Summary
Tet enzymes (Tet1/2/3) convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in various embryonic and adult tissues. Mice mutant for either Tet1 or Tet2 are viable raising the question whether these enzymes have overlapping roles in development. Here, we have generated Tet1 and Tet2 double knockout (DKO) ESCs and mice. DKO ESCs remained pluripotent, but were depleted of 5hmC and caused developmental defects in chimeric embryos. While a fraction of double mutant embryos exhibited mid-gestation abnormalities with perinatal lethality, viable and overtly normal Tet1/Tet2 deficient mice were also obtained. DKO mice had reduced 5hmC and increased 5mC levels and abnormal methylation at various imprinted loci. Nevertheless, animals of both sexes were fertile with females having smaller ovaries and reduced fertility. Our data show that loss of both enzymes is compatible with development but promotes hypermethylation and compromises imprinting. It also suggests a significant contribution of Tet3 to hydroxylation of 5mC during development.
Keywords: Tet1, Tet2, 5-hydroxymethylcytosine, imprinting, DNA methylation, embryonic development
Introduction
DNA methylation is a major epigenetic modification in the eukaryotic genome that regulates gene expression during many biological processes including development. Dynamic changes in the DNA methylation landscape of the genome are essential for proper gene regulation and orchestration of various developmental processes (Ooi and Bestor, 2008; Wu and Zhang, 2010). While the base 5-methylcytosine (5mC) and the DNA methyltransferase (DNMT) family of enzymes that catalyze DNA methylation are well-studied, mechanisms of DNA demethylation are poorly understood. Both replication-dependent “passive” and replication-independent “active” models of DNA demethylation have been proposed. Whereas passive DNA demethylation is caused by inhibition of the maintenance methyltransferase DNMT1, active demethylation requires direct enzymatic removal of methyl groups involving components of the DNA repair machinery (Ooi and Bestor, 2008; Wu and Zhang, 2010).
The Ten eleven translocation (Tet) family of enzymes (Tet1, Tet2 and Tet3) has recently been implicated in DNA demethylation. These enzymes contain a C-terminal catalytic domain that has dioxygenase activity and converts 5mC to 5 hydroxymethylcytosine (5hmC) (Tahiliani et al., 2009). Since 5hmC is poorly recognized by DNMT1, it has been proposed to promote DNA demethylation in a replication-dependent manner or, alternatively, could serve as an intermediate in active DNA demethylation where it is excised from the genome by components of the DNA repair machinery (Wu and Zhang, 2010; Branco et al., 2011). It has also been shown that Tet proteins can convert 5mC and 5hmC to 5-formyl cytosine (5fC) and 5-carboxylcytosine (5caC), two less abundant bases. These bases are also recognized and directly or indirectly removed by the base excision repair (BER) machinery involving thymine DNA glycosylase (TDG) or putative decarboxylases (He et al., 2011; Ito et al., 2011).
Both 5hmC and the Tet enzymes are abundant in various embryonic and adult cell types including the zygote (Wossidlo et al., 2011), primordial germ cells (Hajkova et al., 2010), Purkinje neurons (Kriaucionis and Heintz, 2009) and embryonic stem cells (Ito et al., 2010; Koh et al., 2011). 5hmC is a stable base that is abundant in CpG-rich promoter regions, gene bodies (Ficz et al., 2011; Pastor et al., 2011; Wu et al., 2011; Williams et al., 2011b) and enhancer elements in embryonic stem cells (Stroud et al., 2011) or differentiated tissues (Bocker et al., 2012). It selectively binds Methyl-CpG-Binding Domain protein 3 (MBD3) in ES cells and helps recruit key chromatin regulatory complexes, but is poorly recognized by 5mC-binding proteins (Yildirim et al., 2011). In addition, Tet1 binds to Polycomb target gene promoters and can form chromatin repressive complexes, thereby having a dual role in transcriptional activation and repression in ES cells (Wu et al., 2011; Williams et al., 2011b).
To define the function of the Tet enzymes and of 5hmC in development, mice carrying mutant alleles of the Tet genes have been generated. While Tet1 and Tet2 knockout mice are viable (Dawlaty et al., 2011; Li et al., 2011; Moran-Crusio et al., 2011), deletion of Tet3 leads to neonatal lethality (Gu et al., 2011). Consistent with the high level of Tet3 and 5hmC in the zygote (Wossidlo et al., 2011), conditional deletion of Tet3 in the oocyte leads to aberrant hydroxylation and delayed demethylation of the paternal genome upon fertilization leading to impaired development and reduced fertility (Gu et al., 2011).
Both Tet1 and Tet2 are abundantly expressed in mouse ES cells. Depletion of either of these proteins reduced 5hmC levels but did not affect pluripotency (Dawlaty et al., 2011; Koh et al., 2011). Previously, we have shown that Tet1 knockout ES cells show skewed differentiation toward trophectoderm in vitro, but appear normal in vivo and can support development of viable mice. Tet1 knockout mice are viable and fertile albeit displaying reduced body mass and smaller litter size suggesting a subtle role for Tet1 in animal physiology (Dawlaty et al., 2011). Like Tet1, Tet2 is dispensable for embryonic development and adult Tet2 mutants are viable and fertile. However, Tet2 loss affects hematopoietic stem cells (HSCs) and causes chronic myelomonocytic leukemia (CMML) (Ko et al., 2011; Li et al., 2011; Moran-Crusio et al., 2011; Quivoron et al., 2011). Given the abundance of 5hmC in various tissues and dynamic expression of Tets during various stages of development (Hackett et al., 2012b), it is not known whether Tet1 and Tet2 have different or partially redundant functions in development. In this study we show that combined Tet1 and Tet2 deficiency results in partially penetrant embryonic and neonatal abnormalities associated with perinatal lethality of about half of the mutants with a substantial fraction of mutants surviving to overtly normal and fertile adults.
Results
Generation of Tet1 and Tet2 double mutant ESCs
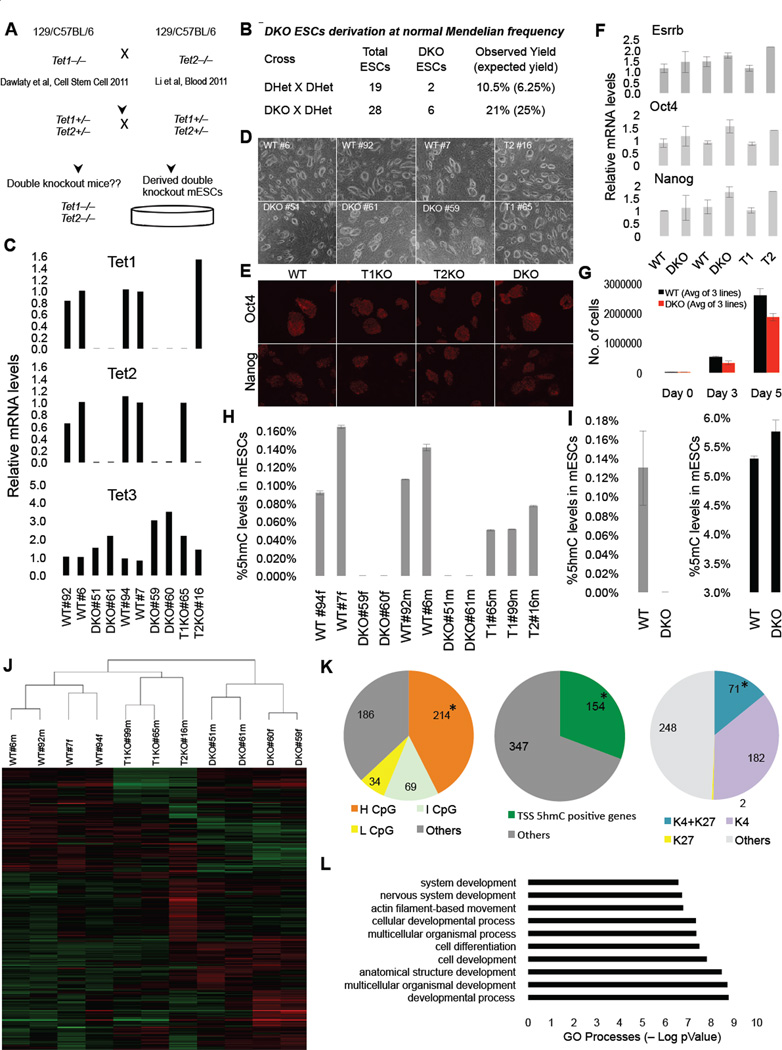
To study the effects of combined deficiency of Tet1 and Tet2 on pluripotency and development we intercrossed Tet1 and Tet2 mice (Dawlaty et al., 2011; Li et al., 2011) and derived double knockout (DKO), Tet1 knockout (T1KO), Tet2 knockout (T2KO) and control wild type (WT) ES cell lines following the scheme in Figure 1A. Genotyping by Southern Blot and PCR (Figure S1A&B) indicated that DKO ES cells were derived at the expected Mendelian frequencies from two independent crosses (Figure 1B). All DKO lines were depleted of both Tet1 and Tet2 transcripts and exhibited an average two fold induction of Tet3 (Figure 1C), which is expressed at a low level in WT ESCs. The cells maintained normal ES cell morphology and expressed the pluripotency markers Oct4, Nanog and Esrrb (Figure 1D–F), but showed a subtle reduction in proliferation (Figure 1G). These results suggest that Tet1 and Tet2 are dispensable for derivation of ESCs and preimplantation development.
Figure 1. Tet1 Tet2 double knockout (DKO) ESCs are depleted of 5hmC and express pluripotency markers.
(A) Schematic of crosses to generate DKO ESCs. (B) Summary of derived ESCs and Mendelian frequency of DKO ESC. (C) RTqPCR for Tet1, Tet2 and Tet3 ESCs of indicated genotypes. Notice that DKO ESCs are depleted of Tet1 and Tet2 mRNA. Data are normalized to Gapdh. (D) Bright field images of ESCs of indicated genotypes. (E) Immunostainning for pluripotency markers Oct4 and Nanog in ESCs. (F) Relative mRNA levels of Esrrb, Nanog and Oct4 in ESCs. Data are normalized to Gapdh. (G) WT and DKO ESCs 5-day cell proliferation count. (H & I) Quantification of 5hmC and 5mC in ESCs of indicated genotypes by mass spectrometry (LC-MS/MS-MRM). Notice in DKO ESCs 5hmC levels are below detection level. (J) Heat map of differentially expressed genes across a panel of wild type, single knockouts and DKOs ESCs. Genes differentially expressed in at least one of the comparisons are displayed. Intensity values are log2 transformed and genes are centered by mean intensity across all arrays. A total of 501 genes are deregulated in DKO ESCs. (K) Pie charts categorizing deregulated genes into sub categories of CpG rich genes (left), TSS 5hmC positive genes (center) and H3K4me and H3K27me bivalent genes (right) in ESCs. Asterisk indicates enriched and statistically significant. (L) Gene ontology analysis of 501 differentially expressed genes in DKO ESCs showing that most GO terms are implicated in development. For all panels m=male cell line and f=female cell line. Error bars indicate s.d. See also Figure S1 and Table S2.
Combined loss of Tet1 and Tet2 leads to depletion of 5hmC and deregulated gene expression
To investigate the role of Tet1 and Tet2 on cytosine modification, we applied mass spectrometry to measure levels of 5mC and 5hmC in genomic DNA isolated from feeder free double knockout, single knockout and wild type ESCs. While individual loss of either enzyme reduced 5hmC levels to about 40 to 60%, DKO ESCs were completely depleted of 5hmC (Figure 1H&I) suggesting that Tet1 and Tet2 are the main 5-methyl-dioxygenases responsible for establishing and maintaining the 5hmC marks in ES cells and that the two-fold induction of Tet3 in DKO ES lines (Figure 1C) did not significantly contribute to the 5hmC level. Also, a subtle increase in global 5mC levels was detected in DKO ESCs (Figure 1I), suggesting that loss of 5hmC leads to increased global hypermethylation.
To determine the effect of Tet1 and Tet2 loss and 5hmC depletion on gene expression, we analyzed mRNA levels from DKO, single knockout and WT ESCs by microarray. We found that a total of 501 genes (327 up and 174 down) were differentially expressed by 1.5 fold or more in DKO ESCs as compared to WT ES cells (Figure 1J, Figure S1C) whereas the expression of fewer genes was changed in single mutant cells (214 up an 110 down in T2KO and 55 up and 88 down in T1KO ESC). The substantial number of genes up regulated in mutant cells are likely indirect downstream effectors of Tet1 and Tet2 or are targets of the transcriptional repressive activity of Tet1. A comparison of the total of 501 differentially expressed genes in DKO ESCs with known CpG-rich genes in ES cells (Bernstein et al., 2006), revealed an enrichment for genes with high CpG content (214/501; 43%, compared to all expressed genes 9533/25512; 37%, p-value=0.009) (Figure 1K, Table S2). A similar analysis of differentially expressed genes for transcriptional start site (TSS) 5hmC positive genes in ES cells (Pastor et al., 2011) showed enrichment for genes with high 5hmC levels (154/501; 31%, compared to all expressed genes 4673/25512; 18%; p-value=7.85×10−12). Most of the 5hmC-rich deregulated genes overlapped with genes with high and intermediate CpG content (Figure S1D). We also found that transcriptionally poised bivalent genes in ES cells (71/501; 14%) were enriched among the deregulated genes as compared to all expressed genes (2133/25512; 8%, p-value=9.25×10−6) (Figure 1K, Table S2). Gene ontology analyses revealed that the majority of the differentially expressed genes are implicated in developmental processes including cellular differentiation and nervous system development (Figure 1L).
Tet1/Tet2 deficient ESCs are pluripotent
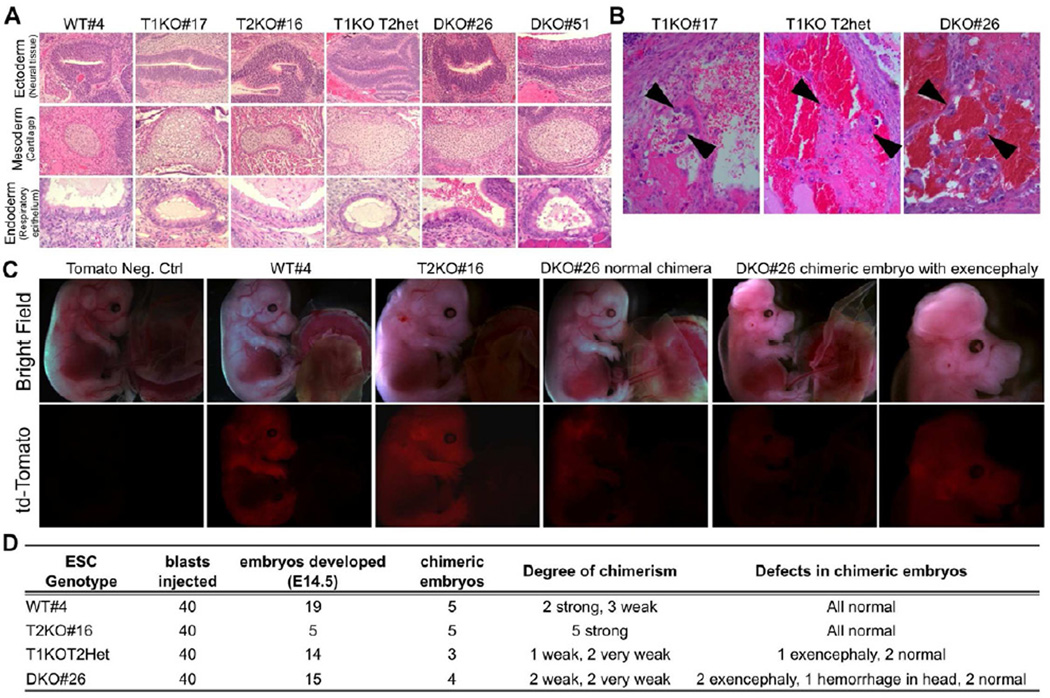
Loss of Tet1 or Tet2 reduced 5hmC levels to half but did not affect pluripotency (Dawlaty et al., 2011; Koh et al., 2011). To assess whether Tet1 and Tet2 are partially redundant and that loss of one could compensate for loss of the other, we tested single and double mutant ESCs for their ability to form tissues of all germ layers in a teratoma assay. All ES cell lines developed teratomas consisting of tissues from the three embryonic layers suggesting that combined deficiency of Tet1 and Tet2 does not affect pluripotency (Figure 2A). However, T1KO, T1KOT2Het and DKO teratomas, but not T2KO teratomas, were hemorrhagic and contained trophoblast-like cells (Figure 2B, Figure S1E). These findings confirm that DKO ES lines are pluripotent but exhibit skewed differentiation defects towards extraembryonic lineage in a teratoma assay similar to what is seen in Tet1 deficient ESCs (Dawlaty et al., 2011; Koh et al., 2011).
Figure 2. Double knockout ESCs remain pluripotent in teratoma and chimera assays.
(A) Representative images of each germ layer from H&E stained sections of teratomas generated from ESCs of indicated genotypes. (B) High magnification images of H&E stained teratoma sections showing hemorrhage and trophoblast-like cells (arrowheads) in tumors derived from DKO and Tet1 deficient ESCs. (C) E14.5 chimeric embryos generated by injecting FUW-td-Tomato transduced ESCs of indicated genotypes into wild type blastocysts. Notice exencephaly and weaker td-tomato signal in DKO ESCs chimeras. (D) Summary table for all chimera injections and defects of DKO chimeric embryos.
To more stringently assess pluripotency, we injected td-tomato labeled WT, T2KO and DKO ES lines into wildtype blastocysts to generate chimeric embryos. Dissection of E14.5 embryos revealed wide contribution of both wild type and T2KO ES cell lines to developing embryos (Figure 2C). While DKO ES cells also contributed to chimeras, half of the DKO and some T1KOT2Het chimeras had exencephaly (Figure 2C&D). Together these findings further ascertain that DKO ES cells remain pluripotent and can contribute to developing embryos, albeit with some defects in differentiation.
Most double mutant mice die perinatally
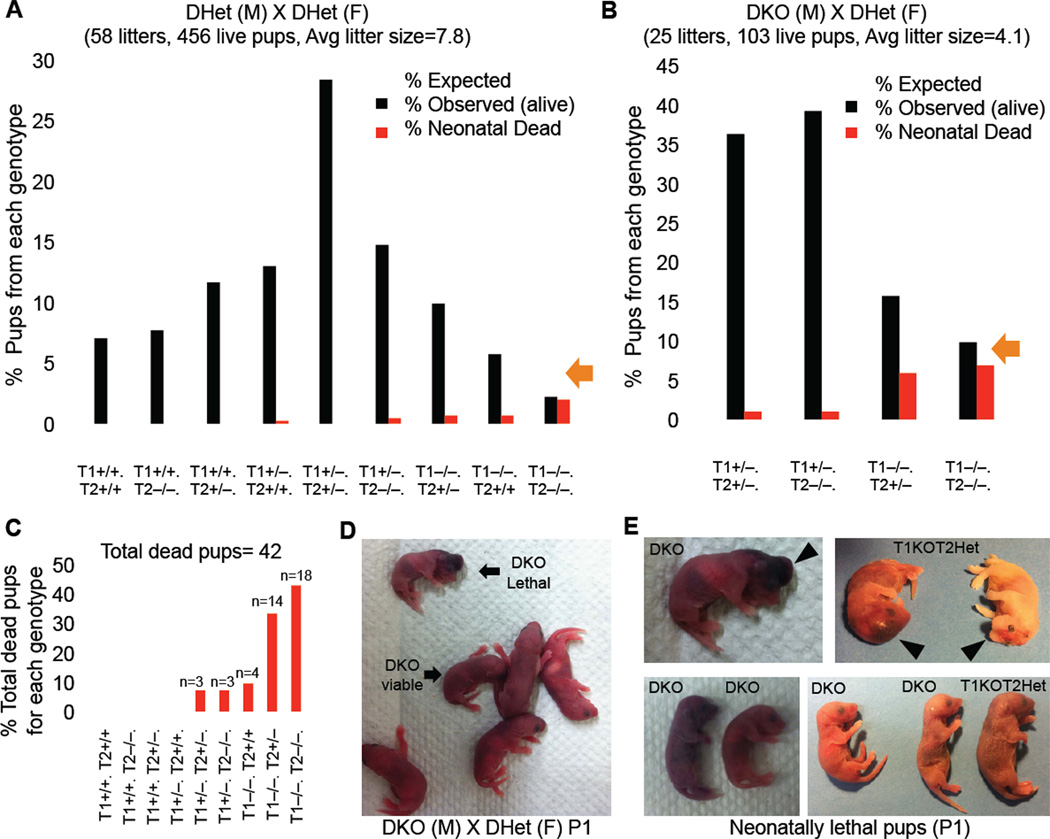
Tet1 and Tet2 single mutant mice are viable (Dawlaty et al., 2011; Ko et al., 2011; Li et al., 2011; Moran-Crusio et al., 2011; Quivoron et al., 2011) consistent with the possibility that deficiency of one gene could be compensated by the other. To investigate whether combined deficiency of Tet1 and Tet2 affected normal development, we generated double knockout mice by intercrossing double heterozygote animals. As summarized in Figure 3A and S2A, a total of 58 litters yielded 456 viable mice. Genotyping by Southern blot and PCR (Figure S2D&E) confirmed animals with the nine predicted genotypes at the expected Mendelian ratios except for DKO animals, which were found at a 3-fold reduced frequency (2.2% vs. expected 6.25%). While DKO mice were born at almost the expected frequency, the majority died within the first 2 days. Reduced survival of DKO pups was confirmed in a cross of DKO males with DHet females yielding double mutant mice at an expected frequency of 25%. As shown in Figure 3B and Figure S2B, a total of 25 litters produced 103 pups, with only 9.8% vs. the expected 25% DKO and 15.7% vs. the expected 25% T1KOT2Het pups surviving to adulthood. Crossing T1HetT2KO males with DHet females yielded similar results (Figure S2C).
Figure 3. Partially penetrant perinatal lethality of Tet1/Tet2 deficient mice.
(A &B) Charts summarizing percent number of pups for each genotype from indicated crosses. Note the increased number of neonatally dead pups with DKO genotype. Dead and alive pups add up close to the expected Mendelian ratio. Average litter sizes shown on top of the charts are calculated based on alive pups. (C) Total dead pups in the study (n=42) are classified based on their genotype. Notice that majority of dead pups are DKO or T1KOT2Het. (D) Gross images of P1 neonates. (E) Representative Images of neonatally lethal pups. See also Figure S2.
The majority of homozygous Tet1/Tet2 mutants died soon after birth or within two days displaying a variety of malformations such as exencephaly, hemorrhage in the head or profound growth retardation (Figure 3C–E). Histopathological examination revealed no overt defects, although smaller organs, lack of milk in the stomach and signs of necrosis were noted in some tissues (Figure S2F). Thus, the partially penetrant perinatal lethality of double mutants in contrast to normal survival of single mutants is consistent with the notion that expression of either enzyme can compensate for deficiency of the other during development
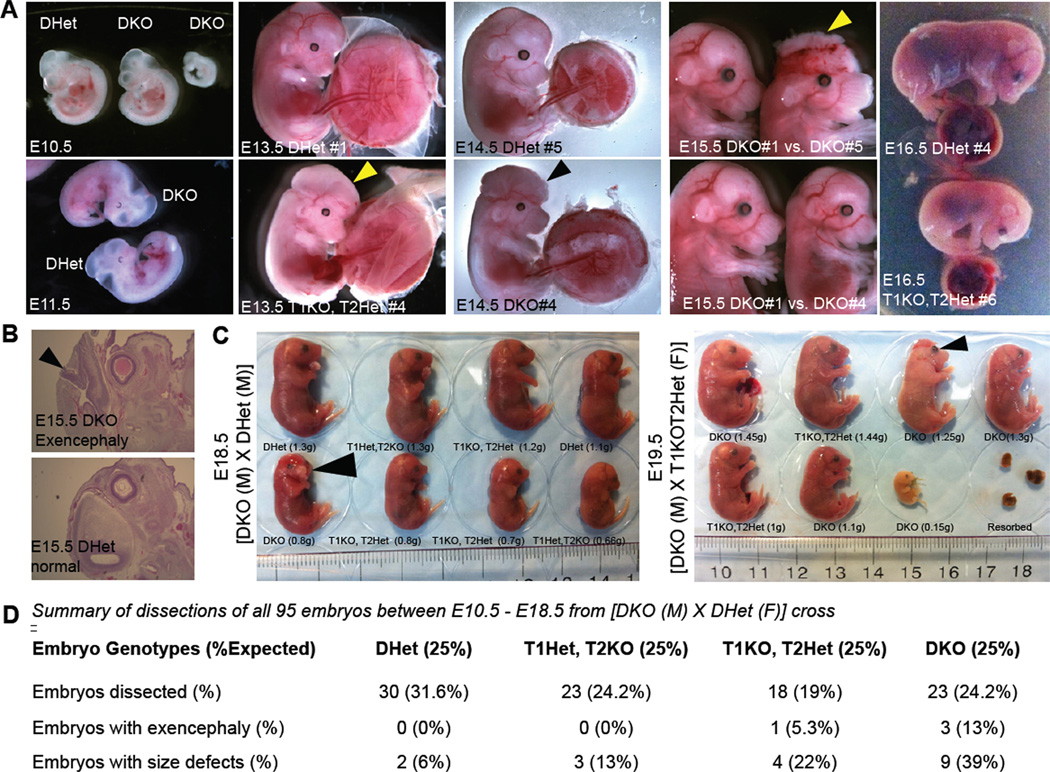
To investigate when in development the combined loss of Tet1 and Tet2 affects the survival of embryos, a total of 95 embryos were isolated between E10.5 and E19.5 and assessed for gross anatomical and growth defects. DKO embryos were present at the expected Mendelian ratio with a substantial fraction (39%) displaying reduced size (Figure 4). Also, similar to the chimeras in Figure 2 and newborn DKO pups in Figure 3, we found an increased incidence of exencephaly (13%) in DKO embryos starting at E13.5 (Figure 4). Our results suggest that the great majority of DKO embryos develop to term but many die perinatally with severe abnormalities. Although a fraction of DKO embryos and newborns were smaller in size, we did not find evidence for any developmental delay as all newborns had fully developed organs.
Figure 4. Defects in midgestation double knockout embryos.
(A) Representative images of mid-gestation E10.5 to E15.5 embryos of indicated genotypes. Notice the presence of normal, smaller size and exencephalic DKO embryos. Arrowheads point to exencephaly/neural tube defects. All embryos are from crosses of DKO male × Dhet female. (B) H&E staining for histological analysis of exencephaly. (C) Images, genotypes and weights of E18.5 and E19.5 litters from the indicated crosses. Arrowheads point to exencephaly/neural tube defects. (D) Summary of genotypes, phenotypes and Mendelian frequency of all mid and late gestation embryos.
Surviving Tet1/Tet2 deficient mice are fertile with females having smaller ovaries and reduced fertility
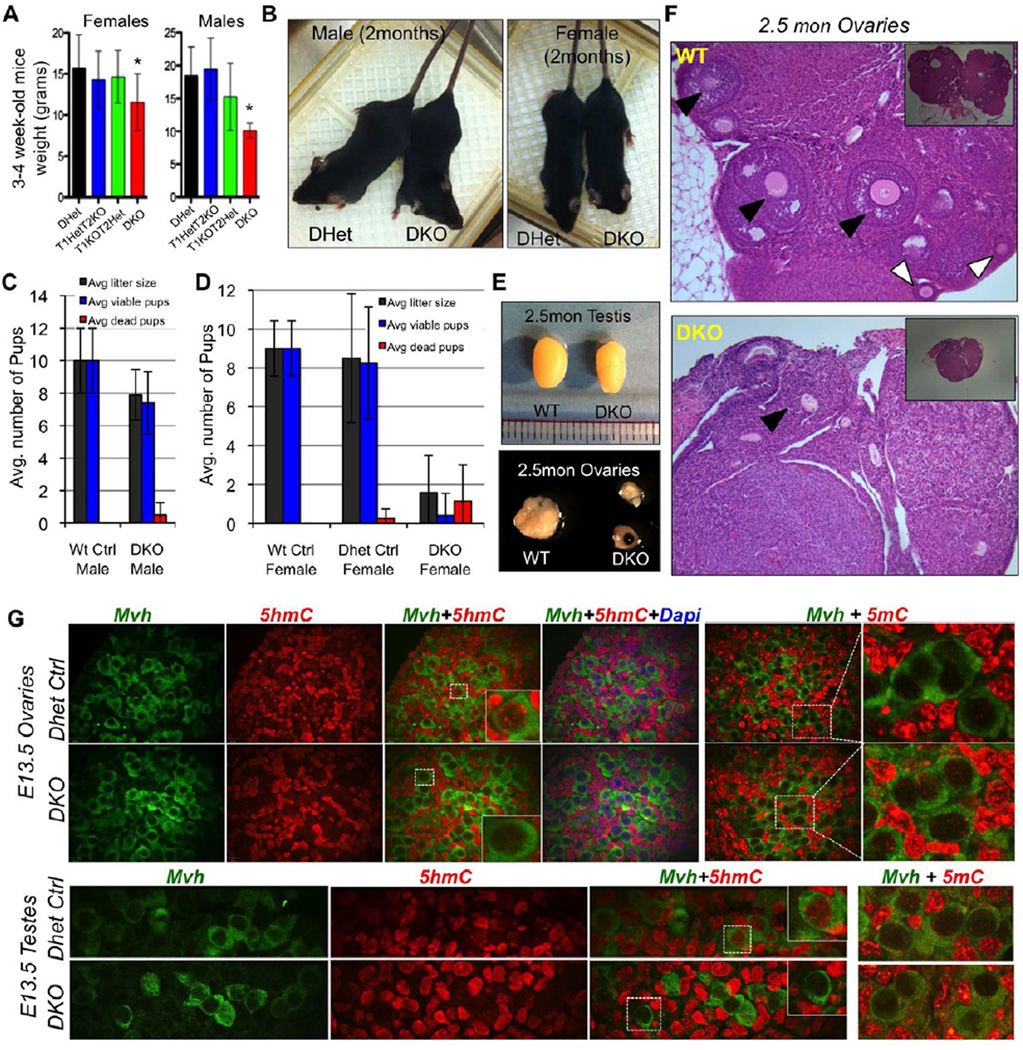
About 40% of DKO newborns survived to adults with a slightly reduced body weight at weaning (Figure 5A) but no significant difference in weight or overall health at two months of age when compared to age matched controls (Figure 5B). Blood analyses, liver and kidney functions showed no major differences except for the presence of few lymphoma cells and a positive hemolysis index in DKO mice, which are likely attributed to Tet2 loss and its role in hematopoietic malignancies (Li et al., 2011; Moran-Crusio et al., 2011) (Figure S2G&H).
Figure 5. Fertility assessment of viable adult double knockout mice.
(A&B) Average weight and gross images of DKO mice. Asterisks indicate p<0.05. (C) Average number of pups per litter from crosses of [DKO male × WT Females] or [Control WT males × WT females]. (D) Average number of pups per litter from crosses of [WT male × Control WT female] or [WT male × Dhet female] or [WT male × DKO female]. (E) Gross images of testis and ovaries. Notice the smaller size of ovaries. (F) H&E stained lateral sections of ovaries. Insets are low magnification images to highlight differences in size of ovaries. Notice the reduced presence of mature follicles and eggs (black arrows) and secondary follicles (white arrows) in DKO ovaries. (G) E13.5 gonad sections stained with anti-Mvh, anti 5hmC and anti 5mC antibodies and Dapi. Notice the lack of 5hmC staining in DKO germ cells. See also Figure S5.
Male DKO mice had normal testes, albeit with some slight variation in size, and, when mated with wild type females, produced normal sized litters (Figure 5C) suggesting that Tet1/Tet2 deficient male mice have normal fertility. In contrast, DKO female mice, while mating normally, showed reduced fecundity when mated with wild type males producing an average litter size of 2 pups as compared to 8 pups for wild type and DHet control females (Figure 5D). Only half of the pups survived to normal adults with the rest dying at P1 or P2. The ovaries of DKO females were substantially smaller than those of wild type mice (Figure 5E) and had, consistent with the reduced fertility, fewer mature follicles (Figure 5F), but contained Mvh-positive germ cells at E13.5 (Figure 5G).
Combined loss of Tet1 and Tet2 reduces 5hmC levels and promotes global hypermethylation
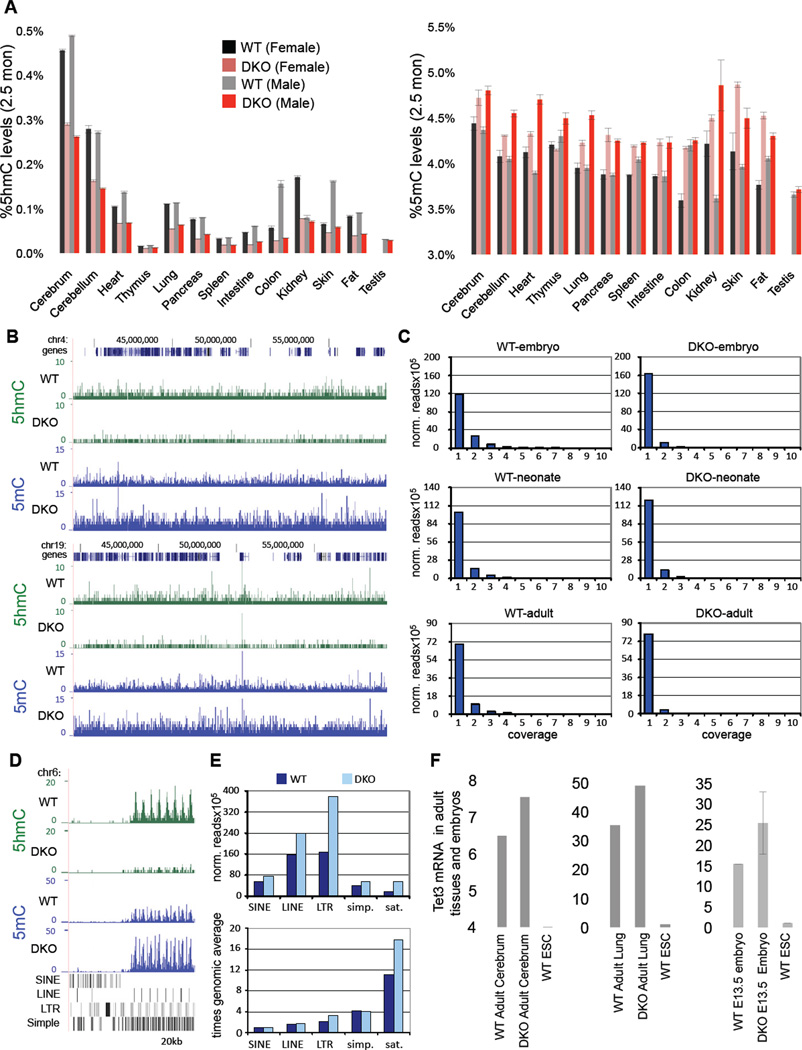
Since Tet proteins are dynamically expressed during development (Hajkova et al., 2010; Ito et al., 2010; Wossidlo et al., 2011), we investigated how the combined loss of Tet1 and Tet2 affects global 5hmC and 5mC levels. DNA was extracted from embryos and multiple organs of adult wild type and DKO mice and 5mC and 5hmC levels were quantified by mass spectrometry. As shown in Figure S3, slightly reduced levels of 5hmC were found in DKO embryos as compared to control embryos, and these were associated with a small but variable increase of 5mC levels. In contrast, the 5hmC levels in most organs of adult DKO mice were significantly reduced, which correlated with increased 5mC levels when compared to controls (Figure 6A). In order to map methylation patterns on a genome wide scale we analyzed DNA samples from a series of wt and DKO embryos, neonates and adults by methylated DNA immunoprecipitation with specific antibodies (Ficz et al., 2011; Wu et al., 2011) against 5mC (MeDIP) and 5hmC (hMeDIP) and massive parallel sequencing. Consistent with the mass spectrometry analysis, we found a general reduction in 5hmC and an increase in 5mC across all chromosomes in DKO samples, which confirmed global hypermethylation of the genome (Figure 6B). As we noticed a substantial increase in baseline levels of 5mC and less well defined peaks, we determined the position-wise coverage of the 5mC sequencing reads in each data set and compared their distribution between WT and DKO samples. As shown in Figure 6C, the distribution of reads showed a consistent change in all three tissues. The fraction with low coverage was increased in DKO samples, whereas peak regions were reduced, suggesting an increase in nonselective methylation in DKO animals. Further data analysis also indicated pronounced differences in the methylation of repetitive elements (Figure 6D), which are well-known targets of DNA methylation. This observation was further confirmed by a quantitative analysis of several retrotransposon classes (LINEs, SINEs, LTRs, satellites and simple repeats), which showed a significant enrichment for methylation and a general increase of 5mC in DKO neonates, in particular on LTRs and satellite repeats (Fig. 6E). The observed global hypermethylation in DKO animals is consistent with the notion that Tet-mediated hydroxymethylation protects against aberrant de novo methylation (Williams et al., 2011b).
Figure 6. Hypermethylation and reduced 5hmC levels in Tet1/Tet2 deficient embryos and adult mice.
(A) Quantification of 5hmC and 5mC in various tissues of adult 2.5-month-old male and female mice by mass spectrometry. (B) hMeDIP-seq (green) and MeDIP-seq (blue) profiles of two 20 MB regions from mouse chromosome 4 (upper panel) and 19 (lower panel). MeDIP was performed on genomic DNA extracted from WT and DKO tails of neonates (pup#7 and #15, respectively). hMeDIP was performed on genomic DNA extracted from WT and DKO adult cerebrum. Enrichments are indicated as normalized read counts. UCSC transcription units are indicated on top in dark blue. Genomic features are viewed as custom tracks in the UCSC genome browser. (C) Quantitative analysis of MeDIP results. Normalized 5mC read counts for each sample were determined for all bases and then distributed into bins with increasing read counts (1–10). The resulting profiles from DKO samples show a consistent increase in the number of bases that are covered by a single read, representing low-level methylation. At the same time, the number of bases with higher coverage becomes decreased in DKO samples. Samples analysed were from WT and DKO neonates (MeDIP performed on DNA from tail/hind leg), embryos (MeDIP performed on DNA from total brain) and adults (MeDIP performed on DNA from cerebrum). (D) hMeDIP-seq and MeDIP-seq profiles (as in B) of the flanks of a CpG-rich repeat region on chromosome 6 (chr6:47,563,923-47,636,951). Genomic features are shown as custom tracks in the UCSC genome browser. (E) Quantitative MeDIP analysis of repetitive elements in WT (dark blue) and DKO (light blue) neonates. The upper panel shows normalized 5mC read counts for several classes of repetitive elements, as indicated. The lower panel shows the enrichment of repetitive element sequencing reads relative to the genome-wide average (indicated by the red line). Abbreviations indicate Long Interspersed Nuclear Elements (LINEs), Short Interspersed Nuclear Elements (SINEs), Long Terminal Repeats (LTRs), simple repeats (simp.) and satellite repeats (sat.). (F) Relative expression of Tet3 in various embryos and adult tissues. Data are normalized to Gapdh. For all panels error bars indicate s.d. See also Figure S3–S4.
The presence of considerable levels of 5hmC in DKO mice suggests that Tet3 may maintain the observed level of 5mC hydroxylation. To assess whether the significant levels of 5hmC in organs of adult DKO animals was due to increased activity of Tet3, we measured Tet3 mRNA levels in wild type and DKO adult lung and brain, two organs with relatively high levels of Tet1 and Tet2, and as well as in E 13.5 embryos. We found a significant increase of Tet3 RNA in these tissues compared to control samples (Figure 6F). These results suggest that combined Tet1 and Tet2 deficiency induces Tet3, which may be responsible for the significant 5hmC levels detected in the double mutants.
5hmC is absent in Tet1/Tet2 deficient germ cells
Since Tet1 and Tet2 are expressed in germ cells and have been proposed to participate in global DNA demethylation and epigenetic reprogramming (Hajkova et al., 2010; Hackett et al., 2012a), we examined the 5hmC levels in germ cells of DHet and DKO E13.5 embryonic gonads by immunofluorscence. Germ cells were identified in gonad sections by staining with mouse vasa homolog (mvh) antibody, which revealed that DHet and DKO gonads of both sexes contained mvh-positive germ cells. However, only DKO germ cells, but not DHet germ cells or the surrounding somatic cells, were exclusively depleted of 5hmC (Figure 5G). This is consistent with specific expression of Tet1 and Tet2 in germ cells, and presence of Tet3 in the surrounding somatic cells in mid-gestation embryonic gonads (Hajkova et al., 2010; Yamaguchi et al., 2012; Hackett et al., 2012a). To examine if absence of 5hmC in DKO germ cells affects the global methylation content of these cells, we stained DHet and DKO gonads for 5mC but failed to detect any overt increase in total 5mC levels. Similarly, quantification of global 5mC and 5hmC levels in sperm DNA by mass spectrometry did not show any significant increase in global 5mC amounts in DKO sperm (Figure S5A). Surprisingly, we found an almost normal level of 5hmC in DKO sperm suggesting that Tet3, which is expressed during spermatogenesis (Figure S5B), achieves 5mC hydroxylation. This is supported by the presence of substantial levels of 5hmC in different sperm cell types in adult DKO testis (Figure S5C).
Imprinting is partially compromised in Tet1/Tet2 deficient mice
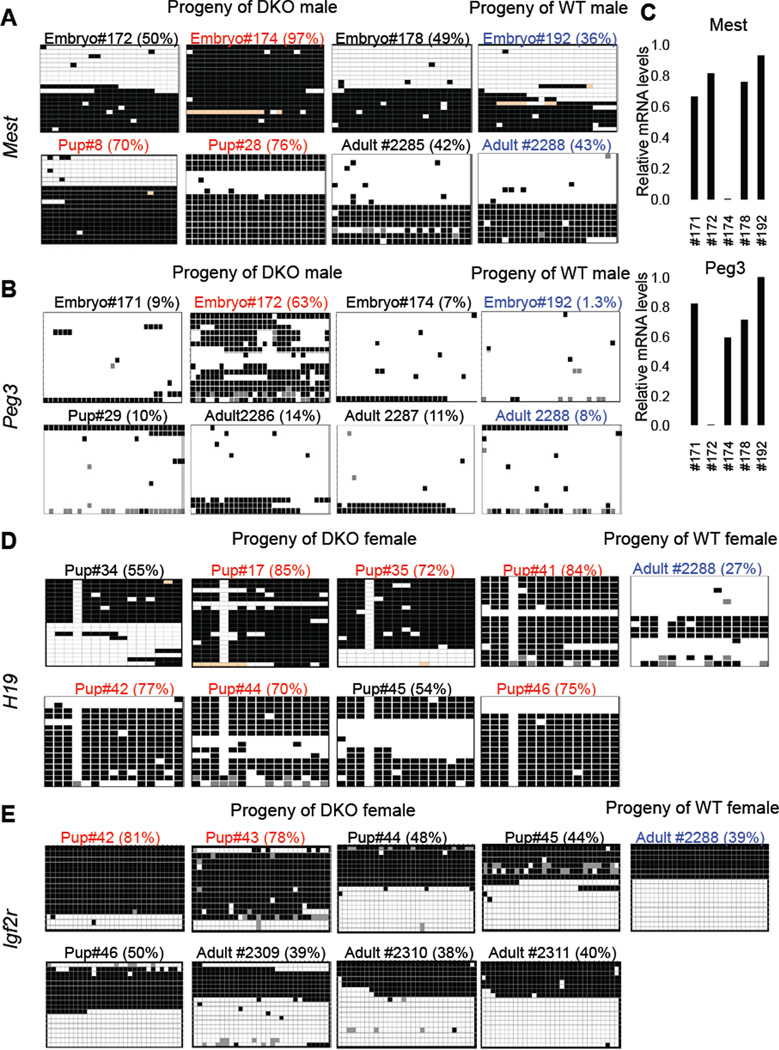
To determine whether loss of Tet1 and Tet2 and absence of 5hmC in germ cells affect erasure and reestablishment of imprinting during development and contribute to increased mortality of DKO mice, we looked for alterations in the methylation status of imprinted loci in genome wide MeDIP data sets generated from tail DNA of selected progeny of DKO mice. We found that heterozygous offspring of DKO mice from crosses of DKO males with WT females and DKO females with WT males showed increased methylation levels across 94 imprinted gene loci (Figure S4), suggesting that proper establishment of imprinting is compromised in DKO gametes. However, this analysis does not have the resolution to reflect the methylation status of imprinted control regions (ICRs) at CpG level. Therefore, to confirm these findings and to obtain more detailed insights, we applied sodium bisulfite sequencing to quantify methylation levels at ICRs of the maternally imprinted genes Mest and Peg3 and the paternally imprinted genes H19 and Igf2r in tail DNA from dead and surviving mutant mice derived from a male or female DKO parent (Figure 7, Table 1 and Table S1). We found that about a quarter of both surviving and dead progeny of DKO males had increased methylation at ICRs of Mest (>60%) and Peg3 (>50%) when compared to the progeny of control WT males (Table 1A, Table S1A, Figure 7 A&B), which also correlated with loss of expression of these genes (Figure 7C). This is consistent with changes in methylation at these ICRs in DKO sperm (Table1C and Figure S5D). In contrast, the majority of the progeny of DKO females harbored more than 65% methylation at the H19 ICR and a substantial number of offspring showed higher methylation levels at the Igf2r ICR as compared to the expected 50% in offspring of WT females (Table 1B and Table S1B and Figure 7 D&E) with three animals having aberrant methylation of both genes (Table 1B). These analyses confirmed imprinting aberrations in Tet1 and Tet2 deficient animals suggesting that these enzymes contribute to proper establishment of imprinting during development. Our results also suggest that Tet1/Tet2 deficiency in oocytes has a more significant effect on methylation of imprinted loci than their combined deficiency in sperm. However, increased methylation at imprinted loci is highly variable allowing apparently normal development in a fraction of embryos.
Figure 7. Tet1/Tet2 deficiency partially compromises imprinting.
(A, B, D, E) Heat map representation of bisulfite sequencing analysis of imprinted genes in selected samples corresponding to table S1. Within each heat map columns represent each CpG position in the sequenced ICR. Rows represent individual clones sequenced. Total of 16 to 20 clones were analyzed for each sample. Black box= methylated, white box=unmethylated and orange or grey box=undetermined due to poor sequence quality. Values on top of each map are % methylated CpG sites. Samples with aberrant (more than 65% methylation) are shown in red. Control samples are shown in blue. Detailed description of the entire analysis and all samples analyzed can be found in Table S1. (C) RTqPCR for Mest and Peg3 in selected E13.5 embryos confirming loss of expression of these genes in embryos with increased methylation shown in A and B. See also Table S1.
Table 1.
Methylation analysis of imprinted genes in DKO mice (see also Table S1)
| A: Analysis of mice from progeny of DKO male × or WT female | |||
|---|---|---|---|
| Mouse Phenotype |
Aberrant Mest Methylation |
Aberrant Peg3 Methylation |
Aberrant Methylation of both genes |
| Dead or abnormal mouse | 2 (out of 8) | 0 (out of 8) | 0 |
| Viable and normal mouse | 1 (out of 8) | 2 (out of 8) | 0 |
| Control (Progeny of WT male × WT or Dhet female) | 0 (out of 3) | 0 (out of 3) | 0 |
| B: Analysis of mice from progeny of WT male × DKO female | |||
|---|---|---|---|
| Mouse Phenotype |
Aberrant H19 Methylation |
Aberrant Igf2r Methylation |
Aberrant Methylation of both genes |
| Dead or abnormal mouse | 8 (out of 10) | 3 (out of 8) | 3 |
| Viable and normal mouse | 2 (out of 3) | 0 (out of 3) | 0 |
| Control (Progeny of WT or DKO male × WT female) | 0 (out of 2) | 0 (out of 1) | 0 |
| C: Analysis of sperm from DKO mouse | |||
|---|---|---|---|
| Genotype of sperm source | % Mest Methylation |
% Peg3 Methylation |
% H19 Methylation |
| DKO | 10.5 | 1.1 | 89 |
| WT | 1.2 | 1.1 | 94 |
Lastly, we also investigated the methylation status of ICRs of the maternally expressed H19 gene in the progeny of DKO males and the paternally expressed Mest gene in the progeny of DKO females. We found that 2 out of 4 offspring of DKO males that had increased Mest or Peg3 methylation also contained increased methylation at the H19 ICR (Table S1A, right column). A similar aberrant methylation was observed for Mest in 3 out of 5 progeny of DKO females that had increased H19 or Igf2r methylation (Table S1B, right column). This suggests that maternally expressed Tet1 and Tet2 is not only important for establishing the normal hypo-methylation state of maternal alleles but may also function in maintaining the hypo-methylated state of paternal alleles.
Discussion
The three enzymes Tet 1, 2 and 3 catalyze 5mC to 5hmC conversion (Tahiliani et al., 2009; Ito et al., 2010) but the dynamic and overlapping expression of the enzymes in various cell types of the embryo and adult poses a challenge to dissecting the function of each of the three proteins. The generation of Tet1 and Tet2 double mutant ESCs and mice has allowed us to investigate how the combined loss of these two enzymes affects pluripotency and development. Our findings can be summarized as follows: (i) Loss of Tet1 and Tet2 is compatible with development to apparently healthy adults although the majority of double mutants die during embryogenesis or perinatally with various developmental abnormalities. (ii) Deficiency of Tet1 and 2 results in complete loss of 5hmC in embryonic stem cells and germ cells and partial loss in tissues of the adult. (iii) Concomitantly with a decreased level of 5hmC the mutants display increased levels of 5mC, consistent with a protective role of the Tet enzymes against de novo methylation (Williams et al., 2011a). (iv) The analysis of DNA methylation levels also revealed abnormal imprinting of some genes in a fraction of embryos, consistent with the notion that disturbed imprinting may contribute to pre and postnatal abnormalities (Reik and Walter, 2001).
Tet1 and Tet2 are the main 5-methylcytosine dioxygenases in ESCs but their loss is dispensable for self-renewal and pluripotency
Tet1 and Tet2 are highly expressed in ES cells in contrast to Tet3, which is barely if at all detectable (Ito et al., 2010; Koh et al., 2011). While deletion of either Tet1 or Tet2 decreased 5hmC levels to 40% or 60% of wt levels, respectively, combined deletion of both genes completely depleted 5hmC from the ES cell genome, consistent with Tet1 and Tet2 being the most important if not the only mediators of hydroxymethylation in ES cells.
Previous work had established that deletion of either Tet1 or Tet2 did not affect self-renewal and pluripotency of ES cells though Tet1 single mutant ES cells displayed increased hemorrhagic tissues and skewed differentiation along trophectoderm lineage in a teratoma assay (Dawlaty et al., 2011; Koh et al., 2011). This raised the question whether complete lack of 5hmC would affect the developmental potential of ES cells. Here we show that Tet1/Tet2 double mutant ES cells can generate cells of all three germ layers in a teratoma assay, though the tumors tend to form an excess of hemorrhagic tissues and exhibit skewed differentiation defects towards the extraembryonic lineage. This was also seen with Tet1−/−/Tet2+/− mutant cells arguing that Tet1 deficiency is responsible for this phenotype. When injected into blastocysts, Tet1/Tet2 double mutant ES cells were able to generate chimeric mice although a significant fraction of chimeric embryos were lost with neural tube defects and exencephaly. This is consistent with an important role of 5hmC in normal development although the severity of the observed embryonic phenotypes may depend on the extent of chimeric contribution. To better define the role of Tet1 and Tet2 in embryos and postnatal mice we generated animals that were deficient for both enzymes.
Combined loss of Tet1 and Tet2 is compatible with development
Previous work has shown that both Tet1 and Tet2 single mutant mice survive at normal Mendelian frequencies (Dawlaty et al., 2011; Ko et al., 2011; Li et al., 2011) suggesting that the two genes may compensate for each other in development. The generation of viable DKO animals established that combined loss of Tet1 and Tet2 is compatible with development and that overtly normal animals can be generated in the absence of these two genes. However, the majority of double mutants die with a variety of developmental abnormalities such as exencephaly, hemorrhage and growth retardation consistent with a crucial role of hydroxymethylation in embryogenesis. Our analysis of mid to late gestation embryos and newborns does not suggest that development of DKO embryos is delayed during gestation as all mutant pups had fully developed organs and lethality was not exclusively associated with animal size or growth retardation. Therefore, embryonic abnormalities observed in DKO mice are unlikely due to a general developmental delay but rather more likely caused by a variable penetrance of epigenetic abnormalities leading to a wide spectrum of defects including, but not limited to, exencephaly.
One possible explanation to the observation that a fraction of double mutant embryos survived to overtly normal and fertile adult mice was that Tet3 compensated in part for combined Tet1/Tet2 deficiency. Indeed, we found increased but variable expression of Tet3 in postnatal double mutant animals resulting in substantial though reduced 5hmC levels suggesting that induction of this enzyme was sufficient to support survival in the absence of Tet1 and Tet2. While adult mutant males had normal gonads and were fertile, double mutant females displayed smaller ovaries, reduced numbers of mature follicles and reduced fertility, which is more pronounced than previously observed in Tet1 single knockout mice (Dawlaty et al., 2011). By a number of criteria, germ cell development appeared to be normal in double mutant embryonic gonads displaying approximately the same number of mvh-positive germ cells as gonads of wild type embryos. This raised the question whether Tet1/Tet2 deficiency could cause epigenetic aberrations in the germ cells, which could possibly explain some of the abnormalities in double mutant embryos.
A recent report suggested lethality of Tet1 KO mice in a 129sv/Ola background and partial lethality when backcrossed to C57BL/6 (Yamaguchi et al., 2012), which contrasts with our previous findings demonstrating full viability of Tet1 mutant mice in a 129/C57BL/6 background (Dawlaty et al., 2011), a result reproduced in this study (figure 3A and supplementary figure S2A). Although variation in background of mice has been suggested as a possible explanation for these differences (Yamaguchi et al., 2012), non-specific DNA binding of the fused CXXC-beta-geo protein, produced from the Tet1 knockout allele in their study, may interfere with the function of Tet3 and possibly explain the lethal phenotype seen by Yamaguchi et al.
Epigenetic abnormalities and stochastic perinatal lethality are associated with combined Tet1 and Tet2 deficiency
It has been suggested that 5hmC protects DNA against de novo methylation (Williams et al., 2011a; 2011b). Consistent with this possibility we found significantly increased levels of genomic 5mC associated with decreased levels of 5hmC in double mutant ES cells and tissues. Tet3 is induced in tissues of adult double mutant mice resulting in significant levels of 5hmC, which likely compensates for Tet1/Tet2 deficiency and assures survival of some mutant embryos. Likewise, the significance of Tet3 is further underscored in a recent study in Xenopus, which does not have Tet1 orthologs, and where deficiency of Tet3 impairs eye and neural development (Xu et al., 2012).
Recent profiling of 5mC and 5hmC distribution in wild type PGCs during different stages of germ cell reprogramming highlights the significance of dynamic changes in DNA methylation for proper germ cell maintenance (Seisenberger et al., 2012; Yamaguchi et al., 2012; Hackett et al., 2012a). These data demonstrate that during the process of global demethylation of PGCs and establishment of imprinting, 5mC erasure is coupled with 5hmC conversion, a process driven by Tet1 and Tet2 (Hackett et al., 2012a). This conclusion is consistent and complementary to our findings that DKO mice harbor increased imprinting defects. Therefore, the variable abnormalities seen in double mutant embryos and pups may be due to epigenetic aberrations caused by Tet1/Tet2 deficiency either during gametogenesis or in somatic tissues. Tet3 is not expressed in early stage germ cells (Hajkova et al., 2010) but is expressed later in gametogenesis (E16.5) (Yamaguchi et al., 2012) as well as during spermatogenesis (Figure S5B) and in the oocytes (Gu et al., 2011; Wossidlo et al., 2011). In agreement with these observations, antibody staining of Tet1/Tet2 deficient embryonic germ cells showed no detectable 5hmC, while sperm of double mutant mice showed nearly normal levels of 5hmC and 5mC (Figure S5A). Furthermore, the deletion of Tet3 in mouse oocytes leads to epigenetic abnormalities in the paternal genome (Gu et al., 2011) arguing that Tet3 expression during early embryogenesis is functionally important. Epigenetic abnormalities seen in the Tet1/Tet2 double mutants may thus be due to a combination of epigenetically aberrant gametes, which would be predicted to lead to abnormal imprinting, and to only partial restoration of normal 5hmC levels by Tet3 expression in somatic tissues. Indeed, genome wide analysis showed widespread changes in methylation levels in genomic DNA. Importantly, a variable fraction of imprinted genes was found to be abnormally methylated suggesting that disturbed imprinting may contribute to the partially penetrant developmental abnormalities of Tet1/Tet2 mutant embryos, although aberrant imprinting was not exclusively linked to lethality (Table 1). Our data also suggest that Tet1/Tet2 deficiency in oocytes has more severe consequences on imprinting than the combined enzyme deficiency in male gametes consistent with the less frequent incidence of imprinting abnormalities in male DKO progeny and the almost normal 5hmC level in mature sperm, which is presumably achieved by Tet3 expression.
Accurate establishment of the role of Tet enzymes in development is further complicated by the findings that Tet enzymes, in addition to the presence of the conserved catalytic domain that drives 5mC to 5hmC conversion, also have functions independent of their catalytic. Tet1 has been implicated in transcriptional repression (Wu et al., 2011; Williams et al., 2011b) and Tet2 has recently been shown to enhance histone glycosylation and gene transcription (Chen et al., 2012). While recent studies demonstrate that Tet1 and Tet2 catalytic activity is essential for germ cell reprogramming (Hackett et al., 2012a), a study of Tet3 in Xenopus shows that the non-catalytic CXXC domain works cooperatively with the catalytic domain to help target Tet3 to its bindings sites during development (Xu et al., 2012). Mutant alleles need to be generated to dissect catalytic domain dependent and independent functions of Tet proteins in development.
Understanding the biological significance of the varying abundance of 5hmC and widespread expression of Tets in different embryonic and adult cell types has been the main focus of establishing the physiological relevance of these enzymes in development. In this study we demonstrate that combined loss of Tet1 and Tet2 is dispensable for pluripotency and compatible with development but leads to epigenetic abnormalities, which cause embryonic defects and impair postnatal survival. Our findings support the notion that combined loss of Tet1 and Tet2 leads to reduced overall Tet activity, which results in reduced hydroxymethylation, increased DNA methylation and aberrant imprinting in embryonic, somatic and germ cells. Our results suggest that these epigenetic aberrations predispose to and then stochastically lead to manifestation of developmental abnormalities in mutant embryos resulting in only a fraction of mutants surviving to adulthood. Adult Tet1/Tet2 deficient animals appear overtly normal and are fertile although we cannot exclude more subtle defects associated with the combined deficiency of these genes that have a late-life onset such as cognitive and neurological dysfunction and hematopoietic disorders given their high expression in hematopoietic (Ko et al., 2011; Li et al., 2011) and neural tissues (Kriaucionis and Heintz, 2009; Ito et al., 2010).
Experimental Procedures
Derivation and culture of ESCs
Mouse embryonic stem cells were derived Tet1/Tet2 double mutant mice as described before (Markoulaki et al., 2008) and briefly explained in the supplemental experimental procedures. Derived ES cell lines were expanded on feeders using regular ESCs media containing LIF. All ESC lines were genotyped by Southern blot using the protocols and probes previously outlined (Dawlaty et al., 2011; Li et al., 2011) and PCR using primers described in supplemental information. For labeling ESCs with td-Tomato, ES cells were transduced with FUW-td-Tomato lentivirus four times over the course of 48 hours and then FAC-sorted to enrich for td-tomato positive cells of equal intensity.
Chimera assays and analysis of midgestation embryos
To generate chimeric embryos, 10–12 td-tomato labeled ES cells were injected in to B6D2F1 × B6D2F1 E3.5 wild-type blastocysts and surgically implanted into 2.5 d.p.c. pseudo-pregnant Swiss Webster female mice following standard procedures. E14.5 embryos were harvested, dissected and imaged under a fluorescence dissecting scope. For analysis of mid-gestation embryos timed pregnancies were set up by crossing DKO males with double heterozygote females. Pregnant mice were sacrificed and dissected at mid gestation days E10.5 to E18.5. Embryo numbers, size and defects were recorded for each time point. Tail or head tissue was used for genotyping.
Generation and maintenance of mouse colony
Tet1/Tet2 Double knockout mice (DKO) were generated by crossing Tet1−/− (Dawlaty et al., 2011) and Tet2−/− (Li et al., 2011) to obtain Tet1+/− | Tet2+/− animals, which were further intercrossed to produce double knockout mice. Additional breeding schemes and details on colony maintenance can be found in the supplemental experimental procedures. For fertility analysis DKO females were crossed with wild type males and DKO males were crossed with wild type females. To ensure mating, vaginal plugs were checked daily. All mice in the study were maintained in a mixed 129 and C57BL/6 background. Animal care was in accordance with institutional guidelines and approved by the Committee on Animal Care, Department of Comparative Medicine, Massachusetts Institute of Technology.
Quantification of 5mC and 5hmC
Combined liquid chromatography-tandem mass spectrometry with multiple reaction monitoring (LC-MS/MS-MRM) was applied to quantify 5hmC and 5hmC levels in DNA extracted from ESCs or tissues as explained before (Le et al., 2011) and described briefly in supplemental experimental procedures.
Sodium bisulfite sequencing of imprinted genes
Two µg of DNA was treated with sodium bisulfite using EpiTect kit (Qiagen). H19, Mest and Peg3 imprinted loci were amplified as explained before (Lucifero et al., 2002), cloned into pcr2.1 TA cloning vector and sequenced with M13rev primer. 16–20 clones were analyzed for each sample.
(h)MeDIPs-Seq
MeDIP-seq was performed as described (Bocker et al., 2012). Briefly, 10 to 50 µg of genomic DNA for each sample was sonicated to 300bp fragments using a Covaris S220 (Covaris) and used for the ligation of barcoded Illumina adapters. Five µg of adapter ligated DNA was used for the immunoprecipitation. Sequencing libraries were generated by pooling immunoprecipitated DNA from 8 samples. Libraries were amplified using Illumina Enrichment Primers. Paired-end 100 bp sequencing was performed on an Illumina HiSeq system. Processing and mapping of the sequencing data were carried out as described in supplemental experimental procedures. Read counts were normalized according to the 5mC and 5hmC levels of each sample DNA, as determined by mass spectroscopy.
Supplementary Material
Highlights.
Tet1/Tet2 double mutant ES cells are depleted of 5hmC and remain pluripotent
Mice with combined loss of Tet1 and Tet2 are viable with reduced fertility
Hypermethylation and partial perinatal lethality are associated with Tet1/Tet2 loss
Deficiency of Tet1 and Tet2 compromises imprinting
Acknowledgments
We thank R. Flannery, K. Ganz, D. Fu and R. Alagappan for help with animal husbandry, histology and blastocyst injections. We are also grateful to S. Sarkar and Y. C. Hu for help with microscopy, J. Cassady for help with FACs analysis, J. Kwon and J. Love from the Whitehead Genome Technology Core for help with microarrays, A. Yoon for help with mass spectrometry, and S. Schmidt from the DKFZ Genomics and Proteomics Core Facility for Illumina sequencing services. We thank members of the Jaenisch laboratory for helpful discussions. We are also thankful to Dr. R. Bronson for analyzing histology slides. M.M.D is a Damon Runyon Postdoctoral Fellow. B.E.P is supported by a PhD fellowship from the Boehringer Ingelheim Fonds. A.W.C. is supported by a Croucher scholarship. T.L. is supported by UCLA Molecular, Cellular and Neurobiology Training Grant, UCLA Mental Retardation Training Grant, and Eugene V. Cota-Robles Fellowship. Work in F.L.’s lab was supported by a grant from the Deutsche Forschungsgemeinschaft (SPP 1463). R.J. is funded by NIH grants 5-RO1-HDO45022 and 5-R37-CA084198.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The (h)MeDIP sequencing data (accession number GSE42396) and gene expression array data sets (accession number GSE42991) have been deposited at the Gene Expression Omnibus (GEO) database, http://www.ncbi.nim.nih.gov/geo
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bocker MT, Tuorto F, Raddatz G, Musch T, Yang F-C, Xu M, Lyko F, Breiling A. Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster. Nature Communications. 2012;3:818–812. doi: 10.1038/ncomms1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011:1–7. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2012 doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu Y-C, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi S-W, Page DC, et al. Tet1 Is Dispensable for Maintaining Pluripotency and Its Loss Is Compatible with Embryonic and Postnatal Development. Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011 doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Gu T-P, Guo F, Yang H, Wu H-P, Xu G-F, Liu W, Xie Z-G, Shi L, He X, Jin S-G, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011:1–7. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA Demethylation Dynamics and Imprint Erasure Through 5-Hydroxymethylcytosine. Science. 2012a doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Zylicz JJ, Surani MA. Parallel mechanisms of epigenetic reprogramming in the germline. Trends in Genetics. 2012b:1–11. doi: 10.1016/j.tig.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-Wide Reprogramming in the Mouse Germ Line Entails the Base Excision Repair Pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science. 2011 doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 Regulate 5-Hydroxymethylcytosine Production and Cell Lineage Specification in Mouse Embryonic Stem Cells. Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T, Kim K-P, Fan G, Faull KF. A sensitive mass spectrometry method for simultaneous quantification of DNA methylation and hydroxymethylation levels in biological samples. Analytical Biochemistry. 2011;412:203–209. doi: 10.1016/j.ab.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai C, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011 doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation Dynamics of Imprinted Genes in Mouse Germ Cells. Genomics. 2002;79:530–538. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- Markoulaki S, Meissner A, Jaenisch R. Somatic cell nuclear transfer and derivation of embryonic stem cells in the mouse. Methods. 2008;45:101–114. doi: 10.1016/j.ymeth.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 Loss Leads to Increased Hematopoietic Stem Cell Self-Renewal and Myeloid Transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SKT, Bestor TH. The Colorful History of Active DNA Demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, Mcloughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011:1–4. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronné L, Valle Della, V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern M-H, et al. TET2 Inactivation Results in Pleiotropic Hematopoietic Abnormalities in Mouse and Is a Recurrent Event during Human Lymphomagenesis. Cancer Cell. 2011:1–14. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The Dynamics of Genome-wide DNA Methylation Reprogramming in Mouse Primordial Germ Cells. Molecular Cell. 2012 doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins—guardians of CpG islands? Nature Publishing Group. 2011a:1–8. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PAC, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011b:1–24. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter JOR. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nature Communications. 2011;2:241–248. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu H, D’alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011:1–18. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nature Publishing Group. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, et al. Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012 doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung J-H, Chen PB, Dong X, Ee L-S, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD Complex Regulates Expression of 5-Hydroxymethylcytosine Marked Genes in Embryonic Stem Cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.