Abstract
Mechanical loading induces both nuclear distortion and alterations in gene expression in a variety of cell types. Mechanotransduction is the process by which extracellular mechanical forces can activate a number of well-studied cytoplasmic signaling cascades. Inevitably such signals are transduced to the nucleus and induce transcription factor-mediated changes in gene expression. However, gene expression can be also regulated through alterations in nuclear architecture, providing direct control of genome function. One putative transduction mechanism for this phenomenon involves alterations in nuclear architecture that result from the mechanical perturbation of the cell. This perturbation is associated with direct mechanical strain or osmotic stress, which is transferred to the nucleus. This review describes the current state of knowledge relating the nuclear architecture and the transfer of mechanical forces to the nucleus mediated by the cytoskeleton, the nucleoskeleton, and the LINC (linker of the nucleoskeleton and cytoskeleton) complex. Moreover, remodeling of the nucleus induces alterations in nuclear stiffness, which may be associated with cell differentiation. These phenomena are discussed in relation to the potential influence of nuclear architecture-mediated mechanoregulation of transcription and cell fate.
Keywords: nucleus, mechanotransduction, chromatin organization, LINC complex, osmotic stress
1. GENERAL INTRODUCTION AND HISTORY
In biological systems, mechanotransduction is defined as the process by which mechanical stimuli are converted to intracellular signals that control cellular physiology in both health and disease. A classic example is shear stress acting on endothelial cells lining the wall of a blood vessel. The force from the blood flowing past an endothelial cell evokes changes in cell morphology; cells reorient their cytoskeletons in the direction of flow and initiate signaling to neighboring cells, resulting in changes to local vessel elasticity (1, 2). In effect, the external stimuli are sensed by the cells, and those signals are transduced across the membrane, where they alter intracellular signaling pathways and result in a cellular response. Similarly, many other cells rely on mechanotransduction for maintenance of their phenotype and metabolic activities. This process is particularly relevant to cells of the musculoskeletal, cardiovascular, and pulmonary systems, which are exposed to significant physical forces under normal conditions (3). Importantly, mechanical factors may be involved in numerous pathological conditions including diseases such as atherosclerosis, osteoarthritis, osteoporosis, and many others (reviewed in 4). Recent years have seen growing evidence of the critical role of mechanical and other physical signals in controlling the fate of stem or progenitor cells (5-7). The mechanisms involved in mechanotransduction are complex and may depend on several cellular pathways involving the cell membrane, ion channels, cytoskeleton, and cell nucleus (reviewed in 8). This review focuses on how the nucleus reacts to extracellular mechanical forces, with specific attention to how these forces can influence nuclear architecture, gene regulation and cell fate.
Conceptually, the idea that development and differentiation are shaped by extrinsic (mechanical) forces is ancient. Aristotle thought that embryonic development was in part directed by the locomotive component of the soul. This so-called Aristotelian soul differed from the more traditional or spiritual concept of a soul in that it is generated through the (physical) mixing of male and female semen (9), producing a teleological driving force that gradually gestates that individual organism to its final form (10). Teleological reasoning such as this persisted for millennia. However, over time, the notion of the Aristotelian soul as a driving force was either replaced by a divine, perfecting force (11) or obviated altogether by the notion that organisms were preformed and grew with no need for ontological development or differentiation (10). In the mid-1800s, Darwin published his theory describing evolution as a slow and gradual process of descent with modification, resulting in stochastic variability capable of conferring selectable (morphological) advantages to promote survival or increased reproductive fitness of an individual. He included many observations of embryos, as he saw them as “picture[s], more or less obscured, of the common parent-form of each great class of animals” (12, p. 450). Along with this, and more central to this work, he was also among the first to promote a movement away from teleological thinking, suggesting that evolution (and embryology) need not be a process seeking to perfect a morphology, per se (11). This point was later elaborated on by Wilhelm Roux and Julius Wolff (13), in Germany, toward the end of the century.
Wolff was an orthopedic surgeon who conducted research into how the structure of bones changed with alteration of function owing to either growth or pathology. His ideas followed from those of Roux; namely, he maintained that life has two periods, an embryonic one characterized by trophic organ growth and differentiation, and an adult one in which growth and remodeling occurs only when stimulated by healing or (cellular) turnover (13). These stimuli, which Roux collectively referred to as developmental mechanics (11), had an overall effect on tissues irrespective of life period, resulting in their remodeling. Wolff postulated that there was a causal relationship between the physical forces acting on bones and the observed changes to both gross morphology and internal architecture. In particular, he predicted that remodeling of bone trabeculae followed the mathematical trajectories of the forces acting on them. He also went a step further, suggesting that these same mechanical signals could provide one plausible mechanism for Darwin’s theory of natural selection (13). Since the publication of Wolff’s work, the accuracy of some of his mathematical predictions have been called into question (reviewed in 14). Another common criticism is that his theory cannot be considered a law because other bones and tissues do not exhibit the universality sufficient to explain the phenomena he described in cancellous bone. Despite these detractions, his observations were made well in advance of the discovery of radiography and modern tissue biology techniques. Wolff’s work is thus deservedly recognized for its pioneering contributions to the burgeoning fields of orthopedics and tissue mechanobiology.
Although an understanding of the complete sequence of biophysical and molecular events involved in mechanotransduction is still incomplete, numerous discoveries in the past decade have added to the evidence for the role of the cell nucleus in this process. Molecular complexes in the nuclear envelope were found to mediate a connection between the cytoskeleton (the cell’s proteinaceous structural support network) and the lamin network (a structural component of the nucleoskeleton), the latter of which is located on the interior of the nuclear membrane. The LINC (linker of the nucleoskeleton and cytoskeleton) complex is of particular interest because lamins and their associated proteins play a role in genomic silencing-opening a hotline of sorts for mechanical communication directly to the genome, potentially literally shaping gene expression. This review examines the current state of nuclear and chromatin biology with a particular focus on how the nucleus responds to mechanical stimuli in terms of both homeostasis and how those stimuli affect gene expression and matters as fundamental as cell fate.
Throughout the 1800s, several prominent scientists theorized about nuclear function, but it was Theodor Boveri’s work near the end of the century that experimentally confirmed the nucleus, and later the chromosomes, as the instruments of heredity in the cell (reviewed in 15). Boveri, who was closely assisted by his wife Marcella, is credited with several major shifts in biological thinking related to cancer and developmental biology. They were the first to report that cancer was clonal in nature, arising from a cell whose chromatin had been damaged (16). More relevant to this review is Boveri’s chromosome theory, which was supported by experimental evidence in parasitic nematodes and sea urchins. The theory, in addition to definitively identifying the nucleus (and more specifically the chromosomes) as the material of heredity, demonstrated that genetic contribution is equal between parents and that all, not just a subset, of the chromosomes are transmitted from germ cells to somatic cells during normal embryonic development (17). This finding was especially important as it overturned the prevailing wisdom that developmental cell fate was specified by an incomplete sharing of the chromosomes among germ cells and the various types of somatic cells. In other words, it was thought that muscle cells received only the “muscle chromosomes,” whereas brain cells inherited only the chromosomes relevant for brain function. Boveri also concluded that normal embryonic development involved a cooperation between the nuclear material and the cytoplasm. Introduced in the early twentieth century, these concepts are highly studied today and pivotal to any stem cell-based regenerative therapy. The importance of cooperation between regionalized factors [usually ribonucleic acids (RNAs) and proteins] in the cytoplasm and lineage-specific gene expression (nuclear factors) in specifying cell fate varies widely among species. The molecular and genomic revolutions of the late twentieth century brought genetic factors and gene-regulatory networks into prominence, especially where mammalian development was concerned.
2. NUCLEAR STRUCTURE AND FUNCTION
2.1. General Structure and Subnuclear Bodies
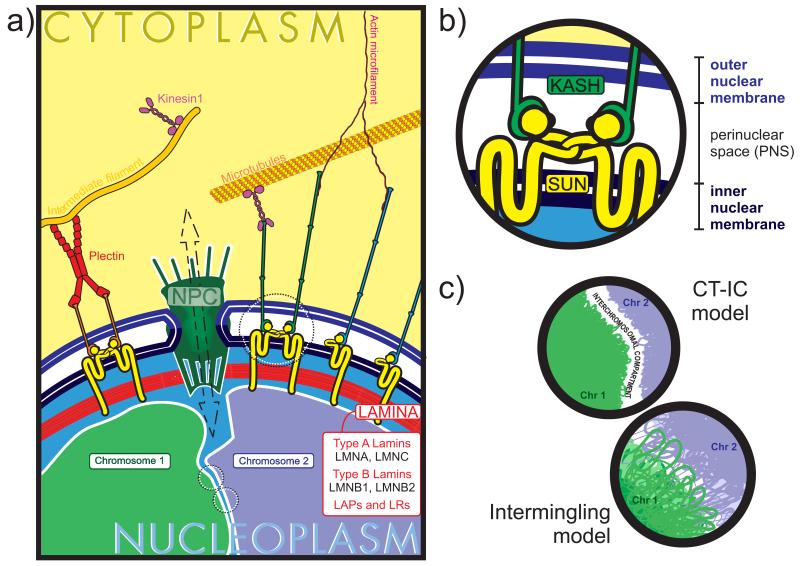
The nucleus houses the nuclear genome and provides a functional environment for the information that will maintain and reproduce that cell, and potentially the whole organism. Eukaryotes, whose word origin is Greek for true kernel or nucleus, are defined by a nucleus that is bound by a membranous envelope. This multilayer, selectively permeable covering is composed of the outer nuclear membrane (ONM) and the inner nuclear membrane (INM) that surround the perinuclear space and of the nuclear lamina on the nucleoplasmic side (Figure 1a,b). Nuclear envelope transmembrane (NET) proteins traverse either one or both of the nuclear membranes and represent a functionally diverse group of nuclear factors. Among these are the nucleoporins, which make up the nuclear pore complexes that bridge the cytoplasmic and nuclear compartments and are embedded ubiquitously throughout the nuclear envelope. These enable bidirectional, passive movement for small molecules and facilitate transport of larger (>40-kDa) ones (reviewed in 18). The innermost compartment of the nucleus is occupied by the nucleoplasm, the (nuclear) genome, and several types of subnuclear bodies. The nucleolus is the largest of these, and nuclei sometimes contain more than one. It is made up of a number of proteins and ribonucleic protein complexes that are involved in ribosomal RNA synthesis and processing. An additional level of the nucleolus’s organization differentiates it from other nuclear bodies; it is surrounded by specific regions of the genome termed nucleolar-associated deoxyribonucleic acid (DNA), which comprise approximately 4% of the genome (19). The other subnuclear bodies are mostly ribonucleic protein complexes that are active in RNA synthesis and processing, such as promyelocytic leukemia bodies, splicing speckles, and Cajal bodies. As none of these subnuclear bodies are membrane bound, it is thought that their structure (and therefore placement) is transient and determined by the electrostatic intermolecular forces that bind the protein and nucleic acid components of these complexes together (20).
Figure 1.
The archetypal SUN-KASH domain association in the LINC complex. (a) The LINC complex is typified by an association between SUN-domain proteins (yellow) on the inner nuclear membrane and KASH-domain proteins (green, blue, and orange) on the outer nuclear membrane. KASH-domain proteins form a functional link with the networks of cytoplasmic intermediate filaments, microtubules, and actin microfilaments, which compose the cytoskeleton. SUN-domain proteins bind to the nuclear lamina, a network of intermediate filaments composed of varying isoforms of A- and B-type lamins, LAPs, and LRs. The lamina also serves as a tethering point for the genome, with associations reported among various lamins, LAPs, and LRs. (b) KASH domains associate with SUN domains in the perinuclear space, and this association maintains the architecture of the nuclear envelope. SUN proteins can associate promiscuously with KASH proteins and can also form homo- and heterodimers with other SUN proteins. (c) Two competing models explain the 3D organization of CTs in the interphase nucleus. The CT-IC model posits that a largely DNA-free compartment of contiguous spaces between adjacent CTs exists. The second model, known as the intermingling model, maintains that whereas CTs occupy nonrandom spaces in the interphase nucleus, there is a large amount of intermingling of the chromatin between adjacent CTs. Abbreviations: CT, chromosome territory; IC, interchromsomal compartment; KASH, Klarsicht/ANC-1/Syne-1 homology; LAP, lamin-associated protein; LINC, linker of the nucleoskeleton and cytoskeleton; LR, lamin receptor; NPC, Nuclear Pore Complex; SUN, Sad1 and UNC84.
2.2. Chromatin
The DNA in eukaryotic nuclei is packaged as chromatin, a complex of DNA and proteins. An organism’s genome contains the total collection of genes and regulatory information, coded as a linear sequence of nucleic acid moieties, needed to maintain and reproduce that organism. Contained within most human nuclei are two copies of the genome, one descendent from each parent. Each copy, which has approximately 3.1 billion nucleotide base pairs (bp) of DNA, containing >22,200 protein-coding genes and >9,900 ribosomal RNA genes, which code for >142,700 different gene products (21). Although the information is encoded linearly, the overall genome is not. Some chromosomes are gene rich, and others are gene poor. Some genes exist in clusters and are functionally coregulated because of their clustering (e.g., Hox genes, globins, protamines), whereas others exist bordered by what are known as gene deserts. With each bp estimated at 330 pm in size (22), the length of each copy of the human genome would be approximately 1 m, if all chromosomes were stretched out and aligned end to end. The diameter of most human nuclei is in the micrometer (to tens-of-micrometers) range. Chromatin proteins serve to compact and provide order to what would be otherwise a chaotic system.
Chromatin is organized into nucleosomes, each consisting of approximately 145 bp of DNA supercoiled around a core complex of four dimeric histone proteins: H2A, H2B, H3, and H4. Approximately 20-80 bp of linker DNA exist between each adjacent nucleosome, and this region is bound by histone H1 (Figure 1b). Although the core’s structure has been mapped to near-atomic resolution (23), there is great debate about how H1 interacts with the nucleosome and whether this interaction is essential for the maintenance of higher chromatin order (24, 25). In low-ionic-strength solutions, a chromatin fiber has been measured to have a thickness of approximately 10 nm that is thought to fold back on itself to form higher-order, compacted chromatin. The interphase nucleus is likely a mixture of several types of folding that could account for an approximate 30-nm-diameter structure, and the manner of folding of a particular region is highly dependent on the internucleosomal linker length and the presence of linker histone H1 (24).
Much of the difficulty in modeling the structure of chromatin in the interphase nucleus comes from the dynamic nature of chromatin and the fact that many of the existing static models rely on experiments conducted in solution, which may not reflect the exact physiological environment. Moreover, the effect of factors such as ion concentration and molecular crowding on nuclear structure is not fully understood (26). Even with this as an open question, investigators have identified hundreds of chromatin and chromatin-associated proteins that either bind to DNA directly or help stabilize chromatin in higher-order, more compacted forms. This hierarchical packing allows the genome to be contained within the available space. However, regions of chromatin must be unpacked before they can be accessed for transcription, and it is now becoming clear that exogenous mechanical factors may influence this process.
Chromatin generally exists in one of two forms: lightly compacted euchromatin or more highly compacted heterochromatin. The former conformation is generally thought to be permissive to transcription, whereas the higher levels of compaction achieved in heterochromatin inhibit transcription. These two forms of chromatin represent two distinct, although dynamic, compartments within the interphase nucleus. Apart from clear differences in levels of compaction, each of these states has fundamental molecular signatures that distinguish one from the other; these signatures involve covalent modifications to DNA and several amino acid residues from the termini of histone proteins from the nucleosome core particle (27). Most organisms, including humans, can methylate cytosine residues in DNA directly in a modification associated with transcriptional silencing in heterochromatin, gene promoters, and repetitive DNA sequences (28). Lysine (K) residues in histone tails, for example, are subject to numerous posttranslational covalent modifications such as methylation, acetylation, phosphorylation, ubiquitination, and SUMOylation. Initially, a so-called histone code was theorized to account for the general trends exhibited by genomic regions baring these marks (29). On one hand, hyperacetylation of K residues in histones H3 and H4 are commonly associated with actively transcribed chromatin, whereas methylation of H3K9 and H4K20 tend to be seen in pericentromeric heterochromatin, which is transcriptionally silent. On the other hand, modifications such as lysine acetylation could influence chromatin topography directly by steric inhibition between neighboring nucleosomes. Additionally, these modifications create new binding epitopes that are recognized by effector proteins, potentially resulting in subsequent activation or silencing. Thus, a modification such as methylated H3K4, which tends to localize to the start of genes, was generally thought to promote transcriptional activation by recruiting a chromatin remodeling complex containing a plant homology domain, a peptide motif that recognizes and binds to the trimethylated H3K4 residue (30). However, plant homology domains are not specific to activating complexes (31). Moreover, if methylated H3K4 marks accumulate at sites other than the starting position of genes (32), this accumulation can signal transcriptional silencing of that locus (33). Thus, the overall impact of any modification requires contextual information including genomic location, cell-cycle stage, the presence of specific histone variants, the expression status of transcriptional coactivators and transcription factors, the net effect of other repressive marks such as DNA methylation, and the expression status of effector proteins (reviewed in 34).
Apart from the canonical histones, a number of histone variant proteins are involved in cellular processes such as chromosome segregation during mitosis, DNA repair, chromatin remodeling, and transcriptional regulation. In many cases, these variants are assembled into the histone core at the time of DNA replication as is H2A.Z in gene promoters and transcription start sites, and thus serve as a biochemical signpost for important genomic features (35). Other histone variants, such as CenH3, are inserted into larger spans of DNA and are localized to highly repetitive centromeric sequences. This histone protein is an essential component of the kinetochore, part of the molecular machinery that segregates chromosomes to daughter cells at the end of cell division (36). The incorporation of an H3 variant, H3.3, into the nucleosome cores was recently reported to be essential to the establishment of heterochromatin in paternal pericentromeric regions of mouse embryos (33). In some cases, histone variants are specific to cell lineages, such as the testes variant H3t, which is important in the histone-to-protamine exchange that takes place in the sperm chromatin of many animals (37). Although the amino acid sequence of the variants may not differ significantly from the canonical protein (33, 35), those small differences often localize to key positions in the nucleosome core particle such that variant-bearing nucleosomes package DNA in a different manner, thereby changing the topology of the chromatin fiber. In some cases, this change stabilizes the chromatin fiber and provides a basic building block for a higher-order structure (e.g., H3.3 and heterochromatin) (33, 35); in other cases, this change produces a less stable structure, like promoting more efficient exchange of the nucleosome structure to the protamine toroids found in sperm nuclei, for example (37).
2.3. Chromosomes and Chromosome Territories
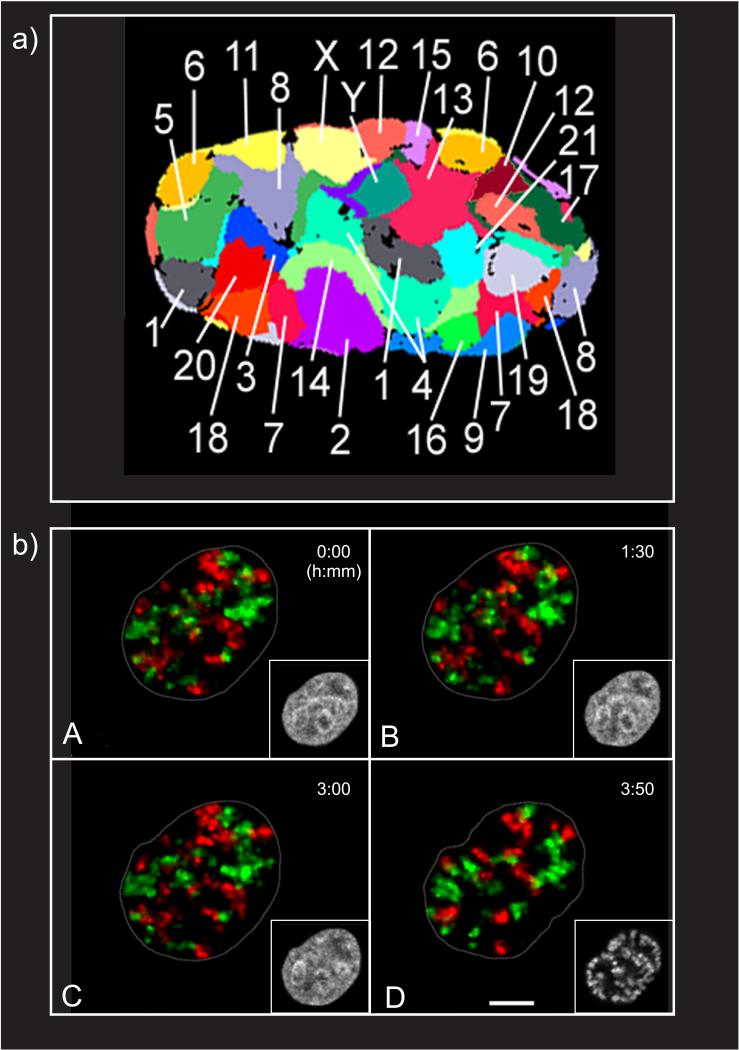
Beyond the transcriptional states of chromatin in the nucleus, there are other degrees of order. Chromatin is organized into chromosomes (whose name derived from the Greek word for colored body, because of the way that chromosomes stained in early cell preparations). Chromosomes condense during metaphase in mitosis, making them visible as discrete bodies by light microscopy. In this form, certain regional domains within a chromosome become obvious-for example, the telomeres and the centromeres. Each of these regions tends to be gene poor and full of repetitive DNA sequences. Centromeric chromatin contains an assortment of centrosome-specific proteins including CenH3, which are essential for chromosome segregation during cell division. Telomeres protect the ends of chromosomes from the shortening that occurs with every round of cell division, ensuring that no critical genes are lost (38). During interphase, the chromosomes decondense (compared with those in metaphase), but telomeres and centromeres maintain a heterochromatic conformation. Overall, however, chromatin maintains a specific nonrandom order within the nucleus, referred to as a chromosome territory (CT) (Figures 1a,c and 2). These CTs were initially described in animal nuclei in the late nineteenth century by Carl Rabl, but it was Boveri who later coined the term in his theory of chromosome individuality (39, 40). It was not until the 1980s that Thomas Cremer’s group provided definitive proof for the nonrandom arrangement of chromosomes in the interphase nucleus, initially using ultraviolet microirradiation (41) and then in situ hybridization (42) techniques.
Figure 2.
Chromosomes occupy nonrandom placement in the cell nucleus. (a) Flattened 3D fluorescent in-situ hybridization representation of all 24 chromosome territories (CTs) in a human G0 fibroblast nucleus (166). (b) Stability of CT neighborhood during interphase of living HeLa cells. HeLa cells were replication-labeled during S-phase of two consecutive cell cycles (first cycle, Cy3-dTUP, red; second cycle, Cy5-dUTP, green). (A) Cells were allowed to complete another two cycles before observation was started. (D) After 3 h, 50 min, the cell entered prophase. Frames of maximum intensity projections from light optical serial sections are displayed for the indicated time point. Insets show H2B-GFP signals representing the chromatin (gray) in confocal nuclear midsections. Images are corrected for translational and rotational nuclear movements. Prophase chromosome condensation is a locally confined process with little change to CT arrangements (compare late G2 in subpanel C with early prophase in subpanel D). Scale bar: 5 μm (167). Abbreviations: Cy, cyanine; dUTP, 2′-deoxyuridine 5′-triphosphate; GFP, green fluorescent protein. Adapted from References 166 and 167 with permission.
Each territory is composed of numerous megabase domains, the architectural units that constitute chromosomes in the mammalian interphase nucleus. Each of these megabase domains has a length of approximately 0.5-1 megabase pairs (Mbp) of DNA (40). Initially, this number was attributed to foci of replicating DNA visualized in the nucleus (43), but recent studies of genomic organization using high-resolution sequencing techniques corroborate this value (19, 44). The radial arrangement of specific CTs in the nucleus is also not random: Gene-dense chromosomes localize to the interior of the nucleus, whereas gene-poor chromosomes are located more peripherally (45-47). This arrangement has been evolutionarily conserved among vertebrates, but other corollaries, such as transcriptional activity and replication timing of certain subregions of chromosomes, have also been reported (reviewed in 40). In addition to radial arrangements in the nucleus, chromosome neighborhoods, which are specific arrangements between a subset of chromosomes, have been described with some tissue or cell-type specificity (48). The majority of these studies used fixed preparations that provided only snapshots in time and space, and, as such, they have not shed any light on how these arrangements are maintained or what mechanism is responsible for their positioning.
Another hotly debated matter with respect to CTs and nuclear architecture centers on the remaining space, or lack thereof, in the nucleus. One model posits that along with the chromatin in CTs that comprise a highly ordered interconnected network, there is an additional interchromosomal compartment (IC), which is largely DNA/chromatin free and contiguous with the nuclear pore complexes (49) (Figure 1c). This IC is the location where splicing speckles, transcription factories, and other subnuclear bodies are found. Transcription factories, as the name implies, are sites that feature several active clusters of RNA polymerase together with all the necessary cofactors that comprise the preassembled transcriptional machinery(50). This notion became quite popular with several reports of gene-dense regions that form discrete chromatin loops outside the periphery of their respective CTs. The human major histocompatibility complex (51) and the mouse HoxB cluster (52) are two such examples. Upon activation-by interferon stimulation in the former case and by developmental timing of cluster expression in the latter case-a decondensation event was triggered at these loci, resulting in their constituent genes being pushed out into the IC to be transcribed. Investigators have also reported other looping phenomena, named chromosome kissing events, that involve the coregulation of disparate genomic loci, sometimes on different chromosomes (53). The competing model suggests a more dynamic picture, in which interactions between neighboring CTs are far more frequent than the rare looping events at a handful of loci. As such, any one snapshot in space and time shows a more intermingled state between adjacent CTs (54).
2.4. The Nuclear Lamina, the LINC Complex, and Their Associations to the Genome
Just interior to the INM, the lamina is a structural layer of intermediate filaments composed largely of lamin protein products from three evolutionarily conserved genes, LMNA/C, LMNB1, and LMNB2, and the proteins that associate to them, the lamin-associated proteins and lamin receptors (Figure 1a). The mechanical and functional properties of the lamina can vary greatly among cell types depending on the relative ratios of lamin splice variants that constitute the intermediate filaments (55). Sometimes referred to as the nuclear scaffold, lamins and their associated proteins can interact with the chromatin (directly at the DNA level or through a chromatin or chromatin-associated protein) by directly binding nuclear scaffold or matrix attachment regions. These regions usually flank the coding sequences of genes and insulate them from the effects of neighboring genomic regulatory elements (56, 57).
Embedded within the nuclear envelope are a series of proteins that facilitate force transfer throughout the cell (58). The LINC complex describes several classes of molecules that are localized to the INM and ONM and provides a functional link between the support structures of the cytoplasmic and nucleoplasmic compartments (Figure 1a,b and Table 1). They were discovered through a series of genetic screens in Caenorhabditis elegans and Drosophila, in which mutants displayed nuclear displacement phenotypes (59). Klarsicht/ANC-1/Syne-1 homology domain proteins (KASH-domain proteins) form the link between the ONM and elements of the cytoskeleton. Nesprins 1 and 2 bind actin filaments as well as several of the motor proteins of the microtubule network (reviewed in 59). Nesprin 3 binds plectin, which connects it to networks of intermediate filaments (60). On the INM side, Sad1 and UNC84 domain proteins (SUN-domain proteins) Sun1, Sun2, and Sun3 bind the lamina in the nucleoplasm (reviewed in 59 and 61). Other Sun proteins, including Sun1η, Sun3, and sperm associated antigen 4 (Spag4) are expressed only in the spermatogentic lineage, and have been reported to be important in the association between the genome and nucleoskeletal manchette microtubes (163, 164). In the perinuclear space, the Nesprins’ KASH domains interacts with the SUN domains, establishing the LINC and maintaining the perinuclear space. Emerin protein (EMD) is a transmembrane protein that localizes to the ONM, where it can form an association with the microtubules and the microtubule organizing center. When localized to the INM, EMD binds the lamina. To date, however, communication between INM- and ONM-bound EMD has not been reported (62). Mutations in lamins and other lamina components have been implicated in a class of pathologies termed laminopathies, such as Hutchinson-Gilford progeria syndrome (63) and Emery-Dreifuss muscular dystrophy (64). Cells in these disorders display a classic nuclear dysmorphology, altered nuclear and cellular stiffness (65), and disorganization of heterochromatin domains in the nucleus (66). As the LINC complex plays an important role in nuclear placement and anchoring and as lamins are important not only for bulk chromatin organization in the nucleus but also for transcriptional regulation, it is a key to how mechanical signals traverse compartments in the cell.
Table 1.
Molecular associations between Sun and Nesprin proteins of the LINC complex
| Nucleoplasmic partners/function |
SUN domain protein |
KASH domain protein |
Cytoplasmic partners/function |
|---|---|---|---|
| Anchorage between lamina and INM Nuclear positioning (158, 159) Maintenance of perinuclear space Anchorage of telomeres (160, 161) |
Sun1 and Sun2 |
Nesprin 1 and nesprin 2 (multiple variants including GIANT isoforms) |
Anchorage between ONM and actin filaments in cytoskeleton Nesprin 2 links nucleus to MTOC and kinesin-1 (162) |
| Nesprin 3 | Anchorage to intermediate filaments via plectin adaptor protein (60) |
||
| Postmeiotic, sperm-specific isoforms (163) |
Sun1rη and Sun3 | Nesprin 1 and nesprin 3 |
Manchette microtubules |
| Sperm-specific isoform (164) | Spag 4 and Spag 4L/2 | Unknown | Unknown |
| Nuclear positioning | Unknown | Nesprin 4 | Connects to kinesin-1 (165) |
Abbreviations: INM, inner nuclear membrane; KASH, Klarsicht/ANC-1/Syne-1 homology; LINC, linker of the nucleoskeleton and cytoskeleton; MTOC, microtubule organizing center; ONM, outer nuclear membrane; Sun, Sad1 and UNC84; Spag4, sperm associated antigen
Domains of highly compacted chromatin have been described at the nuclear periphery and bordering the nucleolus. In higher eukaryotes, associations with the nuclear lamina have been implicated as part of a long-term silencing mechanism for large regions of the genome (44, 67). All nuclear lamins contain domains in their C termini that bind chromatin with a high efficiency through direct association with histone proteins (68) or through interactions with B-type lamin receptor and heterochromatin proteins HP1α and HP1γ (69). However, repression is thought to be cooperative between the C terminus and the rod domain of LNMA only, which targets the promoter regions of genes (70). Through development, as cells differentiate from a more pluripotent (stem cell– or progenitor cell-like) state to a more specified state, the transcriptional demands on the genome change. Genes critical for the maintenance of pluripotency become silenced, and lineage-specific markers become active. This phenomenon has been recently shown to be at least partially mediated through interactions with the nuclear lamina. In mouse embryos, cells that will form placenta were observed with large domains of higher-order chromatin forming adjacent to the nuclear lamina (71), whereas those that retain a pluripotent state-which is necessary to specify all the cell types making up the embryo-maintain their genomes in a state of lower compaction. A dynamic situation is set up whereby segments of the genome that require activation come away from the lamina, whereas the newly silenced loci can become tethered there (40, 72, 73). Three independent 2008 studies used the LacO-LacI system to direct specific loci to the nuclear lamina and reported varying results (67, 74, 75), suggesting that this may be a two-step process involving an initial silencing event followed by a repositioning to the lamina for long-term silencing.
Conventional wisdom has largely held that constrained diffusion is the only means of movement in the interphase nucleus (76, 77). Thus, as with the looping and chromosome kissing events, no definitive mechanisms have been delineated to explain this motion of chromatin with respect to the lamina; however, two groups report interesting findings that shed new light intranuclear repositioning of gene loci and CTs (78, 79). The Belmont group (78), using live-cell time-lapse imaging, observed an actin/myosin-dependent displacement of a green fluorescent protein-tagged transgenic locus from the nuclear periphery to the interior upon induction during interphase. Myosin is an adenosine triphosphate (ATP)-dependent motor protein that tracks along the actin cytoskeleton through the cytoplasmic compartment. Interestingly enough, depriving the cell of ATP leads to changes in chromatin organization in the nucleus, but this has been attributed largely to the ATP dependence of numerous chromatin and nucleosome remodeling complexes that are active in the nucleus (76, 77). The Bridger group (79) reported a nuclear /actin/myosin-dependent mechanism of chromosome and gene locus displacement, but their findings suggest a dependence on entry or exit from the cell cycle (80, 81), as well as an intact lamina (82), as cells with mutations in LMNA linked to Hutchinson-Gilford progeria syndrome fail to show the same chromosome dynamics.
In mitotic cells, changes in intranuclear chromosome position could be explained by a mechanism in which an affinity-based process promotes interactions between mitotic chromosomes and specific NET proteins (83). During mitosis, the nuclear envelope breaks down into smaller vesicles in early prophase, and chromosomes condense, allowing their free movement in the cell. During this time, certain vesicles enriched with chromatin-binding NET proteins have an opportunity to associate with various targets on the mitotic chromosomes, including specific epigenetic marks, and a multitude of chromatin-associated proteins. When the nuclear envelope reforms in the subsequent interphase, the chromosomes that were high-affinity targets for the chromatin-binding NET proteins are tethered to the nuclear periphery, whereas others are forced into the interior of the nucleus by steric hindrance once chromosomes decondense. A recent proteomic analysis identified hundreds of novel NET proteins (84). A screen focused on identifying factors involved in genomic architecture involving just 10 of these yielded two NET proteins that demonstrated the capacity to reposition a gene locus within the nucleus. Given these early results, it is tempting to speculate whether a NET protein-based mechanism could be involved in the changes to 3D genomic architecture observed during differentiative cell divisions.
3. MECHANICAL/STRUCTURAL PROPERTIES OF CELLS
3.1. The Cytoskeleton
The proteins of the cytoskeleton exist in the cytoplasm as a population of dissolved monomers in dynamic equilibrium with large polymeric cytoskeletal structures. As a result of their dynamic nature, cytoskeletal structures are at once robust and easily reconfigured. Tubulin monomers assemble into thick hollow tubes named microtubules that resist bending and are therefore capable of bearing compressive loads. Microtubules radiate throughout the cell from a microtubule organizing center located next to the nucleus. They contribute to the mechanical integrity of the cell and also provide a network of tracks along which organelles are driven by motor proteins (85). Intermediate filament proteins form fibrous structures that are flexible but resistant to fracture and extension (86). Actin monomers remain in solution as long as they are bound to thymosin. However, another actin-binding protein named profilin can displace thymosin and make actin available for assembly. Therefore, the cell can initiate rapid assembly of polymeric actin structures by producing profilin (87). This rapid actin assembly allows the cell to apply forces to its environment. The nature of the assembled structure and the forces generated depend on other actin-associated proteins. Actin filaments associate with one another in parallel to create thick bundles when they are cross-linked by α-actinin (88). Myosin proteins can bind to these filaments and drive them past one another to exert tension through the filament bundle (89). In addition, Filamin cross-links actin filaments at an angle so that they assemble into a gel (90). Taken together, the cell can push on its environment by assembling an actin gel adjacent to the cell membrane, for example, during the extension of a lamellipodium. The mechanical properties of the substrate affect the organization of the actin cytoskeleton. Stiff substrates increase bundling and tension in actin fibers (91). Substrate stiffness influences differentiation in a manner that depends on the activity of the myosin motor proteins that create tension in actin fibers (92). Therefore, physical properties of the extracellular matrix (ECM) influence events occurring within the nucleus via the cytoskeleton.
3.2. Extracellular Matrix: Transfer of External Strain to Cells and Subcellular Compartments
Physical stresses in the cell environment propagate through a series of mechanical connections to the nuclear interior. Bulk compression of cartilage, for example, causes shape changes in the nuclei of chondrocytes in an actin-dependent manner (93). Extracellular mechanical stresses act on the cytoskeleton via integrins, cell-surface proteins that connect to both the cytoskeleton and the ECM. For example, the nucleoli within fibroblast nuclei were found to reorient along the direction of tension that was applied through beads bound to the cell surface via integrins (94). This behavior implies the presence of a mechanical connection spanning the nuclear envelope. As noted above, the LINC complex spans the perinuclear space and connects all three cytoskeletal components (60, 95, 96) with the nuclear lamina (97, 98). Transmission of force from the cytoplasm to the nuclear interior requires intact LINC complexes, although the activation of mechanotransduction processes originating at focal adhesions may not (58). In addition to transmitting cytoskeletal stresses to the nucleus, LINC complexes allow the nuclear lamina to contribute to the overall stiffness of the cell (55, 99). Mechanical activation of nuclear factor κB was compromised in Lmna-deficient mouse embryo fibroblasts, suggesting that the lamina plays an important role in the mechanotransduction of extracellular stresses into gene activation (99). The nuclear lamina not only connects the nucleus to cytoskeletal stresses but also protects it from them. Lmna-deficient cells deformed more during cell-stretching experiments than did their wild-type counterparts and had decreased viability under cyclical mechanical strain (99). EMD is another protein that resides at the interface between the nucleus and the cytoplasm. It binds lamin, actin, and components of the LINC complex as well as several transcriptional repressors. Emd-null cells have approximately normal mechanics but impaired mechanotransduction, suggesting that EMD plays a role in communicating mechanical stresses to the nuclear interior (100). LNMB1’s contribution to the overall mechanical properties of the nucleus is complex. On one hand, its deletion does not impact the overall stiffness of the lamina and only leads to minor aberrations in lamina architecture, as evidenced by the increased incidence of localized blebbing, coinciding with focal reductions of LMNA and C in the nuclei of Lmnb1-deficient cells (101). On the otherhand, cells lacking Lmnb1 display a nuclear rotation phenotype where the nucleus and its contents rotate freely, confined by the cytoskeleton, in an ATP-dependent fashion. It is thought that the loss of LMNB1 disrupts the organization of Sun proteins or other INM proteins required for the localization of the Nesprin proteins to ONM (102).
3.3. Mechanical Properties of Tissues, Cells, and the Nucleus
Biological tissues exhibit complex mechanical properties, and a range of sophisticated mechanical theories have been developed to describe these properties. Of the major tissue types, only bone can reasonably be approximated as a linearly elastic material subject to small strains (103). Large strain effects must be explicitly accounted for in many scenarios, including traumatic brain injury, which occurs at intermediate levels (at strains of 20%) (104). Some tissues, such as the annulus fibrosus of the intervertebral disc or the arterial wall, are anisotropic because they are reinforced by aligned fibers (105, 106). Biological tissues often exhibit complex rate dependence characterized by a spectrum of time constants rather than by a single value (107). In cartilage, rate dependency arises from the flow of fluid through the porous solid matrix, a process that is quantitatively described by biphasic theory (108). The flow of fluid is, in turn, influenced by the presence of fixed electrostatic charges in the matrix, an effect that has been modeled using triphasic theory (109). Stress and strain distributions within biological tissues can be estimated using continuum biomechanics by careful application of the tools described above. The local stress-strain and fluid-flow states within a tissue determine the physical environment of cells at that point. Multiscale modeling can be used to describe this physical environment and how it is modulated by the pericellular domain (110-112).
Cells exhibit viscoelastic material properties determined by their cytoskeletal organization, which is, in turn, influenced by external stimuli. For example, osmotic loading stimulated calcium signaling in chondrocytes, which, in turn, triggered disassembly of the actin cortex and a reduction in cell stiffness (113). Conversely, endothelial cells exposed to shear stress increased actin organization and stiffness (114). Mechanotransduction may alter cell physiology by stimulating conventional biochemical signaling paths via ion channel activation (115) or activation of other membrane-bound receptors (116, 117). Alternatively, however, mechanical stresses are transferred from the ECM directly to the cell nucleus via the LINC complex (58, 118). The consequences of mechanical stresses in the nucleus depend in part on the mechanical properties of the nucleus.
Mechanically, the nucleus is composed of two major structures, the lamina and the nucleoplasm within it. In creep experiments, the nucleoplasm is softer and more viscous than the nuclear lamina. Both structures exhibit power-law rheology under creep loading; i.e., no single time constant dominates the response (119). This type of rheological behavior is common in gels and other materials that exist close to the transition between solid and liquid states (120). Whereas lipid bilayers are commonly described as two-dimensional liquids, the nuclear lamina is a two-dimensional solid (121); the distinction is that the solid lamina can resist shear stress, as evidenced by the buckles that arose in the lamina when it was aspirated into a micropipette in the study by Dahl and colleagues (119). When tested independently of the rest of the nucleus, the nucleoplasm behaves as a Maxwell fluid. The corresponding Young’s modulus was found to be 36 Pa in a study that employed a 100-μm-diameter bead loaded only by Brownian motion (122) and 250 Pa in a study that employed a 500-μm-diameter bead loaded via an externally applied magnetic field (123). This contrast suggests that the mechanical properties of the nucleoplasm may depend on the length scale on which they are measured. The structural stiffness of the nucleus derives primarily from the nuclear lamina. Experiments that loaded the whole nucleus via micropipette aspiration (124) or unconfined compression (125, 126) observed solid viscoelastic behavior, with moduli in the kilopascal range-i.e., at values much higher than those of the nucleoplasm. In summary, much of the behavior of the nucleus under physical stress can be explained by modeling the nucleus as a soft, viscoelastic gel encapsulated in a stiff viscoelastic membrane and connected to the ECM via cytoskeletal structures and integrins.
3.4. Mechanical Perturbation of the Nucleus: Deformation, Osmotic Effects, and Cell Morphology
Mechanical and osmotic stresses can influence the nucleus directly as well as through indirect effects on the cytoskeleton (127). For example, intervertebral disc cells subjected to uniaxial stretch in monolayer culture exhibited nuclear strain that was lower than the applied cytoplasmic strain; this observation was attributed to the fact that the nucleus is typically stiffer than the cytoplasm (128). Epithelial cells experience shear stress in situ because they are loaded by blood flow. Shear stress causes the nucleus to flatten, elongate in the direction of flow, and stiffen (125), probably owing to upregulation and redistribution of A-type lamins (129). The elongated shape persists after the nucleus is isolated from the cell, demonstrating that the nucleoplasm is not a solid capable of elastic rebound (125). When human mesenchymal stem cells were cultured on nanoscale gratings, their nuclei aligned with the direction of the grating because the topography of the grating stimulated rearrangement of the actin and intermediate filament networks in the cytoskeleton (130).
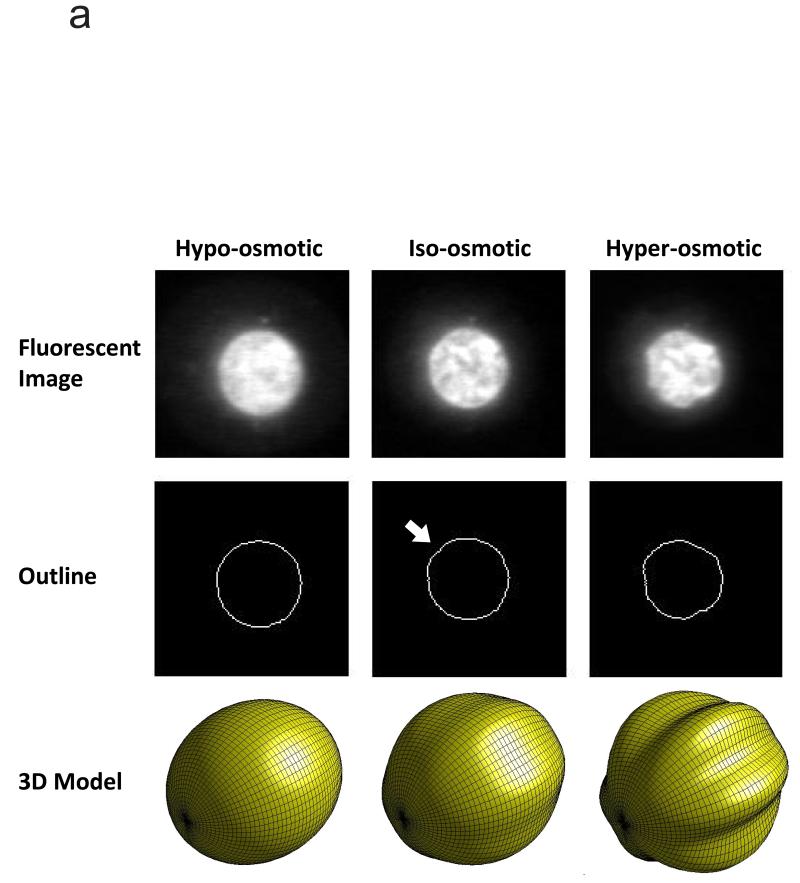
In addition to mechanical stress, osmotic stress is a potent mediator of mechanotransduction in connective tissues such as cartilage and intervertebral disc (131), where it is coupled to compressive stress by triphasic effects (109), and it affects the geometry and other physical properties of the nucleus in several important ways. Osmotic stress alters cell volume (132) and therefore alters the concentration of macromolecules inside the cell. The chromatin inside the nucleus dilates with changes in macromolecule concentration (133-135) as a consequence of the thermodynamics of macromolecular crowding (136, 137). Therefore, under hypo-osmotic stress, the nucleus expands until the nuclear lamina draws taut around the nucleoplasm and prevents further expansion. Under hyperosmotic stress, the nucleoplasm contracts, and the nucleus shrinks. In addition, hyperosmotic stress results in marked chromatin condensation (Figure 3). Since the nuclear lamina resists changes in area, it buckles as the nucleus shrinks, forcing the nucleus into a convoluted shape (134, 138) (Figure 3). The increased macromolecular crowding that accompanies hyperosmotic stress impedes diffusion of macromolecules in the cytoplasm. However, the effect on diffusion within the nucleoplasm is negligible because it is offset by reorganization of the chromatin within the nucleus (135, 138). Chromatin is interdigitated by channels and pores of the IC (139) that accommodate rapid motion of macromolecules (140), and these channels enlarge as the chromatin contracts (49, 135), offsetting the effects of increased macromolecular crowding. The change in nuclear geometry in the absence of changes in intranuclear transport leads to increased nucleocytoplasmic transport of inert macromolecules under hyperosmotic stress (135). This is significant because transport of macromolecules from the cytoplasm to the nucleus is a key activation step in several intracellular signaling pathways. Therefore, osmotic stress may enhance signaling along these pathways through a combination of purely biophysical processes.
Figure 3.
Changes in the nuclear morphology of chondrocytes with osmotic stress. (a) The middle column depicts the iso-osmotic or equilibrium condition (380 mOsm). The osmotic pressurization of the nucleus is barely sufficient to induce mild buckling of the nuclear lamina, as evidenced by the mild undulation in the outline of the nucleus (white arrow). The image in the bottom row of the middle column shows a 3D view of a mildly postbuckled spheroid. The left column depicts the hypo-osmotic condition (180 mOsm). The undulation has disappeared, and the outline of the nucleus is now smooth. The right column depicts the hyperosmotic condition (580 mOsm). Now there are pronounced undulations along the entire circumference of the nuclear outline. The image in the bottom row of this column shows a 3D view of a spheroidal geometry with pronounced longitudinal buckling. These reconstructions are hypothesized to be representative of the nuclear outlines depicted in the middle row. Adapted from Reference 135 with permission. (b) Hyperosmotic challenge on live bovine chondrocytes transfected with H2B-GFP. Chondrocytes were cultured in monolayer initially in an iso-osmotic environment (300 mOsm kg−1) and subjected to hyperosmotic challenge (700 mOsm kg−1) for 15 min, and then brought back to 300 mOsm kg−1 for an additional 15 min while being imaged by time-lapse confocal microscopy. The top row shows an average intensity projection in the xy plane. The middle row shows an average intensity projection in the xz plane. The bottom row shows the middle section of the chondrocyte nuclei. Images were thresholded using the iterative self-organizing data algorithm (134; J. Irianto, R.P. Martins & D.A. Lee, unpublished data). Abbreviations: GFP, green fluorescent protein; mOsm, milliosmole.
3.5. Differentiation and Migration
There is an intriguing relationship between cell fate and compliance of the nucleus. Lmna expression increases as cells differentiate (141), making the nucleus stiffer (Figure 4) (142). Inside the nucleus, chromatin is generally diffuse in embryonic stem cells but condenses into higher-order structures as the cells differentiate (143). This pattern is also evident in the early mouse embryo, as higher-order chromatin structure begins to accumulate at chromocenters and around the periphery of the nucleus in cells that will adopt a trophectodermal fate in contrast to cells on the interior of the embryo that maintain a pluripotent program (71). As differentiation proceeds, epigenetic modifications accumulate throughout the genome in a manner that silences genes associated with self-renewal and pluripotency in favor of terminally differentiated genetic programs (144). This phenomenon suggests that chromatin organization may govern gene expression and hence cell fate, although it is not certain that the association between chromatin condensation and silencing is causal (20). A deformable nucleus facilitates normal stem cell behavior because it accommodates motion of the cell through tortuous three-dimensional environments. The nucleus is the largest, stiffest organelle in the cell and hence is the primary impediment to motion as cells squeeze through tight spaces. Efficient migration requires condensation of chromatin (145), which also helps the nucleus fit through small spaces. Other cells that migrate through the ECM to fulfill their functions also have low levels of A-type lamin expression, which make their nuclei more deformable (101). For example, the human neutrophil has low levels of lamins, lamin-associated proteins, lamin receptors, and EMD expression that allow it to pass through small gaps between endothelial cells in blood vessel walls (146). Similarly, cancer calls invade tissues as they metastasize, and again, nuclear changes that promote deformability occur. Although cancer cells are a heterogeneous population, lamin expression is reduced in many types of cancer (147, 148). The impressions made by individual extracellular collagen fibers can be seen in the nuclei of cancer cells as they invade the ECM, suggesting that the fibers physically compress the nucleus as it passes between them (149). Nuclear deformation during cell migration provides a clear and compelling example of a physiological process that depends on the mechanics of the nucleus, but the interaction is passive. One of the most daunting and important challenges in the field of nuclear mechanics today is demonstrating that a mechanical stress can propagate from the ECM, through the cytoskeleton, across the nuclear envelope, and into the chromatin, where it physically deforms the DNA or associated proteins, thus changing gene expression.
Figure 4.
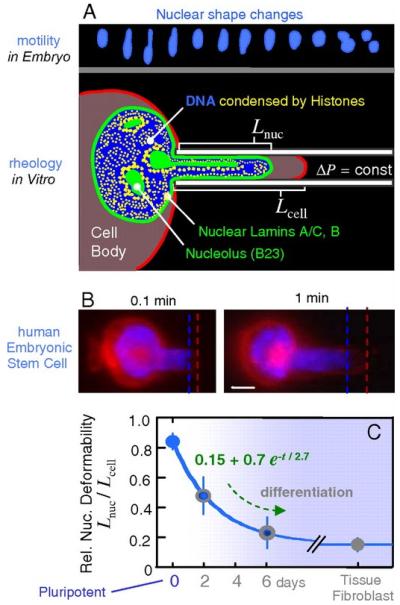
Nuclei of human stem cells are more deformable than nuclei of differentiated cells. (a) Neural progenitor cell nuclei show large deformations during in vivo migration. Micropipette aspiration mimics such distortions. (b) Human embryonic stem cells (ESCs) were aspirated after differentiation in culture, and the ratio of nuclear extension to cytoplasmic extension was measured as Lnuc/Lcell. A day-zero pluripotent ESC is shown with labeling of nuclear DNA (blue) and the cell membrane (red). Scale bar: 3 μm. (c) As differentiation progresses, ESC nuclei stiffen nearly six-fold relative to cytoplasm, and the decrease in relative compliance fits an exponential decay. Differentiated cells such as embryonic fibroblasts also have a nucleus that is stiffer than the cytoplasm. Reproduced from Reference 142 with permission.
3.6. Mechanical Regulation of Gene Expression
Physical stresses can act on the nucleus and nucleoplasm and also cause a change in gene activity, although a causal relationship between mechanical stimuli and a biological response cannot always be shown. For example, the gene CTFR moves from the transcriptionally repressive nuclear periphery into the center of the nucleus upon activation. However, artificially inducing this relocation does not activate the gene, indicating that these are molecularly separate events and that relocation is a consequence of activation rather than a cause (150). Microtubules induce dynamic fluctuations in the nuclear envelope as they push against it, and these fluctuations “agitate” the chromatin within, causing different chromatin domains to move relative to one another (151). Chromatin topology influences gene regulation (139, 152, 153), so these microtubule-pushing forces may alter gene activity as they reorganize the nuclear interior, but direct evidence for this is lacking. Conformational changes occur in several cytoskeletal proteins when a cell is tensed, but none have been reported in chromatin (154). Subnuclear structures such as Cajal bodies and promyelocytic leukemia bodies exist within the IC, and the dynamics and architecture of the surrounding chromatin determine their motion (155). This implies that the RNA-processing interactions between these bodies and the surrounding chromatin are influenced by reorganization of that chromatin under mechanical stress. Mechanical strain rapidly activates tenascin-C gene expression in fibroblasts independent of protein synthesis or paracrine effects, making this a good candidate for investigation as a gene that is directly activated by mechanical stress (21).
4. SUMMARY AND CONCLUSIONS
There is significant evidence that physical signals can be transmitted readily from the ECM to the cell nucleus. In the case of mechanical stresses, there is a direct molecular link between the ECM molecules and the nucleoplasmic compartment. Integrinsspan the cell membrane and connect the ECM molecules to various cytoskeletal elements which in turn associate both directly and indirectly to the LINC complex, whose Sun protein components form the final link to the nucleoplasm and the nuclear lamina (e.g.,93, 94, 156, 157; Fig. 1). Osmotic stress, in contrast, can alter nuclear size, structure, and physical properties through alterations in the physicochemical characteristics of the cytoplasm, potentially circumventing a direct mechanical link (reviewed in 138). In either case, such changes in nuclear and chromatin structure can have a profound effect on biological events such as gene transcription, nucleocytoplasmic transport, and protein synthesis.
The degree of interconnectedness of the cell’s support structures throughout cytoplasmic and nuclear compartments provides a means for direct communication and force transfer. That these forces lead to the agitation of chromatin domains provides tempting grounds for speculation as to how mechanical forces may result in CT movements in an interphase nucleus. Alternatively, the mechanical stimulus may simply provide a signal to the cell, causing it to either enter or exit the cell cycle, with commensurate changes in chromatin and nuclear organization similar to those described by Bridger and colleagues (80, 81). Understanding how NET proteins can specifically target gene loci and entire chromosomes to specific subnuclear regions (83) is also an important open question to be answered. As the tools for studying intracellular biomechanics improve, the connection between physical stresses, nuclear architecture, and gene activity is becoming more apparent, and our increased understanding of this connection opens up the possibility of a new realm of cell signaling pathways comprising biophysical as well as biochemical events. As these discoveries accumulate, they add to the importance of nuclear mechanics in our overall understanding of the cell.
ACKNOWLEDGMENTS
The authors would like to acknowledge Jerome Irianto for his contribution of Figure 3b. FG and JDF were supported in part by NIH grants AR48182, AR50245, AG15768, and AR48852. DAL and RPM were supported in part by an EPSRC platform grant EP/E046975/1 and a Human Frontier of Science Program grant RGP0025-2009.
LIST OF ABBREVIATIONS
- ATP
adenosine triphosphate
- bp
(nucleotide) base pairs
- CT
chromosome territory
- DNA
deoxyribonucleic acid
- ECM
extracellular matrix
- EMD
emerin protein
- H1/H2A/H2B/H3/H4
histone proteins
- IC
interchromosomal compartment
- INM
inner nuclear membrane
- K
amino acid lysine; K20, e.g., represents lysine, residue 20
- LINC
linker of the nucleoskeleton and cytoskeleton
- NET protein
nuclear envelope transmembrane protein
- ONM
outer nuclear membrane
- RNA
ribonucleic acid
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Dewey CF., Jr. Effects of fluid flow on living vascular cells. J. Biomech. Eng. 1984;106:31–35. doi: 10.1115/1.3138453. [DOI] [PubMed] [Google Scholar]
- 2.Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, et al. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–31. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 3.Discher D, Dong C, Fredberg JJ, Guilak F, Ingber D, et al. Biomechanics: cell research and applications for the next decade. Ann. Biomed. Eng. 2009;37:847–59. doi: 10.1007/s10439-009-9661-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann. Med. 2003;35:564–77. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 5.Estes BT, Gimble JM, Guilak F. Mechanical signals as regulators of stem cell fate. Curr. Top. Dev. Biol. 2004;60:91–126. doi: 10.1016/S0070-2153(04)60004-4. [DOI] [PubMed] [Google Scholar]
- 6.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee DA, Knight MM, Campbell JJ, Bader DL. Stem cell mechanobiology. J. Cell. Biochem. 2011;112:1–9. doi: 10.1002/jcb.22758. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–23. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aristotle . Generation of Animals. Harv. Univ. Press; Transl. AL Peck. Cambridge, MA: 1979. p. 607. [Google Scholar]
- 10.Maienschein J. Zalta EN, editor. Epigenesis and preformationism. Stanf. Encycl. Philos. 2005 http://plato.stanford.edu/archives/spr2012/entries/epigenesis/ [Google Scholar]
- 11.Carter DR, Beaupré GS. Skeletal Function and Form. Mechanobiology of Skeletal Development, Aging and Regeneration. : Cambridge Univ. Press; Cambridge, UK: 2001. [Google Scholar]
- 12.Darwin C. On the Origin of Species by Means of Natural Selection, Or the Preservation of Favoured Races in the Struggle for Life. John Murrayp; London: 1859. p. 450. [PMC free article] [PubMed] [Google Scholar]
- 13.Wolff J. The Law of Bone Remodeling. Springer-Verlag; Berlin: 1986. [Google Scholar]
- 14.Barak MM, Lieberman DE, Hublin J-J. A Wolff in sheep’s clothing: trabecular bone adaptation in response to changes in joint loading orientation. Bone. 2011;49:1141–51. doi: 10.1016/j.bone.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Satzinger H. Theodor and Marcella Boveri: chromosomes and cytoplasm in heredity and development. Nat. Rev. Genet. 2008;9:231–38. doi: 10.1038/nrg2311. [DOI] [PubMed] [Google Scholar]
- 16.Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J. Cell Sci. 2008;121:1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- 17.Boveri T. Über mehrpolige Mitosen als Mittel zur Analyse des Zellkerns. Verh. Phys.-Med. Ges. Würzburg. 1902;35:67–90. [Google Scholar]
- 18.Ma J, Yang W. Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots. Proc. Natl. Acad. Sci. USA. 2010;107:7305–10. doi: 10.1073/pnas.0908269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 21.Chiquet M, Gelman L, Lutz R, Maier S. From mechanotransduction to extracellular matrix gene expression in fibroblasts. Biochim. Biophys. Acta. 2009;1793:911–20. doi: 10.1016/j.bbamcr.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Mandelkern M, Elias JG, Eden D, Crothers DM. The dimensions of DNA in solution. J. Mol. Biol. 1981;152:153–61. doi: 10.1016/0022-2836(81)90099-1. [DOI] [PubMed] [Google Scholar]
- 23.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 24.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA. 2008;105:8872–77. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Syed SH, Goutte-Gattat D, Becker N, Meyer S, Shukla MS, et al. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. USA. 2010;107:9620–25. doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 29.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 30.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 31.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 2010;12:853–62. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 35.Talbert PB, Henikoff S. Histone variants-ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010;11:264–75. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 36.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–31. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tachiwana H, Kagawa W, Osakabe A, Kawaguchi K, Shiga T, et al. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc. Natl. Acad. Sci. USA. 2010;107:10454–59. doi: 10.1073/pnas.1003064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–62. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 39.Boveri T. Die Blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenindividualität. Arch. Zellforsch. 1909;3:181–268. [Google Scholar]
- 40.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders M. Analysis of chromosome positions in the interphase nucleus of Chinese hamster cells by laser-UV-microirradiation experiments. Hum. Genet. 1982;62:201–9. doi: 10.1007/BF00333519. [DOI] [PubMed] [Google Scholar]
- 42.Schardin M, Cremer T, Hager HD, Lang M. Specific staining of human chromosomes in Chinese hamster × man hybrid cell lines demonstrates interphase chromosome territories. Hum. Genet. 1985;71:281–87. doi: 10.1007/BF00388452. [DOI] [PubMed] [Google Scholar]
- 43.Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–84. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- 44.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 45.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–31. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cremer M, von Hase J, Volm T, Brero A, Kreth G, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–67. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- 47.Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J, et al. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J. Cell Biol. 2003;162:809–20. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14:707–33. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]
- 50.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat. Rev. Genet. 2009;10:457–66. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 51.Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000;113(Pt. 9):1565–76. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- 52.Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–23. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- 53.Cavalli G. Chromosome kissing. Curr. Opin. Genet. Dev. 2007;17:443–50. doi: 10.1016/j.gde.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 54.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Broers JL, Machiels BM, Kuijpers HJ, Smedts F, van den Kieboom R, et al. A- and B-type lamins are differentially expressed in normal human tissues. Histochem. Cell Biol. 1997;107:505–17. doi: 10.1007/s004180050138. [DOI] [PubMed] [Google Scholar]
- 56.Martins RP, Ostermeier GC, Krawetz SA. Nuclear matrix interactions at the human protamine domain: a working model of potentiation. J. Biol. Chem. 2004;279:51862–68. doi: 10.1074/jbc.M409415200. [DOI] [PubMed] [Google Scholar]
- 57.Martins RP, Krawetz SA. Decondensing the protamine domain for transcription. Proc. Natl. Acad. Sci. USA. 2007;104:8340–45. doi: 10.1073/pnas.0700076104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 2011;286:26743–53. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 2010;26:421–44. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koizumi H, Gleeson JG. Sun proteins enlighten nuclear movement in development. Neuron. 2009;64:147–49. doi: 10.1016/j.neuron.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, et al. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys. J. 2008;95:5462–75. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–98. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manilal S, Nguyen TM, Sewry CA, Morris GE. The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet. 1996;5:801–8. doi: 10.1093/hmg/5.6.801. [DOI] [PubMed] [Google Scholar]
- 65.Rowat AC, Lammerding J, Ipsen JH. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys. J. 2006;91:4649–64. doi: 10.1529/biophysj.106.086454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chaturvedi P, Parnaik VK. Lamin A rod domain mutants target heterochromatin protein 1α and β for proteasomal degradation by activation of F-box protein, FBXW10. PLoS ONE. 2010;5:e10620. doi: 10.1371/journal.pone.0010620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–47. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 68.Taniura H, Glass C, Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J. Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Makatsori D, Kourmouli N, Polioudaki H, Shultz LD, McLean K, et al. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J. Biol. Chem. 2004;279:25567–73. doi: 10.1074/jbc.M313606200. [DOI] [PubMed] [Google Scholar]
- 70.Lee DC, Welton KL, Smith ED, Kennedy BK. A-type nuclear lamins act as transcriptional repressors when targeted to promoters. Exp. Cell Res. 2009;315:996–1007. doi: 10.1016/j.yexcr.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS ONE. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 2010;38:603–13. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deniaud E, Bickmore WA. Transcription and the nuclear periphery: edge of darkness? Curr. Opin. Genet. Dev. 2009;19:187–91. doi: 10.1016/j.gde.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 74.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 2002;12:439–45. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- 77.Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 1997;7:930–39. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- 78.Chuang C-H, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr. Biol. 2006;16:825–31. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 79.Mehta IS, Elcock LS, Amira M, Kill IR, Bridger JM. Nuclear motors and nuclear structures containing A-type lamins and emerin: Is there a functional link? Biochem. Soc. Trans. 2008;36:1384–88. doi: 10.1042/BST0361384. [DOI] [PubMed] [Google Scholar]
- 80.Szczerbal I, Foster HA, Bridger JM. The spatial repositioning of adipogenesis genes is correlated with their expression status in a porcine mesenchymal stem cell adipogenesis model system. Chromosoma. 2009;118:647–63. doi: 10.1007/s00412-009-0225-5. [DOI] [PubMed] [Google Scholar]
- 81.Mehta IS, Amira M, Harvey AJ, Bridger JM. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 2010;11:R5. doi: 10.1186/gb-2010-11-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mehta IS, Eskiw CH, Arican HD, Kill IR, Bridger JM. Farnesyltransferase inhibitor treatment restores chromosome territory positions and active chromosome dynamics in Hutchinson-Gilford progeria syndrome cells. Genome Biol. 2011;12:R74. doi: 10.1186/gb-2011-12-8-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zuleger N, Robson MI, Schirmer EC. The nuclear envelope as a chromatin organizer. Nucleus. 2011;2:339–49. doi: 10.4161/nucl.2.5.17846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Korfali N, Wilkie GS, Swanson SK, Srsen V, Batrakou DG, et al. The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture. Mol. Cell. Proteomics. 2010;9:2571–85. doi: 10.1074/mcp.M110.002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olmsted JB, Borisy GG. Microtubules. Annu. Rev. Biochem. 1973;42:507–40. doi: 10.1146/annurev.bi.42.070173.002451. [DOI] [PubMed] [Google Scholar]
- 86.Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. 1994;63:345–82. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 87.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–76. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 88.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–99. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 89.Lazarides E. Actin, α-actinin, and tropomyosin interaction in structural organization of actin-filaments in nonmuscle cells. J. Cell Biol. 1976;68:202–19. doi: 10.1083/jcb.68.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA. 1998;95:6181–86. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 92.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 93.Guilak F. Compression-induced changes in the shape and volume of the chondrocyte nucleus. J. Biomech. 1995;28:1529–41. doi: 10.1016/0021-9290(95)00100-x. [DOI] [PubMed] [Google Scholar]
- 94.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA. 1997;94:849–54. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, et al. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp. Cell Res. 2004;295:330–39. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 96.King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–38. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, et al. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 2006;26:3738–51. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lammerding J, Kamm RD, Lee RT. Mechanotransduction in cardiac myocytes. Ann. N. Y. Acad. Sci. 2004;1015:53–70. doi: 10.1196/annals.1302.005. [DOI] [PubMed] [Google Scholar]
- 100.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 2005;170:781–91. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, et al. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006;281:25768–80. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 102.Ji JY, Lee RT, Vergnes L, Fong LG, Stewart CL, Reue K, Young SG, Zhang Q, Shanahan CM, Lammerding J. Cell nuclei spin in the absence of Lamin B1. J. Biol. Chem. 2007;282:20015–20026. doi: 10.1074/jbc.M611094200. [DOI] [PubMed] [Google Scholar]
- 103.McElhaney JH, Fogle JL, Melvin JW, Haynes RR, Roberts VL, Alem NM. Mechanical properties on cranial bone. J. Biomech. 1970;3:495–511. doi: 10.1016/0021-9290(70)90059-x. [DOI] [PubMed] [Google Scholar]
- 104.Morrison B, III, Cater HL, Wang CC, Thomas FC, Hung CT, et al. A tissue level tolerance criterion for living brain developed with an in vitro model of traumatic mechanical loading. Stapp Car Crash J. 2003;47:93–105. doi: 10.4271/2003-22-0006. [DOI] [PubMed] [Google Scholar]
- 105.Elliott DM, Setton LA. Anisotropic and inhomogeneous tensile behavior of the human anulus fibrosus: experimental measurement and material model predictions. J. Biomech. Eng. 2001;123:256–63. doi: 10.1115/1.1374202. [DOI] [PubMed] [Google Scholar]
- 106.Schulze-Bauer CA, Morth C, Holzapfel GA. Passive biaxial mechanical response of aged human iliac arteries. J. Biomech. Eng. 2003;125:395–406. doi: 10.1115/1.1574331. [DOI] [PubMed] [Google Scholar]
- 107.Tripathy S, Berger EJ. Measuring viscoelasticity of soft samples using atomic force microscopy. J. Biomech. Eng. 2009;131:094507. doi: 10.1115/1.3194752. [DOI] [PubMed] [Google Scholar]
- 108.Mow VC, Kuei SC, Lai WM, Armstrong CG. Biphasic creep and stress relaxation of articular cartilage in compression? Theory and experiments. J. Biomech. Eng. 1980;102:73–84. doi: 10.1115/1.3138202. [DOI] [PubMed] [Google Scholar]
- 109.Lai WM, Hou JS, Mow VC. A triphasic theory for the swelling and deformation behaviors of articular cartilage. J. Biomech. Eng. 1991;113:245–58. doi: 10.1115/1.2894880. [DOI] [PubMed] [Google Scholar]
- 110.Guilak F, Mow VC. The mechanical environment of the chondrocyte: a biphasic finite element model of cell-matrix interactions in articular cartilage. J. Biomech. 2000;33:1663–73. [PubMed] [Google Scholar]
- 111.Alexopoulos LG, Haider MA, Vail TP, Guilak F. Alterations in the mechanical properties of the human chondrocyte pericellular matrix with osteoarthritis. J. Biomech. Eng. 2003;125:323–33. doi: 10.1115/1.1579047. [DOI] [PubMed] [Google Scholar]
- 112.Alexopoulos LG, Setton LA, Guilak F. The biomechanical role of the chondrocyte pericellular matrix in articular cartilage. Acta Biomater. 2005;1:317–25. doi: 10.1016/j.actbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 113.Guilak F, Erickson GR, Ting-Beall HP. The effects of osmotic stress on the viscoelastic and physical properties of articular chondrocytes. Biophys. J. 2002;82:720–27. doi: 10.1016/S0006-3495(02)75434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sato M, Nagayama K, Kataoka N, Sasaki M, Hane K. Local mechanical properties measured by atomic force microscopy for cultured bovine endothelial cells exposed to shear stress. J. Biomech. 2000;33:127–35. doi: 10.1016/s0021-9290(99)00178-5. [DOI] [PubMed] [Google Scholar]
- 115.Guharay F, Sachs F. Stretch-activated single ion channel currents in tissue-cultured embryonic chick skeletal muscle. J. Physiol. 1984;352:685–701. doi: 10.1113/jphysiol.1984.sp015317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen KD, Li YS, Kim M, Li S, Yuan S, et al. Mechanotransduction in response to shear stress: roles of receptor tyrosine kinases, integrins, and Shc. J. Biol. Chem. 1999;274:18393–400. doi: 10.1074/jbc.274.26.18393. [DOI] [PubMed] [Google Scholar]
- 117.Correa-Meyer E, Pesce L, Guerrero C, Sznajder JI. Cyclic stretch activates ERK1/2 via G proteins and EGFR in alveolar epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;282:L883–91. doi: 10.1152/ajplung.00203.2001. [DOI] [PubMed] [Google Scholar]
- 118.Chen CS, Ingber DE. Tensegrity and mechanoregulation: from skeleton to cytoskeleton. Osteoarthr. Cartil. 1999;7:81–94. doi: 10.1053/joca.1998.0164. [DOI] [PubMed] [Google Scholar]
- 119.Dahl KN, Engler AJ, Pajerowski JD, Discher DE. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J. 2005;89:2855–64. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sollich P. Rheological constitutive equation for a model of soft glassy materials. Phys. Rev. E. 1998;58:738–59. [Google Scholar]
- 121.Rowat AC, Foster LJ, Neilson MM, Weiss M, Ipsen JH. Characterizing the elastic properties of the nuclear envelope. J. R. Soc. Interface. 2005;22:63–9. doi: 10.1098/rsif.2004.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tseng Y, Lee JS, Kole TP, Jiang I, Wirtz D. Micro-organization and viscoelasticity of the interphase nucleus revealed by particle nanotracking. J. Cell Sci. 2004;117:2159–67. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
- 123.de Vries AH, Krenn BE, van Driel R, Subramaniam V, Kanger JS. Direct observation of nanomechanical properties of chromatin in living cells. Nano Lett. 2007;7:1424–27. doi: 10.1021/nl070603+. [DOI] [PubMed] [Google Scholar]
- 124.Guilak F, Tedrow JR, Burgkart R. Viscoelastic properties of the cell nucleus. Biochem. Biophys. Res. Commun. 2000;269:781–86. doi: 10.1006/bbrc.2000.2360. [DOI] [PubMed] [Google Scholar]
- 125.Deguchi S, Maeda K, Ohashi T, Sato M. Flow-induced hardening of endothelial nucleus as an intracellular stress-bearing organelle. J. Biomech. 2005;38:1751–59. doi: 10.1016/j.jbiomech.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 126.Caille N, Thoumine O, Tardy Y, Meister JJ. Contribution of the nucleus to the mechanical properties of endothelial cells. J. Biomech. 2002;35:177–87. doi: 10.1016/s0021-9290(01)00201-9. [DOI] [PubMed] [Google Scholar]
- 127.Dahl KN, Booth-Gauthier EA, Ladoux B. In the middle of it all: mutual mechanical regulation between the nucleus and the cytoskeleton. J. Biomech. 2010;43:2–8. doi: 10.1016/j.jbiomech.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 128.Gilchrist CL, Witvoet-Braam SW, Guilak F, Setton LA. Measurement of intracellular strain on deformable substrates with texture correlation. J. Biomech. 2007;40:786–94. doi: 10.1016/j.jbiomech.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 129.Philip JT, Dahl KN. Nuclear mechanotransduction: response of the lamina to extracellular stress with implications in aging. J. Biomech. 2008;41:3164–70. doi: 10.1016/j.jbiomech.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 130.Chalut KJ, Kulangara K, Giacomelli MG, Wax A, Leong KW. Deformation of stem cell nuclei by nanotopographical cues. Soft Matter. 2010;6:1675–81. doi: 10.1039/B921206J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chao PH, West AC, Hung CT. Chondrocyte intracellular calcium, cytoskeletal organization, and gene expression responses to dynamic osmotic loading. Am. J. Physiol. Cell Physiol. 2006;291:C718–25. doi: 10.1152/ajpcell.00127.2005. [DOI] [PubMed] [Google Scholar]
- 132.Lucke B, McCutcheon M. The living cell as an osmotic system and its permeability to water. Physiol. Rev. 1932;12:68–139. [Google Scholar]
- 133.Richter K, Nessling M, Lichter P. Experimental evidence for the influence of molecular crowding on nuclear architecture. J. Cell Sci. 2007;120:1673–80. doi: 10.1242/jcs.03440. [DOI] [PubMed] [Google Scholar]
- 134.Finan JD, Chalut KJ, Wax A, Guilak F. Nonlinear osmotic properties of the cell nucleus. Ann. Biomed. Eng. 2009;37:477–91. doi: 10.1007/s10439-008-9618-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Finan JD, Leddy HA, Guilak F. Osmotic stress alters chromatin condensation and nucleocytoplasmic transport. Biochem. Biophys. Res. Commun. 2011;408:230–35. doi: 10.1016/j.bbrc.2011.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marenduzzo D, Finan K, Cook PR. The depletion attraction: an underappreciated force driving cellular organization. J. Cell Biol. 2006;175:681–86. doi: 10.1083/jcb.200609066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marenduzzo D, Micheletti C, Cook PR. Entropy-driven genome organization. Biophys. J. 2006;90:3712–21. doi: 10.1529/biophysj.105.077685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Finan JD, Guilak F. The effects of osmotic stress on the structure and function of the cell nucleus. J. Cell. Biochem. 2010;109:460–67. doi: 10.1002/jcb.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 140.Bacher CP, Reichenzeller M, Athale C, Herrmann H, Eils R. 4-D single particle tracking of synthetic and proteinaceous microspheres reveals preferential movement of nuclear particles along chromatin-poor tracks. BMC Cell Biol. 2004;5:45. doi: 10.1186/1471-2121-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–85. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- 142.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA. 2007;104:15619–24. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–16. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 145.Gerlitz G, Bustin M. Efficient cell migration requires global chromatin condensation. J. Cell Sci. 2010;123:2207–17. doi: 10.1242/jcs.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Olins AL, Zwerger M, Herrmann H, Zentgraf H, Simon AJ, et al. The human granulocyte nucleus: unusual nuclear envelope and heterochromatin composition. Eur. J. Cell Biol. 2008;87:279–90. doi: 10.1016/j.ejcb.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Prokocimer M, Margalit A, Gruenbaum Y. The nuclear lamina and its proposed roles in tumorigenesis: projection on the hematologic malignancies and future targeted therapy. J. Struct. Biol. 2006;155:351–60. doi: 10.1016/j.jsb.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 148.Konety BR, Getzenberg RH. Nuclear structural proteins as biomarkers of cancer. J. Cell. Biochem. 1999;32-33(Suppl):183–91. doi: 10.1002/(sici)1097-4644(1999)75:32+<183::aid-jcb22>3.3.co;2-1. [DOI] [PubMed] [Google Scholar]
- 149.Wolf K, Friedl P. Mapping proteolytic cancer cell-extracellular matrix interfaces. Clin. Exp. Metastasis. 2009;26:289–98. doi: 10.1007/s10585-008-9190-2. [DOI] [PubMed] [Google Scholar]
- 150.Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 2004;166:815–25. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hampoelz B, Azou-Gros Y, Fabre R, Markova O, Puech PH, Lecuit T. Microtubule-induced nuclear envelope fluctuations control chromatin dynamics in Drosophila embryos. Development. 2011;138:3377–86. doi: 10.1242/dev.065706. [DOI] [PubMed] [Google Scholar]
- 152.Lanctôt C, Kaspar C, Cremer T. Positioning of the mouse Hox gene clusters in the nuclei of developing embryos and differentiating embryoid bodies. Exp. Cell Res. 2007;313:1449–59. doi: 10.1016/j.yexcr.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 153.Goetze S, Mateos-Langerak J, Gierman HJ, de Leeuw W, Giromus O, et al. The three-dimensional structure of human interphase chromosomes is related to the transcriptome map. Mol. Cell. Biol. 2007;27:4475–87. doi: 10.1128/MCB.00208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–66. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gorisch SM, Wachsmuth M, Ittrich C, Bacher CP, Rippe K, Lichter P. Nuclear body movement is determined by chromatin accessibility and dynamics. Proc. Natl. Acad. Sci. USA. 2004;101:13221–26. doi: 10.1073/pnas.0402958101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Knight MM, van de Breevaart Bravenboer J, Lee DA, van Osch GJ, Weinans H, Bader DL. Cell and nucleus deformation in compressed chondrocyte-alginate constructs: temporal changes and calculation of cell modulus. Biochim. Biophys. Acta. 2002;1570:1–8. doi: 10.1016/s0304-4165(02)00144-7. [DOI] [PubMed] [Google Scholar]
- 157.Lee DA, Knight MM, Bolton JF, Idowu BD, Kayser MV, Bader DL. Chondrocyte deformation within compressed agarose constructs at the cellular and sub-cellular levels. J. Biomech. 2000;33:81–95. doi: 10.1016/s0021-9290(99)00160-8. [DOI] [PubMed] [Google Scholar]
- 158.Lei K, Zhang X, Ding X, Guo X, Chen M, et al. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc. Natl. Acad. Sci. USA. 2009;106:10207–12. doi: 10.1073/pnas.0812037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yu J, Lei K, Zhou M, Craft CM, Xu G, et al. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum. Mol. Genet. 2011;20:1061–73. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell. 2007;12:863–72. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 161.Schmitt J, Benavente R, Hodzic D, Hoog C, Stewart CL, Alsheimer M. Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc. Natl. Acad. Sci. USA. 2007;104:7426–31. doi: 10.1073/pnas.0609198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schneider M, Lu W, Neumann S, Brachner A, Gotzmann J, et al. Molecular mechanisms of centrosome and cytoskeleton anchorage at the nuclear envelope. Cell. Mol. Life Sci. 2011;68:1593–610. doi: 10.1007/s00018-010-0535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Göb E, Schmitt J, Benavente R, Alsheimer M. Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS ONE. 2010;5:e12072. doi: 10.1371/journal.pone.0012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Frohnert C, Schweizer S, Hoyer-Fender S. SPAG4L/SPAG4L-2 are testis-specific SUN domain proteins restricted to the apical nuclear envelope of round spermatids facing the acrosome. Mol. Hum. Reprod. 2011;17:207–18. doi: 10.1093/molehr/gaq099. [DOI] [PubMed] [Google Scholar]
- 165.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, et al. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA. 2009;106:2194–99. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 2003;160:685–97. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]