A functional screen identified MARK4 as a positive regulator of axonemal extension and ciliogenesis via its interaction with the mother centriolar protein ODF2.
Abstract
Despite the critical contributions of cilia to embryonic development and human health, key regulators of cilia formation await identification. In this paper, a functional RNA interference–based screen linked 30 novel protein kinases with ciliogenesis. Of them, we have studied the role of the microtubule (MT)-associated protein/MT affinity regulating kinase 4 (MARK4) in depth. MARK4 associated with the basal body and ciliary axoneme in human and murine cell lines. Ultrastructural and functional analyses established that MARK4 kinase activity was required for initiation of axoneme extension. We identified the mother centriolar protein ODF2 as an interaction partner of MARK4 and showed that ODF2 localization to the centriole partially depended on MARK4. Our data indicated that, upon MARK4 or ODF2 knockdown, the ciliary program arrested before the complete removal of the CP110–Cep97 inhibitory complex from the mother centriole, suggesting that these proteins act at this level of axonemal extension. We propose that MARK4 is a critical positive regulator of early steps in ciliogenesis.
Introduction
The primary cilium is a conserved microtubule (MT)-based structure that plays important roles in coordinating key signaling pathways in both embryonic and adult tissues (D’Angelo and Franco, 2009). The physiological significance of primary cilia for human health has been highlighted by the association of several human genetic diseases with ciliary dysfunction (Fliegauf et al., 2007).
The primary cilium is formed by an MT-based core structure (named the axoneme), which consists of nine MT doublets that are surrounded by the ciliary membrane. The MTs of the axoneme are nucleated by the basal body, an MT-based cylindrical structure derived from the mother centriole of the centrosome. The assembly of the primary cilium is initiated at the G0/G1 phase of the cell cycle, and it follows an ordered sequence of steps (Sorokin, 1962, 1968). At the earliest stages of cilia formation, Golgi-derived vesicles are recruited to the distal end of the mother centriole. This step is followed by the extension of the axoneme and its associated ciliary membrane and the subsequent docking of this complex to the plasma membrane (the intracellular pathway). Alternatively, depending on the cell type, the mother centriole docks with the plasma membrane, and the cilium elongates directly out into the extracellular environment (the extracellular pathway; Ghossoub et al., 2011). Cilia extension depends on targeted vesicle transport regulated by the conserved Rab family of GTPases and their associated protein complexes (Yoshimura et al., 2007; Knödler et al., 2010; Westlake et al., 2011). In addition, components of the intraflagellar transport (IFT) machinery, together with motor proteins, contribute to the retrograde and anterograde transport of cargoes along the forming axoneme (Ishikawa and Marshall, 2011).
The primary cilium is formed at the mother but not at the daughter centriole (Nigg and Raff, 2009). The mother centriole possesses electron-dense, spikelike structures at its subdistal and distal ends, referred to as appendages. Many of the appendage proteins, including ODF2 (outer dense fiber protein 2), centriolin, ninein, and Cep164, are required for cilia assembly (Ishikawa et al., 2005; Graser et al., 2007; Mikule et al., 2007; Schmidt et al., 2012). In addition, in cycling cells, the protein CP110 and its interaction partner Cep97 localize at the distal ends of both mother and daughter centrioles to block inappropriate cilia formation. Remarkably, CP110 and Cep97 disappear from the mature basal body, whereas they still persist at the daughter centriole in ciliated cells (Spektor et al., 2007).
Although a large number of cilia-associated components have been identified in the past decade, the identity of the key regulators that control the initial steps of ciliogenesis awaits definition. Centriolar/basal body proteins and other molecules that promote cilia elongation are subjected to phosphorylation (Guarguaglini et al., 2005; Graser et al., 2007; Boesger et al., 2009; Soung et al., 2009). It is therefore conceivable that phosphorylation of centriolar components may regulate the transition between the distinct steps of ciliogenesis. However, we have a very limited understanding of the kinases that may govern these changes.
Here, we performed an RNAi-based screen using human telomerase-immortalized retinal pigment epithelial (RPE1) cells to search for kinases required for ciliogenesis. Among the kinases that we identified, we focused our analysis on the role of the MT-associated protein (MAP)/MT affinity regulating kinase 4 (MARK4; Kato et al., 2001; Trinczek et al., 2004). We show that MARK4 associates with the basal body and is required for the initiation of axoneme extension after the docking of ciliary vesicles to the mother centriole. These data therefore establish MARK4 as a critical kinase in cilia biogenesis. We anticipate that the characterization of the role played by the other kinases identified here will shed further light on the molecular basis for the spatial and temporal control of ciliogenesis.
Results
A kinome-wide siRNA screen for new regulators of primary cilium formation
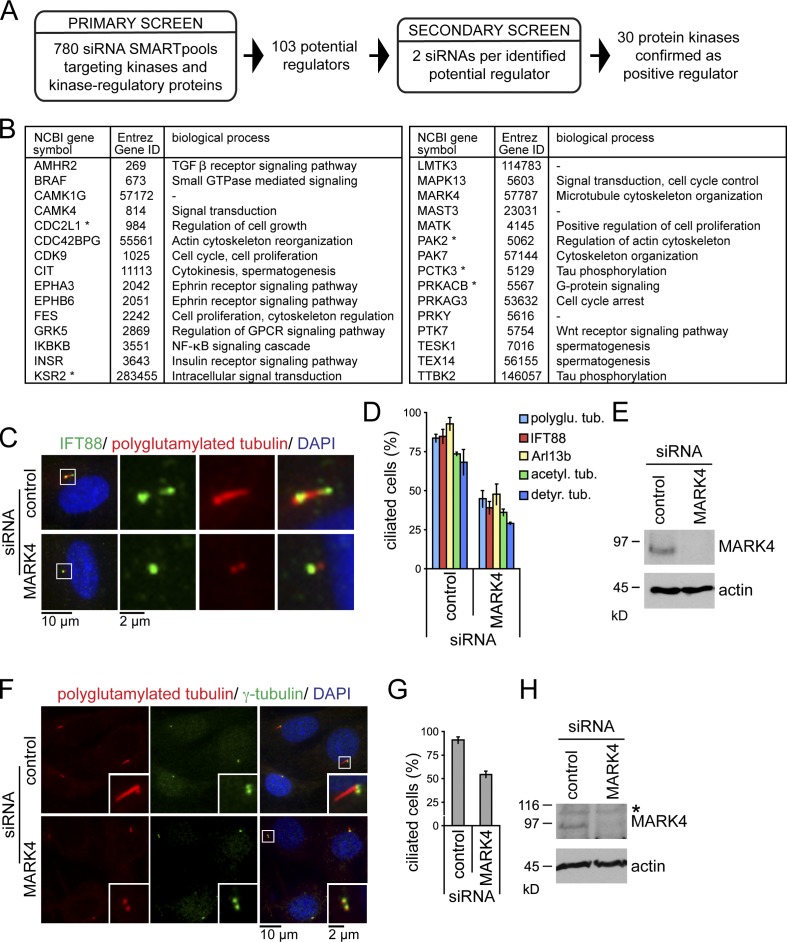
To identify new regulators of ciliogenesis, we performed an siRNA-based screen using RPE1 cells, in which ciliation can be induced by serum withdrawal. For high-throughput screening, we established a simple protocol for siRNA transfection under reduced serum conditions and an automated microscope-based readout. The latter was based on staining for acetylated tubulin, a posttranslational modification of tubulin that is highly enriched in the cilium (Figs. 1 A and S1; Janke and Bulinski, 2011). The primary screen was performed using a human siRNA kinome library consisting of 780 pools of four siRNAs targeting kinases and kinase-regulatory proteins. 103 putative regulators of ciliogenesis were identified. To minimize false positives arising from off-target effects, we conducted a secondary screen using two distinct siRNAs that confirmed, for 30 protein kinases, a reduction in ciliogenesis with at least one siRNA (Figs. 1 B and S1 D). Although five of the identified kinases were annotated in the ciliome database (a compilation of cilia-related studies using genomic and proteome-based analyses; Inglis et al., 2006), only TTBK2 has been reported to influence ciliogenesis (Kim et al., 2010). As a proof of principle, for the significance of our screen, we subsequently focused our investigations on MARK4, a conserved kinase that localizes at the centrosome but has not been linked to ciliogenesis (Kato et al., 2001; Trinczek et al., 2004).
Figure 1.
A kinome-wide siRNA screen identifies MARK4 as positive regulator of ciliogenesis. (A) Screen flowchart. (B) Kinases positively regulating ciliogenesis. Functional notes are obtained from the Entrez Gene database. Asterisks indicate kinases presented in the ciliome database. (C) RPE1 cells, treated with nontargeting (control) or MARK4 siRNA, were serum starved for 48 h and stained for IFT88, polyglutamylated tubulin, and DNA (DAPI). The left images show merged images. Regions within the white boxes are shown at higher magnification on the right. (D) Percentages of ciliated cells. polyglu. tub., polyglutamylated tubulin; acetyl., acetylated; detyr., detyrosinated. (E) Total cell extracts of control and MARK4-depleted RPE1 cells were analyzed by immunoblotting. Actin served as a loading control. (F) NIH 3T3 cells treated with control or MARK4 siRNA were serum starved for 24 h and stained for polyglutamylated and γ-tubulin and DNA. Insets show higher magnification of the centrosome region. (G) Percentages of ciliated cells determined using polyglutamylated tubulin. (H) Same as in E, except using extracts of NIH 3T3 cells. The asterisk indicates an unspecific band. Data are means ± SD of three independent experiments.
MARK4 depletion leads to loss of primary cilia
Using RPE1 cells and complete serum withdrawal, we confirmed that depletion of MARK4 (Figs. 1 E and S2, A–D) led to a significant decrease in ciliogenesis assessed by staining for cilia-associated components (IFT88 and Arl13b; Figs. 1, C and D; and S1 E) or ciliary MT modifications (Figs. 1, C and D; and S1, F and G). As primary cilia form in G0/G1 phase of the cell cycle (Plotnikova et al., 2008), we used FACS analysis and Ki67 staining (a marker for proliferating cells) to confirm that MARK4-depleted cells arrested upon serum starvation with a 2N DNA content (Fig. S1 H) and low levels of Ki67 (Fig. S1 I). This excluded the possibility that the defective ciliogenesis was an indirect consequence of an inappropriate cell cycle arrest. Remaining cilia in MARK4-depleted cells that may be a result of residual MARK4 activity (see Discussion) displayed no alteration in cilia length in comparison to control cells (Fig. S1 J). More importantly, depletion of MARK4 in mouse fibroblast NIH 3T3 cells (mMARK4) also hindered ciliogenesis (Figs. 1, F–H; and S1 K), indicating functional conservation across species. We conclude that MARK4 is required for efficient cilia formation in human and murine cells.
MARK4 localizes to the basal body
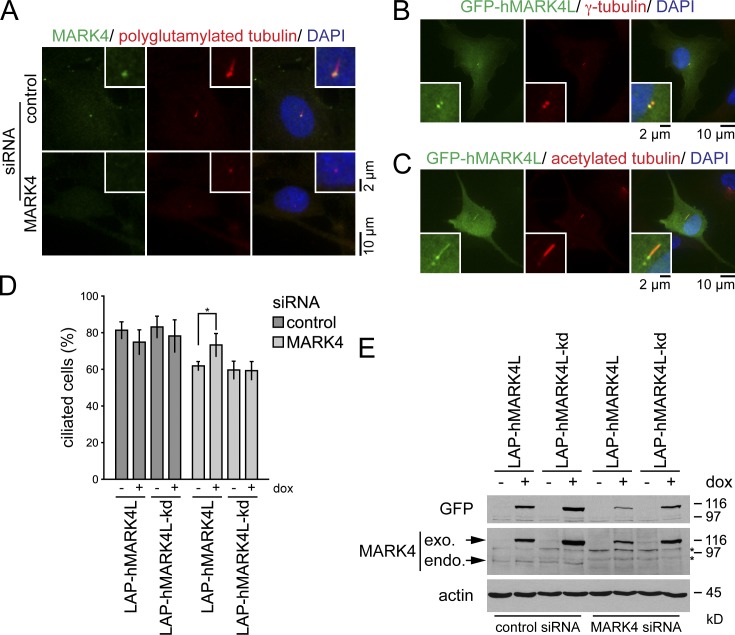
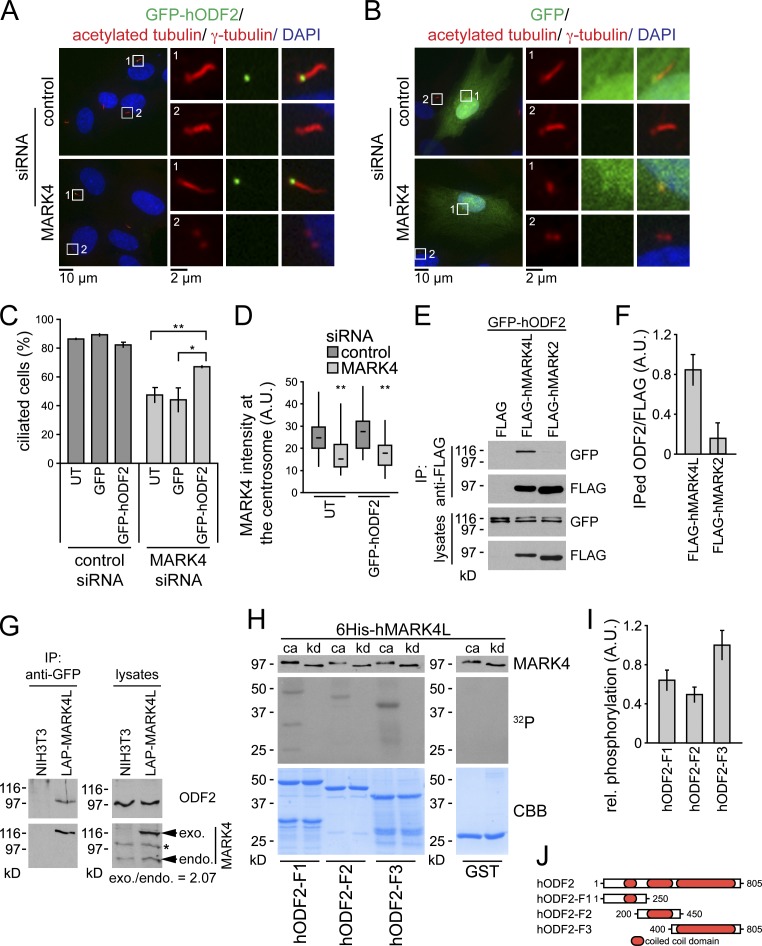
A previous study had localized MARK4 to the centrosomes of glioma cell lines and the midbody during cytokinesis (Trinczek et al., 2004). Using MARK4 antibodies, we found that MARK4 colocalized with polyglutamylated tubulin at the centrioles in a vast majority (>99%) of the analyzed ciliated RPE1 and NIH 3T3 cells (Figs. 2 A and S2 E). The centriolar staining was specific to MARK4, as it was strongly reduced upon MARK4 depletion or preabsorption of MARK4 antibodies with blocking peptides (Figs. 2 A and S2, E–G). To confirm MARK4 localization by an independent approach, we transiently transfected RPE1 cells with a GFP-tagged fusion of MARK4 large isoform (GFP-hMARK4L), which is the most abundant MARK4 isoform expressed in tissues other than the brain (Moroni et al., 2006). GFP-hMARK4L decorated both centrioles (Fig. 2 B) and, in addition, was seen along the cilium in 36% of transfected ciliated cells (Fig. 2 C). The lack of ciliary localization of the endogenous protein might be caused by the overexpression, or alternatively, it could be explained by the inaccessibility of the antibody to the cilia compartment, as previously documented for IFT20 (Follit et al., 2006). Collectively, these data indicate that MARK4 associates with both the basal body and the daughter centriole in ciliated cells and along the ciliary axoneme upon overexpression.
Figure 2.
MARK4 localizes to the basal body in ciliated cells. (A) RPE1 cells treated with control or MARK4 siRNA were serum starved for 48 h and stained for MARK4, polyglutamylated tubulin, and DNA. (B and C) RPE1 cells were transiently transfected with GFP-hMARK4L and stained for γ-tubulin (B) or acetylated tubulin (C) and DNA. (D) After control or MARK4 depletion, LAP-hMARK4L or LAP-hMARK4L-kd (kinase-dead mutant) expression was induced by the addition of 100 ng/ml doxycycline (dox), the cells were serum starved for 24 h, and percentages of ciliated cells were determined using acetylated tubulin as a cilia marker. Data are means ± SD of four independent experiments. *, P < 0.05. (E) Immunoblot analysis of the indicated NIH 3T3 cell extracts using anti-GFP and anti-MARK4 antibodies. Actin served as a loading control. Asterisks indicate unspecific bands. exo., exogenous; endo., endogenous. In A–C, the right images show merged images. Insets show higher magnification.
MARK4 catalytic activity is required to promote ciliogenesis
To investigate whether MARK4 catalytic activity is required for its function in promoting ciliogenesis, we generated NIH 3T3 cell lines stably expressing localization and affinity purification (LAP; Cheeseman and Desai, 2005)–tagged wild-type human MARK4L (LAP-hMARK4L) or an established kinase-dead (kd) mutant, in which amino acids 214 and 218 in the kinase domain were substituted to alanine (LAP-hMARK4L-kd; Lizcano et al., 2004; Trinczek et al., 2004). Both constructs were expressed under the control of the inducible tetracycline (TET)-on promoter. NIH 3T3 cells were treated with mouse-specific MARK4 siRNA to selectively deplete mMARK4 followed by serum withdrawal in the presence or absence of doxycycline. In doxycycline-deprived samples, mMARK4 depletion led to a 20% reduction in the number of ciliated cells (Fig. 2 D, compare first and fifth bars). The expression of low levels of active LAP-hMARK4L, but not LAP-hMARK4L-kd, restored the deficiency in cilia formation that arose from mMARK4 depletion (Fig. 2, D and E). These data therefore indicate that the kinase activity of MARK4 is important for its role in ciliogenesis.
MARK4 regulates the centrosomal localization of ODF2
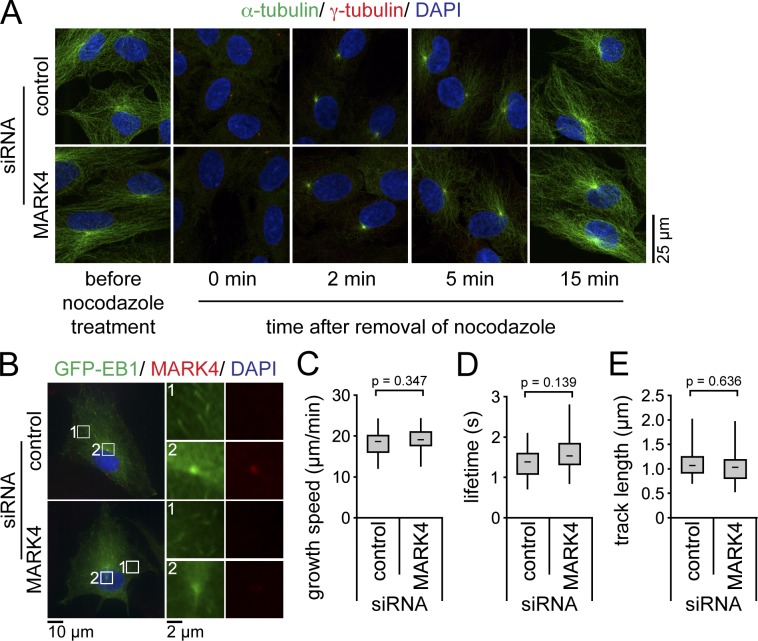
In neuroblastoma cells, MARK4 function was reported to influence MT stability through phosphorylation of the MT-associated proteins tau, MAP2, and MAP4 (Trinczek et al., 2004). We thus asked whether the defective ciliogenesis observed in RPE1 cells lacking MARK4 could arise from malfunction of the MT cytoskeleton. For this, we analyzed the ability of the centrosome to renucleate MT after nocodazole treatment. We observed no difference in the timing of MT regrowth at the centrosome after nocodazole washout between control and MARK4-depleted serum-starved cells (Fig. 3 A). To more precisely examine MT dynamics, we monitored the behavior of the MT end-binding protein EB1 tagged to GFP by time-lapse cell imaging. GFP-EB1 served as a marker for the growing ends of MTs and accurately allowed the determination of MT growth rates. As previously described (Piehl et al., 2004), GFP-EB1 foci emanated from the centrosome (Fig. 3 B, inset 2; and Video 1). The growth speed, lifetime, and length of GFP-EB1 tracks did not significantly differ between control and MARK4-depleted cells (Fig. 3, B–E; and Videos 1 and 2). This indicated that MT dynamics were not affected by MARK4 depletion. Consistently, the centriolar satellite proteins PCM-1, Cep290, BBS4, and OFD1, which are transported to the centrosome/basal body in an MT-dependent manner (Dammermann and Merdes, 2002; Lopes et al., 2011), were all recruited to the centrosome in cells depleted for MARK4 (Fig. S3, A–D). However, we noticed a slight decrease in the amount of interphase acetylated and detyrosinated MTs in cells lacking MARK4 (Fig. S3, E–I). Nevertheless, there was no correlation between the levels of these modifications and the loss of cilia, as we detected ciliated cells with reduced MT modification as well as nonciliated cells in which detyrosination or acetylation levels were comparable to wild-type cells (Fig. S3, E and H). Interestingly, the treatment of MARK4-depleted cells with the histone deacetylase inhibitor Trichostatin A (TSA) was able to restore the levels of acetylation but not ciliation (Fig. S3, E–G), implying that MARK4 regulates ciliogenesis by some other means.
Figure 3.
MARK4 depletion does not affect MT dynamics. (A) After control or MARK4 depletion, RPE1 cells were serum starved for 48 h and treated with 8 µM nocodazole at 37°C for 90 min to depolymerize MTs. Cells were stained for α- and γ-tubulin and DNA. (B–E) After control or MARK4 depletion, GFP-EB1–expressing RPE1 cells were serum starved for 24 h and either stained for MARK4 and DNA (B) or analyzed by live-cell imaging (Videos 1 and 2). The left images in B show merged images. Regions within the white boxes are depicted at higher magnification on the right showing GFP-EB1 tracks in the cytoplasm (1) or the centrosome (2). Time-lapse sequences were analyzed using automated tracking software. There is no significant difference in the mean growth speeds (C), lifetimes (D), and track lengths (E) between cells treated with control or MARK4 siRNA (control, n = 15; MARK4, n = 22). Boxes show the top and bottom quartiles (25–75%) with a line at the median, and whiskers extend from the minimum to the maximum of all data.
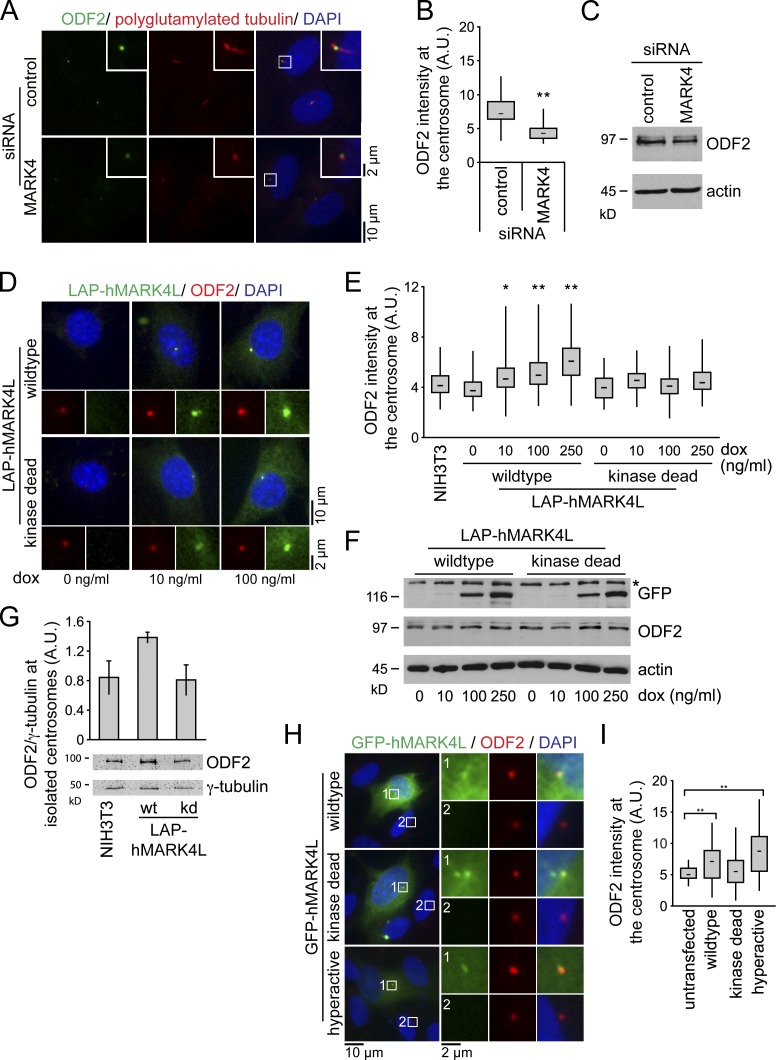
Given the localization of MARK4 to the centrosome and basal body in ciliated cells, we wondered whether MARK4 specifically regulated centrosomal components, as many of them are required for ciliogenesis, especially the appendage proteins of the mother centriole. Quantitative fluorescence microscopy revealed that MARK4 depletion significantly reduced the amount of ODF2 that associated with centrioles by over 40% compared with control levels (Fig. 4, A and B). Total ODF2 protein levels were not affected (Fig. 4 C). Identical results were obtained for MARK4-depleted NIH 3T3 cells (unpublished data). In contrast, the centrosomal levels of the pericentriolar marker γ-tubulin, the subdistal appendage proteins centriolin and ninein, and the distal components Cep164 and NPHP1 were similar in control and MARK4 siRNA–treated cells (Fig. S4, A–E; and not depicted). Thus, MARK4 depletion did not affect the overall composition of the centrosome but specifically caused a mislocalization of ODF2. Importantly, ODF2 localization was not influenced by perturbations of the MT cytoskeleton (Fig. S4 F), indicating that the association of ODF2 with centrosomes in RPE1 cells is independent of MT integrity, as previously established for mouse F9 cells (Ishikawa et al., 2005). We therefore concluded that ODF2 localization is partially controlled by MARK4.
Figure 4.
MARK4 regulates centrosomal localization of ODF2. (A) After control or MARK4 depletion, RPE1 cells were serum starved for 48 h and stained with the indicated antibodies. The right images show merged images. Insets show higher magnification of the regions within the white boxes. (B) Box and whisker plots show the relative ODF2 intensity at the centrosome after control and MARK4 depletion in RPE1 cells. One representative experiment out of three is shown. **, P < 0.001. A.U., arbitrary units. (C) Immunoblot analysis of the indicated RPE1 cell extracts using anti-ODF2 antibodies. Actin served as a loading control. (D) NIH 3T3 cells stably expressing either LAP-hMARK4L wild-type or kd mutant under the control of the TET-on promoter were treated with doxycycline (dox) and stained for ODF2 and DNA. The LAP tag enabled visualization of MARK4 constructs via a GFP moiety. (E) Box and whisker plots show the relative ODF2 intensity at the centrosome of untransfected cells (NIH 3T3) and cells expressing LAP-hMARK4L wild-type or kd mutant. One representative experiment out of three is shown. *, P < 0.05; **, P < 0.001 with respect to untransfected cells. (F) Immunoblot analysis of NIH 3T3 cells carrying the indicated constructs and treated with variable amounts of doxycycline as in D. ODF2 was detected with ODF2-specific antibodies. Anti-GFP antibodies were used to detect the LAP-hMARK4L constructs. Actin served as a loading control. The asterisk indicates an unspecific band. (G) Centrosomes from NIH 3T3 cells and NIH 3T3 cells expressing either LAP-hMARK4L wild-type (wt) or kinase-dead (kd) mutant were isolated by centrifugation using a discontinuous sucrose gradient and immunoblotted for ODF2 and γ-tubulin. The graph shows the quantification of centrosome-associated ODF2 normalized to γ-tubulin. Data are means ± SD of three independent experiments. (H) RPE1 cells were transiently transfected with the indicated constructs for 24 h, serum starved for 24 h, and stained for ODF2 and DNA. The left images show merged images. Regions within the white boxes are shown at a higher magnification to the right. (I) Box and whisker plots show the relative ODF2 fluorescence intensity at the centrosome in untransfected cells and after overexpression of the indicated constructs. One representative experiment out of three is shown. **, P < 0.001 with respect to untransfected cells. Boxes show the top and bottom quartiles (25–75%) with a line at the median, and whiskers extend from the minimum to the maximum of all data.
If MARK4 promoted ODF2 centrosomal association, one would expect that elevation of MARK4 levels would increase the levels of ODF2 at the centrosome. To test this notion, we used NIH 3T3 cells stably expressing either LAP-hMARK4L or the kd mutant under the inducible TET-on promoter (Fig. 4, D–F). Increased expression of LAP-hMARK4L enhanced the centrosomal localization of ODF2 (Fig. 4, D–E), without interfering with ODF2 protein levels (Fig. 4 F). The centrosomal accumulation of ODF2 required MARK4 catalytic activity, as ODF2 centrosomal levels did not change after overexpression of the kd mutant (Fig. 4, D and E). An increase in ODF2 centrosomal levels could also be detected in isolated centrosomes (Figs. 4 G and S4 G). In addition, we obtained similar results in RPE1 cells transiently expressing GFP-hMARK4L (Fig. 4, H and I). The centrosomal accumulation of ODF2 in RPE1 cells was even more pronounced after overproduction of hyperactive GFP-hMARK4L, in which threonine 214 was substituted to glutamic acid (Fig. 4, H and I; Lizcano et al., 2004). Together, these data suggest that MARK4 kinase activity contributes to the centrosomal recruitment of ODF2.
ODF2 overexpression rescues cilia loss upon MARK4 depletion
The suggestion that MARK4 contributes to ciliogenesis by promoting ODF2 centrosomal association prompted us to ask whether the ciliogenesis defect that arises from MARK4 depletion could be compensated by the ectopic overproduction of ODF2. This suppression effect would be expected if MARK4 increases the binding affinity of ODF2 for centrioles. To address this question, GFP-hODF2 was transiently overexpressed in RPE1 cells before subjecting the cells to control or MARK4 depletion and subsequent serum withdrawal (Fig. 5 A). In untransfected RPE1 cells, MARK4 depletion led to a 40% reduction in the number of ciliated cells (Fig. 5 C). Ectopic expression of GFP-hODF2 significantly increased the number of ciliated cells upon MARK4 knockdown (Fig. 5, A and C), whereas expression of GFP alone did not (Fig. 5, B and C). Importantly, the depletion of MARK4 was comparable between untransfected and GFP-hODF2–expressing cells (Fig. 5 D). These data indicate that enhancement of ODF2 levels compensates for the defective ciliogenesis that occurs after a reduction in MARK4 activity. Similar results were obtained for NIH 3T3 cells stably expressing LAP-hODF2 (unpublished data). Collectively, our data support the model that MARK4 promotes ciliogenesis by acting upstream of ODF2.
Figure 5.
ODF2 overexpression rescues cilia loss after MARK4 depletion. (A and B) RPE1 cells were transiently transfected with GFP-hODF2 (A) or GFP (B) and subsequently treated with control or MARK4 siRNA for 24 h before serum withdrawal and incubation for another 24 h. Cells were stained for DNA, γ-tubulin, and acetylated tubulin. The left images show merged images. Regions within the white boxes are shown at a higher magnification on the right. (C) Percentages of ciliated cells based on acetylated tubulin as a cilia marker. UT, untransfected. (D) Efficiency of MARK4 depletion in untransfected and GFP-hODF2–expressing cells were determined by quantitative fluorescence microscopy using MARK4-specific antibodies. Boxes show the top and bottom quartiles (25–75%) with a line at the median, and whiskers extend from the minimum to the maximum of all data. (E and F) HEK293T cells were transiently transfected with the indicated constructs. Immunoprecipitations (IP) were performed using anti-FLAG agarose. (E) Interacting proteins were detected by immunoblotting. (F) Quantification of E. (G) NIH 3T3 cells and NIH 3T3 cells expressing low levels of LAP-MARK4L were serum starved for 24 h, and immunoprecipitations were performed on the lysates using anti-GFP beads and probed for endogenous ODF2 and MARK4. exo., exogenous; endo., endogenous. The asterisk indicates an unspecific band. (H) For in vitro kinase assays, purified 6His-MARK4L, either catalytic active (ca) or kinase dead (kd), was incubated with recombinant ODF2 truncations. GST was used as a negative control. Samples were subjected to SDS-PAGE followed by autoradiography (32P) and Coomassie Brilliant blue staining (CBB). An aliquot of each reaction was analyzed by immunoblotting with anti-MARK4. (I) Relative phosphorylation of ODF2-F1, ODF2-F2, and ODF2-F3 normalized to protein amounts. (J) Schematic representation of ODF2 truncations. A.U., arbitrary unit. Data are means ± SD of three independent experiments. *, P < 0.05; **, P < 0.001.
These results raised the question as to whether MARK4 interacts with ODF2 in vivo. To investigate this possibility, we performed coimmunoprecipitation assays from human embryonic kidney 293T cells (HEK293T) ectopically expressing GFP-hODF2 alongside FLAG-hMARK4. As a negative control, we used MARK2, which shares 68% homology to MARK4 (Timm et al., 2008; Marx et al., 2010). GFP-hODF2 coimmunoprecipitated with FLAG-hMARK4 but not with FLAG-hMARK2 or FLAG peptide (Fig. 5, E and F). Endogenous ODF2 also coimmunoprecipitated with LAP-MARK4L, which was expressed at low levels in serum-starved NIH 3T3 cells (Fig. 5 G). Together, these data strongly suggest that ODF2 is able to associate with MARK4 in vivo.
To test whether MARK4 is able to phosphorylate ODF2, we performed in vitro kinase assays using bacterially purified catalytic-active (ca) or kd 6His-hMARK4L and ODF2. Full-length ODF2 was highly insoluble; however we were able to purify ODF2 truncated forms comprising the N-terminal, central, and C-terminal domains (ODF2-F1, -F2, and -F3, respectively; Fig. 5, H and J). 6His-hMARK4L-ca, but not the kd mutant, preferentially phosphorylated ODF2-F3 (Fig. 5 H). Collectively, our data indicate that MARK4 interacts with ODF2 in vivo and phosphorylates ODF2 in vitro.
MARK4 depletion blocks axoneme extension
To gain insight into how MARK4 regulates ciliogenesis, we used transmission EM (TEM) to analyze the basal body and/or ciliary structure of serum-starved RPE1 cells lacking MARK4. Ciliogenesis is a multistep process that begins with the docking of Golgi-derived vesicles to the mother centriole, after which the axoneme elongates (Fig. 6 A; Sorokin, 1962, 1968). When we analyzed our RPE1 cell line grown in the presence of serum, we observed that, in 50% of the G1 cells, the mother centriole was deprived of any vesicle (Fig. 6, A [step 0], B, and M). In the remaining cells, the mother centriole was either associated with a small vesicle (17%; Fig. 6, A [step 1], C, and M), a capping vesicle (11%; Fig. 6, A [step 2], D, and M), or an elongated cilium (22%; Fig. 6, A [step 4] and M). To enrich for the early stages of ciliogenesis, we investigated control depleted cells that had been subjected to serum starvation for a brief period of time (24 h). Under such growth conditions, 47% of mother centrioles were associated with ciliary vesicles (Fig. 6, D, E, and M). After longer periods of serum withdrawal (48 h), all mother centrioles were associated with an elongated cilium in control depleted cells (Fig. 6, F and M). In contrast, 46% of MARK4-depleted cells were unciliated. In 38% of these cells, the distal end of the mother centriole was capped by the ciliary vesicle, but extended axonemal MTs were absent (Fig. 6, G, H, and M). Interestingly, no gross alteration was detected in the ultrastructure of the basal body. We therefore concluded that MARK4 is not required for the initial docking of ciliary vesicles but is regulating an initial step of axoneme extension,
Figure 6.
Ultrastructural analysis of MARK4 depletion phenotypes. (A) A schematic model depicting the different stages of ciliogenesis, according to Sorokin (1962, 1968). The mother centriole is characterized by the presence of distal (DA) and subdistal appendages (SDA; 0). The initial step of ciliogenesis is the docking of ciliary vesicles (CV) to the mother centriole (1). The CVs increase in size by fusion with nearby vesicles (V; 2) and become invaginated by the ciliary bud (CB), an accumulation of electron-dense material at the distal end of the mother centriole (3). MT doublets start to elongate from the basal body giving rise to the axonemal shaft (AS) and ciliary pocket (CP; 4). The elongated ciliary vesicle fuses with the plasma membrane (PM), and the cilium emerges in the extracellular environment (5). (B and C) Electron micrographs showing serial sections of unstarved RPE1 cells. Cells were seeded 24 h before fixation and cultivated in medium with 10% serum. (D and E) Electron micrographs showing serial sections of RPE1 cells treated with control siRNA. Cells were starved for 24 h to enrich early stages of ciliogenesis. (F–H) Electron micrographs showing serial sections of RPE1 cells treated with control (F) or MARK4 siRNA (G and H). Cells were serum starved for 48 h before fixation for TEM analysis. (I) After control or ODF2 depletion, RPE1 cells were serum starved for 48 h and analyzed by immunofluorescence. Percentages of ciliated cells were determined using polyglutamylated tubulin. Data are means ± SD of three independent experiments. (J) Immunostaining with anti-ODF2 revealed residual low levels of ODF2 at the centrosome in cells treated with ODF2 siRNA. Box and whisker plots show the relative ODF2 intensity at the centrosome after control and ODF2 depletion. One representative experiment out of three is shown. Boxes show the top and bottom quartiles (25–75%) with a line at the median, and whiskers extend from the minimum to the maximum of all data. A.U., arbitrary unit; polyglu. tub., polyglutamylated tubulin. (K and L) Electron micrographs showing serial sections of RPE1 cells treated with ODF2 siRNA. Cells were serum starved for 48 h before fixation for TEM analysis. (M) Quantification of ultrastructural phenotypes in control, MARK4-, and ODF2-depleted RPE1 cells after the indicated time of serum starvation. For unstarved control cells (0 h), only cells in G1 with unduplicated centrioles were considered. The asterisk indicates that the ODF2-depleted RPE1 cells showed an additional defect in the subdistal appendage formation; subdistal appendages were either completely absent (77%) or grossly impaired (23%).
We next asked whether lowering the levels of ODF2 would influence ciliogenesis in a way similar to MARK4 depletion. Treatment of RPE1 with an established ODF2 siRNA impaired ciliogenesis as previously described (Fig. 6 I; Soung et al., 2009) and significantly diminished the levels of ODF2 at centrosomes (Fig. 6 J). TEM analysis revealed that 48% of ODF2-depleted cells possessed mother centrioles that were associated with a capping ciliary vesicle but no extended axoneme (Fig. 6, K and L). This phenotype is highly reminiscent of that arising from MARK4 depletion (Fig. 6, G and H), but unlike MARK4-depleted cells and in agreement with a previous study using murine Odf2−/− knockout cells (Ishikawa et al., 2005), depletion of ODF2 greatly impaired subdistal appendage formation. This was observed by TEM (Fig. 6, K and L; and S5 C) and quantification of the subdistal marker protein centriolin at the centrosome (Fig. S5, D and E). This lack of subdistal appendages was also observed in cells with an elongated cilium (unpublished data), indicating that it was not causative for the ciliary defect. However, in contrast to F9 Odf2−/− cells, in which distal appendages were not formed (Ishikawa et al., 2005), we observed no difference in distal appendage formation upon ODF2 depletion by TEM (Fig. S5 C) and quantitative analysis of the levels of the distal appendage protein Cep164 at the centrosome (Fig. S5, F and G). This may be explained by the presence of residual levels of ODF2, which may be sufficient to support the formation of distal but not subdistal appendages. Importantly, similarly to MARK4, all ODF2-depleted nonciliated cells were defective in the initiation of axoneme extension but not in the initial docking of the ciliary vesicle.
To further investigate whether the ciliogenesis program had initiated in MARK4- and ODF2-depleted cells, we analyzed the localization of the Rab family GTPase Rab8a, which was reported to be absent from the centrosome in cycling cells but to be recruited to this location after serum withdrawal (Westlake et al., 2011). Accordingly, no centrosomal localization of GFP-Rab8a was observed in the majority (72%) of interphase RPE1 cells grown in the presence of serum (Fig. 7, control unstarved). However, upon serum starvation, GFP-Rab8a accumulated at the centrosome in a large percentage of nonciliated cells after MARK4 or ODF2 depletion (Fig. 7), indicating that the ciliary program had initiated in those cells. Together, we concluded that MARK4 and ODF2 are not required for the initiation of the ciliary program, but instead, they play a role in promoting axoneme extension after the docking of Rab8a-positive vesicles to the mother centriole.
Figure 7.
MARK4 or ODF2 depletion leads to accumulation of cells in early stages of ciliogenesis. (A) RPE1 cells stably expressing GFP-Rab8a were treated with the indicated siRNAs. Cells, grown in medium with 10% serum (unstarved) or serum starved for 24 h were stained for γ-tubulin, acetylated tubulin (acetyl. tub.), and DNA. Representative fluorescence micrographs are shown for the localization patterns of GFP-Rab8a. The left images show merged images. Centrosomal regions are shown at higher magnification on the right. (B) Quantification of A. One representative experiment out of three is shown.
Depletion of MARK4 blocks ciliogenesis before the complete displacement of CP110 and Cep97 from the mother centriole
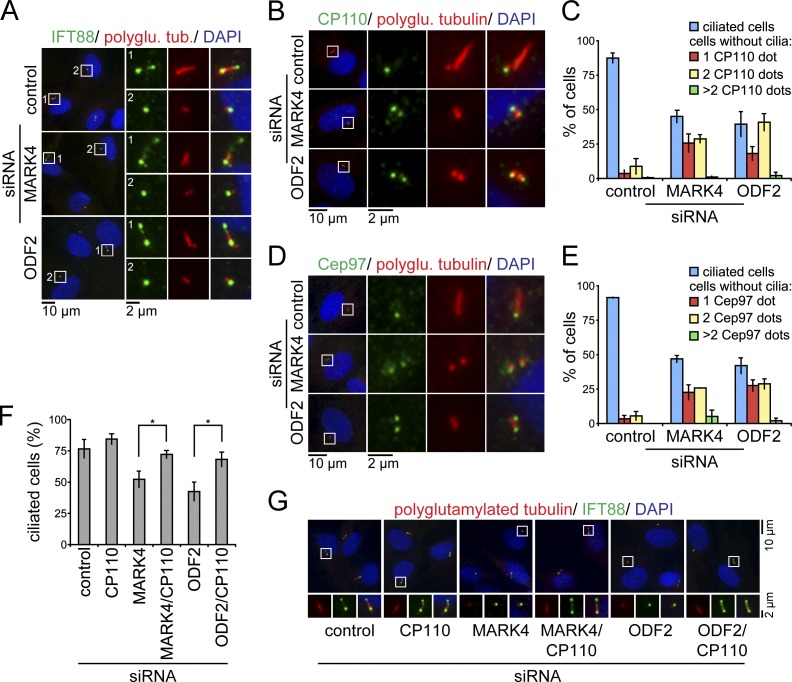
The depletion of MARK4 or ODF2 blocks the assembly of the ciliary axoneme. Several deficiencies may account for this phenotype. The defect in axoneme extension may be a consequence of defective IFT transport, or it may arise from impaired control of the CP110–Cep97 complex (Spektor et al., 2007; Pedersen and Rosenbaum, 2008). To discern between these two possibilities, we analyzed the localization of IFT88 and CP110–Cep97 in serum-starved RPE1 cells after depletion of MARK4 or ODF2. The IFT protein IFT88 is found at the mother centriole and, additionally, at the axoneme in ciliated cells (Fig. 8 A) and is required for axoneme extension in a variety of organisms (Pazour et al., 2000; Jurczyk et al., 2004; Pedersen and Rosenbaum, 2008). This mother centriole/basal body recruitment of IFT88 was not affected by MARK4 or ODF2 depletion (Fig. 8 A), excluding the possibility that the centrosomal association of IFT88 is under MARK4 or ODF2 control.
Figure 8.
CP110 persists at the mother centriole after MARK4 depletion. (A) RPE1 cells, treated with the indicated siRNAs, were serum starved for 48 h and stained for IFT88, polyglutamylated tubulin (polyglu. tub.), and DNA. (B–E) Same as in A; however, cells were stained for CP110 (B) or Cep97 (D), polyglutamylated tubulin, and DNA. (C and E) Quantification of B and D, respectively. The number of ciliated cells and CP110 (C) or Cep97 dots (E) per centrosome are indicated. (F and G) Codepletion of CP110 rescues the loss of cilia in MARK4- or ODF2-depleted RPE1 cells. (F) After treatment with control, MARK4, or ODF2 siRNA for 24 h, RPE1 cells were subjected to control or CP110 depletion for 24 h and subsequent serum starvation for 24 h. Percentages of ciliated cells based on polyglutamylated tubulin as a cilia marker. *, P < 0.05. (G) RPE1 cells were treated as in F and stained for IFT88, polyglutamylated tubulin, and DNA. In A, B, D, and G, merged images are shown. Regions within the white boxes are shown at higher magnification at the left (A, B, and D) or in the bottom images (G). Data are means ± SD of three independent experiments.
We next asked whether the inhibitory protein CP110 remained at the mother centrioles of cells from which MARK4 had been depleted. As previously reported (Spektor et al., 2007; Schmidt et al., 2009), CP110 decorates the daughter centriole but not the basal body in cells with an elongated cilium (Fig. 8 B). Remarkably, in cells that lacked MARK4 or ODF2, CP110 persisted at the mother centriole after serum starvation (Fig. 8, B and C). Similar results were obtained for the CP110-interacting protein Cep97 (Fig. 8, D and E). We thus postulated that if the reason for the lack of axoneme extension was related to the inhibitory role of CP110, we would be able to rescue ciliogenesis in MARK4- or ODF2-deficient cells by decreasing CP110 protein levels. The depletion of CP110 also interfered with the centrosomal localization of Cep97 (unpublished data), which depends on CP110 (Spektor et al., 2007). The percentage of ciliation in cells lacking MARK4 or ODF2 was significantly increased upon the subsequent knockdown of CP110 (Fig. 8 F). As CP110 depletion may also induce elongation of centrioles (Spektor et al., 2007; Schmidt et al., 2009), we confirmed by IFT88 labeling that the observed structures were genuine primary cilia (Fig. 8 G). Collectively, the data indicate that reduced levels of MARK4 or ODF2 block the ciliary program before the complete exclusion of CP110–Cep97 from the mother centriole, a prerequisite for cilia extension.
Discussion
Kinases involved in cilia formation
Our understanding of the molecular basis for the remarkable conversion of the mother centriole into the basal body that nucleates the primary cilium remains in its infancy. Several constitutive centriolar/basal body proteins and other molecules that promote cilia elongation and/or maintenance are subjected to phosphorylation (Guarguaglini et al., 2005; Graser et al., 2007; Boesger et al., 2009; Soung et al., 2009). It is therefore reasonable to assume that protein kinases may play a critical role in the regulation of distinct steps in cilia assembly. However, despite the analysis of a variety of ciliated organisms, only a few kinases have actually been functionally linked to ciliogenesis (Quarmby and Parker, 2005; Santos and Reiter, 2008; Cao et al., 2009). Here, we performed an RNAi-based screen in human RPE1 cells that enabled us to identify novel protein kinases involved in ciliogenesis. Our approach was designed to minimize false positives related to off-target effects of the siRNA probes used. However, false negatives remain possible, implying that the number of ciliogenesis-promoting kinases described here is most likely underestimated. In total, we functionally linked 30 protein kinases to cilia formation. Importantly, with the exception of TTBK2 and five other kinases that were annotated in the ciliome database (Inglis et al., 2006), none had been linked to ciliogenesis previously.
The role of MARK4 in ciliogenesis
In human cells, four MARK kinases (MARK1–4) have been identified. They belong to a conserved family of serine/threonine protein kinases represented by PAR-1 (portioning-defective 1) in nematodes and flies, and the Kin1–4 kinases in budding yeast (Tassan and Le Goff, 2004; Marx et al., 2010). Kinases of the MARK family are involved in the control of the MT cytoskeleton, cell polarity, and/or asymmetric cell division (Drewes et al., 1998; Hurov and Piwnica-Worms, 2007; Matenia and Mandelkow, 2009). The high degree of evolutionary conservation, the association of MARK4 with centrosomes in neuroblastoma cell lines (Trinczek et al., 2004), and the identification of MARK4 in our screen motivated us to pursue the role performed by MARK4 in ciliogenesis in greater depth.
The depletion of MARK4 using multiple siRNA sequences led to a reduction in cilia formation in both human and murine cells. The reduced ciliogenesis upon MARK4 depletion was not caused by off-target effects, as it could be rescued by expression of an exogenous siRNA-resistant MARK4 construct. Considering that MARK4 is a kinase, a residual level of MARK4 activity might be sufficient to promote ciliogenesis. Alternatively, the activity of kinases working in parallel to MARK4 during ciliogenesis could explain the incomplete penetrance of cilia loss. Members of the MARK family would be the most obvious candidates for molecules that act alongside MARK4 in ciliogenesis. However, we did not observe any further reduction in ciliogenesis after coknockdown of MARK4 with MARK1, MARK2, or MARK3 (unpublished data). We therefore consider functional redundancies in ciliogenesis between MARK kinases as less likely. However, we are currently unable to rule out the possibility that another kinase functionally overlaps with MARK4’s role in promoting ciliogenesis.
How does MARK4 affect ciliogenesis in RPE1 cells? Cilia formation starts with the docking of Golgi-derived vesicles to the apical part of the mother centriole, a step that is regulated by components of the distal appendages (Fig. 9; Sorokin, 1968; Schmidt et al., 2012). Our data indicate that MARK4 influenced neither the localization of the distal appendage components NPHP1 and Cep164 nor the initiation of the ciliary program, i.e., the initial process of vesicle docking, which is under Cep164 control (Schmidt et al., 2012). Remarkably, in nonciliated MARK4-depleted cells, the initiation of axoneme extension was impaired, implying that MARK4 plays a critical role at this step of ciliation.
Figure 9.
Model for the role of MARK4 in primary cilium formation. Primary cilium formation is initiated by the fusion of vesicles to the mother centriole, which thus becomes a basal body. Subsequently, the ciliary axoneme elongates from the basal body. MARK4 is not involved in vesicle docking but is crucial for the initiation of axoneme extension. MARK4 most likely regulates ODF2 functions but may also act on other targets to promote ciliogenesis. A decrease in MARK4 activity blocks ciliogenesis after the initial vesicle docking and before the complete displacement of the inhibitory proteins CP110 and Cep97.
In the search for centriolar targets being affected by MARK4, we observed that MARK4 knockdown reduced the centrosomal levels of ODF2 by 40%. This effect was specific to ODF2, as the localization of centriolin and other subdistal components remained unchanged (Fig. S4 and unpublished data). Interestingly, overexpression of ODF2 rescued ciliation of cells from which MARK4 had been depleted, implying that the reduced centrosomal level of ODF2 upon MARK4 knockdown limits ciliogenesis. Consistently, our TEM analysis revealed that the reduction of ODF2 protein levels in RPE1 cells led to a phenotype that was remarkably similar to the one observed for MARK4 depletion, i.e., block of axoneme extension. It is therefore feasible that MARK4 contributes to ciliogenesis by promoting the accumulation of a critical level of ODF2 at the mother centriole, for example, by promoting or stabilizing complex formation between ODF2 and centriolar components. This assumption is supported by the fact that overproduced MARK4 triggered ODF2 accumulation to the centrosome (Fig. 4), ODF2 interacted with MARK4 in vivo (Fig. 5), and MARK4 phosphorylated ODF2 in vitro (Fig. 5). It is therefore possible that ODF2 may be an in vivo substrate of MARK4, although a final proof for this assumption awaits clarification. Interestingly, a study addressing the impact of centriole age upon ciliogenesis showed that, after mitosis, the oldest mother centriole possesses higher levels of ODF2 (Anderson and Stearns, 2009). This older centriole is the first to form the primary cilium (Anderson and Stearns, 2009), thereby suggesting that recruitment and/or maintenance of a threshold level of ODF2 at the mother centriole may indeed constitute a rate-limiting step in cilia formation.
In addition to the block of axoneme extension, we also observed that ODF2 depletion additionally impaired subdistal appendage formation in RPE1 cells. This defect is in agreement with a previous study showing that murine F9 Odf2−/− cells are deprived of subdistal appendages (Ishikawa et al., 2005). Importantly, whereas distal appendages were also lost in F9 Odf2−/− cells, we observed no loss of distal appendage components when comparing wild-type and ODF2-depleted cells. This may be explained by the presence of residual levels of ODF2, which may still be sufficient to support formation of distal but not subdistal appendages in RPE1 cells. Indeed, ODF2 was reported to localize to both distal and subdistal appendages (Lange and Gull, 1995; Nakagawa et al., 2001), and a recent study revealed that Odf2Δ/Δ mouse embryonic fibroblast cells expressing a truncated form of ODF2 were able to form distal but not subdistal appendages (Kunimoto et al., 2012). In contrast to ODF2-depleted RPE1 cells, Odf2Δ/Δ mouse embryonic fibroblast cells were able to form cilia because of the expression of the ODF2 truncation (Kunimoto et al., 2012), implying that distinct subdomains of ODF2 might independently promote subdistal and distal appendage formation.
We found that in the absence of either MARK4 or ODF2, the proteins CP110 and Cep97 persisted at the mother centriole after serum withdrawal. The CP110–Cep97 complex was reported to have an inhibitory function at the distal end of the centriole, where they prevent axoneme assembly (Spektor et al., 2007). Interestingly, the depletion of CP110 was able to rescue ciliogenesis in cells depleted for MARK4 or ODF2. It is thus possible that MARK4 and ODF2 are part of a regulatory mechanism leading to the specific removal of the CP110–Cep97 complex from the mother centriole during early steps of ciliogenesis. It is also tempting to speculate that the confinement of ODF2 to the mother centriole would explain why CP110 and Cep97 would be selectively displaced from this location.
The tumor-suppressor protein kinase LKB1/STK11, which works as an upstream activating kinase for MARK4 (Brajenovic et al., 2004; Lizcano et al., 2004), has been reported to localize at the cilium, to regulate cilia length, and to locally regulate mTOR (mammalian target of rapamycin) signaling and cell size through AMP-activated protein kinase phosphorylation (Boehlke et al., 2010; Jacob et al., 2011). This observation opens the exciting possibility that LKB1 may locally activate MARK4 to promote cilia formation. The investigation of MARK4 regulation by LKB1 and its physiological contribution for ciliogenesis will constitute an exciting aspect of future research. Furthermore, we are confident that the molecular characterization of the additional kinases identified here will constitute an important step toward the molecular understanding of the mechanisms of primary cilia formation.
Materials and methods
Plasmids and reagents
Full-length cDNA for MARK4L was provided by A. Beghini (University of Milan, Milan, Italy), and full-length cDNA for ODF2 isoform 3, also known as Cenexin1, was obtained from the German Resource Center for Genome Research (clone: IRATp970A059D6). Full-length cDNA for MARK2 and Rab8a was amplified from total cDNA of RPE1 cells. Expression constructs comprising an N-terminal GFP, FLAG, LAP, GST, or 6His tag were generated by PCR cloning into pEGFP-C1 (Takara Bio Inc.), pCMV-3Tag-1 (Agilent Technologies), pIC113 (LAP construct; gift from I. Cheeseman, Whitehead Institute, Cambridge, MA; Cheeseman and Desai, 2005), pGEX-5X-1 (GE Healthcare), or pET28 (EMD Millipore). Point mutations in MARK4 were generated by site-directed mutagenesis. All constructs were confirmed by DNA sequencing. Retroviral expression vectors, which contained the TET-on inducible promoter (Gossen and Bujard, 1992), were generated by subcloning into pMOWSIN-TREt (TET response element tight; Pfeifer et al., 2010). For generation of stable NIH 3T3 cells, retroviral expression vectors were cotransduced into NIH 3T3 cells together with the cDNA for the transactivator protein contained in pMOWS-rtTAM2 (Pfeifer et al., 2010). Expression of TET-on–inducible constructs was induced by the addition of doxycycline (Sigma-Aldrich) at a concentration of 10–250 ng/ml during serum starvation. For perturbation of the MT cytoskeleton, cells were treated at 37°C with nocodazole (Sigma-Aldrich) at a final concentration of 8 µM, with taxol (Sigma-Aldrich) at a final concentration of 50 nM, or TSA (Sigma-Aldrich) at a final concentration of 0.5 µM.
Cell culture and transfection
RPE1 cells and RPE1 cells stably expressing GFP-EB1 (pEGFP-C1-EB1; Rosa et al., 2006) were gifts from S. Doxsey (University of Massachusetts Medical School, Worcester, MA). Cells were grown in DMEM/F12 medium supplemented with 10% FCS, 2 mM l-glutamine, and 0.348% sodium bicarbonate. HEK293T cells (gift from T. Benzing, University of Cologne, Cologne, Germany) and Phoenix Eco cells were cultured in DMEM supplemented with 10% FCS. NIH 3T3 cells were grown in DMEM supplemented with 10% newborn calf serum. All cell lines were grown at 37°C under 5% CO2. HEK293T and Phoenix Eco cells were transiently transfected with plasmid DNA using the calcium phosphate precipitation method. RPE1 cells were transiently transfected with plasmid DNA using Fugene 6 (Roche) and fixed 24–48 h after transfection. To generate stable RPE1 cell lines, cells were transfected with plasmid DNA using Fugene 6 and subsequently selected with 800 µg/ml G418 (Invitrogen) for 2 wk. Stable NIH 3T3 cells were produced as previously described (Ketteler et al., 2002; Pfeifer et al., 2010). In brief, retroviral expression vectors were transiently transfected in Phoenix Eco cells. 24 h later, transducing supernatants were harvested and added to NIH 3T3 cells. Plates with cells were centrifuged for 3 h at 340 g at 37°C. Cells were then cultivated with normal growth media for 24 h and selected with 3 µg/ml puromycin (GE Healthcare). NIH 3T3 and RPE1 cells were incubated in serum-free medium for 24–48 h to induce cilia formation. The percentages of ciliated cells were calculated from the observation of 100–150 cells per experimental condition.
RNAi
Synthetic siRNA oligonucleotides were obtained from Ambion and targeted to the following sequences: human MARK4, 5′-GCATCATGAAGGGCCTAAA-3′ and 5′-CTGCAGCCTGTTGCCCAATAA-3′; mouse MARK4, 5′-GGGATCTAAAGGCTGAAAA-3′, 5′-GGTCGCTATTAAGATCATT-3′, and 5′-GGCCAACATCAAAATCGCC-3′; human CP110, 5′-AAGCAGCATGAGTATGCCAGT-3′ (Spektor et al., 2007); human ODF2, 5′-AAAGACTAATGGAGCAACAAG-3′ (Soung et al., 2009); and nontargeting control siRNA, 5′-AACGTACGCGGAATACTTCGA-3′. DharmaFECT 3 (Thermo Fisher Scientific) or Lipofectamine RNAiMAX (Invitrogen) were used for transfection of siRNAs in RPE1 and NIH 3T3 cells.
Screening procedure
For high-throughput screening, RPE1 cells were reverse transfected in 384-well plates using DharmaFECT 3. In brief, siRNAs were prealiquoted in 384-well plates to a final concentration of 50 nM. DharmaFECT 3 was diluted in Opti-MEM and incubated with the siRNAs for 30 min. RPE1 cells (2,000 cells/well) were seeded in medium with 3% FCS (low serum medium) onto the 384-well plates containing siRNAs. After 24 h, serum-free medium was added to enhance the induction of ciliogenesis. Cells were further incubated for 96 h. Before fixation, 384-well plates were placed on ice for 45 min to depolymerize cytoplasmic MTs. Cells were fixed with 3% PFA for 20 min and permeabilized with 0.1% Triton X-100 in PBS for 10 min. Cilia were stained using mouse monoclonal anti–acetylated tubulin antibodies (C3B9; Sigma-Aldrich) coupled to Cy3. The coupling of the antibodies with Cy3 monoreactive dye (GE Healthcare) was performed according to the manufacturer’s instructions. DNA was stained using DAPI (Sigma-Aldrich). Cells were washed with PBS and either analyzed immediately or stored in PBS at 4°C for ≤5 d.
The high-throughput screen was performed using a human SMARTpool siRNA library (Thermo Fisher Scientific) consisting of a pool of four siRNAs per gene, targeting kinases, and kinase-regulatory proteins. Two independent screens were performed in duplicate, resulting in four replicates. Only results that increased or reduced the percentage of ciliated cells from the sample median by at least two median absolute deviations were considered as hits. A gene was considered as a potential regulator of ciliogenesis when at least two of the four replicates were scored as hits.
Image analysis of screening data
Image acquisition was performed at two wavelengths (DAPI and Cy3) using an automated scanning microscope station (IX81 ScanR; Olympus) equipped with a 20×, 0.4 NA air objective lens (Olympus). Automated focusing was performed on DAPI-stained DNA. Four fields per well were acquired as z stacks, and maximum projections of z stacks were used for the image analysis.
To identify nuclei and cilia and to determine the percentage of ciliated cells, we developed a cilia analysis software. The morphological characteristics of the objects were extracted via several image-processing steps using MATLAB (MathWorks) and DIPimage (DIPlib 1.6 Scientific Image Processing Library; Quantitative Imaging Group, Delft University of Technology, Dleft, Netherlands). From the raw image (TIF file; 12 bit), a smoothing function corresponding to the radii of the objects of interest (nuclei: DAPI channel; cilia: Cy3 channel) was used to obtain background-corrected images. The global threshold was calculated using Otsu’s method, which returned normalized intensity values lying in the range [0, 1]. Binary images were then generated, and only objects not touching the image boarder (edge objects) were considered for subsequent evaluation. Edge objects were discarded. After labeling the binary image, objects were classified and identified as nuclei/cilia if they fulfilled defined criteria, such as object size, shape (eccentricity, perimeter, and Feret diameter), and intensity. Objects featuring parameters lying outside of these classifications, i.e., objects that were too large or small, with atypical shape, too weak, or high intensities were excluded from further analysis. To correlate cilia and nuclei, the border pixels of the identified nuclei were calculated, resulting in corresponding polygons for each of the nuclei. Cilia belonging to a certain nuclei were calculated via the distance between the center of mass of the cilia and the nearest pixel of the polygon. If this distance was smaller than or equal to two thirds of the minor axis length of the nuclei, the cilia was regarded as belonging to the nuclei, and consequently, the object was counted as one ciliated cell. The percentage of ciliated cells for each well was determined by correlating the numbers of ciliated cells and all nuclei.
Immunofluorescence staining and microscopy
Cells were grown on coverslips and either fixed in ice-cold methanol for 5 min and permeabilized in ice-cold acetone for 20 s or fixed in 3% PFA for 20 min. For cilia staining with anti–acetylated tubulin antibodies, cells were incubated on ice for 10–30 min before fixation to depolymerize cytoplasmic MTs. PFA fixation was followed by a 10-min quenching step in 50 mM NH4Cl and a 5-min permeabilization step in 0.1% Triton X-100. After blocking with 0.5% BSA in 0.1% Triton X-100/PBS for 30 min, cells were incubated with primary antibodies at 37°C for 1 h. Primary antibodies were detected with Alexa Fluor–conjugated secondary antibodies for 1 h at room temperature. DAPI was included with the secondary antibodies for DNA staining. All antibodies were diluted in blocking solution. Coverslips were mounted on glass slides in Mowiol (EMD Millipore). Confocal images were taken using a microscope (TCS SP5; Leica) equipped with an HCX Plan Apochromat λ blue 63.0×, 1.4 NA oil objective. Other images were acquired as z stacks (0.3-µm steps spacing 3 µm) using a microscope (Axiophot; Carl Zeiss) equipped with a 63×, 1.4 NA or 100×, 1.45 NA Plan Fluor oil immersion objective lens (Carl Zeiss), a charge-coupled device camera (Cascade 1K; Photometrics), and MetaMorph software (Universal Imaging Corp.). Color balance was adjusted to the same level in MetaMorph. Image brightness and contrast were adjusted equally in Photoshop CS3 (Adobe). The z plane of focus was selected for images.
Intensity quantification of centrosomal proteins
Quantification of fluorescence intensity of centrosomal proteins was performed using ImageJ software (National Institutes of Health). An area of 25 pixels encompassing the centrosome was measured using summed projections of acquired z stacks. Background signals were measured in the near proximity of each centrosome and subtracted from the centrosomal measurement. 50–100 cells were measured for each experimental condition. Distribution of fluorescence intensity measurements is displayed in box and whiskers plots. Boxes show the top and bottom quartiles (25–75%) with a line at the median, and whiskers extend from the minimum to the maximum of all data. Statistical analyses of fluorescence intensity measurements and ciliation assays were performed using two-tailed Student’s t test. P < 0.05 was considered statistically significant.
Analysis of MT dynamics using live-cell time-lapse imaging
After siRNA transfection, GFP-EB1–expressing RPE1 cells were reseeded onto 35-mm µ-dishes (Ibidi GmbH). After 24 h, the cells were washed twice with serum-free DMEM/F12 medium without phenol red and incubated for additional 24 h under serum starvation conditions before image acquisition. Time-lapse images were recorded every 400 ms for 1 min using a spinning-disk microscope (UltraVIEW ERS; PerkinElmer) equipped with a 100×, 1.4 NA Plan Apochromat oil objective and an electron multiplying charge-coupled device (C9100-50; Hamamatsu Photonics) camera. Cells were incubated on the microscope at 37°C. MT dynamics were analyzed from GFP-EB1 time-lapse videos using a published multiple particle-tracking software (Sironi et al., 2011). The software automatically detects GFP-EB1–labeled MT ends, reconstitutes MT tracks over consecutive time frames, and calculates for each track the mean growth speed, lifetime, and track length. To compare different cells, the median of the mean growth speeds was calculated, and the distributions of lifetimes and track lengths were fitted with exponential decay to calculate the characteristic lifetime and track length for each cell as previously described (Sironi et al., 2011).
TEM
RPE1 cells were grown on coverslips and fixed with 2.5% glutaraldehyde for 20 min starting at 30°C with a temperature decrease to 4°C. Glutaraldehyde was washed out with 50 mM cacodylate buffer. After postfixation in 2% OsO4 for 1–2 h at 4°C, cells were incubated in 0.5% aqueous uranyl acetate overnight and then dehydrated at 4°C using ascending ethanol concentrations and propylene oxide. After overnight incubation in an 1:1 mixture of Epon and propylene oxide, the samples were infiltrated with Epon for 2 h and polymerized at 60°C for 48 h. Serial ultrathin sections (70 nm) were stained with uranyl acetate and lead citrate and examined under a microscope (Philips EM 410; FEI) equipped with a digital camera (BioScan 792; Gatan) and DigitalMicrograph software (Gatan) or a microscope (EM 910; Carl Zeiss) equipped with a charge-coupled device camera (2,048 × 2,048 pixels; TRS Sharpeye; Tröndle) and Image SP software (Sys-Prog). Image brightness and contrast were adjusted in Photoshop CS3 (Adobe).
Centrosome purification
NIH 3T3 cells and NIH 3T3 cells stably carrying inducible LAP-hMARK4L wild-type or kd constructs were grown to a density of ∼80%. The TET-on constructs were induced by addition of 250 ng/ml of doxycycline in serum-free medium overnight. After washing with ice-cold PBS, the cells were harvested by scraping, pelleted for 5 min at 4°C and 1,200 g, washed with 10 ml of 8% sucrose (BDH) in PBS, and sedimented at 1,200 g for 5 min. The cell pellet was resuspended in 250 µl of 8% sucrose and frozen in liquid nitrogen for further processing. Centrosomes were isolated by centrifugation using a discontinuous sucrose gradient (Mitchison and Kirschner, 1984). In brief, cells were lysed for 10 min in 1.5 ml lysis buffer (1 mM Hepes-NaOH, pH 7.2, 0.5% NP-40, 0.5 mM MgCl2, 1 mM DTT, 1 mM PMSF, 10 µg/ml cytochalasin D, 200 µM sodium vanadate, and 50 mM sodium fluoride) supplemented with complete EDTA-free protease inhibitor cocktail (Roche). Swollen nuclei and cell debris were sedimented at 1,200 g for 10 min, and 1.2 ml of the supernatant was supplemented with 1 M Hepes-NaOH, pH 7.2, and 2 mg/ml DNase I to a final concentration of 10 mM and 10 µg/ml, respectively. After incubation at 4°C for 30 min, 1 ml lysis supernatant was overlaid on a discontinuous sucrose gradient set in one 11 × 34–mm polyallomer tube (Beckman Coulter), with 300 µl of 70% sucrose (wt/wt), 200 µl of 50% sucrose (wt/wt), and 400 µl of 40% sucrose (wt/wt) prepared in 10 mM Pipes-KOH, pH 7.2, 0.1% Triton X-100, 1 mM DTT, 200 µM sodium vanadate, and 50 mM sodium fluoride. Gradients were run at 100,000 g at 4°C for 1 h in a rotor (TLS-55; Beckman Coulter). Fractions of four drops (∼100 µl) were collected from the bottom of the gradient, and the protein concentration was determined using the Advanced Protein Assay (Cytoskeleton). The protein content of each fraction was precipitated with TCA before SDS-PAGE and immunoblot analysis. For protein quantification, samples corresponding to the centrosomal fractions were separated by SDS-PAGE and analyzed using anti–γ-tubulin and anti-ODF2 primary antibodies and IRDye 800–conjugated secondary antibody. Membranes were scanned with an imaging system (Odyssey Imager; LI-COR Biosciences), and band intensities were quantified using ImageJ software. The background measured in an empty area of the membrane was subtracted from all measured values. The amount of ODF2 in centrosomal fractions was normalized to the γ-tubulin amount, and the ratio of ODF2/γ-tubulin in each individual sample was calculated as the percentage within each experimental set to allow the comparison of independent experiments.
Immunoprecipitation
Transfected HEK293T cells were lysed for 20 min at 4°C in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 10% glycerol, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, and 1 mM DTT) supplemented with complete EDTA-free protease inhibitor cocktail. Lysates were clarified by centrifugation, and FLAG-tagged proteins were immunoprecipitated for 2 h using anti–FLAG M2 beads (Sigma-Aldrich). The beads were washed five times with radioimmunoprecipitation assay buffer, and bound proteins were analyzed by SDS-PAGE and immunoblotting. For immunoprecipitations from NIH 3T3 cells, lysis was performed at 4°C for 30 min with ELB buffer (50 mM Hepes, pH 7, 250 mM NaCl, 5 mM EDTA, 10% glycerol, 0.1% NP-40, 10 mM NaF, 50 mM β-glycerophosphate, 1 mM PMSF, and 1 mM DTT) supplemented with complete EDTA-free protease inhibitor cocktail. After centrifugation, 2 mg of the resulting supernatant was incubated with anti-GFP beads for 2 h. The beads were washed five times with ELB buffer, and immunoprecipitates were probed for endogenous ODF2 and MARK4 after SDS-PAGE and immunoblotting.
In vitro kinase assay
GST, GST-ODF2-F1, GST-ODF2-F2, and 6His-ODF2-F3 as well as 6His-MARK4L-ca and -kd were expressed in BL21 (DE3) Codon Plus cells and purified by affinity chromatography according to manufacturer’s instructions using glutathione Sepharose (GE Healthcare) or Ni-nitrilotriacetic acid agarose (QIAGEN). In the 6His-MARK4L-ca, the autophosphorylation sites (S26 and T214; Wissing et al., 2007) were substituted to glutamic acid to increase the kinase activity and to avoid background signals caused by autophosphorylation of the kinase. For in vitro kinase assays, the substrate and kinase were mixed in kinase buffer (50 mM Hepes, pH 7.4, 100 mM NaCl, 1 mM DTT, 10 mM MgCl2, and 200 µM ATP) supplemented with 5 µCi γ-[32P]ATP and incubated for 20 min at 30°C. Kinase reactions were stopped in sample buffer and subjected to SDS-PAGE followed by autoradiography and Coomassie Brilliant blue staining. Radioactivity was detected using an imaging system (Bas 1800 II; Fujifilm). Band intensities on protein gels detected with autoradiography or Coomassie staining were quantified using ImageJ software. Signal intensities were corrected against the gel background. The radioactive signal was normalized to the protein amount, and the relative phosphorylation in each individual sample was calculated as the percentage within each experimental set to allow the comparison of independent experiments.
Antibodies
Polyclonal MARK4 antibodies were raised in rabbit using the synthetic peptides TEEGGDRGAPLC (1) and CTLVTRISNDLEL (2; Peptide Specialty Laboratories GmbH) and purified from preabsorbed serum by affinity purification using peptide 2. Polyclonal ODF2 antibodies were raised in rabbit against amino acids 250–632 using a bacterially purified His-tagged ODF2 truncation (Peptide Specialty Laboratories GmbH). Polyclonal IFT88 antibodies were raised in rabbit as previously described (residues 365–824 of mouse IFT88; Pazour et al., 2002). Antibodies were purified from preabsorbed serum by affinity purification using the immobilized antigen.
Antibodies from commercial sources were obtained as follows: mouse anti–acetylated tubulin C3B9, rat anti–α-tubulin YL1/2, mouse anti–FLAG M2, mouse anti–γ-tubulin GTU88, or rabbit anti–γ-tubulin T5192 (Sigma-Aldrich); mouse antiactin (EMD Millipore); rabbit anti–detyrosinated tubulin (EMD Millipore); rabbit anti-Ki67 (Novocastra Laboratories); rabbit anti-Cep97, rabbit anti-CP110, or rabbit anti-Cep290 (Bethyl Laboratories, Inc.; kind gift of I. Hoffmann, DKFZ, Heidelberg, Germany); rabbit anti-Arl13b (Proteintech); and species-specific secondary antibodies coupled to Alexa Fluor dyes for immunofluorescence (Molecular Probes) or to horseradish peroxidase for immunoblotting (Dianova). Additional antibodies used in this study were rabbit anti-Cep164 (residues 1–298; Schmidt et al., 2012); rabbit anti-PCM1 (residues 665–2,024; gift of A. Merdes, Centre National de la Recherche Scientifique, Toulouse, France; Dammermann and Merdes, 2002); rabbit anti-BBS4 (residues 235–519) and rabbit anti-OFD1 (residues 145–1,012; gifts from A. Fry, University of Leicester, Leicester, England, UK; Lopes et al., 2011); mouse anti–polyglutamylated tubulin GT335 (synthetic peptide mimicking the structure of polyglutamylated sites of α-tubulin was used as an antigen; gift of C. Janke, Institute Curie, Orsay/Paris, France; Wolff et al., 1994); rabbit anti-GFP and rabbit anticentriolin (residues 938–2,711; gifts from E. Schiebel, ZMBH, University of Heidelberg, Heidelberg, Germany); and rabbit anti-Odf2 (residues 41–266 of mouse Odf2; gift of S. Tsukita, Osaka University, Osaka, Japan). Anti-GFP beads were generated as previously described (Rothbauer et al., 2008). In brief, the 6His-tagged GFP binding protein (6His-GBP) was expressed in BL21 (DE3) Codon Plus cells, purified by affinity chromatography using Ni-nitrilotriacetic acid agarose, and coupled to N-hydroxysuccinimide–activated Sepharose beads (Sigma-Aldrich) according to manufacturer’s instructions.
FACS analysis
Cell cycle profiles of serum-starved control or MARK4-depleted cells were determined by analyzing total DNA content using propidium iodide staining (30-min incubation of ethanol-fixed cells in PBS containing 50 µg/ml propidium iodide, 3.8 mM sodium citrate, 100 mg/ml RNase A, and 0.1% Triton X-100). Cells were subjected to flow cytometry by using a flow cytometer (FACScan; BD).
Online supplemental material
Fig. S1 illustrates the siRNA screening and microscope-based readout procedures used for the kinome-wide screen and shows the characterization of the MARK4 depletion phenotype with additional cilia markers. Fig. S2 shows the characterization of the anti-MARK4 antibody. Fig. S3 demonstrates that MARK4 depletion does not affect the MT-dependent transport of centriolar satellite proteins to the centrosome and shows the effect of MARK4 depletion on MT modifications. Fig. S4 shows the effect of MARK4 (depletion or overexpression) on the localization of centrosomal proteins determined by fluorescence microscopy and biochemical fractionation. Fig. S5 demonstrates that ODF2 depletion impairs subdistal but not distal appendage formation. Videos 1 and 2 show the MT dynamics in control-treated (Video 1) and in MARK4-treated (Video 2) RPE1 cells stably expressing GFP-EB1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201206013/DC1.
Supplementary Material
Acknowledgments
We thank Stephen Doxsey, Thomas Benzing, Alessandro Beghini, Elmar Schiebel, Sachiko Tsukita, Andreas Merdes, Andrew Fry, Carsten Janke, Iain Cheeseman, and Ingrid Hoffmann for reagents; Astrid Hofmann for excellent technical support; Alwin Krämer for sharing equipment; Karsten Richter for support with EM; Maren Klinger and Oliver Gruss for support with MT dynamics analysis; and Elmar Schiebel, Iain Hagan, Fouzia Ahmad, and laboratory members for comments on the manuscript. The ZMBH and DKFZ Light Microscopy facilities are gratefully acknowledged.
The ViroQuant-CellNetworks RNAi screening facility is supported by CellNetworks-Cluster of Excellence (EXC81). S. Kuhns and K.N. Schmidt were supported by the NG-HZ-111 grant and Helmholtz International Graduate School. M. Boutros received funding from the Helmholtz Alliance for Systems Biology. R. Carvalho is a fellow of the cooperative graduate program Disease Models and Drugs funded by the Landesgraduiertenförderung (University of Heidelberg) and a member of the Hartmut Hoffmann-Berling International Graduate School. This work was funded by the Helmholtz Association Grant NG-HZ-111 granted to G. Pereira.
Footnotes
Abbreviations used in this paper:
- ca
- catalytic active
- IFT
- intraflagellar transport
- kd
- kinase dead
- LAP
- localization and affinity purification
- MAP
- MT-associated protein
- MARK
- MT affinity regulating kinase
- MT
- microtubule
- TEM
- transmission EM
- TET
- tetracycline
- TSA
- Trichostatin A
References
- Anderson C.T., Stearns T. 2009. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr. Biol. 19:1498–1502 10.1016/j.cub.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehlke C., Kotsis F., Patel V., Braeg S., Voelker H., Bredt S., Beyer T., Janusch H., Hamann C., Gödel M., et al. 2010. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat. Cell Biol. 12:1115–1122 10.1038/ncb2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesger J., Wagner V., Weisheit W., Mittag M. 2009. Analysis of flagellar phosphoproteins from Chlamydomonas reinhardtii. Eukaryot. Cell. 8:922–932 10.1128/EC.00067-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brajenovic M., Joberty G., Küster B., Bouwmeester T., Drewes G. 2004. Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J. Biol. Chem. 279:12804–12811 10.1074/jbc.M312171200 [DOI] [PubMed] [Google Scholar]
- Cao M., Li G., Pan J. 2009. Regulation of cilia assembly, disassembly, and length by protein phosphorylation. Methods Cell Biol. 94:333–346 10.1016/S0091-679X(08)94017-6 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1 10.1126/stke.2662005pl1 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Merdes A. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo A., Franco B. 2009. The dynamic cilium in human diseases. Pathogenetics. 2:3 10.1186/1755-8417-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G., Ebneth A., Mandelkow E.M. 1998. MAPs, MARKs and microtubule dynamics. Trends Biochem. Sci. 23:307–311 10.1016/S0968-0004(98)01245-6 [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- Follit J.A., Tuft R.A., Fogarty K.E., Pazour G.J. 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 17:3781–3792 10.1091/mbc.E06-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghossoub R., Molla-Herman A., Bastin P., Benmerah A. 2011. The ciliary pocket: a once-forgotten membrane domain at the base of cilia. Biol. Cell. 103:131–144 10.1042/BC20100128 [DOI] [PubMed] [Google Scholar]
- Gossen M., Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA. 89:5547–5551 10.1073/pnas.89.12.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179:321–330 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarguaglini G., Duncan P.I., Stierhof Y.D., Holmström T., Duensing S., Nigg E.A. 2005. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol. Biol. Cell. 16:1095–1107 10.1091/mbc.E04-10-0939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurov J., Piwnica-Worms H. 2007. The Par-1/MARK family of protein kinases: from polarity to metabolism. Cell Cycle. 6:1966–1969 10.4161/cc.6.16.4576 [DOI] [PubMed] [Google Scholar]
- Inglis P.N., Boroevich K.A., Leroux M.R. 2006. Piecing together a ciliome. Trends Genet. 22:491–500 10.1016/j.tig.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Marshall W.F. 2011. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12:222–234 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Kubo A., Tsukita S., Tsukita S. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7:517–524 10.1038/ncb1251 [DOI] [PubMed] [Google Scholar]
- Jacob L.S., Wu X., Dodge M.E., Fan C.W., Kulak O., Chen B., Tang W., Wang B., Amatruda J.F., Lum L. 2011. Genome-wide RNAi screen reveals disease-associated genes that are common to Hedgehog and Wnt signaling. Sci. Signal. 4:ra4 10.1126/scisignal.2001225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Bulinski J.C. 2011. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 12:773–786 10.1038/nrm3227 [DOI] [PubMed] [Google Scholar]
- Jurczyk A., Gromley A., Redick S., San Agustin J., Witman G., Pazour G.J., Peters D.J., Doxsey S. 2004. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J. Cell Biol. 166:637–643 10.1083/jcb.200405023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Satoh S., Okabe H., Kitahara O., Ono K., Kihara C., Tanaka T., Tsunoda T., Yamaoka Y., Nakamura Y., Furukawa Y. 2001. Isolation of a novel human gene, MARKL1, homologous to MARK3 and its involvement in hepatocellular carcinogenesis. Neoplasia. 3:4–9 10.1038/sj.neo.7900132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketteler R., Glaser S., Sandra O., Martens U.M., Klingmüller U. 2002. Enhanced transgene expression in primitive hematopoietic progenitor cells and embryonic stem cells efficiently transduced by optimized retroviral hybrid vectors. Gene Ther. 9:477–487 10.1038/sj.gt.3301653 [DOI] [PubMed] [Google Scholar]
- Kim J., Lee J.E., Heynen-Genel S., Suyama E., Ono K., Lee K., Ideker T., Aza-Blanc P., Gleeson J.G. 2010. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 464:1048–1051 10.1038/nature08895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., Guo W. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA. 107:6346–6351 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto K., Yamazaki Y., Nishida T., Shinohara K., Ishikawa H., Hasegawa T., Okanoue T., Hamada H., Noda T., Tamura A., et al. 2012. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell. 148:189–200 10.1016/j.cell.2011.10.052 [DOI] [PubMed] [Google Scholar]
- Lange B.M., Gull K. 1995. A molecular marker for centriole maturation in the mammalian cell cycle. J. Cell Biol. 130:919–927 10.1083/jcb.130.4.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano J.M., Göransson O., Toth R., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Mäkelä T.P., Hardie D.G., Alessi D.R. 2004. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23:833–843 10.1038/sj.emboj.7600110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes C.A., Prosser S.L., Romio L., Hirst R.A., O’Callaghan C., Woolf A.S., Fry A.M. 2011. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J. Cell Sci. 124:600–612 10.1242/jcs.077156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A., Nugoor C., Panneerselvam S., Mandelkow E. 2010. Structure and function of polarity-inducing kinase family MARK/Par-1 within the branch of AMPK/Snf1-related kinases. FASEB J. 24:1637–1648 10.1096/fj.09-148064 [DOI] [PubMed] [Google Scholar]
- Matenia D., Mandelkow E.M. 2009. The tau of MARK: a polarized view of the cytoskeleton. Trends Biochem. Sci. 34:332–342 10.1016/j.tibs.2009.03.008 [DOI] [PubMed] [Google Scholar]
- Mikule K., Delaval B., Kaldis P., Jurcyzk A., Hergert P., Doxsey S. 2007. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat. Cell Biol. 9:160–170 10.1038/ncb1529 [DOI] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M. 1984. Microtubule assembly nucleated by isolated centrosomes. Nature. 312:232–237 10.1038/312232a0 [DOI] [PubMed] [Google Scholar]
- Moroni R.F., De Biasi S., Colapietro P., Larizza L., Beghini A. 2006. Distinct expression pattern of microtubule-associated protein/microtubule affinity-regulating kinase 4 in differentiated neurons. Neuroscience. 143:83–94 10.1016/j.neuroscience.2006.07.052 [DOI] [PubMed] [Google Scholar]
- Nakagawa Y., Yamane Y., Okanoue T., Tsukita S., Tsukita S. 2001. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol. Biol. Cell. 12:1687–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., Raff J.W. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Dickert B.L., Vucica Y., Seeley E.S., Rosenbaum J.L., Witman G.B., Cole D.G. 2000. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151:709–718 10.1083/jcb.151.3.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Baker S.A., Deane J.A., Cole D.G., Dickert B.L., Rosenbaum J.L., Witman G.B., Besharse J.C. 2002. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 157:103–113 10.1083/jcb.200107108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L.B., Rosenbaum J.L. 2008. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr. Top. Dev. Biol. 85:23–61 10.1016/S0070-2153(08)00802-8 [DOI] [PubMed] [Google Scholar]
- Pfeifer A.C., Kaschek D., Bachmann J., Klingmüller U., Timmer J. 2010. Model-based extension of high-throughput to high-content data. BMC Syst. Biol. 4:106 10.1186/1752-0509-4-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehl M., Tulu U.S., Wadsworth P., Cassimeris L. 2004. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc. Natl. Acad. Sci. USA. 101:1584–1588 10.1073/pnas.0308205100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova O.V., Golemis E.A., Pugacheva E.N. 2008. Cell cycle-dependent ciliogenesis and cancer. Cancer Res. 68:2058–2061 10.1158/0008-5472.CAN-07-5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby L.M., Parker J.D. 2005. Cilia and the cell cycle? J. Cell Biol. 169:707–710 10.1083/jcb.200503053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa J., Canovas P., Islam A., Altieri D.C., Doxsey S.J. 2006. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol. Biol. Cell. 17:1483–1493 10.1091/mbc.E05-08-0723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., Leonhardt H. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 7:282–289 [DOI] [PubMed] [Google Scholar]
- Santos N., Reiter J.F. 2008. Building it up and taking it down: the regulation of vertebrate ciliogenesis. Dev. Dyn. 237:1972–1981 10.1002/dvdy.21540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.D., Nigg E.A. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Schmidt K.N., Kuhns S., Neuner A., Hub B., Zentgraf H., Pereira G. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199:1083–1101 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Solon J., Conrad C., Mayer T.U., Brunner D., Ellenberg J. 2011. Automatic quantification of microtubule dynamics enables RNAi-screening of new mitotic spindle regulators. Cytoskeleton (Hoboken). 68:266–278 10.1002/cm.20510 [DOI] [PubMed] [Google Scholar]
- Sorokin S. 1962. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15:363–377 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S.P. 1968. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 3:207–230 [DOI] [PubMed] [Google Scholar]
- Soung N.K., Park J.E., Yu L.R., Lee K.H., Lee J.M., Bang J.K., Veenstra T.D., Rhee K., Lee K.S. 2009. Plk1-dependent and -independent roles of an ODF2 splice variant, hCenexin1, at the centrosome of somatic cells. Dev. Cell. 16:539–550 10.1016/j.devcel.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A., Tsang W.Y., Khoo D., Dynlacht B.D. 2007. Cep97 and CP110 suppress a cilia assembly program. Cell. 130:678–690 10.1016/j.cell.2007.06.027 [DOI] [PubMed] [Google Scholar]
- Tassan J.P., Le Goff X. 2004. An overview of the KIN1/PAR-1/MARK kinase family. Biol. Cell. 96:193–199 10.1016/j.biolcel.2003.10.009 [DOI] [PubMed] [Google Scholar]
- Timm T., Marx A., Panneerselvam S., Mandelkow E., Mandelkow E.M. 2008. Structure and regulation of MARK, a kinase involved in abnormal phosphorylation of Tau protein. BMC Neurosci. 9(Suppl. 2):S9 10.1186/1471-2202-9-S2-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinczek B., Brajenovic M., Ebneth A., Drewes G. 2004. MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes. J. Biol. Chem. 279:5915–5923 10.1074/jbc.M304528200 [DOI] [PubMed] [Google Scholar]
- Westlake C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA. 108:2759–2764 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing J., Jänsch L., Nimtz M., Dieterich G., Hornberger R., Kéri G., Wehland J., Daub H. 2007. Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Mol. Cell. Proteomics. 6:537–547 [DOI] [PubMed] [Google Scholar]
- Wolff A., Houdayer M., Chillet D., de Néchaud B., Denoulet P. 1994. Structure of the polyglutamyl chain of tubulin: occurrence of alpha and gamma linkages between glutamyl units revealed by monoreactive polyclonal antibodies. Biol. Cell. 81:11–16 10.1016/0248-4900(94)90049-3 [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Egerer J., Fuchs E., Haas A.K., Barr F.A. 2007. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 178:363–369 10.1083/jcb.200703047 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.