Abstract
Microfluidic circuits are characterized by fluidic channels and chambers with a linear dimension on the order of tens to hundreds of micrometers. Components of this size enable lab-on-a-chip technology that has much promise, for example, in the development of point-of-care diagnostics. Micro-scale fluidic circuits also yield practical, physical, and technological advantages for studying biological systems, enhancing the ability of researchers to make more precise quantitative measurements. Microfluidic technology has thus become a powerful tool in the life science research laboratory over the past decade. Here we focus on chip-in-a-lab applications of microfluidics and survey some examples of how small fluidic components have provided researchers with new tools for life science research.
INTRODUCTION
Integrated microfluidic devices have been used in research labs for over twenty years1 and within the last decade their use in life science research has increased dramatically in large part because of the invention of soft lithography2 and microfluidic large scale integration.3 These technological advances have increased the throughput of device production, improved rapid prototyping efforts, and have enabled researchers to enhance the complexity and sophistication of experiments that can be performed on a microfluidic chip. As a result, microfluidic technology is beginning to realize its immense potential for research in life science and medicine.
The realm of microfluidic technology is home to a wide variety of embodiments and applications, but the motivation for its use in medicine and life science research can be reduced to two essential themes. One reason that microfluidic solutions are desired is because the size of the device itself is small. The other motivation to use microfluidic technology comes from the advantages gained from the small size of individual fluidic components. The small device size makes microfluidic technology the ideal platform for portable, point-of-care diagnostic devices. In addition to being small, microfluidic devices can be easy to use, cheap to fabricate and operate, require very little sample, and they can be easily disposed of. For these reasons and more the handheld diagnostic device has been recognized as a potential killer application of microfluidics and is the motivation for much of the research effort in microfluidic technology development.
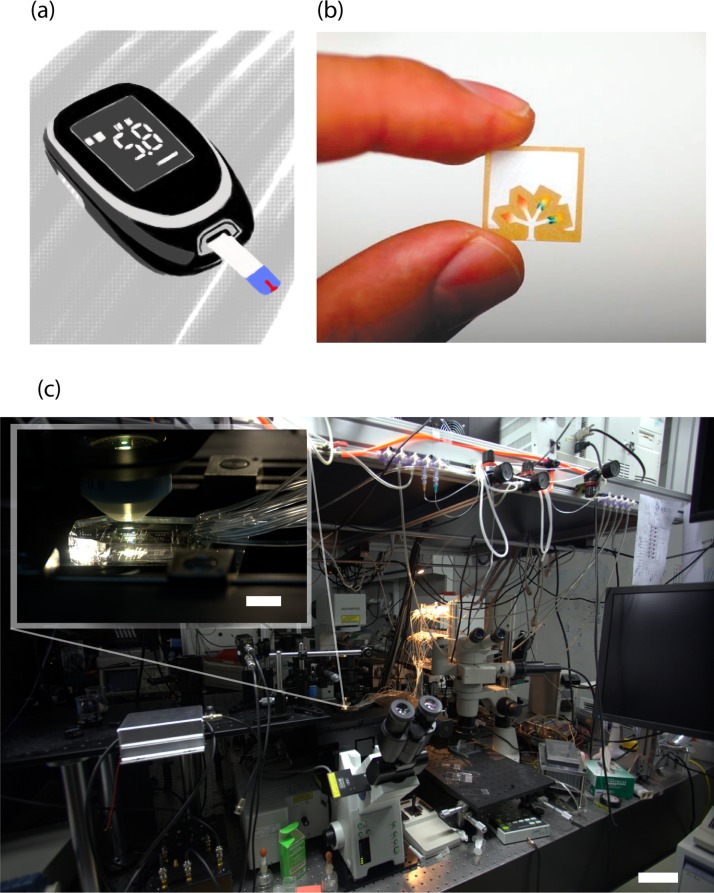
The commercially available glucose meter, for example, is often regarded as the archetype for a handheld diagnostic device4 (Figure 1a). It is cheap ($10–$20 US), easy to operate, provides a clear digital readout of blood glucose level, and uses disposable paper strips for sample delivery. By combining a miniature bio-sensor with a simple and passive microfluidic delivery system the glucose meter requires only a small drop of blood from a finger prick of the operator. The use of paper in microfluidic device fabrication is emerging as a popular strategy and is a promising format for point-of-care diagnostic devices.5 Microfluidic paper-based analytical devices or μPADs as coined by Whitesides6 (Figure 1b) are not only small, they are also thin, cheap and disposable and thus would be ideal for healthcare applications in the developing world. Meanwhile, much work has been done to incorporate microfluidic pumps and actuators into handheld devices for sophisticated and multifunctional diagnostics. For example, a consortium headed by Paul Yager developed a prototype device called the DxBox, a handheld device capable of differential diagnostics of disease specific antigens and antibodies.7 For a more comprehensive survey of current developments towards point-of-care microfluidic immunoassay based diagnostic technology, see the review by Gervais and colleagues.8
Figure 1.
(a) Illustration of generic hand held digital glucose meter. (b) A microfluidic paper-based analytical device (μPAD) Reprinted with permission from A. W. Martinez, S. T. Phillips, G. M. Whitesides, and E. Carrilho, Anal. Chem. 82(1), 3–10 (2010). Copyright 2010 American Chemical Society. (c) A microfluidic chip (inset) in a laboratory. The scale bars are 10 cm and 1 cm (inset).
While much of the research effort in the development of microfluidic technology has been driven by the potential to use small “lab-on-a-chip” technology in point-of-care diagnostic applications, the private sector has yet to reap the full benefits of small microfluidic-based devices. The small size of the individual fluidic circuit components in microfluidic devices, however, provides an arsenal of experimental advantages which make microfluidic solutions desirable, particularly in life science research laboratories. Small components enable researchers to perform experiments which are not possible or practical to execute with more traditional bench top techniques. When evaluating tools of measurement, the size of the probe often determines the resolution and thus precision of the measurement. This is true for physical measurements like atomic force microscopy which can achieve measurement resolution down to the angstrom scale because of a probe tip which is sharpened to a single atom, and it is true for optical measurement, where resolution is proportional to the inverse of the wavelength of the probing light. This concept is also true in biological measurement in many respects. With fluidic components that approach the cellular scale, microfluidic circuits offer exquisite liquid handling capabilities allowing manipulation of single cells and even single molecules in vitro. The size of the components of a microfluidic circuit also allows for high density and parallel architecture thereby increasing the throughput of biological and chemical assays for more precise characterization of heterogeneous systems. Additionally, microfluidic devices are compatible with integration into experimental setups ranging from custom built optical analysis apparatuses to commercial imaging systems and can execute computer automated protocols increasing the efficiency and reproducibility of experimentation. Because of these advantages, microfluidic devices have also been increasingly used not only as a platform for chemical and biological analysis but also as a tool in larger experimental pipelines which require microfluidic solutions to sample preparation or quantification. All of these virtues come from micrometer-scale fluidic components which yield an increased ability to make precise quantitative biological measurements.
These two genres of microfluidic research collectively embody the great potential of microfluidic technology in medicine. On one hand, point-of-care diagnostic device development is centered on coming up with elegant solutions to practical problems to create accessible technology for healthcare applications in resource limited locations. On the other hand, microfluidic platforms are also routinely used in resource-rich research laboratories to provide innovative solutions to “non-practical” problems which may yield fundamental insight into human biology. Here, we focus on this second application of microfluidics and examine the roll of the integrated microfluidic chip in life science research labs or “chip-in-a-lab” applications. Instead of offering a comprehensive review of the state-of-the-art in microfluidic technology, we survey some examples of how researchers have been able to exploit the small size of microfluidic components and incorporate these devices into larger experimental pipelines to make high precision biological measurements. Here, we focus particularly on the miniaturization of fluidic manipulation achieved with droplets and valves in devices made of polydimethylsiloxane (PDMS). There is a large body of work based on electrophoretic manipulation in microfluidic channels which is not covered here but is reviewed elsewhere.9 This work is organized by the unique advantages that microfluidic technology offers in order to highlight ways in which recent microfluidic innovations have revolutionized biology research.
SMALL VOLUMES
Fluidic channels and chambers that are tens to hundreds of micrometers wide are the defining characteristic of microfluidic devices. The small volume of these fluidic circuit components, loosely on the order of nanoliters, enables experiments on-chip that have no bench top equivalent. There are not only practical advantages of small reaction volumes in chemical and biological analysis but there are also fundamental physical and chemical scaling effects that can be exploited in microchannels and compartments. This section presents some old and some new work to summarize the principle advantage of microfluidics.
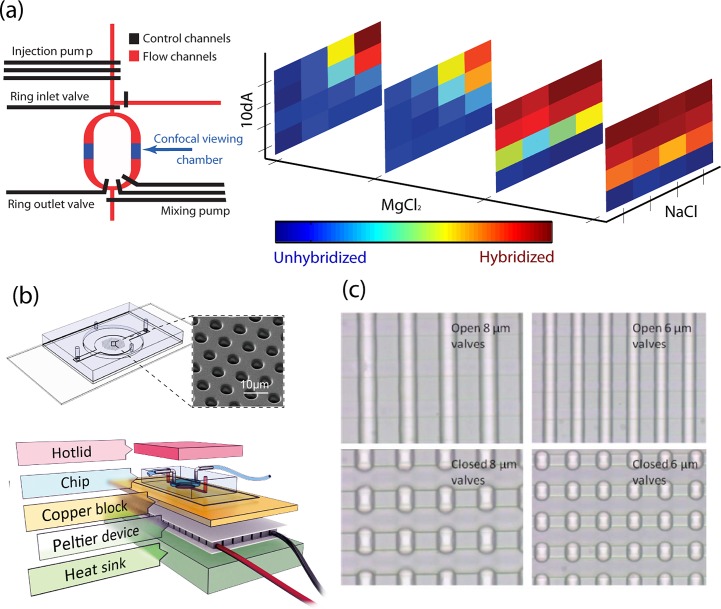
One of the most basic benefits of performing analysis in small volumes is that very little sample is needed. Low-sample consumption is one of the heralded virtues of microfluidic applications and an early demonstration of this was the use of microfluidic devices for protein crystallization screening studies. One of the many challenges of protein crystallization is often the low yield of protein purification. Many of the most coveted protein structures are in part unsolved because of the precious little amount of undegraded product which comes off of purification columns, sometimes with as little as microgram yield for a standard purification protocol. Commercial screens, however, which are used to find crystallization conditions, often require on the order of milligrams of protein to screen hundreds of different potential conditions. Seasoned crystallographers have bags full of tricks to deal with the many obstacles of protein crystallization but more often than not brute force is the solution of choice and to combat low purification yield researchers commonly undergo large volume protein preparation protocols, starting with up to tens of liters of growth media to harvest the bacteria needed for protein expression. In an early application of microfluidic large scale integration, Hansen et al. demonstrated an elegant solution to this problem by using a microfluidic platform for high-throughput screening of crystallization reagents.10, 11 They used two devices; one was a microfluidic formulation device (Figure 2a) based on a ring-shaped mixing chamber which used peristaltic pumps to actively mix protein and reagent in order to characterize the solubility of the protein against combinations of various precipitants and buffers.10 With this automated device thousands of conditions could be coarsely screened with less than 10 μl of protein solution. The second device (Figure 2b) incorporated a passive diffusion-based mixing scheme and was used to screen high potential conditions identified in the formulation device.11 With less than three microliters of protein this chip could perform crystal screens on 244 conditions demonstrating a screening efficiency of over 100-fold better than commercial screens. This paradigm has been used to solve novel protein structures12 and was appropriated into a commercial crystal screening platform.
Figure 2.
(a) A microfluidic formulation device for high throughput solubility screening of proteins. The primary element is a mixing ring. Peristaltic pumps (in red) inject protein and precipitant into ring and yellow pumps mix the contents of the ring. Reprinted with permission from C. L. Hansen, M. O. A. Sommer, and S. R. Quake, Proc. Natl. Acad. Sci. USA 101(40), 14431–14436 (2004). Copyright 2004 National Academy of Sciences, USA. (b) A free interface diffusion based mixing array for protein crystal screening similar to the device used in Refs. 11, 12. (c) A simplified illustration of two microfluidic valves creating a chamber. Typical channels are on the order of 100 μm. Chambers defined in the flow channel by two such valves can be on the order of 100 pl. (d) An emulsion generator using cross flow to shear droplets from the “Y” junction. The two reagents will eventually mix diffusively. (e) A diagram and micrograph showing bacterial confinement in droplets. Green arrows point to bacteria which initiated quorum sensing. Reprinted with permission from J. Q. Boedicker, M. E. Vincent, and R. F. Ismagilov, Angew. Chem., Int. Ed. Engl. 48(32), 5908–5911 (2009). Copyright 2009 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (f) A two-layered microfluidic device for trapping, lysing, and amplifying the genetic material ofsingle cells. Reprinted with permission from Y. Marcy, C. Ouverney, E. M. Bik, T. Losekann, N. Ivanova, H. G. Martin, E. Szeto, D. Platt, P. Hugenholtz, D. A. Relman, and S. R. Quake, Proc. Natl. Acad. Sci. USA 104(29), 11889–11894 (2007). Copyright 2007 National Academy of Sciences, USA.
This crystallization platform is an example of how micro-chambers separated by microfluidic valves can been used for miniature compartmentalization (Figure 2c). Another effective way to achieve small, isolated volumes on chip is with liquid emulsions or droplets (Figure 2d). Stable emulsions can be formed when one fluid is sheared into another immiscible fluid, for example water in oil, using cross flow microfluidic channels.13 These droplets can be used as miniature reactors by introducing multiphase flow to the immiscible medium.14 Droplet based microfluidics occupies a large portion of the field and has been used for countless life science research applications as well as in many commercial laboratory tools, some of which we discuss in following sections. Zheng et al. demonstrated the droplet equivalent of the protein crystallization screening chip15 which can screen hundreds of conditions with microliters of protein. This technology has also evolved into a commercial crystal screening product.16
Droplets created in microfluidic devices, which are most commonly in the range of a few picoliters to hundreds of nanoliters, provide an important demonstration of how small volume confinement can effectively amplify the concentration of molecules or cells under investigation. An elegant example of this is the work by Boedicker et al. on quorum sensing in single bacterial cells.17 When cells reach certain critical density, the concentration of secreted auto inducers reaches a level that can be detected by neighboring cells and activates quorum sensing. At low cell density, these auto inducers rapidly diffuse and delocalize. In a small confined volume, however, the concentration of one or few cells is effectively increased and secreted auto inducers remain localized in the droplet, initiating quorum sensing with low numbers of cells or even a single cell (Figure 2e).
In this work, droplets were created in a different manner18 than cross flow shearing but the principle can be extended to any small volume confinement. We will see that variations on this approach are exploited for a wide variety of applications. Marcy et al. took advantage of nanoliter compartmentalization in a two-layer valve-based microfluidic chip for genomic amplification of single cells. They developed a platform to isolate single cells in a series of chambers for sequential lysis and genomic amplification using multiple displacement amplification and used it to sequence the genomes of uncultivated bacteria found in the human oral cavity19 (Figure 2f). They showed that multiple displacement amplification benefits from the 60-nl reaction chambers, which both increases the effective concentration of genetic material from a single bacterial cell, and reduces amplification bias.20 Further examples of devices which take advantage of this particular small volume asset are discussed later.
The physical properties of fluids change drastically as length scale is reduced and many microfluidic devices take advantage of the unique fluidic physics in small volumes. These physical properties are expressed by dimensionless numbers which define ratios between various fluidic parameters such as viscosity and buoyancy. For example, the Reynolds number is defined as the ratio of inertial force to viscous force and is notoriously low in microfluidic channels. When the Reynolds number is low (≪1) viscosity overpowers inertia and thus particles or molecules, which are not actively set into motion are limited to diffusive movement. The Grashof number relates inertial to viscous forces for buoyancy driven flow and is helpful for understanding the extent of advective mixing in two fluids. The low Grashof number in microfluidic channels is particularly advantageous in the case of protein crystallization on chip. In the passive protein crystal screening device developed by Hansen et al.,11 the precipitating reagent is introduced into a micro-chamber containing protein through a process called free-interface diffusion. This is a technique used in bench top crystallography in which protein and precipitant solution are carefully prepared such that a free interface between the two liquids is created and mixing is purely diffusive creating a concentration gradient to form between protein and precipitant. In a macro-scale environment, however, advective mixing is only avoided by performing the crystal reaction in low-gravity settings, or in a vertical capillary creating a horizontal interface, perpendicular to the direction of gravity. Protein crystals nucleate at some specific point along the chemical gradient but as they gain mass they can fall out of the region conducive to crystal growth and dissolve. In the low Grashof number environment of microfluidic channels a vertical free-interface can be created by “dead-end” filling two opposing channels separated by a microfluidic valve. When the valve is opened, mixing of the two liquids is dominated purely by diffusion and an ideal configuration for protein crystallization is created. Incidentally it is the gas permeability of PDMS which allows the channels to be primed in this manner.
In general, small length scale fluid physics gives microfluidic devices unique and unexpected characteristics. Sometimes low Reynolds number or low Grashof number poses obstacles to researchers, particularly when mixing reagents on chip. A significant body of work negotiates the obstacles imposed by low Reynolds number flow by using surface acoustic waves to drive transport in microfluidic channels.21 This technique can be used to pump fluid through microchannels achieving orders of magnitude increase in flow rates21, 22 and has also been used for active cell sorting applications.23, 24 Often times the aspects of small volumes which would otherwise pose obstacles in microfluidic manipulation, such as low Reynolds number, are exploited for benefits which are unavailable in larger volumes. Cho et al., for example, developed a clever method for separating motile sperm from a heterogeneous sample in a single layer device by concentrating passive particles along laminar flow lines and relying on the active cells to self separate.25 Additionally, Wang et al. exploited the high surface area-to-volume ratio in microfluidic channels to capture circulating tumor cells with high efficiency.26
Low Reynolds number flow also becomes advantageous when it is necessary to manipulate fluid in microfluidic channels with high precision. When inertia is negligible, the microfluidic valve quantizes fluid manipulation the way an electric switch digitizes electronic signals. A single valve is a three-terminal component in which a pressure level on one terminal, the control line, determines the binary fluidic current on the flow line—on or off (Figure 2c). In this sense, the valve is much like a transistor in an integrated electronic circuit, offering an added layer of complexity in circuit architecture. With a minimum of three consecutive valves a peristaltic pump can be created in which a cyclic input signal on the control lines moves discrete amounts of fluid on the flow lines. The quantum of fluid that can be manipulated in a single pump cycle is related to the volume beneath the valve. The pump thus offers exquisite metering resolution and can be used to titrate chemicals with high reproducibility.
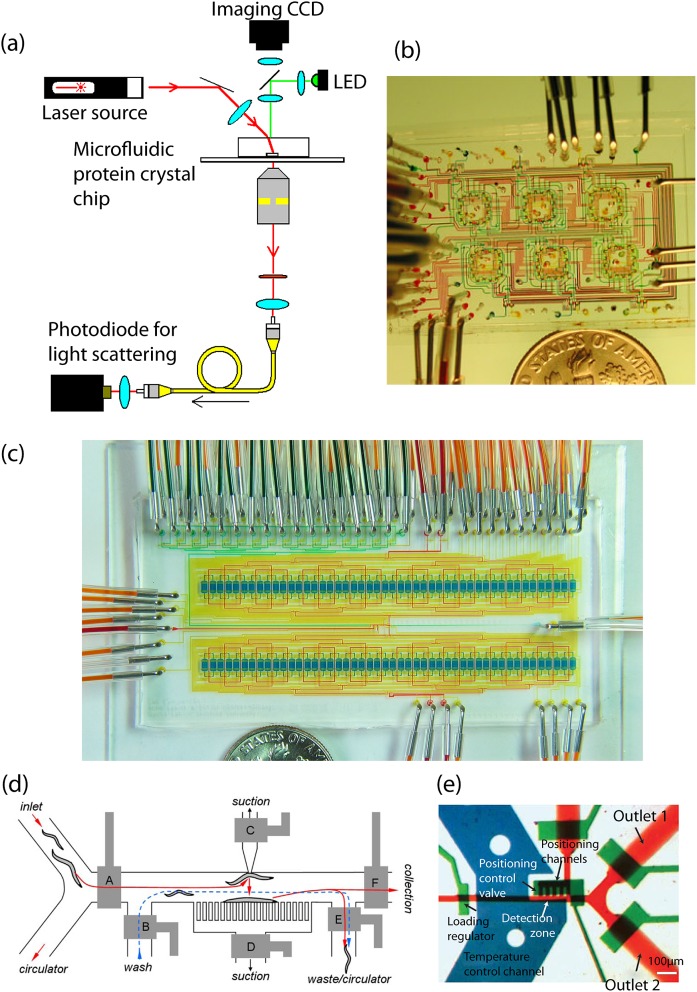
In collaboration with the Weiss group, we took advantage of microfluidic chemical metering to map the conformation of single-stranded DNA molecules in a high density, multi-dimensional chemical space.27 A microfluidic mixing circuit capable of performing arbitrary combinatorial reactions of chemical inputs was integrated with a confocal microscope in order to make fluorescence resonance energy transfer measurements on single DNA molecules. The mixing circuit, powered by a peristaltic pump, could perform serial chemical titrations with resolution of tens of picoliters. With this automated device we constructed two- and three- dimensional conformational maps of the end-to-end distance of ssDNA in response to changing environmental conditions by performing up to 64 sequential single molecule experiments (Figure 3a). On a practical level, this work demonstrates how microfluidic automation can save a researcher time as well as many tedious manual pipetting steps. However, it also shows how the advantage of small volume fluidic manipulation can lead to fundamentally higher precision measurements. While typical pipetting error can be around 0.5%, or 50 nanoliters for microliter reactions, the peristaltic pump injection error is about 0.0001% or in this embodiment 10 femtoliters for 10 nanoliter injections.
Figure 3.
(a) A microfluidic mixing device for high throughput single molecule measurements (left). FRET labeled single stranded DNA is mixed with its complimentary molecule or various salts to measure conformational response. A 3D conformation map is constructed through sequential titration of reagents and automated data collection (right). Reprinted with permission from S. Kim, A. M. Streets, R. R. Lin, S. R. Quake, S. Weiss, and D. S. Majumdar, Nat. Methods 8(3), 242–245 (2011). (b)Ultra-high density digital PCR chip with 36-femtoliter microwells. The illustration depicts the experimental setup for thermal cycling the device. Reprinted with permission from Y. Men, Y. Fu, Z. Chen, P. A. Sims, W. J. Greenleaf, and Y. Huang, Anal. Chem. 84(10), 4262–4266 (2012). Copyright 2012 American Chemical Society. (c) Optical micrograph of microfluidic very large scale integration. These valve arrays are made of channels with cross section of 8 and 6 μm. Reprinted with permission from I. E. Araci and S. R. Quake, Lab Chip 12(16), 2803–2806 (2012). Copyright 2012 by the Royal Society of Chemistry.
While the typical valve can be used to define compartments of tens to hundreds of picoliters, femtoliter confinements have also been incorporated into integrated microfluidic devices using micro-wells fabricated with soft lithography. Our group patterned micro wells of 3 μm diameter onto a deformable PDMS membrane in order to construct a chip capable of performing digital polymerase chain reaction in isolated microreactors packed at a density of greater than 20 000/mm2 (Ref. 28) (Figure 3b). Microfluidic digital PCR is an established technique in which template DNA is diluted and distributed into independent chambers such that on average each chamber has either zero or one molecule. The contents of each chamber are then amplified in parallel by PCR and single starting molecules are quantified by positive amplification events detected with fluorescence of a TaqMan probe.29 This technique allows absolute quantification of the starting genetic material without amplification noise or bias. The 36-fl reaction chambers are particularly advantageous for digital PCR because the increased effective concentration of template molecules allows for fewer thermal cycles. Additionally the high reactor density yields a quantification dynamic range of almost 105. Microfluidic digital PCR is a powerful technique for quantitative measurement of genetic material. Some applications and new approaches will be discussed further in the following sections.
The digital PCR chip is an example of semi-active confinement of femtoliter chambers as a pneumatic press is used to isolate the microreactors. Femtoliter confinement with two-layer valves however has recently been demonstrated by Araci and Quake.30 They achieved microfluidic very large scale integration (mVLSI) by fabricating valves as small as 6 × 6 × 1.5 μm (Figure 3c). Typical microfluidic large scale integration circuits achieve density of thousands of valves per cm2. mVLSI beats this benchmark by two orders of magnitude approaching one million valves per cm2. A peristaltic pump fabricated with these valves could achieve chemical metering with a resolution close to a hundred femtoliters but this has not yet been demonstrated.
These are some first-order examples of how the small volume of microfluidic chambers and channels has enabled researchers to develop new types of research tools. In the proceeding sections, we examine how these elements are compounded, layered, and exploited with integrated microfluidic approaches and used to solve compelling biological questions.
HIGH THROUGHPUT
A natural extension of the small volume advantage is high density. The dense and parallel architecture of microfluidic circuits is the cornerstone of another marquee attribute of the technology; high throughput analysis. In some cases, a high throughput microfluidic solution proves to be a practical advantage, saving a researcher time and resources by scaling up the number of parallel reactions on chip or incorporating automated protocols. In other cases, the scale of an experiment can be prohibitive in a bench top setting and a microfluidic solution fundamentally enables the collection of previously unattainable data. Here, a few examples of the extremely wide range of high throughput microfluidic technologies are presented in order to give a sense for the diversity of approaches to high throughput data collection on chip.
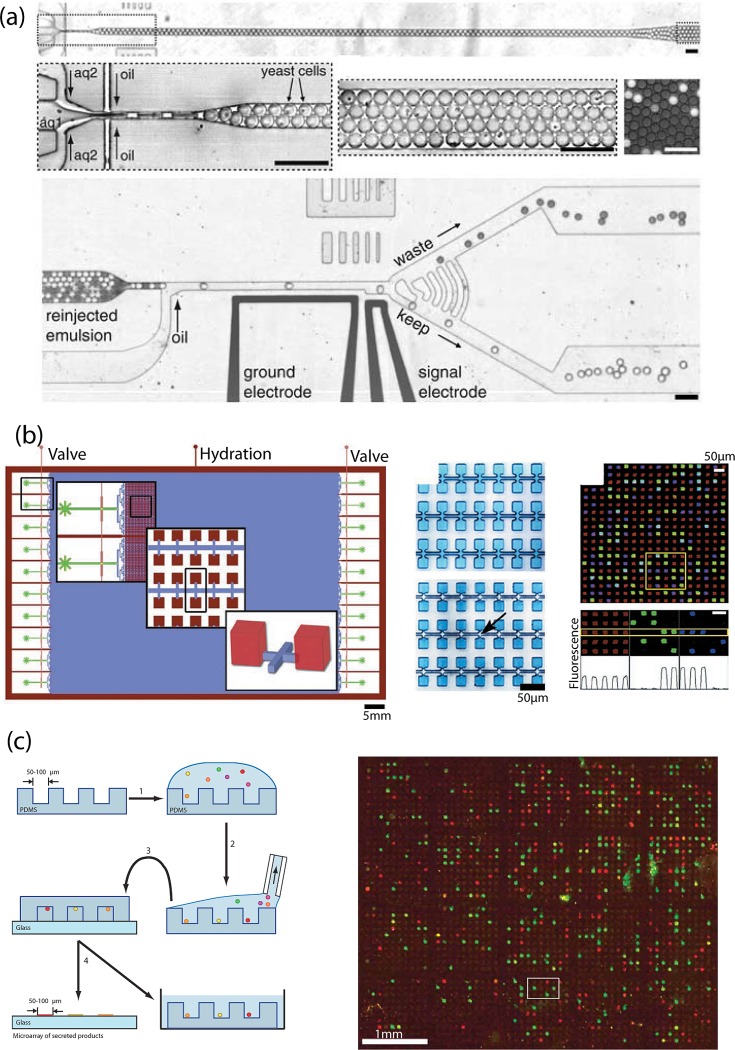
Directed evolution is a powerful technique in which researchers harness the fitness optimization of natural evolution in order to generate potentially useful but unnatural proteins.31 Genetic mutation is artificially induced in cells expressing a protein of potential interest in order to produce new variants. Then, a criterion is used to select cells which tend towards a desired phenotype. Cyclic implementation of this strategy can generate new proteins with desired function. Because natural evolution is an extremely slow process, a necessary quality of directed evolution experiments is the ability to screen variants in a high throughput fashion. Robotic screening tools which use micoilter well array plates can perform assays in excess of 100 000 per day. Agresti et al. used a droplet-based microfluidic device to screen 108 assays in about 10 h, a 1000 fold increase over the robotic platforms32 (Figure 4a). The device is capable of rapidly mixing yeast cells with a fluorogenic substrate into uniform, 6 picoliter droplets. The droplets are packed and stored in a transfer line for incubation. Another device then receives the droplets, refocuses them to a linear stream, and then sorts them using dielectrophoresis based on fluorescence which indicates substrate binding.
Figure 4.
(a) Picoliter emulsion generator, transfer line, and sorter. Reprinted with permission from J. J. Agresti, E. Antipov, A. R. Abate, K. Ahn, A. C. Rowat, J. C. Baret, M. Marquez, A. M. Klibanov, A. D. Griffiths, and D. A. Weitz, Proc. Natl. Acad. Sci. USA 107(9), 4004–4009 (2010). (b) Megapixel digital PCR array. After chambers are filled, they are isolated by flowing an immiscible liquid down the connecting fluid lines preventing molecules from diffusing between chambers. The micrograph shows the imaged array after amplification. Reprinted with permission from K. A. Heyries, C. Tropini, M. Vaninsberghe, C. Doolin, O. I. Petriv, A. Singhal, K. Leung, C. B. Hughesman, and C. L. Hansen, Nat. Methods 8(8), 649–651 (2011). Copyright 2011 Macmillan Publishers Ltd. (c) Process for microengraving (left). A dilute cell suspension is dropped onto the microwell array for cell sequestering. This array is used to print secreted antibodies onto a pre-coated slide. The scanned micrograph displays the resulting microengraved array with fluorescently detected spots (right). Reprinted with permission from J. C. Love, J. L. Ronan, G. M. Grotenbreg, A. G. van der Veen, and H. L. Ploegh, Nat. Biotechnol. 24(6), 703–707 (2006). Copyright 2006 Macmillan Publishers Ltd.
In general, the high throughput capability of microfluidic platforms is useful for chemical screening applications. Droplet microfluidics is well suited for ultra high throughput applications because density and scale limitations of device fabrication are avoided and instead the “chambers” are produced on the fly. Additionally, there is a variety of robust droplet-based circuits for screening assays, including combinatorial mixing14 and serial dilution.33
Chemical screening on chip can also take advantage of automation. The rotary mixer10 is a versatile tool for automated combinatorial mixing of reagents. In addition to the examples mentioned above,10, 27 the microfluidic mixing circuits have been used in applications ranging from automated chromatin immunoprecipitation based drug screening34 to in situ click chemistry screening.35, 36
In array based devices, high throughput is attained by minimizing the size of reaction chambers and increasing density. Heyries et al. increased the throughput of valve-based digital PCR by 100-fold by employing a novel partitioning method to replace the valve in order to achieve a density of 440 000 reaction chambers per cm2 (Figure 4b).37 They performed digital PCR in 1 000 000 ten picoliter chambers to quantify DNA samples with a dynamic range of 107, and single nucleotide variant detection sensitivity below one copy per 100 000 wild type molecules.
Discovery of monoclonal antibodies is a pursuit which has benefits in both biomedical research and in medicine. This task involves screening, identifying, and recovering hybridoma which are cell lines produced by fusing an antibody producing lymphocyte with a myeloma cell to produce antigen-specific antibodies. Success in finding the desired antibody is proportional to the number of screened hybridoma which must be isolated by rounds of serial dilution in order to achieve monoclonality using conventional microwell plates. Love et al. developed a high throughput solution to hybridoma screening and clonal expansion using a PDMS micro-well array that was used to engrave a microarray of secreted antibodies.38, 39 In their technique, a dilute suspension of a polyclonal mixture of hybridomas was dispensed onto a PDMS device containing 25 000 arrayed wells of 100-μm diameter. Single cells settled into the wells, excess media was removed and the device was sealed with a glass slide pretreated with secondary antibodies (Figure 4c). After incubation, secreted antibodies were “engraved” onto the glass slide creating a microarray which was then stained with fluorescently labeled antigens and screened for secreted antigen-specific antibody. The cells remained viable in the micro wells for recovery and clonal expansion.
This technique, termed microengraving, is a powerful tool for characterization and recovery of cells and has been used for investigation into T cell behavior. Varadarajan et al. used microengraving to demonstrate efficient characterization of clinically obtained T cells requiring up to 1000 times fewer cells than traditional approaches.40 After antigen-specific CD8+ T cells were identified they could be recovered for further in vitro characterization or to establish monoclonal cell lines. Han et al. used sequential microengraving to measure temporal cytokine secretion profiles in T cells.41 In a study of stem cells with surface receptors engineered for fluorescent detection of signaling, microengraving was used to mediate cell-to-cell interactions, and assess the spatial-temporal characteristics of the probe.42
Recent work from Shi et al. using a single-cell barcoding chip (SCBC)43 provides another example of how single cell proteomics has and will likely continue to benefit from high throughput microfluidic solutions. The technique takes advantage of two microfluidic devices. The first utilized a concept similar to micro-engraving in which a single-layered passive flow chip was used to pattern a DNA barcode array onto a glass slide.44 The second device, the SCBC, was then aligned to the glass slide containing the DNA barcode array such that each of the 120 cell chambers contained a DNA barcode (Figure 5a). DNA-antibody conjugates were flowed through the chip converting the DNA microarray into a multiplexed antibody array. Single or few cells were then flowed into these chambers, segregated by microfluidic valves, and lysed via diffusive mixing of a lysis buffer. Quantitative single cell expression profiling was achieved for nine cytoplasmic proteins via staining with biotinylated detection antibodies and fluorescent labels43 (Figure 5a).
Figure 5.
(a) Single cell barcoding chip (SCBC) (left). The chip is aligned with a pre-printed DNA bar code (not shown). After conversion to antibody bar code and incubation with single cells the chip is removed and the bar code array is scanned (right). Reprinted with permission from C. Ma, R. Fan, H. Ahmad, Q. Shi, B. Comin-Anduix, T. Chodon, R. C. Koya, C. C. Liu, G. A. Kwong, C. G. Radu, A. Ribas, and J. R. Heath, Nat. Med. 17(6), 738–743 (2011). Copyright 2011 Macmillan Publishers Ltd. (b) Mechanically induced trapping of molecular interactions (MITOMI) chip (top). Optical micrograph inset shows the button and incubation chamber pair. The chip preparation pipeline and button action are illustrated below. Reprinted with permission from S. J. Maerkl and S. R. Quake, Science 315(5809), 233–237 (2007). Copyright 2007 AAAS.
The two nanoliter single cell chambers effectively enhanced the local concentration of cellular protein improving antibody capture efficiency and enabled low abundance expression profiling. The SCBC chip was scaled up to include 1000 cell chambers and used to quantify heterogeneity in cytokine secretion levels from clinical samples of cytotoxic T lymphocytes from both healthy donors and melanoma patients.45
These examples of high throughput microfluidic analysis systems are based on the principle of a well known and mature high throughput technique; DNA microarrays. Maerkl and Quake took advantage of this pre-existing technology and coupled a parallel microfluidic circuit with microarray printing in order to systematically measure transcription factor binding sites.46 The technique, termed “mechanically induced trapping of molecular interactions” (MITOMI), used a microfluidic valve like “button” to mechanically immobilize DNA motif-transcription factor binding events, while unbound molecules were washed away allowing for low background measurement of low affinity interactions (Figure 5b). The button unit cell was arrayed so that 2400 combinations of variants of the basic helix-loop-helix transcription factor family and permutations of its typical DNA binding site could be assessed on a single chip. With 17 chips, they measured 41 000 binding reactions with 464 target DNA sequences. Combining these measurements with bioinformatic analysis of the yeast genome they predicted the biological function of two yeast transcription factors.
MITOMI has proven to be a versatile technology. It has since been used to examine the evolutionary plasticity of transcription factors47 and map protein-protein interaction networks.48, 49, 50 Einav et al. used MITOMI to discover a binding target for a hepatitis C virus transmembrane protein.51 And recently Martin et al. demonstrated RNA-MITOMI,52 a systematic approach to examine the sequence-structure-affinity relationship in RNA-protein interactions.
In an era where proteomics and genomics are handling more and more complex biological networks, high throughput microfluidic platforms have demonstrated effective solutions to systems biology investigations. In addition to the increase in experimental efficiency, high throughput approaches significantly increase experimental precision. This is in part because when making measurements of a single value that are subject to random error, the mean of the measurements approach the actual value of interest as the number of measurements approaches infinity according to the law of large numbers. However, the measurement precision gained with the use of microfluidics does not come only from high throughput data collection. In many instances, the precise liquid handling and technical capabilities of microfluidic circuits enable single or few high quality measurements that qualitatively improve upon the type of experiments that are possible off chip. As we continue, we explore experiments and measurements that are uniquely made possible by microfluidic integration.
INTEGRATION
When conducting on-site measurements in the field, the ideal microfluidic device should be packaged in as small and simple a platform as possible. Hand-held devices are by definition self-sufficient with minimal or passive fluid driving mechanisms and integrated detection systems for easy readout either by eye or by smart phone camera.53 When conducting experiments in the lab, researchers have access to a gambit of external control options and mature table-top analysis techniques. The small size and planar format of microfluidic devices combined with the optical transparency of PDMS makes the technology compatible with many methods of optical detection including microscopic imaging and spectroscopy. In this sense, a microfluidic device is not a research end in and of itself but instead should be considered a research tool, like a pipette or a 96-well plate, to be used in concert with other laboratory technology. As multi-layer soft lithography has become more routine and computer controlled valving more accessible via open source web resources,54, 55 it is now more common to find microfluidic platforms integrated into sophisticated experimental setups for synchronized protocol execution and data collection. It has arguably taken some time for this approach to transcend proof of principle demonstrations, but recently these efforts have begun to bear fruit of scientific significance.
X-ray crystallography is the primary tool for protein structure determination but growing diffraction quality protein crystals is a challenging task. There have been enormous strides in high throughput screening technology, x-ray science, and computational approaches to structure reconstruction from diffraction data. However, the fundamental theory of macromolecule crystal nucleation and growth is relatively incomplete compared to inorganic crystal growth, and experimental observations are rarely predicted by classical theory. Thus building a complete theory of protein crystal nucleation and phase behavior is of fundamental and practical interest.56 We investigated crystal nucleation and growth with multimodal optical analysis of protein crystals grown in a microfluidic environment. Combing dynamic light scattering and imaging with a microfluidic free-interface diffusion device (Figure 6a) similar to the chip described earlier,11 crystal nucleation and growth were monitored in time, enabling crystal size determination with a dynamic range of nanometers to hundreds of micrometers.57 This multimodal approach allowed us to determine for the first time that pre-crystal protein clusters exhibit classical Ostwald ripening after the nucleation of a stable protein crystal. This provides one of many examples of how integration of quantitative analysis with small fluidic components provides an ideal platform for making physical measurements of biological systems.
Figure 6.
(a) Schematic diagram of integrated dynamic light scattering and microscopy for microfluidic-based protein crystal growth studies from Ref. 57. (b) Photograph of the microchemostat. Reprinted with permission from F. K. Balagadde, L. You, C. L. Hansen, F. H. Arnold, and S. R. Quake, Science 309(5731), 137–140 (2005). Copyright 2005 AAAS. (c) Photograph of the cell culture chip used in Ref. 62 courtesy of R. Gomez-Sjoberg. (d) Diagram of chip for C. elegan trapping, imaging, and sorting used in Ref. 64. Reprinted with permission from C. B. Rohde, F. Zeng, R. Gonzalez-Rubio, M. Angel, and M. F. Yanik, Proc. Natl. Acad. Sci. USA 104(35), 13891–13895 (2007). Copyright 2007 National Academy of Sciences, USA. (e) Optical micrograph of C. elegan immobilization chamber in the device described in Refs. 67, 68. Reprinted with permission from K. Chung, M. M. Crane, and H. Lu, Nat. Methods 5(7), 637–643 (2008). Copyright 2008 Macmillan Publishers Ltd.
Burg et al. built a micro-resonator into a microfluidic chip to make high sensitivity measurements of particle mass in an aqueous environment.58 By suspending an enclosed fluidic microchannel in a vacuum, their device was able to measure the mass of particle suspensions in the microchannel with sub-femtogram resolution. Grover et al. used this device, called the suspended microchannel resonator, to make high throughput measurements of single live cell density59 which showed significantly less variability than cell mass or volume. Son et al. combined the suspended microchannel resonator with a fluorescent microscope in order to monitor cell-cycle with an ubiquitination-based cell cycle indicator.60 They measured the mass of single mammalian cells while monitoring cell cycle progression in order to characterize cell growth.
The microscope is arguably one of the most powerful and ubiquitous research tool in biology and it is a fundamental companion of microfluidic devices in the laboratory setting, acting as the premier chip-to-world mediator. Microfluidic platforms possess a handful of attributes that enhance microscope capability as well. For one, microfluidic chips often have regular, arrayed chambers which make automated image collection a very straight forward task. This is particularly useful for multiplexed time lapsed cellular imaging. Additionally, because microfluidic valves and pumps can be computer controlled, it is possible to incorporate machine vision based feedback control loops for active fluidic manipulation.
Two early microfluidic platforms paved the way for automated live cell imaging on a microfluidic platform. Blagadde et al. constructed a microfluidic bioreactor called the microchemostat which could culture and observe bacterial populations for hundreds of hours.61 The device contained six independently controlled reactors (Figure 6b) and actively maintained conditions for bacterial growth by replacing media and maintaining temperature, humidity and nitrogen levels within the microscope housing. The six colonies were constantly monitored with automated microscope stage translation and image acquisition, to create time-lapsed images with single cell resolution. The platform was used to characterize oscillations in cell population density engineered by a population control signaling circuit.
Gomez-Sjoberg et al. developed a general platform for microfluidic cell culture.62 The system contained 96 independently addressable cell culture chambers to which customized media and reagents could be delivered at preset schedules (Figure 6c). Again time lapse imaging was incorporated so that the individual chambers could be automatically maintained and monitored for weeks. Tay et al. used this platform to monitor NF-κB signaling dynamics in single cells responding to TNF-α.63 With active fluidic control, temporal TNF-α stimulation could be delivered to cell chambers independently. They found that at low levels of TNF-α, single cells demonstrated a digital response to the signaling molecule based on analogue processing of parameters like dose intensity or timing.
Whole organism investigation is becoming a growing trend in microfluidic applications as new strategies to manipulate and probe animals in micro channels are emerging. Rohde et al. developed a suite of devices for manipulating, sorting, and screening Caenorhabditi elegans.64 They first presented a strategy for trapping and then immobilizing single C. elegans using microfluidic suction channels (Figure 6d). The worm was flattened against a suction array so that they could be analyzed with bright field and fluoresce imaging. From there, the trapped animal was delivered to an independently addressable incubation chamber where it was again immobilized with suction for time-lapse imaging. A third device was designed to interface the screening chip with reagents from a multi-well plate so that drug and RNAi screening could be carried out in vitro.
There has since been a handful of applications of on-chip C. elegan studies including long term, live organism monitoring for studies of neurotoxin induced response65 and passive age-based sorting.66 Other groups have built upon microfluidic C. elegans screening by implementing an automated active sorting scheme67, 68 (Figure 6e). Crane et al. demonstrate automated screening of a C. elegan mutant library and identified phenotypes related to synapse formation.68 In order to achieve completely autonomous screening and sorting they implemented a machine vision based control loop which operated fluidic valves to introduce a worm into an imaging chamber, capture a pseudo-three-dimensional image, process the image and classify the worm as mutant or wild type, then deliver the worm to respective sorting chambers. They screened worms at a rate of over 220 per hour, out performing manual screening by a factor of ten. Additionally, they achieved ten-fold higher sensitivity to phenotypic variance by applying a sophisticated multi-stage machine vision and learning algorithm that could assess an image using over 30 qualifiers. The platform was robust and has been used to screen up to 40 000 animals and identify novel phenotypic variants, previously undetected by manual classification.
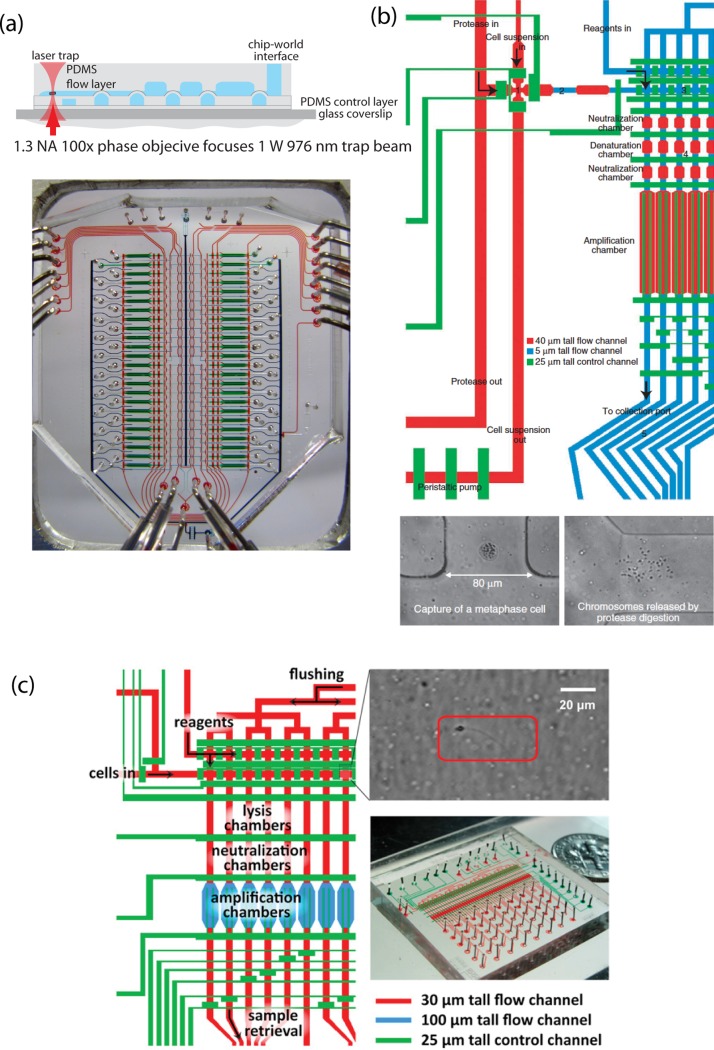
Manipulation of single cells is the area in which microfluidic technology has arguably made some of its most notable progress. This is especially true for application in single cell genomic studies. Cells are particularly easy to handle in microfluidic environments compared to Petri dishes and well plates because the scale of microfluidic components and their moving parts approach the size of cells and because the low Reynolds number environment keeps suspended particles like cells relatively stationary. Handling of cells becomes even more precise when optical manipulation is applied. Blainey et al. used a laser trap to separate single ammonia-oxidizing archaea from a bulk sample and sort them into individual micro-chambers69 (Figure 7a). They used a device similar to the one described earlier by Marcy and colleagues19 to lyse the isolated single cells and amplify their genetic material using MDA (Figure 7a). The genetic material from five single archaea was collected from the chip and sequenced independently. With single cell genomic data and metagenomic data they assembled a draft genome of this low-salinity ammonia-oxidizing archaea. The laser sorting single cell platform is critical for species which cannot be easily cultured in the lab and has been used to assemble a majority of the genome of Thiovulum sp, an un-cultivated genus of sulfur bacteria and the second fasted bacterium ever observed.70 The same approach was used to partially assemble the genome of a member of an un-cultured bacterial lineage labeled candidate division OP11 which are found ubiquitously in nature but whose ecological interactions are largely unknown.71 This platform has also been applied to the study of mammalian microbiome. Pamp et al. sequenced the genome of single segmented filamentous bacterial cells, an intestinal symbiont found in mammals and revealed a complex interdependent relationship of the microbe and its host.72
Figure 7.
(a) Diagram of laser trap sorting of single cells. This is a two layer “push-up” device and the laser trap drags the cell past two open valves and then into the lysis chamber. (Below)An optical micrograph of the whole device which was used in Refs. 69, 70, 71, 72. Device was fabricated by the Stanford microfluidic foundry staff. Figure courtesy of P. Blainey. (b) Device schematic for single chromosome sequencing. Red and blue lines represent the flow channels and green are the control. Single cells are trapped in the cross-junction, lysed in the following chamber and then chromosomes are separated into the sequential channels. Optical micrographs (bottom) depict a single metaphase cell in the channel and its chromosomes after lysis.73 Reprinted with permission from H. C. Fan, J. Wang, A. Potanina, and S. R. Quake, Nat. Biotechnol. 29(1), 51–57 (2011). Copyright 2011 Macmillan Publishers Ltd. (c) Device schematic for the single sperm sequencing chip used in Ref. 74. Optical micrograph depicts a single sperm cell trapped a flow channel. Reprinted with permission from J.Wang, H. C. Fan, B. Behr, and S. R. Quake, Cell 150(2), 402–412 (2012). Copyright 2012 Elsevier.
The single cell laser trap-based sorting platform in fact conceptually combines two types of microfluidic integration. On one hand, the device is integrated into a microscope platform combining laser tweezing with a semi-automated stage for manipulation and imaging. Meanwhile, these experiments provide an example of systems level integration of microfluidic technology where the whole apparatus is used as a single tool and integrated into a larger experimental pipeline. In this case, the microfluidic device is used simply to collect a sample, which is then prepared for high throughput sequencing that yields reads to be further processed by bioinformatic computing resources.
Microfluidic systems level integration has been particularly effective for single cell genome sequencing studies. Permutations of the single cell isolation and genomic amplification device developed by Marcy et al. have been used in a variety of single human cell sequencing applications. Fan et al. sorted individual human metaphase cells and then further separated homologous copies of chromosomal molecules for amplification and sequencing in order to perform whole genome personal haplotyping73 (Figure 7b). Wang et al. used a similar device to isolate the genetic material from single human sperm cells for whole-genome sequencing in order to measure genomic variation caused by recombination and de novo mutation during gametogenisis74 (Figure 7c). These works provide a clear indication that microfluidic technology has much to offer in the field of single cell genomic analysis.
HETEROGENEITY AND STOCHASTICITY
From both a genotypic and phenotypic standpoint, cellular systems display significant cell to cell variability, even among the same type of cell. In fact, this heterogeneity may not only be a characteristic of cellular networks but may also be critical to their function.75 In recent years, microfluidic platforms have emerged as powerful tools for characterizing the nature and origin of variation in cellular populations through single cell analysis.76, 77, 78 Here, we look at a few such examples of an already large and growing field. For a more comprehensive survey, see the recent reviews by Lecault et al.76 and Ryan et al.79
Growth rate is an important cellular parameter which displays significant heterogeneity in cell populations. Leung et al. characterized heterogeneity in proliferation of monoclonal bacterial colonies with varying initial cell number, ranging between one and 1000.80 These experiments were performed on a powerful and versatile microfluidic platform that took advantage of emulsion-based compartmentalization and valve-based fluid routing (Figure 8a). The device was capable of formulating droplet reactions containing single cells and combinations of up to seven input reagents with ten-fold concentration metering per reagent. These droplets were stored in one of 95 individually addressable chambers for incubation and could be later recovered for further off-chip analysis. The chip architecture allows for generalized experimental protocol ranging from on chip monoclonal cell culture to single or multiple cell lysis, whole genome amplification, and recovery for sequencing. They demonstrated the versatility of this generalized cell manipulation platform by conducting PCR based genotyping of single bacteria, whole genome single bacterial sequencing, and simultaneous whole genome amplification, sequencing, and genomic comparison of naturally occurring bacterial colonies from three different environments. In another cell proliferation study, Lecault et al. successfully cultured non-adherent hematopoietic stem cells in a microfluidic chip containing densely arrayed 4 nanoliter chambers81 (Figure 8b). The device was engineered to maintain culture conditions which facilitated growth rates comparable to a traditional Petri dish culture environment. With automated media exchange and time-lapse imaging, they monitored proliferation of over 6000 colonies of primary HSCs for several days with single cell resolution and assessed how growth factor deprivation affects cell fate.
Figure 8.
(a) Device schematic of the programmable droplet based reaction array used in Ref. 80. Droplets from 8 reagent inputs are directed to an array address and merged with other droplets to form a desired reaction. Reprinted with permission from K. Leung, H. Zahn, T. Leaver, K. M. Konwar, N. W. Hanson, A. P. Page, C. C. Lo, P. S. Chain, S. J. Hallam, and C. L. Hansen, Proc. Natl. Acad. Sci. USA 109(20), 7665–7670 (2012). (b) Chamber array chip for high density culture of non-adherent mammalian cells. Micrographs (top) show chamber array with single cells. Reprinted with permission from V. Lecault, M. Vaninsberghe, S. Sekulovic, D. J. Knapp, S. Wohrer, W. Bowden, F. Viel, T.McLaughlin, A. Jarandehei, M. Miller, D. Falconnet, A. K. White, D. G. Kent, M. R. Copley, F. Taghipour, C. J. Eaves, R. K. Humphries, J. M. Piret, and C. L. Hansen, Nat. Methods 8(7), 581–586 (2011). Copyright 2011 Macmillan Publishers Ltd. (c) Experimental setup for single molecule gene expression studies in single cells.83 Two control channels define nanoliter cell confinement. Reprinted with permission from L. Cai, N. Friedman, and X. S. Xie, Nature 440(7082), 358–362 (2006). Copyright 2006 Macmillan Publishers Ltd. (d) Experimental setup for proteome and transcriptome profiling.84 Different strains of engineered bacteria are monitored in parallel flow channels. Micrograph (right) depicts gene expression observation in single bacteria. Reprinted with permission from Y. Taniguchi, P. J. Choi, G. W. Li, H. Chen, M. Babu, J. Hearn, A. Emili, and X. S. Xie, Science 329(5991), 533–538 (2010). Copyright 2010 AAAS.
Stochastic gene expression is one of the primary contributors to cellular heterogeneity in isogenetic populations.82 Cai et al. measured gene expression of β-galactosidase in single E. coli cells with single molecule sensitivity by detecting a synthetic fluorogenic substrate which is hydrolyzed by the protein upon expression.83 The fluorescent product of this hydrolysis reaction is released through the cell membrane and would typically diffuse quickly into the surrounding medium. In order to combat this loss of signal, these experiments were performed in a microfluidic device, wherein cells were confined, cultured, and interrogated in 100-pl chambers enclosed with microfluidic valves (Figure 8c). Confining the cell to these small volumes localized the secreted product preserving the fluorescent signal associated with enzymatic amplification. The microfluidic platform offered the added benefit of arraying these confinements for parallel observation of multiple single cells, addressed by translating the microscope stage. The authors directly measured protein expression burst amplitude and frequency in real time and showed that these parameters can also be calculated from steady-state copy number distribution measurements. They went on to demonstrate the versatility of technique with measurements of low-level expression in yeast and mammalian cells.
Taniguchi et al. measured stochastic variability in gene expression in single E. coli cells at the transcriptome wide level.84 They used a yellow fluorescent protein fusion library and fluorescence in situ hybridization to count protein and mRNA expression for 1000 genes with single molecule sensitivity. A single layer high density microfluidic flow channel array was used to spatially organize and separate bacterial strains in order to facilitate high throughput image collection (Figure 8d). Coupling the flow cell array with an automated fluorescence excitation and image acquisition system the authors screened 96 strains per device at a rate of 160 cells/second per strain. By measuring steady-state population level copy number distributions of protein and mRNA molecules, they were able to characterize transcription rates and protein expression burst amplitudes, as demonstrated by Cai et al.,83 with single cell resolution.
Single cell gene expression analysis is especially useful for the study of heterogeneous cell populations, which is often the case when investigating tissue or environmental samples where populations do not necessarily contain a single cell type. In such a case population wide gene expression analysis with single cell resolution is critical for defining subsets of cells or identifying rare cells. In order to differentiate cell type amidst a noisy gene expression profile, many genes and many cells must be examined and microfluidics offers significant practical advantage for this task.85 Dalerba et al. used fluorescence activated cell sorting and microfluidic gene expression arrays to identify distinct subpopulations of cells in colon cancer tissue.86 The microfluidic device used here was a commercial chip sold by Fluidigm Corp. which enables quantitative PCR of 96 target genes in 96 samples. With nanoliter reaction chambers, a low number of mRNA copies can be quantified allowing for multiplexed expression analysis of material from single cells. Normal and cancer tumor tissues were dispersed to single cell suspensions and sorted by flow cytometry based on markers which identify cells of specific lineage, function or location in the colon epithelial. Expression of up to 53 genes, selected for differential expression from a pool of over 200 genes was then quantified in hundreds of single cells from these populations using the microfluidic array. The authors used statistical clustering algorithms to characterize the transcriptional identity of tumor subpopulations in order to explain the nature of heterogeneity in cancer tissue.
Microfluidic quantitative PCR has proven to be a powerful tool for characterization of cellular heterogeneity. Buganim et al. studied gene expression at various stages of cellular reprogramming and by virtue of the single cell resolution of their approach, revealed two characteristic stages distinguished by transcriptional profile.87 Using microfluidic qPCR, they quantified expression of 48 genes in 7000 cells and showed that transcription during early stages of cell reprogramming is a stochastic process, however, in later stages gene activation mechanisms that lead to pluripotency can follow hierarchical trajectory.
The ability of these platforms to perform experiments in a high throughput and parallel fashion increases both the reproducibility and the quantity of each single measurement. This significantly increases measurement precision allowing for more accurate estimation of the variation in these stochastic systems. These last few examples demonstrated the potential for more regular application of robust microfluidic platforms in life science laboratories. While it is clear that microfluidic technology has great potential as a tool for single cell analysis, whether it becomes a common and practical tool in the research lab depends on the extent to which experimental platforms can develop into robust, user friendly technologies that can be exploited by the larger life science community. There have been a handful of examples which point towards this paradigm. In the next section, we look at microfluidic devices which have found their way into commercial platforms.
CHIP IN A BOX
In the introduction, we discussed how point-of-care diagnostics was identified early on as a primary commercial application of microfluidic technology. Another area in which microfluidics has begun to demonstrate commercial impact is in laboratory research equipment. Many modern high throughput life science research tools now regularly take advantage of the “small component” aspect of microfluidic circuits and sell microfluidic platforms powered by highly engineered control infrastructure and analysis technology, packaged into a black box for the research scientist end user.
Microfluidic based bench top machines provide a practical alternative to traditional wet lab protocols. A virtue of many of these products is that they are simple to operate. The user only has to load their sample and some reagents from a kit and press go while the control machinery routes the samples through the chip, performs the assay, makes the measurement and then prints the read out on a screen. In addition to the small component advantages which have been reviewed, the microfluidic element of these commercial machines is often a disposable cartridge, thus avoiding contamination between experiments.
Caliper Life Sciences88 offers a series of platforms called the LabChip Systems which perform capillary electrophoresis in a microfluidic chip. A robotic sample delivery mechanism loads a single layer microfluidic device with sample and labeling reagents and then performs electrophoretic separation in the microfluidic channel detecting bands of protein or nucleic acid with inline fluorescent excitation and detection. The Agillent 2100 Bioanalyzer is another versatile system for separation, sizing, and quantification of DNA, RNA, and protein. It is a one-box system which also uses a disposable cartridge with a microfluidic circuit and a reagent kit designed for electrophoretic quantification and whole cell flow cytometry. Commercial microfluidic machines like these take a variety of traditional wet lab protocols like gel electrophoresis and reduce them to a single easy-to-use platform and have been used in countless published works.89, 90
Other commercial platforms take advantage of microfluidic technology to present qualitatively new approaches to quantifying biological systems. Fluidigm offers a suite of microfluidic platforms for genetic analysis from high throughput gene expression to sample quantitation and sequencing preparation. Their BioMark system uses dense microfluidic chamber arrays to perform quantitative or digital PCR on small volumes of genetic material. In addition to allowing densely packed arrays for high throughput analysis, the small volume reaction chambers yield higher sensitivity enabling detection of single cell sample quantities. A few applications of the BioMark to single cell gene expression were discussed in the previous section. Digital PCR is also powerful tool for quantitation of genetic material. Because microfluidics has proved to be an ideal way to implement the technique,29 the technology has found many uses as a research tool. The single molecule sensitivity of digital PCR makes for extremely precise counting of DNA and groups have used the platform for quantitation of viral load,91 viral detection in bacterial communities,92 detection of human aneuploidy,93 and as a novel platform for noninvasive detection of organ transplant rejection,94 to cite a few examples.
Droplet microfluidics presents another elegant solution to digital PCR. Genetic material can be diluted into droplets contained in an immiscible medium creating microreactors for PCR. Droplets are created sequentially then sorted into a densely packed storage array where they undergo thermal cycling. Afterwards, they can be recovered sequentially for sorting or detection as Agresti et al. demonstrated.32 Two commercial platforms take advantage of this approach to digital PCR; the QX100™ Droplet Digital™ PCR system from BIO-RAD Laboratories Inc., and the RainDrop™Digital PCR system from RainDance technologies. With these platforms, millions of PCR reactions can be created and analyzed providing a powerful platform for high throughput DNA quantitation. For example, droplet based PCR has been used for copy number variant counting in human evolution studies,95 genomic enrichment for targeted sequencing,96 and quantitative detection of rare mutated DNA molecules.97, 98
The marketing departments of many of the companies mentioned embrace the microfluidic components in their machines because incorporation of microfluidic plumbing highlights the new capacity and capability of these platforms. Indeed, it is the small fluidic components which make these products useful to researchers in the lab. However, there is a whole class of transformative life science technology which regularly relies on microfluidic components but does not necessarily need to advertise this aspect in order to sell units. Next generation sequencing has revolutionized life science research99 and with the plummeting time and cost required for DNA sequencing more labs have gained access to this powerful technology. Just about every next generation sequencing platform on the market incorporates microfluidic technology for reagent handling. A passive microfluidic flow cell array or microwell array is used in most machines to handle reactions and align the sample with detection optics. Many of the microfluidic advantages covered in the preceding sections are exploited in these machines, particularly low sample consumption, high density compartmentalization, and increased effective concentration in small volumes. Additionally, there are a handful of commercial sequencing sample preparation devices that also take advantage of microfluidic technology. Table TABLE I. organizes some notable commercial products which incorporate microfluidic technology.
TABLE I.
A list of commercial laboratory machines that incorporate microfluidic technology.
| Machine | Company | Application | Microfluidic element | URL Reference |
|---|---|---|---|---|
| LabChip | Caliper/Perkin Elmer | DNA, RNA, and Protein quantification | Microchip electrophoresis | 89 |
| 2100 Bioanalyzer | Agilent | DNA, RNA, and Protein quantification | Microchip electrophoresis | 90 |
| MPCS | Emerald BioSystems | Protein crystal screening | Emulsion generator | 16 |
| BioMark | Fluidigm | Gene expression analysis Digital PCR | Integrated fluidic circuit | 101 |
| C1 | Fluidigm | Single cell processing | Integrated fluidic circuit | 101 |
| QX100 ddPCR | Bio-Rad | Digital PCR | Emulsion generator | 102 |
| RainDrop | RainDance | Digital PCR | Emulsion generator | 103 |
| Mondrain SP | Nugen | Sequencing sample prep | Digital microfluidics | 104 |
| One Touch | Life technologies | Sequencing sample prep | Emulsion generator | 105 |
| Ion Torrent PGM/Proton | Life technologies | Next generation Sequencing | Microfluidic flowcell/microwell array | 105 |
| Genome Analyzer/HiSeq/MiSeq | Illumina | Next generation Sequencing | Microfluidic flowcell | 106 |
| Heliscope | Helicos | Next generation Sequencing | Microfluidic flowcell | 107 |
CONCLUSION
The advantages of small fluidic circuit components that have been covered here enable quantitatively more and qualitatively new measurements of biological systems. For the last decade, however, the question has been whether microfluidic approaches will have major and lasting impact on life science. The integrated circuit analogy is popular in the microfluidic community to illustrate the evolution of microfluidic applications. There are of course some aspects of the respective technologies for which the analogy breaks down, for example, we may be approaching the valve density limit for integrated microfluidic circuits.30 However, in lieu of recent applications of the technology, many presented here, it is not clear that this limit will have the same effect on microfluidic technology development as it is projected to have in micro-electronics. The analogy between the microfluidic integrated circuit and the microelectronic integrated circuit is still, however, quite insightful and is especially useful for contextualizing the potential of microfluidic technology. As electronic components have become smaller and smaller, consumer electronics have become smaller, cheaper, and more useful. Twenty years ago not every household had a personal computer but now it seems just about everyone has a smart phone. Equally, as smaller and more efficient microfluidic elements are successfully integrated into hand held devices, there is no doubt that we will see the impact all over the world, from home medical diagnostics to point-of-care diagnostics in the developing world. At the same time, as computer processors have steadily increased transistor density, large scale computational servers with immense computing power are now able to fit into a box the size of a refrigerator or small room. We now have machines that can process data of a volume and complexity that was hard to imagine even a decade ago and these machines are revolutionizing how we manage information and in the case of bioinformatics, changing how we fundamentally conduct life science research. The same might be said for microfluidic technology. As we have reviewed here some microfluidic chips are being integrated into commercial and experimental research tools that are fundamentally changing the way we do science. As the technology matures these powerful research platforms are slowly being adopted by a wider scope of users. If six years ago microfluidic technology was adolescent,100 then now it is coming of age in life science research. The challenge now, as is always the case with growing up, will be how the field handles new responsibility. For research applications, microfluidic technologies should not be evaluated anymore on their extensive properties like size, throughput, complexity, etc. Instead, the technology should be evaluated by its scientific contribution. There will always be technology developers who can produce impressive machines, but their usefulness will now be judged by the extent to which they yield new science. This means that developers need to either become more equipped to assess the needs of life science research and apply their inventions to real problems, or there should be more and closer collaborations between life science research labs and technology development labs. Fortunately, this appears to be the trend.
ACKNOWLEDGMENTS
The authors would like to thank Chen Tao for illustrations and helpful discussions. They would also like to thank Paul Blainey, Rafael Gomez-Sjoberg, Brian Yu, Stephen Quake and the Stanford microfluidic foundry for images and figures of their microfluidic devices. A.M.S. was supported by the Whitaker International Biomedical Engineering Fellowship and Y.H. by the Ministry of Science and Technology of China (2011CB809106), the National Natural Science Foundation of China (20890020, 91913011, 21222501), and the Fok Ying Tung Education Foundation.
References
- Manz A., Graber N., and Widmer H. M., Sens. Actuators B 1(1–6), 244–248 (1990). 10.1016/0925-4005(90)80209-I [DOI] [Google Scholar]
- Xia Y. and Whitesides G. M., Angew. Chem., Int. Ed. 37(5), 550–575 (1998). [DOI] [PubMed] [Google Scholar]
- Thorsen T., Maerkl S. J., and Quake S. R., Science 298(5593), 580–584 (2002). 10.1126/science.1076996 [DOI] [PubMed] [Google Scholar]
- Whitesides G. M., Lab Chip 13, 11–13 (2013). 10.1039/c2lc90109a [DOI] [PubMed] [Google Scholar]
- Li X., Ballerini D. R., and Shen W., Biomicrofluidics 6(1), 11301–1130113 (2012). 10.1063/1.3687398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. W., Phillips S. T., Whitesides G. M., and Carrilho E., Anal. Chem. 82(1), 3–10 (2010). 10.1021/ac9013989 [DOI] [PubMed] [Google Scholar]
- Lafleur L., Stevens D., McKenzie K., Ramachandran S., Spicar-Mihalic P., Singhal M., Arjyal A., Osborn J., Kauffman P., Yager P., and Lutz B., Lab Chip 12(6), 1119–1127 (2012). 10.1039/c2lc20751f [DOI] [PubMed] [Google Scholar]
- Gervais L., de Rooij N., and Delamarche E., Adv. Mater. 23(24), H151–176 (2011). 10.1002/adma.201100464 [DOI] [PubMed] [Google Scholar]
- Pethig R., Biomicrofluidics 4(2), 022811–022835 (2010). 10.1063/1.3456626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C. L., Sommer M. O. A., and Quake S. R., Proc. Natl. Acad. Sci. USA 101(40), 14431–14436 (2004). 10.1073/pnas.0405847101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C. L., Skordalakes E., Berger J. M., and Quake S. R., Proc. Natl. Acad. Sci. USA 99(26), 16531–16536 (2002). 10.1073/pnas.262485199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. J., Hansen C. L., and Quake S. R., Proc. Natl. Acad. Sci. USA 103(45), 16746–16751 (2006). 10.1073/pnas.0605293103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen T., Roberts R. W., Arnold F. H., and Quake S. R., Phys. Rev. Lett. 86(18), 4163–4166 (2001). 10.1103/PhysRevLett.86.4163 [DOI] [PubMed] [Google Scholar]
- Tice J. D., Song H., Lyon A. D., and Ismagilov R. F., Langmuir 19(22), 9127–9133 (2003). 10.1021/la030090w [DOI] [Google Scholar]
- Zheng B., Roach L. S., and Ismagilov R. F., J. Am. Chem. Soc. 125(37), 11170–11171 (2003). 10.1021/ja037166v [DOI] [PubMed] [Google Scholar]
- See http://www.emeraldbiosystems.com/t-mpcsplugmaker.aspx for Emerald Biosystems' Microcapillary Protein Crystallization System.
- Boedicker J. Q., Vincent M. E., and Ismagilov R. F., Angew. Chem., Int. Ed. Engl. 48(32), 5908–5911 (2009). 10.1002/anie.200901550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. C., Hur J. Y., Kwon K. W., Park S.-H., and Suh K. Y., Lab Chip 6(8), 988–994 (2006). 10.1039/b602961b [DOI] [PubMed] [Google Scholar]
- Marcy Y., Ouverney C., Bik E. M., Losekann T., Ivanova N., Martin H. G., Szeto E., Platt D., Hugenholtz P., Relman D. A., and Quake S. R., Proc. Natl. Acad. Sci. USA 104(29), 11889–11894 (2007). 10.1073/pnas.0704662104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y., Ishoey T., Lasken R. S., Stockwell T. B., Walenz B. P., Halpern A. L., Beeson K. Y., Goldberg S. M., and Quake S. R., PLoS Genet. 3(9), 1702–1708 (2007). 10.1371/journal.pgen.0030155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo L. Y. and Friend J. R., Biomicrofluidics 3(1), 012002–012023 (2009). 10.1063/1.3056040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardo S., Cecchini M., Beltram F., Cingolani R., and Pisignano D., Lab Chip 8(9), 1557–1563 (2008). 10.1039/b803967d [DOI] [PubMed] [Google Scholar]
- Ding X., Lin S.-C. S., Lapsley M. I., Li S., Guo X., Chan C. Y., Chiang I. K., Wang L., McCoy J. P., and Huang T. J., Lab Chip 12(21), 4228–4231 (2012). 10.1039/c2lc40751e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J., Lim H., Kim C., Kang J. Y., and Shin S., Biomicrofluidics 6(2), 024120-1–024120-10 (2012). 10.1063/1.4718719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B. S., Schuster T. G., Zhu X. Y., Chang D., Smith G. D., and Takayama S., Anal. Chem. 75(7), 1671–1675 (2003). 10.1021/ac020579e [DOI] [PubMed] [Google Scholar]
- Wang S., Liu K., Liu J., Yu Z. T. F., Xu X., Zhao L., Lee T., Lee E. K., Reiss J., Lee Y.-K., Chung L. W. K., Huang J., Rettig M., Seligson D., Duraiswamy K. N., Shen C. K. F., and Tseng H.-R., Angew. Chem., Int. Ed. 50(13), 3084–3088 (2011). 10.1002/anie.201005853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Streets A. M., Lin R. R., Quake S. R., Weiss S., and Majumdar D. S., Nat. Methods 8(3), 242–U283 (2011). 10.1038/nmeth.1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men Y., Fu Y., Chen Z., Sims P. A., Greenleaf W. J., and Huang Y., Anal. Chem. 84(10), 4262–4266 (2012). 10.1021/ac300761n [DOI] [PubMed] [Google Scholar]
- Ottesen E. A., Hong J. W., Quake S. R., and Leadbetter J. R., Science 314(5804), 1464–1467 (2006). 10.1126/science.1131370 [DOI] [PubMed] [Google Scholar]
- Araci I. E. and Quake S. R., Lab Chip 12(16), 2803–2806 (2012). 10.1039/c2lc40258k [DOI] [PubMed] [Google Scholar]
- Jäckel C., Kast P., and Hilvert D., Annu. Rev. Biophys. 37(1), 153–173 (2008). 10.1146/annurev.biophys.37.032807.125832 [DOI] [PubMed] [Google Scholar]
- Agresti J. J., Antipov E., Abate A. R., Ahn K., Rowat A. C., Baret J. C., Marquez M., Klibanov A. M., Griffiths A. D., and Weitz D. A., Proc. Natl. Acad. Sci. USA 107(9), 4004–4009 (2010). 10.1073/pnas.0910781107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X. Z., Gielen F., Edel J. B., and deMello A. J., Nat. Chem. 3(6), 437–442 (2011). 10.1038/nchem.1046 [DOI] [PubMed] [Google Scholar]
- Wu A. R., Kawahara T. L. A., Rapicavoli N. A., van Riggelen J., Shroff E. H., Xu L. W., Felsher D. W., Chang H. Y., and Quake S. R., Lab Chip 12(12), 2190–2198 (2012). 10.1039/c2lc21290k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sui G., Mocharla V. P., Lin R. J., Phelps M. E., Kolb H. C., and Tseng H.-R., Angew. Chem., Int. Ed. 45(32), 5276–5281 (2006). 10.1002/anie.200601677 [DOI] [PubMed] [Google Scholar]
- Wang Y., Lin W. Y., Liu K., Lin R. J., Selke M., Kolb H. C., Zhang N., Zhao X. Z., Phelps M. E., Shen C. K., Faull K. F., and Tseng H. R., Lab Chip 9(16), 2281–2285 (2009). 10.1039/b907430a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyries K. A., Tropini C., Vaninsberghe M., Doolin C., Petriv O. I., Singhal A., Leung K., Hughesman C. B., and Hansen C. L., Nat. Methods 8(8), 649–651 (2011). 10.1038/nmeth.1640 [DOI] [PubMed] [Google Scholar]
- Love J. C., Ronan J. L., Grotenbreg G. M., van der Veen A. G., and Ploegh H. L., Nat. Biotechnol. 24(6), 703–707 (2006). 10.1038/nbt1210 [DOI] [PubMed] [Google Scholar]
- Ogunniyi A. O., Story C. M., Papa E., Guillen E., and Love J. C., Nat. Protoc. 4(5), 767–782 (2009). 10.1038/nprot.2009.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan N., Kwon D. S., Law K. M., Ogunniyi A. O., Anahtar M. N., Richter J. M., Walker B. D., and Love J. C., Proc. Natl. Acad. Sci. USA 109(10), 3885–3890 (2012). 10.1073/pnas.1111205109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q., Bagheri N., Bradshaw E. M., Hafler D. A., Lauffenburger D. A., and Love J. C., Proc. Natl. Acad. Sci. USA 109(5), 1607–1612 (2012). 10.1073/pnas.1117194109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Schafer S., Choi J., Yamanaka Y. J., Lombardi M. L., Bose S., Carlson A. L., Phillips J. A., Teo W., Droujinine I. A., Cui C. H., Jain R. K., Lammerding J., Love J. C., Lin C. P., Sarkar D., Karnik R., and Karp J. M., Nat. Nanotechnol. 6(8), 524–531 (2011). 10.1038/nnano.2011.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Qin L., Wei W., Geng F., Fan R., Shin Y. S., Guo D., Hood L., Mischel P. S., and Heath J. R., Proc. Natl. Acad. Sci. USA 109(2), 419–424 (2012). 10.1073/pnas.1110865109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y. S., Ahmad H., Shi Q., Kim H., Pascal T. A., Fan R., W. A.GoddardIII, and Heath J. R., ChemPhysChem 11(14), 3063–3069 (2010). 10.1002/cphc.201000528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Fan R., Ahmad H., Shi Q., Comin-Anduix B., Chodon T., Koya R. C., Liu C. C., Kwong G. A., Radu C. G., Ribas A., and Heath J. R., Nat. Med. 17(6), 738–743 (2011). 10.1038/nm.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerkl S. J. and Quake S. R., Science 315(5809), 233–237 (2007). 10.1126/science.1131007 [DOI] [PubMed] [Google Scholar]
- Maerkl S. J. and Quake S. R., Proc. Natl. Acad. Sci. USA 106(44), 18650–18655 (2009). 10.1073/pnas.0907688106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber D., Maerkl S. J., and Quake S. R., Nat. Methods 6(1), 71–74 (2009). 10.1038/nmeth.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M., Sit R., Pan W., and Quake S. R., Anal. Chem. 84(21), 9572–9578 (2012). 10.1021/ac302436y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M., Sit R. V., and Quake S. R., Proc. Natl. Acad. Sci. USA 110(2), 477–482 (2013). 10.1073/pnas.1210634110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einav S., Gerber D., Bryson P. D., Sklan E. H., Elazar M., Maerkl S. J., Glenn J. S., and Quake S. R., Nat. Biotechnol. 26(9), 1019–1027 (2008). 10.1038/nbt.1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L., Meier M., Lyons S. M., Sit R. V., Marzluff W. F., Quake S. R., and Chang H. Y., Nat. Methods 9, 1192–1194 (2012). 10.1038/nmeth.2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. W., Phillips S. T., Carrilho E., S. W.ThomasIII, Sindi H., and Whitesides G. M., Anal. Chem. 80(10), 3699–3707 (2008). 10.1021/ac800112r [DOI] [PMC free article] [PubMed] [Google Scholar]
- See http://www.stanford.edu/group/foundry/ for Stanford Microfluidic Foundry website.
- See http://microfluidics.lbl.gov/ for Lawrence Berkeley National Lab Microfluidics Lab website (R. Gómez-Sjöberg).
- Vekilov P. G. and Chernov A. A., in Solid State Physics, edited by Henry E. and Frans S. (Academic, 2003), Vol. 57, pp. 1–147. [Google Scholar]
- Streets A. M. and Quake S. R., Phys. Rev. Lett. 104(17), 178102 (2010). 10.1103/PhysRevLett.104.178102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg T. P., Godin M., Knudsen S. M., Shen W., Carlson G., Foster J. S., Babcock K., and Manalis S. R., Nature 446(7139), 1066–1069 (2007). 10.1038/nature05741 [DOI] [PubMed] [Google Scholar]
- Grover W. H., Bryan A. K., Diez-Silva M., Suresh S., Higgins J. M., and Manalis S. R., Proc. Natl. Acad. Sci. USA 108(27), 10992–10996 (2011). 10.1073/pnas.1104651108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S., Tzur A., Weng Y., Jorgensen P., Kim J., Kirschner M. W., and Manalis S. R., Nat. Methods 9(9), 910–912 (2012). 10.1038/nmeth.2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagadde F. K., You L., Hansen C. L., Arnold F. H., and Quake S. R., Science 309(5731), 137–140 (2005). 10.1126/science.1109173 [DOI] [PubMed] [Google Scholar]
- Gomez-Sjoberg R., Leyrat A. A., Pirone D. M., Chen C. S., and Quake S. R., Anal. Chem. 79(22), 8557–8563 (2007). 10.1021/ac071311w [DOI] [PubMed] [Google Scholar]
- Tay S., Hughey J. J., Lee T. K., Lipniacki T., Quake S. R., and Covert M. W., Nature 466(7303), 267–271 (2010). 10.1038/nature09145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde C. B., Zeng F., Gonzalez-Rubio R., Angel M., and Yanik M. F., Proc. Natl. Acad. Sci. USA 104(35), 13891–13895 (2007). 10.1073/pnas.0706513104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Jiang L., Shi W., Qin J., and Lin B., Biomicrofluidics 3(4), 044114–044118 (2009). 10.1063/1.3274313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- i Solvas X. C., Geier F. M., Leroi A. M., Bundy J. G., Edel J. B., and deMello A. J., Chem. Commun. 47(35), 9801–9803 (2011). 10.1039/c1cc14076k [DOI] [PubMed] [Google Scholar]
- Chung K., Crane M. M., and Lu H., Nat. Methods 5(7), 637–643 (2008). 10.1038/nmeth.1227 [DOI] [PubMed] [Google Scholar]
- Crane M. M., Stirman J. N., Ou C. Y., Kurshan P. T., Rehg J. M., Shen K., and Lu H., Nat. Methods 9(10), 977–980 (2012). 10.1038/nmeth.2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey P. C., Mosier A. C., Potanina A., Francis C. A., and Quake S. R., PLoS ONE 6(2), e16626 (2011). 10.1371/journal.pone.0016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall I. P., Blainey P. C., Spormann A. M., and Quake S. R., Appl. Environ. Microbiol. 78(24), 8555–8563 (2012). 10.1128/AEM.02314-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef N. H., Blainey P. C., Quake S. R., and Elshahed M. S., Applied Appl. Environ. Microbiol. 77(21), 7804–7814 (2011). 10.1128/AEM.06059-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp S. J., Harrington E. D., Quake S. R., Relman D. A., and Blainey P. C., Genome Res. 22(6), 1107–1119 (2012). 10.1101/gr.131482.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H. C., Wang J., Potanina A., and Quake S. R., Nat. Biotechnol. 29(1), 51–57 (2011). 10.1038/nbt.1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Fan H. C., Behr B., and Quake S. R., Cell 150(2), 402–412 (2012). 10.1016/j.cell.2012.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken R. S., Nat. Rev. Microbiol. 10(9), 631–640 (2012). 10.1038/nrmicro2857 [DOI] [PubMed] [Google Scholar]
- Lecault V., White A. K., Singhal A., and Hansen C. L., Curr. Opin. Chem. Biol. 16(3–4), 381–390 (2012). 10.1016/j.cbpa.2012.03.022 [DOI] [PubMed] [Google Scholar]
- Yin H. and Marshall D., Curr. Opin Biotechnol. 23(1), 110–119 (2012). 10.1016/j.copbio.2011.11.002 [DOI] [PubMed] [Google Scholar]
- Zare R. N. and Kim S., Annu. Rev. Biomed. Eng. 12, 187–201 (2010). 10.1146/annurev-bioeng-070909-105238 [DOI] [PubMed] [Google Scholar]
- Ryan D., Ren K., and Wu H., Biomicrofluidics 5(2), 021501–021509 (2011). 10.1063/1.3574448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K., Zahn H., Leaver T., Konwar K. M., Hanson N. W., Page A. P., Lo C. C., Chain P. S., Hallam S. J., and Hansen C. L., Proc. Natl. Acad. Sci. USA 109(20), 7665–7670 (2012). 10.1073/pnas.1106752109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecault V., Vaninsberghe M., Sekulovic S., Knapp D. J., Wohrer S., Bowden W., Viel F., McLaughlin T., Jarandehei A., Miller M., Falconnet D., White A. K., Kent D. G., Copley M. R., Taghipour F., Eaves C. J., Humphries R. K., Piret J. M., and Hansen C. L., Nat. Methods 8(7), 581–586 (2011). 10.1038/nmeth.1614 [DOI] [PubMed] [Google Scholar]
- Li G. W. and Xie X. S., Nature 475(7356), 308–315 (2011). 10.1038/nature10315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Friedman N., and Xie X. S., Nature 440(7082), 358–362 (2006). 10.1038/nature04599 [DOI] [PubMed] [Google Scholar]
- Taniguchi Y., Choi P. J., Li G. W., Chen H., Babu M., Hearn J., Emili A., and Xie X. S., Science 329(5991), 533–538 (2010). 10.1126/science.1188308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisky T. and Quake S. R., Nat. Methods 8(4), 311–314 (2011). 10.1038/nmeth0411-311 [DOI] [PubMed] [Google Scholar]
- Dalerba P., Kalisky T., Sahoo D., Rajendran P. S., Rothenberg M. E., Leyrat A. A., Sim S., Okamoto J., Johnston D. M., Qian D., Zabala M., Bueno J., Neff N. F., Wang J., Shelton A. A., Visser B., Hisamori S., Shimono Y., van de Wetering M., Clevers H., Clarke M. F., and Quake S. R., Nat. Biotechnol. 29(12), 1120–1127 (2011). 10.1038/nbt.2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D. A., Cheng A. W., Itskovich E., Markoulaki S., Ganz K., Klemm S. L., van Oudenaarden A., and Jaenisch R., Cell 150(6), 1209–1222 (2012). 10.1016/j.cell.2012.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recently acquired by PerkinElmer.
- See http://www.perkinelmer.com/catalog/category/id/labchip%20systems for PerkinElmer LabChip.
- See http://www.genomics.agilent.com for Agilent.
- R. A.WhiteIII, Quake S. R., and Curr K., J. Virol. Methods 179(1), 45–50 (2012). 10.1016/j.jviromet.2011.09.017 [DOI] [PubMed] [Google Scholar]
- Tadmor A. D., Ottesen E. A., Leadbetter J. R., and Phillips R., Science 333(6038), 58–62 (2011). 10.1126/science.1200758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H. C. and Quake S. R., Anal. Chem. 79(19), 7576–7579 (2007). 10.1021/ac0709394 [DOI] [PubMed] [Google Scholar]
- Snyder T. M., Khush K. K., Valantine H. A., and Quake S. R., Proc. Natl. Acad. Sci. USA 108(15), 6229–6234 (2011). 10.1073/pnas.1013924108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettger L. M., Handsaker R. E., Zody M. C., and McCarroll S. A., Nat. Genet. 44(8), 881–885 (2012). 10.1038/ng.2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R., Warner J. B., Nakano M., Libby B., Medkova M., David P. H., Kotsopoulos S. K., Samuels M. L., Hutchison J. B., Larson J. W., Topol E. J., Weiner M. P., Harismendy O., Olson J., Link D. R., and Frazer K. A., Nat. Biotechnol. 27(11), 1025–1031 (2009). 10.1038/nbt.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson B. J., Ness K. D., Masquelier D. A., Belgrader P., Heredia N. J., Makarewicz A. J., Bright I. J., Lucero M. Y., Hiddessen A. L., Legler T. C., Kitano T. K., Hodel M. R., Petersen J. F., Wyatt P. W., Steenblock E. R., Shah P. H., Bousse L. J., Troup C. B., Mellen J. C., Wittmann D. K., Erndt N. G., Cauley T. H., Koehler R. T., So A. P., Dube S., Rose K. A., Montesclaros L., Wang S., Stumbo D. P., Hodges S. P., Romine S., Milanovich F. P., White H. E., Regan J. F., Karlin–Neumann G. A., Hindson C. M., Saxonov S., and Colston B. W., Anal. Chem. 83(22), 8604–8610 (2011). 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekin D., Skhiri Y., Baret J. C., Le Corre D., Mazutis L., Salem C. B., Millot F., El Harrak A., Hutchison J. B., Larson J. W., Link D. R., Laurent-Puig P., Griffiths A. D., and Taly V., Lab Chip 11(13), 2156–2166 (2011). 10.1039/c1lc20128j [DOI] [PubMed] [Google Scholar]
- Niedringhaus T. P., Milanova D., Kerby M. B., Snyder M. P., and Barron A. E., Anal. Chem. 83(12), 4327–4341 (2011). 10.1021/ac2010857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesides G. M., Nature 442(7101), 368–373 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- See http://www.fluidigm.com/ for information about Fluidigm Corporation.
- See http://www.bio-rad.com/prd/en/US/LSR/PDP/M9HE3XE8Z/QX100-Droplet-Digital-PCR-System for Bio-Rad QX 100 Droplet Digital PCR system.
- See http://raindancetech.com/ for RainDance Technologies website.
- See http://www.nugeninc.com/nugen/index.cfm/products/msp/ for Nugen.
- See http://www.iontorrent.com/ for Life Technologies iontorrent system.
- See http://www.illumina.com/ for Illumina Incorporated website.
- See http://www.helicosbio.com/ for Helicos Biosciences corporation website.