Abstract
Almost all primary transcripts in higher eukaryotes undergo several splicing events and alternative splicing is a major factor in generating proteomic diversity. Thus, the spliceosome, the ribonucleoprotein assembly that performs splicing, is a highly critical cellular machine and as expected, a very complex one. Indeed, the spliceosome is one of the largest, if not the largest, molecular machine in the cell with over 150 different components in human. A large fraction of the spliceosomal proteome is organized into ribonucleoprotein particles (snRNPs) by associating with one of the small nuclear RNAs (snRNAs), and the function of many spliceosomal proteins revolve around their association or interaction with the spliceosomal RNAs or the substrate pre-messenger RNAs. In addition to the complex web of protein-RNA interactions, an equally complex network of protein-protein interactions exists in the spliceosome which includes a number of large, conserved proteins with critical functions in the spliceosomal catalytic core. These include the largest conserved nuclear protein, Prp8, which plays a critical role in spliceosomal function in a hitherto unknown manner. Taken together, the large spliceosomal proteome and its dynamic nature has made it a highly challenging system to study, and at the same time, provides an exciting example of the evolution of a proteome around a backbone of primordial RNAs likely dating from the RNA World.
Keywords: Proteome, Prp8, RNA helicases, Spliceosome, Splicing
Introduction
The last decade has witnessed major advances in our understanding of spliceosomal function and has heightened our appreciation of its daunting complexity. Indeed, the spliceosome is a very large ribonucleoprotein assembly that performs the splicing reaction in eukaryotes with over 150 different components in human [1–3]. In addition, it is also highly dynamic and undergoes several major conformational rearrangements during its complicated assembly process. On the other hand, its critical function in the expression of almost all eukaryotic genes, which often undergo several splicing events, and its role in generating proteomic diversity via alternative splicing, highlights the need for functional accuracy. While the role of spliceosomal components, their mode of interaction and function, and the way the spliceosome ensures splicing fidelity are largely unknown, recent progress has started to provide insights into the function of this highly interesting and complex cellular machine. This review is by no means a comprehensive account of the function of the spliceosomal proteome, but rather attempts to provide a brief overview of our knowledge of the major spliceosomal proteome as a whole, and subsequently focus on a small number of exciting new discoveries on the role of several proteins whose function directly impacts the spliceosomal active site.
The evolution of a large RNP enzyme
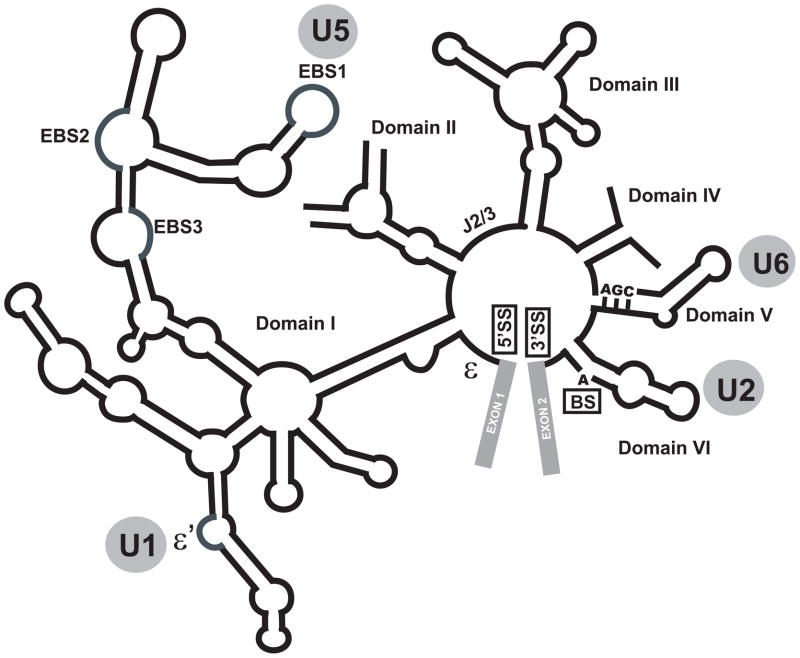
The close mechanistic similarities between the spliceosome and group II self-splicing introns, RNA enzymes found in all three kingdoms of life (Fig. 1), has raised the possibility that the two may be evolutionarily related [4,5]. Both systems are complex enzymes which catalyze a mechanistically identical two-step splicing reaction [6,7]. In both systems, the splicing reaction occurs in two steps entailing SN2-type transesterification reactions. In the first step, the 2′OH of the conserved branch site adenosine attacks the 5′ splice site and generates a free 5′ exon and a lariat intron that is still attached to the 3′ exon. In the second step, the 3′OH of the free 5′ exon launches an attach on the 3′ splice site, leading to a transesterification reaction that ligates the two exons together [1–5]. In addition to these mechanistic similarities, the snRNAs, the RNA components of the spliceosome, closely resemble a number of critical RNA domains of group II self-splicing introns. The major spliceosomes, which are the focus of this review, contain five snRNAs, named U1, U2, U4, U5 and U6. All except U4 contain functional counterparts in group II introns (Fig. 1, 2), and sequence swapping experiments have indicated the functional equivalence of U5 and U6 snRNAs with the EBS1 and DV elements in group II introns [6]. In the case of U6 snRNA, the similarity to the corresponding group II intron domain extends to both the primary sequence and the secondary structure, in addition to the extensive functional similarities (Fig. 1, 2) [4,6,7]. In addition, the snRNAs are highly conserved, and even point mutations in them, especially in the case of U6, are often incompatible with splicing [6,7]. These have led to the hypothesis that the spliceosomes may have evolved from group II intron-like ancestors and thus the snRNAs may be direct descendants of group II introns [8,9].
Fig. 1. Schematic representation of a group II self-splicing intron, from which spliceosomes may have evolved.
The exons are shown in gray rectangles. The identity of each RNA domain is indicated close to each domain. The sites of 5′ and 3′ splice sites and branch site are marked as 5′SS, 3′SS and BS, respectively. The functional and/or structural homologue of each RNA element in the spliceosome is shown in gray circles.
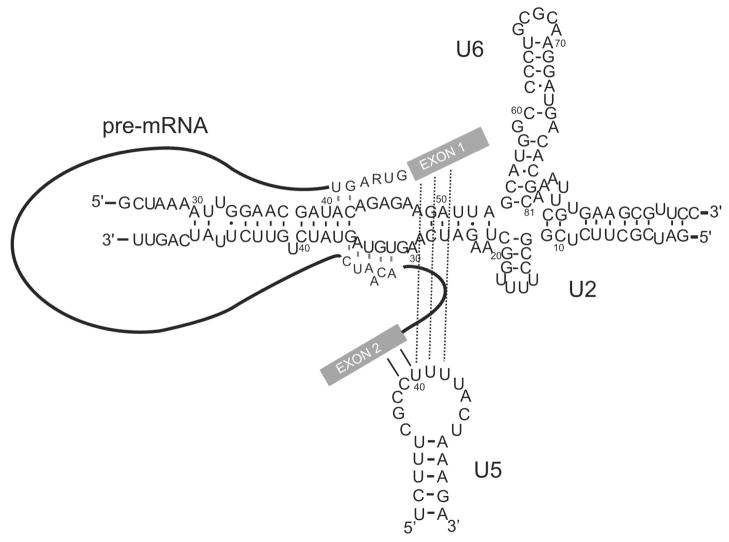
Fig. 2. The interaction of U6, U5 and U2 snRNAs with the pre-mRNA before the first step of splicing.
The basepairing interactions within each snRNA, between U6 and U2 snRNAs and between snRNAs and the pre-mRNA are shown. Numbers reflect the human numbering system. The exons in pre-mRNA are shown as solid rectangles, with the intron shown as a line. The sequence of the 5′ splice site and branch site are shown.
An extensive body of data support this possibility [6,10]. The snRNAs play critical roles in splicing by recognition of the 5′ splice site and branch site through basepairing (U1 and U2), regulating the catalytic activity of the spliceosome by basepairing to a catalytically critical snRNA (U4), binding to the reacting groups of the splicing reaction and juxtaposing them (U2 and U6), maintaining the positioning of the splicing intermediates for the second step of splicing (U5), keeping the reactive groups in a constrained conformation required for optimal catalysis (U2) and binding of functionally critical metal ions (U6)[4,6,7,10]. In the spliceosomal catalytic core, three snRNAs, U2, U5 and U6, maintain close contact with the splice sites and branch site and indeed, the first step of splicing occurs in the vicinity of an evolutionarily invariant sequence in U6, which may form part of the spliceosomal active site [6,11,12]. Finally, in vitro synthesized, protein-free U2 and U6 snRNAs can catalyze a two-step splicing reaction on small RNA oligonucleotides which is indistinguishable from the group II intron splicing and the second step of spliceosomal catalysis [13].
These evidence suggest that the snRNAs play a central role in many aspects of spliceosomal function. Comparing the size of snRNAs with the much larger group II introns raises the possibility that many group II intron domains are replaced by proteins in the spliceosomes during evolution, giving rise to the modern ribonucleoprotein eukaryotic splicing machines. While no one-to-one tally yet exists, it is easily possible to identify proteins which perform a function mediated by RNA elements in group II introns. For example, several U2-associated proteins function in initiating and stabilizing the U2 snRNA-branch site interaction (Fig. 2, see below). In group II introns, the U2 equivalent is covalently linked to the branch site via a hyperstable hairpin loop, which ensures the formation and stability of their interaction (Fig. 1). From this evolutionarily perspective, it is not surprising that a large fraction of spliceosomal proteins exist in association with an snRNA, often assisting it in its function (Fig. 3). However, as detailed below, many spliceosomal proteins are involved in regulatory functions not required in the context of a self-splicing intron and are likely more recent additions to the spliceosomal protein complement.
Fig. 3. The RNA and protein composition of each snRNP particle.
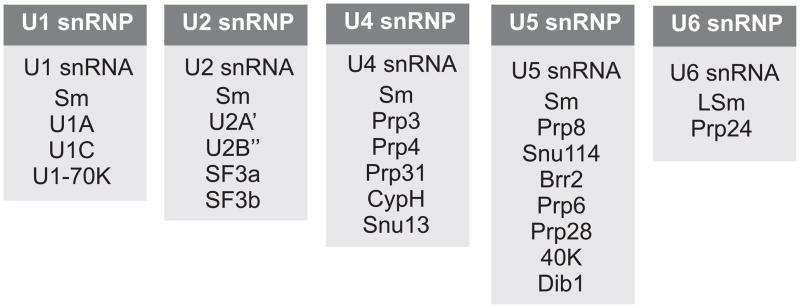
Note that all snRNPs except U6 carry a set of Sm proteins, whereas U6 instead contains LSm proteins.
How many spliceosomal proteins?
It is thought that the last common ancestor of eukaryotes had a highly evolved, fully functional spliceosome which resembled those found in modern eukaryotes. Although it likely contained significantly fewer components compared to even the smallest of modern spliceosomes, data indicate that most of the key components were present in even the earliest versions of the spliceosomes [14,15]. Indeed, a subset of the spliceosomal proteome shows a significant level of conservation among different eukaryotic species, with a number of spliceosomal protein being among the most conserved cellular proteins.
The study of spliceosomal proteome is complicated by the dynamic nature of the spliceosome, which undergoes a large number of compositional rearrangements each involving recruitment and discard of several proteins [Ref. 1 and see below]. Initial attempts using a mixture of spliceosomes at various stages of assembly placed the total number of spliceosomal proteins in a range of 150–300 distinct proteins [16,17]. More recent analyses, however, have taken advantage of the technical advances allowing the purification of individual spliceosomal complexes to near homogeneity, resulting in much more informative proteomic pictures [18–24]. Depending on the species, the latest studies indicate that the spliceosomes contain between nearly 80 proteins in the budding yeast to ~ 170 different proteins in human, with clear counterparts in human for almost all yeast spliceosomal proteins [1].
The protein complement of snRNPs and the non-snRNP complexes
1) The U1 snRNP and recognition of the 5′ splice site
The association of U1 snRNP with the 5′ splice site is one of the earliest events in the spliceosomal assembly pathway [1,3]. The recognition and binding of 5′ splice site is mediated both by basepairing of U1 snRNA to the 5′ splice site and through an intricate web of interactions between the pre-mRNA and U1C, a U1-specific protein [25]. U1, similar to all other spliceosomal snRNAs except U6, contains seven Sm proteins, namely B/B′, D1, D2, D3, E, F and G, which form a ring structure with a hole in the middle, through which a U-rich sequence in the snRNA passes [26–29]. In addition, it contains three U1-specific proteins, namely the RNA-recognition motif (RRM)-containing U1A and U1-70K and the zinc-finger domain containing U1C. The structure of an almost complete U1 snRNP particle, which contains the U1 snRNA and the ten associated proteins, has been determined by both cryo-EM and X-ray crystallography at 5.5 A resolution [30,31]. In addition, the structure of several of its components including U1A, U1C and the Sm ring either in part or in their entirety has been determined at higher resolution [for a review, see Ref. 32], which allowed a detailed interpretation of the low resolution structure of the entire particle. Importantly, this high-resolution structure indicated the presence of a number of interactions between U1C and the nucleotides at the 5′ end of U1, which basepair to the 5′ splice site, thus providing a structural basis for the dual RNA-protein recognition of the 5′ splice site [31].
2) The U2 snRNP and recognition of the branch site and 3′ splice site
Similar to the binding of U1 to the 5′ splice site, the recognition and interaction of U2 snRNP with the branch site and 3′ splice site is one of the earliest events in the spliceosomal assembly pathway (Fig. 4) [33,34]. U2 was initially described as a 12S particle consisting of a set of seven Sm proteins and only two additional proteins, the U2 specific proteins A′ and B″. The functionally active 17S U2 snRNP, however, contains several additional proteins including SF3a (itself composed of SF3a 120, 66 and 60 subunits) and SF3b (which contains SF3b155, 145, 130, 49, 14a, 14b and 10 subunits) [1,34]. The RNA-protein interactions within the 17S particle were studied by Dybkov and colleagues [35], which indicated a significant level of dynamic RNA-protein interactions within the particle. SF3a and b are required for the formation of the spliceosomal commitment complex and function in stabilizing the functionally critical basepairing interaction between the U2 snRNA and the branch site of introns, with at least one of the subunits (SF3b14a) directly contacting the branch site adenosine [1,36].
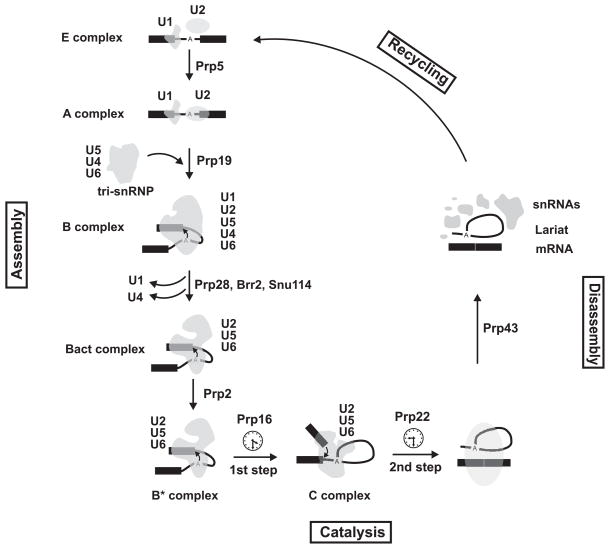
Fig. 4. The spliceosomal cycle.
The pre-mRNA is shown in black, and the location of the branch site is marked by an A. The exons are shown as solid rectangles and the intron is drawn as a thin line. The spliceosomal complexes and snRNPs are shown in gray. The identity of each spliceosomal complex and snRNP particle is shown. The main steps of spliceosomal cycle, assembly, catalysis, disassembly and recycling are marked on the graph. The main proofreading steps of spliceosomal cycle are marked by clock signs and the main RNA helicases involved in each conformational rearrangement are indicated. The snRNAs within each spliceosomal complex are shown.
In addition to the essential role of U2 snRNA and the associated proteins in recognition of the branch site of introns, a non-snRNP protein, the U2 auxiliary factor (U2AF), also significantly contributes to both branch site and 3′ splice site recognition during the early steps of spliceosomal assembly. In human, U2AF contains a large subunit, U2AF65, and a small subunit, U2AF35. U2AF65 contains an RNA binding domain through which it binds the polypyrimidine track at the 3′ end of introns, and an activation domain which mediates recruitment of the rest of the U2 snRNP particle to the vicinity of the branch site and stabilization of the basepairing interaction between U2 snRNA and the branch site nucleotides. U2AF35 interacts with the 3′ splice site and plays an important role in its recognition, especially in introns with weak splice site consensus sequences [1,34].
3) The U5 snRNP and its complement of regulatory proteins
In the spliceosomal catalytic core, the U5 snRNA is involved in aligning the exons to ensure their optimal positioning for the second step of splicing, which as mentioned above involves a nucleophilic attack by a hydroxyl group at the end of the newly cleaved 5′ exon on the 3′ splice site (Fig. 4) [34]. In addition, U5 binds several functionally critical U5-specific regulatory proteins, including Prp8/P220K, Brr2/200K, Snu114/116K, Prp6/102K, Prp28/100K, 40K, 15K and 52K in addition to the set of the seven common Sm proteins. Together, the U5 snRNA and its associated proteins form the largest RNP in the spliceosome and make up a large part of the fully assembled spliceosomes [34,37]. The structure of the isolated U5 snRNP has been determined using cryo electron microscopy (cryo-EM)[38]. Using a labeling approach, Sander and colleagues could show that the conserved loop I of U5 snRNA is located in a central position in the U5 snRNP particle, with its 5′ end located nearby, suggesting that the snRNA forms the center of the particle, with the associated proteins on the periphery [38]. Since loop I in U5 snRNA plays an important functional role in the spliceosomal catalytic center, these studies together with cryo-EM studies of the tri-snRNP particle (see below) suggest that the other catalytic center components likely contact the U5 snRNP particle at this site during the assembly of the spliceosome.
4) The U4/U6 di-snRNP: keeping the catalyst in check?
U6 is the most conserved spliceosomal snRNA and contains two invariant domains which play a critical functional role in splicing. Further, it contains a stemloop which is almost identical to the catalytic domain of group II introns (Fig. 1) and similar to its group II intron counterpart, binds a functionally required divalent cation [6,7,10,39–41]. As mentioned above, in the spliceosomal catalytic core, U6 closely interacts with the 5′ splice site at the time of the first step of splicing and along with U2 snRNA can catalyze a two-step splicing reaction in the absence of proteins in vitro ([11–13]. Perhaps in order to prevent it from prematurely forming a catalytically active spliceosomal complex, U6 is kept in an inactive conformation with the formation of its group II intron-like stemloop disrupted through a mutually exclusive basepairing interaction with U4. Current data do not indicate any additional functions for U4 snRNA except acting as a negative chaperon for U6. Once the basepaired U4/U6 complex joins the spliceosome in association with U5, the U4/U6 duplex is unwound in a tightly controlled manner and U2 snRNA replaces U4 as the basepairing partner of U6 (Fig. 4) [33,34].
Several additional characteristics make U6 an unusual snRNP particle: unlike the other snRNAs which are transcripts of RNA polymerase II, are transported to the cytoplasm during their assembly into snRNPs and receive a set of seven Sm proteins, U6 is transcribed by RNA polymerase III and is assembled into an RNP within the nucleus [33,34]. Further, it contains a set of LSm (like Sm) proteins composed of LSm proteins 2–8, which are thought to be an evolutionarily ancient paralog of Sm proteins [14]. In addition to the LSm proteins, U6 contains only one additional protein, Prp24, which contains four RNA recognition motif (RRM) domains, of which at least two contribute to binding to U6 snRNA [for a review, see Ref. 1]. It has been suggested that the LSm ring and Prp24 may act as chaperons, preparing the U6 snRNA for interacting with its binding partner, U4. The cryo-EM structures for the U6 and U4/U6 snRNP particles have been described [38,42]. It is interesting to note that while the proteins associated with other snRNAs at least partly remain bound to the RNA and perform functions in the spliceosome which in many cases are related to that of the snRNA, the U6 associated proteins leave the spliceosome after the integration of U6 into the assembling spliceosomes and do not seem to have a function beyond escorting U6 to the assembling spliceosomal complexes (see below).
5) The tri-snRNP: U5.U4/U6 complex
The U4/U6 di-snRNP and U5 snRNP form a ternary complex termed the “tri-snRNP” which is the form that can integrate into the assembling spliceosomes. Sander and colleagues determined the three dimensional structure of the 1.8 Mega Dalton tri-snRNP by cryo-EM [38] and could identify the position of the U5 and U4/U6 di-snRNPs within the tri-snRNP complex. The U5 snRNP accounted for ~ 60% of the total mass of the particle, and determined the overall shape of the complex. Labeling of different positions of the snRNAs and proteins within the complex permitted the identification of the location of the RNAs and several critical protein component, including Prp8, Brr2, Snu114, Prp6, Prp3 and Prp31 within the tri-snRNP. These studies indicated that the C-terminal domain of Brr2 was localized in the head domain where as that of prp8 and snu114 were localized in the neighboring so-called body of the particle [37,43], and provided a first glimpse into the structural organization of this important snRNP particle.
6) The SR proteins
A large portion of the non-snRNP spliceosomal proteins belong to the SR protein family, including ASF/SF2, SC35 and both subunits of U2AF. These typically consist of one or two copies of an N-terminal RRM and a serine and arginine-rich C-terminal domain (the RS domain). While several of the snRNP-associated proteins are related to SR proteins (e.g., U1-70K, Snu66, Srm160 and U5-100K/Prp28), many of them fall into the category of non-snRNP proteins [44–46]. SR proteins in general stimulate splicing by binding to exonic sequences and recruiting spliceosomal proteins and stabilizing RNA-RNA interactions. However it has been shown that the binding of SR proteins within an intron can be inhibitory [47,48]. The RS domains mediate protein-protein or RNA-protein interactions and their function is often regulated by phosphorylation. Several kinases with a high specificity for the serine residues of the RS domains have been described, including Clk/Sty and related kinases, DNA topoisomerase I and the SRPK family of kinases [1]. The RS domains have also been shown to interact with the pre-mRNA in the branch site region in a tethering experiment [49]. A functionally unusual SR protein, SRp38, has all the features of a classical SR protein with an N-terminal RRM and a C-terminal RS domain, however, it acts as a potent inhibitor, rather than activator, of splicing once dephosphorylated [50,51]. SRp38 is dephosphorylated upon heat shock treatment of nuclear extracts and during M phase in cell cycle, suggesting that it plays a role in global regulation of splicing under these conditions [52]. It should be mentioned that the spliceosome contains many non-snRNP, non-SR-family protein components, a detailed description of which has been recently reviewed in detail [1–3]. The remainder of this review focuses on the subset of spliceosomal proteins which directly impact the assembly of the active site and catalytic function of the spliceosome.
The proteome of spliceosomal complexes
As mentioned above, spliceosomes assemble on the pre-messenger RNAs in a highly elaborate, stepwise fashion. However, a complex containing all five snRNPs, termed the penta-snRNP, has been purified from yeast [53]. This finding suggests that the spliceosome can exist in a pre-assembled form which undergoes a myriad of rearrangements during the different stages of spliceosomal assembly by remodeling and stabilization of a subset of interactions. The first stage in the spliceosomal assembly pathway is the formation of the ATP-independent E or early complex, in which the 5′ splice site is recognized and bound by U1 snRNP, and the branch site and 3′ splice site are loosely recognized by U2 snRNPs (Fig. 4). In the next stage, the A complex, the association of U2 with the branch site/3′ splice site region is remodeled in an ATP-dependent fashion to stabilize its binding to this region. The proteomic analysis and cryo-EM structure of the human A complex has been described by Behzadnia and colleagues [21] at ~40Å. As expected, the complex contains nearly all U1 and U2 snRNP protein components and the pre-mRNA, in addition to close to 50 non-snRNP proteins [21]. These include members of the Prp19/CDC5 complex (see below), which were previously thought to be added to the spliceosomes at later stages.
Next, the U4/U6.U5 tri-snRNP joins the assembling spliceosome to form the B complex (Fig. 4). The B complex undergoes several conformational rearrangements, several of which are regulated by phosphorylation. In one, a tri-snRNP associated protein kinase, SRPK2, phosphorylates the RS domain of the RNA helicase Prp28. Prp28 phosphorylation is required for its stable association with the tri-snRNP and also for the tri-snRNP association with the assembling spliceosomes during B complex formation [54]. It has been shown that in yeast, Prp28 is an RNA helicase which mediates the dissociation of U1 snRNP from the 5′ splice site, however the yeast protein lacks the N-terminus RS domain which may explain its lack of stable association with the tri-snRNP [55,56]. The functional significance of the lack of RS domain in yeast Prp28 in B complex formation in yeast remains to be understood.
After the dissociation of U1 snRNP, the U4/U6 basepairing interaction is unwound by Brr2, a U5-associated RNA helicase [1,34]. U1 and U4 leave the assembling spliceosome, and U6 replaces U1 at the 5′ splice site as the B complex progresses toward the B* complex, which is catalytically active and is competent to perform the first step of splicing. A set of proteins including Prp19 stably associate with the assembling spliceosomes subsequent to U4 dissociation and likely play a role in the transition of the B complex to the catalytically active B* complex [1,19]. Two additional tri-snRNP components, Prp6 and Prp31, are phosphorylated by Prp4 kinase (Prp4K) as they incorporate into the B complex to form stable, functional assemblies [57]. Prp4K also interacts with Prp8 and Brr2, and thus may have additional roles in spliceosomal function [58]. The proteomic analysis and cryo-EM structure of the pre-catalytic complex B, which contains ~130 proteins in human and Drosophila and ~60 proteins in the yeast S. cerevisiae, have been described [59–61]. Comparison of the B complex proteome in the yeast and human has indicated a high level of evolutionary conservation. While over 85% of the yeast splicing factors found in purified spliceosomal B and B* complexes had clear evolutionary counterpart in human, a large fraction of the proteins found in human spliceosomal complexes have no yeast homologs, including several SR and hnRNP proteins [61]. Using antibodies containing colloidal gold, Wolf and colleagues [62] used electron microscopy to identify the location of the two exons, as well as SF3B155, a component of U2 snRNP, in B complexes. While this was a promising first step, further use of such molecular mapping approaches, combined with high resolution study of the individual components, are needed to provide a thorough understanding of this large spliceosomal assembly.
The B*Complex, in which the spliceosomes become catalytically competent, follows the B complex. Fabrizio and colleagues purified the B, Bact (which is a pre-catalytic complex formed immediately prior to B*) and C complexes from the budding yeast S. cerevisiae, and performed a thorough proteomic analysis. These studies indicated that conversion of B to Bact involves a dramatic compositional change [61]. In yeast, B and Bact complexes contain 60 and 40 proteins, respectively. This contrasts with the ~130 proteins found in B complexes in human and Drosophila [20,60]. As expected, U1 and U4/U6-specific proteins are present in the B complex but absent in Bact complex, in addition to the U6 LSm proteins [61]. 12 proteins that were not present in the B complex were recruited to the Bact complex, including the NTC proteins (see below). How all the above conformational changes will lead to the activation of the spliceosomes for performing the first step of splicing is poorly understood. Recent evidence suggest that the addition of Cwc25, a protein previously identified to be associated with the Prp19-associated complex, to spliceosomes that are fully assembled but not catalytically active can result in catalysis of the first step of splicing [63]. Two other factors, Yju2 and an unknown heat-resistant factor, likely function in the same final step of spliceosomal activation, although their function is again poorly understood [1,64]. Finally, ATP hydrolysis by the RNA helicase Prp2 and its dissociation from the spliceosome marks the very last known step before the catalysis of the first step of splicing and conversion of the Bact to the B* complex.
Recently, using a temperature-sensitive mutant of the RNA helicase Prp2, Warkocki and colleagues [65] succeeded in purifying spliceosomes stalled before the first step of splicing, which likely correspond to an inactive Bact complex, and determined their protein composition and structure using mass spectroscopy and cryo-EM studies. They could show that by the addition of Prp2, Spp2 and Cwc25, the stalled spliceosomes could undergo the first step of splicing and that the U2 snRNP proteins SF3a and SF3b were dissociated during this process, likely as a result of remodeling activity of Prp2. Once purified Prp16, Slu7, Prp18 and Prp22 were also added, the spliceosomes transitioned to the C complex and became competent to also perform the second splicing step [65].
Comparison of purified Bact and C complexes also indicated that further rearrangements occur during the conversion of these two complexes. In yeast this includes the recruitment of nine additional proteins including Prp22, Slu7, Cwc23, Cwc25, Prp43, Spp382/Ntr1, Ntr2 and Prp18 [61]. This is a longer list compared to the minimal set of proteins mentioned above which are absolutely required for the spliceosomes to undergo the second splicing step [65]. However, all proteins present in Bact were also present in the C complex in yeast.
In metazoans, the C complex contains 110–150 proteins compared to the 40–50 in yeast, which likely reflects the more complex splicing regulation in higher eukaryotes [18,20,60,61]. Proteomic analysis of an isolated but functionally active human C complex, in which the second step of splicing was shown to occur, indicated that it contains 150 proteins, of which 105 were also present in the B complex [18]. These included most of the U5 and U2 snRNP protein complement plus the Prp19-CDC5 complex and related factors, in addition to the components of the Retention and Splicing (RES) complex [18]. As expected, the U1 and U4 snRNP components were almost completely absent in the human complex C, in addition to U6 LSm proteins and a number of non-snRNP factors. Interestingly, Prp16, a helicase which plays a central role in the conformational transition between the first and second step spliceosomes, is absent in this complex, suggesting that it is either loosely associated with spliceosomes or that the purified complexes belong to a stage after the one in which Prp16 functions. On the other hand, several proteins in complex C seemed specific to this complex and were not found in other spliceosomal assemblies. These included proteins known to be important in the second step of splicing such as a number of DEAD-box helicases and peptidyl-prolyl isomerases, which likely help facilitate the conformational changes associated with the second step of splicing. Importantly, the core components of the exon junction complex (EJC), an assembly of proteins deposited ~20 nucleotides upstream of the exon-exon junctions in spliced mRNAs, are also significantly enriched in the C complex [18].
Further analysis of the human C complex showed that it contained a salt-stable RNP core which consisted of a number of critical components of the spliceosome, including members of the Prp19 complex and related factors as well as Prp8 and Snu114 and the U5-40K protein, in addition to equimolar amounts of the snRNAs U2, U5 and U6 and splicing intermediates [18]. Although this salt-stable complex was not functionally active, it may correspond to a stalled version of the catalytic core of the spliceosome [18]. Interestingly, the pre-mRNA splicing intermediates may play a role in stabilization of the C complex, since removal of part of the 3′ exon by nucleolytic cleavage in pre-assembled C complexes resulted in loss of a fraction of the particle in cryo-EM studies [66]. While a complete understanding of the significance of each conformational rearrangement in the spliceosome awaits future studies, a number of interesting findings have elucidated the functional importance of at least some of the spliceosomal rearrangements. The following passages discuss a select number of these findings which involve the spliceosomal catalytic core in detail.
A central role for helicases: the timers and the remodelers
At least eight DExD/H-box proteins function at various steps of the spliceosomal cycle and several show weak helicase activity in vitro [44,67]. The spliceosomal helicases play central roles in remodeling of RNA-RNA, RNA-protein and protein-protein interactions and in a number of cases, their remodeling activity seem to be tied to the spliceosomal quality control mechanisms [2,68,69]. UAP56, a DExD/H-box helicase originally discovered as a U2AF65-associated protein, is known to promote the early steps of spliceosome assembly through interaction with U2AF65 in an ATP-dependent way [71]. Further, it has been shown that UAP56 is also required in later steps of spliceosomal assembly, when it contacts U4 and U6 snRNAs and likely help in the unwinding of the U4/U6 basepaired duplexes, a step previously shown to be dependent on Brr2 (see below) [71]. Interestingly, UAP56 is also involved in a number of other aspects of cellular RNA metabolism, including mRNA nuclear export and cytoplasmic localization [72–75].
A major remodeling helicase in the spliceosome is the U5-associated DExD/H-box protein Brr2, a large protein (~2100 amino acids in yeast and human) which disrupts the interaction of U6 with its basepairing partners in both pre-catalytic and post-catalytic spliceosomes [34]. The protein contains two helicase-like domains followed by a Sec63 domain of unknown function. High resolution structural studies indicate that the second Sec63 domain is closely similar to the DNA helicase Hel308, which also contains similar helicase-like domains, suggesting mechanistic similarities between Brr2 and Hel308 [76,77]. The ATPase and helicase activity of Brr2, at least in vitro, is modulated by Snu114 and the C-terminal domain of Prp8, which also improves the binding of Brr2 to one of its targets, the U4/U6 basepaired complex [78,79].
The remodeling function of a number of spliceosomal helicases is tied to the spliceosomal quality control. The activation of this class of helicases leads to remodeling of the existing spliceosomal conformation to the next stage in the spliceosomal cycle and thus limits the amount of time the spliceosome dwells in that particular conformation. If this conformation is associated with a given function, for example performing the first step of splicing, the remodeling activity of the helicase would limit the amount of time available for catalyzing the first step. While the allocated amount of time is more than enough for optimal substrates to react, it is not sufficient to allow the suboptimal splice sites, which react more slowly, to go through the first step. Thus, the helicase-mediated remodeling would prevent suboptimal splice sites and branch sites from undergoing splicing, providing a quality control mechanism [68–70].
Perhaps the best-studied example of this class of helicases is Prp16, which associates with the spliceosomes before the first step of splicing; however, it functions in proofreading of the first step of splicing by performing its remodeling action after the first step of splicing on optimal substrates has occurred [1,33,34,70]. Mutations that impair the ATPase or helicase activity of Prp16 would result in splicing of suboptimal branch site and 5′ splice site substrates which would have been otherwise discarded. Interestingly, the Prp16-mediated rejection of erroneous splicing events is reversible, requiring a downstream discard pathway which involves Prp43, a helicase that mediates spliceosomal disassembly [80]. The direct molecular target of Prp16 is not yet determined, however, it has been shown to directly interact with the pre-messenger RNA in the 3′ splice site region and also to have a role in remodeling of one of the U6/U2 basepaired helices [81,82].
Another RNA helicase, Prp22, interacts with a region immediately downstream of the 3′ splice site in the fully spliced mRNAs after the second step of splicing [81,83]. Through its 3′ to 5′ helicase activity, it likely disrupts the mRNA/U5 snRNP contacts, thus leading to the release of the spliced mRNA at the end of the splicing cycle [83]. Mayas and colleagues [84] studied the effect of mutations in Prp22 that impaired its ATPase or RNA unwindase activity, and showed that these mutations resulted in loss of splicing fidelity, since pre-mRNAs with mutations in 3′ splice site could be spliced with much higher efficiency. These data indicated that the Prp22-mediated step in spliceosomal disassembly is yet another quality control checkpoint which controls the fidelity of the second step of splicing.
Prp43, a DExD/H-box RNA helicase which is involved in ribosomal biogenesis [85,86] also plays an important role in the disassembly of spliceosomes. Together with Ntr1/Spp382 and Ntr2, Prp43 forms the NTR complex which mediates spliceosomal disassembly. Ntr2 interacts with Brr2 and U5, suggesting that it may be involved in the recruitment of the NTR complex to the post-catalytic spliceosomes [87,88]. Ntr1 functions by activating the inherently weak helicase activity of Prp43, which upon activation triggers the release of the lariat intron from the post-catalytic spliceosomes [70, 89]. Mutations in Ntr1 and Prp43 suppressed the splicing defects of Prp8 and Prp38 mutants, suggesting that Prp43 is yet another quality control checkpoint in the spliceosomal cycle [90]. More recently, it has been shown that Prp43 is also involved in rejection of suboptimal 5′ exon and lariat intermediates which are blocked from undergoing splicing in a Prp16-dependent way (see above). This data suggests that similar to a normal splicing reaction, aberrant, stalled spliceosomes also depend on Prp43 for disassembly [91].
In addition to the examples discussed above, several other helicases play important roles at various stages of spliceosomal assembly, for example, Prp28 mediates the release of U1 snRNA from the 5′ splice site in the B complex (see above) and Prp5, also a DExD/H-box protein, functions in recruiting the U2 snRNP to the pre-mRNA in early steps of spliceosomal assembly ([1,3,92].
A G-protein in control
As mentioned above, Brr2 is involved in two critical remodeling steps in the spliceosomal cycle, namely, the unwinding of U4/U6 duplex as the B complex is transitioning to the B* complex, and the release of U6 from its interaction with U2 after the second step of splicing. These two functions of Brr2 are in turn regulated by Snu114, a U5 snRNP-associated GTPase with significant homology to the ribosomal translocase EF-G [79]. In an elegant series of experiments, Small and colleagues showed that Snu114 functions as a classic regulatory G protein, serving as a signal-dependent switch [79]. Based on their studies, Snu114 transduces signals to Brr2, thus regulating its molecular remodeler function. Binding of Snu114 to GDP prevented Brr2 from performing its function in both spliceosomal assembly and disassembly, while replacing GDP with GTP or a non-hydrolyzable GTP analog removed this block [79]. These findings are consistent with previous observations, which indicated that Snu114 extensively interacts with U5-specific proteins Prp8, Prp28 and Brr2 [93,94]. Further, Brenner and colleagues [93] showed that truncation mutations in the C-terminal domain of Snu114 allows the initial stages of spliceosomal assembly to happen but blocks the release of U4 during the conversion of B to B* complex. Finally, mutations in the GTPase domain of Snu114 prevent its interaction with Prp8 and U5 snRNA and blocks the assembly of U5 snRNP particle [93]. It is plausible that the interaction of Snu114 with Prp8 through its GTPase domain may be one of the mechanisms by which Prp8 and other factors regulate the spliceosomal cycle.
Activating the spliceosome: the role of Prp19 complex
A set of 7 proteins in human and 8–11 proteins in yeast associate with Prp19 through a complex set of interactions to form the Prp19/CDC5 complex (in humans) or the so-called NineTeen Complex (NTC) in yeast (Table 1) [95,96]. The NTC complex is required for stable association of U5 and U6 with the exonic sequences and the 5′ splice site in the assembling spliceosomes after the dissociation of U4 and U1 snRNAs [96–98]. It has been shown that NTC is involved in mediating a conformational change in U6 which involves remodeling of its interaction with the 5′ splice site and removal of the LSm proteins [19,96]. In human, Prp19 is also found together with CDC5 in a larger complex containing ~30 proteins and is likely to play a role in the second step of splicing [99]. Recently, it has been shown that Cwc21, a member of the NTC, binds directly to both Prp8 and Snu114 and thus, may mediate the interaction of the NTC with the U5 snRNP [100,101]. Another study has shown that even in the very divergent trypanosomes, the interaction of Cwc21 and U5 snRNP is conserved and essential for splicing, which further underscores its functional importance [102]. Taken together, the existing data suggest that NTC is involved in stabilizing a number of RNA-RNA interactions which are among the latest steps before formation of the catalytically competent spliceosomes [1,63].
Table 1.
The protein composition of the Prp19 complex in human and yeast S. cervisiae. The first four yeast proteins are the homologues of the first four human proteins. The last five components in the yeast column are written in the order of discovery. Only the 8 characterized components of the NineTeen complex have been listed, although a number of additional currently uncharacterized components have also been co-purified with this complex in yeast [94, 122, 123].
| NineTeen Complex (yeast) | Prp19/CDC5 complex (Human) |
|---|---|
| Prp19p | hPrp19 |
| Cef1p/NTC 85 | CDC5L |
| Prp46p* | PRL1 |
| Snt309p/NTC25 | SPF27 |
| NTC 30 | AD002 |
| NTC 20 | CTNNBL1 |
| NTC 90 | HSP73 |
| NTC 77 | |
| NTC 31 |
The special case of Prp8
Prp8 is arguably the most interesting of the spliceosomal proteins. It is an unusually conserved protein with 61% identity between yeast and human. However, it lacks clear functional motifs in its ~ 2300 amino acid length and the few functional domains that have been discerned are degenerate and likely perform functions unrelated to the original cellular role of the motif [94]. On the other hand, Prp8 clearly plays a very critical role in the spliceosomal active site. It has been shown that Prp8 interacts with several spliceosomal proteins including Snu114 and Brr2 [1,94,103]. Further, it has been crosslinked to the 5′ and 3′ splice sites and the branch site of the pre-messenger RNAs, in addition to crosslinks to U5 and U6 snRNAs, which indicates that it is present in the spliceosomal catalytic core and in direct contact with all critical players in the splicing reaction [94,104]. Mutations in Prp8 are associated with a wide range of spliceosomal defects including alterations in the ability of spliceosomes to reject suboptimal splice sites and suppression of defects caused by mutations in other spliceosomal components, for example mutations in Prp28, Brr2, U4 and U6 snRNAs [94,105,106]. A subset of these phenotypes, which are observed in aggregate with certain Prp8 alleles, are likely to be caused by abnormal stabilization of a particular spliceosomal conformation by the Prp8 mutant alleles. As mentioned above, the spliceosomal fidelity mechanism depends on timely conversion of one conformation to the next one in the spliceosomal cycle. Thus, hyperstabilization of one conformation, for example the one which is conducive to catalysis of the first step of splicing, would result in splicing of substrates with suboptimal 5′ splice sites [2,68,69,107].
Very little is known about the way Prp8 performs its function. Transposon-mediated screening assays indicated that large regions of Prp8 are highly sensitive to insertion of transposons and likely function as a single structural unit [104,108]. Apart from a degenerate nuclear localization signal close to its N terminus and a degenerate RRM motif in the middle of the protein, only two other functional domains have been identified in the ~2300 amino acid-long Prp8. A degenerate variant of the MPN/Jab1 domain found in deubiquitinating enzymes is found near the C terminus of Prp8 [94]. A high resolution structure of this domain has shown that the metal binding site of the isopeptidase center is impaired and thus, it likely does not function as a deubiquitinating enzyme [109,110]. In vitro, a fragment of Prp8 containing this domain could directly bind to ubiquitin with an affinity that was comparable to other known ubiquitin binding proteins [111]. A number of known Prp8 mutations that disrupted splicing also disrupted ubiquitin binding, raising the possibility of a functional role for ubiquitination in splicing regulation. Interestingly, proteomic analyses have indicated that a number of spliceosomal factors, including Sad1, Snu114, Rse1 and Prp8 itself are ubiquitinated in vivo, and further, Prp19 exhibits E3 ubiquitin ligase activity in vitro [111–115]. Further, ubiquitin plays a role in the formation and maintenance of tri-snRNP, likely by modulating the Prp8-mediated regulation of the function of Brr2 [115]. Alternatively, the MPN/Jab1 domain may function as a protein-protein interaction platform. A number of mutations in Prp8 which fall in this domain result in the hereditary blindness Retinitis Pigmentosa [94], and these mutations, once introduced into Prp8, result in weakening of the interaction of Prp8 with Brr2 and Snu114 [109].
High resolution studies of another fragment of Prp8 which encompasses a highly conserved region (69% amino acid identity between yeast and human) close to its C-terminal domain, indicated the presence of a β–hairpin finger resembling those found in ribosomal proteins and a degenerate RNase H-like domain [116–118]. The overall geometry of the RNase H-like motif at the level of secondary and tertiary structure was well conserved but the primary sequence showed a much lower level of conservation, for example, only one of the active site residues was present. Interestingly, amino acids located adjacent to the active site in this degenerate RNase H domain had been previously shown to be close to the 5′ splice site in precatalytic spliceosomes [119], suggesting that the degenerate active site may nonetheless form part of the catalytic core of the spliceosome. However, mutation of the single conserved amino acid at the RNase H-like active site either did not have a phenotype, which could indicate redundancy or lack of a critical function [118]; or its effect could not be examined since the mutation induced misfolding of the protein fragment [116, see also 117]. Interestingly, the fragment demonstrated an affinity for duplex RNAs containing four-way junction conformations [118]. In activated spliceosomes, U2 and U6 snRNAs are predicted to form such a structure [120,121]. While these results are intriguing, more in-depth analyses are needed to elucidate the role of this domain of Prp8 in spliceosomal function.
Concluding remarks
In its heart, the spliceosome is ultimately an enzyme with an RNA substrate. It is also a ribonucleoprotein complex evolved around a core of five snRNAs which most likely are descendants of an ancient catalytic RNA. Unlike the self-splicing ribozymes, the spliceosome has evolved to splice a diverse variety of substrates in trans in a tightly regulated way and thus, in addition to proteins which have replaced the function of the lost RNA elements, many of the spliceosomal proteins function in substrate recognition and regulation of splicing. As expected from an RNP enzyme with an RNA substrate, a large fraction of spliceosomal proteins are RNA-binding proteins or function in remodeling of RNA-RNA and RNA-protein interactions. Although our knowledge of the organization and function of the many spliceosomal complexes is rudimentary at present, recent proteomic and structural biology studies combined with genetic and biochemical approaches have started to provide major insights into the way spliceosomes function. The last decade has seen a vast increase in our knowledge of all aspects of spliceosomal function. The next decade is likely to witness the emergence of a detailed working model for this large ribonucleoprotein cellular machine.
Acknowledgments
This work was supported by the NIH grant RO1GM078572 and the accompanying ARRA administrative supplement to S.V.
Abbreviations
- RNP
ribonucleoprotein
- snRNA
small nuclear RNA
Footnotes
Conflict of interest statement
The author declares no conflict of interest.
References
- 1.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Smith DJ, Query CC, Konarska MM. “nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman AJ, Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr Opin Struct Biol. 2010;20:82–89. doi: 10.1016/j.sbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Dayie KT, Padgett RA. A glimpse into the active site of a group II intron and maybe the spliceosome, too. RNA. 2008;14:1697–1703. doi: 10.1261/rna.1154408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keating KS, Toor N, Perlman PS, Pyle AM. A structural analysis of the group II intron active site and implications for the spliceosome. RNA. 2010;16:1–9. doi: 10.1261/rna.1791310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valadkhan S. Role of the snRNAs in spliceosomal active site. RNA Biol. 2010;7 doi: 10.4161/rna.7.3.12089. ePublication. [DOI] [PubMed] [Google Scholar]
- 7.Michel F, Costa M, Westhof E. The ribozyme core of group II introns: a structure in want of partners. Trends Biochem Sci. 2009;34:189–199. doi: 10.1016/j.tibs.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Sharp PA. On the origin of RNA splicing and introns. Cell. 1985;42:397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 9.Cech TR. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986;44:207–10. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- 10.Valadkhan S. The spliceosome: a ribozyme at heart? Biol Chem. 2007;388:693–697. doi: 10.1515/BC.2007.080. [DOI] [PubMed] [Google Scholar]
- 11.Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 13.Valadkhan S, Mohammadi A, Jaladat Y, Geisler S. Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc Natl Acad Sci USA. 2009;106:11901–11906. doi: 10.1073/pnas.0902020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veretnik S, Wills C, Youkharibache P, Valas RE, et al. Sm/LSm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comput Biol. 2009;5:e1000315. doi: 10.1371/journal.pcbi.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins L, Penny D. Complex spliceosomal organization ancestral to extant eukaryotes. Mol Biol Evol. 2005;22:1053–1066. doi: 10.1093/molbev/msi091. [DOI] [PubMed] [Google Scholar]
- 16.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 18.Bessonov S, Anokhina M, Will CL, Urlaub H, et al. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 19.Makarova OV, Makarov EM, Urlaub H, Will CL, et al. A subset of human 35S U5 proteins, including prp19, function prior to catalytic step 1 of splicing. EMBO J. 2004;23:2381–2391. doi: 10.1038/sj.emboj.7600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stark H, Lührmann R, Behzadnia N, Golas MM, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartmuth K, Urlaub H, Vornlocher H, Will CL, et al. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, et al. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarov EM, Makarova OV, Urlaub H, Gentzel M, et al. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 25.Du H, Rosbash M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- 26.Achsel T, Brahms H, Kastner B, Bachi A, et al. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mura C, Cascio D, Sawaya MR, Eisenberg DS. The crystal structure of a heptameric archaeal Sm protein: implications for the eukaryotic snRNP core. Proc Natl Acad Sci USA. 2001;98:5532–5537. doi: 10.1073/pnas.091102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaric B, Chami M, Rémigy H, Engel A, et al. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J Biol Chem. 2005;280:16066–16075. doi: 10.1074/jbc.M414481200. [DOI] [PubMed] [Google Scholar]
- 29.Kambach C, Walke S, Young R, Avis JM, et al. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 30.Stark H, Dube P, Lührmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- 31.Pomeranz Krummel DA, Oubridge C, Leung AKW, Li J, et al. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie DB, Schellenberg MJ, MacMillan AM. Spliceosome structure: piece by piece. Biochim Biophys Acta. 2009;1789:624–633. doi: 10.1016/j.bbagrm.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Nilsen TW. RNA-RNA interactions in nuclear pre-mRNA splicing. In: Simons R, Grunberg-Manago M, editors. RNA structure and function. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1998. pp. 279–307. [Google Scholar]
- 34.Will CL, Luhrmann R. Spliceosome structure and function. In: Gesteland R, Cech T, Atkins J, editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 369–400. [Google Scholar]
- 35.Dybkov O, Will CL, Deckert J, Behzadnia N, et al. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol Cell Biol. 2006;26:2803–2816. doi: 10.1128/MCB.26.7.2803-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sattler M, Spadaccini R, Reidt U, Dybkov O, et al. Biochemical and NMR analyses of an SF3b155-p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA. 2006;12:410–425. doi: 10.1261/rna.2271406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lührmann R, Stark H. Structural mapping of spliceosomes by electron microscopy. Curr Opin Struct Biol. 2009;19:96–102. doi: 10.1016/j.sbi.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Sander B, Golas MM, Makarov EM, Brahms H, et al. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 di-snRNP within U4/U6.U5 tri-snRNP as revealed by electron cryomicroscopy. Mol Cell. 2006;24:267–278. doi: 10.1016/j.molcel.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 39.Toor N, Keating KS, Pyle AM. Structural insights into RNA splicing. Curr Opin Struct Biol. 2009;19:260–266. doi: 10.1016/j.sbi.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huppler A, Nikstad LJ, Allmann AM, Brow DA, et al. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat Struct Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- 41.Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karaduman R, Dube P, Stark H, Fabrizio P, et al. Structure of yeast U6 snRNPs: arrangement of prp24p and the LSm complex as revealed by electron microscopy. RNA. 2008;14:2528–2537. doi: 10.1261/rna.1369808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Häcker I, Sander B, Golas MM, Wolf E, et al. Localization of prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat Struct Mol Biol. 2008;15:1206–1212. doi: 10.1038/nsmb.1506. [DOI] [PubMed] [Google Scholar]
- 44.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors clocks, springs and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 45.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin S, Fu X. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- 47.Ibrahim EC, Schaal TD, Hertel KJ, Reed R, et al. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci USA. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 49.Shen H, Kan JLC, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 50.Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 51.Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 52.Nilsen TW. Too hot to splice. Nat Struct Mol Biol. 2004;11:208–209. doi: 10.1038/nsmb0304-208. [DOI] [PubMed] [Google Scholar]
- 53.Stevens SW, Ryan DE, Ge HY, Moore RE, et al. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 54.Mathew R, Hartmuth K, Möhlmann S, Urlaub H, et al. Phosphorylation of human prp28 by SRpk2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat Struct Mol Biol. 2008;15:435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 55.Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 56.Chen JY, Stands L, Staley JP, Jackups RR, et al. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of prp28p, an essential DEAD box splicing factor. Mol Cell. 2001;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 57.Schneider M, Hsiao H, Will CL, Giet R, et al. Human prp4 kinase is required for stable tri-snRNP association during spliceosomal B complex formation. Nat Struct Mol Biol. 2010;17:216–221. doi: 10.1038/nsmb.1718. [DOI] [PubMed] [Google Scholar]
- 58.Bottner CA, Schmidt H, Vogel S, Michele M, et al. Multiple genetic and biochemical interactions of Brr2, Prp8, Prp31, Prp1 and Prp4 kinase suggest a function in the control of the activation of spliceosomes in Schizosaccharomyces pombe. Curr Genet. 2005;48:151–161. doi: 10.1007/s00294-005-0013-6. [DOI] [PubMed] [Google Scholar]
- 59.Boehringer D, Makarov EM, Sander B, Makarova OV, et al. Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat Struct Mol Biol. 2004;11:463–468. doi: 10.1038/nsmb761. [DOI] [PubMed] [Google Scholar]
- 60.Herold N, Will CL, Wolf E, Kastner B, et al. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol. 2009;29:281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fabrizio P, Dannenberg J, Dube P, Kastner B, et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 62.Wolf E, Kastner B, Deckert J, Merz C, et al. Exon, intron and splice site locations in the spliceosomal B complex. EMBO J. 2009;28:2283–2292. doi: 10.1038/emboj.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chiu Y, Liu Y, Chiang T, Yeh T, et al. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol Cell Biol. 2009;29:5671–5678. doi: 10.1128/MCB.00773-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, Chen H, Wu N, Cheng S. A novel splicing factor, Yju2, is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol Cell Biol. 2007;27:5403–5413. doi: 10.1128/MCB.00346-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, et al. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol. 2009;16:1237–1243. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 66.Ilagan J, Yuh P, Chalkley RJ, Burlingame AL, et al. The role of exon sequences in C complex spliceosome structure. J Mol Biol. 2009;394:363–375. doi: 10.1016/j.jmb.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jankowsky E, Fairman ME. RNA helicases--one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 68.Konarska MM, Query CC. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- 69.Query CC, Konarska MM. Splicing fidelity revisited. Nat Struct Mol Biol. 2006;13:472–474. doi: 10.1038/nsmb0606-472. [DOI] [PubMed] [Google Scholar]
- 70.Boon KL, Auchynnikava T, Edwalds-Gilbert G, Barrass JD, et al. Yeast Ntr1/Spp382 mediates Prp43 function in postspliceosomes. Mol Cell Biol. 2006;26:6016–23. doi: 10.1128/MCB.02347-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen H, Zheng X, Shen J, Zhang L, et al. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev. 2008;22:1796–1803. doi: 10.1101/gad.1657308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gatfield D, Le Hir H, Schmitt C, Braun IC, et al. The DExH/D box protein Hel/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol. 2001;11:1716–1721. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- 73.Luo ML, Zhou Z, Magni K, Christoforides C, et al. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 74.Herold A, Teixeira L, Izaurralde E. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J. 2003;22:2472–2483. doi: 10.1093/emboj/cdg233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meignin C, Davis I. UAP56 RNA helicase is required for axis specification and cytoplasmic mRNA localization in Drosophila. Dev Biol. 2008;315:89–98. doi: 10.1016/j.ydbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 76.Zhang L, Xu T, Maeder C, Bud L, et al. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009;16:731–739. doi: 10.1038/nsmb.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pena V, Jovin SM, Fabrizio P, Orlowski J, et al. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell. 2009;35:454–466. doi: 10.1016/j.molcel.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 78.Maeder C, Kutach AK, Guthrie C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol. 2009;16:42–48. doi: 10.1038/nsmb.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell. 2010;39:385–95. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McPheeters DS, Muhlenkamp P. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol Cell Biol. 2003;23:4174–4186. doi: 10.1128/MCB.23.12.4174-4186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mefford MA, Staley JP. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA. 2009;15:1386–1397. doi: 10.1261/rna.1582609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Combs DJ, Nagel RJ, Ares M, Stevens SW. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leeds NB, Small EC, Hiley SL, Hughes TR, et al. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsai R, Fu R, Yeh F, Tseng C, et al. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai R, Tseng C, Lee P, Chen H, et al. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol Cell Biol. 2007;27:8027–8037. doi: 10.1128/MCB.01213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tanaka N, Aronova A, Schwer B. Ntr1 activates the prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pandit S, Lynn B, Rymond BC. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci USA. 2006;103:13700–13705. doi: 10.1073/pnas.0603188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mayas RM, Maita H, Semlow DR, Staley JP. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0906022107. Epublication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kosowski TR, Keys HR, Quan TK, Ruby SW. DExD/H-box Prp5 protein is in the spliceosome during most of the splicing cycle. RNA. 2009;15:1345–1362. doi: 10.1261/rna.1065209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brenner TJ, Guthrie C. Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain. RNA. 2006;12:862–871. doi: 10.1261/rna.2319806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grote M, Wolf E, Will CL, Lemm I, et al. Molecular architecture of the human Prp19/Cdc5l complex. Mol Cell Biol. 2010;30:2105–2119. doi: 10.1128/MCB.01505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chan S, Kao D, Tsai W, Cheng S. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 97.Chan S, Cheng S. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem. 2005;280:31190–31199. doi: 10.1074/jbc.M505060200. [DOI] [PubMed] [Google Scholar]
- 98.McGrail JC, Krause A, O’Keefe RT. The RNA binding protein Cwc2 interacts directly with the U6 snRNA to link the nineteen complex to the spliceosome during pre-mRNA splicing. Nucleic Acids Res. 2009;37:4205–4217. doi: 10.1093/nar/gkp341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ajuh P, Kuster B, Panov K, Zomerdijk JC, et al. Functional analysis of the human Cdc5l complex and identification of its components by mass spectrometry. EMBO J. 2000;19:6569–6581. doi: 10.1093/emboj/19.23.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grainger RJ, Barrass JD, Jacquier A, Rain J, et al. Physical and genetic interactions of yeast Cwc21p, an ortholog of human Srm300/Srrm2, suggest a role at the catalytic center of the spliceosome. RNA. 2009;15:2161–2173. doi: 10.1261/rna.1908309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khanna M, Van Bakel H, Tang X, Calarco JA, et al. A systematic characterization of Cwc21, the yeast ortholog of the human spliceosomal protein Srm300. RNA. 2009;15:2174–85. doi: 10.1261/rna.1790509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Luz Ambrósio D, Lee JH, Panigrahi AK, Nguyen TN, et al. Spliceosomal proteomics in Trypanosoma brucei reveal new RNA splicing factors. Eukaryot Cell. 2009;8:990–1000. doi: 10.1128/EC.00075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu S, Rauhut R, Vornlocher H, Lührmann R. The network of protein-protein interactions within the human U4/U6. U5 tri-snRNP. RNA. 2006;12:1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Turner IA, Norman CM, Churcher MJ, Newman AJ. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA. 2006;12:375–386. doi: 10.1261/rna.2229706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kuhn AN, Brow DA. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics. 2000;155:1667–1682. doi: 10.1093/genetics/155.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuhn AN, Reichl EM, Brow DA. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc Natl Acad Sci USA. 2002;99:9145–9149. doi: 10.1073/pnas.102304299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal Prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 108.Boon K, Norman CM, Grainger RJ, Newman AJ, et al. Prp8p dissection reveals domain structure and protein interaction sites. RNA. 2006;12:198–205. doi: 10.1261/rna.2281306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pena V, Liu S, Bujnicki JM, Lührmann R, et al. Structure of a multipartite protein-protein interaction domain in splicing factor Prp8 and its link to retinitis pigmentosa. Mol Cell. 2007;25:615–624. doi: 10.1016/j.molcel.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 110.Zhang L, Shen J, Guarnieri MT, Heroux A, et al. Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants. Protein Sci. 2007;16:1024–1031. doi: 10.1110/ps.072872007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bellare P, Kutach AK, Rines AK, Guthrie C, et al. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA. 2006;12:292–302. doi: 10.1261/rna.2152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hatakeyama S, Yada M, Matsumoto M, Ishida N, et al. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 113.Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, et al. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bellare P, Small EC, Huang X, Wohlschlegel JA, et al. A role for ubiquitin in the spliceosome assembly pathway. Nat Struct Mol Biol. 2008;15:444–451. doi: 10.1038/nsmb.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang K, Zhang L, Xu T, Heroux A, et al. Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci USA. 2008;105:13817–13822. doi: 10.1073/pnas.0805960105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pena V, Rozov A, Fabrizio P, Lührmann R, et al. Structure and function of an RNAse H domain at the heart of the spliceosome. EMBO J. 2008;27:2929–2940. doi: 10.1038/emboj.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ritchie DB, Schellenberg MJ, Gesner EM, Raithatha SA, et al. Structural elucidation of a Prp8 core domain from the heart of the spliceosome. Nat Struct Mol Biol. 2008;15:1199–1205. doi: 10.1038/nsmb.1505. [DOI] [PubMed] [Google Scholar]
- 119.Reyes JL, Gustafson EH, Luo HR, Moore MJ, et al. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA. 1999;5:167–179. doi: 10.1017/s1355838299981785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sun JS, Manley JL. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- 121.Sashital DG, Cornilescu G, McManus CJ, Brow DA, et al. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol. 2004;11:1237–42. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- 122.Hogg R, McGrail JC, O’Keefe RT. The function of the NineTeen Complex (NTC) in regulating spliceosome conformations fidelity during pre-mRNA splicing. Biochem Soc Trans. 2010;38:1110– 1115. doi: 10.1042/BST0381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]