Abstract
Asthma is a chronic inflammatory disease of the airways that leads to various degrees of recurrent respiratory symptoms affecting patients globally. Specific subgroups of asthma patients have severe disease leading to increased healthcare costs and socioeconomic burden. Despite the overwhelming prevalence of the asthma, there are limitations in predicting response to therapy and identifying patients who are at increased risk of morbidity. This syndrome presents with common clinical signs and symptoms; however, awareness of subgroups of asthma patients with distinct characteristics has surfaced in recent years. Investigators attempt to describe the phenotypes of asthma to ultimately assist with diagnostic and therapeutic applications. Approaches to asthma phenotyping are multifold; however, it can be partitioned into 2 essential groups, clinical phenotyping and molecular phenotyping. Innovative techniques such as bipartite network analysis and visual analytics introduce a new dimension of data analysis to identify underlying mechanistic pathways.
Keywords: Asthma, Allergic, Exercise induced, Molecular, Bipartite, Phenotype, Network, Logistic regression, Multivariate adaptive regression splines (MARS), Cytokines, Cluster analysis, Classification regression trees (CART), Heterogeneous, Airway, Inflammation
Introduction
Asthma is a chronic inflammatory disease characterized by inflammation of the airways leading to variable airflow obstruction and airway hyper-responsiveness affecting close to 300 million people globally [1]. Severe asthma, accounting for up to 15% of asthmatic patients, consumes extensive health care costs and is an immense economic burden to society [2]. In industrialized countries, despite 1% to 2% of health care expenditure in management of asthma, clinicians continue to struggle with treatment of severe asthma because response to current therapy is often variable and unpredictable [3, 4].
The mechanism of airway inflammation in asthma has been extensively studied; however, the underlying pathophysiology of asthma requires further investigation [5], as it is heterogeneous. Such mechanistic heterogeneity may underlie important clinical phenotypes, sometimes called ‘endotypes’. Technological advances in proteomics, genomics, and computer science in the recent era have led to ongoing studies to explore the inflammatory pathways and signaling mechanisms associated with asthma. Numerous research techniques have been utilized to characterize the multiple phenotypes of asthma to assist with therapy and prognostication. The latest investigations attempt to use methods of classification that rely less on a priori assumptions in an attempt to create distinct subsets of patients. Ultimately, the objective of defining the clear phenotypes of asthma includes prediction of response to therapy, calculation of the clinical trajectory, and reduction of clinical heterogeneity in clinical trials.
Asthma is a heterogeneous clinical disorder with variable ages of onset, duration of disease process and extent of airway obstruction [6]. Over the past decade, despite the commonality of some clinical signs and symptoms, an awareness of subgroups of asthma patients with distinct characteristics has surfaced [7]. The ‘phenotype’ represents the patients’ observed characteristics and, given the heterogeneity of asthma, this becomes a complex task to undertake. Approaches to asthma phenotyping are multifold; however, it can be partitioned into 2 essential groups, clinical phenotyping and molecular phenotyping. Clinical phenotyping attempts to utilize the clinical presentations, characteristics, and results of common diagnostic tests to subgroup patients. In contrast, molecular phenotyping attempts to demystify the complex pathways in an attempt to classify the pathophysiologic process at a molecular level, based in patterns of expression of proteins or nucleic acids.
Clinical Phenotypes
Allergic Asthma
In the spectrum of clinical phenotypes, several subtypes are noteworthy. First, the allergic asthma phenotype is a syndrome used frequently by clinicians to describe the constellation of airway hyperesponsiveness to various stimuli, excessive mucus production, airway eosinophilia, positive skin test responses to aeroallergens, and elevated serum immunoglobulin E (IgE) [8, 9]. The airway inflammation is mediated by the T cell differentiation to the TH2 cellular pathway and production of inflammatory mediators, specifically interleukin 4 (IL-4), interleukin 5 (IL 5), and Interleukin 13 (IL-13) [10]. The term allergic asthma gained popularity in the clinical arena by general practitioners and subspecialists to describe the seasonal respiratory decompensation associated with allergens, or persistent disease associated with perennial allergens.
Exercise Induced Asthma
Second, exercise induced asthma (EIA) is well described entity, but the pathophysiology is controversial. The presentation of EIA is focused on the sudden onset of asthma symptoms, typically shortness of breath and wheezing, which occurs after the onset of exercise [11]. Patients will exhibit dyspnea and bronchoconstriction, often manifested to the greatest extent after 10 minutes of exercise, or shortly following its cessation. Most patients recover within 60 minutes of cessation of exercise [12]. Both the thermal and osmotic theories have attempted to uncover the mechanism of EIA, but this is yet to be entirely clarified. The thermal theory suggests that the airway cooling and subsequent rewarming sets off an inflammatory cascade. On the other hand, the osmotic theory proposes cellular volume changes as the trigger for inflammatory mediator release [13]. Clinicians have used exercise induced asthma to describe the dyspnea associated with exercise in known asthmatics, and this entity can be observed in close to half of the patients with asthma [14]. It is often important to distinguish patients with exercise as one of several triggers of asthma symptoms from those whose exclusive trigger is exercise.
Despite the heterogeneity of asthma, underlying chronic airway inflammation is the hallmark of this disease process. Thus, therapy with inhaled corticosteroids has been the standard of care for symptom control, often leading to reduction of inflammatory mediators and reduced airway obstruction [7], improvements in quality of life, and reduction in exacerbations of asthma. Interestingly, clinicians have recognized a subset of patients with severe or refractory asthma, accounting for about 5%–7% of patients with asthma [6], who have a decreased response to steroids and have increased symptoms. Although this clinical phenotype is small in number, these patients utilize 50% of health care cost for asthma secondary to the frequent exacerbations, uncontrolled symptoms, and requirement for hospitalization [15, 16].
Clinical Clusters
Given the significant morbidity and lack of understanding of this subset of asthma patients, the National Heart, Lung, and Blood Institute (NHLBI) established the Severe Asthma Research Program (SARP) made up of several academic institutions to further investigate this group of patients. Several seminal papers have emerged from this effort, but relevant to the question of phenotyping, SARP studies revealed 5 clinical phenotypes of asthma by using unsupervised modeling suggesting that different pathophysiologic mechanisms likely leads to the clinical presentations. The recent SARP publication [17••] notes that close to 1600 asthma patients and over 500 severe asthma patients have been evaluated with the SARP collaborative network.
Using the extensive researched clinical variables recorded on the patients, agglomerative cluster analysis was used to classify patients into 5 distinct clusters [18]. The first cluster encompasses patients with mild allergic asthma with early onset and history of atopy. These patients generally have normal lung function and have less health care utilization. The largest group of asthma patients are in Cluster 2, who have mild to moderate allergic asthma. This group exhibits atopy and early onset but are different than Cluster 1 due to the borderline low FEV1 values. Cluster 3 patients are older, very late onset, and have a higher BMI. They are distinct from Clusters 1 and 2 secondary to the less atopic nature, frequent need for systemic corticosteroids, and greater than 3 controller medications. One-third of the patients belong to Clusters 4 and 5, who have longer durations of disease compared with the other clusters. These 2 clusters differ in pulmonary function tests and response to bronchodilators. Patients in Cluster 4 have a less reduced FEV1 with reversibility to almost normal range with bronchodilators (albuterol). However, Cluster 5 patients have more severely reduced FEV1 with much reduced response to bronchodilators.
Induced Phenotype
These 5 clinical phenotypes give clinicians a framework to categorize patients, but do not provide much help in predicting response to therapy. The presence of comorbidities or modifying factors such as smoking, gastroesophageal reflux disease and sinusitis frequently encountered in clinical practice leads to constraints in classifying patients into these clusters. Inhaled corticosteroids continue to be the mainstay of chronic therapy for asthma; however, the response to this therapy remains inconsistent. Response to corticosteroids is an “induced phenotype,” and physicians use the degree of response to tailor therapy. Early identification of these induced phenotypes is vital as corticosteroid-resistant patients have increased morbidity and mortality [17••]. Further investigations in genetic analysis, molecular science, and imaging attempt to expose the mechanisms underlying airway inflammation and, furthermore, to understand the variable response to therapy.
One common theme in the methods to classify asthma thus far is that divisions are based on grouping of patients with similar characteristics, be it molecular or clinical data. These independent divisions do not necessarily correlate across the spectrum for a universal asthma classification schema. This disconnect has prompted investigators to develop novel methods to classify asthma phenotypes, based on molecular markers or inflammatory pathways.
Molecular Phenotype
Molecular phenotypes of asthma have been investigated with great vigor in an attempt to shed light on the fundamental pathways leading to the clinical presentations of asthma. The putative link between the distinct mechanism and the variation in clinical presentation needs further exploration. Molecular phenotyping has the advantage of incorporating knowledge of the fundamental pathways of disease. This understanding is essential to development biomarkers, diagnostic strategies and therapeutics. Historically, airway inflammation in asthma is thought to be T-helper type 2 (Th2) mediated and the cells involved include basophils, eosinophils, and mast cells [19]. Cytokines, including IL-4, IL-5, and IL13 play a prominent role in the inflammatory cascade [20, 21].
Cellular Phenotype
In this subdivision of molecular phenotypes, the cellular phenotypes are noteworthy. As early as the 1990s, Wenzel et al conducted sputum analysis and bronchoscopic evaluation on severe asthmatics, and 2 subtypes of SA were identified: eosinophilic and non-eosinophilic [21]. Further work continued to focus on the cellular component of airway inflammation. Eosinophilic airway inflammation in generally associated with airway hyper-responsiveness and chronic asthma. With continued investigation of induced sputum, elevated numbers of neutrophils were noted in asthmatic patients with increased airflow obstruction [21]. Subsequent studies supported these results that a neutrophilic phenotype correlates with the chronic narrowing of the airways [22], lower eosinophilic counts, and poor response to inhaled steroids [23]. In addition, it is well known that during acute asthma exacerbations, airway neutrophilia is present [24]. Ongoing studies continue and infectious etiologies of the neutrophilic phenotype have been suggested.
Gene Expression
Woodruff and colleagues extended the molecular investigations to gene expression profiling of airway epithelial brushings. They noted that a group of up-regulated genes were enhanced by IL 13 suggesting that IL-13 likely is an activator of epithelial cells in asthma. However, there was variability in IL-13 expression in patients with asthma, and there is a subgroup of asthmatics with low levels of IL-13 despite similar symptoms. Their work revealed a distinct group of up-regulated genes in asthmatics and additionally noted that elevated expression of a set of genes, CLCA1, periostin, and serpinB2, was associated with enhanced response to corticosteroids [25]. Further investigations elucidated that asthmatic patients can be classified into “Th2 high” and “Th2 low” groups suggesting different degrees of airway inflammation [26]. The 2 observations were key because of the concept that a panel of genes or proteins may be a better index of molecular phenotype than any single molecule could be.
Studies conducted by Brasier et al [27] explored phenotypes at a molecular level in depth using the assistance of the SARP database and biobank. The clinical clusters described by SARP suggest that severe asthma patients have different inflammatory processes and likely have distinct airway inflammatory mediators. This observation led to the hypothesis that cytokine expression in the airway might discriminate severe from non-severe asthma. Thus airway cytokine expression patterns in bronchoalveolar lavage (BAL) from matched group of patients with non-severe and severe asthma were analyzed using multiplex cytokine arrays. The data revealed 18 cytokines that have measurable concentrations in the BAL. Unsupervised agglomerative hierarchical clustering was performed and this identified 4 asthma phenotypes. Importantly, Group 1 had a significantly reduced FEV1, FVC, and FEV1 improvement after bronchodilator therapy compared with the other groups, and 60% of these patients were classified as severe asthma by SARP investigators. Group 2, interestingly, had the best preservation of lung function and predominantly composed of non-severe asthma patients. The findings revealed that BAL cytokine patterns were informative distinguishing asthma phenotypes, but did not per se provide much information about the underlying mechanisms of inflammation. Hence, further studies at the proteomic and advanced analytic levels were required to understand the relationship between protein expression patterns and variable asthma presentations.
Machine Learning Methods to Predict Asthma Phenotypes
Brasier et al [27] undertook an analysis of BAL protein expression patterns with the intermediate phenotypes of bronchial responsiveness to methacholine, beta-agonist bronchodilator response, airway eosinophilia, and airway neutrophilia. In the SARP dataset, these intermediate phenotypes had little overlap. Additional analysis of the protein expression patterns demonstrated that each the intermediate phenotype was associated with a distinct protein expression pattern. It is perhaps not surprising that cellular inflammation (neutrophilic or eosinophilic) was related to cytokine and chemokine expression. However, it was of considerable interest that protein expression patterns also related to physiologic variables of methacholine responsiveness and β-2 agonist bronchodilator responses. The observations hint at the power of protein expression profiling.
Brasier et al [28] have tested the accuracy of four different statistical (machine) learning methods to predict each intermediate phenotype. The authors classified 1048 subjects enrolled in the U.S. severe Asthma Research Program (SARP) into 4 distinct clinicopathologic subsets, heuristically derived from pathophysiologic phenotypes of these patients. BAL cellularity determined 2 groups based on ‘eosinophil rich’ or ‘neutrophil rich’ pathology while physiologic response to albuterol (bronchodilators) and methacholine (hyper-responders) comprised of the other two groups. Using logistic regression (LR), multivariate adaptive regression splines (MARS), classification and regression trees (CART) and random forest (RF) methods, cytokine data from the BAL fluid was analyzed for prediction of the above defined subtypes.
This approach led to the identification of LR and MARS as optimal statistical learning approaches for phenotyping of asthma. The method described in the article attempts to find the optimal statistical technique capable of working with molecular data (cytokines) and predicting a clinical phenotype.
The advantage of this method allows for prediction of asthma type based on molecular data. Another advantage was the molecular cytokine data supported the empiric grouping and strengthened classification architecture by finding cytokine profiles that were unique enough to distinguish the a priori classified patient subsets. Moreover, one could surmise that this method is clonally applicable to a similar data set in other disease conditions.
One drawback that this method has in common with others is the heuristic grouping of patients into defined phenotypes. A change in the defining conditions could lead to a different set of patient subgroups, leading to a new classification, which may then have its own unique statistical method capable of predicting the phenotype. Thus, the emergent classification would be ‘condition based’. Moreover, subsets based on the assumption that the defining characteristic is a ‘fixed attribute’ for that group may not apply in situations with high fluxes. Patients with rapidly evolving pathology or in the case where there is change from eosinophilic to neutrophilic infiltrate and vice-versa would not fall into any one category. Another difficulty of this methodology is that just as patient subgroups are predefined, so are the choices of statistical models used, leading to lack of nonexclusivity in choice of machine learning methods.
One way to let the information reveal patterns of disease is to use Network analysis. Information gleaned from statistical learning approaches can be supplemented with a network based approach.
Network Analysis Methods
Network Basics
Network analysis is a discipline of network science and a part of graph theory in mathematics. A network is formed by a set of nodes connected in pairs by “edges” (Fig. 1). Networks have been used to analyze a wide range of datasets, including gene–disease interactions [29] and disease–gene associations [30]. A molecular unipartite (single variable) association network of asthma with 129 SNP has been published by Renkonen et al [31]. Though asthma classification was not attempted, important associations between subgroups of protein classes like Toll-like receptors and chemokines were demonstrated. Recently, networks have also been used to determine if quantitative proteomics of bronchial biopsies from asthmatics can distinguish biological functions [32]. Though networks analyses are finding increasing use in the published literature to analyze a wide range of scientific data such as social networks, occurrence of comorbidities and gene–gene interactions [33•], these unipartite analyses show only one part of a bipartite (eg, subject–cytokine) relationship.
Fig. 1.
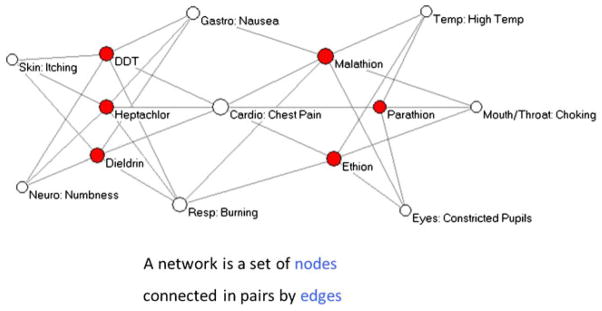
Network basics. A simple network is composed of nodes and edges. Here the network attempts to describe the relationship between the drug and clinical reaction. Red nodes denote the drug. The white nodes denote the reaction. The edges connect the nodes. White nodes (reaction) are distributed in space according to their degree of association to the red nodes (chemical agent). By this representation subsets of reactions and drugs can be identified. For example, it appears that DDT, Hepatachor, and Dieldrin can lead to similar side effects
Bipartite Networks
In contrast to the unipartite networks used in previous studies, bipartite networks contain 2 sets of nodes such as subjects and cytokines, and edges can connect only nodes from different sets. This approach is ideal for the representation of bipartite relationships, which is both more powerful and considerably more complex. Bhavnani et al [34] used this representation to conduct a secondary analysis of the SARP cytokine data. This analysis of the SARP data differed from previous attempts to classify asthma patients, as it did not assume an a priori classification of either patients based on phenotypic (severe vs non-severe or hyper-responsive vs normal), or molecular information.
Application of Bipartite Networks
For a bipartite network analysis to reveal patterns of meaningful relations between variables being studied, the tabular data need to be converted into a network representation. In this case, asthma patients and cytokines were represented as nodes and their normalized cytokine expression values, (Fig. 2), represented as edges connected each patient to each cytokine. Furthermore, in common with network principles, graphical elements like node size (proportional to total expression value of the connecting edges, in this study) and edge thickness (proportional to normalized cytokine expression values in this study) allowed for an information-rich representation in addition to affecting the network architecture. Application of force-directed algorithms (eg, Kamada-Kawai algorithm [35] used in this case) to the layout pushes together nodes that have a similar profile, and pushes apart nodes that do not. Thus, patients who had a higher cytokine expression value for a particular cytokine were spatially closer compared with those who have lower cytokine expression value for the same cytokine. Once the topological relationships are established, the next step is to contrast the real network to random permutations of the network. This is done by comparing the variance, skewness, and kurtosis of the dissimilarities (either between patients, or between cytokines) in the ‘real’ network, to 1000 permutations of the same data. Validation of a pattern is based on the principle that the probability of the observed pattern in the network could not have occurred by random chance.
Fig. 2.
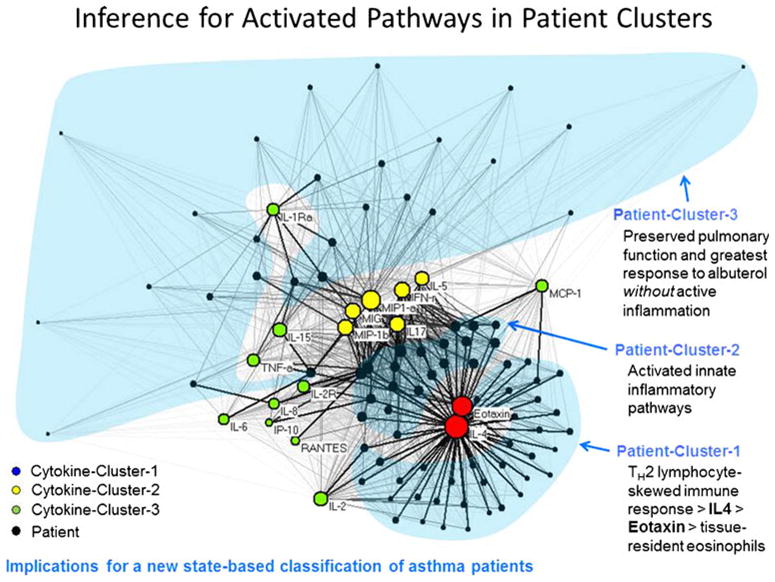
Network analysis of asthma patients and cytokines. The network reveals cytokine and patient clusters
Bipartite Networks in Asthma
As an example (Fig. 2) using this technique, the authors demonstrated that eotaxin and IL-4 (cytokine nodes) were placed close to each other. Exploratory visual analysis of the force-directed layout revealed topological characteristics, which can denote important relationships. Exploratory visual analysis in the SARP bipartite network also revealed 3 patient clusters based on their cytokine profiles and the nature of their relationship with the rest of the patient–cytokine network. The next step was to quantitatively verify the boundaries and the members of the individual clusters, which was done by using agglomerative hierarchical clustering.
Biologic Implications of Bipartite Networks in Severe Asthma
The significance of such exploratory visual and bipartite network analysis comes from the biological implications of clustering effect of cytokines with patients. In this study, the finding of co-occurrence of eotaxin and IL4 (cytokine cluster 1) with patient cluster 1 allowed for authors to infer the pathology of the pathways responsible for the phenotype. Similarly, the specific grouping of cytokines in cluster 2 suggested the presence of a shared NFkB mediated innate response (IL5, IFNγ, MIP1a, MIG, IL-17, and MIP-1b), which is distinct from the cluster 1-Th2 mediated inflammatory pathways. The third cluster of patients, with the weakest cytokine expressions, also had the largest differences in obstructive measures of lung functions and lowest responsiveness to methacholine. By analyzing the clinical variable associated with this subgroup, the authors inferred that this subgroup had preserved lung function and less inflammatory pathway activation. Thus, this molecular information-based patient classification using bipartite networks revealed groups of asthma patients with distinct pathologic mechanisms. Moreover, the comparison of the above classification of patients to the severe/non-severe classification did not show a concordance, suggesting a shortcoming in using a single variable to classify patients.
Application of this methodology is still in its early stages when it comes to analysis of disease patterns in patient phenotypes. Using this method for asthma classification presents an exciting precedent for other diseases and pathologic states. Although bipartite networks are a powerful technique, they are limited in the number of variables that can be simultaneously represented through graphical properties such as color, size, and shape. However, the strengths include a unified representation of both sides of a bipartite relationship (for example, patients and cytokines) in addition to the nature of each relationship through the edge weights. In addition, visualizing the data using 3-dimensional layouts is possible [36], allowing for an extension of analytical capabilities. Another advantage of this method is that it guides the selection of appropriate quantitative measures (eg, cluster analysis if such a pattern exists in the data) that match the underlying structure of the data.
There are other instances where bipartite networks have proven to be useful. Lu et al have done bipartite analysis of human microRNAs and disease associations [37] and discovered patterns of microRNA associated with disease, while other authors have extended network analysis of microRNA data to oncogenic pathways [38] and even protein complexes and drug interactions [39].
Conclusions
Asthma is a chronic disease affecting a large segment of the population. There has been an explosion of information, processes, and data regarding asthma pathogenesis. Discovery of new information permits characterization of the disease in a new light. Apt classification and characterization of asthma will help in providing the most appropriate care to patients with diverse underlying pathophysiology. Network analysis of biomedical data is a relatively new technique of analyzing data. This robust method allows for visualization of emergent patterns in seemingly complex information dense data sets.
Acknowledgments
The authors acknowledge support from the UTMB CTSA grant.
Footnotes
Disclosure Dr. Brasier has received grant support from the National Institutes of Health. Dr. Calhoun has received grant and travel support from the NHLBI/Asthma Clinical Research Network. Drs. Pillai, Divekar, and Bhavnani reported no potential conflicts of interest relevant to this article.
Contributor Information
Regina R. Pillai, Email: rrpillai@utmb.edu, Department of Internal Medicine, Division of Pulmonary and Critical Care Medicine, University of Texas Medical Branch, 5.140 John Sealy Annex, 301 University Blvd, Galveston, TX 77555-0561, USA
Rohit Divekar, Email: rddiveka@utmb.edu, Department of Internal Medicine, Division of Allergy and Clinical Immunology, University of Texas Medical Branch, 5.140 John Sealy Annex, 301 University Blvd, Galveston, TX 77555-0561, USA.
Allan Brasier, Email: arbrasie@utmb.edu, Institute for Translational Sciences, University of Texas Medical Branch, MRB 8.122, 301 University Blvd, Galveston, TX 77555-1060, USA.
Suresh Bhavnani, Email: subhavna@utmb.edu, Institute for Translational Sciences, University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77555-0129, USA.
William J. Calhoun, Email: William.Calhoun@utmb.edu, Department of Internal Medicine, Division of Pulmonary and Critical Care Medicine, University of Texas Medical Branch, 4.118 John Sealy Annex, 301 University Blvd, Galveston, TX 77555-0568, USA
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.National Institutes of Health. National Heart, Lung, and Blood Institute. National Asthma Education and Prevention Program. Expert Panel Report 3: guidelines for the diagnosis and management of asthma. [Accessed March 2012];NIH publication no 07-4051. 2007 Aug; Available at: http://www.nhlbi.nih.gov/guidelines/asthma/index.htm.
- 2.Bousquet J, Khaltaev N. Global Alliance against Chronic Respiratory Disease. Geneva: World Health Organization; 2007. Global surveillance, prevention, and control of chronic respiratory diseases: a comprehensive approach. [Google Scholar]
- 3.Calhoun WJ, Sutton LB, Emmett A, Dorinsky PM. Asthma variability in patients previously treated with B2agonists alone. J Allergy Clin Immunol. 2003;112:1088–94. doi: 10.1016/j.jaci.2003.09.044. [DOI] [PubMed] [Google Scholar]
- 4.Weiss KB, Gergen PJ, Hodgson TA. An economic evaluation of asthma in the United States. N Engl J Med. 1992;326:862–6. doi: 10.1056/NEJM199203263261304. [DOI] [PubMed] [Google Scholar]
- 5.Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–3. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore WC, Peters SP. Severe asthma: an overview. J Allergy Clin Immunol. 2006;117:487–94. doi: 10.1016/j.jaci.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 7.Busse WW, Lemanske RF., Jr Asthma. N Engl J Med. 2001;344:350–62. doi: 10.1056/NEJM200102013440507. [DOI] [PubMed] [Google Scholar]
- 8.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 9.Walker C, Bode E, Boer L, Hansel TT, Blaser K, Virchow JC., Jr Allergic and nonallergic asthmatics have distinct patterns of T-cell activation and cytokine production in the peripheral blood and bronchoalveolar lavage. Am Rev Respir Dis. 1992;146:109–15. doi: 10.1164/ajrccm/146.1.109. [DOI] [PubMed] [Google Scholar]
- 10.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, et al. Interleukin-13: central mediator of allergic asthma. Science. 1998;282(5397):2258–61. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 11.Storms WW. Review of exercise-induced asthma. Med Sci Sports Exercise. 2003;35:1464–70. doi: 10.1249/01.MSS.0000084533.75912.B4. [DOI] [PubMed] [Google Scholar]
- 12.Edmunds AT, Tooley M, Godfrey S. The refractory period after exercise-induced asthma: its duration and relation to the severity of exercise. Am Rev Respir Dis. 1978;117:247–54. doi: 10.1164/arrd.1978.117.2.247. [DOI] [PubMed] [Google Scholar]
- 13.Anderson SD, Daviskas E. The mechanism of exercise-induced asthma is…. J Allergy Clin Immunol. 2000;106:453–9. doi: 10.1067/mai.2000.109822. [DOI] [PubMed] [Google Scholar]
- 14.Hallstrand TS, Curtis JR, Koepsell TD, Martin DP, Schoene RB, Sullivan SD, et al. Effectiveness of screening examinations to detect unrecognized exercise-induced bronchoconstriction. J Pediatr. 2002;141:343–8. doi: 10.1067/mpd.2002.125729. [DOI] [PubMed] [Google Scholar]
- 15.Godard P, Chanez P, Siraudin L, et al. Costs of asthma are correlated with severity. Eur Respir J. 2002;19:61–7. doi: 10.1183/09031936.02.00232001. [DOI] [PubMed] [Google Scholar]
- 16.Serra-Batlles J, Plaza V, Morejón E, Comella A, Brugués J. Costs of asthma according to the degree of severity. Eur Respir J. 1998;12:1322–6. doi: 10.1183/09031936.98.12061322. [DOI] [PubMed] [Google Scholar]
- 17••.Jarjour NN, Erzurum SC, Bleecker ER, Calhoun WJ, Castro M, Comhair SA, et al. Am J Respir Crit Care Med. 2012;185:356–62. doi: 10.1164/rccm.201107-1317PP. This is the latest summary of the research conducted by the Severe Asthma Research Program “SARP”. It provides clinicians the basis of the clinical clusters and the latest ongoing research strategies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, D’Agostino R, Jr, et al. Am J Respir Crit Care Med. 2010;181:315–23. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenzel SE, Schwartz LB, Langmack EL, Halliday JL, Trudeau JB, Gibbs RL, et al. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J RespirCrit Care Med. 1999;160:1001–8. doi: 10.1164/ajrccm.160.3.9812110. [DOI] [PubMed] [Google Scholar]
- 20.Chanez P, Wenzel SE, Anderson GP, Anto JM, Bel EH, Boulet LP, et al. Severe asthma in adults: what are the important questions? J Allergy Clin Immunol. 2007;119:1337–48. doi: 10.1016/j.jaci.2006.11.702. [DOI] [PubMed] [Google Scholar]
- 21.Wenzel SE, Szefler SJ, Leung DY, Sloan SI, Rex MD, Martin RJ. Bronchoscopic evaluation of severe asthma: persistent inflammation associated with high dose glucocorticoids. Am J Respir Crit Care Med. 1997;156:737–43. doi: 10.1164/ajrccm.156.3.9610046. [DOI] [PubMed] [Google Scholar]
- 22.Shaw DE, Berry MA, Hargadon B, McKenna S, Shelley MJ, Green RH, et al. Association between neutrophilic airway inflammation and airflow limitation in adults with asthma. Chest. 2007;132:1871–5. doi: 10.1378/chest.07-1047. [DOI] [PubMed] [Google Scholar]
- 23.Green RH, Brightling CE, Woltmann G, Parker D, Wardlaw AJ, Pavord ID, et al. Analysis of induced sputum in adults with asthma: identifi cation of subgroup with isolated sputum neutrophilia and poor response to inhaled corticoids. Thorax. 2002;57:875–9. doi: 10.1136/thorax.57.10.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sur S, Crotty TB, Kephart GM, Hyma BA, Colby TV, Reed CE, et al. Sudden-onset fatal asthma. A distinct entity with few eosinophils and relatively more neutrophils in the airway submucosa. 1993;148(3):713–9. doi: 10.1164/ajrccm/148.3.713. [DOI] [PubMed] [Google Scholar]
- 25.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA. 2007;104:15858–63. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–95. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brasier AR, Victor S, Boetticher G, Ju H, Lee C, Bleecker ER, et al. Molecular phenotyping of severe asthma using pattern recognition of bronchoalveolar lavage-derived cytokines. J Allergy Clin Immunol. 2008;121:30–7. doi: 10.1016/j.jaci.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brasier AR, Victor S, Ju H, Busse WW, Curran-Everett D, Bleecker E, et al. Predicting intermediate phenotypes in asthma using bronchoalveolar lavage-derived cytokines. Clin Transl Sci. 2010;3(4):147–5. doi: 10.1111/j.1752-8062.2010.00204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–52. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renkonen J, Joenväärä S, Parviainen V, et al. Network analysis of single nucleotide polymorphisms in asthma. J Asthma Allergy. 2010;9(3):177–86. doi: 10.2147/JAA.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Neil SE, Sitkauskiene B, Babusyte A, Krisiukeniene A, Stravinskaite-Bieksiene K, Sakalauskas R, et al. Network analysis of quantitative proteomics on asthmatic bronchi: effects of inhaled glucocorticoid treatment. Respir Res. 2011;22:12:124. doi: 10.1186/1465-9921-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Newman MEJ. Networks: an introduction. Oxford: Oxford University Press; 2010. This text provides insight into network analysis for clinicians and its importance in clinical arena. [Google Scholar]
- 34.Bhavnani SK, Victor S, Calhoun WJ, et al. How cytokines co-occur across asthma patients: from bipartite network analysis to a molecular-based classification. J Biomed Inform. 2011;44 (Suppl 1):S24–30. doi: 10.1016/j.jbi.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman M. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 36.Bhavnani SK, Ganesan A, Hall T, et al. Discovering hidden relationships between renal diseases and regulated genes through 3D network visualizations. BMC Res Notes. 2010;11;3:296. doi: 10.1186/1756-0500-3-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu M, Zhang Q, Deng M, Miao J, et al. An analysis of human microRNA and disease associations. PLoS One. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ooi CH, Oh HK, Wang HZ, et al. A densely interconnected genome-wide network of microRNAs and oncogenic pathways revealed using gene expression signatures. PLoS Genet. 2011;7(12):e1002415. doi: 10.1371/journal.pgen.1002415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nacher JC, Schwartz JM. Modularity in protein complex and drug interactions reveals new polypharmacological properties. PLoS One. 2012;7:e30028. doi: 10.1371/journal.pone.0030028. [DOI] [PMC free article] [PubMed] [Google Scholar]


