Summary
Clonal evolution is a key feature of cancer progression and relapse. We studied intratumoral heterogeneity in 149 chronic lymphocytic leukemia (CLL) cases by integrating whole-exome sequence and copy number to measure the fraction of cancer cells harboring each somatic mutation. We identified driver mutations as predominantly clonal (e.g., MYD88, trisomy 12 and del(13q)) or subclonal (e.g., SF3B1, TP53), corresponding to earlier and later events in CLL evolution. We sampled leukemia cells from 18 patients at two timepoints. Ten of 12 CLL cases treated with chemotherapy (but only 1 of 6 without treatment) underwent clonal evolution, predominantly involving subclones with driver mutations (e.g., SF3B1, TP53) that expanded over time. Furthermore, presence of a subclonal driver mutation was an independent risk factor for rapid disease progression. Our study thus uncovers patterns of clonal evolution in CLL, providing insights into its stepwise transformation, and links the presence of subclones with adverse clinical outcome.
INTRODUCTION
Recent genomic studies have revealed that individual cancer samples are genetically heterogeneous and contain subclonal populations (Carter et al., 2012; Ding et al., 2012; Gerlinger et al., 2012; Mullighan et al., 2008; Navin et al., 2011; Nik-Zainal et al., 2012; Shah et al., 2012). Indeed, tumors likely evolve through competition and interactions between genetically diverse clones (Snuderl et al., 2011). While the existence of intratumoral subclones has long been appreciated, little is known about the frequency, identity and evolution of subclonal genetic alterations or their impact on clinical course.
To examine the evolution and impact of subclonal mutations, we focused on chronic lymphocytic leukemia (CLL), a slow-growing B cell malignancy with disease onset in older individuals. CLL shows a highly variable disease course, partly explained by the diverse combinations of somatic mutations uncovered by sequencing studies (Quesada et al., 2012; Wang et al., 2011). We hypothesized that the presence, diversity and evolutionary dynamics of subclonal mutations in CLL also contribute to the variations observed in disease tempo and response to therapy (Schuh et al., 2012; Stilgenbauer et al., 2007). Importantly, the slow growth of CLL-B cells (relative to other malignancies) provides an extended window for observing the process of clonal evolution, as it may take months to years for a new clone to fully replace previous clones (Schuh et al., 2012; Wu, 2012).
Subclonal mutations in CLL have been detected by fluorescence in situ hybridization (FISH)(Shanafelt et al., 2008) and microarrays (Grubor et al., 2009), showing that they harbor driver lesions and evolve over time. Since these methods can only be used to detect a limited number of genetic alterations, more recent studies have used whole-genome sequencing to quantify thousands of somatic mutations per sample and track subclones by clustering alterations of similar allelic frequency (Ding et al., 2012; Egan et al., 2012; Nik-Zainal et al., 2012; Schuh et al., 2012; Shah et al., 2012; Walter et al., 2012). However, because genome-wide sequencing is currently not feasibly applied to large sample collections, the patterns of clonal evolution and their effects on disease course have not been fully elucidated.
Whole-exome sequencing (WES) (Gnirke et al., 2009) of tumors is an affordable, rapid and comprehensive technology for detecting somatic coding mutations. We sought to refine and apply a method for analysis of subclonal mutations using WES since: (i) the high sequencing depth obtained by WES (typically ~100-150X) enables reliable detection of subclonal mutations required for defining subclones and tracking them over time (Cibulskis et al, under review); (ii) coding mutations likely encompass many of the important driver events that provide fitness advantage for specific clones; and finally, (iii) the relatively low cost of WES permits studies of large cohorts, which is key for understanding the relative fitness and temporal order of driver mutations and for assessing the impact of clonal heterogeneity on disease outcome.
To this end, we performed large-scale WES of 160 CLL tumor/normal pairs that represented the broad clinical spectrum of CLL. In particular, we examined the roles of CLL subclones and the mutations that they harbor by integrative analysis of coding mutations and somatic copy number alterations, which enabled estimation of the cancer cell fraction. This was performed in samples from 149 CLL patients, including 18 patients sampled at two timepoints for which both exome sequencing data and copy number data were available. This analysis allowed us to study mutation frequencies, observe clonal evolution and link subclonal mutations to clinical outcome.
RESULTS
Large-scale WES analysis of CLL expands the compendium of CLL drivers and pathways
We performed whole-exome sequencing (WES) of 160 matched CLL and germline DNA samples (including 82 of the 91 samples previously reported (Wang et al., 2011)). This cohort included patients with both low- and high-risk features based on established prognostic risk factors (Table S1). We applied MuTect (a highly sensitive and specific mutation-calling algorithm; Cibulskis et al., under review) to the WES data to detect somatic single nucleotide variations (sSNVs) present in as few as 10% of cancer cells. Average sequencing depth of WES across samples was ~130X (see Extended Experimental Procedures). In total, we detected 2,444 nonsynonymous and 837 synonymous mutations in protein-coding sequences, corresponding to a mean (±SD) somatic mutation rate of 0.6±0.28 per megabase (range, 0.03 to 2.3), and an average of 15.3 nonsynonymous mutations per patient (range, 2 to 53) (Table S2).
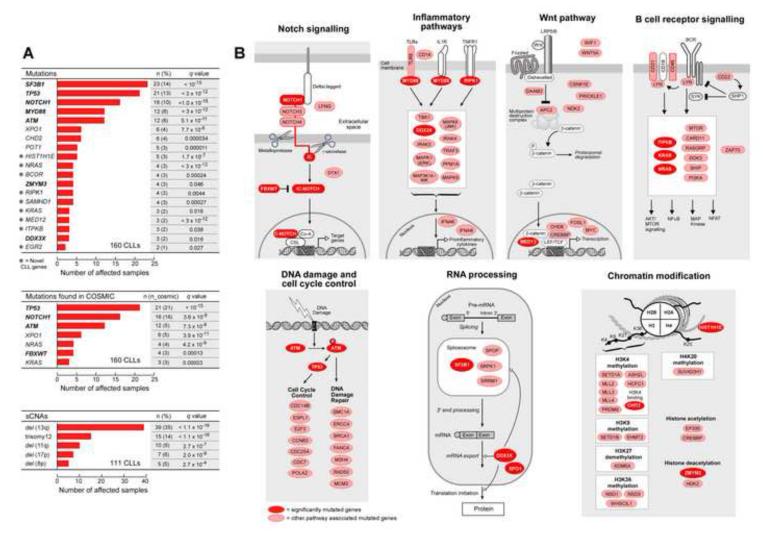
Expansion of our sample cohort provided us with the sensitivity to detect 20 putative CLL cancer genes (q<0.1), which was accomplished through recurrence analysis to detect genes enriched with mutations beyond the background mutation rate (Figure 1A-top, Figure S1A) or genes with mutations that overlap with previously reported mutated sites (from COSMIC (Forbes et al., 2010); Figure 1A-middle) (see Experimental Procedures). These included 8 of the 9 genes identified in our initial report (TP53, ATM, MYD88, SF3B1, NOTCH1, DDX3X, ZMYM3, FBXW7) (Wang et al., 2011). The missing gene, MAPK1, did not harbor additional mutations in the increased sample set and therefore its overall mutation frequency now fell below our significance threshold. The 12 newly identified genes were mutated at lower frequencies, and hence were not detected in the previously reported subset of samples. Three of the 12 additional candidate driver genes were identified in recent CLL sequencing efforts (XPO1, CHD2, and POT1) (Fabbri et al., 2011; Puente et al., 2011). The 9 remaining genes represent novel candidate CLL drivers, with mutations occurring at highly conserved sites (Figure S1B). These included six genes with known roles in cancer biology (NRAS, KRAS (Bos, 1989), BCOR (Grossmann et al., 2011), EGR2 (Unoki and Nakamura, 2003), MED12 (Makinen et al., 2011) and RIPK1 (Hosgood et al., 2009)), two genes that affect immune pathways (SAMHD1 (Rice et al., 2009), ITPKB (Marechal et al., 2011)) and a histone gene (HIST1H1E (Alami et al., 2003)).
Figure 1. Significantly mutated genes and associated gene pathways in 160 CLL samples.
(A) Mutation significance analysis, using the MutSig2.0 and GISTIC2.0 algorithms identifies recurrently mutated genes and recurrent sCNAs in CLL, respectively. Bold – significantly mutated genes identified in a previous CLL sequencing effort (Wang et al., 2011). * - novel CLL genes identified in the present analysis (Figure S1). ‘n’ – number of samples out of 160 CLLs harboring a mutation in a specific gene; ‘n_cosmic’ – number of samples harboring a mutation in a specific gene at a site previously observed in the COSMIC database. (B) The significantly mutated genes fall into seven core signaling pathways, in which the genes play well-established roles. Red - genes with significant mutation frequencies; pink - additional pathway genes with mutations.
Together, the 20 candidate CLL driver genes appeared to fall into 7 core signaling pathways, in which the genes play well-established roles. These include all five pathways that we previously reported to play a role in CLL (DNA repair and cell-cycle control, Notch signaling, inflammatory pathways, Wnt signaling, RNA splicing and processing). Two new pathways were implicated by our analysis: B cell receptor signaling and chromatin modification (Figure 1B). We also noted that the CLL samples contained additional mutations in the genes that form these pathways, some of which are known drivers in other malignancies.
Because recurrent chromosomal abnormalities have defined roles in CLL biology (Döhner et al., 2000; Klein et al., 2010), we further searched for loci that were significantly amplified or deleted by analyzing somatic copy-number alterations (sCNAs). We applied GISTIC2.0 (Mermel et al., 2011) to 111 matched tumor and normal samples which were analyzed by SNP6.0 arrays (Brown et al., 2012). Through this analysis, we identified deletions in chromosomes 8p, 13q, 11q, and 17p and trisomy of chromosome 12 as significantly recurrent events (Figure 1A-bottom). Thus, based on WES and copy number analysis, we altogether identified 20 mutated genes and 5 cytogenetic alterations as putative CLL driver events.
Inference of genetic evolution with whole-exome sequencing data
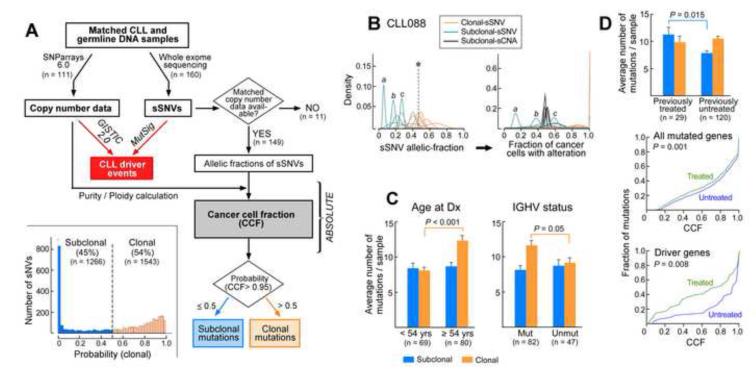
In order to study clonal evolution in CLL, we performed integrative analysis of sCNAs and sSNVs using a recently reported algorithm ABSOLUTE (Carter et al., 2012), which jointly estimated the purity of the sample (fraction of cancer nuclei) and the average ploidy of the cancer cells. All samples were estimated to have near-diploid DNA content; these estimates were confirmed by FACS analysis of 7 CLL samples (Figure S2A). Our data were sufficient for resolution of these quantities in 149 of the 160 samples (Table S3A), allowing for discrimination of subclonal from clonal alterations, including sCNAs, sSNVs, and selected indels (see Extended Experimental Procedures). Our analysis approach is outlined in Fig 2A. For each sSNV, we estimated its allelic fraction by calculating the ratio of alternate to total number of reads covering the mutation site in the WES data. These estimates were consistent with independent deeper sequencing and RNA sequencing (Figure S2B-C, Tables S4-5). Next, we used ABSOLUTE to estimate the cancer cell fraction (CCF) harboring the mutation by correcting for sample purity and local copy-number at the sSNV sites (Experimental Procedures, Table S3B, Figure 2B). We classified a mutation as clonal if the CCF harboring it was >0.95 with probability > 0.5, and subclonal otherwise (Figure 2A inset). The results remained unchanged when more stringent cutoffs were used (Extended Experimental Procedures). For sSNVs designated as subclonal, median CCF was 0.49 with a range of 0.11 to 0.89.
Figure 2. Subclonal and clonal somatic single nucleotide variants (sSNVs) are detected in CLL in varying quantities based on age at diagnosis, IGHV mutation status, and treatment status (also see Figure S2).
(A) The analysis workflow: ‘CLL driver events’ (red box) were identified by mutation significance analysis using WES and SNP array data collected from matched germline and tumor DNA. For the 149 samples that had matched WES and copy number data, ABSOLUTE was applied to estimate the cancer cell fraction (CCF). Mutations were classified as subclonal (blue) or clonal (orange), based on the probability that their CCF is greater than 0.95 (clonal). Inset - Histogram of the probability of being clonal for all sSNVs across 149 CLL samples. (B) A representative example of the transformations generated by ABSOLUTE (for sample CLL088). First, probability density distributions of allelic fractions for each mutation are plotted (representative peaks for sSNVs a, b and c shown). Second, these data are converted to CCF (right panel), incorporating purity and local copy number information. The probability of the event being clonal (i.e affecting >0.95 of cells) is represented by a color spectrum: orange-high probability; blue-low probability. *- allelic fraction of a clonal mutation at multiplicity of 1 (for example, a heterozygous mutation in a diploid region). (C) Comparison of the number of subclonal and clonal sSNVs/sample based on patient age at diagnosis and IGHV mutation status. (D) Comparison of the number of subclonal and clonal sSNVs/sample based on treatment status at time of sample collection (top panel). Cumulative distribution of the sSNVs by CCF is shown for samples from treated and untreated patients for all (middle panel) and only driver sSNVs (bottom panel).
Overall, we identified 1,543 clonal mutations (54% of all detected mutations, average of 10.3±5.5 mutations per sample, Table S1). These mutations were likely acquired either before or during the most recent complete selective sweep. This set therefore includes both neutral somatic mutations that preceded transformation and the driver and passenger event(s) present in each complete clonal sweep. A total of 1,266 subclonal sSNVs were detected in 146 of 149 samples called by ABSOLUTE (46%; average of 8.5±5.8 subclonal mutations per sample). These subclonal sSNVs exist in only a fraction of leukemic cells, and hence occurred after the emergence of the “most-recent common ancestor”, and by definition, also after disease initiation. The mutational spectra were similar in clonal and subclonal sSNVs (Figure S2D), consistent with a common set of mutational processes giving rise to both groups.
Age and mutated IGHV status are associated with an increased number of clonal somatic mutations
The identification of subclones enabled us to analyze several aspects of leukemia progression. We first addressed how clonal and subclonal mutations relate to the salient clinical characteristics of CLL. CLL is generally a disease of the elderly with established prognostic factors, such as the IGHV mutation (Döhner, 2005) and ZAP70 expression. Patients with a high number of IGHV mutations (mutated IGHV) tend to have better prognosis than those with a low number (unmutated IGHV) (Döhner, 2005). This marker distinguishes between leukemias originating from B cells that have or have not yet, respectively, undergone the process of somatic hypermutation that occurs as part of normal B cell development. We examined the association of these factors, as well as patient age at diagnosis, with the prevalence of clonal and subclonal mutations. Age and mutated IGHV status (but not ZAP70 expression) were found to associate with greater numbers of clonal (but not subclonal) mutations (age, P<0.001; mutated vs unmutated IGHV, P=0.05; Figure 2C, Table S1). Since CLL samples with mutated IGHV derive from B-cells that have experienced a burst of mutagenesis as part of normal B cell somatic hypermutation, the increased number of clonal somatic mutations is likely related to aberrant mutagenesis that preceded clonal transformation (Deutsch et al., 2007; McCarthy et al., 2003). Furthermore, the higher number of clonal sSNVs in older individuals is consistent with the expectation that more neutral somatic mutations accumulate over the patient’s lifetime prior to the onset of cancer later in life (Stephens et al., 2012; Welch et al., 2012).
Subclonal mutations are increased with treatment
The effect of treatment on subclonal heterogeneity in CLL is unknown. In samples from 29 patients treated with chemotherapy prior to sample collection, we observed a significantly higher number of subclonal (but not clonal) sSNVs per sample than in the 120 patients who were chemotherapy-naïve at time of sample (Figure 2D-top and middle panels). Using an analysis of covariance model, we observed that receipt of treatment prior to sample among the 149 patients was statistically significant (P=0.048) but time from diagnosis to sample was not (P=0.31). Because patients that do not require treatment in the long-term may have a distinct subtype of CLL, we also restricted the comparison of the 29 pre-treated CLLs to only the 42 that were eventually treated after sample collection and again confirmed this finding (P=0.02). In these 42 patients, a higher number of subclonal mutations was not correlated with a shorter time to treatment (correlation coefficient =0.03; P=0.87). Thus, therapy prior to sample was associated with a higher number of subclonal mutations, and furthermore, the number of subclonal sSNVs detected increased with the number of prior therapies (P=0.011, Table S1).
Cancer therapy has been theorized to be an evolutionary bottleneck, in which a massive reduction in malignant cell numbers results in reduced genetic variation in the cell population (Gerlinger and Swanton, 2010). It is likely that the overall diversity in CLL is diminished after therapeutic bottlenecks as well. Because most of the genetic heterogeneity within a cancer is present at very low frequencies (Gerstung et al., 2012) --below the level of detection afforded by the ~130X sequence coverage we generated -- we were unable to directly assess reduction in overall genetic variation.
However, in the range of larger subclones that were observable by our methods, (>10% of malignant cells), we witnessed increased diversity after therapy (Figure 2D). Although, the available data cannot definitively rule out extensive diversification following therapy, this increase likely results, at least in part, from outgrowth of pre-existing minor subclones (Schuh et al., 2012; Wu, 2012). This may result from the removal of dominant clones by cytotoxic treatment, eliminating competition for growth and allowing the expansion of one or more fit subclones to frequencies above our detection threshold. Further supporting our interpretation that fitter clones grow more effectively and become detectable after treatment, we observed an increased frequency of subclonal driver events (which are presumably fitter) in treated relative to untreated patients (Figure 2D-bottom) (note that driver events include CLL driver mutations (Figure 1A) and sSNVs in highly conserved sites of genes in the Cancer Gene Census (Futreal et al., 2004)).
Inferring the order of genetic changes underlying CLL
While general aspects of temporal evolution could not be completely resolved in single timepoint WES samples, the order of driver mutation acquisition could be partially inferred from the aggregate frequencies at which they are found to be clonal or subclonal. We considered the 149 samples as a series of “snapshots” taken along a temporal axis. Clonal status in all or most mutations affecting a specific gene or chromosomal lesion would suggest that this alteration was acquired at or prior to the most recent selective sweep before sampling and hence could be defined as a stereotypically early event. Conversely, predominantly subclonal status in a specific genetic alteration implies a likely later event that is tolerated and selected for only in the presence of an additional mutation.
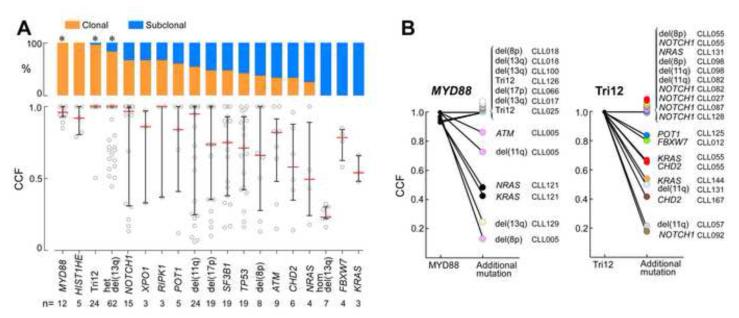
This strategy was used to infer temporal ordering of the recurrent sSNVs and sCNAs (Fig 3A, Figure S3). We focused on alterations found in at least 3 samples within the cohort of 149 CLL samples. We found that three driver mutations – MYD88 (n=12), trisomy 12 (n=24), and hemizygous del(13q) (n=70) – were clonal in 80-100% of samples harboring these alterations, a significantly higher level than for other driver events (q<0.1, Fisher exact test with Benjamini-Hochberg FDR (Benjamini and Hochberg, 1995)), implying that they arise earlier in typical CLL development. Mutations in HIST1H1E, although clonal in 5 of 5 affected samples, did not reach statistical significance. Other recurrent CLL drivers – for example, ATM, TP53 and SF3B1 (9, 19 and 19 mutations in 6, 17 and 19 samples, respectively) -- were more often subclonal, indicating that they tend to arise later in leukemic development and contribute to disease progression. We note that the above approach assumed that different CLL samples evolve along a common temporal progression axis. We therefore examined specifically CLL samples that harbored one ‘early’ driver mutation and any additional driver alteration(s). As expected, the ‘early’ events had either similar or a higher CCF compared to ‘later’ events (examples for trisomy 12 and MYD88 given in Figure 3B).
Figure 3. Identification of earlier and later CLL driver mutations (also see Figure S3).
(A) Distribution of estimated cancer cell fraction (CCF) (bottom panel) and percent of the mutations classified as clonal (orange) or subclonal (blue) for CLL drivers (top panel); * - drivers with q-values <0.1 for a higher proportion of clonal mutations compared with the entire CLL drivers set. Het - heterozygous deletion; Hom - homozygous deletion. The analysis includes all recurrently mutated genes (Figure 1A) with 3 or more events in the 149 samples, excluding sSNVs affecting the X chromosome currently not analyzable by ABSOLUTE, and also excluding indels in genes other than NOTCH1. (B) All CLL samples with MYD88 (left) or trisomy 12 (right) and at least 1 additional defined CLL driver (i.e. 9 of 12 samples with mutated MYD88; 14 of 16 tumors with trisomy 12) are depicted. Each dot color denotes separate individual CLL samples.
Direct observation of clonal evolution by longitudinal data analysis of chemotherapy-treated CLL
To directly assess the evolution of somatic mutations in a subset of patients, we compared CCF for each alteration across two clinical timepoints in 18 of the 149 samples (median years between timepoints was 3.5; range 3.1-4.5). Six patients (‘untreated’) did not receive treatment throughout the time of study. The remaining 12 patients (‘treated’) received intervening chemotherapy (primarily fludarabine and/or rituxan-based) (Table S6). The two patient groups were not significantly different in terms of elapsed time between first and second sample (median 3.7 years for the 6 untreated patients compared to 3.5 years for the 12 treated patients, P=0.62; exact Wilcoxon rank-sum test), nor did it differ between time of diagnosis to first sample (P=0.29).
Analysis of the 18 sets of data revealed that 11% of mutations increased (34 sSNVs, 15 sCNAs), 2% decreased (6 sSNVs, 2 sCNAs) and 87% did not change their CCF over time (q <0.1 for significant change in CCF, Table S7). As suggested by our single timepoint analysis, we observed a shift of subclonal driver mutations (e.g., del(11q), SF3B1 and TP53) towards clonality over time. Changes in the genetic composition of CLL cells with clonal evolution were associated with network level changes in gene expression related to emergence of specific subclonal populations (e.g. changes in signatures associated with SF3B1 or NRAS mutation, Figure S4D-E, Table S10)). Finally, expanding sSNVs were enriched in genes included in the Cancer Gene Census (Futreal et al., 2004) (P=0.021) and in CLL drivers (P=0.028), consistent with the expected positive selection for the subclones harboring them.
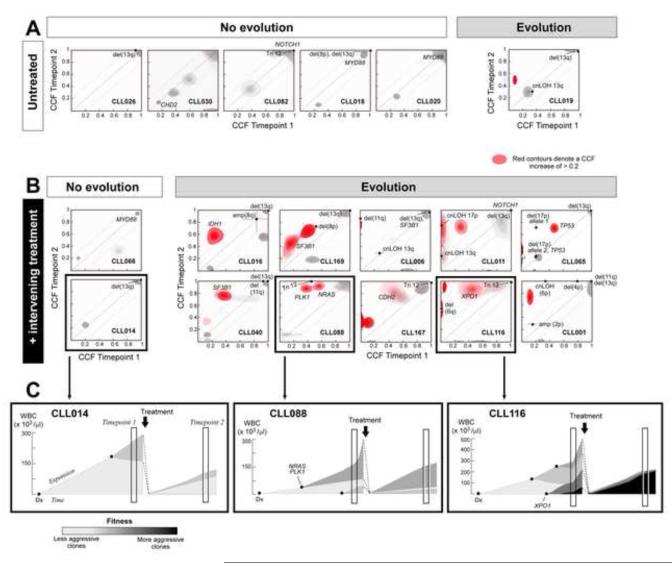
Clustering analysis of CCF distributions of individual genetic events over the two timepoints (Extended Experimental Procedures), revealed clear clonal evolution in 11 of 18 CLL sample pairs. We observed clonal evolution in 10 of 12 sample pairs which had undergone intervening treatment between timepoints 1 and 2 (Figure 4B, Figure S4A-C). This was contrasted with the 6 untreated CLLs, 5 of which demonstrated equilibrium between subpopulations that was maintained over several years (Figure 4A; P=0.012, Fisher exact test). Of the 11 patients with subclonal evolution across the sampling interval, 5 followed a branched evolution pattern as indicated by the disappearance of mutations with high CCF co-occurring with the expansion of other subclones (Figure 4B). This finding demonstrates that co-existing sibling subclones are at least as common in CLL as are linear nested subclones, as demonstrated in other hematological malignancies (Ding et al., 2012; Egan et al., 2012). We conclude that chemotherapy-treated CLLs often undergo clonal evolution resulting in the expansion of previously minor subclones. Thus, these longitudinal data validate the insights obtained in the cross-sectional analysis, namely that (i) ‘later’ driver events expand over time (Figure 3A) and (ii) treatment results in the expansion of subclones enriched with drivers (and thus presumably have higher fitness) (Figure 2D).
Figure 4. Longitudinal analysis of subclonal evolution in CLL and its relation to therapy (also see Figure S4).
Joint distributions of cancer cell fraction (CCF) values across two timepoints were estimated using clustering analysis (see Expanded Experimental Procedures). Red - a mutation with an increase in CCF of greater than 0.2 (with probability >0.5). The dotted diagonal line represents y=x, or where identical CCF values across the two timepoints fall; the dotted parallel lines denote the 0.2 CCF interval on either side. Likely driver mutations were labeled. Six CLLs with no intervening treatment (A) and 12 CLLs with intervening treatment (B) were classified according to clonal evolution status, based on the presence of mutations with an increase of CCF > 0.2. (C) Hypothesized sequence of evolution, inferred from the patients’ WBC counts, treatment dates, and changes in CCF for 3 representative examples.
Presence of subclonal drivers adversely impacts clinical outcome
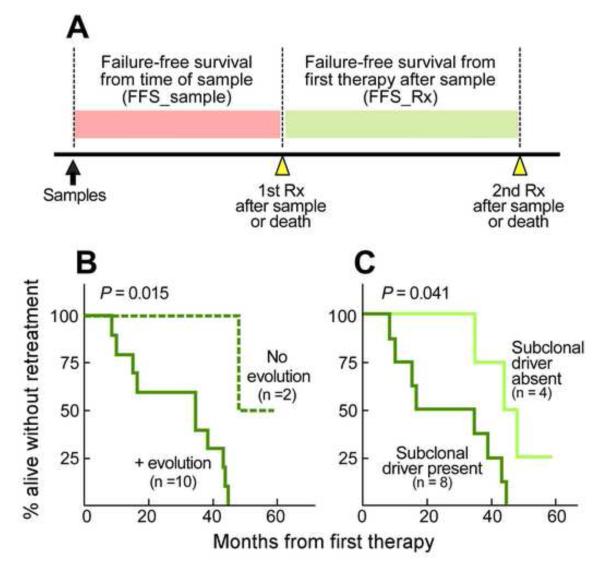
We observed treatment-associated clonal evolution to lead to the replacement of the incumbent clone by a fitter pre-existing subclone (Figure 4B). Therefore, we would expect a shorter time to relapse in individuals with evidence of clonal evolution following treatment. As a measure of relapse, we assessed failure-free survival from time of sample (‘FFS_Sample’) and failure-free survival from time of next therapy (‘FFS_Rx’, Figure 5A), where failure is defined as retreatment (a recognized endpoint in slow growing lymphomas (Cheson et al., 2007)) or death. For the study of clonal evolution in CLL, retreatment as an endpoint is preferable to other measures such as progression alone, as this is a well-defined event that reflects CLL disease aggressiveness. For example, disease progression alone in CLL may be asymptomatic without necessitating treatment; conversely, treatment is administered only in the setting of symptomatic disease or active disease relapse (Hallek et al., 2008).
Figure 5. Genetic evolution and clonal heterogeneity results in altered clinical outcome.
(A) Schema of the main clinical outcome measures that were analyzed: failure free survival from time of sample (FFS_Sample) and from initiation of first treatment after sampling (FFS_Rx). Within the longitudinally followed CLLs that received intervening treatment (12 of 18), shorter FFS_Rx was observed in CLL samples that (B) had evidence of genetic evolution (n=10) compared to samples with absent or minimal evolution (n=2; Fisher exact test), and that (C) harbored a detectable subclonal driver in the pretreatment sample (n=8) compared to samples with absent subclonal driver (n=4).
Within the 12 of 18 longitudinally analyzed samples that received intervening treatment, we observed that the 10 samples with clonal evolution exhibited shortened FFS_Rx (log-rank test; P=0.015, Figure 5B). Importantly, the somatic driver mutations that expanded to take over the entire population upon relapse (‘timepoint-2’), were often already detectable in the pre-treatment (‘timepoint-1’) sample (Figures 4B and S4B). Our results thus suggested that presence of detectable subclonal drivers in pre-treatment samples can anticipate clonal evolution in association with treatment. Indeed, the 8 of 12 samples with presence of subclonal drivers in pretreatment samples exhibited shorter FFS_Rx than the 4 samples with subclonal drivers absent (p=0.041; Figure 5C). Together, the results of our longitudinally studied patient samples suggested that the presence of driver events within subclones may impact prognosis and clinical outcome.
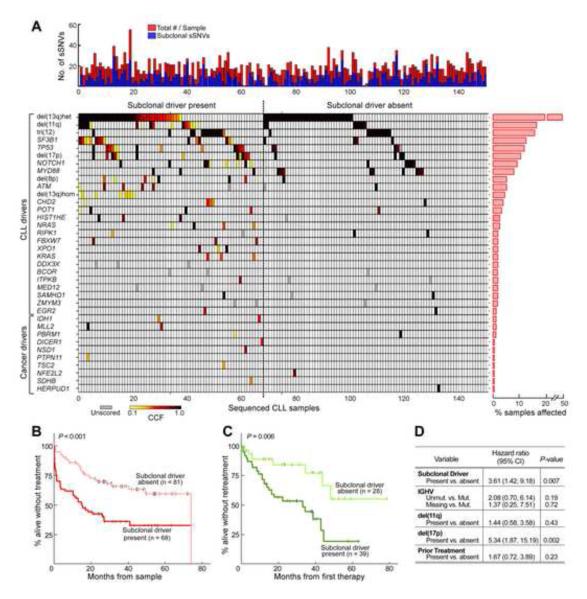
We tested this hypothesis in the set of 149 patient samples, of which subclonal driver mutations were detected in 46% (Figure 6A; Extended Experimental Procedures; Table S8). Indeed, we found that CLL samples with subclonal driver mutations were associated with a shorter time from sample collection to tratment or death (‘FFS_Sample’, P<0.001, Figure 6B, Table S9A,C), that seemed to be independent of established markers of poor prognosis (i.e. unmutated IGHV, or presence of del(11q) or del(17p), Figure S5). Moreover, we tested specifically whether the presence of pre-treatment subclonal drivers was associated with a shorter FFS_Rx, as we observed in the longitudinal data. Therefore, we focused on the 67 patients who were treated after sample collection (median time to first therapy from time of sample was 11 months [range 1-45]). These patients could be divided into two groups based on the presence (n=39) or absence (n=29) of a subclonal driver (62% and 64%, respectively, were treated with fludarabine-based immunochemotherapy, P=0.4). The 39 of these patients in which subclonal CLL drivers were detected required earlier retreatment or died (shorter FFS_Rx; log-rank test, P=0.006; Figure 6C, Table S9A), indicative of a more rapid disease course.
Figure 6. Presence of subclonal drivers mutations adversely impacts clinical outcome.
(A) Analysis of genetic evolution and clonal heterogeneity in 149 CLL samples. Top panel - total number (red) and the number of subclonal (blue) mutations per sample. Bottom panel - co-occurring CLL or cancer drivers (sSNVs in highly conserved sites in Cancer Gene Census genes) detected in the 149 samples. Color spectrum (light yellow to black) corresponds to estimated cancer cell fraction (CCF); white boxes - not detected; grey – CCF not estimated (genes on the X chromosome and indels other than in NOTCH1). (B-C) Subclonal drivers are associated with adverse clinical outcome. (B) CLL samples containing a detectable subclonal driver (n=68) exhibited shorter FFS_Sample compared to samples with absent subclonal drivers (n=81) (also see Figure S5). (C) Subclonal drivers were associated with shorter FFS_Rx in 67 samples which were treated after sampling. (D) A Cox multivariable regression model designed to test for prognostic factors contributing to shorter FFS_Rx showed that presence of a subclonal driver was an independent predictor of outcome.
Regression models adjusting for multiple CLL prognostic factors (IGHV status, prior therapy and high risk cytogenetics) supported the presence of a subclonal driver as an independent risk factor for earlier retreatment (adjusted hazard ratio (HR) of 3.61 (CI 1.42-9.18), Cox P=0.007; unadjusted HR, 3.20 (CI 1.35-7.60); Figure 6D), comparable to the strongest known CLL risk factors. In similar modeling within a subset of 62 patients who had at least one driver (clonal or subclonal), the association of the presence of a subclonal driver with a shorter time to retreatment or death was also significant (P=0.012, Table S9B) reflecting that this difference is not merely attributable to the presence of a driver. Additionally, an increased number of subclonal driver mutations per sample (but not clonal drivers) was also associated with a stronger HR for shorter FFS_Rx (Table S9D). Finally, this association retained significance (Cox P=0.033, Table S9E) after adjusting for the presence of mutations previously associated with poor prognosis (ATM, TP53, SF3B1), suggesting that in addition to the driver’s identity, its subclonal status also affects clinical outcome.
DISCUSSION
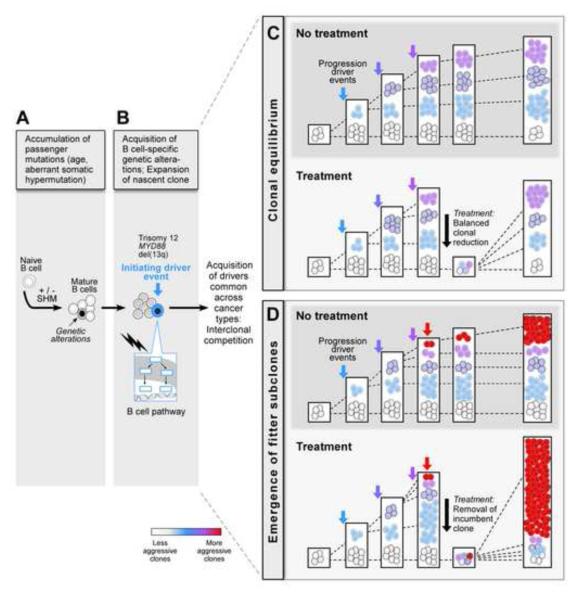
While intertumoral (Quesada et al., 2012; Wang et al., 2011) and intra-tumoral (Schuh et al., 2012; Stilgenbauer et al., 2007) genetic heterogeneity had been previously demonstrated in CLL, our use of novel WES-based algorithms enabled a more comprehensive study of clonal evolution and its clinical impact. We propose the existence of distinct periods in CLL progression. In the first period prior to transformation, passenger events accumulate in the cell that will eventually be the founder of the leukemia (in proportion to the age of the patient; Figure 2C), and are thus clonal mutations (Figure 7A). In the second period, the founding CLL mutation appears in a single cell and leads to transformation (Figure 7B); these are also clonal mutations, but unlike passenger mutations, these are recurrent across patients. We identified driver mutations that were consistently clonal (del(13q), MYD88 and trisomy 12; Figure 3A) and which appear to be relatively specific drivers of CLL or B cell malignancies (Beroukhim et al., 2010; Döhner et al., 2000; Ngo et al., 2010). In the third period of disease progression, subclonal mutations expand over time as a function of their fitness integrating intrinsic factors (e.g. proliferation and apoptosis) and extrinsic pressures (e.g. interclonal competition and therapy) (Figure 7C-D). The subclonal drivers include ubiquitous cancer genes, such as ATM, TP53 or RAS mutations (Figure 3A). These data suggest that mutations that selectively affect B cells may contribute more to the initiation of disease and precede selection of more generic cancer drivers that underlie disease progression – providing predictions that can be tested in human B cells or animal models of CLL.
Figure 7. A model for the stepwise transformation of CLL.
Our data suggest distinct periods in the history of CLL. We observed an increase in clonal mutations in older patients and in the IGHV mutated subtype, likely corresponding to pre-transformation mutagenesis (A). We identified earlier and later mutations in CLL, consistent with B cell-specific (B) and ubiquitous cancer events (C-D), respectively. Finally, clonal evolution and treatment show a complex relationship. Most untreated CLLs and a minority of treated CLLs maintain stable clonal equilibrium over years (C). However, in the presence of a subclone containing a strong driver, treatment may disrupt interclonal equilibrium and hasten clonal evolution (D).
An important question addressed here is how treatment affects clonal evolution in CLL. In the 18 patients monitored at 2 timepoints, we observed two general patterns – clonal equilibrium in which the relative sizes of each subclone were maintained and clonal evolution in which some subclones emerge as dominant (Figure 4). We propose that in untreated samples, more time is needed for a new fit clone to take over the population in the presence of existing dominant clones (Figure 7D-top). In contrast, in treated samples, cytotoxic therapy typically removes the incumbent clones (Jablonski, 2001) -- acting like a ‘mass extinction’ event (Jablonski, 2001) -- and shifts the evolutionary landscape (Nowak and Sigmund, 2004; Vincent and Gatenby, 2008) in favor of one or more aggressive subclones (Maley et al., 2006) (Figure 7D-bottom). Thus, highly fit subclones likely benefit from treatment and exhibit rapid outgrowth (Greaves and Maley, 2012).
CLL is an incurable disease with a prolonged course of remissions and relapses. It has been long recognized that relapsed disease responds increasingly less well to therapy over time. We now show an association between increased clinical aggressiveness and genetic evolution, which has therapeutic implications. We found that the presence of pre-treatment subclonal driver mutations anticipated the dominant genetic composition of the relapsing tumor. Such information may eventually guide the selection of therapies to prevent the expansion of highly fit subclones. In addition, the potential hastening of the evolutionary process with treatment provides a mechanistic justification for the empirical practice of ‘watch and wait’ as the CLL treatment paradigm (CLL Trialists Collaborative Group, 1999). The detection of driver mutations in subclones (a testimony to an active evolutionary process) may thus provide a new prognostic approach in CLL, which can now be rigorously tested in larger clinical trials.
In conclusion, we demonstrate the ability to study tumor heterogeneity and clonal evolution with standard WES. These innovations will allow characterization of the subclonal mutation spectrum in large, publically available datasets (Masica and Karchin, 2011). The implementation described here may also be readily adopted for clinical applications. Even more importantly, our studies underscore the importance of evolutionary development as the engine driving cancer relapse. This new knowledge challenges us to develop novel therapeutic paradigms that not only target specific drivers (i.e ‘targeted therapy’) but also the evolutionary landscape (Nowak and Sigmund, 2004) of these drivers.
Experimental procedures
149 patients with CLL provided tumor and normal DNA for exome-sequencing and copy number assessment in this study. Tumor and normal DNA from 11 additional patients were also analyzed by DNA sequencing alone (a total of 160 CLL samples). 82 CLL samples were previously reported (Wang et al., 2011), and the raw BAM files for these samples were re-processed and re-analyzed together with the new data, to ensure the consistency of the results and to enable the detection of smaller subclones made possible with a newer version of the mutation caller. Written informed consent was obtained prior to sample collection according to the Declaration of Helsinki. DNA was extracted from blood- or marrow-derived lymphocytes (tumor) and autologous epithelial cells (saliva), fibroblasts or granulocytes (normal).
Libraries for WES were constructed and sequenced on either an Illumina HiSeq 2000 or GA-IIX using 76 bp paired-end reads (Berger et al., 2011; Chapman et al., 2011). Output from Illumina software was processed by the Picard data processing pipeline to yield BAM files containing well calibrated, aligned reads (Chapman et al., 2011; DePristo et al., 2011). sSNVs and indels were identified using MuTect [V119, (Cibulskis et al., under review)].and indelocator [V61, (Wang et al., 2011)], respectively. Recurrent sSNV and indels in 160 CLLs were identified using MutSig2.0 (Lohr et al., 2012). For 111 of 149 matched CLL-normal DNA samples, copy number profiles were obtained using the Affymetrix Genome-wide Human SNP Array 6.0, with allele-specific analysis [HAPSEG (Carter, 2011)]. Recurrent sCNAs were identified using the GISTIC2.0 algorithm (Mermel et al., 2011), after excluding germline copy number variants. For CLL samples with no available SNP arrays (38 of 149 CLLs), sCNAs were estimated directly from the WES data, based on the ratio of CLL sample read-depth to the average read-depth observed in normal samples for that region. We applied ABSOLUTE, to estimate sample purity, ploidy, and absolute somatic copy numbers. These were used to infer the CCFs of point mutations from the WES data. Following the framework previously described (Carter et al., 2012), we computed the posterior probability distribution over CCF c as follows. Consider a somatic mutation observed in a of N sequencing reads on a locus of absolute somatic copy-number q in a sample of purity α. The expected allele-fraction f of a mutation present in one copy in a fraction c of cancer cells is calculated by Then Binom assuming f(c) = αc/(2(1 - α) + αq), with c ∈ [0.01,1]. Then P(c) α Binom(a|N,f(c)), assuming a uniform prior on c. The CCF over was then obtained by calculating these values over a regular grid of 100 c values and normalizing by dividing them by their sum, which is the constant of proportionality in the above equation. Mutations were thereafter classified as clonal based on the posterior probability that the CCF exceeded 0.95, and subclonal otherwise. Validation of allelic fraction was performed by using deep sequencing with indexed libraries recovered on a Fluidigm chip. Resulting normalized libraries were loaded on a MiSeq instrument (Illumina) and sequenced using pairedend 150bp sequencing reads to an average coverage depth of 4200X.
Associations between mutation rates and clinical features were assessed by the Wilcoxon rank-sum test, Fisher exact test, or the Kruskal–Wallis test, as appropriate. Time-to-event data were estimated by the method of Kaplan and Meier, and differences between groups were assessed using the log-rank test. Unadjusted and adjusted Cox modeling was performed to assess the impact of the presence of a subclonal driver on clinical outcome measures alone and in the presence of clinical features known to impact outcome. A chi-square test with 1 degree of freedom and the -2 Log-likelihood statistic were used to test the prognostic independence of subclonal status in Cox modeling.
A complete description of the Materials and Methods is provided in the Extended Experimental Procedures.
Supplementary Material
Highlights.
Whole exome analysis of clonal heterogeneity in 149 chronic lymphocytic leukemias
Earlier and later mutations in the temporal evolution of CLL identified
Clonal evolution is commonly seen with treatment, typically in a branched pattern
A subclonal driver in a pre-treatment sample is associated with adverse outcome
Acknowledgments
DAL dedicates this manuscript to the loving memory of his mother Nina, who passed away during the final stages of this work. We thank all members of the Broad Institute’s Biological Samples, Genetic Analysis and Genome Sequencing Platforms, who made this work possible (NHGRI-U54HG003067). DAL is supported by an American Society of Hematology (ASH) Research Award for Fellows-in-Training, and an ACS Post-Doctoral Fellowship. JRB is supported by NIH K23 CA115682, the Melton and Rosenbach Funds, and is an ASH Scholar and a LLS Clinical Research Scholar. CJW acknowledges support from the Blavatnik Family Foundation, AACR (SU2C Innovative Research Grant), NHLBI (1RO1HL103532-01), NCI (1R01CA155010-01A1) and is a Clinical Investigator supported in part by the Damon-Runyon Cancer Research Foundation (CI-38-07).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alami R, Fan Y, Pack S, Sonbuchner TM, Besse A, Lin Q, Greally JM, Skoultchi AI, Bouhassira EE. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci U S A. 2003;100:5920–5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Brown JR, Hanna M, Tesar B, Werner L, Pochet N, Asara JM, Wang YE, Dal Cin P, Fernandes SM, Thompson C, et al. Integrative genomic analysis implicates gain of PIK3CA at 3q26 and MYC at 8q24 in chronic lymphocytic leukemia. Clin Cancer Res. 2012;18:3791–3802. doi: 10.1158/1078-0432.CCR-11-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Meyerson M, Getz G. Accurate estimation of homologue-specific DNA concentration ratios in cancer samples allows long-range haplotyping. 2011 Available from Nature Precedings < http://hdlhandlenet/10101/npre201164941%3E. [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheson BD, Pfistner B, Juweid ME, Gascoyne RD, Specht L, Horning SJ, Coiffier B, Fisher RI, Hagenbeek A, Zucca E, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007;25:579–586. doi: 10.1200/JCO.2006.09.2403. [DOI] [PubMed] [Google Scholar]
- CLL Trialists Collaborative Group Chemotherapeutic options in chronic lymphocytic leukemia: a meta-analysis of the randomized trials. J Natl Cancer Inst. 1999;91:861–868. doi: 10.1093/jnci/91.10.861. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch AJ, Aigelsreiter A, Staber PB, Beham A, Linkesch W, Guelly C, Brezinschek RI, Fruhwirth M, Emberger W, Buettner M, et al. MALT lymphoma and extranodal diffuse large B-cell lymphoma are targeted by aberrant somatic hypermutation. Blood. 2007;109:3500–3504. doi: 10.1182/blood-2006-06-030494. [DOI] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner H. The use of molecular markers in selecting therapy for CLL. Clin Adv Hematol Oncol. 2005;3:103–104. [PubMed] [Google Scholar]
- Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- Egan JB, Shi CX, Tembe W, Christoforides A, Kurdoglu A, Sinari S, Middha S, Asmann Y, Schmidt J, Braggio E, et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012;120:1060–1066. doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103:1139–1143. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstung M, Beisel C, Rechsteiner M, Wild P, Schraml P, Moch H, Beerenwinkel N. Reliable detection of subclonal single-nucleotide variants in tumour cell populations. Nat Commun. 2012;3:811. doi: 10.1038/ncomms1814. [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust E, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann V, Tiacci E, Holmes AB, Kohlmann A, Martelli MP, Kern W, Spanhol-Rosseto A, Klein HU, Dugas M, Schindela S, et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 2011;118:6153–6163. doi: 10.1182/blood-2011-07-365320. [DOI] [PubMed] [Google Scholar]
- Grubor V, Krasnitz A, Troge J, Meth J, Lakshmi B, Kendall J, Yamrom B, Alex G, Pai D, Navin N, et al. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA) Blood. 2009;113:1294–1303. doi: 10.1182/blood-2008-05-158865. [DOI] [PubMed] [Google Scholar]
- Hallek M, Cheson B, Catovsky D, Caligaris-Cappio F, Dighiero G, Döhner H, Hillmen P, Keating M, Montserrat E, Rai K, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosgood HD, 3rd, Baris D, Zhang Y, Berndt SI, Menashe I, Morton LM, Lee KM, Yeager M, Zahm SH, Chanock S, et al. Genetic variation in cell cycle and apoptosis related genes and multiple myeloma risk. Leuk Res. 2009;33:1609–1614. doi: 10.1016/j.leukres.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski D. Lessons from the past: evolutionary impacts of mass extinctions. Proc Natl Acad Sci U S A. 2001;98:5393–5398. doi: 10.1073/pnas.101092598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, Gentile M, Yan J, Enge M, Taipale M, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, Paulson TG, Blount PL, Risques RA, Rabinovitch PS, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- Marechal Y, Queant S, Polizzi S, Pouillon V, Schurmans S. Inositol 1,4,5-trisphosphate 3-kinase B controls survival and prevents anergy in B cells. Immunobiology. 2011;216:103–109. doi: 10.1016/j.imbio.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Masica DL, Karchin R. Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer Res. 2011;71:4550–4561. doi: 10.1158/0008-5472.CAN-11-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy H, Wierda WG, Barron LL, Cromwell CC, Wang J, Coombes KR, Rangel R, Elenitoba-Johnson KS, Keating MJ, Abruzzo LV. High expression of activation-induced cytidine deaminase (AID) and splice variants is a distinctive feature of poor-prognosis chronic lymphocytic leukemia. Blood. 2003;101:4903–4908. doi: 10.1182/blood-2002-09-2906. [DOI] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, Downing JR. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al. Oncogenically active MYD88 mutations in human lymphoma. 2010 doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Sigmund K. Evolutionary dynamics of biological games. Science. 2004;303:793–799. doi: 10.1126/science.1093411. [DOI] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, Ramsay AJ, Bea S, Pinyol M, Martinez-Trillos A, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R, Feller SM, Grocock R, Henderson S, Khrebtukova I, et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012 doi: 10.1182/blood-2012-05-433540. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanafelt TD, Hanson C, Dewald GW, Witzig TE, LaPlant B, Abrahamzon J, Jelinek DF, Kay NE. Karyotype evolution on fluorescent in situ hybridization analysis is associated with short survival in patients with chronic lymphocytic leukemia and is related to CD49d expression. J Clin Oncol. 2008;26:e5–6. doi: 10.1200/JCO.2008.16.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilgenbauer S, Sander S, Bullinger L, Benner A, Leupolt E, Winkler D, Krober A, Kienle D, Lichter P, Dohner H. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92:1242–1245. doi: 10.3324/haematol.10720. [DOI] [PubMed] [Google Scholar]
- Unoki M, Nakamura Y. EGR2 induces apoptosis in various cancer cell lines by direct transactivation of BNIP3L and BAK. Oncogene. 2003;22:2172–2185. doi: 10.1038/sj.onc.1206222. [DOI] [PubMed] [Google Scholar]
- Vincent TL, Gatenby RA. An evolutionary model for initiation, promotion, and progression in carcinogenesis. Int J Oncol. 2008;32:729–737. [PubMed] [Google Scholar]
- Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, Larson DE, McLellan MD, Dooling D, Abbott R, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CJ. CLL clonal heterogeneity: an ecology of competing subpopulations. Blood. 2012;120:4117–4118. doi: 10.1182/blood-2012-09-452805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.