Abstract
Type VI protein secretion system (T6SS) is important for bacterial competition through contact-dependent killing of competitors. T6SS delivers effectors to neighboring cells and corresponding antagonistic proteins confer immunity against effectors that are delivered by sister cells. Although T6SS has been found in more than 100 gram-negative bacteria including many important human pathogens, few T6SS-dependent effector and immunity proteins have been experimentally determined. Here we report a high-throughput approach using transposon mutagenesis and deep sequencing (Tn-seq) to identify T6SS immunity proteins in Vibrio cholerae. Saturating transposon mutagenesis was performed in wild type and a T6SS null mutant. Genes encoding immunity proteins were predicted to be essential in the wild type but dispensable in the T6SS mutant. By comparing the relative abundance of each transposon mutant in the mutant library using deep sequencing, we identified three immunity proteins that render protection against killing by T6SS predatory cells. We also identified their three cognate T6SS-secreted effectors and show these are important for not only antibacterial and antieukaryotic activities but also assembly of T6SS apparatus. The lipase and muramidase T6SS effectors identified in this study underscore the diversity of T6SS-secreted substrates and the distinctly different mechanisms that target these for secretion by the dynamic T6SS organelle.
Keywords: Dictyostelium discoideum, T6SS assembly, actin cross-linking, lipase, VgrG
Microbes often exist in complex communities in nature and compete with other species for limited resources. To gain relative fitness, microbes have evolved diverse attack mechanisms that include secretion of toxins, injection of effector proteins into target cells, and production of small molecule antibiotics. Many gram-negative bacteria also encode a cytotoxic organelle called the type VI protein secretion system (T6SS). T6SS has been found in more than 25% of gram-negative bacteria (1), including human pathogens Vibrio cholerae (2) and Pseudomonas aeruginosa (3). Several T6SS proteins are structurally related to proteins associated with bacteriophage tails (4, 5), and indeed the T6SS apparatus appears morphologically similar to contractile phage tails within the bacterial cytoplasm (6). These structures are nearly 10 times larger than bacteriophage tails, attached to the cytoplasmic membrane, and highly dynamic (6, 7). Type T6SS proteins have been identified that correspond to orthologous components in phage tails that include the tail spike, tube, sheath, and base plate (4, 6). Inner- and outer-membrane-associated T6SS proteins likely anchor the T6SS organelle to the cell envelope (8–10). Because the functionality of the T6SS apparatus has been correlated with its ability to kill or inhibit both eukaryotic as well as prokaryotic cells, most models for T6SS function postulate that the contraction of the sheath minimally ejects a complex corresponding to the T6SS spike/tube complex out of the T6SS+ cells and into target cells in close contact (4, 6).
V. cholerae employs T6SS to kill Escherichia coli, Dictyostelium discoideum amoebae, and macrophage cell lines (2, 11, 12). Three proteins secreted by the V. cholerae T6SS belong to the VgrG family of proteins and one of these (VgrG1) carries an enzymatically active domain that is capable of cross-linking actin in eukaryotic cells (5, 11, 13). VgrG proteins are thought to constitute a trimeric structure located at the very tip of a T6SS tube structure with the Hcp protein forming the tube (4). V. cholerae can also use its T6SS apparatus to secrete the protein VasX, which contributes to the killing of amoebae, but it is unclear how this protein is recognized or secreted by the T6SS apparatus (14). Based on homology of the C-terminal domain of VgrG3 to peptidoglycan-binding domains, Pukatzki and coworkers (5) first suggested that T6SS might also target prokaryotic cells. This prediction was shown to be the case for P. aeruginosa, which secretes three T6SS-dependent effectors that are inhibitory to other Pseudomonas species and E. coli (15, 16). These effectors have corresponding antagonistic immunity proteins that prevent self-killing (15, 16). Indeed, microscopic analysis of P. aeruginosa cultures suggest that sister cells are often found battling each other and it seems that dynamic T6SS activity directed at one cell triggers corresponding T6SS activity in the cell that is directed against the attacker (7). Such “T6SS dueling” behavior suggests that immunity proteins are needed because of repeated sister cell attacks rather than for protection against the toxicity exhibited by effectors before or during the secretion process per se (7, 15, 16). T6SS dueling activity has also been recently shown to be critical to the selection and targeting of T6SS+ prey by P. aeruginosa (17), suggesting immunity proteins may also be important for resistance to heterologous attack by related T6SS+ organisms.
Despite the large number of T6SS-possessing bacteria, T6SS effector–immunity pairs have only been experimentally identified in P. aeruginosa (15), Burkholderia thailandensis (18), and Serratia marcescens (19), and they share little homology in primary sequence. A heuristic approach based on physical and contextual properties of known effector–immunity pairs predicts that they are widespread in T6SS-possessing bacteria (18). Nonetheless, no effector–immunity protein has been identified in V. cholerae using this approach (18), which suggests that other classes of T6SS effector–immunity pairs remain to be discovered.
Transposon mutagenesis coupled with next generation sequencing (Tn-seq) is a powerful tool to uncover gene functions (20–23). The relative abundance of each mutant in the transposon library pool can be tracked by massively parallel sequencing, which can be used to determine the relative fitness of each mutant under specific conditions (21, 22, 24). Here we use a Tn-seq approach to identify genes whose essentiality is dependent on the presence of a functional T6SS. We predicted that if immunity proteins confer protection against the T6SS-attack of neighboring sister cells, then these immunity proteins would be required for survival in a T6SS+ strain but dispensable in a T6SS mutant. Using Tn-seq, we report three immunity proteins and their closely linked, corresponding effectors in V. cholerae. This approach is generally applicable to studying immunity–effector pairs in other T6SS-possessing bacteria and may also define gene products that may be required for surviving damage done by the dynamic T6SS organelle.
Results
Identification of Immunity Proteins Required for Self-Protection.
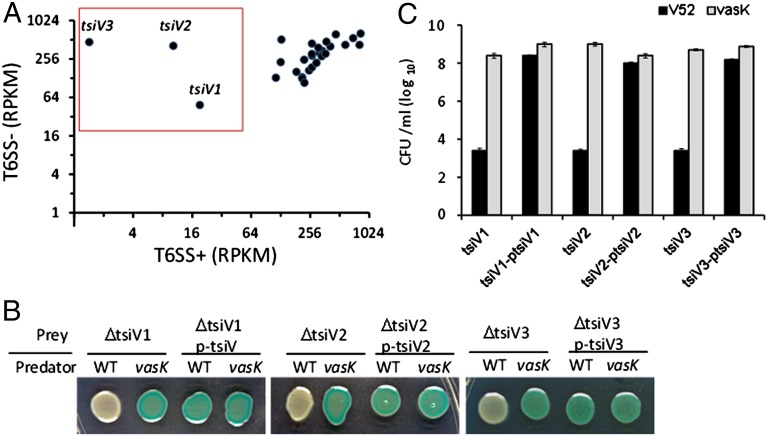
We designed a selection/screening scheme for T6SS immunity proteins based on our hypothesis that cells are protected from the T6SS activity of neighboring sister cells by immunity proteins (Fig. 1). Such immunity proteins would be crucial for survival only in wild type but not in T6SS mutants. To identify T6SS-dependent essential genes, we constructed saturating transposon libraries in wild-type V. cholerae V52 and a T6SS double hcp mutant missing the key component that assembles the T6SS tube (4, 25). Each library contained ∼1 million mutants that were cultured on agar as a pool to maximize interaction of different mutant cells with each other; we envisioned such interactions would deplete cells carrying mutations in immunity genes in the T6SS+ population selectively. The mutant cells surviving these conditions were then subjected to deep sequencing to identify specific transposon locations and those genes whose inactivation occurred more frequently in the T6SS− background than in the T6SS+ background. In total, there were 18.7 million and 14.5 million mapped unique reads for the wild-type and the hcp mutant libraries, respectively. We used a normalized value reads per kilobase per million (RPKM) mapped reads to represent the relative number of insertions in each gene in the library pool, with low RPKM values indicating genes required for survival and high values indicating genes dispensable under the testing conditions. We identified eight genes with substantially lower RPKM values in the wild type compared with the T6SS null mutant (Table S1). Mutants in seven of the genes identified were available in our defined V. cholerae mutant library (26). Because the parental strain of these mutants (C6706) is not phenotypically T6SS+ (27), these mutants were not subject to counter selection during construction of the ordered library (26). By mixing these mutants with the T6SS+ wild-type strain V52, we confirmed that two mutants were sensitive to T6SS-dependent killing (VCA0021 and VCA0124). Because the defined library lacks a specific transposon mutant for VC1419, we used a previously constructed V52 multiple mutant VC1417-21 (13) to test survival of a VC1419 mutant. This mutant was highly sensitive to killing by V52, which suggests that our Tn-seq analysis was likely identifying genes encoding T6SS immunity. By comparing the RPKM values of these three genes (VC1419, VCA0021, and VCA0124) with the other T6SS genes, we found that they showed considerably higher RPKM values in the T6SS mutant than in the wild type (Fig. 2A), which suggests that they are important for survival in the wild type but not in the T6SS hcp double mutant. We name these three putative T6SS immunity genes in V. cholerae tsiV1 (VC1419), tsiV2 (VCA0021), and tsiV3 (VCA0124). Although single mutants for tsiV1–3 genes are likely not viable in V52, we have previously constructed two V52 multiple mutants VC1417-21 (lacking TsiV1) and VCA0019-21 (lacking TsiV2) (13), and made a V52 double mutant VCA0123-24 (lacking TsiV3) in this study. To confirm the protective role of tsiV1–3, we tested whether complementing these tsiV1–3 mutants with corresponding tsiV genes could improve survival when challenged with wild-type T6SS+ V52 strain. We found that complementation in trans increased survival of these tsiV1–3 mutants by 10,000-fold (Fig. 2 B and C). These results confirm that tsiV1–3 genes encode immunity proteins required for self-protection against T6SS.
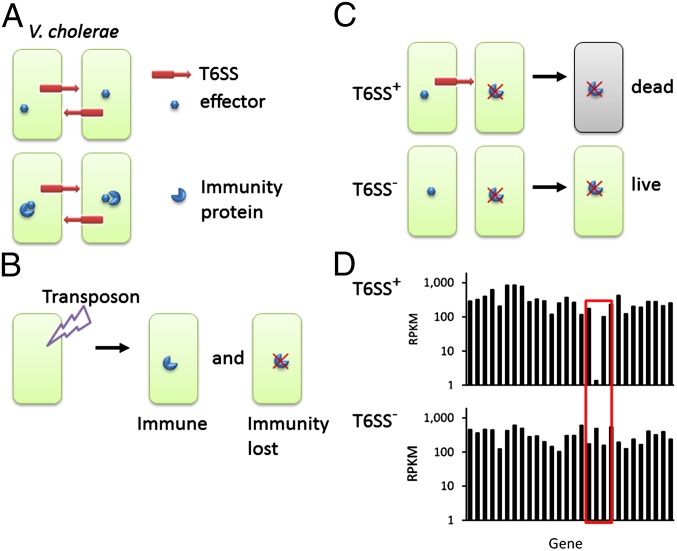
Fig. 1.
Schematic diagram of the selection method for immunity genes. (A) T6SS organelles of two V. cholerae sister cells actively attack each other and inject a potentially lethal effector protein. Because a cognate immunity protein binds to and neutralizes the effector, the cells survive the attack. A corollary to this model is that injection of the effector protein per se should not be a lethal event in sister cell–sister cell interactions. (B) Saturating transposon mutagenesis generates two populations of mutants, one still immune and the other lost immunity due to transposon inactivation of a T6SS effector–immunity protein. (C) Mutants that lost immunity were killed by T6SS of neighboring cells and diminished in the mutant pool. However, such mutants would survive if constructed in a strain lacking T6SS apparatus. (D) Deep sequencing of transposon insertion junctions (Tn-seq) is used to quantify the relative abundance of transposon insertions in every gene in the context of the T6SS+ and T6SS− backgrounds. The genes for T6SS immunity proteins correspond to those genes that show a lower normalized value of mapped DNA sequence reads expressed as RPKM in the T6SS+ strain than in the T6SS− strain.
Fig. 2.
Immunity proteins confer self-protection. (A) Comparison of the RPKM values for insertions in various genes associated with T6SS function and immunity. RPKM vales for the same genes were determined in either wild type (T6SS+) or the hcp double mutant (T6SS−) of V. cholerae V52. The RPMK values of most T6SS genes formed a cluster located at the upper right of the graph because transposons inserted in these genes at the same normalized frequency in the T6SS+ and T6SS− strain. In contrast, three genes (highlighted in the red box), named tsiV1–3, showed considerably lower number of insertions in the T6SS+ wild type, which suggests that they are important for T6SS-dependent survival. (B) Confirmation of immunity genes by complementation. Mutants (lacZ+) of immunity genes (tsiV1–3) were coincubated with the wild-type strain V52 (lacZ−) and the vasK mutant (lacZ−) on LB with 40 µg/mL X gal. Blue or yellow color indicates survival or killing of tsiV mutants, respectively. (C) Survival of tsiV mutants was quantified by serial plating on LB medium. Note that the tsiV mutants used in the killing assay lack the corresponding tsiV genes as well as neighboring genes (see Table S3 for detailed genotypes). The vasK mutant is a known T6SS null mutant (2).
Identification of Corresponding Effector Proteins.
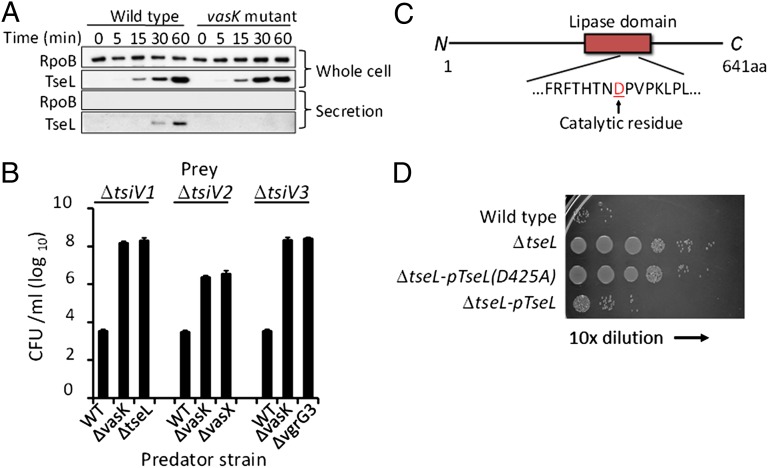
Upstream of two of the immunity genes tsiV2 and tsiV3 are known T6SS-secreted proteins encoded by VCA0020 (VasX) (14) and VCA0123 (VgrG3) (2). We hypothesized that the gene product of VC1418 (located upstream of tsiV1) may also be secreted by T6SS. To test this, we expressed the VC1418 product in wild-type V52 and the vasK mutant (T6SS−). VC1418 was secreted in the wild type but not in the T6SS mutant, indicating its secretion is T6SS-dependent (Fig. 3A). We have named this gene product TseL to reflect the likely nature of its biological activity (see below). We then tested if these three T6SS-secreted proteins (VgrG3, VasX, and TseL) are required for killing corresponding immunity gene mutants. We found that mutants lacking individually each of these three secreted proteins were unable to kill mutants lacking the corresponding immunity genes (Fig. 3B). Thus, VasX (VCA0020), VgrG3 (VCA0123), and TseL (VC1418) are T6SS-dependent, antibacterial effectors of V. cholerae.
Fig. 3.
Identification of effectors required for killing the corresponding immunity gene mutants. (A) The secretion of TseL (the VC1418 gene product) is T6SS dependent. Samples were taken at different time points after induction for TseL expression and analyzed by Western blot analysis. The RNA polymerase subunit RpoB was used as a loading control. (B) Deletions in tseL, vasX, or vgrG3 abolished killing activity against the corresponding tsiV mutants. Survival of prey cells was quantified by serial plating after coincubation with predatory cells on LB medium for 3 h. (C) TseL is a 641 residue protein that carries a putative lipase domain. (D) A tsiV1 mutant of V52 was incubated with various predator strains as indicated and then titered for survival. The tsiV1 mutant was killed by wild-type V52 and the tseL mutant complemented with TseL but not by the tseL mutant complemented with TseLD425A, a mutation in the putative lipase domain active site.
TseL is predicted to carry a lipase domain (Fig. 3C). We asked if this lipase domain is important for its antibacterial activity. Using site-directed mutagenesis, we constructed a mutant of TseL that was predicted to change a conserved catalytic residue aspartate-425 to alanine. This mutant construct TseLD425A was unable to complement the knockout mutant of TseL in assays for killing the tsiV1 mutant (Fig. 3D), suggesting the antibacterial effect of TseL is dependent on its lipase domain. It should be emphasized that we attempted to detect the lipase activity associated with TseL by using several standard substrates but were unsuccessful, which suggests that this protein might require undefined cofactors to activate its lipase activity.
Killing of E. coli by V. cholerae Effector Mutants.
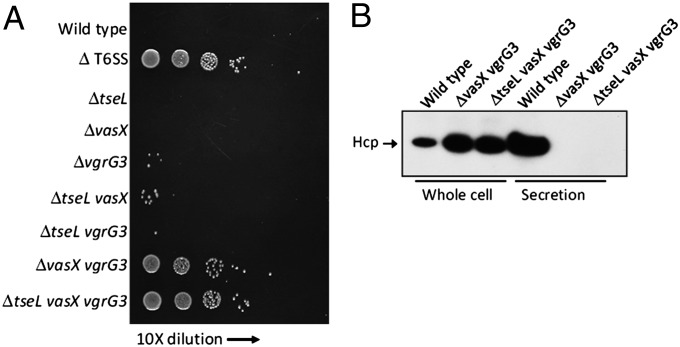
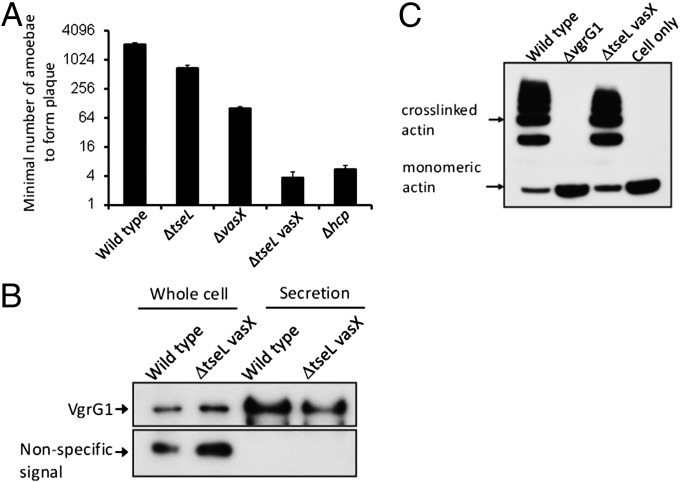
To test whether these effectors are required for T6SS assembly or competition with other bacteria, we mixed effector mutants with E. coli on agar and quantified survival of E. coli after coincubation. Single mutants lacking each of the effectors killed E. coli as efficiently as wild type (Fig. 4A), indicating that these effectors individually are not essential for T6SS secretion and lethal antibacterial activity. Mutants missing tseL and vasX or tseL and vgrG3 also killed E. coli. By contrast, a mutant missing vasX and vgrG3 or a triple mutant lacking tseL, vasX, and vgrG3 failed to kill E. coli. This loss of killing may result from loss of effector activities, or, alternatively, from loss of a T6SS functional apparatus. To test the latter, we checked the secretion of Hcp, a hallmark for T6SS organelle function (2). We found that Hcp was not secreted in the mutant lacking vasX and vgrG3 nor the triple mutant lacking tseL, vasX, and vgrG3 (Fig. 4B). These results suggest that these two double and triple effector mutants do not produce a functional T6SS apparatus.
Fig. 4.
Effects of T6SS effectors on killing E. coli and Hcp secretion. (A) Different V. cholerae strains were individually mixed with E. coli CC114 (tetracycline resistant) on LB medium at 37 °C for 3 h and then titered for viable counts on LB containing tetracycline. (B) Western blot analysis to compare Hcp protein levels in cells and supernatant fluids of different V. cholerae strains. Hcp secretion was abolished in mutants missing both vasX and vgrG3.
VgrG3 Targets the Cell Wall.
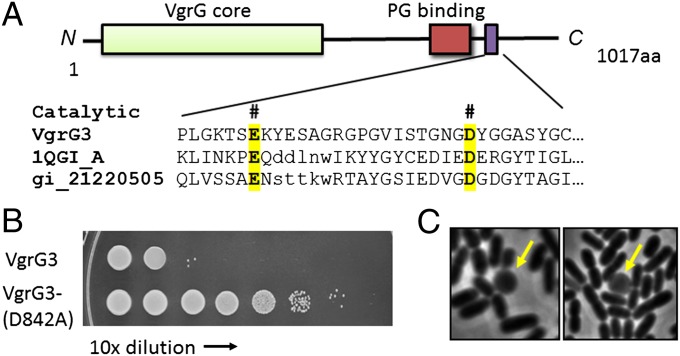
The C terminus of VgrG3 contains a peptidoglycan-binding domain (PBD) (2) suggesting that VgrG3 likely targets the cell wall of prey cells for degradation (Fig. 5A). Using the homology detection program HHpred (28), we found that the sequence following the PBD domain of VgrG3 shares conserved residues with the catalytic sites of lysozyme-like chitosanases (29–31). When cloning VgrG3 into the pBAD-expression vector, we noticed that induced expression of this protein leads to lysis of E. coli culture. To test if this lysis phenotype was caused by the activity of this lysozyme-like domain, we performed site-directed mutagenesis to change the predicted catalytic residue aspartate-842 to alanine (Fig. 5A). The point mutation D842A abolished the toxicity of VgrG3 upon arabinose induction in E. coli (Fig. 5B). Microscopic analysis showed that cells expressing wild-type VgrG3 turned from rod to spherical shape, consistent with the hypothesis that the PBD/lysozyme-like C-terminal domain of VgrG3 is indeed capable of disrupting the integrity of the bacterial cell wall (Fig. 5C).
Fig. 5.
VgrG3 targets the cell wall. (A) Domain structure of VgrG3 and the alignment of the putative catalytic site of VgrG3 with identified lysozymelike chitosanase homologs. The conserved catalytic residues are highlighted. (B) The catalytic residue aspartate-842 is critical for VgrG3 function. Induced expression of VgrG3 in E. coli resulted in considerable cell death although expression of VgrG3D842A mutant construct allowed nearly 10,000-fold better survival. (C) Light microscopic imaging of E. coli expressing VgrG3. Yellow arrows highlight cells with a spherical shape that are seen frequently when VgrG3 is expressed in E. coli and seldom seen when VgrG3D842A is expressed.
TseL and VgrG3 Directly Interact.
When screening for T6SS genes important for secretion of TseL, we found that an VgrG3 mutant was unable to secrete TseL (Fig. 6A). This was surprising because VgrG3 mutants kill E. coli, which indicates a functional T6SS organelle (13). We hypothesized that if TseL depends on VgrG3 for its secretion, then these two proteins might interact. To test if there is direct interaction between TseL and VgrG3, we performed an immunoprecipitation analysis using a monoclonal antibody to VgrG3 to pull down VgrG3-bound proteins and then used an antibody to TseL to detect its presence in the immune complex. This analysis demonstrated that TseL and VgrG3 directly interact (Fig. 6B).
Fig. 6.
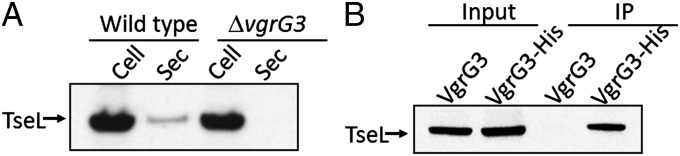
Direct interaction between TseL and VgrG3. (A) Secretion of TseL requires VgrG3. One milliliter of exponential phase culture was collected by centrifugation, and the resultant pellet (Cell) and supernatant fluids (Sec) were tested for TseL levels by Western blot analysis. (B) Immunoprecipitation of TseL and VgrG3. Cell lysates (Input) were incubated with anti-His antibody-conjugated beads for 2 h at 4 °C. Bound proteins (IP) were eluted with SDS/PAGE loading buffer and detected by Western blot analysis using an antibody against the 3 x V5 epitope tag fused to the C terminus of TseL.
TseL and VasX Are Required for Killing D. discoideum Amoebae.
Previous results have shown that mutations in tseL (13) or vasX (14) reduced the virulence of V. cholerae V52 against D. discoideum, a phenotype that infers both the ability of the T6SS to kill amoeba as well as resist killing themselves by amoebae predation (2, 5, 13, 14). To test if TseL and VasX function synergistically, we quantified the predation of D. discoideum against mutants lacking tseL and/or vasX. As expected, tseL and vasX individual mutants exhibited impaired amoebae virulence (Fig. 7A). However, the double mutant defective in both tseL and vasX was found to be completely avirulent and thus similar to a T6SS-null mutant. In addition, this loss of virulence is not due to changes in secretion of VgrG1, which is known to be required for virulence in D. discoideum by causing actin cross-linking (5). We found that VgrG1 was secreted in the tseL vasX double mutant (Fig. 7B). To test if the double mutant can still cause actin cross-linking, we infected macrophage J774 cells with the corresponding mutant and wild-type V. cholerae strains (5, 11). As expected, wild-type V. cholerae resulted in actin cross-linking in J774 cells that is dependent on VgrG1. The tseL vasX double mutant also caused actin cross-linking similar to wild-type levels (Fig. 7C). These results indicate that V. cholerae requires TseL and VasX to kill D. discoideum by a mechanism that depends on actin cross-linking but does not require these effectors to either secrete or deliver the actin cross-linking domain of VgrG1 to the cytosol of mammalian macrophages such as J774 cells.
Fig. 7.
Role of TseL and VasX in killing eukaryotic cells. (A) Determination of the minimal number of amoebae required to form a plaque on lawns of different strains of V. cholerae. (B) Western blot analysis of VgrG1 in the tseL and vasX double mutant. (C) Determination of the level of actin cross-linking in J774 cells exposed to various V. cholerae mutants. All strains are defective in RtxA toxin, HlyA hemolysin, and HapA hemagglutinin/protease to eliminate background toxicity and actin cross-linking that are not associated with the T6SS system of V52 (11).
Discussion
Time-lapse microscopy analysis has revealed that T6SS organelles in V. cholerae cells are very dynamic and likely secrete their T6SS spike/tube VgrG/Hcp complex in multiple directions over a period of minutes of observation (6). Such constitutive activity is likely to result in the attack of neighboring sister cells under conditions of growth on solid media or within biofilms. Therefore, it is not surprising that immunity proteins would evolve in T6SS+ species to resist the toxicity associated with the attack of sister cells.
In this study, we report three immunity genes and their corresponding effectors in V. cholerae. Inactivation of any of three immunity genes results in susceptibility to T6SS-dependent killing by T6SS+ strain V52. Deletion of any of the three cognate effectors in V52 abolishes killing of the mutant defective in the corresponding immunity protein. The identified effector–immunity proteins share no homology with other known T6SS effector–immunity pairs, indicating the diversity of T6SS toxic effector substrates and immunity proteins.
Our data demonstrate that Tn-seq is an effective approach to identifying T6SS immunity genes in V. cholerae and likely in other T6SS-possessing bacteria as well. It is worth noting that, as a screening tool, Tn-seq results require validation by other assays. In our study, we confirmed that three putative immunity genes potentiated sensitivity to a heterologous and homologous V. cholerae T6SS+ strain although the other five candidate genes did not confer sensitivity when inactivated. It is possible that these latter five genes are not directly involved in anti-effector activities but are important for mitigating damage done by the dynamic T6SS organelle within the predator cell. T6SS forms multiple long (∼0.5–1 µm) tubular structures in the cell that correspond to extended and contracted organelles (6). T6SS sheath contraction likely inserts half of the internal inner Hcp/VgrG tube/spike complex through the inner and outer membrane of the predator cell in less than 5 ms (6). This sudden traversal of the bacterial cell envelope may cause damage to the T6SS+ cell that requires specialized repair proteins. Thus, these five postulated T6SS repair proteins may be important for survival in T6SS+ cells but not in T6SS mutants or strains such as C6706 that do not express T6SS. We further propose that the damage controlled by these five proteins is distinctly different from the T6SS-mediated damage caused to prey cells by T6SS attack; the latter likely involves lesions caused by highly active enzymatic effectors delivered to the prey cell and the former may be transient but repeated mechanical perturbations to the envelope of the predator cell. This hypothesis will be tested through future work in our laboratory.
In these studies we also identified three effectors that can kill neighboring cells lacking their cognate immunity proteins. All three effectors lack an N-terminal secretion signal. Although VasX and VgrG3 were known to be T6SS secreted proteins (2, 14), their function as effectors directed at bacterial cells was not previously defined. Previously, the C-terminal domain of VgrG3 was predicted to share similarity to other peptidoglycan-binding domains prompting speculation that this VgrG might be an antibacterial effector (5). Here we show that expression of VgrG3 inside E. coli causes cell rounding and lysis. These results are consistent with the conclusion that VgrG3 is a lysozyme-like T6SS effector and, like other effectors described in P. aeruginosa, likely targets the cell wall of bacterial prey for enzymatic digestion (16).
It is interesting to note that VgrG3 causes cell lysis even when expressed in the cytosol of E. coli. Because VgrG3 lacks a canonical signal that would target its secretion to the periplasm or outer membrane, we hypothesize that its transport to the periplasm may depend on a property yet to be defined. For example, overexpression of VgrG3 may result in formation of VgrG3 trimers that spontaneously insert themselves into the plasma membrane and thus deliver its lysozyme-like domain to the periplasm. E. coli is also known to constitutively secrete outer membrane vesicles (OMV) that contain proteins from the outer membrane, periplasm, and the cytosol (32). It is possible that overexpressed VgrG3 gets incorporated into secreted OMVs, which later fuse with the outer membrane of sister cells releasing VgrG3 to the periplasm. Finally, VgrG3 translocation may be similar to some phage-related muramidases that have no secretion signal sequences and rely on accessory proteins such as phage holins to gain access to the periplasm (33). Regardless of the mechanism of its transit to the periplasm, the fact that VgrG3 is toxic when expressed in the cytosol of E. coli suggests that delivery of this effector into the cytosol of prey cells by T6SS may still result in the lysis of prey.
The in vivo bacterial targets of TseL and VasX were not determined in this study. TseL possesses a lipase domain, which suggests it may target membrane-associated lipids; and indeed inactivating a conserved residue located in this putative lipase's activity site abolished its toxic activity as a T6SS effector. VasX has been shown to interact with phospholipids in vitro (14). In addition to their antibacterial activities, TseL and VasX also play a role in antieukaryotic predation. Deletion of both tseL and vasX completely abolishes the virulence of V. cholerae toward D. discoideum amoebae, without affecting T6SS-dependent delivery of VgrG1 into these cells as evidenced by VgrG1-dependent actin cross-linking in these cells. This result suggests that killing of D. discoideum amoebae by V. cholerae results from effector functions of TseL and VasX in addition to actin cross-linking. Because amoebae killing also requires the VgrG1 actin cross-linking domain (5, 11), we propose that the cytotoxic effects of TseL and VasX likely occur at a step that follows disruption of the actin cytoskeleton by VgrG1. These effectors could block repair of VgrG1-mediated damage to the cytoskeleton or potentiate other lethal cell biological events (e.g., lysis) that do not occur efficiently as a result of actin cross-linking alone.
Although many of the key components T6SS proteins have been determined by genetic, biochemical, and structural analyses (2, 5, 6, 34, 35), it remains unclear how exactly the T6SS structure is assembled and effectors are recognized for secretion. Derived from structural information on contractile phage tails and imaging analysis (4, 6), the current T6SS model postulates that Hcp polymerizes and forms an inner tube that is wrapped with an outer tube consisting of T6SS sheath proteins VipA and VipB (6). Sheath contraction pushes the inner VgrG/Hcp spike tube complex out of the cell and into target cells. VgrG-based effectors can be translocated into target cells by this mechanism alone (6, 11). However, other effectors such as the Tse proteins of P. aeruginosa (15, 16) are not VgrG orthologs and thus must be recognized and secreted by another mechanism. It has been proposed that effectors may be delivered through the inner channel of the Hcp tube (∼4 nm in diameter), which might allow transport of folded proteins less than 50 kDa (3, 36). Consistent with this, known T6SS effectors in P. aeruginosa and S. marcescens all have molecular weight less than 50 kDa (Table S2). However, V. cholerae effectors are much larger (>70 kDa). If these proteins were transported through the Hcp tube they would need to be folded into an extended axial structure (e.g., alpha helix) to fit in the lumen of the Hcp tube. In this study we provide evidence for another mechanism of transport that does not require transit through the Hcp tube. We show that TseL and VgrG3 directly interact and that TseL secretion depends on VgrG3. These results provide strong evidence that TseL is likely secreted in a complex that includes VgrG3. We speculate that other T6SS effectors may also bind to cognate VgrG orthologs or specific heterotrimers of these proteins. Evidence has been previously reported that heterotrimers of VgrG proteins can be secreted by V. cholerae (5).
Structural analysis of VgrG and Hcp proteins suggests that these proteins will form a trimer at the tip of the Hcp tube (4, 5). However, it is unknown whether these trimers are homotrimers or heterotrimers within the functional organelle because three distinct VgrG proteins are expressed and secreted by V. cholerae (5). It is also unclear if these trimers are in a complex with other proteins in the T6SS base plate complex just before secretion driven by T6SS sheath contraction (6). The interaction between VgrG3 and TseL becomes more interesting given that deletion of both vasX and vgrG3 results in loss of Hcp secretion, a hallmark for T6SS function. These results suggest that these two effectors likely form a complex that is essential for proper assembly and function of a T6SS organelle. Future research will focus on identifying which step in the T6SS organelle biogenesis requires the presence of these two effector proteins.
Given its antibacterial and antieukaryotic activities, T6SS likely plays an important ecological role in survival and competition with other species in the natural environment. T6SS-associated antibacterial effector–immunity proteins are predicted to be widespread in gram-negative bacteria (18). However, the three V. cholerae effector–immunity pairs we identified in this study differ greatly from previously predicted families of effector/immunity proteins (18). The Tn-seq approach we describe here will provide an effective tool for experimentally screening for immunity proteins and their cognate effectors in other T6SS+ species that display antibacterial activity.
Materials and Methods
Strains and plasmids are listed in Table S3. Mutants and expression vectors were constructed as previously described (37–40). The mariner-transposon–based vector pSAMDGm used for the transposon library construction is a gift from Stephen Lory (Harvard Medical School, Boston). To compare transposon insertions per gene, we use a normalized RPKM value (41), which represents the relative abundance of transposon insertions in each gene in the transposon library. Assays for T6SS-dependent killing (12), amoebae survival (2, 13), and actin cross-linking (5), were performed as previously described. Details for data analysis and methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Marek Basler, Bryan Davies, Stephen Lory, and Deepa Patel for technical assistance and the J.J.M. group for helpful discussion. We also thank Bryan Davies for critical reading of this manuscript. This work was supported by National Institute of Allergy and Infectious Diseases Grant AI-01845 (to J.J.M.). T.D. was supported by a Banting Postdoctoral Fellowship awarded by the Canadian Institutes of Health Research (CIHR).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222783110/-/DCSupplemental.
References
- 1.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: A beginner’s guide. Curr Opin Microbiol. 2008;11(1):3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Pukatzki S, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103(5):1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312(5779):1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leiman PG, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106(11):4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. 2007;104(39):15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483(7388):182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basler M, Mekalanos JJ. Type 6 secretion dynamics within and between bacterial cells. Science. 2012;337(6096):815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aschtgen MS, Bernard CS, De Bentzmann S, Lloubès R, Cascales E. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol. 2008;190(22):7523–7531. doi: 10.1128/JB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aschtgen MS, Thomas MS, Cascales E. Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP... what else? Virulence. 2010;1(6):535–540. doi: 10.4161/viru.1.6.13732. [DOI] [PubMed] [Google Scholar]
- 10.Cascales E, Cambillau C. Structural biology of type VI secretion systems. Philos Trans R Soc, B. 2012;367(1592):1102–1111. doi: 10.1098/rstb.2011.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 2009;5(3):234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci USA. 2010;107(45):19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS ONE. 2011;6(8):e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyata ST, Kitaoka M, Brooks TM, McAuley SB, Pukatzki S. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun. 2011;79(7):2941–2949. doi: 10.1128/IAI.01266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hood RD, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7(1):25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell AB, et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475(7356):343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basler M, Ho B, Mekalanos JJ. Tit-for-tat: Type VI secretion system counterattack during bacterial cell-cell interactions. Cell. 2013 doi: 10.1016/j.cell.2013.01.042. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell AB, et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe. 2012;11(5):538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.English G, et al. New secreted toxins and immunity proteins encoded within the Type VI secretion system gene cluster of Serratia marcescens. Mol Microbiol. 2012;86(4):921–936. doi: 10.1111/mmi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6(3):279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman AL, Wu M, Gordon JI. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat Protoc. 2011;6(12):1969–1980. doi: 10.1038/nprot.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106(38):16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallagher LA, Shendure J, Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2011;2(1):e00315–e10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Opijnen T, Bodi KL, Camilli A. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6(10):767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA. 2009;106(11):4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron DE, Urbach JM, Mekalanos JJ. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci USA. 2008;105(25):8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 2010;107(49):21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 29.Saito J, et al. Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-A resolution and its substrate recognition mechanism. J Biol Chem. 1999;274(43):30818–30825. doi: 10.1074/jbc.274.43.30818. [DOI] [PubMed] [Google Scholar]
- 30.Boucher I, et al. Site-directed mutagenesis of evolutionary conserved carboxylic amino acids in the chitosanase from Streptomyces sp. N174 reveals two residues essential for catalysis. J Biol Chem. 1995;270(52):31077–31082. doi: 10.1074/jbc.270.52.31077. [DOI] [PubMed] [Google Scholar]
- 31.Monzingo AF, Marcotte EM, Hart PJ, Robertus JD. Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat Struct Biol. 1996;3(2):133–140. doi: 10.1038/nsb0296-133. [DOI] [PubMed] [Google Scholar]
- 32.Lee EY, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7(17):3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- 33.Young R. Bacteriophage holins: Deadly diversity. J Mol Microbiol Biotechnol. 2002;4(1):21–36. [PubMed] [Google Scholar]
- 34.Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28(4):315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J, Leung KY. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol. 2007;66(5):1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- 36. Silverman JM, Brunet YR, Cascales E, Mougous JD (2012) Structure and regulation of the type VI secretion system. Annu Rev Microbiol 66:453-472. [DOI] [PMC free article] [PubMed]
- 37.Metcalf WW, et al. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35(1):1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 38.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies BW, Bogard RW, Mekalanos JJ. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc Natl Acad Sci USA. 2011;108(30):12467–12472. doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong TG, Mekalanos JJ. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 2012;40(16):7766–7775. doi: 10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.