Abstract
The influenza virus fusion peptide is the N-terminal ~20 residues of the HA2 subunit of the hemagglutinin protein and this peptide plays a key role in the fusion of the viral and endosomal membranes during initial infection of a cell. The fusion peptide adopts N-helix/turn/C-helix structure in both detergent and membranes with reports of both open and closed interhelical topologies. In the present study, backbone 13CO-15N REDOR solid-state NMR was applied to the membrane-associated fusion peptide to detect the distribution of interhelical distances. The data clearly showed a large fraction of closed and semi-closed topologies and were best-fitted to a mixture of two structures that do not exchange. One of the earlier open structural models may have incorrect G13 dihedral angles derived from TALOS analysis of experimentally correct 13C shifts.
Keywords: REDOR, influenza virus, fusion peptide, membranes, solid-state NMR, hemagglutinin
The influenza virus fusion peptide is the ~20 N-terminal residues of the HA2 subunit of the hemagglutinin fusion protein which is a single-pass integral membrane protein of the viral membrane that assembles as molecular trimers (White et al. 2008). The virus is first endocytosed into host respiratory epithelial cells and the subsequent lowering of the pH of the endosome to the 5–6 range leads to a large change in the HA2 structure and binding of the fusion peptide to the endosomal membrane (Durrer et al. 1996; Durell et al. 1997). The subsequent fusion of the viral and endosomal membranes allows the virus nucleocapsid to enter the cytoplasm and this entry is a requirement for infection of the cell. Peptides with the fusion peptide sequence but lacking the rest of hemagglutinin have been studied to better understand the role of the fusion peptide in membrane fusion (Epand 2003). In this paper, we also refer to these peptides as “fusion peptides” which is consistent with the nomenclature of earlier papers. Many of these fusion peptides induce fusion of membrane vesicles and there is more rapid and extensive vesicle fusion at pH 5.0 than at pH 7.4 (Han and Tamm 2000; Korte et al. 2001). These pHs respectively correspond to the lowest endosomal pH and to typical physiological pH.
The present study aims to understand significant differences between the structure of the fusion peptide in membranes derived from an earlier solid-state NMR study and various structures in detergent derived from several earlier liquid-state NMR studies. To our knowledge, the fusion peptide does not induce fusion between detergent micelles although it does induce fusion between membrane vesicles. Most of the high-resolution fusion peptide structural models are in detergent and it is therefore important to understand the similarities and differences between the detergent and membrane structures.
Human cell membranes typically contain ~0.3 mole fraction cholesterol although this fraction may be lower in the endosomal membrane (Worman et al. 1986). For membranes containing ~0.3 mole fraction cholesterol and for fusion peptide: lipid mole ratio ≈ 0.01, the fusion peptide forms dominant β sheet structure that is likely oligomeric (Wasniewski et al. 2004). For membranes that lack cholesterol, the dominant structure is α helical and likely monomeric (Bodner et al. 2004; Bodner et al. 2008; Sun and Weliky 2009). The fusion peptide induces fusion both between vesicles whose membranes contain cholesterol and between vesicles that lack cholesterol (Han and Tamm 2000; Korte et al. 2001). The present study only considers membranes that lack cholesterol for which α helical structure is dominant.
There are several differences among the fusion peptide sequences used in the detergent and membrane studies and we provide a list of these sequences and nomenclatures: H1_23; GLFGAIAGFIEGGWTGMIDGWYG; H1_23_G8A, GLFGAIAAFIEGGWTGMIDGWYG; H1_20; GLFGAIAGFIEGGWTGMIDG; and H3_20, GLFGAIAGFIENGWEGMIDG. The “H1” and “H3” refer to the different viral serotypes for which sequence differences are G12 and T15 for the H1 serotype vs N12 and E15 for the H3 serotype. The “23” and “20” refer to the number of fusion peptide residues. Residues 21-23, ie. WYG, are conserved for both the H1 and H3 serotypes (Curtis-Fisk et al. 2007; Lorieau et al. 2010). For most studies, the fusion peptide sequence also had a non-native C-terminal tag to increase aqueous solubility, eg. GGKKKKG.
The first liquid-state NMR studies were done on a H3_20 fusion peptide in detergent at pH 5.0 and supported a ~10-residue N-terminal helix/2-residue defined turn/~5-residue C-terminal helix structure (Dubovskii et al. 2000; Han et al. 2001). The structure was “open”, ie. the two helices had a relatively oblique topology. One metric of interhelical topology is: (1) representing each helix axis as a vector from the N-terminal to the C-terminal helix residue; and (2) calculating the interhelical angle using the scalar product between the two helices (Lee et al. 2007). For this H3_20 open structure in detergent at pH 5.0, the interhelical angle is 103° by this calculation method. There was also a liquid-state NMR study of this H3_20 fusion peptide in detergent at pH 7.4 that was consistent with a different structure composed of a N-helix/turn/extended C-terminal structure (Han et al. 2001). Accompanying EPR studies for spin-labeled H3_20 fusion peptides in membranes were interpreted to mean that at pH 5.0, the N-helix axis makes a 52° angle with respect to the bilayer normal whereas at pH 7.4, this angle is 67°, ie. the insertion is less oblique at pH 5.0. In addition, at pH 5.0, the spin label of the most deeply inserted F3 residue was ~4 Å from the bilayer center whereas at pH 7.4, this label was ~10 Å from the bilayer center, ie. the fusion peptide was more deeply inserted at the lower pH. These findings were integrated into a model that explained the higher vesicle fusion at pH 5.0 with coupled: (1) helical rather than extended structure in the C-terminal region of the fusion peptide; (2) change in N-helix orientation relative to the bilayer normal with consequent deeper insertion of the N-helix; and (3) greater perturbation of the lipids particularly those close to the hydrophobic pocket on the interior of the “boomerang” structure. The importance of the open structure was further supported by subsequent liquid-state NMR in detergent and EPR in membranes of mutants of the H3_20 peptide (Lai et al. 2006). These studies correlated disruption of the open structure with slower membrane fusion.
More recently, liquid-state NMR was applied to the H1_23 fusion peptide in detergent at pH 7.4 and showed a “closed” N-helix/turn/C-helix structure, ie. tightly packed antiparallel N- and C-helices with interhelical angle of 158° (Lorieau et al. 2010). Additional liquid-state NMR studies were carried out on the H1_23_G8A peptide mutant in detergent at pH 7 and were interpreted in terms of a N-helix/turn/C-helix structure in which there was rapid exchange between different turn conformations and consequent exchange between closed structures with typical interhelical angle of ~159° and very open structures with typical angle of ~73° (Lorieau et al. 2012). Relaxation data for wild-type H1_23 peptide in detergent at pH 7.4 were consistent with only closed structure. By contrast, relaxation data at pH 4 for H1_23 were interpreted in terms of two rapidly exchanging peptide populations. There was ~0.8 fraction with closed structure and ~0.2 fraction with open structures similar to those of the H1_23_G8A mutant. For vesicles with ~0.3 fraction cholesterol at pH 7.2, there was greater fusion induced by wild-type H1_23 peptide than the H1_23_G8A mutant. This observation coupled with the structures were interpreted to signify the importance of the closed structure in lipid mixing which is an early step of membrane fusion. The open structures were thought to be important in the later step of fusion pore formation (Durrer et al. 1996).
Additional studies compared the 1H-15N HSQC spectrum of the H1_20 peptide to the spectrum of the H3_20 peptide where both peptides were in detergent at pH 7.3 (Lorieau et al. 2013). The peak positions were generally very similar between the two peptides although there were additional peaks for H3_20 that were attributed to deamidation of N12. Such peaks had not been reported in the earlier liquid-state NMR studies of H3_20 in detergent (Han et al. 2001; Lai et al. 2006). Liquid-state NMR studies of H1_20 peptide in detergent at pH 7.3 were interpreted as two rapidly interchanging populations of N-helix/turn/C-helix structures. The minor population with ~0.11 fraction was the closed structure and the major population with ~0.89 fraction was a distribution of open structures.
These detergent results provide some context for a previous solid-state NMR study of the H3_20 fusion peptide in membranes without cholesterol with samples prepared at both pH 5.0 and pH 7.4 (Sun and Weliky 2009). The experimental data were unambiguously assigned 13C chemical shifts in the N-helix and turn regions as well as rotational-echo double-resonance (REDOR) measurements on samples that were 13CO labeled at residue i and 15N labeled at residue i + 4 (Gullion and Schaefer 1989; Zheng et al. 2006). The REDOR-derived A5 13CO-F9 15N distance was 4.0 Å for both pH 5.0 and pH 7.4 samples, the F9 13CO-G13 15N distance was 3.6 Å at pH 5.0 and 3.7 Å at pH 7.4 and the G13 13CO-M17 15N distance was 4.5 Å at pH 5.0 and 4.6 Å at pH 7.4. The typical uncertainty in each distance was 0.1 Å. These distances and the 13C secondary shifts were consistent with N-helix/turn/C-helix structure at both pH 5.0 and pH 7.4.
There was a single set of 13C shifts for all residues at pH 7.4 and these shifts were also observed typically observed at pH 5.0. We denote these shifts to be associated with “structure A”. There was an additional set of 13C shifts only for residue E11 and only at pH 5.0. There was ~0.8 fraction of the H3_20 peptides with structure A E11 13C shifts and a ~0.2 fraction with “structure B” E11 shifts that differed by ~4 ppm from the structure A shifts. These structure B E11 shifts were observed in both fluid-phase and gel-phase membranes. The two structures did not appear to be exchanging on the NMR timescale.
The TALOS program analysis of the major population 13C shifts yielded an open N-helix/turn/C-helix structure A with an interhelical angle of 94°, cf. Fig. 1a (Cornilescu et al. 1999). Structure A was consistent with the REDOR-determined residue i to i + 4 distances. TALOS provides a distribution of values for each φ or ψ angle and the typical standard deviation of a distribution was 10° (Sun and Weliky 2009). TALOS analysis using the minor population B shifts also yielded a N-helix/turn/C-helix structure and the major difference between the A and B structures was the turn conformation. There were no interhelical distance constraints in the development of these structural models.
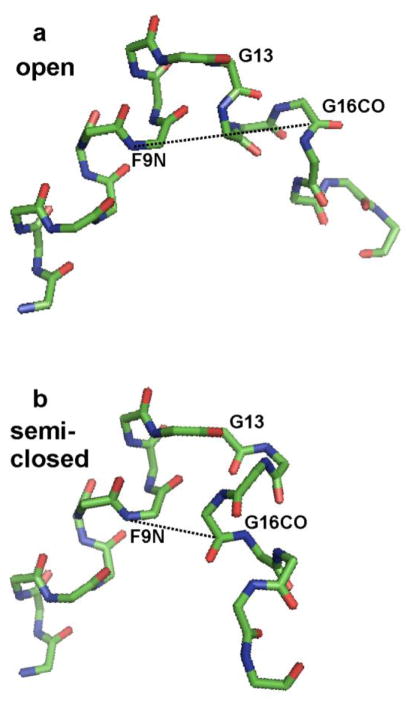
Figure 1.
Heavy-atom backbone structural models of residues L2 to G20 of membrane-associated H3_20 fusion peptide based on solid-state NMR data. Carbon atoms are represented by green vertices, nitrogen atoms are represented by blue vertices, and oxygen atoms are represented by red segments. The (φ, ψ) dihedral angles were the same for the panel a and b structures except that in the a structure, the G13 (φ, ψ) = (87°, 10°) were TALOS-derived and based on solid-state NMR 13C shifts and in the b structure, the G13 (φ, ψ) = (45°, −146°) were those of the closed structure in detergent. The structures were aligned to have the same N-helix orientation. The panel a structure has open interhelical topology and the dashed-line G16 CO-F9N internuclear distance is 10.5 Å. The panel b structure has semi-closed topology, and the internuclear distance is 5.2 Å. For reference, the G16 CO-F9N distance is 3.7 Å for the closed structure in detergent.
Structure A for membrane-associated H3_20 fusion peptide at pH 7.4 and 5.0 is similar to the open N-helix/turn/C-helix structure of H3_20 peptide in detergent at pH 5.0 (Han et al. 2001). Structure A is different from the H3_20 peptide structure in detergent at pH 7.4 which has extended rather than helical C-terminal structure. However, there is similarity to the open N-helix/turn/C-helix structures of the H1_20 peptide in detergent at pH 7.3 (Lorieau et al. 2013). Structure A was also compared to the closed N-helix/turn/C-helix structure of the H1_23 peptide in detergent at either pH 7.4 or pH 4.0 (Lorieau et al. 2010). The REDOR-derived F9 13CO-G13 15N distance of 3.6 Å agrees quantitatively with the distance of the closed structure and less well with the distance distribution of the open structure of the H3_20 peptide in detergent at pH 5.0. There is also a potential correlation between the ~0.2 fraction of structure B for membrane-associated H3_20 peptide at pH 5.0 and the ~0.2 fraction of open structures for detergent-associated H1_23 peptide at pH 4.0.
The key difference between the open and closed peptide structures lies in the G13 (φ, ψ) angles. For the open solid-state NMR structure in membranes, the average values of these angles are (87°, 10°) whereas for the closed structure in detergent, they are (45°, −146°). This effect is clearly seen in Fig. 1 where Fig. 1a has solid-state NMR-derived (φ, ψ) for all residues whereas Fig. 1b has G13 closed structure (φ, ψ) while retaining the solid-state NMR (φ, ψ) for the other residues. The interhelical angle for the “semi-closed” Fig. 1b structure is 136° and contrasts with the 94° angle in Fig. 1a. The solid-state NMR-derived (φ, ψ) for G13 were based on TALOS analysis of the 13CO and 13Cα shifts for G13 and the 13CO, 13Cα, and 13Cβ shifts for N12. The solid-state NMR G13 13CO and 13Cα shifts of 174.5 and 45.8 ppm were similar to the respective shifts of 174.1 ppm and 45.3 ppm for the closed structure in detergent (Lorieau et al. 2010). These similar shifts suggested that the solid-state NMR derived G13 angles that lead to open structure may have been incorrect. In addition, the G13 13C shifts do not appear to be strongly dependent on conformation. For example, the open H1_20 peptide in detergent at pH 7.3 has respective G13 13CO and 13Cα shifts of 174.2 ppm and 45.7 ppm and the open H1_23_G8A peptide has a G13 13Cα shift of 45.6 ppm (Lorieau et al. 2012; Lorieau et al. 2013).
In order to design experiments to distinguish between the models in Fig. 1, we looked for specific interhelical internuclear distances that differed greatly between them. For example, in the Fig. 1a structural model, the G16 CO – F9 N distance (r) was 10.5 Å whereas in Fig. 1b, the distance was 5.2 Å. The H3_20 peptide sequence GLFGAIAGFIENGWEGMIDGGGKKKKG was then chemically synthesized with a specific 13CO label at G16 and a specific 15N label at F9. The G16 CO-F9 N distance was probed using the REDOR experiment (Gullion and Schaefer 1989; Gullion 1998). This H3_20 sequence had been used to obtain the solid-state NMR-derived structure in membranes, cf. Fig. 1a, and we decided it was the best sequence to begin study of the relative populations of open vs closed fusion peptide structures in membranes. Future studies might examine the H1_23 sequence that was used to obtain the closed structure in detergent. We also chose to consider the structure near the endosomal pH of 5. Future studies might examine the pH dependence of closed vs open peptide populations and the possible relationship between this dependence and increased vesicle fusion at lower pH.
Sample preparation included: (1) ~30 mL of pH 5.1 buffer containing 2.0 μmole of peptide; (2) ~2 mL of pH 5.1 buffer containing extruded ~100 nm diameter vesicles made from 40 μmole of 1,2-di-O-tetradecyl-sn-glycero-3-phosphocholine (DTPC) and 10 μmole 1,2-di-O-tetradecyl-sn-glycero-3-[phospho-(1′-rac-glycerol)] (DTPG); (3) overnight mixing of solutions 1 and 2 followed by centrifugation at ~100000g to pellet the proteoliposome complexes; (4) lyophilization of the pellet, packing in a 4 mm diameter magic angle spinning (MAS) rotor, and hydration of the pellet with 20 μL of pH 5.1 buffer. The DTPC:DTPG (4:1) composition reflects aspects of the composition of the membranes of host respiratory epithelial cells including the significant fraction of phosphatidylcholine (Worman et al. 1986). Ether- rather than ester-linked lipids were used to reduce the natural abundance 13CO signal in the solid-state NMR spectra. Previous studies have compared the spectra of the H3_20 peptide with DTPC:DTPG (4:1) lipids to the spectrum of the peptide with the ester-linked lipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-(1′-rac-glycerol)] (POPG) in 4:1 mole ratio. There was little variation in 13C shifts and presumably peptide structure between samples containing either lipid type and also little variation between DTPC:DTPG and POPC:POPG samples at ~ − 30 °C with lipids in the gel-phase and the POPC:POPG sample at +10 °C with lipids in the fluid-phase (Sun and Weliky 2009). The solid-state NMR measurements were carried out using a 9.4 T spectrometer and triple resonance magic angle spinning (MAS) probe and further experimental details are given in Fig. 2 caption.
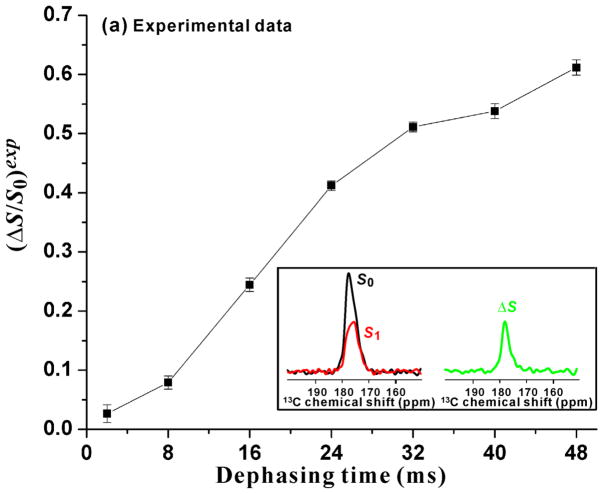
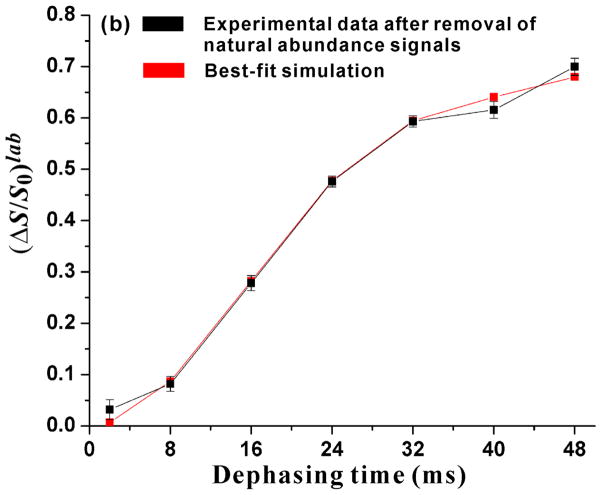
Figure 2.
Plots of (a) experimental (ΔS/S0)exp vs τ and (b) labeled G16 13CO (ΔS/S0)lab and best-fit-simulated (ΔS/S0)sim vs τ. The sample was the membrane-associated H3_20 fusion peptide at pH 5.1. Panel a inset displays the 13CO regions of the S0 (black), S1 (red), and ΔS ≡ S0 − S1 (green) spectra for τ = 32 ms. Each spectrum was processed with 20 Hz exponential line broadening and baseline correction. The (ΔS/S0)exp were calculated from the S0 and S1 13CO intensities integrated in 3.0 ppm windows centered at the peak 13CO shift = 177.1 ppm. The displayed ±σ uncertainties in ΔS/S0 were calculated using the amplitudes of the spectral noise (Zheng et al. 2006). The REDOR experiment consisted of cross-polarization (CP) to generate 13C magnetization followed by the dephasing period of duration τ and then 13C acquisition. During the dephasing period of the S0 acquisition, 13C π pulses were applied at the end of every rotor cycle except the last one. During the dephasing period of the S1 acquisition, there were additional 15N π pulses at the center of each rotor cycle. XY-8 phase cycling was applied to the 13C π pulses and was also applied to the 15N π pulses (Gullion et al. 1990). The NMR experimental conditions included 10.0 kHz MAS frequency, sample cooling gas temperature of −50 °C with sample temperature of approximately −30 °C, 50 kHz 1H π/2 and CP fields, 80 kHz 1H decoupling field applied during the dephasing and acquisition periods, 61–65 kHz ramped 13C CP field, 8.0 μs 13C π pulses, and 10.0 μs 15N π pulses. The pulses were calibrated using a lyophilized helical peptide as described previously (Zheng et al. 2006). For each τ, the numbers of S0 and S1 acquisitions were equal. The specific number of acquisitions was τ-dependent, varied between ~3000 and ~55000, and was set so that the uncertainty in (ΔS/S0)exp was about 0.01. Panel b displays (ΔS/S0)lab (black squares) calculated after removal of the natural abundance contributions from (ΔS/S0)exp. The (ΔS/S0)lab were largely due to the labeled G16 13CO-F9 15N dipolar coupling. The red squares are the simulated (ΔS/S0)sim calculated using the closed/semi-closed model and the best-fit parameters fc = 0.41, dc = 65.4 Hz, fs = 0.59, and ds = 21.2 Hz.
In the REDOR experiment, two 13CO spectra are obtained that are denoted by S0 and S1. Spectra are acquired for a series of dephasing times (τ) and are specifically sensitive to the 13CO-15N dipolar coupling parameter (d ) where for d in Hz and interspin distance (r) in Å, d = 3066/r 3. The value of d is reflected in reduced 13CO signal intensity in the S1 spectrum relative to the S0 spectrum and in the buildup of this intensity reduction with increased τ. The reduction is quantified as dephasing ≡ ΔS/S0 = (S0 − S1)/S0 where S0 and S1 are the 13CO intensities of the respective spectra. For semi-quantitative analysis of REDOR data, a useful model is based on the approximations: (1) the 13CO signal is only due to the labeled G16 13CO nuclei; (2) there is a single peptide structure; and (3) each G16 13CO nucleus is only strongly coupled to the F9 15N nucleus in the same peptide molecule. Because of approximations (1-3), there are only isolated intramolecular 13CO-15N spin pairs and all pairs in the sample have a single value of r and therefore single value of d. For this model, each τ has a corresponding unitless λ = dτ and there is a single universal function for ΔS/S0 (λ) that has approximately sigmoidal shape (Gullion 1998). For λ ≈ 0.2, ΔS/S0 ≈ 0.05 and for l » 0.7, ΔS/S0 » 0.5 while for λ ≈ 1.5, ΔS/S0 reaches its asymptotic value of ~1. The value of d can be estimated using d ≈ 0.7/τ1/2 where τ1/2 is the τ value for which ΔS/S0 has half its maximal value.
We first consider this model for a single peptide structure with a single r and corresponding single d. For the Fig. 1a open structure, the r = 10.5 Å with corresponding d = 3 Hz and for the Fig. 1b semi-closed structure, the r = 5.2 Å and d = 22 Hz. For the closed structure in detergent, the r = 3.7 Å and d = 61 Hz. The experimental τ vary between 2 and 48 ms so for the open structure with d = 3 Hz, the maximum λmax ≤ 0.2 and the expected (ΔS/S0)max ≤ 0.05. For the Fig. 1b semi-closed structure, there would be substantial buildup of ΔS/S0 with τ and τ1/2 ≈ 32 ms. For the closed structure, there would be the most rapid buildup of ΔS/S0 with τ and τ1/2 ≈ 12 ms.
Fig. 2a displays the experimental (ΔS/S0)exp vs τ and the S0, S1, and ΔS spectra for τ = 32 ms are displayed in the inset. The ΔS/S0 increase rapidly with τ with (ΔS/S0)max ≈ 0.6 for τ = 48 ms. One potential reason why (ΔS/S0)max < 1 is that the 13CO signal is a sum of the labeled (lab) signal from the labeled G16 site and the natural abundance (na) signals from the unlabeled 13CO sites. The distances between some of these natural abundance sites and the F9 15N are likely larger than the distance for the G16 13CO nucleus. Relative to the earlier model where all 13CO-15N pairs have the same d, this heterogeneity could lead to reduced (ΔS/S0)exp. Spin counting supports a ~0.75 fractional contribution to the 13CO intensity from G16. However, the G16 contribution to the (ΔS/S0) in Fig. 2a is likely larger because the intensities are integrated in a 3.0 ppm interval centered at the S0 peak shift. More detailed consideration of the peak shift and the labeled vs natural abundance contributions are presented later in this paper. At this point, we consider an approximate model where the data represent a ~0.6 fractional population of the peptide with a single r and d and this population completely dephases, ie. (ΔS/S0) ≈ 1 for τ = 48 ms. For this population, the (ΔS/S0)max ≈ 0.6 with consequent τ1/2 ≈ 20 ms and corresponding d ≈ 35 Hz which is intermediate between the calculated d values of the closed and semi-closed structures. This qualitative analysis does support a large population of some type of closed structure.
For Fig. 2a inset, the respective peak S0 and ΔS shifts of 177.1 and 177.2 ppm support a dominant contribution to each spectrum of labeled G16 13CO in helical conformation. These shifts are similar to the G16 13CO shift of 177.0 ppm for closed H1_23 peptide in detergent with G16 in helical conformation in this closed structure (Lorieau et al. 2010). The 177.1 ppm shift also matches much better database shifts of Gly 13CO nuclei in helical conformation than in β strand conformation (Zhang et al. 2003). Relative to the ΔS0 spectrum, the narrower linewidth of the ΔS spectrum correlates with a larger contribution to ΔS from G16 13CO nuclei and a smaller contribution from natural abundance 13CO nuclei. This is consistent with the proximity of G16 13CO and F9 15N in a closed or semi-closed structure.
The quantitative analysis of the REDOR data started with SIMPSON program calculation of (ΔS/S0) vs τ for each natural abundance 13CO site (Bak et al. 2000). Because the qualitative analysis of (ΔS/S0)exp vs τ supported a large fraction of peptide in some type of closed structure, the input d of the calculation for a particular natural abundance site was based on the r between this natural abundance CO nucleus and the F9 N nucleus in the closed structure in detergent (Lorieau et al. 2010). For each τ, these individual site (ΔS/S0) were then averaged to yield (ΔS/S0)na. Fig. 2b displays the labeled (ΔS/S0)lab calculated from (ΔS/S0)exp = flab(ΔS/S0)lab + fna(ΔS/S0)na where flab = 0.78 and fna = 0.22 were estimated from spin counting. Because the effects of natural abundance 13CO nuclei were removed to obtain (ΔS/S0)lab, these (ΔS/S0)lab could then be analyzed considering only the G16 13CO-F9 15N spin pairs in the sample. As expected from the dominant contribution of the labeled G16 to the experimental spectra, the (ΔS/S0)lab and (ΔS/S0)exp for a given τ typically differed by less than 15%.
The asymptotic value of (ΔS/S0)lab at large τ is ~0.7 and is not consistent with a single d for all G16 13CO-F9 15N spin pairs. A “closed/open” model was therefore considered with a fc fraction of peptides with closed structure and a fo fraction of peptides with open structure. The fc + fo = 1 and the closed and open structures were non-exchanging. The approximation do = 0 was made with consequent (ΔS/S0)o = 0. SIMPSON calculations of (ΔS/S0)c(dc, τ) were then done for a range of dc values. For each pair of fc and dc values, the χ2(fc,dc) was then computed using [(ΔS/S0)lab(τ) − fc × (ΔS/S0)c(dc, τ)]2/σ(τ)2 with summing over the seven τ values and the σ(τ) calculated from the spectral noise amplitudes (Zheng et al. 2006). For a grid of fc and dc values, the smallest χ2(fc,dc) yielded best-fit fc = 0.64 and dc = 51 Hz which corresponded to rc = 3.9 Å. However, the best-fit χ2 = 33 for the closed/open model is significantly larger than the number of degrees of fitting (ν) = 5. The poor fitting with this model is specific to these data. For 13CO-15N REDOR of other membrane-associated fusion peptides, statistically reasonable χ2 were obtained from analysis of (ΔS/S0)lab with a model with a single d (Zheng et al. 2006).
A more general “closed/semi-closed” model was then considered in which there were peptide populations with either closed or semi-closed structure and the closed and semi-closed structures were non-exchanging. The respective fractional populations were fc and fs with fc + fs = 1 and the dipolar couplings were dc and ds. The fc, dc, and ds were fitted by a procedure analogous to that previously described for the closed/open model. The best-fit values for the closed structure were fc = 0.41 and dc = 65 Hz (rc = 3.6 Å) and those for the semi-closed structure were fs = 0.59 and ds = 21 Hz (rs = 5.2 Å). The best-fit χ2 = 5.7 is similar to ν = 4 of the closed/semi-closed model and supports the validity of this model. The (ΔS/S0) from these best-fit values are plotted as red squares in Fig. 2b. One estimate of uncertainty in a best-fit value is the deviation that leads to a +1 increase in χ2. For this approach, the other parameters are kept at their best-fit values. Application of this method yielded fc = 0.41 ± 0.01, dc = 65.4 ± 1.7 Hz with corresponding rc = 3.61 ± 0.03 Å, and ds = 21.2 ± 0.5 Hz with rs = 5.25 ± 0.04 Å. Sample-to-sample uncertainties were estimated by preparing a second sample, acquiring new REDOR data, and fitting these new data. For this second sample, the best-fit χ2 = 0.83 and the best-fit parameter values were fc = 0.38, dc = 63.2 Hz with rc = 3.65 Å, and ds = 19.0 Hz with rs = 5.44 Å.
These results support the dominance of closed and semi-closed structures for membrane-associated H3_20 peptide at pH 5.1. The best-fit rc = 3.61 Å is very close to the 3.67 Å G16 CO-F9 N distance of the closed structure in detergent while the best-fit rs = 5.2 Å matches the 5.2 Å G16 CO-F9 N distance of the Fig. 1b semi-closed structural model. We note that the interhelical geometry of the Fig. 1b model is based on the turn dihedral angles derived primarily from 13C shifts. Additional interhelical distances will help to validate/refine this semi-closed structural model.
There are several lines of evidence that support assignment to intra- rather than intermolecular distances. First, when fitting the data with two distances, ~0.4 fraction of the molecules in membranes have a distance that is the same as that of the closed structure in detergent. Second, the earlier solid-state NMR study supported the helix-turn-helix structure and large dephasing through intermolecular contacts of this structure would probably require a large aggregate of molecules (Sun and Weliky 2009). Earlier solid-state NMR studies are consistent with significant motion of the molecules near ambient temperature and are inconsistent with an aggregate (Bodner et al. 2004). Third, although fully antiparallel homodimers could be consistent with our data, there is strong evidence for a turn, as noted earlier. In addition, this model has been experimentally ruled out for the H1_23 peptide in detergent (Lorieau et al. 2010). Fourth, earlier EPR studies were carried out on a construct that consisted of residues 1-127 of the HA2 subunit of hemagglutinin (Macosko et al. 1997). This construct forms molecular trimers like HA2 and was spin-labeled in the N-terminal fusion peptide region. The EPR spectra of the membrane-bound construct were consistent with three non-interacting fusion peptides that splayed apart from one another.
We note that there are other reports of multiple structures for a fusion peptide including: (1) the H1_23 peptide in detergent at pH 4.0 which has ~0.8 fraction closed structure and ~0.2 fraction open structures in fast exchange (Lorieau et al. 2012); (2) two different continuous helical structures for the HIV fusion peptide in detergent, perhaps with different curvatures (Gabrys and Weliky 2007); (3) helical and β sheet structure for both the membrane-associated influenza fusion peptide and the membrane-associated HIV fusion peptide, with β structure favored in membranes containing cholesterol (Wasniewski et al. 2004; Qiang and Weliky 2009); and (4) multiple antiparallel registries of the β sheet membrane-associated HIV fusion peptide (Schmick and Weliky 2010). In our view, it is not yet clear whether/how this structural plasticity is important in fusion catalysis.
The (ΔS/S0)lab could be well-fitted by a model without any population of open structure. Open structure was predominant in detergent with the H3_20 sequence also used for the present study in membranes (Han et al. 2001). These results therefore suggest that the fractional population of open structure is very different in detergent vs membranes. Other differences between the detergent and membrane-associated H3_20 peptide have been previously reported including respective loss and retention of the C-helix at pH 7.4 (Sun and Weliky 2009). In our view, the present data do not rule out a relatively small (fo < 0.3) open population characterized by do < 4 Hz and (ΔS/S0)o < 0.05 for τ ≤ 48 ms. Future studies could probe for open structure and would likely require extending the τ range to τ ≈ 100 ms so that λo ≈ 0.4 with corresponding (ΔS/S0)o ≈ 0.2.
The earlier solid-state NMR study also showed the minor “B structure” population of membrane-associated H3_20 peptide with very different E11 13C shifts. The TALOS-derived minor peptide structure has a turn that extended over E11, N12, and G13 and differed from the shorter N12 and G13 turn of the major structure. This minor structure is open with r ≈ 13 Å and d ≈ 1 Hz and consequent (ΔS/S0) ≈ 0. An alternate minor structure was also considered, analogous to Fig. 1b, in which the TALOS-derived (φ, ψ) of G13 = (87°, 10°) were replaced by the closed structure (φ, ψ) = (45°, −146°). This structure is also open with r ≈ 8 Å and d ≈ 6 Hz and consequent (ΔS/S0) ≈ 0.1 for τ = 48 ms. As noted above, detection of a small population of such open structures will likely require measurement of (ΔS/S0) at larger τ.
The present study illustrates how solid-state NMR experiments can distinguish between structural models of membrane-associated peptides and can quantitatively determine the distribution of structures including fractional populations. Detection of closed and semi-closed structures supports earlier incorrect determination of the G13 (φ, ψ) based on accurate and correct 13C shifts. The interhelical topology was consequently incorrect. Relative to the open structure of detergent-associated H3_20 peptide, the closed/semi-closed structures of membrane-associated H3_20 peptide may reflect a real difference between fusion peptide/detergent and fusion peptide/membrane interactions.
Distinguishing between the open and closed structures is important because each structure leads to a different model for membrane interaction and for consequent catalysis of the lipid mixing step of membrane fusion. For the fixed-angle open “boomerang” structure, the hydrophobic sidechains are predominantly on the interior of the inserted boomerang and this hydrophobic pocket is hypothesized to perturb the surrounding membrane (Han et al. 2001). By contrast in the closed structure, these hydrophobic sidechains are predominantly on one outside face of the structure and this face likely contacts the membrane (Lorieau et al. 2010).
In addition to these experimental studies, several different groups have done molecular dynamics simulations for the fusion peptide with varied results about the extents of helicity and ranges of interhelical angles (Vaccaro et al. 2005; Jang et al. 2008; Panahi and Feig 2010; Legare and Lague 2012). There was one study which showed predominant closed structure for the H3_20 peptide in implicit membranes (Panahi and Feig 2010).
Acknowledgments
The research was supported by NIH AI47153.
References
- Bak M, Rasmussen JT, Nielsen NC. SIMPSON: A general simulation program for solid-state NMR spectroscopy. J Magn Reson. 2000;147:296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- Bodner ML, Gabrys CM, Parkanzky PD, Yang J, Duskin CA, Weliky DP. Temperature dependence and resonance assignment of 13C NMR spectra of selectively and uniformly labeled fusion peptides associated with membranes. Magn Reson Chem. 2004;42:187–194. doi: 10.1002/mrc.1331. [DOI] [PubMed] [Google Scholar]
- Bodner ML, Gabrys CM, Struppe JO, Weliky DP. 13C-13C and 15N-13C correlation spectroscopy of membrane-associated and uniformly labeled HIV and influenza fusion peptides: Amino acid-type assignments and evidence for multiple conformations. J Chem Phys. 2008;128:052319. doi: 10.1063/1.2829984. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Curtis-Fisk J, Preston C, Zheng ZX, Worden RM, Weliky DP. Solid-state NMR structural measurements on the membrane-associated influenza fusion protein ectodomain. J Am Chem Soc. 2007;129:11320–11321. doi: 10.1021/ja073644g. [DOI] [PubMed] [Google Scholar]
- Dubovskii PV, Li H, Takahashi S, Arseniev AS, Akasaka K. Structure of an analog of fusion peptide from hemagglutinin. Prot Sci. 2000;9:786–798. doi: 10.1110/ps.9.4.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell SR, Martin I, Ruysschaert JM, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol Membr Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- Durrer P, Galli C, Hoenke S, Corti C, Gluck R, Vorherr T, Brunner J. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J Biol Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- Epand RM. Fusion peptides and the mechanism of viral fusion. Biochim Biophys Acta. 2003;1614:116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- Gabrys CM, Weliky DP. Chemical shift assignment and structural plasticity of a HIV fusion peptide derivative in dodecylphosphocholine micelles. Biochim Biophys Acta. 2007;1768:3225–3234. doi: 10.1016/j.bbamem.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullion T. Introduction to rotational-echo, double-resonance NMR. Concepts Magn Reson. 1998;10:277–289. [Google Scholar]
- Gullion T, Baker DB, Conradi MS. New, compensated Carr-Purcell sequences. J Magn Reson. 1990;89:479–484. [Google Scholar]
- Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J Magn Reson. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Han X, Bushweller JH, Cafiso DS, Tamm LK. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nature Struct Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- Han X, Tamm LK. A host-guest system to study structure-function relationships of membrane fusion peptides. Proc Natl Acad Sci USA. 2000;97:13097–13102. doi: 10.1073/pnas.230212097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Michaud-Agrawal N, Johnston JM, Woolf TB. How to lose a kink and gain a helix: pH independent conformational changes of the fusion domains from influenza hemagglutinin in heterogeneous lipid bilayers. Proteins-Struct Funct Bioinform. 2008;72:299–312. doi: 10.1002/prot.21925. [DOI] [PubMed] [Google Scholar]
- Korte T, Epand RF, Epand RM, Blumenthal R. Role of the Glu residues of the influenza hemagglutinin fusion peptide in the pH dependence of fusion activity. Virology. 2001;289:353–361. doi: 10.1006/viro.2001.1108. [DOI] [PubMed] [Google Scholar]
- Lai AL, Park H, White JM, Tamm LK. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J Biol Chem. 2006;281:5760–5770. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- Lee HS, Choi J, Yoon S. QHELIX: A computational tool for the improved measurement of inter-helical angles in proteins. Protein J. 2007;26:556–561. doi: 10.1007/s10930-007-9097-9. [DOI] [PubMed] [Google Scholar]
- Legare S, Lague P. The Influenza fusion peptide adopts a flexible flat V conformation in membranes. Biophys J. 2012;102:2270–2278. doi: 10.1016/j.bpj.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieau JL, Louis JM, Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid: water interface. Proc Natl Acad Sci USA. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieau JL, Louis JM, Bax A. The impact of influenza hemagglutinin fusion peptide length and viral subtype on its structure and dynamics. Biopolymers. 2013;99:189–195. doi: 10.1002/bip.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieau JL, Louis JM, Schwieters CD, Bax A. pH-triggered, activated-state conformations of the influenza hemagglutinin fusion peptide revealed by NMR. Proc Natl Acad Sci USA. 2012;109:19994–19999. doi: 10.1073/pnas.1213801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko JC, Kim CH, Shin YK. The membrane topology of the fusion peptide region of influenza hemagglutinin determined by spin-labeling EPR. J Mol Biol. 1997;267:1139–1148. doi: 10.1006/jmbi.1997.0931. [DOI] [PubMed] [Google Scholar]
- Panahi A, Feig M. Conformational sampling of Influenza fusion peptide in membrane bilayers as a function of termini and protonation states. J Phys Chem B. 2010;114:1407–1416. doi: 10.1021/jp907366g. [DOI] [PubMed] [Google Scholar]
- Qiang W, Weliky DP. HIV fusion peptide and its cross-linked oligomers: efficient syntheses, significance of the trimer in fusion activity, correlation of β strand conformation with membrane cholesterol, and proximity to lipid headgroups. Biochemistry. 2009;48:289–301. doi: 10.1021/bi8015668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmick SD, Weliky DP. Major antiparallel and minor parallel beta sheet populations detected in the membrane-associated Human Immunodeficiency Virus fusion peptide. Biochemistry. 2010;49:10623–10635. doi: 10.1021/bi101389r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Weliky DP. 13C-13C Correlation spectroscopy of membrane-associated Influenza virus fusion peptide strongly supports a helix-turn-helix motif and two turn conformations. J Am Chem Soc. 2009;131:13228–13229. doi: 10.1021/ja905198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro L, Cross KJ, Kleinjung J, Straus SK, Thomas DJ, Wharton SA, Skehel JJ, Fraternali F. Plasticity of influenza haemagglutinin fusion peptides and their interaction with lipid bilayers. Biophys J. 2005;88:25–36. doi: 10.1529/biophysj.104.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasniewski CM, Parkanzky PD, Bodner ML, Weliky DP. Solid-state nuclear magnetic resonance studies of HIV and influenza fusion peptide orientations in membrane bilayers using stacked glass plate samples. Chem Phys Lipids. 2004;132:89–100. doi: 10.1016/j.chemphyslip.2004.09.008. [DOI] [PubMed] [Google Scholar]
- White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Brasitus TA, Dudeja PK, Fozzard HA, Field M. Relationship between lipid fluidity and water permeability of bovine tracheal epithelial cell apical membranes. Biochemistry. 1986;25:1549–1555. doi: 10.1021/bi00355a014. [DOI] [PubMed] [Google Scholar]
- Zhang HY, Neal S, Wishart DS. RefDB: A database of uniformly referenced protein chemical shifts. J Biomol NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Yang R, Bodner ML, Weliky DP. Conformational flexibility and strand arrangements of the membrane-associated HIV fusion peptide trimer probed by solid-state NMR spectroscopy. Biochemistry. 2006;45:12960–12975. doi: 10.1021/bi0615902. [DOI] [PMC free article] [PubMed] [Google Scholar]