbred strains, is the reference strain for the mouse genome sequence and breed well in the laboratory. Thus, the study of mutant alleles in a pure C57BL/6 genetic background is considered to be ideal for large-scale phenotyping efforts that will follow. Highly germline-competent ES cell lines from the C57BL/6N substrain of mice have been established for this project15–17. A common web portal providing information and access to the resource has been established18, with links to designated repositories for ordering vectors, ES cell clones and mice.
Here we describe a pipeline for the design and mass parallel construction of conditional targeting vectors by serial 96-well BAC recombineering and high-throughput gene targeting in C57BL/6 ES cells. Our pipeline is configured to create a number of useful resources en route to the generation of targeted ES cells (Supplementary Fig. 1). Ongoing large-scale production of targeted ES cell lines demonstrates rates of homologous recombination in C57BL/6 ES cells well above the historical average. Our pipeline forms the basis for the generation of thousands of lacZ-tagged conditional alleles for the European Conditional Mouse Mutatgenesis (EUCOMM) and the National Institutes of Health Knockout Mouse (KOMP) programs as part of the international knockout effort14.
Computer-assisted design of alleles
Conditional alleles permit the analysis of gene function in a tissue-specific or temporal manner during embryonic and postnatal development10,19. Our conditional allele isbased on the ‘knockout-first’ design20, a strategy that combines the advantages of both a reporter-tagged and a conditional mutation (Fig. 1 and Supplementary Fig. 2). In contrast to standard conditional designs, the initial unmodified allele is predicted to generate a null allele through splicing to a lacZ trapping element contained in the targeting cassette. Our trapping cassettes include the mouse En2 splice acceptor and the SV40 polyadenylation sequences, signals that have proven to be highly effective in creating null alleles in mice2,21.
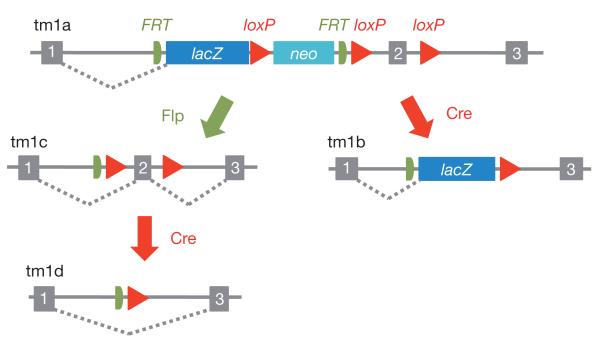
Figure 1. Schematic of the ‘knockout-first’ conditional allele.
The ‘knockout-first’ allele (tm1a) contains an IRES:lacZ trapping cassette and a floxed promoter-driven neo cassette inserted into the intron of a gene, disrupting gene function. Flp converts the ‘knockout-first’ allele to a conditional allele (tm1c), restoring gene activity. Cre deletes the promoter-driven selection cassette and floxed exon of the tm1a allele to generate a lacZ-tagged allele (tm1b) or deletes the floxed exon of the tm1c allele to generate a frameshift mutation (tm1d), triggering nonsense mediated decay of the deleted transcript.
The knockout-first allele can be easily modified in ES cells or in crosses to transgenic FLP and cre mice. Conditional alleles are generated by removal of the gene-trap cassette by Flp recombinase, which reverts the mutation to wild type, leaving loxP sites on either side of a critical exon. Subsequent exposure to Cre deletes the critical exon to induce a frameshift mutation and trigger nonsense-mediated decay of the mutant transcript. Many cre transgenic strains are available for the study of gene function in specific tissues and developmental time points (see http://www.creline.org).
Typically, loxP sites are placed in introns of genes to avoid disrupting normal transcription, processing and translation of the target gene. The loxP and FRT sites are positioned to minimize possible interference with the splice sites of the critical exon. In some cases, the presence of the recombinase sites may perturb normal splicing patterns22. This caveat notwithstanding, knockout first alleles are very useful for proving the causality of gene disruptions and observed phenotypes. Reversion of the phenotype with Flp, or conversely, induction of the phenotype with Cre, rule out potential effects of secondary linked mutations that can arise in cultured ES cells23. Furthermore, removal of the FRT-flanked stop cassette is particularly useful for further studies of genes that present heterozygous lethal phenotypes.
The vector design process ideally begins with high-quality manual annotation of gene structures24. Manual annotation identifies and resolves errors in automated gene predictions and captures all known transcript variants from available messenger RNA evidence. However, manual annotation of genes is a time-consuming process and proved rate-limiting in our high-throughput pipeline. Although the accuracy of automated gene prediction is improving, vector designs for Ensembl gene structures must be approached with caution.
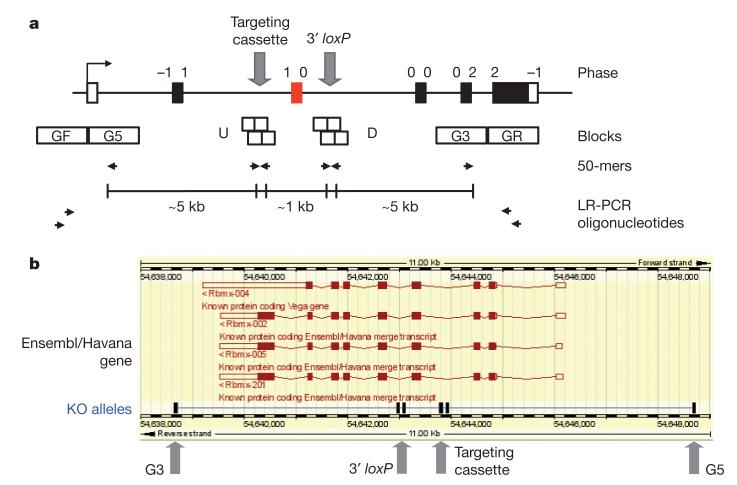
To assist in the design of conditional alleles, we developed a computational tool to identify oligonucleotide sequences (50-mers) suitable for recombineering. These sequences are used to insert a selection cassette and loxP site around the critical exon and to recover homologous sequence from the BAC required for gene targeting (Fig. 2a). More generally, these computational tools can be applied to any other mammalian or non-mammalian genome for which the construction of large numbers of recombineered DNA constructs is desired. Each design is displayed on the genome browser (Fig. 2b) and manually inspected to choose the optimal design. Valid designs are selected for the 5′-most critical exon(s) that is common to all known transcript variants and disrupts at least 50% of the protein-coding sequence. Designs are rejected if the deleted region contains highly conserved intronic sequence as these elements are likely to correspond to regulatory elements and complicate the interpretation of the mutant phenotype in mice12,13.
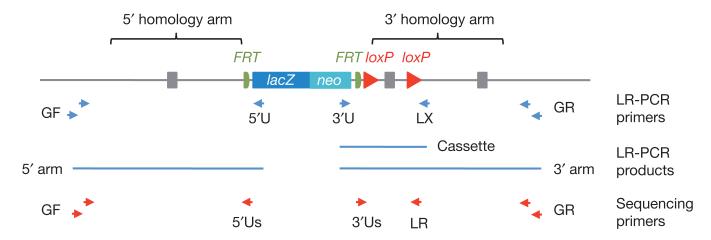
Figure 2. Computational design of oligonucleotides for recombineering and LR-PCR genotyping.
a, A critical exon(s) common to all transcript variants (red box) is identified. Recombineering oligonucleotides (50-mers) are identified by ArrayOligoSelector46 within pre-defined blocks (G5, U, D, G3) of genomic sequence for insertions of the targeting cassette and 3′ loxP site and for plasmid rescue of the 5′ and 3′ homology arms by gap repair. For LR-PCR genotyping, multiple primers (25 to 30-mers) are then selected from 1-kb blocks of genomic sequence (GF, GR) outside the homology arms. b, Display of conditional alleles on the Ensembl genome browser (Distributed Annotation System (DAS) source = KO alleles). A conditional design for the merged Ensembl/Havana Rbmx gene on the reverse strand is shown.
Approximately 40% of protein-coding genes do not fit our design criteria, most commonly, small transcription units composed of one or two exons. Genes with alternative 5′ end transcripts are also problematical. In some cases, it is not possible to remove a single exon or cluster of exons that disrupts all isoforms. These genes have been set aside for other partners within the international knockout consortium to generate standard lacZ-tagged deletion alleles using, for example, Velocigene technology11.
Construction of modular targeting vectors
For the generation of conditional gene-targeting vectors, we developed a strategy for high-throughput, serial, liquid BAC recombineering in 96-well format (Fig. 3) similar to that reported for transgene production25,26. We adopted a modular strategy for the construction of targeting vectors using recombineering to create Gateway-adapted intermediate vectors (Fig. 4a) that are later assembled into the final targeting construct through in vitro Gateway reactions (Fig. 4b). For targeting in C57BL/6N ES cells16, we made use of indexed C57BL/6J BAC libraries27 for the construction of targeting vectors.
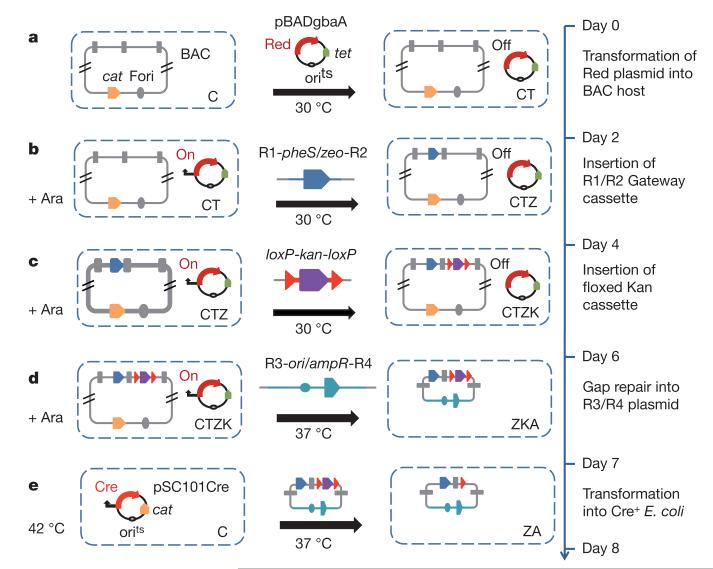
Figure 3. Construction of Gateway-adapted intermediate targeting vectors by 96-well BAC recombineering.
Recombineering steps and elapsed time are shown. a, BAC clones, arrayed in 96-well format and electroporated with a plasmid expressing arabinose-inducible Red proteins (pBADgbaA)47.
b–d, After arabinose induction, cells are electroporated with PCR fragments containing R1-pheS/zeo-R2 Gateway element (b), loxP-kan-loxP cassette
(c) and R3-ori/ampR-R4 subcloning plasmid (d). e, After gap repair, plasmid DNA is prepared and transformed into Cre-expressing bacteria to remove the kanR cassette, leaving a single loxP site downstream of the critical exon. Antibiotics used at each step are: A, ampicillin; C, chloramphenicol; K, kanamycin; T, tetracycline; Z, zeocin.
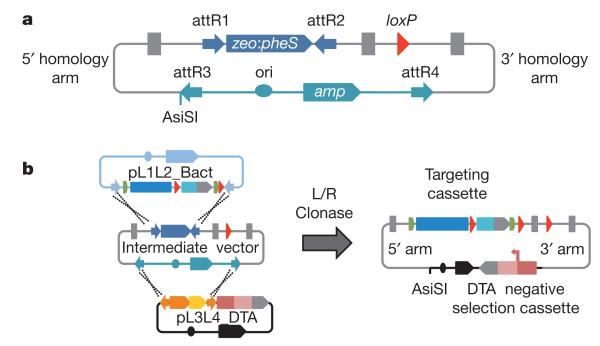
Figure 4. Intermediate and final targeting constructs.
a, Schematic showing the structure of the Gateway-adapted intermediate plasmid. A rare AsiSI restriction site is included in the gap repair plasmid for linearizing the final targeting vector before electroporation of ES cells. b, Assembly of final targeting vectors in a multi-Gateway reaction. See Supplementary Fig. 4 for a full description of the custom Gateway-adapted plasmids used for vector construction.
The construction of Gateway-adapted intermediate targeting vectors from BACs involves three consecutive recombineering steps: insertion of an attR1/attR2 zeo-pheS Gateway element upstream of the critical exon (Fig. 3b and Supplementary Fig. 3); insertion of a floxed kanR cassette downstream of a critical exon (Fig. 3c); and subcloning of the modified region of genomic DNA (8–10 kb) into a Gateway-adapted plasmid backbone by gap repair (Fig. 3d and Supplementary Fig. 3). Heterologous attR3/attR4 sites are included to enable switching of the plasmid backbone to introduce a negative selection cassette for positive–negative targeting in ES cells. The exquisite efficiency and nucleotide precision of Red operon-induced recombination in bacteria permitted the assembly of DNA constructs in 96-well format through three rounds of recombineering with an 80% overall efficiency (Supplementary Table 1). This efficiency of vector production readily accommodates the needs of the global mouse gene-targeting projects that aim to knock out thousands of genes per year14.
Assembly of the final targeting constructs
Gateway technology has been successfully used for the construction of large-scale genomic resources28,29. The use of Gateway technology minimizes the potential for deleterious mutations common to polymerase chain reaction (PCR)-based cloning methods. We developed a series of promoterless and promoter-driven selection cassettes flanked by attL1/attL2 sites (Supplementary Fig. 4). To use positive–negative selection for gene targeting30, a plasmid backbone was constructed that contains attL3/attL4 Gateway elements and a diphtheria-toxin-A-chain31 (DTA) expression cassette. Final targeting constructs were assembled in vitro in a three-part Gateway reaction (Fig. 4b) in 96-well format and sequence-confirmed across all recombineered junctions. Final targeting vectors were recovered from 95% of the intermediate plasmids (Supplementary Table 1). Thus, the overall efficiency of vector construction is 75% and, so far, we have constructed more than 12,000 final targeting vectors.
The intermediate vectors themselves (Fig. 4a) represent an important modular resource that can be re-used to generate alternative vector designs oradditional mutant alleles in the future. For example, targeting cassettes containing specialized reporters, such as alkaline phosphatase or green fluorescent protein, can be rapidly assembled to provide alternative visualization of gene expression. Furthermore, targeting vectors with different selectable markers can be readily constructed to knock out the second allele of genes for functional studies in homozygous ES cells. Finally, knock-ins of wild-type and mutant cDNAs provide an avenue for detailed structure–function studies or to explore human variation. Thus, a permanent library of intermediate targeting plasmids will permit the further exploitation of targeting technology in the future.
High-throughput ES cell production
To scale targeting experiments to high throughput, we optimized electroporation conditions for C57BL/6N ES cells16 in multi-well cuvettes. Here we aimed to minimize the number of cells and amount of plasmid DNA required to obtain sufficient drug-resistant colonies for screening (Table 1). After selection, expansion and freezing, most (65%) ES cell clones retained their ability to colonize the germ line of mice16.
Table 1.
Targeting efficiency using promoterless and promoter-driven cassettes
| Vector type | Number of unique targeting vectors |
Number of successful electroporations |
Number of colonies* |
Number of genes targeted |
Genes targeted (%) |
Targeting efficiency (%) |
Number of colonies screened* |
Number of targeted clones* |
Number of targeted clones with 39 loxP site* |
|---|---|---|---|---|---|---|---|---|---|
| Promoterless | 1,285 | 778 | 224 | 621 | 48 | 51 | 24 | 12 | 6 |
| Promoter | 1,811 | 1,671 | 348 | 1,440 | 80 | 35 | 29 | 10 | 3.5 |
| Promoter (–DTA) | 87 | 87 | 729 | 49 | 56 | 12 | 34 | 4 | 1 |
Average values.
Homologous recombinants generated with targeting vectors are usually identified by Southern blotting. However, this method is not practical for large-scale screening. Long-range PCR (LR-PCR) is an alternative method32 which is better-suited to high-throughput genotyping of ES cell clones. We developed a 384-well LR-PCR method to identify correctly targeted events (Fig. 5). PCR fragments, amplified with gene-specific primers outside the homology arms in combination with primers in the targeting cassette, were sequence-verified. In general, LR-PCR was performed across the 3′ homology arm. Because the targeted clones are genotyped at one end, non-homologous events within the opposite arm will occur in rare cases. Furthermore, mixed clones composed of targeted and non-targeted cells are not detected by our high-throughput genotyping protocol. For these reasons, further validation of targeted alleles using standard Southern blot assays is highly recommended before use.
Figure 5. Genotyping ES clones by LR-PCR sequencing.
Five LR-PCR reactions are carried out: two 5′ arm (GF/5′U), two 3′ arm (3′U/GR) and one cassette (3′U/LX). Sequence verification of LR-PCR products is carried out with gene-specific primers (GF and GR) and with nested primers in the targeting cassette (5′Us and 3′Us). To confirm the presence or absence of the 3′ loxP site, 3′ arm LR-PCR products are sequenced with a primer adjacent to the loxP site (LR). In cases where 3′ arm LR-PCR fails to generate a product, the 3′ loxP site is confirmed by sequencing the cassette product.
Owing to frequent crossover events between the selectable marker and 3′ loxP site, many of the targeted ES cell clones lose the 3′ loxP site and cannot be converted to a conditional allele. To distinguish between these two alternative products of homologous recombination, LR-PCR products amplified from the 3′ homology arm were sequenced with a primer at the loxP site. Where 3′ LR-PCR failed to generate a product, LR-PCR was performed across the 5′ homology arm (5′ LR-PCR). For these cases, the retention of the 3′ loxP site was confirmed by PCR between the cassette and 3′ loxP site.
Gene targeting is highly efficient
High-throughput gene targeting depends on achieving high targeting efficiencies. For genes expressed in ES cells, a promoterless targeting strategy (referred to as ‘targeted trapping’)33 has been shown to yield targeting efficiencies averaging above 50%. By design, promoterless vectors effectively suppress the recovery of random non-homologous events in the genome as only insertions in transcribed loci, in the correct orientation and reading frame, will confer drug resistance. We electroporated 1,285 different promoterless constructs and obtained targeted clones from nearly half of these constructs with an average targeting efficiency of 50% (Table 1). These data confirm and extend the results of ref. 33, demonstrating that targeted trapping is a highly efficient method for genes expressed in ES cells.
Only half of the promoterless targeting vectors were effective in producing targeted clones. Electroporation of these vectors produced variable numbers of drug-resistant colonies. In general, high colony numbers were predictive of successful targeting experiments, whereas low colony numbers usually indicated a failure to target the locus (Supplementary Table 2). The success or failure of a construct correlated with the number of clones with gene-trap events in the International Gene Trap Consortium database (Supplementary Table 3). Thus, gene-trapping data serve as a useful guide to identify the subset of genes that are amenable to a promoterless targeting strategy34. Correlation with classes of gene was also observed. For instance, targeted trapping was less effective with secreted proteins compared to non-secreted proteins, indicating that our cassette designed for trapping secreted proteins (pL1L2_ST, see Supplementary Fig. 4)35 is not optimal for this class of gene36.
Given that only half of all genes are expressed at a sufficient level in ES cells to support a targeted trapping strategy, we switched to using a promoter-driven cassette for positive selection for non-expressed genes combined with negative DTA selection to select against random insertions. We electroporated different positive–negative targeting cassettes and from the analysis of approximately 30 ES cell clones per unique construct, we recovered targeted events for 80% of genes with an average targeting efficiency of 35% (Table 1; for a complete list of targeted genes see Supplementary Data). A combination of factors probably contribute to our high targeting efficiencies, including the use of isogenic DNA, relatively long recombineered homology arms and DTA negative selection.
Gene targeting is dependent on both the length and the extent of homology between the targeting vector and the target locus37–39. Our vectors typically contain 10 kb of homology to the endogenous locus and originate from a C57BL/6J BAC library. Although the ES cells are derived from the C57BL/6N sub-strain, the Jackson (J) and NIH (N) substrains of C57BL/6 are very closely related16, thus our targeting vectors will have identical sequence with the ES cell genome in the great majority of cases. Negative selection was introduced to improve targeting efficiencies30,31. Overall we observed a threefold enrichment of targeted clones with DTA counter-selection, consistent with previous observations30,31,40 (Table 1).
In a high-throughput pipeline, projects inevitably fail at one or more steps and overall pipeline efficiency depends on effective recovery of these failures. In our experience, most failures are technical in nature and are most efficiently recovered by repeating the procedure. For example, 70% of targeting experiments are rescued after re-electroporation of cells with an alternative preparation of vector DNA (Supplementary Data). Similarly, re-synthesis of oligonucleotides for recombineering or repeating the Gateway reaction recovers a majority of intermediate and final targeting vectors (data not shown). Thus, completion of the mutant resource will require iterative rounds of recovery. Whether some genes are refractory to targeting will become apparent once all technical issues have been ruled out.
Discussion
Our targeting pipeline is the major contributor to the international mouse knockout programmes that aim to generate lacZ-tagged null mutations in every protein-coding gene in mouse. With the technology described here, more than 9,000 genes have been successfully targeted in C57BL/6N ES cells to date. The value of our knockout ES cell resource critically depends on the germline potential of individual targeted C57BL/6N ES cell clones. In a separate study16, hundreds of targeted cell lines generated in our pipeline were assessed for contribution to the germline after blastocyst injection. At least 65% of targeted clones colonized the germ line of chimaeric mice. Thus, our library of mutant C57BL/6N ES cells is robust and will support the production of mutant mice for future large-scale phenotyping programmes.
The scale of mass parallel vector construction and gene targeting described here has implications for functional genomics and proteomics in many model systems. New systematic, genome-scale programmes can now be contemplated. Using available BAC or fosmid genome resources, the high-throughput production of complex transgenes and/or targeting constructs will facilitate the generation of sophisticated, physiologically accurate, cell and animal models. For example, tagging all proteins in the mouse genome by knock-in targeting to establish a proteomic mapping programme equivalent to the highly successful yeast TAP-tagging programmes41 is now feasible.
In the coming years, it is likely that the genome engineering technologies pioneered in the mouse will be also applicable to other model systems such as the rat42,43 and human pluripotent stem cells44,45. The capacity for fluent gene targeting also permits the systematic generation of doubly targeted ES cell lines for functional studies by conditional mutagenesis, which will serve to complement and extend RNA interference studies by providing complete genetic knockouts. Coupled with the power to differentiate ES cells into many cell types, such resources will not only provide means to gaining unique functional insights but will also reduce animal experimentation. With pioneering methodologies, we have overcome the considerable technical challenges involved in establishing the most complex and accurate high-throughput functional genomics platform yet attempted. We believe that our work raises the standards of achievement and expectation for future genome-scale programmes.
METHODS
Computational design of conditional alleles
Gene structures to be targeted are first extracted from a current release of the Ensembl (NCBIM37 assembly) or Vega database. Critical exons, which when deleted induce a frameshift, are chosen computationally (start phase – end phase ≠ 0) or manually (exon length not divisible by 3). Primers (50-mer oligonucleotides) for recombineering are then selected from overlapping blocks of sequence (typically 120 bp) flanking the critical exons at a predefined distance from the splice sites (300 bp from the splice acceptor and 100 bp from the splice donor). Primers for gap repair were chosen from sequence blocks (typically 1 kb) at the ends of the desired homology arms (4–6 kb). Each block was analysed by ArrayOligoSelector46 (http://sourceforge.net/projects/arrayoligosel/) generating one or more candidate primers inside each sequence block with a minimum of 28% G+C content. Candidate primers were rejected if they were repetitive inside a region spanning 100 kb either side of the critical exon(s), and gap repair primers at the ends of the homology arms were also rejected if they shared sequences of 6 bp or more. The final recombineering primer sequences were mapped to the current NCBI assembly, recorded with their genomic coordinates in a database, and displayed in an Ensembl DAS-track. After manual inspection, complete sets of recombineering primers were selected from the database, automatically reverse complemented (where appropriate) and appended with 20–23 bp of sequence homology to the appropriate recombineering cassettes before ordering. In parallel, BACs from the RP23/RP24 indexed library were chosen based on end-mappings of the clones. A vector design interface (Custom Design Tool; http://www.sanger.ac.uk/htgt) is available online.
96-well recombineering
BACs from the RPCI-23/RPCI-24 indexed C57BL/6J libraries27 were arrayed in 96-well format to match the corresponding 96-well plates of 70-mer oligonucleotides (desalted; Illumina/Invitrogen) used to PCR amplify the cassettes used for recombineering. PCR amplifications were performed using the FastStart High Fidelity PCR System (Roche) and the products were desalted using High Pure 96UF Cleanup kits (Roche). The arrayed BAC clones were initially grown at 37 °C inLuria broth(LB) containing chloramphenicol (12.5 μg ml−1) to early log phase and made electrocompetent by washing three times with ice-cold HPLC grade water and the cells are transformed with pBADgbaA plasmid DNA47 using an ECM 630 96-well electroporator/HT-200 automatic plate handler (BTX Harvard Apparatus; pulse conditions of 2,400 V, 700 Ω, 25 μF) followed by growth at 30 °C in liquid medium containing tetracycline (5 μg ml−1) and chloramphenicol (12.5 μg ml−1). The BAC cultures underwent three rounds of recombineering, changing only the PCR products used for each electroporation and the antibiotic selection applied after each step, using the following standard procedure: early log phase cultures were induced to express the red operon following addition of 0.1% arabinose and incubated for 40 min at 37 °C; electrocompetent cells were electroporated in 96-well format (as above) with 1–2 μg of desalted PCR products and allowed to recover at 37 °C for 90 min; an aliquot was then inoculated into a new 96-well box containing media plus the appropriate antibiotics and grown at 30 °C for 2 days. The PCR cassette and antibiotic cocktail used at each step shown in Fig. 3 was as follows. (1) R1-pheS/zeo-R2, zeocin (4 μg ml−1), tetracycline (5 μg ml−1), chloramphenicol (12.5 μg ml−1); (2) loxP-kan-loxP, kanamycin (15 μg ml−1), zeocin (6.5 μg ml−1), tetracycline (5 μg ml−1), chloramphenicol (12.5 μg ml−1); and (3) pR3R4, zeocin (6.5 μg ml−1), kanamycin (15 μg ml−1) carbenicillin (50 μg ml−1). After the gap repair step, the temperature was shifted to 37 °C to eliminate the recombineering plasmid. Intermediate plasmid DNA was purified using standard procedures from saturated cultures (1.5 ml) grown in 96-well blocks. Approximately 50 ng was transformed into electro-competent DH10B E. coli carrying the 705-Cre plasmid (Gene Bridges), pre-induced at 42 °C to express Cre recombinase from the λPR promoter, and selected in liquid culture containing carbenicillin (50 μg ml−1) and zeocin (10 μg ml−1). After overnight growth at 37 °C, individual colonies were streaked out on ampicillin/zeocin plates to isolate individual clones and were sequence-verified.
Gateway exchange reaction
Three-way Gateway reactions were carried out in 96-well format using LR Clonase II Plus enzyme mix (Invitrogen) essentially as described by the manufacturer. In an overnight reaction at 25 °C, 100–200 ng of intermediate targeting vector (prepared from 1.5-ml cultures in 96-well blocks using the Qiagen Turboprep kit) was combined with 60 ng of L1/L2 targeting cassette vector and 60 ng of L3/L4 DTA plasmid backbone in a 10 μl volume. After treatment with Proteinase K, 2 μl of the reaction was transformed into 30 μl of chemically competent Escherichia coli (DH10B, Invitrogen) and plated onto YEG agar plates containing 4-chlorophenylalanine48 and the appropriate antibiotics. Individual colonies were picked and sequenced across all recombineered junctions. Reads were automatically aligned against the synthetic vector sequences and assigned pass levels based on the number and position of matching reads.
ES cell culture and electroporation
The final targeting constructs were prepared for ES cell electroporation from 2 ml of culture (2X LB plus antibiotics) in 96-well format using the Qiagen Turboprep kit. Before electroporation, vectors were linearized with AsiSI and examined by gel electrophoresis. For most clones, the digested DNA migrated as a single high-molecular-mass band of the expected size (Supplementary Fig. 5). Occasionally, contaminating smaller molecular mass bands were also observed on the gel (DNA quality failures).
JM8 mouse ES cell lines derived from the C57BL/6N strain were grown either on a feeder layer of SNL6/7 fibroblasts (neomycin and/or puromycin resistant) or on gelatinized tissue culture plates16. Both feeder-independent and feeder-dependent lines were maintained in Knockout DMEM (500 ml, Gibco) supplemented with 2 mM glutamine, 5 ml 100× β-mercaptoethanol (360 μl in 500 ml PBS, filter sterilized), 10–15% fetal calf serum respectively (Invitrogen) and 500 U ml−1 leukaemia-inhibitory factor (ESGRO, Millipore). Trypsin solution was prepared by adding 20 ml of 2.5% trypsin solution (Gibco) and 5 ml chicken serum (Gibco) to 500 ml filter-sterilized PBS containing 0.1 g EDTA (Sigma) and 0.5 g d-glucose (Sigma).
Electroporations of ES cells were carried out in a 25-well cuvette using the ECM 630 96-well electroporator /HT-200 automatic plate handler (BTX Harvard Apparatus; set at 700 V, 400 Ω, 25 μF). Immediately before electroporation, cell suspensions of ~1 × 107 cells and ~2 μg of linearized targeting vector DNA were mixed in a final volume of 120 μl PBS. Cells were seeded onto a 10-cm dish (with feeders or gelatin) and colonies were picked after 10 d of selection in 100 μg (active) per ml Geneticin (Invitrogen). To expand cells into duplicate wells for archiving and preparation of genomic DNA, confluent cultures of JM8 ES cells grown on feeder cells were washed twice with pre-warmed PBS and trypsinized for 15 min at 37 °C. Five volumes of pre-warmed media were added and the cells were gently dispersed by tituration and passed at a dilution of 1:4 into new plates containing feeder cells. Passage of cells grown on gelatinized plates was carried out in a similar manner except that the cells were trypsinized for 10 min and passed at a dilution of 1:6 into freshly gelatin-coated plates (0.1% gelatin, Sigma G1393). Culture medium was replaced daily and cells reached confluence 2 days after passage. To archive ES cell clones, trypsinized cells from confluent 96-well plates were transferred in 200 μl freezing medium (Knockout DMEM, 15% serum/ 10% DMSO) to 96-well cryovials (Matrix) and overlayed with sterile mineral oil. The cells were placed at −80 °C overnight and then transferred to liquid nitrogen.
Computational design of primers for long-range PCR
To identify targeted ES cell clones, we developed a robust LR-PCR system that uses one set of reaction conditions for every targeted allele screened. In addition, we used an in-house primer generation program (“Primer Brain”) to generate genome-specific primers for the LR-PCR. Primers were selected from 2-kb blocks of sequence upstream of the 5′ homology arm (GF) and downstream of the 3′ homology arm (GR) and from a variable-sized region that contains the critical exon (EX). Primers were first extracted by a single-base-pair tiling of each region into 24- to 30-mers that end in G/C, have at least 10 G/C bases and have a melting temperature of at least 64 °C. Primer choice was weighted negatively to avoid both ‘runs’ of nucleotides (for example, ‘AAA’) and self-annealing ends. The top 100 high-scoring primers in each region were aligned against the current mouse genome (NCBIM37) with Exonerate software (http://www.ebi.ac.uk/~guy/exonerate) and were weighted negatively based on the number of alignments to the genome, with added negative weight given to alignments close to the 3′ end of primers. The two best-scoring primers from each block (GF1 and GF2; GR1 and GR2; EX1 and EX2) were grouped and primer combinations (for example, GF1 and EX1) were screened to eliminate pairs with a 4-bp overlap at their 3′ ends. The resulting GF, GR and EX primers were stored in an Oracle database.
LR-PCR genotyping
ES cell genomic DNA was isolated by digesting the cells with Proteinase K and RNase A. Each well of a confluent 96-well plate was lysed with 30 μl of lysis buffer (10 mM Tris/HCl ph 8, 1 mM EDTA, 50 mM KCl, 2 mM MgCl2) containing 200 μg ml−1 RNase A (Sigma) and 0.67 mg ml−1 proteinase K (Life Technologies). After overnight digestion at 60 °C, the samples were heated to 90 °C (2 min) and 1–2 μl of the lysate was used in a 10 μl LR-PCR reaction. To generate LR-PCR amplicons, two genomic-specific primers outside each end of the 5′ and 3′ homology arms (GF and GR, respectively) were used in combination with the appropriate universal cassette primers (5U (5′-CACAACGGGTTC TTCTGTTAGTCC-3′) and 3U (5′-ATCCGGGGGTACCGCGTCGAG-3′)) (Fig. 5).
Using the SequalPrep kit (0.1 μl 100% v/v DMSO, 0.5 μl 10× enhancer A, 0.5 μl 10× enhancer B, 1.0 μl 10× buffer, 0.2 μl Taq Enzyme/dNTPs; Life Technologies) or LongAMP Taq mix (0.2 μl 100% v/v DMSO (Sigma), 0.3 μl 10 mM dNTPs (Thermo Fisher Scientific), 2.0 μl 5× LongAMP buffer (NEB), 0.4 μl LongAMP Taq (NEB)), 10 μl reactions were set up in 384-well format with ~30–50 ng (1–2 μl) genomic DNA and 12 pmol of each primer. Thermal cycling was performed using the following conditions: 1 cycle 93 °C for 3 min; 8 cycles 92 °C for 15 s, 65 °C for 30 s decreasing by 1 °C per cycle, 65 °C (LongAMP) or 68 °C (SequalPrep) for 8 min; 30 cycles 92 °C for 15 s, 55 °C for 30 s, 65 °C (LongAMP) or 68 °C (SequalPrep) for 8 min increasing 20 s per cycle; 1 cycle 65 °C (LongAMP) or 68 °C (SequalPrep) for 9 min. The PCR products were visualized on 1% E-gels (Life Technologies) and scored for the presence of high-molecular-mass fragments (Supplementary Fig. 6). The LR-PCR products were treated with exonuclease I and shrimp alkaline phosphatase (0.3 U μl−1 and 0.19 U μl−1, respectively; NEB) in 20 mM Tris/HCl, 10 mM MgCl2 for 1 h at 37 °C followed by 80 °C for 15 min. PCR products were sequenced with the genomic primers used for amplification and universal primers to the targeting cassette (5′Us (5′-CGTGGTATCGT TATGCGCCT-3′) and 3′Us (5′-TCTATAGTCGCAGTAGGCGG-3′)) and 3′ loxP (LR (5′-ACTGATGGCGAGCTCAGACC-3′)). Sequence reads were compared by BLAST against synthetic sequences for each targeted allele and clones with correctly aligned sequences were marked as valid. Clones that retained the 3′ loxP site and have 3′ or 5′ sequence-verified LR-PCR bands are marked for distribution and clones that have lost the 3′ loxP are marked as targeted, non-conditional events.
Supplementary Material
Acknowledgements
We thank the following people for technical assistance: D. Klose, D. Oakley, W. Yang and L. Stebbings for informatics/vector design; R. Bennett, A. Horton and A. van Brunt for manual gene annotation/vector design; L. Cho, R. Li, J.-F. Popoff, M.Sharma andY.Zhang for recombineering; G. Belteki, P.Tate, Y.Bekele and S. Borchia for targeting vectors; D. Fraser, J. Greystrong, N. Gueorguieva, M. Jackson, P. Ramagiri, I. Walczak, J. Woodward, E. Stebbings, M. Martinez, A. Tsang and Y. Yoshinaga for vector/ES quality control; and D. Edwards, S. Harris, N. Krishnappa, R. Leah and A. Tait for ES cells. We are grateful for advice on the Gateway system from J. Chesnut of Invitrogen. Finally, we wish to thank W. Wurst, K. Lloyd, and our EUCOMM and KOMP colleagues who are contributing to the production and distribution of the conditional knockout resource. This work was funded by the Wellcome Trust Sanger Institute, grants from the National Institutes of Health (KOMP, U01-HG004080 to W.C.S., P.J.d.J. and A.B.) from the EU Sixth Framework Programme (EUCOMM, to W.C.S., A.F.S. and A.B.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Detailed information on targeted genes is available from the IKMC web portal (http://www.knockoutmouse.org). Targeting constructs and mutant ES cells are available upon request from the EUCOMM (http://www.eummcr.org) and KOMP (http://www.komp.org) repositories.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Gossler A, Joyner AL, Rossant J, Skarnes WC. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989;244:463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- 2.Skarnes WC, Auerbach BA, Joyner AL. A gene trap approach in mouse embryonic stem cells:the lacZ reported is activated bysplicing, reflectsendogenous gene expression, and is mutagenic in mice. Genes Dev. 1992;6:903–918. doi: 10.1101/gad.6.6.903. [DOI] [PubMed] [Google Scholar]
- 3.Zambrowicz BP, et al. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl Acad. Sci. USA. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Gene Trap Consortium A public gene trap resource for mouse functional genomics. Nature Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen GM, et al. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 2008;18:1670–1679. doi: 10.1101/gr.078352.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 7.Robertson E, Bradley A, Kuehn M, Evans M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 8.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 9.van der Weyden L, Adams DJ, Bradley A. Tools for targeted manipulation of the mouse genome. Physiol. Genomics. 2002;11:133–164. doi: 10.1152/physiolgenomics.00074.2002. [DOI] [PubMed] [Google Scholar]
- 10.Glaser S, Anastasiadis K, Stewart AF. Current issues in mouse genome engineering. Nature Genet. 2005;37:1187–1193. doi: 10.1038/ng1668. [DOI] [PubMed] [Google Scholar]
- 11.Valenzuela DM, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nature Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 12.Lettice LA, et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl Acad. Sci. USA. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuniga A, et al. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev. 2004;18:1553–1564. doi: 10.1101/gad.299904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Mouse Knockout Consortium A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Poueymirou WT, et al. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nature Biotechnol. 2007;25:91–99. doi: 10.1038/nbt1263. [DOI] [PubMed] [Google Scholar]
- 16.Pettitt SJ, et al. Agouti C57BL/6N embryonic stem cells for mouse genetics resources. Nature Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gertsenstein M, et al. Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS ONE. 2010;5:e11260. doi: 10.1371/journal.pone.0011260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ringwald M, et al. The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res. 2011;39(Database issue):D849–D855. doi: 10.1093/nar/gkq879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 20.Testa G, et al. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell KJ, et al. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nature Genet. 2001;28:241–249. doi: 10.1038/90074. [DOI] [PubMed] [Google Scholar]
- 22.Yang SH, et al. Caution! Analyze transcripts from conditional knockout alleles. Transgenic Res. 2009;18:483–489. doi: 10.1007/s11248-008-9237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang Q, Conte N, Skarnes WC, Bradley A. Extensive genomic copy number variation in embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105:17453–17456. doi: 10.1073/pnas.0805638105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilming LG, et al. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarov M, et al. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nature Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- 26.Poser I, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nature Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osoegawa K, et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- 28.Hope IA, et al. Feasibility of genome-scale construction of promoter:reporter gene fusions for expression in Caenorhabditis elegans using a multisite gateway recombination system. Genome Res. 2004;14:2070–2075. doi: 10.1101/gr.2463804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rual JF, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 30.Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 31.Yagi T, et al. Homologous recombination at c-fyn locus of mouse embryonic stem cells with use of diphtheria toxin A-fragment gene in negative selection. Proc. Natl Acad. Sci. USA. 1990;87:9918–9922. doi: 10.1073/pnas.87.24.9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lay JM, Friis-Hansen L, Gillespie PJ, Samuelson LC. Rapid confirmation of gene targeting in embryonic stem cells using two long-range PCR techniques. Transgenic Res. 1998;7:135–140. doi: 10.1023/a:1008876526826. [DOI] [PubMed] [Google Scholar]
- 33.Friedel RH, et al. Gene targeting using a promoterless gene trap vector (“targeted trapping”) is an efficient method to mutate a large fraction of genes. Proc. Natl Acad. Sci. USA. 2005;102:13188–13193. doi: 10.1073/pnas.0505474102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skarnes WC. Two ways to trap a gene in mice. Proc. Natl Acad. Sci. USA. 2005;102:13001–13002. doi: 10.1073/pnas.0506279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skarnes WC, Moss JE, Hurtley SM, Beddington RS. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc. Natl Acad. Sci. USA. 1995;92:6592–6596. doi: 10.1073/pnas.92.14.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Felipe P, Luke GA, Brown JD, Ryan MD. Inhibition of 2A-mediated ‘cleavage’ of certain artificial polyproteins bearing N-terminal signal sequences. Biotechnol. J. 2010;5:213–223. doi: 10.1002/biot.200900134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasty P, Rivera-Perez J, Bradley A. The length of homology required for gene targeting in embryonic stem cells. Mol. Cell. Biol. 1991;11:5586–5591. doi: 10.1128/mcb.11.11.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng C, Capecchi MR. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell. Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.te Riele H, Maanday ER, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc. Natl Acad. Sci. USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanagawa Y, et al. Enrichment and efficient screening of ES cells containing a targeted mutation: the use of DT-A gene with the polyadenylation signal as a negative selection maker. Transgenic Res. 1999;8:215–221. doi: 10.1023/a:1008914020843. [DOI] [PubMed] [Google Scholar]
- 41.Collins SR, et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Buehr M, et al. Captureof authentic embryonic stem cellsfrom rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 43.Li P, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Bozdech Z, et al. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, et al. An improved recombineering approach by adding RecA to lambda Red recombination. Mol. Biotechnol. 2006;32:43–53. doi: 10.1385/mb:32:1:043. [DOI] [PubMed] [Google Scholar]
- 48.Kast P. pKSS—a second-generation general purpose cloning vector for efficient positive selection of recombinant clones. Gene. 1994;138:109–114. doi: 10.1016/0378-1119(94)90790-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.