Abstract
In many DNA viruses, genome packaging is initiated by the small subunit of the packaging terminase, which specifically binds to the packaging signal on viral DNA and directs assembly of the terminase holoenzyme. We have experimentally mapped the DNA-interacting region on Shigella virus Sf6 terminase small subunit gp1, which occupies extended surface areas encircling the gp1 octamer, indicating that DNA wraps around gp1 through extensive contacts. High resolution structures reveal large-scale motions of the gp1 DNA-binding domain mediated by the curved helix formed by residues 54-81 and an intermolecular salt bridge formed by residues Arg67 and Glu73, indicating remarkable structural plasticity underlying multivalent, pleomorphic gp1:DNA interactions. These results provide spatial restraints for protein:DNA interactions, which enable construction of a three-dimensional pseudo-atomic model for a DNA-packaging initiation complex assembled from the terminase small subunit and the packaging region on viral DNA. Our results suggest that gp1 functions as a DNA-spooling device, which may transform DNA into a specific architecture appropriate for interaction with and cleavage by the terminase large subunit prior to DNA translocation into viral procapsid. This may represent a common mechanism for the initiation step of DNA packaging in tailed dsDNA bacterial viruses.
Keywords: terminase, DNA packaging, bacteriophage, virus assembly, nucleoprotein complex
Introduction
In many tailed double-stranded DNA (dsDNA) bacteriophages, the phage DNA newly synthesized in the infected host cell exists as concatemers, each comprised of multiple copies of unit-length genome. During virus morphogenesis, the DNA is packaged into a preformed capsid precursor termed procapsid through a channel formed by a portal protein complex embedded in a unique vertex of the procapsid. The DNA-packaging process is carried out by a molecular machine called terminase that consists of two proteins: a DNA-recognition component and a catalytic component, also known as the “small” and “large” subunit, respectively. The terminase small subunit specifically binds to concatemeric viral DNA, allowing the large subunit to cleave DNA and thread it through the portal protein channel into the procapsid powered by ATP hydrolysis. Several features of phage DNA packaging, e.g., selectivity for viral DNA through specific protein:DNA interaction and the presence of a virally encoded ATPase to fuel the packaging process, appear to be shared by herpesvirus 3, adenoviruses, poxvirus, and some small single-stranded DNA viruses such as parvoviruses and adeno-associated virus 10.
The terminase small subunits play central roles in DNA packaging by recognizing viral DNA, regulating ATPase and nuclease activities of terminase large subunits, and directing assembly of the packaging-competent terminase holoenzymes. The high resolution X-ray structure of the full-length terminase small subunit gp1 of Shigella phage Sf6 showed a ring-like octameric assembly formed by monomers each consisting of an N-terminal domain surrounding the assembly, a central α-helical domain embracing a molecular channel, and a C-terminal domain forming a β-barrel 14. The terminase small subunits of bacteriophages T4, T7, SPP1 and P22 were reported to form oligomers as deduced from biochemical, mass spectrometry, electron microscopic and crystallographic analysis 15-20. X-ray structures of the oligomerization core of an SPP1-like Bacillus phage terminase small subunit and a low-resolution model of the full-length protein 21, as well as the terminase small subunit oligomerization core of a T4-like phage 22, showed similar ring-like arrangement, suggesting a common architecture for terminase small subunits. The oligomerization states of structurally characterized ring-like terminase small subunits appear diverse, which range from octamer for Sf6 gp1 14, nonamer for P22 gp3, nonamer and decamer for Siphovirus SF6 G1P 21, and 11-mer and 12-mer for a T4-like phage terminase small subunit 22. Additionally, it was reported that phage T4 terminase small subunit formed 22-mer double rings as well as 11-mer single rings. In bacteriophage lambda, the terminase subunits gpNu1 and gpA formed a hetero-oligomer with a 8:4 stoichiometry 23.
In phages such as P22, SPP1 and lambda, the terminase small subunits are responsible for selectivity of DNA packaging by recognizing specific regions on phage DNA that usually harbor sequence-specific motifs called pac signals. The pac regions have been identified in several phages as well as in herpesvirus. A common feature of those pac regions is that they span a range of a hundred to a few hundred base pairs. In phage lambda, the pac region spreads over a 120-bp site on phage DNA called cosB, which contains three repeats of an essential DNA motif. In phage P1, a 161-bp region on phage DNA was identified as the pac site, which contained the centrally located packaging cleavage sites with seven repeats of a conserved hexanucleotide motif 29, and binding of the terminase small subunit to the pac DNA and subsequent cleavage by the terminase large subunit is dependent on methylation on those motifs. A 22-bp region of phage DNA was identified as the pac signal of phage P22, which was centered in a 120-bp DNA region where the initiation cleavage sites by the terminase large subunit were distributed 32. In Bacillus phage SPP1, the terminase small subunit specifically binds to an extended pac region spanning 270 bps on the phage DNA 33. In herpes simplex virus type 1, an approximately 200-bp terminal redundant region of viral genome have been shown to harbor the two conserved DNA-packaging signals named pac1 and pac2. However, how these extended DNA regions interact with terminase small subunits remains elusive. It was proposed that binding of terminase small subunits induced bending or looping of phage DNA.
Bacteriophage Sf6 belongs to the Podoviridae family of tailed dsDNA bacteriophage 36. Previously we determined the high resolution X-ray structure of the full-length Sf6 terminase small subunit gp1 14. Here, we show that gp1 binds to DNA in a cooperative manner, indicating multivalent protein:DNA interactions. Using structure-directed mutagenesis studies, we have mapped the DNA-interacting region on gp1, which occupies an extended surface area encircling the gp1 octamer. Comparative structural analysis of gp1 and its mutants at high resolution reveals remarkable domain-wise motion for the DNA-binding domain mediated by the curved helix formed by residues 54-81 and an intermolecular salt bridge formed by residues Arg67 and Glu73. These indicate remarkable structural plasticity of the DNA-binding domain, which provides a structural basis underlying cooperative DNA-binding of gp1. The spatial restraints derived from these experimental results have enabled construction of a three-dimensional pseudo-atomic model for the DNA-packaging initiation complex, in which the phage DNA wraps around the gp1 octamer through multivalent protein:DNA interactions to form a nucleosome-like complex, facilitated by the structural plasticity of gp1. These suggest that gp1 functions as a DNA-spooling device, which may transform the DNA into a specific three-dimensional architecture that is suitable for interaction with and cleavage by the terminase large subunit prior to translocation of DNA into the phage procapsid.
Results
The gp1 binds to DNA cooperatively
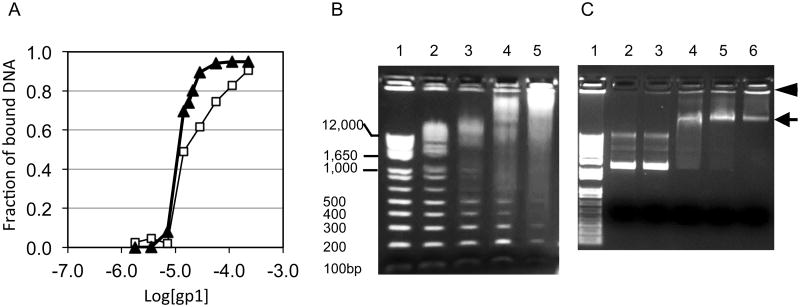
We analyzed the DNA-binding activity of gp1 semi-quantitatively using electrophoretic mobility shift assay. The ∼1800-bp region on the Sf6 genome corresponding to the gp1 and gp2 coding sequences was used. At a protein concentration lower than 3.5 μM, no binding was observed. At higher protein concentration, the dependence of DNA-binding on the protein concentration shows a characteristic sigmoid curve (Figure 1A), indicating a cooperative manner of the gp1:DNA binding. The degree of cooperativity is estimated to be approximately 5.6, suggesting that more than five gp1 molecules bind simultaneously to one single DNA molecule in a cooperative manner.
Figure 1.
Cooperative binding of Sf6 gp1 to DNA.
(A) Fractions of bound DNA plotted against logarithmic gp1 concentrations. Filled triangles and open squares are for 1836-bp and 200-bp linear DNA, respectively.
(B) Binding of gp1 to a DNA ladder. Lane 1, 1kb plus DNA ladder (Invitrogen). Lanes 2-5, the DNA ladder incubated with gp1 at 28.19, 56.39, 112.78 and 225.56 μM respectively.
(C) Binding of gp1 to supercoiled plasmid DNA. Lane 1, 1kb plus DNA ladder (Invitrogen). Lane 2, the plasmid DNA alone. Lanes 3-6, the plasmid DNA incubated with gp1 at 2.82, 28.19, 56.39 and 112.78 μM respectively. The gp1:DNA complex is indicated with an arrow. The very slowly migrating diffusive bands near or in the wells in (B) and (C) may represent higher-molecular-weight protein:DNA complexes or networks and areindicated with an arrowhead.
We then tested the binding of gp1 to 200-bp DNA, and the analysis showed that the binding is cooperative (Figure 1A; Figure S1). The degree of cooperativity is estimated to be approximately 5.5. These data indicate that gp1 binds to 200-bp DNA in a multivalent manner, and each DNA molecule may be bound by more than five gp1 monomers. Gp1 exists as an octameric assembly in solution 14. It is unlikely that multiple gp1 octamers binds to a single 200-bp DNA molecule simultaneously at lower concentrations of gp1 (Figure S1, Lanes 3-8). Thus, these data indicate a multivalent interaction between the gp1 octamer and DNA, i.e., multiple gp1 monomers in one single octamer simultaneously interact with a DNA molecule. Taking into consideration the approximate diameter of 90 Å of the gp1 octamer (thus a perimeter of ∼280 Å), the length of the 200-bp DNA and the persistence length of double-helix DNA of ∼500 Å (140 bps) 37, these results strongly suggest that the DNA wraps around the gp1 assembly through extensive and contiguous molecular contact. Accumulation of DNA in very slowly migrating bands and in wells were observed (Figure 1B and C), suggesting further formation of heterogeneous, higher-molecular-weight complexes or networks by gp1 and DNA.
Correlation of the binding activity of gp1 with the length of linear DNA
We analyzed the gp1 DNA-binding activity using linear dsDNA of various lengths. Analysis of DNA-binding of gp1 using a DNA ladder clearly shows a correlation between gp1 DNA-binding affinity and the DNA length (Figure 1B). At a concentration of ∼28 μM, gp1 selectively binds to DNA that is longer than 1200 bps, while other DNA molecules remains largely unbound (Figure 1B, Lane 2). When the gp1 concentration increases, it begins to bind to DNA of 400-1000 bps (Figure 1B, Lanes 3 and 4). At a higher concentration, gp1 begins to bind to 100- and 200-bp DNA (Figure 1B, Lane 5). These data demonstrate that gp1 selectively binds to longer DNA, indicating that gp1 has higher binding affinity for longer DNA. This may reflect the greater capability of longer DNA to adopt a bent conformation that is needed for multivalent gp1:DNA interactions. As the persistence length of double-helix DNA is ∼500 Å (140 bps) 37, DNA molecules with a length nearer to the persistence length are more likely to adopt extended, elastic-rod-like conformation in solution, thus is energetically less favorable for gp1 binding. In contrast, longer DNA molecules more likely adopt bent conformations, which favors gp1 binding.
The binding of gp1 to supercoiled plasmid DNA
Previous electrophoretic mobility shift assay of gp1 with long, linear DNA showed diffusive bands for potential gp1:DNA complexes 14, suggesting considerable dynamics for the interactions of gp1 with long, linear DNA, i.e., gp1 and DNA undergoes fast association and dissociation. We analyzed the binding of gp1 with supercoiled plasmid DNA. Interestingly, we observed a clear, sharp band (Figure 1C). This suggests that gp1 forms a more stable protein:DNA complex with the supercoiled plasmid DNA, which is in contrast to the dynamic protein:DNA association observed in gp1 binding to long linear DNA. As the DNA binding observed in Figure 1 B and C is not sequence-specific, we infer that formation of a more stable complex by gp1 with the plasmid DNA may be attributed to supercoiling of the substrate DNA, which is present in the plasmid DNA but absent in linear DNA.
Identification of an extensive DNA-binding region on the gp1 assembly by structure-directed mutagenesis
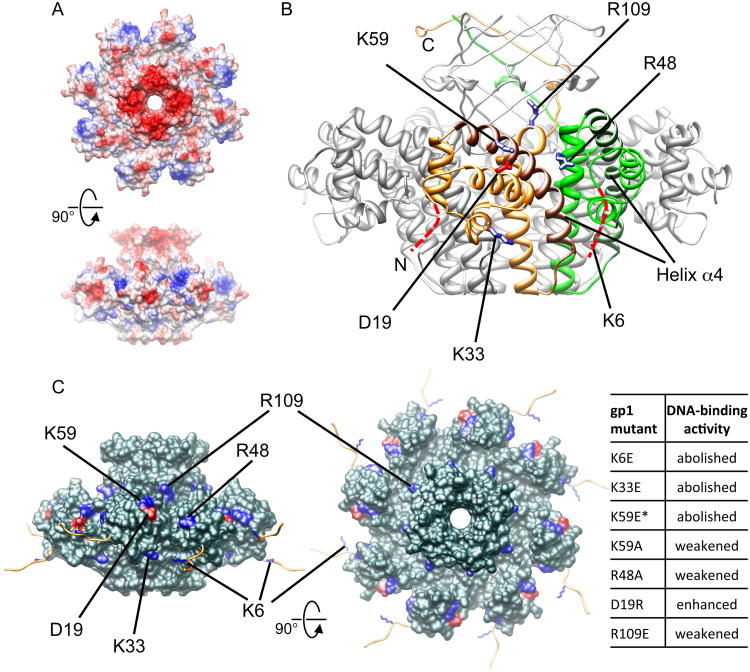
Based on the previously determined X-ray structure of Sf6 gp1 14, we designed mutations of surface residues to experimentally examine their effects on gp1 DNA-binding without interfering with protein folding. The gp1 structure shows a ring-like octameric assembly, with the N-terminal domains hanging at the periphery about the elongated, curved α-helix formed by residues 54-81, referred to as helix α4 hereafter (Figure 2). The C-proximal β-barrel region and the bottom of the gp1 octamer both have strong negative electrostatic potential, thus is unlikely to be the site for DNA binding (Figure 2A). The N-terminal domain is located in the surrounding region of the octamer and is largely exposed to the solvent, serving as a potential candidate for the DNA-binding domain. To gain insights into the gp1:DNA interactions, we selected a series of charged residues in the N-terminal domain, i.e., K59, K33, R48, D19, R109 and K6, for mutagenesis studies. The mutant proteins were expressed in E. coli, purified to homogeneity, and subject to electrophoretic mobility shift assay (EMSA) to examine their DNA-binding activities.
Figure 2.
Mutagenesis analysis of gp1:DNA binding.
(A). The electrostatic surface potential of the Sf6 gp1 octamer (PDB ID 3HEF) viewed down the 8-fold axis (top panel) and from the side (bottom panel). Blue, positively charged molecular surface. Red, negatively charged molecular surface.
(B) Locations of mutated residues on gp1. The gp1 octamer (PDB ID 3HEF) is shown as a ribbon diagram in grey. Two neighboring gp1 monomers are colored in gold and green respectively, in which the curved helices α4 are in brown and dark green respectively. Side chains of the residues subject to mutations are shown in blue for K6, K33, R48, K59 and R109 and red for D19. The N-terminal nine residues that are disordered in X-ray structures are schematically shown as dashed curves in red. The N- and C-termini are labeled for the monomer in yellow.
(C) Distribution of mutated residues in the context of the gp1 octamer (molecular surface in dark cyan; PDB ID 4DYQ). The residues subject to mutagenesis analysis are indicated. The computationally modeled N-terminal nine residues of gp1 are shown as loops in gold, and the side chains of K6 are blue.
The mutations K59E 14 and K33E completely abolished DNA-binding of gp1 (Figure S2 A and B; Figure 2). The K59A mutant showed considerably reduced DNA-binding activity (Figure S2B). These suggest that reduced DNA binding of the K59A mutant was attributed to loss of contact between residue 59 and DNA, whereas loss of DNA binding in the K59E mutant was due to a combined effect of loss of contact and repellent electrostatic interactions between the DNA phosphate backbone and the glutamic acid residues in the mutant protein. The mutation of R48A binds to DNA at a considerably lower affinity than wt-gp1 (Figure S2C). The DNA-binding activity of the R109E mutant is detectable but considerably reduced compared to the wt-gp1 (Figure S2C). This indicates that the R109E mutation shows less effect on DNA-binding than the K33E and K59E mutations, presumably due to the greater distance between residue R109 and the bound DNA phosphate backbones. The N-terminal 9 residues are disordered in the previous X-ray structure (Figure 2). However, as this N-terminal polypeptide is located at the periphery of the gp1 octamer thus is readily accessible for interactions with DNA, we conducted mutagenesis studies on residue K6 within this polypeptide. Interestingly, the K6E mutation abolishes the DNA binding of gp1 (Figure S2C), showing a drastic effect similar to those of the K33E and K59E mutations. This suggests that residue K6 may be at the close proximity to the bound DNA phosphate backbone. The D19R mutation apparently enhances the DNA-binding affinity (Figure S2D). The effects of these mutations on gp1 DNA-binding activity result from the mutations per se instead of structural or folding changes caused by the mutations, as X-ray structures for K59E, D19R and R48A mutants are essentially the same as that of wt-gp1 except for the mutated residues (Table 1).
Table 1. X-ray data collection and structure refinement statistics.
| Protein | gp1-FL* | gp1-R48A | gp1-D19R | gp1-K59E | gp1-FL-CAPS** | |
|---|---|---|---|---|---|---|
| PDB accession code | 4DYQ | 4DZP | 4DYC | 4DZJ | 4DYR | |
| Beamline | SSRL 7-1 | SSRL 12-2 | SSRL 12-2 | SSRL 14-1 | SSRL 14-1 | |
| Resolution/Å | 1.5(1.55-1.5) | 1.8(1.85-1.8) | 1.8(1.85-1.8) | 1.9(1.97-1.9) | 1.65(1.71-1.65) | |
| Completeness(%) | 97.9(97.6) | 99.6(98.9) | 99.6(97.1) | 99.9(99.4) | 97.9(94.5) | |
| I/σ | 50.2(2.80) | 31.7(7.78) | 30.6(4.45) | 60.6(6.73) | 48.1(4.06) | |
| Rmerge | 0.044(0.503) | 0.034(0.203) | 0.042(0.384) | 0.067(0.484) | 0.042(0.381) | |
| Rwork/Rfree | 0.18/0.20 | 0.21/0.25 | 0.21/0.23 | 0.20/0.23 | 0.20/0.22 | |
| RMSD, Bond lengths/Å | 0.005 | 0.006 | 0.007 | 0.006 | 0.006 | |
| RMSD, Bond angles/° | 0.891 | 0.859 | 0.948 | 0.845 | 0.899 | |
| Conformations of DNA-binding domains | Mol. A | “Up” | “Up” | “Up” | “Up” | Intermediate |
| Mol. B | “Down” conformation | |||||
gp1-FL, full-length gp1.
Crystals grown with addition of the CAPS pH10.5 buffer. See Materials and Methods for details.
These data demonstrate that the gp1 N-terminal domain provides the site for DNA binding, therefore experimentally establish its role as the DNA-binding domain. Moreover, these results indicate that residues K6, K33, K59, R48 and D19 must be located in proximity of bound DNA. The drastic effects of mutations at K6, K33 and K59 on the gp1 DNA-binding activity suggest that these residues are in the close proximity of DNA and are presumably involved in electrostatic interactions with the DNA phosphate backbones. The mutation at residue 48 from a positively charged residue Arg to a neutral residue Ala considerably reduces the DNA-binding activity of gp1, suggesting that this residue also makes close contact with the bound DNA. The R109E mutation shows only modest effect, suggesting that this residue is less proximal to the bound DNA. The residues K6, K33, K59, R48 and D19 occupy an extensive surface region that forms a circular zone surrounding the gp1 assembly (Figure 2C). Residues K59 and K33 are located above and beneath the DBD respectively, suggesting that the bound DNA must make contact with gp1 through sites both on the top and bottom of the DBDs. K6 is located in a N-terminal 9-residue polypeptide that is disordered in the X-ray structure (Figure 2C). Computational modeling of this polypeptide shows that eight copies of it emanate from the gp1 assembly, forming a group of arms that can potentially make intimate interactions with DNA (Figure 2C). Residues D19 and R48 are situated on DBDs at the periphery of the gp1 assembly. The distribution of these residues indicates that the bound DNA surrounds the gp1 assembly, making extensive contact with the gp1 DBDs. Furthermore, the distribution of these residues and the differential effects of these mutations on DNA-binding activity of gp1 provide spatial restraints for positioning the bound DNA with respect to the gp1 assembly, albeit at low resolution, suggestive of a wrap-around mode for DNA binding of gp1 (see Discussion). These results demonstrate that the gp1 octamer assembles with DNA into a high-order nucleoprotein complex.
The DNA-binding domain of gp1 exhibits remarkable structural plasticity
We have determined high resolution X-ray structures of full-length gp1 and its mutants at up to 1.5 Å resolution with excellent refinement statistics (Table 1). The crystallographic asymmetric unit contains two gp1 molecules, designated as Molecules A and B. No non-crystallographic symmetry restraints were imposed, as the data-to-parameter ratios were sufficient for bias-free refinement. The refined structures are of high quality (Table 1). Comparison of these structures reveals differential conformations for DBDs.
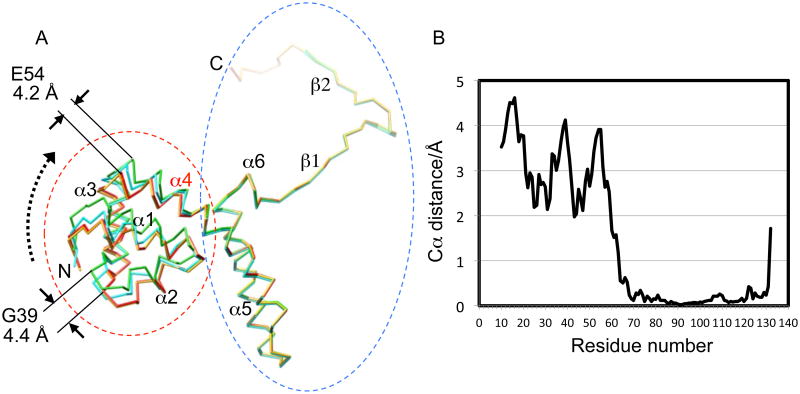
The structure of the gp1 monomer in the 1.5 Å resolution structure, hereafter designated as gp1-FL (Table 1), shows an overall architecture of an N-terminal DBD followed by two a-helices forming an α-barrel and a C-terminal β-strand region forming a β-barrel (Figure 3A). However, least-square superimposition of the two molecules in the crystallographic asymmetric unit shows a root-mean-square (rms) deviation of 1.34 Å for Cα, which indicates significant structural differences for the two molecules. A careful comparison of the two molecules in the asymmetric unit reveals that the N-terminal DNA-binding domain undergoes a domain-wise movement such that it appears to swing upward from the position in molecule B to that in molecule A (Figure 3), which are hereafter termed the “down” and “up” conformations respectively. Superimposition conducted upon residues 70-132 resulted in an rms deviation of 0.21 Å for Cα, indicating that the two molecules are essentially identical in this region. Likewise, the two N-terminal DBDs (residues 10-65) alone are readily superimposable, with an rms deviation of 0.51 Å for Cα. Thus, the internal structures of the DBDs and the rest of the gp1 molecule remain the same respectively, but the position/orientation of the DBD with respect to the rest of the gp1 molecule differs in Molecules A and B. Indeed, after molecule B is superimposed on A by least-square fitting for residues 70-132, an offset for the DBDs is clearly visible with a 2.45 Å distance between centroids of the two domains plus a rotation of 12.9°. The average and maximal distances between main chain atoms of the DBDs of Molecule A and B are 3.2 and 4.6 Å, respectively. The overall motion of the DBDs from Molecule B to Molecule A can be explained as a domain-wise oscillation upward when viewed from the side of the gp1 assembly, plus an additional oscillation clockwise about the octameric axis (Figures 3 and 4A). In the region of residues 60-70 through which the DBD is connected to the rest of the molecule, pairwise distances between main chain atoms of the two molecules decrease gradually (Figure 3), leading to a contiguous change in the curvature of helix α4 that encompasses residues 54-81. No sharp kink is present in this helix in either gp1 monomer. It is likely that helix α4 possesses intrinsic flexibility, which enables curving of this helix to various extents, thus leading to the “up” and “down” conformations of the DBDs in the two gp1 molecules.
Figure 3.
Structural plasticity of the gp1 DNA-binding domain.
(A). Superimposition of Molecules A and B from the structures of gp1-FL and gp1-FL-CAPS (see Table 1). Molecules A and B of gp1-FL are green (“up” conformation) and red (“down” conformation), respectively. Molecules A and B of gp1-FL-CAPS are cyan (“intermediate” conformation) and yellow (“down” conformation), respectively. Superimposition was conducted upon residues 70-132 as indicated by a blue dashed ellipsoid. The DNA-binding domains are indicated by a red dashed ellipsoid. The view is from the side of the gp1 octamer. The putative movement of the DNA-binding domain is indicated by a dashed arrow. Distances of Cα atoms after superimposition are indicated for residues E54 and G39. The secondary structural elements and the N- and C-termini of the modeled gp1 structure corresponding to residue 10 and 139 respectively are labeled.
(B) Cα distances for the two DNA-binding domains in gp1-FL after superimposition upon residues 70-132.
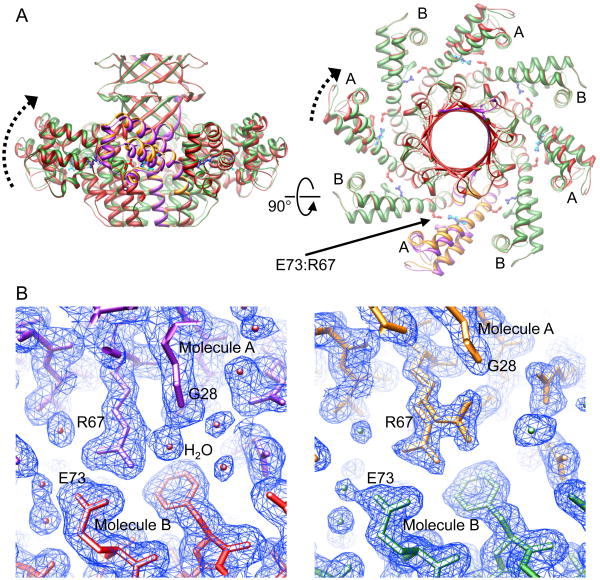
Figure 4.
The “up” and “down” conformations of the gp1 DNA binding domains are mediated by the intermolecular salt bridge between residues E73 and R67.
(A). Locations of the E73:R67 salt bridges in the context of the gp1 octamers. The gp1 octamers are shown as ribbon diagrams in green and red for the superimposed structures of gp1-FL and gp1-FL-CAPS respectively. One gp1 molecule is highlighted in gold and magenta in the two structures respectively. The B molecules in both structures are superimposable, whose DNA-binding domains all adopt the “down” conformation (marked with B), whereas the DNA-binding domains of the A molecules (marked with A) adopt “up” and “intermediate” conformations in gp1-FL and gp1-FL-CAPS structures respectively. The putative movements of the DNA-binding domains from the “down” to the “up” conformations are indicated by dashed arrows. Side chains of residues E73 and R67 are shown in red and blue respectively. Side chains of residues R67 in the four A molecules of the gp1-FL structure that adopt dual conformations are shown in cyan.
(B) Local structures of the E73:R67 salt bridges in structures of gp1-FL-CAPS (left panel)and gp1-FL (right panel) showing the “intermediate” and “up” conformations respectively. Molecules A and B are magenta and red in the gp1-FL-CAPS structure, and gold and green in the gp1-FL structure, respectively. In the left panel, the water molecule that is H-bonded to residues R67 and G28 is labeled. The 2mFo-DFc electron density maps contoured at 0.65 (left panel) and 1.0 σ (right panel) calculated with the final refined models are superimposed (blue chicken wire).
An “intermediate” conformation of the DNA-binding domain
Comparison of the gp1-FL structure with that of the gp1-FL-CAPS whose crystal was grown under slightly higher pH in the presence of the CAPS pH10.8 buffer (Table 1) shows essentially identical structures for Molecules B. However, the conformations of the DBDs of Molecules A are different in the two crystals (Figure 3). In the gp1-FL-CAPS structure, the DBD of Molecule A adopts an “intermediate” conformation that is between the “up” and “down” conformations observed in the present 1.5 Å structure (Figure 3). Superimposition by residues 70-132 of Molecules A and B in the gp1-FL-CAPS structure shows a clear offset for the two DBDs, and it requires an additional 1.59 Å translation and 6.33° rotation to superimpose the DBD of Molecule B upon that of Molecule A (Figure 3). The average and maximal distances between main chain atoms of the two DBDs are 1.81 and 2.95 Å, respectively. While these inter-molecular structural differences are smaller than those in the gp1-FL structure as described above, they are significant at the current resolutions and reflect true conformational differences.
Thus, three distinct conformations have been observed for the gp1 DBD, that is, the “down” conformation in Molecule B in both crystals, the “up” conformation in Molecule A in the gp1-FL structure, and the “intermediate” conformation in Molecule A in the gp1-FL-CAPS structure. In solution, the eight DBDs in a gp1 octamer may toggle between the two conformations randomly and perhaps continuously. These results suggest that gp1 DBD bears considerable conformational plasticity, enabling the gp1 octamer to allosterically adjust its conformation for optimal DNA binding.
Structural plasticity of the DNA-binding domain is mediated by the intermolecular salt bridge between residues R67 and E73
We have inspected intermolecular interactions that may contribute to the differential conformations of the DBDs. The DBDs are engaged in few intermolecular interactions, which is evident by buried solvent accessible areas of as small as 147.0 and 177.2 Å2 for the DBDs of Molecules A and B respectively. Notably, a major intermolecular interaction for the DBD involves a salt bridge between residue R67 of the DBD from one gp1 molecule and residue E73 from a neighboring gp1 molecule in the same octamer. In the “intermediate” or “down” conformations, R67 from Molecule A and E73 from an adjacent Molecule B forms a salt bridge, and a water molecule forms H-bonds with two amide nitrogen atoms of R67 and with the main chain oxygen atom of G28 from the same molecule A (Figure 4). The “intermediate” or “down” conformations differ slightly as the water molecule bridging R67 and G28 moves by 1.0 Å, causing a displacement of 1.2 Å for the G28 main chain oxygen atom. In contrast, in the “up” conformation, R67 adopts dual conformations for its side chain: one participates in the salt bridge with E73, while the alternative conformation flips over and is H-bonded to the main chain oxygen atom of G28 from the same Molecule A. The water molecule between R67 and G28 is absent in the “up” conformation. Flipping of the R67 side chain causes a displacement of 2.7 Å for the G28 main chain oxygen atom, which pushes the DBD of Molecule A upwards, leading to the “up” conformation (Figure 4). As the movement of the DBD is hinged about the curved helix α4, a positional change of 2.7 Å for G28 results in a displacement of as much as 4.6 Å in the distal region of the DBD. In the context of the gp1 octamer, residues R67 from four molecules adopt dual conformations, thus, DBDs of four gp1 monomers adopt the “up” conformation while the other four adopt the “down” conformation (Figure 4A). These data show that the intermolecular salt bridge R67:E73 mediates the formation of variable conformations of the DBDs.
The salt bridge between residues R67 and E73 is required for the optimal DNA-binding activity of gp1
We have over-expressed and purified the E73A mutant of gp1 using the same procedure as for wt-gp1. Size-exclusion chromatography shows that gp1-E73A elutes at the same volume as wt-gp1, indicating that it exists in the solution as an octamer as wt-gp1 (data not shown). Electrophoretic mobility shift assay shows considerably reduced DNA binding of gp1-E73A compared to wt-gp1 (Figure S3). These data demonstrate that the E73A mutation reduces the DNA-binding activity of gp1. Attempt to crystallize gp1-E73A using the same condition for wt-gp1 failed and generated clear drops in the crystallization trays. It is worth pointing out that the condition for wt-gp1 has been used to crystallize K59E, R48A and D19R to yield crystals belonging to the same space group with essentially the same unit cell dimensions and resulting in high resolution structures (Table 1). These imply that the E73A mutation may reduce the conformational dynamics of the gp1 DBD and restrain it to a conformation that is considerably different from that of wt-gp1 so that the crystal packing as in wt-gp1 crystals was not maintained. These data indicate that the R67:E73 salt bridge is required for the optimal DNA binding of gp1 and the role of this salt bridge may be to modulate the DBD conformations between the “down” and “up” conformation.
Discussion
Multivalent binding of Sf6 gp1 and DNA
Previous works indicate that initiation of DNA packaging in many tailed bacteriophages and herpesviruses involves extended pac regions on viral DNA that span over a hundred base pairs. However, it has been elusive how terminase small subunits bind to these pac regions. In this work, we have shown that Sf6 terminase small subunit gp1 binds to DNA in a cooperative manner, indicating multivalent protein:DNA interactions. Gp1 binds to a 200-bp DNA cooperatively, demonstrating that gp1 is capable of drastically altering the DNA conformation and leading to DNA bending or looping, and that the DNA may wrap around the gp1 assembly via extensive and contiguous molecular contact to form a high-order nucleoprotein complex. The gp1:DNA interaction upon each monovalent site may be weak, but simultaneous gp1:DNA interactions at multiple sites may energetically favor the stable gp1:DNA binding. Gp1 preferentially binds to supercoiled plasmid DNA, and forms a sharp band in contrast to diffusive bands of complexes of gp1 and linear DNA, indicating that gp1 exhibits preference for a specific conformation, or the capability to form such conformation, of the substrate DNA that is present in the plasmid DNA but absent in linear DNA. Plasmid DNA has supercoils as a result of DNA replication. Thus, it is likely that these supercoiled regions are where gp1 prefers to bind. Cooperative DNA binding was observed for terminase small subunits from phages SPP1 33 and lambda 27. In SPP1, it was shown that binding of its terminase small subunit lead to DNA bending or looping 33. Thus, alteration of DNA conformation through multivalent binding may be a common feature of terminase small subunits.
Extended regions encircling the gp1 assembly are responsible for gp1:DNA interactions
Our structure-directed mutagenesis studies show that extended regions surrounding the gp1 assembly are engaged in protein:DNA contact. The locations of those mutated residues and their differential effects on gp1 DNA-binding activity define a pattern of molecular contact between gp1 and DNA. The bound DNA must make close contact with three types of regions on the surface of the gp1 assembly, i.e., above, beneath and on the side of the DNA-binding domains, where residues K59, K33, and K6, D19 and R48 are located respectively (Figure 2). It is important to note that DNA must make contact with all three types of regions simultaneously. Contact of DNA solely with any one of the three types of regions may not explain the mutagenesis results. The R109E mutation only modestly reduces gp1:DNA binding, suggesting that this residue is at a greater distance from the bound DNA than other mutated residues. Additionally, the bound DNA may not interact with the C-terminal beta-barrel region or the bottom area of the gp1 assembly, as these regions are abundant in negatively charged residues (Figure 2A). The distribution of these regions confers spatial and conformational restraints for the bound DNA that converges to a high-order nucleoprotein complex via a “wrap-around” DNA-binding mode for gp1 (see below).
A molecular model for the gp1:DNA nucleoprotein complex
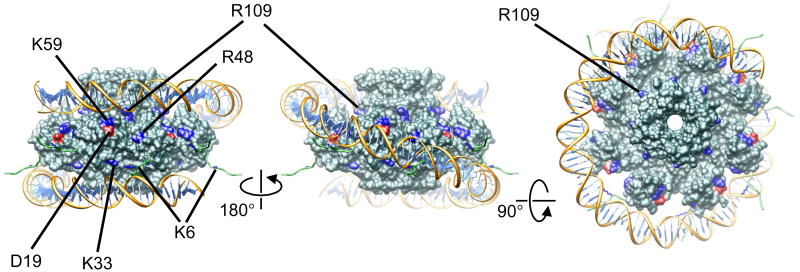
The multivalency of gp1:DNA binding and the spatial restraints for gp1:DNA interactions provided by our structure-directed mutagenesis analysis have enabled construction of a pseudo-atomic model for the gp1:DNA nucleoprotein complex (Figure 5). In this model, the bound DNA wraps around the gp1 octameric assembly. One and a quarter turns, or 120 bps, of DNA superhelix are modeled so that the upper half of the DNA makes multiple contacts with the top regions of DNA-binding domains where residues K59 are located, while the other half of the DNA makes contact with the bottom regions of the DNA-binding domains where residues K33 are located. Residues R48 are also in proximity with the DNA, contributing to protein:DNA interactions. The role of negatively charged residues D19 may be to help precisely position the DNA with respect to gp1. The N-terminal polypeptides (residues 1 to 9) harboring residue K6 form extended arms that may interact with DNA backbones or insert into DNA grooves, helping hold DNA in place. The role of the N-terminal arms is reminiscent of those of basic-residue-rich tails of histone proteins in the nucleosome core particle 38. This model of the gp1:DNA complex sheds light on the overall conformation of DNA as induced by gp1 binding and locates the sites on gp1 that makes physical contact with DNA, although the atomic details of protein:DNA interactions are hypothetical and are to be ultimately unveiled in a high-resolution structure of the gp1:DNA complex.
Figure 5.
The pseudo-atomic model of the nucleoprotein complex formed by gp1 (molecular surface in dark cyan) and the putative pac DNA (ribbon diagram with backbones in gold and base pairs in light blue). The amino acid residues that were subject to mutagenesis analysis are shown in blue for K6, K33, R48, K59 and R109 and in red for D19. The computationally modeled N-terminal nine residues of gp1 are shown as loops in green, and the side chains of K6 are shown in blue. One and a quarter turns, or 120 bps, of a DNA superhelix are modeled. The left and middle panels are side views, and the right panel is viewed down the 8-fold axis of the gp1 octamer.
This model is in good agreement with previous genetic and biochemical results on interactions of terminase small subunits with DNA. In phage SPP1, DNA-protection assay on the pac region bound by the terminase small subunit showed that hypersensitive sites for DNase digestion were distributed along the pac DNA with approximately 10-bp periodicity, which indicates that these hypersensitivity sites were all located on one side of the DNA double helix while the other side was protected presumably by bound proteins 33. Based on this, a nucleosome-like wrap-around mode for DNA binding was proposed 33. The length of the left pac region of SPP1 is 100 bps, which agrees well with the 96 bps per turn of the wrapped DNA superhelix in our model, as the length (thus the number of base pairs) per wrap of the DNA in such a binding mode is largely determined by the diameter of the terminase small subunit assembly, which is 92 Å and 90 Å in SPP1 G1P and Sf6 gp1 respectively. Interestingly, in phage P22, the initiation cleavage sites by the terminase large subunit were distributed over a 120-bp DNA region, and the high-frequency initiation cleavage sites showed an approximate periodicity of ∼20 base pairs 32, which may reflect the periodicity of the interactions of the terminase small subunit with the pac DNA, assuming the initiation cleavage by the terminase large subunit is directed by the interaction of the small subunit and DNA. Our gp1:DNA complex is consistent with the observation in P22, taking into consideration that the terminase large subunit that actually conducts the cleavage could be positioned with a periodicity directed by the small subunit. Recently, a wrap-around mode of DNA:terminase interaction was proposed based on the X-ray structure of the full-length terminase small subunit from a Siphovirus, although the limited resolution and anisotropy of X-ray diffraction data as well as ultra-high B-factors (average B-factor of 220 Å2 for the whole structure and 300.6 for one of the two modeled DNA-binding domains) indicated elusive DNA-binding domains and resulted in speculative positioning of DNA with respect to the terminase small subunit oligomer 21. While differences are present regarding the details of the positions and orientations of terminase small subunit DNA-binding domains with respect to bound DNA, our gp1:DNA complex model and the model proposed by Buttner et al 21 are consistent in formation of nucleosome-like complexes directed by terminase small subunit oligomers. The wrap-around mode is also supported by the ring-like structures of the terminase small subunit oligomerization domain of a T4-like phage 22. Thus, the nucleosome-like model may represent a common architecture for the mode of interaction between pac DNA and terminase small subunits that display ring-like oligomers in phages such as Sf6, P22, T4 and SPP1. In phage lambda, the terminase small subunit gpNu1 N-terminal DNA-binding domain formed a tight dimer, and the specific binding of gpNu1 with two DNA motifs in the cosB region was proposed to induce sharp DNA bending 35. It was reported that gpNu1 formed a heterotrimeric complex with the terminase large subunit gpA with a 2:1 molar ratio 23, and this complex further assembled into a larger complex with a gpNu1(8):gpA(4) stoichiometry 39. However, formation of high-order complexes of terminase small subunits with the cos DNA was speculated to occur in phage lambda 27. It would be interesting to examine if formation of the nucleosome-like complex by terminase small subunit and DNA is also applicable for phage lambda, which cleaves DNA at a precise site.
Multiple conformations have been observed for the gp1 DNA-binding domain in our structural studies. This structural plasticity is mediated by the curved helix α4 and the intermolecular interactions such as the intermolecular salt bridge R67:E73. The structural plasticity allows movement of gp1 DNA-binding domains that may be needed for formation of the gp1:DNA complex, as our pseudo-atomic model suggests pleomorphic interactions between DNA and the eight gp1 molecules. The recent X-ray structure of the full-length Siphovirus terminase small subunit shows disordered DNA-binding domains putatively due to flexible linkers with the oligomerization core 21. This supports structural dynamics of terminase small subunits, which may facilitate DNA binding through pleomorphic protein:DNA interaction.
Implications for assembly of the gp1:DNA nucleoprotein complex
Our results on the DNA-binding behavior of gp1 suggest a mechanism for the formation of the gp1:pac nucleoprotein complex. Assembly of the gp1:DNA complex may begin with binding of the gp1 octamer to one or a few high affinity sites in the packaging region on viral DNA, which promotes binding to multiple secondary sites in the proximity, leading to wrapping of DNA around the gp1 octamer. Each occurrence of DNA:gp1 binding event would bend the DNA only slightly (∼45° assuming an octamer is surrounded by a complete wrap of DNA). A combination of multiple DNA:gp1 binding events scaffolded by the gp1 octamer would lead to an overall drastic conformational change of DNA. This mode of protein:DNA interaction is energetically more favorable than, for instance, making sharper DNA bending such as a DNA U-turn using only two occurrences of protein:DNA contacts in a mechanism proposed for the phage lambda gpNu1 protein 35.
The 3D model of the gp1:DNA complex provides insights into the pac signals on viral DNA. Sf6 uses a headful DNA packaging mechanism 36 similar to phage P22 but unlike phage lambda in which DNA is cut at a precise site upon initiation and completion of each DNA packaging cycle. A DNA fragment in Sf6 genomic DNA was predicted as the potential pac signal based on the homology to the P22 pac, and is located within the gp1 coding region 36. The pac signal of P22 is also located in the coding region of the terminase small subunit. In phage T4, a 24-bp fragment within the terminase small subunit gp16 coding region was suggested to be the pac site. Hence, the DNA interactions observed in our electrophoretic mobility shift assay using DNA containing the gp1 coding sequence may at least partially reflect the binding of gp1 with the actual pac region of Sf6. However, readily recognizable, repeating DNA sequence motifs are absent in the pac regions in Sf6 and P22. On the other hand, our model of gp1:DNA binding implies sequence periodicity in bound DNA as imposed by the repeating gp1 molecules, although this periodicity may be promiscuous. These results suggest that the pac signals may not be present as readily discernible DNA sequence motif repeats, but may be present as specific patterns buried in the DNA sequence in a way similar to, for instance, the nucleosome-positioning sequence, which possess the capacity to bend and form specific, high-order nucleoprotein complexes with terminase small subunits. The pac fragments in P22 and Sf6 may represent a primary, high-affinity binding site, and binding to this site may promote further binding of the terminase small subunit to adjacent secondary sites in a cooperative manner as demonstrated in our in vitro studies of Sf6 gp1:DNA interactions. Such implications for the pac region organization are consistent with observations in several other phages. In phage SPP1 pac, three different DNA sequence motifs of 7 or 10 base pairs are present and each is repeated only twice, and there are no readily discernible DNA sequence motifs that are repeated with a frequency that matches the oligomerization stoichiometry of the terminase small subunit 33. The phage lambda pac region contains three repeats of a conserved DNA sequence motif, but exactly how this motif physically binds to the terminase small subunit gpNu1 hasn't been well established 35. In phage P1, the 161-bp pac region contains seven repeats of a nearly conserved hexanucleotide motif, four on the right side and three on the left side of the packaging cleavage sites 29. It would be interesting to inspect the viral DNA pac regions to detect potential buried DNA sequence patterns that are responsible for the interactions with the terminase small subunits, and this can help understand sequence-dependent DNA curvature and bending in general, whose mechanisms remain to be understood 37. Nevertheless, understanding this type of molecular determinants of protein:DNA interaction may require three-dimensional structural information.
Implications for assembly of the viral DNA-packaging molecular apparatus
Our 3D model of the gp1:DNA complex indicates that the bound DNA may adopt a specific 3D architecture that significantly deviates from an extended linear conformation. Such a DNA conformation is formed only when it binds to gp1, and may provide the binding site for the terminase large subunit gp2. This is consistent with previous work in which it was proposed that binding of terminase small subunits to phage DNA induced DNA bending or looping, which transformed the DNA into a mature form for cleavage by the terminase large subunits. Our results delineate a detailed three-dimensional picture for initiation of DNA packaging. The terminase small subunit forms a ring-like oligomer, which presents multiple DNA-binding domains at the periphery and functions as a DNA-spooling device. The pac region of the viral DNA then wraps around through multivalent protein:nucleic acid interactions to form a nucleosome-like complex. This step may involve cellular factors such as the integration host factor and/or the HU protein in the cases of phages lambda and P1 41, which can help modulate the DNA conformation to facilitate DNA wrapping. The complex of the terminase small subunit and DNA possesses the binding site for the terminase large subunit, which together form a ternary molecular apparatus consisting the two terminase subunits and DNA. The terminase large subunit is then activated to make the initial cleavage on DNA and generate the new DNA end that is to be threaded into the procapsid through the portal channel. Thus, the terminase small subunit plays a central role in initiation of viral DNA packaging in that it directs the assembly of the ternary pre-packaging complex at specific sites on viral DNA. Phage terminase small subunits may play additional crucial roles during the course of DNA translocation, for instance, stimulating the ATPase activity and prohibiting the nuclease activity of the large subunit. Whether those additional roles involve interactions of terminase small subunits with DNA, and if so, the nature of those interactions, is not known. It was proposed that two types of gp1:DNA interactions may exist, which involve the N-terminal DNA-binding domain during packaging initiation and the central channel of the gp1 ring-like assembly during DNA translocation, respectively 14. A recent in vitro study reported failure to form a complex of terminase small subunit with DNA inserted in the channel 21. However, the channels of the T4-like phage terminase small subunit oligomerization core 11-mer and 12-mer 22 and the SPP1-like phage oligomerization core 10-mer 21 are sufficiently wide to accommodate DNA. Thus, additional in vivo data are required to determine whether the channels are used during DNA translocation. Nevertheless, it is clear that the terminase small subunit is a versatile molecular regulator that plays multiple roles throughout the DNA packaging process.
Materials and Methods
Protein expression and purification
The pET28b constructs containing inserts for wt-gp1 and gp1 mutants (K59E, K59A, K6E, D19R, R48A, K33E, R109E and E73A) were transformed into E. coli strain BL21(DE3) pLysS (Novagen). The recombinant cells were grown to OD600 of 0.5 and induced with 1mM IPTG at 30°C for 3 hours to over-express proteins. Cells were harvested by centrifugation at 6,000 g for 10 min, washed and frozen at -20°C. Cells were thawed, lysed with a French press and centrifuged, and the supernatant was passed through a Ni-NTA column. The proteins were eluted with a gradient of 50-500 mM imidazole, the peak fractions were collected and concentrated to 4 ml and loaded onto a size-exclusion chromatographic column (Sephacryl S- 300, GE Healthcare). The peak fractions were combined, concentrated to ∼12 mg/ml, and stored at 4°C.
Electrophoretic mobility shift assay
Two linear DNA fragments were used for electrophoretic mobility shift assay (EMSA). A 1,836-bp DNA fragment corresponding to the coding regions of gp1 and gp2 in the Sf6 genome which may harbor the packaging signal 36 was generated by polymerase chain reaction (PCR) using oligonucleotides 5′-CCA TGG CGA CTG AAC CAA AAG-3′ and 5′- TTA CCA ACC GGA GGA TGA GGG-3′ as primers, and the PCR product was purified by Qiaquick PCR purification kit (Qiagen). The 200-bp DNA fragment at the 5′ end of the gp1 gene was generated with the same method. For the 200-bp DNA, the purified gp1 at concentrations of 1.76, 3.52, 7.05, 14.10, 28.19, 56.39, 112.78 and 225.56 μM was incubated with DNA at approximately 150 nM in 12 μl reaction solution containing 10 mM Tris-HCl pH 7.8, 100 mM NaCl and 10mM MgCl2 at room temperature for 30 minutes, and was loaded onto 1% (w/v) agarose gel followed by ethidium bromide staining. For the 1836-bp DNA, gp1 at concentrations of 1.76, 3.52, 7.05, 14.10, 17.59, 21.15, 28.19, 56.39, 112.78 and 225.56 μM was incubated with DNA at approximately 25 nM. For EMSA of gp1 with plasmid DNA, the plasmid DNA pET20b were purchased from Novagen and amplified in NovaBlue cells (Novagen) and purified by plasmid maxi kit (Omega Biotek). The concentration of plasmid DNA used in EMSA was roughly 15nM. All the gp1 concentrations were calculated as a monomer. The bands of free DNA in gels were quantified with an AlphaImager EP gel documentation system (Cell Biosciences) for calculation of the fractions of bound DNA. The degree of cooperativity was calculated using the Hill equation as described in Lehninger's Principles of Biochemistry, by Nelson and Cox, fifth edition, pp.162-165.
Crystallization, data collection and structure determination
The crystals of gp1-FL, gp1-FL-CAPS and gp1 mutants (K59E, D19R and R48A) were grown at 20°C by vapor diffusion in hanging drops containing 1ul of protein solution at ∼12 mg/ml and an equal volume of well solution containing 0.2M Li2SO4, 1.2M NaH2PO4 and 0.8M K2HPO4 with (for gp1-FL-CAPS) or without (for all other proteins) 100mM CAPS pH10.8, representing a final pH of 5.92 and 6.05 in the crystal droplets, respectively. The space group of all crystals is P4212. Freezing of crystals in cryogenic protectants and X-ray data collection and processing were performed essentially as described 14. The structures were determined with the molecular replacement method using the previous Sf6 gp1 structure (RCSB PDB accession code 3HEF) as the initial model 14. Model building and refinement were performed with the programs COOT 43 and PHENIX 44, respectively. X-ray data collection and structure refinement statistics are summarized in Table 1. For the 1.5 Å resolution structure of gp1-FL, the anisotropic B-factor refinement, the TLS refinement and the “riding hydrogen” model were used for protein atoms, whereas only isotropic B-factor refinement and the TLS refinement were used for other structures. The 8-fold rotational axis of the gp1 octamer coincides with the crystallographic 4fold axis, and the crystallographic asymmetric unit contains two molecules of gp1, namely A and B. No non-crystallographic symmetry restraints were imposed, as the data-to-parameter ratio was sufficiently large. The refined models exhibit excellent electron density. In all structures, residues 1-9 in both molecules, residues 133-140 in Molecule A and the C-terminal residue 140 in Molecule B are disordered, thus not modeled.
Modeling of the protein:DNA complex
A left-handed DNA superhelix was generated computationally with the aid of the program X3DNA 45 using standard B-form dsDNA parameters. The DNA model was docked onto the gp1 octamer manually, taking into consideration the spatial restraints derived from the mutagenesis and EMSA results and avoiding steric clashes between DNA and protein. The pitch of the DNA superhelix was adjusted to yield the optimal docking so that DNA phosphate backbones were within interacting distance with those DNA-interacting residues as defined in the mutagenesis studies. In the final pseudo-atomic model of the gp1:DNA complex, the DNA superhelix has a radius of 51.5 Å and a pitch of 35 Å with 96 basepairs per turn, corresponding to 12 basepairs per gp1 molecule. The N-terminal arms of gp1 comprising residues 1-9 that were disordered in the X-ray structures but harbors residue K6 were modeled with COOT 43. There were no steric clashes between DNA and gp1.
Supplementary Material
Supplemental Figures: Figure S1. Binding of Sf6 gp1 with 200-bp linear DNA. Lane 1, 1kb plus DNA ladder (Invitrogen). Lane 2, 200-bp DNA alone. Lanes 3-10, DNA incubated with gp1 at a concentration of 1.76, 3.52, 7.05, 14.10, 28.19, 56.39, 112.78 and 225.56 μM respectively.
Figure S2. Binding of Sf6 gp1 mutants with linear DNA. Lane 1 in all gels is 1kb plus DNA ladder (Invitrogen).
(A) Lane 2, 1836-bp DNA alone. Lanes 3-6, DNA incubated with wt-gp1 at 28.19, 56.39, 84.58 and 112.78 μM respectively. Lanes 7-10, the same as Lanes 3-6 except that gp1-K33E was used.
(B) Lane 2, 1836-bp DNA alone. Lanes 3-4, DNA incubated with wt-gp1 at 56.39 and 112.78 μM respectively. Lanes 5-6, 7-8 and 9-10, the same as Lanes 3-4 except that gp1-R48A, gp1-K6E and gp1-R109E were used respectively.
(C) Binding of gp1-D19R with DNA. Lanes 2, 423-bp DNA alone (DNA1), which corresponds to the coding sequence of gp1. Lanes 3-6, DNA incubated with gp1-D19R at 28.19, 56.39, 84.58 and 112.78 μM respectively. Lanes 7-11, the same as Lanes 2-6 except that the 1413-bp gp2 coding DNA (DNA2) was used. Lanes 12-16, the same as Lanes 2-6 except that the 1836-bp gp1 and gp2 coding DNA (DNA3) was used. Lane 17-19, the three DNA molecules were incubated with wt-gp1 at 56.39 μM respectively. Notice a fraction of DNA remains unbound in Lanes 17-19, whereas unbound DNA is significantly less in Lanes 4, 9 and 14 in which protein concentrations were the same.
Figure S3. Binding of Sf6 wt-gp1 and gp1-E73A with linear DNA. Lanes 1 and 11, 1kb plus DNA ladder (Invitrogen). Lanes 2 and 12, DNA alone. Lanes 3-10, DNA incubated with wt-gp1 at a concentration of 1.76, 3.52, 7.05, 14.10, 28.19, 56.39, 112.78 and 225.56 μM respectively. Lanes 13-16, DNA incubated with gp-E73A at a concentration of 28.19, 56.39, 112.78 and 225.56 μM respectively. Notice that the vast majority of DNA are bound in Lane 7, whereas a significant fraction of DNA remain unbound in Lane 13.
Highlights.
The terminase small subunit gp1 directs initiation of viral DNA packaging;
DNA-interacting regions were mapped on the gp1 octameric assembly;
DNA wraps around the gp1 octamer to form a nucleosome-like complex;
Structural plasticity of gp1 underlies pleomorphic protein:DNA interactions;
The gp1 serves as a DNA-spooling device during initiation of DNA packaging.
Acknowledgments
We thank the staff at the Stanford Synchrotron Radiation Laboratory for assistance in X-ray data collection. This work was supported by the NIH grant R01GM090010 to L.T.
Abbreviations
- dsDNA
double-stranded DNA
- DBD
DNA-binding domain
- bp
base pair
- wt
wild-type
- 3D
three-dimensional
- IPTG
isopropyl-beta-D-thiogalactopyranoside
Footnotes
Accession Numbers: The coordinates and structure factors have been deposited with the Worldwide Protein Data Bank under the accession codes of 4DYQ, 4DZP, 4DYC, 4DZJ and 4DYR for gp1-FL, gp1-R48A, gp1-D19R, gp1-K59E and gp1-FL-CAPS, respectively.
Supplemental data including 3 figures are available online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 2.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 3.Homa FL, Brown JC. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 4.Ostapchuk P, Hearing P. Control of adenovirus packaging. J Cell Biochem. 2005;96:25–35. doi: 10.1002/jcb.20523. [DOI] [PubMed] [Google Scholar]
- 5.Ewing SG, Byrd SA, Christensen JB, Tyler RE, Imperiale MJ. Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins. J Virol. 2007;81:12450–12457. doi: 10.1128/JVI.01470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koonin EV, Senkevich TG, Chernos VI. Gene A32 product of vaccinia virus may be an ATPase involved in viral DNA packaging as indicated by sequence comparisons with other putative viral ATPases. Virus Genes. 1993;7:89–94. doi: 10.1007/BF01702351. [DOI] [PubMed] [Google Scholar]
- 7.Cassetti MC, Merchlinsky M, Wolffe EJ, Weisberg AS, Moss B. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson GM, Jindal HK, Yeung DE, Chen W, Astell CR. Expression of minute virus of mice major nonstructural protein in insect cells: purification and identification of ATPase and helicase activities. Virology. 1991;185:90–98. doi: 10.1016/0042-6822(91)90757-3. [DOI] [PubMed] [Google Scholar]
- 9.Gorbalenya AE, Koonin EV, Wolf YI. A new superfamily of putative NTP- binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 10.King JA, Dubielzig R, Grimm D, Kleinschmidt JA. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 2001;20:3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catalano CE. Viral genome packaging machines: genetics, structure, and mechanism. Landes Bioscience/Eurekah.com; Kluwer Academic/Plenum Publishers, Georgetown, Tex; New York, N.Y: 2005. [Google Scholar]
- 12.Ghosh-Kumar M, Alam TI, Draper B, Stack JD, Rao VB. Regulation by interdomain communication of a headful packaging nuclease from bacteriophage T4. Nucleic acids research. 2011;39:2742–2755. doi: 10.1093/nar/gkq1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S, Rao VB. Specificity of interactions among the DNA-packaging machine components of T4-related bacteriophages. The Journal of biological chemistry. 2011;286:3944–3956. doi: 10.1074/jbc.M110.196907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, Casjens SR, Tang L. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc Natl Acad Sci U S A. 2010;107:1971–1976. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J Biol Chem. 1997;272:3495–3501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- 16.Nemecek D, Lander GC, Johnson JE, Casjens SR, Thomas GJ., Jr Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J Mol Biol. 2008;383:494–501. doi: 10.1016/j.jmb.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camacho AG, Gual A, Lurz R, Tavares P, Alonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J Biol Chem. 2003;278:23251–23259. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- 18.White JH, Richardson CC. Gene 18 protein of bacteriophage T7. Overproduction, purification, and characterization. J Biol Chem. 1987;262:8845–8850. [PubMed] [Google Scholar]
- 19.Roy A, Bhardwaj A, Cingolani G. Crystallization of the nonameric small terminase subunit of bacteriophage P22. Acta crystallographica Section F, Structural biology and crystallization communications. 2011;67:104–110. doi: 10.1107/S174430911004697X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Duijn E. Current limitations in native mass spectrometry based structural biology. J Am Soc Mass Spectrom. 2010;21:971–978. doi: 10.1016/j.jasms.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 21.Buttner CR, Chechik M, Ortiz-Lombardia M, Smits C, Ebong IO, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, Alonso JC, Antson AA. Structural basis for DNA recognition and loading into a viral packaging motor. Proc Natl Acad Sci U S A. 2012;109:811–816. doi: 10.1073/pnas.1110270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc Natl Acad Sci U S A. 2012;109:817–822. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maluf NK, Yang Q, Catalano CE. Self-association properties of the bacteriophage lambda terminase holoenzyme: implications for the DNA packaging motor. J Mol Biol. 2005;347:523–542. doi: 10.1016/j.jmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 25.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The Bacteriophages. Vol. 1. Plenum Press; New York City: 1988. pp. 15–91. [Google Scholar]
- 26.Adelman K, Salmon B, Baines JD. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc Natl Acad Sci U S A. 2001;98:3086–3091. doi: 10.1073/pnas.061555698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker A, Murialdo H. Bacteriophage lambda DNA: the beginning of the end. J Bacteriol. 1990;172:2819–2824. doi: 10.1128/jb.172.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Catalano CE. The terminase enzyme from bacteriophage lambda: a DNA- packaging machine. Cell Mol Life Sci. 2000;57:128–148. doi: 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sternberg N, Coulby J. Recognition and cleavage of the bacteriophage P1 packaging site (pac). II. Functional limits of pac and location of pac cleavage termini. Journal of molecular biology. 1987;194:469–479. doi: 10.1016/0022-2836(87)90675-9. [DOI] [PubMed] [Google Scholar]
- 30.Skorupski K, Sternberg N, Sauer B. Purification and DNA-binding activity of the PacA subunit of the bacteriophage P1 pacase enzyme. Journal of molecular biology. 1994;243:258–267. doi: 10.1006/jmbi.1994.1652. [DOI] [PubMed] [Google Scholar]
- 31.Sternberg N, Coulby J. Cleavage of the bacteriophage P1 packaging site (pac) is regulated by adenine methylation. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8070–8074. doi: 10.1073/pnas.87.20.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Sampson L, Parr R, Casjens S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol Microbiol. 2002;45:1631–1646. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- 33.Chai S, Lurz R, Alonso JC. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. J Mol Biol. 1995;252:386–398. doi: 10.1006/jmbi.1995.0505. [DOI] [PubMed] [Google Scholar]
- 34.Roizman B, Knipe DM. Herpes simplex virus. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields' virology 5 edit. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2501–2601. [Google Scholar]
- 35.de Beer T, Fang J, Ortega M, Yang Q, Maes L, Duffy C, Berton N, Sippy J, Overduin M, Feiss M, Catalano CE. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol Cell. 2002;9:981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 36.Casjens S, Winn-Stapley DA, Gilcrease EB, Morona R, Kuhlewein C, Chua JE, Manning PA, Inwood W, Clark AJ. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J Mol Biol. 2004;339:379–394. doi: 10.1016/j.jmb.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 37.Peters JP, 3rd, Maher LJ. DNA curvature and flexibility in vitro and in vivo. Q Rev Biophys. 2010;43:23–63. doi: 10.1017/S0033583510000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 39.Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- 40.Lin H, Black LW. DNA requirements in vivo for phage T4 packaging. Virology. 1998;242:118–127. doi: 10.1006/viro.1997.9019. [DOI] [PubMed] [Google Scholar]
- 41.Skorupski K, Sauer B, Sternberg N. Faithful cleavage of the P1 packaging site (pac) requires two phage proteins, PacA and PacB, and two Escherichia coli proteins, IHF and HU. Journal of molecular biology. 1994;243:268–282. doi: 10.1006/jmbi.1994.1653. [DOI] [PubMed] [Google Scholar]
- 42.Al-Zahrani AS, Kondabagil K, Gao S, Kelly N, Ghosh-Kumar M, Rao VB. The small terminase, gp16, of bacteriophage T4 is a regulator of the DNA packaging motor. The Journal of biological chemistry. 2009;284:24490–24500. doi: 10.1074/jbc.M109.025007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 45.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures: Figure S1. Binding of Sf6 gp1 with 200-bp linear DNA. Lane 1, 1kb plus DNA ladder (Invitrogen). Lane 2, 200-bp DNA alone. Lanes 3-10, DNA incubated with gp1 at a concentration of 1.76, 3.52, 7.05, 14.10, 28.19, 56.39, 112.78 and 225.56 μM respectively.
Figure S2. Binding of Sf6 gp1 mutants with linear DNA. Lane 1 in all gels is 1kb plus DNA ladder (Invitrogen).
(A) Lane 2, 1836-bp DNA alone. Lanes 3-6, DNA incubated with wt-gp1 at 28.19, 56.39, 84.58 and 112.78 μM respectively. Lanes 7-10, the same as Lanes 3-6 except that gp1-K33E was used.
(B) Lane 2, 1836-bp DNA alone. Lanes 3-4, DNA incubated with wt-gp1 at 56.39 and 112.78 μM respectively. Lanes 5-6, 7-8 and 9-10, the same as Lanes 3-4 except that gp1-R48A, gp1-K6E and gp1-R109E were used respectively.
(C) Binding of gp1-D19R with DNA. Lanes 2, 423-bp DNA alone (DNA1), which corresponds to the coding sequence of gp1. Lanes 3-6, DNA incubated with gp1-D19R at 28.19, 56.39, 84.58 and 112.78 μM respectively. Lanes 7-11, the same as Lanes 2-6 except that the 1413-bp gp2 coding DNA (DNA2) was used. Lanes 12-16, the same as Lanes 2-6 except that the 1836-bp gp1 and gp2 coding DNA (DNA3) was used. Lane 17-19, the three DNA molecules were incubated with wt-gp1 at 56.39 μM respectively. Notice a fraction of DNA remains unbound in Lanes 17-19, whereas unbound DNA is significantly less in Lanes 4, 9 and 14 in which protein concentrations were the same.
Figure S3. Binding of Sf6 wt-gp1 and gp1-E73A with linear DNA. Lanes 1 and 11, 1kb plus DNA ladder (Invitrogen). Lanes 2 and 12, DNA alone. Lanes 3-10, DNA incubated with wt-gp1 at a concentration of 1.76, 3.52, 7.05, 14.10, 28.19, 56.39, 112.78 and 225.56 μM respectively. Lanes 13-16, DNA incubated with gp-E73A at a concentration of 28.19, 56.39, 112.78 and 225.56 μM respectively. Notice that the vast majority of DNA are bound in Lane 7, whereas a significant fraction of DNA remain unbound in Lane 13.