Summary
Histone H3K4 methylation is associated with active genes and, along with H3K27 methylation, is part of a bivalent chromatin mark that typifies poised developmental genes in embryonic stem cells (ESCs). However, its functional roles in ESC maintenance and differentiation are not established. Here we show that mammalian Dpy-30, a core subunit of the SET1/MLL histone methyltransferase complexes, modulates H3K4 methylation in vitro, and directly regulates chromosomal H3K4 trimethylation (H3K4me3) throughout the mammalian genome. Depletion of Dpy-30 does not affect ESC self-renewal, but significantly alters the differentiation potential of ESCs, particularly along the neural lineage. The differentiation defect is accompanied by defects in gene induction and in H3K4 methylation at key developmental loci. Our results strongly indicate an essential functional role for Dpy-30 and SET1/MLL complex-mediated H3K4 methylation, as a component of the bivalent mark, at developmental genes during the ESC fate transitions.
Introduction
Embryonic stem (ES) cells have two key properties: self-renewal, the capability of maintaining cellular identity after each division, and pluripotency, the capacity to differentiate into all cell types. How pluripotency is maintained and executed at the molecular level remains a central question in ESC biology (Jaenisch and Young, 2008; Niwa, 2007). Posttranslational modifications of histone proteins are thought to be important epigenetic events intimately associated with transcription regulation for both of these processes (Jenuwein and Allis, 2001; Spivakov and Fisher, 2007; Szutorisz and Dillon, 2005; Niwa, 2007). Prominent histone modifications include H3K4 methylation, implicated in transcriptional activation and deposited by Trithorax group proteins, and H3K27 methylation, implicated in transcriptional repression and deposited by Polycomb group proteins (reviewed in Kouzarides, 2007).
In undifferentiated ESCs, pluripotency maintenance genes (e.g., Nanog, Oct4, and Sox2) are marked with high levels of H3K4 methylation at their transcriptional start sites (TSSs) (Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007). Many developmental regulatory gene loci, however, are marked with both H3K4 and H3K27 methylation, the so-called “bivalent marks” (Azuara et al., 2006; Bernstein et al., 2006; Pan et al., 2007). The combination of the seemingly “conflicting” marks suggests that these genes are kept silenced by H3K27 methylation in ESCs, while remaining “poised” for expression events that are presumably dependent upon H3K4 methylation. This poised state was proposed to be central both for the maintenance of the ground state and for the developmental potential of ESCs. The repressive function of H3K27 methylation at the lineage-specific loci is supported by the aberrant expression of these target genes in ESCs lacking key subunits of the PRC2 H3K27 methyltransferase complex (Azuara et al., 2006; Boyer et al., 2006; Lee et al., 2006). However, functional roles for H3K4 methylation in ESCs lack experimental support, despite the association of H3K4 methylation, particularly trimethylation, with active gene expression (Sims et al., 2003). Specifically, it remains unknown whether efficient H3K4 methylation is important either for maintaining expression of stemness genes or for induction of lineage-specific genes during differentiation of ESCs.
In mammalian cells, SET1/MLL family complexes (hereafter, MLL complexes) are important enzymes catalyzing H3K4 methylation. Apart from some specialized subunits, they contain either hSET1, MLL1, MLL2, MLL3, or MLL4 as the catalytic subunit and WDR5, RbBP5, and Ash2L as integral core subunits that are necessary for the methylation activity of the complexes (Dou et al., 2006). Deletion of any one of the MLL family members usually has minimal effects on the global levels of H3K4 methylation likely due to redundancy among the MLL complexes (Lubitz et al., 2007; Wang et al., 2009). In this regard, loss of MLL2 in mouse ESCs leads to skewed differentiation, but evidence for a connection to H3K4 methylation is weak (Lubitz et al., 2007). MLL1-deficient ESCs are defective in hematopoiesis but, for similar reasons, it is not known if H3K4 methylation is directly involved (Ernst et al., 2004). There are no reports regarding ESCs deficient in MLL3, MLL4, or SET1. In contrast, depletion of any of the core subunits effectively reduces the global level of H3K4 methylation (Dou et al., 2006). However, severe loss of H3K4 methylation could potentially affect cell viability and make it difficult to proceed with further biological analyses or to interpret the results. In order to facilitate a genetic approach, we sought a subunit of MLL complexes whose loss would significantly reduce, but not eliminate, H3K4 methylation activity. Dpy-30 emerged as a good candidate in this sense. Originally discovered as a gene essential for dosage compensation in C. elegans (Hsu and Meyer, 1994), Dpy-30 also plays important roles in worm development and behavior (Hsu et al., 1995) through mechanisms that remain unknown. The Dpy-30 homolog in fission yeast S. pombe, SDC1, encodes an integral subunit of the Set1 complex and is important for global H3K4 methylation, but its deletion has less severe effects than deletions of other core subunits (Dehe et al., 2006). In mammals, the homolog of Dpy-30 directly binds to Ash2L and is a common subunit of all of the MLL complexes (Cho et al., 2007), but its function has never been reported.
Here, we show that mammalian Dpy-30 directly regulates H3K4 methylation by MLL family complexes both in vitro and genome-wide in vivo. We then demonstrate that depletion of Dpy-30 or RbBP5 in mouse ESCs leads to a defect in lineage specification that is accompanied by reduced H3K4 methylation and impaired plasticity in transcriptional reprogramming, but does not significantly affect ESC self-renewal. Our data provide strong experimental evidence in support of critical and relatively specific functional roles of MLL complexes and the associated H3K4 methylation in activating the poised developmental genes during ESC fate transitions.
Results
Mammalian Dpy-30 Enhances H3K4 Methylation by MLL Complexes In Vitro
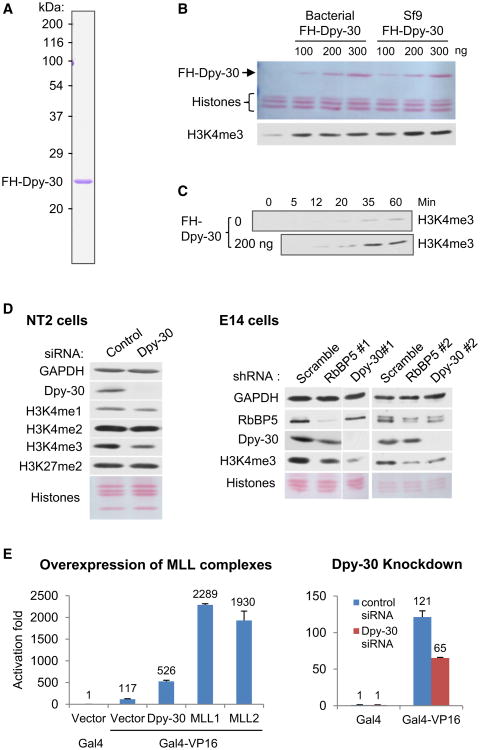
To assess the biochemical activity of Dpy-30, we performed in vitro histone methylation assays with a purified MLL2 core complex. FLAG-HA-tagged human Dpy-30 (FH-Dpy-30) was purified to near homogeneity from bacteria (data not shown) or baculovirus-infected Sf9 cells (Figure 1A). A saturating level of Dpy-30 stimulated H3K4 methylation several fold (Figure 1B), indicating that Dpy-30 has a significant yet limited activity in stimulating methylation by the MLL2 complex. This contrasts with more critical roles for WDR5 and RbBP5 in H3K4 methylation by MLL1 (Dou et al., 2006). A kinetic analysis demonstrated that Dpy-30 significantly enhances the rate of H3K4 trimethylation by the MLL2 core complex (Figure 1C). Because Dpy-30 directly binds to Ash2L and is shared by all MLL complexes (Cho et al., 2007), it is likely that Dpy-30 regulates H3K4 methylation by all MLL complexes. The general composition of MLL complexes was also confirmed in mammalian ESCs, of interest here, by co-immunoprecipitation of RbBP5 with other common core subunits and two catalytic subunits, MLL1 and MLL2, from the nuclear extract of the mouse ESC line E14TG2A (E14) (Figure S1A, available online).
Figure 1. Human Dpy-30 Is Important for Efficient H3K4 Methylation In Vitro and In Vivo.
(A) Coomassie staining of FH-Dpy-30 purified from virally infected Sf9 cells.
(B) Effect of Dpy-30 on H3K4me3 by an MLL2 core complex. An increasing amount of FH-Dpy-30 purified from either bacteria or Sf9 cells was added as indicated. Histones and FH-Dpy-30 were detected by Ponceau S staining, while methylation signals in B, C, and D were detected by immunoblot.
(C) Kinetic analysis of in vitro methylation by an MLL2 core complex in the absence or presence of Dpy-30 purified from Sf9 cells.
(D) Effect of RNAi-mediated Dpy-30 or RbBP5 knockdown on global H3K4 methylation level in NT2 (left) and E14 (right) cell lines. Proteins or histone modifications were detected by immunoblot.
(E) Effects of overexpression of components of MLL complexes (left panel) or effects of Dpy-30 knockdown (right panel) on Gal4-VP16 mediated activation of an integrated reporter. Averages ± SD from duplicate samples are plotted.
See also Figure S1.
Mammalian Dpy30 Is Required for Efficient H3K4 Methylation and Reporter Gene Expression in Cells
Knockdown of Dpy-30 by small interfering RNAs (siRNAs) in MCF7 (Figure S1B), a human breast cancer cell line, and in NT2 (Figure 1D, left), a human embryonic carcinoma (EC) cell line resulted in a significant reduction of H3K4 di- and tri-methylation (H3K4me3), indicating that Dpy-30 is important for maximal global methylation of H3K4 in human cells. Consistent with the observations in yeast (Dehe et al., 2006), this effect of Dpy-30 depletion on H3K4me3 appears less significant than that of RbBP5 or WDR5 depletion (Dou et al., 2006).
We extended these observations to ESCs via Dpy-30 depletion by two lenti-viruses expressing short hairpin RNAs (shRNAs) against two different sequences of mouse Dpy-30 (Dpy-30#1 and #2). Viruses expressing nonhairpin (NH) and scrambled control shRNA sequences, as well as shRNAs against mouse RbBP5 (two sequences: RbBP5#1 and #2), were also constructed for control and later functional studies. Dpy-30#1/#2 or RbBP5#1/#2 shRNAs effectively knocked down expression of the shRNA target genes and correspondingly reduced global H3K4me3 (Figure 1D, right) in E14 cells. Among the two shRNAs for each gene, the more effective ones, Dpy-30#1 and RbBP5#1 (Figure S1C) (hereafter, Dpy-30 and RbBP5, respectively), were selected for most further studies. The knockdown efficiency was significantly higher for Dpy-30 than for RbBP5 (Figure S1C), such that the reduction of methylation also appeared more effective for Dpy-30 (Figure 1D, right).
To investigate the basic effect of MLL complex-associated H3K4 methylation on transcription in mammalian cells, we employed a 293T cell line that contains a chromosomally integrated luciferase reporter gene downstream of five tandem Gal4-binding sites. In control cells, expression of a Gal4-VP16 fusion protein consisting of the Gal4 DNA binding and VP16 transactivation domains strongly activated luciferase expression. Overexpression of MLL complex components including Dpy-30, MLL1 or MLL2 all significantly enhanced the reporter expression, while siRNA-mediated knockdown of Dpy-30 significantly reduced activator-dependent luciferase expression (Figure 1E). These results implicate a positive role of Dpy-30 and MLL complex-mediated H3K4 methylation in chromosomal gene transcription.
Dpy-30 Regulates Chromosomal H3K4me3 throughout the Genome of Mouse ESCs
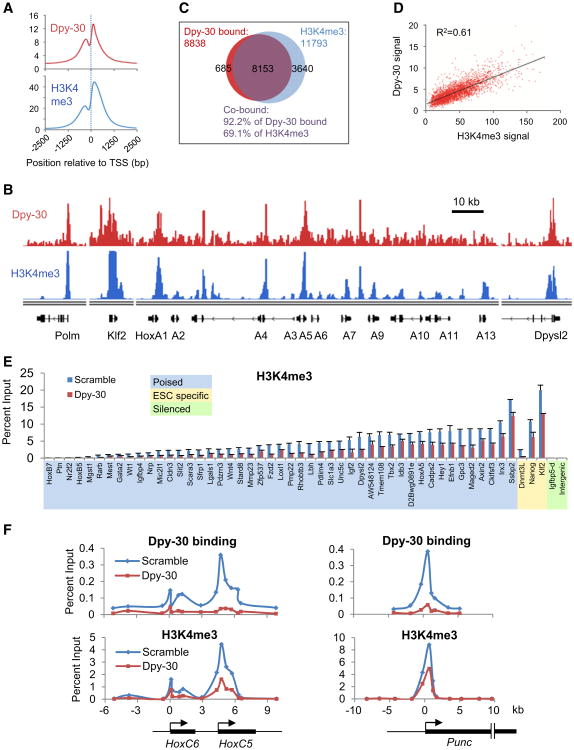
To firmly establish a role for Dpy-30 in regulating genomic H3K4 methylation, we wished to examine (1) the relationship of Dpy-30 and H3K4me3 enrichment across the ESC genome, and (2) the changes in H3K4me3 levels at individual genes upon depletion of Dpy-30 in ESCs. These goals were achieved through a combination of genome-wide analysis by ChIP-seq and more quantitative analysis at specific loci by ChIP-qPCR. The specificity of our anti-Dpy-30 antibody in ChIP assays was first validated by the significant reduction of the Dpy-30 ChIP signals on all of the monitored gene loci upon Dpy-30 depletion (Figure S2A).
Dpy-30 ChIP-seq results revealed that Dpy-30 is highly enriched in gene promoter regions and 5′UTRs, but not in downstream regions of genes or 3′UTRs (Figure S2B). It is also evident from the parallel H3K4me3 ChIP-seq that Dpy-30 binding and H3K4me3 enrichment share an almost identical profile on a composite gene representing the average of all of the marked loci in the whole genome (Figure 2A). Both signals are enriched within 1kb regions upstream and downstream of TSSs, show a major dip slightly upstream of TSSs (coincident with sites of nucleosome depletion), and peak at the same site around 150 bp downstream of TSSs (Figure 2A). A more detailed comparison on the genome browser further showed their strong and genome-wide overlap in both peak distribution and relative heights (Figure 2B and Figure S2C). A statistical analysis revealed that the vast majority (92.2%) of the Dpy-30-occupied regions were also marked with significant levels of H3K4me3 and, conversely, that most (69.1%) H3K4me3-enriched regions were occupied by Dpy-30 (Figure 2C). The detected Dpy-30-bound regions are presumably under-estimated due to the relative technical difficulty in cross-linking of Dpy-30 (which most likely does not directly contact DNA). A scatter plot analysis has further revealed a strong yet quantitatively imperfect correlation of the magnitude of the Dpy-30 occupancy versus H3K4me3 enrichment on all of the marked genes in ESC genome, including the bivalently marked genes (Figure 2D) and the genes marked with H3K4me3 but not H3K27me3 (Figure S2D).
Figure 2. Dpy-30 Regulates Chromosomal H3K4me3 throughout the Genome of Mouse ESCs.
(A) Composite profiling of Dpy-30 (top) and H3K4me3 (bottom) ChIP signals around TSSs as determined by ChIP-seq. Average ChIP-seq signals of all of the H3K4me3 enriched genes are depicted.
(B) ChIP-seq signals of Dpy-30 and H3K4me3 in the genome browser for representative gene loci that include a house-keeping gene (polm, DNA polymerase μ), an ESC-specific gene (Klf2), and some bivalent developmental genes (HoxA cluster and Dpysl2).
(C) Venn diagram showing the overlap of genes occupied by Dpy-30 and genes enriched with H3K4me3 at high confidence in ESCs as determined by ChIP-seq.
(D) Quantitative correlation of signal strength of Dpy-30 binding and H3K4me3 enrichment at TSSs of all of the bivalent genes as determined by ChIP-seq. The linear regression trendline and the correlation coefficient-square are displayed.
(E) Dpy-30 dependence of H3K4me3 at a large panel of selected gene TSSs or genomic regions. Igfbp5-d is a region downstream of the Igfbp5 gene locus. The poised developmental genes are sorted by their H3K4me3 levels. H3K4me3 was determined by ChIP-qPCR in control (Scramble) or Dpy-30-depleted (Dpy-30) ESCs. Averages ± SD from duplicate reactions are plotted.
(F) In situ impact of Dpy-30 binding on H3K4me3 at the broad regions of HoxC6, HoxC5 and Punc genes. Dpy-30 binding and H3K4me3 were determined by ChIP-qPCR in control or Dpy-30-depleted ESCs.
See also Figure S2.
To quantitatively confirm the ChIP-seq results, we performed ChIP-qPCR for Dpy-30 and H3K4me3 on genomic regions that included TSSs of several highly expressed house-keeping genes and ESC-specific genes, a few silent regions and, of most relevance in this work, TSSs of a large panel of poised developmental genes (randomly picked from the group of genes that were highly induced by RA-mediated differentiation as described later in this work).The results (Figure S2E) confirmed the overall correlation between Dpy-30 binding and H3K4me3 as seen in ChIP-seq assays.
To determine a causal relationship of Dpy-30 binding and chromosomal H3K4me3, we next examined by ChIP-qPCR the effect of Dpy-30 depletion on H3K4me3 at the entire panel of developmental genes mentioned above and a subset of the ESC-specific TSSs and silent regions. On all of the monitored genes, H3K4me3 was significantly reduced upon depletion of Dpy-30 (Figure 2E); and on the vast majority of the developmental genes, the reduction ranged from 1.5 to 4 fold. This indicates a relatively universal, yet quantitatively variable, dependence of H3K4me3 on Dpy-30 for all marked genes. When the broad TSS-proximal regions of some developmental genes, such as HoxC6, HoxC5, and Punc, were examined by ChIP-qPCR with primer series, it immediately became obvious that the H3K4me3 profiles are true and direct footprints of Dpy-30, as indicated by their almost identical patterns across these regions (Figure 2F). Importantly, H3K4me3 was most significantly affected at precisely the positions where the most profound depletion of the bound Dpy-30 occurred around the TSSs of these genes (Figure 2F). These results demonstrate that Dpy-30 in situ (therefore, directly) impacts H3K4me3 level in the ESC genome.
The collective evidence of the biochemical activity and the genome-wide distribution and importance of Dpy-30 for H3K4me3 establishes a direct and causal role for Dpy-30 in the regulation of MLL complex-mediated H3K4 methylation throughout the ESC genome. Having laid this foundation, we then employed Dpy-30 depletion, sometimes in conjunction with RbBP5 depletion, to examine the role of H3K4 methylation in the maintenance and execution of the pluripotency of ESCs.
Normal Levels of Mammalian Dpy-30 and RbBP5 Are Not Essential for ESC Self-Renewal or Stress Responses
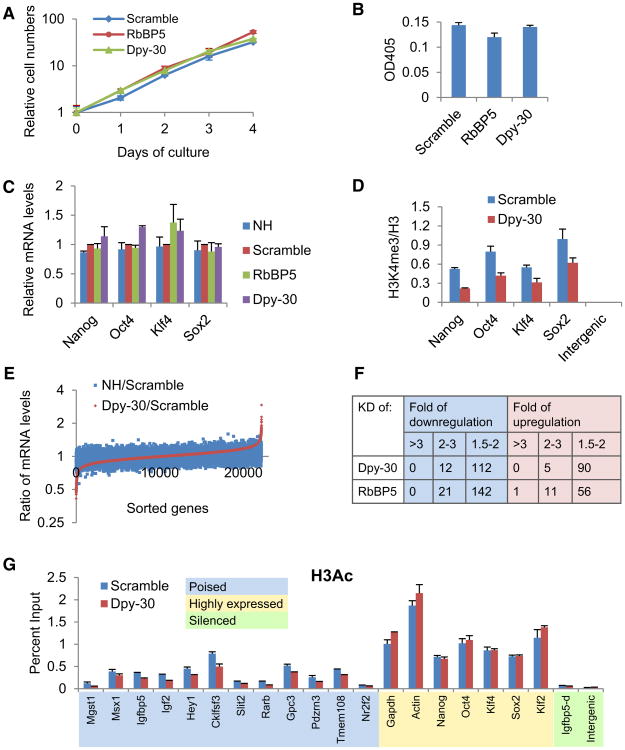
Several observations suggest that ESCs are able to maintain largely normal properties of self-renewal after partial depletion of RbBP5 or Dpy-30. First, the depleted cells exhibited the aggregated morphology typical of undifferentiated ESCs (data not shown). Second, the proliferation rate was not significantly changed (Figure 3A and Figure S3A). Third, the level of alkaline phosphatase (AP), a typical ESC marker (Pease et al., 1990), was not affected (Figure 3B and Figure S3B). Fourth, the expression levels of most genes that are critical for ESC self-renewal (including Nanog, Oct4, Klf4, and Sox2) were not significantly affected (Figure 3C and Figure S3C), despite a significant reduction of local H3K4me3 upon knockdown of Dpy-30 (Figure 3D and Figure S3D). To ensure sufficient time of impact by the depletions, all of these analyses were performed after culturing the cells for more than ten days following viral infection. Based on the entirety of these results, we conclude that self-renewal of ESCs does not require the normal high level of H3K4 methylation, at least under the culture conditions employed here.
Figure 3. Depletion of RbBP5 or Dpy-30 Reduces H3K4 Methylation without Affecting Self-Renewal of ESCs.
(A) Proliferation curves of control and RbBP5- or Dpy-30-depleted ESCs. In (A), (B), (C), (D), and (G), averages ± SD from triplicate measurements are plotted.
(B) AP levels of control and RbBP5- or Dpy-30-depleted ESCs.
(C) Expression of selected key stemness genes in control and RbBP5- or Dpy-30-depleted ESCs.
(D) Relative H3K4 methylation levels at TSSs of key stemness genes or an intergenic region in control and Dpy-30-depleted ESCs.
(E) Microarray analysis of genes affected by Dpy-30 depletion in ESCs. Genes were sorted on the basis of effects of Dpy-30 depletion. The ratio of expression levels between the two different controls (blue) and that between Dpy-30-depleted and scramble control ESCs (red) are shown.
(F) Microarray analysis of genes affected by Dpy-30 or RbBP5 depletion in ESCs. The numbers of affected gene probes (out of a total of 22155) are shown with indicated fold of down- or upregulation.
(G) Differential effects on H3Ac by Dpy-30 depletion, as determined by ChIP-qPCR in control and Dpy-30-depleted ESCs.
See also Figure S3.
Consistent with the largely unaffected self-renewal, microarray analyses revealed that the reduction of global H3K4 methylation had minimal effects on expression of most genes in ESCs (Figure 3E). For the vast majority of genes, expression differences between Dpy-30 depletion and scramble shRNA control were limited to a small range equivalent to the expression differences between the nonhairpin and scramble shRNA control sets (Figure 3E) and, hence, are considered insignificant. Out of 22,155 analyzed gene probes, only a very small portion showed significant down- or upregulation upon RbBP5 or Dpy30 depletion (Figure 3F). A gene ontology analysis found no significant functional clustering of the up- or downregulated genes.
A possible effect of Dpy-30 depletion on local H3 acetylation (H3Ac) was also examined by ChIP-qPCR, since H3Ac is believed to have positive effect on gene expression. Interestingly, Dpy-30 depletion significantly, although not profoundly, affected H3Ac at the TSSs of almost all monitored developmental genes that are poised in ESCs, but did not affect H3Ac at the TSSs of genes that are highly expressed in ESCs (including house-keeping genes and ESC maintenance genes) (Figure 3G).
We next asked whether active gene induction in the ESCs was affected by Dpy-30 or RbBP5 depletion. To this end, stress responses to heat shock and DNA damage were tested. Both control and RbBP5- or Dpy-30-depleted ESCs showed similar levels of induction of the heat shock protein Hsp70 gene upon incubation at elevated temperature (Figure S3E) or comparable upregulation of p21 and Mdm2 genes when treated with the DNA-damaging reagent doxorubicin (Figure S3F), indicating that gene induction is not generally affected by depletion of RbBP5 and Dpy-30 in ESCs. ChIP analyses showed that the H3K4me3 level at the TSS of Hsp70 was moderately decreased upon heat shock (Figure S3G), and that the H3K4me3 levels at the TSSs of p21 and Mdm2 were increased after doxorubicin treatment (Figure S3H). In each of these two different stress responses, Dpy-30 depletion significantly reduced the H3K4me3 levels at relevant TSSs in both control and treated cells (Figures S3G and S3H). Therefore, we conclude that maintenance or induction of gene expression in ESCs does not necessarily require the full level of H3K4me3.
Dpy-30 and RbBP5 Are Required for ESC Differentiation upon LIF Withdrawal
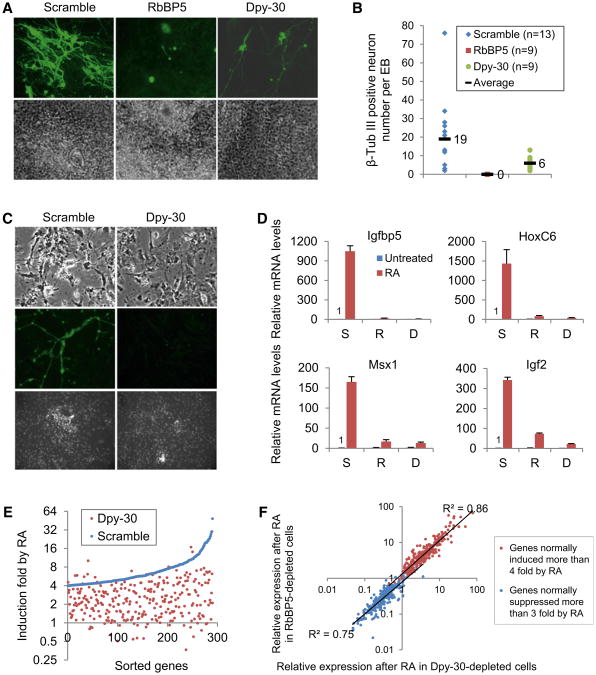
We next examined whether the differentiation capacity of ESCs was affected when H3K4 methylation was reduced. ESCs readily differentiate in the absence of leukemia inhibitory factor (LIF) (Smith et al., 1988). After culturing control and knockdown ESCs without LIF for eight days, we found (1) that control ESCs differentiated to cells with fibroblast-like morphology, whereas most RbBP5- or Dpy-30-depleted ESCs still exhibited aggregated ESC-like morphology (Figure 4A); (2) that the AP level was higher in the RbBP5- or Dpy-30-depleted cells than in control cells (Figure 4B); and (3) that early developmental genes like lgfbp5 and Mest were activated to significant levels in control cells, but not in RbBP5- or Dpy-30-depleted cells (Figure 4C). These findings all indicate a blockade of differentiation in cells depleted of RbBP5 or Dpy-30.
Figure 4. Inefficient Differentiation of Rbbp5- or Dpy-30-Depleted ESCs upon LIF Withdrawal.
(A) Morphology of control, RbBP5- or Dpy-30-depleted cells after being cultured without LIF for 8 days.
(B) AP levels of scramble control (S) and RbBP5-(R) or Dpy-30-depleted (D) ESCs after being cultured without LIF for eight days. In (B) and (C), averages ± SD from triplicate measurements are plotted.
(C) Expression of developmental genes in control and RbBP5- or Dpy-30-depleted ESCs before and after being cultured without LIF for 8 days.
Dpy-30 and RbBP5 Are Important for Conveying Plasticity in Global Transcription during ESC Differentiation into a Neural Lineage
We next focused on the effect of Dpy-30 or RbBP5 depletion on the retinoic acid (RA)-induced neural differentiation of either ESCs-derived embryoid bodies (EBs) or ESCs in monolayer culture. Dpy-30- or RbBP5- depleted ESCs were able to initiate aggregation into EB-like structures with no major morphological abnormalities compared to control ESCs. When treated with RA, certain areas in control EBs differentiated into neuronal fiber structures that were positive for β-tubulin III, a classical neuronal marker (Figure 5A). Such structures were completely missing or far less extensive (and with a lower staining intensity) in the RbBP5- or Dpy-30-depleted EBs, respectively. Moreover, the number of β-tubulin III-positive neurons in Dpy-30-depleted EBs was significantly less than that in control EBs (Figure 5B). These observations are not likely due to off-target effects of the Dpy-30 shRNA sequence, as similar effects were seen with Dpy-30#2 (data not shown). These results indicate that core subunits of MLL complexes are essential for neural specification of ESCs.
Figure 5. RbBP5 and Dpy-30 Are Crucial for ESC Differentiation into a Neural Lineage.
(A) Neuronal structures in EBs derived from control and RbBP5- or Dpy-30-depleted ESCs. EBs were treated with RA for 14days and stained for β-tubulin III (top). Phase contrast microscopic images of the corresponding EBs were shown at the bottom.
(B) Quantitation of β-tubulin III positive neurons. Positive neurons in each EB were counted and the means are indicated.
(C) RA-induced neural differentiation of control or Dpy-30-depleted ESCs in monolayer culture. Cell morphologies were shown two days after the cells were transferred to uncoated plates (top). Cells were stained for β-tubulin III (middle) or for DNA by Hoechst staining (bottom) four days after the transfer.
(D) Expression of developmental genes before and after RA-mediated differentiation in control (S), RbBP5- (R), or Dpy-30-depleted (D) cells in monolayer culture. mRNA levels were measured by qPCR and averages ± SD from triplicate reactions are plotted.
(E) Microarray analysis of genes whose expression was induced more than 4 fold after RA treatment in control cells. Genes were sorted according to induction folds by RA treatment (as compared to expression levels right before RA treatment) in the control cells.
(F) Correlation of the effects of RbBP5 depletion and Dpy-30 depletion on post-RA gene expression. Genes whose expression was normally induced more than 4 fold after RA treatment (red) and genes whose expression was normally suppressed more than 3 fold after RA treatment (blue) were plotted by their post-RA expression levels in RbBP5-and Dpy-30-depleted cells. The linear regression trendlines and the correlation coefficient-squares are displayed.
See also Figure S4.
To obtain more cellular materials for mechanistic analyses, ESCs were cultured in monolayer form and treated with RA to induce differentiation into neural precursor cells. A few days after transfer of these precursor cells to nongelatin-coated plates, β-tubulin III-positive fiber structures typical of neurons could be observed in control cells, but were much less frequently found in Dpy-30-depleted cells (Figure 5C). An analysis of samples at the end of RA treatment revealed that early developmental genes such as lgfbp5, Msx1 and HoxC6, and neurotrophic factors such as lgf2 were induced significantly in control cells, but not in RbBP5- or Dpy-30-depleted cells (Figure 5D).
As revealed by microarray analysis and consistent with previous findings (Walker et al., 2007), RA treatment of control ESCs induced many lineage-associated genes, including many Hox genes and genes involved in early neural differentiation, while silencing many ESC specific genes. Strikingly, depletion of Dpy-30 or RbBP5 significantly repressed the upregulation of the vast majority of genes normally induced by RA (Figure 5E and data not shown). As the same pattern of effects was also observed as early as 1 hr after RA treatment (Figure S4A), it suggests that most of the direct RA-responding genes depend on efficient H3K4 methylation for full induction mediated by RA. Consistent with a blockade of differentiation, depletion of Dpy-30 also antagonized the downregulation of genes normally suppressed by RA, as revealed both by microarray analysis on global genes (Figure S4B) and by qPCR on selected ESC-specific genes, including Upp1, Klf4, Oct4, and Dnmt3L (Figure S4C). These results indicate that core subunits of MLL complexes play an important role in mediating the plasticity of the expression program during the ESC fate transitions. Importantly, a strong positive correlation of RA-induced expression changes resulting from RbBP5 and Dpy-30 depletion, as indicated by a scatter plot with a diagonal distribution of near straight line and a correlation coefficient value close to 1 (Figure 5F), further supports the notion that RbBP5 and Dpy-30 function in the same complexes to regulate transcription of the same genes and in the same direction.
To assess whether depletions of core subunits of MLL complexes have similar effects in human cells, we chose the human EC cell line NT2, which also differentiates into the neural lineage upon RA treatment, as a convenient alternative to human ESCs. Knockdown of Dpy-30 (Figure 1D, left) or WDR5 (Figure S4D) in NT2 cells significantly reduced the global H3K4me3 level. Similar to the observations made with mouse ESCs, lineage-specific genes including Hand1 and Msx1 were strongly induced after RA treatment in control NT2 cells, whereas induction was significantly impaired in WDR5- or Dpy-30-depleted NT2 cells (Figure S4E). These results indicate that RA-mediated induction of the lineage genes is critically dependent on normal levels of MLL complex core subunits in human EC cells.
Dpy-30 Is Important for Normal H3K4me3 Increase at Developmental Genes for RA-Mediated Cell-Fate Transition
Having established the importance of RbBP5 and Dpy-30 in mediating the plasticity of gene expression during ESC differentiation, we next focused on the relevant chromatin-related molecular mechanisms underlying the observed phenotypes. As revealed by ChIP-qPCR on TSSs of lgfbp5, HoxC6, and Msx1, the representative genes that were shown (in Figure 5D) to be significantly induced by RA-mediated differentiation in control cells but not in RbBP5- or Dpy-30-depleted cells, a strong increase of H3K4me3 at these TSSs was observed following RA-mediated differentiation, but such increase was significantly affected by the knockdown of RbBP5 or Dpy-30 (Figure 6A). We then extended this observation to the TSSs of a large panel of randomly picked developmental genes that were significantly induced (over 4-fold) in control cells, but much less so in Dpy-30 knockdown cells, by RA treatment (Figure 6B). H3K4me3 levels were found to be significantly enhanced at the TSSs of the majority of the developmental genes after RA-mediated differentiation in the control cells (this general trend is apparent when comparing Figure 6B and Figure 2E), and this increase was significantly impaired by Dpy-30 depletion (Figure 6B). A few ESC-specific genes and silenced genes were also examined for local H3K4me3. In contrast to the developmental genes, H3K4me3 levels dropped significantly at ESC-specific genes after RA-mediated differentiation in both control and Dpy-30-depleted cells (Figure 6B and Figure S5A). No significant change of H3K4me3 by RA treatment was found at silenced regions (Figures 6A and 6B). The full picture of the chromatin state around broad TSS-proximal regions of several developmental genes, including HoxC6, HoxC5, and Punc, clearly confirmed the significant and Dpy-30-dependent increases in the H3K4me3 peaks at all of the TSS-proximal regions after RA treatment (Figure 6C). The H3K27me3 levels at the TSSs of some bivalent developmental genes were also monitored and found to drop after RA-mediated differentiation in both control and RbBP5- or Dpy-30-depleted cells (Figure S5B).
Figure 6. Dpy-30 Is Important for Normal H3K4me3 Increases at Developmental Genes for RA-Mediated Cell-Fate Transitions.
(A) H3K4me3 levels at the representative developmental genes and an intergenic region in control (S), RbBP5- (R), or Dpy-30-depleted (D) cells before and after RA-mediated differentiation in monolayer culture. In A-D, signals were determined by ChIP-qPCR. InA and D, averages ± SD from triplicate reactions are plotted.
(B) H3K4me3 levels at a large panel of genomic regions after RA-mediated differentiation in monolayer culture. Note the gene list is exactly the same as that in Figure 2E, and a general increase of H3K4me3 is apparent in the comparison to Figure 2E. Averages ± SD from duplicate reactions are plotted.
(C) H3K4me3 levels at the broad TSS-proximal regions of HoxC5, HoxC6 and Punc genes before and after RA-mediated differentiation in monolayer culture.
(D) Dpy-30 (left) and RNA Pol II (right) recruitment at some RA-inducible genes and an intergenic region before and after RA-mediated differentiation in monolayer culture.
See also Figure S5.
To adequately examine Dpy-30 involvement in orchestrating the differentiation program of ESCs, we performed ChIP-chip analyses to determine H3K4me3 levels at large promoter regions (TSS −8.2 to +3 kb) of the whole genome in both control and Dpy-30 depleted cells after RA-mediated differentiation. Although quantitative cross-sample comparison of ChIP-chip results is difficult, genes on which Dpy-30 depletion had a particularly significant effect could still be revealed. Indeed, the broad TSS-proximal regions at many RA-inducible genes such as Igf2 and Mgst1 (Figure S5C) were found to be marked with significantly higher levels of H3K4me3 in the control cells than in the Dpy-30-depleted cells after RA-mediated differentiation. Ontology analysis has revealed that the promoter regions with largest post-RA H3K4me3 defect by Dpy-30 depletion were highly enriched in genes involved in transcriptional regulation in neuron differentiation, axon guidance, and pattern specification process (Figure S5D), perfectly consistent with the actual differentiation program of ESCs after RA treatment. These results demonstrate, at a genome-wide scale in the post-differentiation setting, that Dpy-30 depletion most significantly impacts the promoter H3K4me3 of the RA-mediated lineage specification genes.
Consistent with a previous report of increased recruitment of core subunits of MLL complexes to certain Hox genes upon RA treatment in NT2 cells (Lee et al., 2007), Dpy-30 binding at the TSSs of several monitored developmental genes, but not at an intergenic region, was significantly enhanced after RA treatment in control cells (Figure 6D, left). Depletion of Dpy-30 dramatically crippled such elevated binding, consistent with the RA-mediated increase of H3K4me3 at those developmental genes in control, but not in Dpy-30 depleted cells. Similarly, RNA Pol II recruitment increased significantly at the TSSs of these developmental genes in the control cells after RA treatment; and this increase was severely hampered in the Dpy-30-depleted cells after the same RA treatment (Figure 6D, right). These results are consistent with the idea that recruitment of a regulatory subunit of MLL complexes contributes to the control of H3K4 methylation and gene expression levels in a specific biological process.
To investigate mechanisms by which depletion of RbBP5 or Dpy-30 leads to inefficient downregulation of many ESC-specific genes during differentiation, we checked histone methylation on Upp1, Klf4 and Oct4 by ChIP before and after RA-induced differentiation. As expected, H3K4 methylation at the TSSs of these genes dropped after RA treatment in both control and RbBP5- or Dpy-30-depleted cells (Figure S5A). Strikingly, whereas the H3K27me3 levels at the TSSs of these ESC-specific genes increased significantly after RA treatment in the control cells, the increases were markedly lower in RbBP5- or Dpy-30-depleted ES cells (Figure S5E). Therefore, MLL complexes are important not only for increasing H3K4 methylation on lineage-specific genes, but also for establishing the repressive H3K27me3 mark on ESC-specific genes for efficient silencing during ES cell differentiation.
Discussion
The recent boom in genome-wide studies of histone modifications has provided rich information about the distinctive chromatin states at a global scale in ES and lineage progenitor cells (Bernstein et al., 2006; Cui et al., 2009; Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007), including provocative observations regarding the bivalent marks on many developmental genes. However, this body of information is to a large extent descriptive and correlative in nature, and the actual functions of some of the modifications, including H3K4 methylation, in the biology of those cells have remained unclear.
By a combination of biochemical, cellular and genomic approaches, we establish a direct and causal role for Dpy-30, a core subunit of MLL complexes, in efficient chromosomal H3K4 methylation the ESC genome. We then demonstrate essential roles for normal levels of Dpy-30 and RbBP5 in ESC differentiation. Our data provide strong experimental evidence for the hypothesis that MLL complexes and their corresponding H3K4 methylation marks within bivalent chromatin domains keep silenced developmental genes poised for expression in mammalian ESCs and are functionally important in powering the fate transitions of ESCs to specific lineages. A recent profiling of H3K4me3 during zebrafish embryonic development suggests that this modification may set the stage for genomic activation during the maternal-zygotic transition (Vastenhouw et al., 2010). Apart from being consistent with these findings, our results in a related mammalian (ESC) system also provide strong support for the functional significance of such observations. Our results are also in full agreement with a recent report of an important role for MLL2 in activation of the mouse embryonic genome (Andreu-Vieyra et al., 2010). These findings offer at least partial mechanistic explanations for the requirement of Dpy-30 in the development of C. elegans (Hsu et al., 1995), and are in concord with an essential role of WDR5 in early development of X. laevis tadpoles (Wysocka et al., 2005).
A Direct and Causal Role for Dpy-30 in Enhancing MLL Complex-Mediated H3K4 Methylation throughout the Mammalian Genome
In addition to the dependence of cellular H3K4 methylation levels on Dpy-30, the full extent of Dpy-30 s impact on chromosomal H3K4 methylation is highlighted by (1) the strong genome-wide overlap of the Dpy-30 occupancy and H3K4me3 profiles both in signal positions and in magnitudes and (2) the dependence of chromosomal H3K4me3 on Dpy-30 binding at individual TSSs. The directness of the effect is indicated by the combination of (1) the biochemical activity of purified Dpy-30 in enhancing H3K4 methylation by one of the MLL complexes and (2) the in situ effect on chromosomal H3K4me3 upon loss of Dpy-30 occupancy. Thus, these results collectively provide a physical and functional picture of Dpy-30 and its associated MLL complexes in the mammalian genome, and firmly establish a direct and causal role for Dpy-30 in facilitating chromosomal H3K4 methylation across the entire ESC genome. However, we note that in addition to the recruitment per se of the H3K4 methyltransferase complexes, other factors such as H2B ubiquitylation (Kim et al., 2009; Sun and Allis, 2002), transcription status (Pavri et al., 2006), and stability of the methyl mark may all contribute to the final methylation level at a particular genomic site in a specific cell state. This is consistent with the imperfect correlation of signal magnitudes for Dpy-30 binding and local H3K4me3 (Figure 2D and Figures S2D and S2E) and with quantitative variations in effects of Dpy-30 depletion on H3K4me3 levels at different gene loci (Figure 2E).
Impact of Reduced H3K4 Methylation on Global Gene Expression
The demonstration of direct (causal) effects of specific histone modifications on the transcription of specific target genes in animal cells represents a challenging problem (Kouzarides, 2007). However, recent studies have linked H3K4m3 to the regulation of gene expression through its specific recognition by various domains–including the PHD finger, the chromodomain, and the double tudor domain–that are found in many proteins that are implicated in transcription (reviewed in Ruthenburg et al., 2007). It is likely, therefore, that H3K4 methylation does directly affect expression of many genes on which it is found. In this regard, our ongoing studies of transcription in reconstituted cell free systems have shown a direct (causal) role for H3K4 methylation in the transcription of recombinant chromatin templates (our unpublished results). These results are also consistent with the causal effects of both the Dpy-30 and the catalytic subunits of MLL complexes on expression of a chromosomal reporter gene (Figure 1E). However, compared to the relatively simple and straightforward mode of transcriptional regulation in these model systems, expression of most endogenous genes is probably regulated by multiple layers of factors and diverse histone modifications (Suganuma and Workman, 2008), with complex and interactive relationships that make it difficult to establish unequivocally the direct effect of a single modification/factor on a specific gene.
Our results in ESC differentiation are most consistent with a crucial role of MLL complex-mediated H3K4 methylation in priming the developmental genes for efficient induction upon differentiation, and are also consistent with the biochemical capability of H3K4 methylation to enhance transcription in model systems. In undifferentiated ESCs, however, the normal level of H3K4 methylation appears less essential either for expression of the vast majority of genes in the genome or for gene induction in stress responses. Because H3K4 methylation is not completely lost in the RbBP5- or Dpy-30-depleted ESCs, the possibility remains that expression of stemness genes may rely on low levels of cellular H3K4 methylation. However, the clear differentiation-deficient phenotypes of cells with reduced H3K4 methylation levels sufficient for ESC maintenance and stress responses unambiguously demonstrate a relatively specific role for high level function of MLL complexes in mediating cell-fate transitions of ESCs. We consider two possible explanations for the differential effects of the depletion of MLL complex components.
First, these results may suggest the importance of the detailed physiological setting for the impact of a specific regulatory modification such as H3K4me3. As chromatin is overall less compact and transcription more permissive in ESCs than in differentiated cells (Efroni et al., 2008), it seems plausible that ESCs may tolerate the reduction in H3K4 methylation because of the presence of sufficient amounts of other stimulatory regulators (and/or lack of repressive regulators) that keep chromatin hyperdynamic and accessible to the transcription machineries (Meshorer and Misteli, 2006; Meshorer et al., 2006). In this respect, yeast cells may superficially resemble mammalian ESCs in that (i) they also have a relatively loose chromatin organization largely devoid of heterochromatin and (ii) the loss of H3K4 methylation also has very little effect on global gene transcription in yeast (Miller et al., 2001). As mammalian ESCs go through their differentiation programs, the overall nuclear environment becomes more restrictive (Meshorer and Misteli, 2006) and thus may necessitate maintenance of sufficiently high levels of H3K4 methylation to allow for dynamic re-organization of transcriptional programs.
Alternatively, the poised developmental gene loci may have intrinsically different chromatin structures and hence a higher dependence on H3K4 methylation compared to the ESC specific gene loci. This speculation is supported by the finding that H3 acetylation is selectively lowered at the poised developmental genes, but not at the highly expressed genes (including the stemness genes), in Dpy-30-depleted ESCs (Figure 3G). As histone acetylation is thought to neutralize the charge on histones and/or to stabilize binding of chromatin remodeling factors, and may reflect a chromatin state more accessible to transcription machineries, these differential H3Ac effects are consistent with both the unaffected expression of those genes by Dpy-30 depletion and a role of Dpy-30 in helping establish a potentially activated chromatin state (priming) for a more efficient induction of the poised developmental genes. The mechanisms underlying the differential H3Ac effects (and possibly different chromatin architectures) at the different types of genes remain an interesting question.
Although enhanced H3K4 methylation is the major function established for the core subunits of MLL complexes, we cannot formally exclude the possibility that they may share some other functions that contribute to ESC differentiation. As these core subunits have only been reported in association with MLL complexes, any of their shared functions are most likely related to the function of MLLs. In this regard, we note that the cellular phenotypes resulting from depletion of the core subunits partially resemble those for loss of MLL2 in mouse ESCs (Lubitz et al., 2007).
Model for Regulation of Pluripotency by H3K4 Methylation
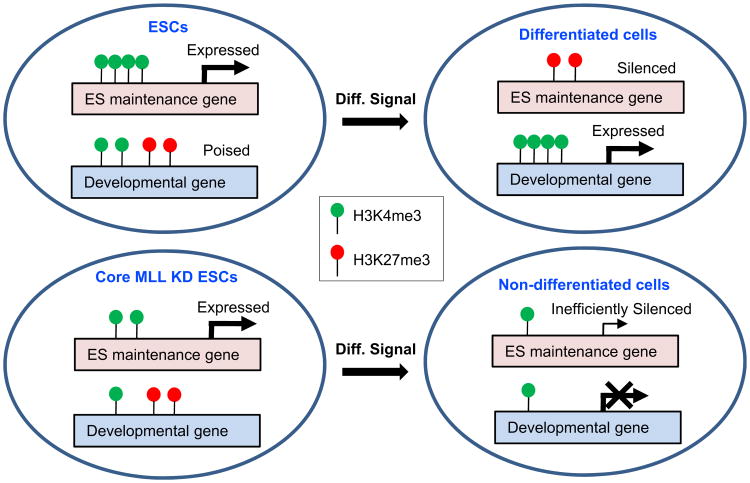
Based on our results, we refine earlier proposals (Azuara et al., 2006; Bernstein et al., 2006; Spivakov and Fisher, 2007) and present a revised model (Figure 7) for how the two opposing methylation activities coordinate the maintenance and fulfillment of pluripotency potentials of ESCs. As described, genes critical for self-renewal and pluripotency maintenance are selectively marked with H3K4 methylation relative to H3K27 methylation and are highly expressed in ESCs, while many developmental genes carry both H3K4 and H3K27 methylation marks and remain poised. Upon differentiation, ESC-specific genes begin to lose K4 methylation and gain K27 methylation, while induced lineage-specific genes gain K4 methylation at the expense K27 methylation. As shown here, ESCs depleted of core MLL complex subunits exhibit a global deficit in H3K4 methylation that appears to leave expression of stemness genes largely unaffected but has a major effect on the induction potential of lineage genes. Together with a compromised enhancement of the repressive H3K27 methylation of the ESC-specific genes, which may involve indirect mechanisms, these effects result in a blockade of differentiation and inefficient silencing of ESC-specific genes.
Figure 7. A Model for How MLL Complexes Might Affect ESC Maintenance and Differentiation.
See Discussion for details. The numbers of the lollipops on the depicted genes are not meant to be quantitatively meaningful.
We note a contrast between the defective differentiation shown here for ESCs depleted of core subunits of MLL complexes, and the relatively higher tendency of spontaneous differentiation seen in ESCs deficient for subunits of Polycomb repressive complexes (Azuara et al., 2006; Boyer et al., 2006). These contrasting phenotypes confirm that established principles of antagonism between Trithorax- and Polycomb-group proteins (Hanson et al., 1999; Klymenko and Muller, 2004; Ring-rose and Paro, 2004) are evident in a model of mammalian embryonic development.
Experimental Procedures
Antibodies
Details of antibodies can be found in Supplemental Information.
Recombinant Proteins and Histone Methylation Assay
Details of recombinant proteins and histone methylation assay can be found in Supplemental Information.
ESC Culture and Differentiation
E14 cells were cultured on 0.1% gelatinized tissue-culture plates in complete ESC growth medium supplemented with LIF (Kindly provided by David Allis's lab). Embryoid bodies were derived using the hanging drop method (Wang and Yang, 2008) with modifications. For details see Supplemental Information. For neural differentiation under monolayer culture conditions, ESCs were plated on gelatinized 6-well tissue-culture dishes at 30,000 cells per well in complete growth medium with LIF. Next day cells were washed and incubated in growth medium without LIF for4 days before incubation with 1 μM all-trans RA (Sigma) for 4 additional days. Cells were either subjected to RNA extraction, fixation for ChIP analyses, or transfer in growth medium (without LIF or RA) to uncoated 24-well plates for further development of neural morphology.
RNA Interference
Lentiviral constructs expressing shRNAs (listed in Table S1) including the controls were purchased from OpenBiosystems. Viral particles were produced by following the recommended protocols (Addgene). Two days after infection of ESCs with viruses, puromycin was added at 2 μg/ml to select for pooled populations of stably-infected cells. MCF7 and NT2 cells were transfected with siRNA duplexes (listed in Table S1) or an ON-TARGETplus nontargeting pool as the negative control (Dharmacon) using Lipofectamine (Invitrogen) according to manufacturer’s instructions.
ChIP, ChIP-seq, and ChIP-chip
For Dpy-30 ChIP, cells were fixed by double crosslinking method, except for Figure 6D, where single crosslinking was used. For all other ChIP assays cells were fixed by the single crosslinking method. Remaining steps in ChIP were performed essentially following the protocol in the ChIP Assay Kit (Upstate) except that, for ChIP-qPCR, DNA was eluted using Chelex 100 resin following the fast ChIP protocol (Nelson et al., 2006).
For Dpy-30 ChIP-seq, sonicated chromatin derived from ∼3 × 108 E14cells was subjected to immunoprecipitation by 36 μg anti-Dpy-30 antibody following the protocol in the ChIP Assay Kit (Upstate) in the absence of salmon sperm DNA. Purified DNA was submitted to ArrayStarInc. (Rockeville, MD) for library construction, sequencing and basic data analyses. ChIP-seq for H3K4me3 was carried out as described (Bernstein et al., 2006; Mikkelsen et al., 2007).
For details on ChIP assays including H3K4me3 ChIP-chip see Supplemental Information.
RT-PCR and qPCR
Total RNAs were extracted using the RNeasy kit (QIAGEN) and reverse-transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen). Fold differences in gene expression levels were normalized against Gapdh. More details and the primer sequences can be found in Extended Experimental Procedures and in Table S2.
Gene Expression Microarray Analysis
Microarray analysis for global gene expression was performed using standard methods on Illumina MouseRef-8 v2.0 expression beadchip. For details see Supplemental Information.
Supplementary Material
Acknowledgments
We are grateful to Yali Dou for baculoviruses encoding RbBP5, WDR5 and Ash2L, Cristina Hughes (David Allis lab), and Eric McIntush (Bethyl Laboratories) for Dpy-30 antibody, Joanna Wysocka for WDR5 antibody, Aaron Goldberg (David Allis lab) for LIF, Shannon Lauberth and Zhanyun Tang for providing supporting data, Zheng-Yuan Fu for technical assistance, Esther Rheinbay, Jing Wang and Xiaojian Sun for data analyses, and Bing Ren (UCSD) and Xiangdong Lu for insightful discussions of the project. H.J. and W.-Y.C. are Leukemia and Lymphoma Society Fellows. X.W. is a recipient of a Tri-Institutional Starr Stem Cell Scholars fellowship. This work was supported by NIH (DK071900), Starr Cancer Consortium (I2-A88), and Leukemia and Lymphoma Society (7132-08) grants to R.G.R.
Footnotes
Accession Numbers: Our microarray and ChIP-seq datasets have been deposited in the GEO data-base under GSE26136.
Supplemental Information: Supplemental Information includes Extended Experimental Procedures, five figures, and two tables and can be found with this article online at doi:10. 1016/j.cell.2011.01.020.
References
- Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, Stewart AF, Matzuk MM. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 2010;8:e1000453. doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehe PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodriguez-Gil A, Mkandawire M, Landsberg K, Shevchenko A, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem. 2006;281:35404–35412. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004;6:437–443. doi: 10.1016/s1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- Hanson RD, Hess JL, Yu BD, Ernst P, van Lohuizen M, Berns A, van der Lugt NM, Shashikant CS, Ruddle FH, Seto M, et al. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci USA. 1999;96:14372–14377. doi: 10.1073/pnas.96.25.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DR, Chuang PT, Meyer BJ. DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development. 1995;121:3323–3334. doi: 10.1242/dev.121.10.3323. [DOI] [PubMed] [Google Scholar]
- Hsu DR, Meyer BJ. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics. 1994;137:999–1018. doi: 10.1093/genetics/137.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol Biol Cell. 2007;18:2356–2366. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pease S, Braghetta P, Gearing D, Grail D, Williams RL. Isolation of embryonic stem (ES) cells in media supplemented with recombinant leukemia inhibitory factor (LIF) Dev Biol. 1990;141:344–352. doi: 10.1016/0012-1606(90)90390-5. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Crosstalk among Histone Modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Dillon N. The epigenetic basis for embryonic stem cell pluripotency. Bioessays. 2005;27:1286–1293. doi: 10.1002/bies.20330. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E, Ohishi M, Davey RE, Zhang W, Cassar PA, Tanaka TS, Der SD, Morris Q, Hughes TR, Zandstra PW, et al. Prediction and testing of novel transcriptional networks regulating embryonic stem cell self-renewal and commitment. Cell Stem Cell. 2007;1:71–86. doi: 10.1016/j.stem.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang P. In vitro differentiation of mouse embryonic stem (mES) cells using the hanging drop method. J Vis Exp. 2008 doi: 10.3791/825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.