Summary
DEADSouth mRNA is a component of germ plasm in Xenopus laevis and encodes a DDX25 DEAD-box RNA helicase. To determine the intracellular localization of DEADSouth protein, we injected mRNA encoding DEADSouth tagged with mCherry fluorescent protein into fertilized eggs from transgenic Xenopus expressing EGFP fused with a mitochondrial targeting signal. The DEADSouth-mCherry fusion protein was localized to the germ plasm, a mitochondria-rich region in primordial germ cells (PGCs). DEADSouth overexpression resulted in a reduction of PGC numbers after stage 20. Conversely, DEADSouth knockdown using an antisense locked nucleic acid gapmer inhibited movement of the germ plasm from the cortex to the perinuclear region, resulting in inhibition of PGC division at stage 12 and a decrease in PGC numbers at later stages. The knockdown phenotype was rescued by intact DEADSouth mRNA, but not mutant mRNA encoding inactive DEADSouth helicase. Surprisingly, it was also rescued by mouse vasa homolog and Xenopus vasa-like gene 1 mRNAs that encode DDX4 RNA helicases. The rescue was dependent on the 3′ untranslated region (3′UTR) of DEADSouth mRNA, which was used for PGC-specific expression. The 3′UTR contributed to localization of the injected mRNA to the germ plasm, resulting in effective localization of DEADSouth protein. These results demonstrate that localization of DEADSouth helicase to the germ plasm is required for proper PGC development in Xenopus laevis.
Keywords: RNA helicase, Germline, LNA gapmer
Introduction
In most animals, germline cells are generated apart from the somatic lineage in early development. In contrast to mice, in which the germline is generated by inductive signals from the surrounding tissues, the Xenopus germline is established by inheriting specialized cytoplasm localized in the vegetal cortex of the egg. Such cytoplasm in Xenopus is also observed in various animals including zebrafish, nematode and fly, which is termed ‘germ plasm’ in general, the P-body in Caenorhabditis elegans and ‘polar plasm’ in Drosophila melanogaster (Ikenishi, 1998). It has been demonstrated that germ plasm contains determinants required and sufficient for germline differentiation in Drosophila (Illmensee and Mahowald, 1974; Okada et al., 1974) and Xenopus (Buehr and Blackler, 1970; Tada et al., 2012).
In early Xenopus development, germ plasm at the vegetal cortex of fertilized eggs is divided into about four blastomeres through the first two cleavages, and then distributes to only one side of two daughter cells because it is present in a particular region of the cortex of blastomeres (Whitington and Dixon, 1975). Thus, the number of primordial germ cells (PGCs) harboring germ plasm remains at about four, although the size of PGCs become gradually smaller through repeated cell division until the late blastula at stage 9. Early in gastrulation, germ plasm moves from the cortex to the perinuclear region in PGCs and divides equally into two daughter PGCs in later cell divisions. PGCs incorporating germ plasm divide about three times and migrate to the genital ridge (Dziadek and Dixon, 1977). The germ plasm is composed of electron dense granules, many mitochondria, and specific mRNAs and proteins. Although many molecules in germ plasm have been identified and investigated, the mechanisms of germline development remain unknown (Cuykendall and Houston, 2010; King et al., 2005).
VASA/DDX4 of the DEAD-box RNA helicase family is a component of germ plasm and widely used as a germline marker in various animals because of its germline-specific expression (Gustafson and Wessel, 2010). It has been reported that vasa is required for germ cell development. In fly and nematode, loss-of-function of vasa results in a defect of oogenesis (Kuznicki et al., 2000; Styhler et al., 1998). MVH (mouse vasa homolog) is present in the chromatoid body that is observable during spermatogenesis and resembles germ plasm (Toyooka et al., 2000). Mvh-null mutants show defects in spermatogenesis (Tanaka et al., 2000). Recently, it was also revealed that vasa is involved in cell cycle progression in fly (Pek and Kai, 2011a) and sea urchin (Yajima and Wessel, 2011), and piRNA production in mice (Kuramochi-Miyagawa et al., 2010). In contrast, XVLG1 (Xenopus vasa-like gene 1) is expressed in somatic and germ cells in Xenopus, although XVLG1 is certainly a homolog of vasa/Ddx4 (Ikenishi and Tanaka, 2000; Komiya et al., 1994). Functional inhibition of XVLG1 by an antibody results in aberrant morphogenesis of somatic cells and the loss of germ cells, suggesting that XVLG1 is involved in the differentiation of both somatic and germline cells (Ikenishi and Tanaka, 1997; Ikenishi and Tanaka, 2000).
DEADSouth is a DEAD-box RNA helicase belonging to DDX25, but not the VASA/DDX4 family, and its transcript is an RNA component of germ plasm in Xenopus (MacArthur et al., 2000). DEADSouth transcripts were initially detected prior to mitochondrial cloud formation, accumulate in the mitochondrial cloud and then co-localize to the germ plasm during early development. It is also expressed in spermatogonia, spermatocytes and spermatids in testes. In mammals, DDX25 has been identified as a gonadotropin-regulated testicular RNA helicase (GRTH) (Tang et al., 1999). Knockout of the gene results in remarkably diminished sizes of chromatoid bodies in spermatids and incomplete spermatogenesis in mice (Tsai-Morris et al., 2004). It has been suggested that GRTH/DDX25 may be required to maintain chromatoid bodies in spermatogonia (Sato et al., 2010).
In this study, we focus on the function of DEADSouth in Xenopus germline development in relation to VASA/DDX4. By expression of mRNA encoding DEADSouth fused with mCherry fluorescent protein, we show that the DEADSouth protein is localized to the germ plasm. We show that PGCs decrease in number both in DEADSouth-overexpressing and -depleted embryos. The depleted embryos are deficient for translocation of germ plasm from the cortex to the perinuclear region in PGCs before midblastula transition (MBT), causing inhibited PGC division. Surprisingly, such a knockdown phenotype is rescued by expression of MVH and XVLG1, but not inactive DEADSouth helicases. The rescue is dependent on the 3′ untranslated region (3′UTR) of DEADSouth mRNA, which is required for localization of the injected mRNA to the germ plasm. Together, we demonstrate here that localization of DEADSouth protein is required for proper development of Xenopus PGCs.
Materials and Methods
Xenopus embryos
Adult male and female wild-type Xenopus laevis were purchased commercially and maintained at 22°C in circulatory tanks. Female transgenic Xenopus laevis expressing EGFP fused with a mitochondrial targeting signal (mito-EGFP Xenopus) was also used in this study (Taguchi et al., 2012). Because embryos from the transgenic frog had maternally supplied mitochondria with EGFP, the mitochondria-rich germ plasm was clearly visible until stage 20 under a fluorescence microscope. Embryos were obtained as described previously (Kataoka et al., 2006), allowed to develop at 18°C, and staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1994).
Preparation of constructs
Mvh cDNA was a gift from Drs Kuniya Abe (RIKEN BioResource Center) and Toshiaki Noce (Keio University). DEADSouth and XVLG1 cDNAs were amplified by PCR using Xenopus laevis ovarian cDNA as a template, and cloned into pCR2.1 or pCRII vectors. Then, the ORFs and 3′UTRs were amplified by PCR using PrimeStar polymerase (TaKaRa), and inserted into appropriate regions of pCS2-based vectors such as pCS2-Venus-DEADSouth 3′UTR and pCS2-mCherry-DEADSouth 3′UTR (Kataoka et al., 2006) by In-Fusion technology (Clontech). pCS2-DEADSouth-DEADSouth 3′UTR and pCS2-DEADSouth-AAA-DEADSouth 3′UTR were constructed by inverse PCR with mutated primers and In-Fusion technology. The coding regions of all constructs were confirmed by sequencing. XVLG1Δ-DS 3′UTR and XVLG1Δ-XVLG1 3′UTR were generated by removing the EcoRV fragment of the XVLG1 ORF from pCS2-XVLG1-DS 3′UTR and pCS2-XVLG1-XVLG1 3′UTR plasmids, respectively.
Locked nucleic acid (LNA) gapmers
According to previous reports (Kurreck et al., 2002; Braasch et al., 2002), antisense and mismatch LNA gapmers (19 bases each) were designed. Sequences of antisense and mismatch LNA gapmers were TTAGCGGCCATCGTTCCTT and TATCCGGCCATCGTTGGAT (modification with LNA underlined), respectively (Nippon EGT). LNA gapmers were dissolved in water at 20 µM. Ninety-two femtomoles of gapmer were injected into each embryo.
Preparation and microinjection of mRNA
Template plasmids were linearized by digestion with XhoI, and used as templates for in vitro mRNA synthesis with an mMESSAGE mMACHINE SP6 kit (Ambion). mRNAs of XVLG1Δ-DS 3′UTR and XVLG1Δ-XVLG1 3′UTR were labeled using a Label IT Cy3 labeling kit (Mirus). mRNAs and LNA gapmers were microinjected into the cortical region at the vegetal pole of a fertilized egg with a Nonoject II microinjector (Drummond Scientific Company). The amount of each injected mRNA per egg was determined empirically as follows. Four-hundred and sixty picograms of Venus-DEADSouth 3′UTR (v-DS), DS-mCherry-DS 3′UTR, DS-mCherry-XVLG1 3′UTR or mCherry-DS 3′UTR; 46 pg (for overexpression) or 92 pg (for rescue) of DS-full; 46 pg of DS-stop; 92 pg of DS-AAA, Mvh-DS 3′UTR or DS-XVLG1 3′UTR; 276 pg of XVLG1-DS 3′UTR or XVLG1-XVLG1 3′UTR; 3.7 ng of Cy3-XVLG1Δ-DS 3′UTR or Cy3-XVLG1Δ-XVLG1 3′UTR were injected. Injection at these doses did not affect normal development.
PGC isolation and observation
Mito-EGFP Xenopus or v-DS-injected embryos at the indicated stages were dissected in 70% Dulbecco's phosphate-buffered saline without Ca2+ and Mg2+ (PBS−) and incubated for 1 hour. Then, PGCs were collected manually with a micropipette and observed under an MZ16F fluorescence stereomicroscope (Leica) equipped with a DS-5Mc digital camera (Nikon).
The diameter of PGCs was measured in images with a scale. To visualize mCherry protein, isolated PGCs were fixed in 2% paraformaldehyde/0.1 M MOPS (pH 7.5)/0.5 M NaCl at 4°C for at least 2 days, stained with 2 µg/mL Hoechst 33342 (Sigma) in PBS−, washed with PBS− and mounted in 50% glycerol/PBS−. Cells were observed under a BX60 fluorescence microscope (Olympus) equipped with a Nuance Fx digital camera (Caliper PerkinElmer).
Quantitative real-time RT-PCR
Total RNA was extracted from 10 embryos using TRIzol (Invitrogen). One microgram of total RNA was subjected to cDNA synthesis using Ready-To-Go You-Prime First-Strand Beads with random hexamers (GE healthcare Life Science), according to the manufacturer's protocol. mRNA quantification was performed by a PRISM7000 Sequence Detection System (Applied Biosystems) using SYBR Premix Ex Taq (TaKaRa). Primer sequences and annealing temperatures were as follows. DEADSouth forward ATGGGCTTCAACAGACCTTC and reverse TCCACACGACTCAGCATAGC, 60°C; Xpat forward GAGGAAATTGTTGCAAGTGCTCTAAACAGG and reverse TGTCAGGAAGCATAAAATAGCCATCTGTAG, 63°C; EF1α forward CAGGCCAGATTGGTGCTGGATATGC and reverse GCTCTCCACGCACATTGGCTTTCCT, 66°C. Standard curves were generated for each gene by serial dilution of cDNA from uninjected stage 8 embryos. EF1α gene expression was used as an internal control. Each experiment was repeated three times and RNA quantification was performed twice for each experiment.
Immunostaining
Embryos were fixed in 4% paraformaldehyde/PBS− at 4°C overnight, washed in TPBS− (0.1% Triton X-100 in PBS−), and then incubated in blocking buffer (10% goat serum in TPBS−) at room temperature for 2 hours. Samples were treated with blocking buffer containing mouse anti-Xdazl (1:1000 dilution) (Mita and Yamashita, 2000) and rabbit anti-caspase-3 (1:1000 dilution) (Abcam) antibodies at 4°C for 3 days, washed in TPBS−, incubated in blocking buffer containing Alexa Fluor 488-conjugated anti-mouse IgG (H+L) (1:1000 dilution) (Molecular Probes) and Cy3-conjugated anti-rabbit IgG (1:1000 dilution) (Rockland) at 4°C overnight and then washed in TPBS−. Specimens were dissociated manually in PBS− and then observed under an MZ16F fluorescence stereomicroscope (Leica).
Statistical analyses
The Student's t-test was carried out for all statistical analyses. One-tailed P-values were used for significance.
Results
Localization of DEADSouth protein in Xenopus PGCs
Although DEADSouth mRNA is localized to the germ plasm during oogenesis and early development (MacArthur et al., 2000), localization of the protein is unknown. Because an antibody against DEADSouth protein was not available, we expressed DEADSouth protein tagged with a fluorescent protein. To reveal the relative location of DEADSouth protein in germ plasm, we also used mito-EGFP Xenopus (Taguchi et al., 2012). Germplasm is a PGC-specific organelle enriched with mitochondria that are a useful marker of germ plasm (Venkatarama et al., 2010; Elinson et al., 2011). We prepared mRNA encoding DEADSouth protein and mCherry fluorescent protein, followed by the DEADSouth 3′UTR (DS-mCherry-DS 3′UTR), and injected it into fertilized eggs obtained by crossing a mito-EGFP female with a wild-type male. At stage 12, the germ plasm was observed in the perinuclear region of PGCs (Fig. 1A–E). The mCherry signal clearly overlapped with the germ plasm and appeared to be observable in the nuclei of PGCs. This observation suggested that DEADSouth protein played a role in the nucleus and the germ plasm, but required further analysis. As a control, mCherry signal from mCherry-DS 3′UTR was observed throughout PGCs, but not localized to the germ plasm (Fig. 1F–J). This result indicates that DEADSouth protein is localized to the germ plasm.
Fig. 1. DEADSouth-mCherry fusion protein is localized to the germ plasm in PGCs.
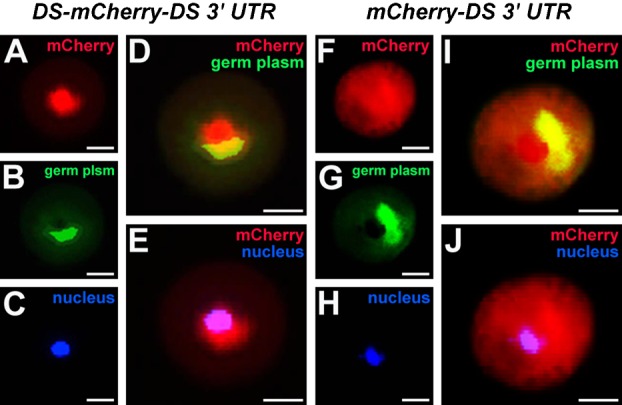
mRNA of DEADSouth-mCherry-DS 3′UTR (A–E) or mCherry-DS 3′UTR (F–J) was injected at the vegetal pole of fertilized eggs by crossing wild-type male and mito-EGFP transgenic female Xenopus. PGCs were isolated from embryos at stage 12, fixed and stained with Hoechst 33342. Signals for mCherry, germ plasm (mitochondria) and nuclei are red, green and blue, respectively. Scale bars: 10 µm.
Overexpression effects of the DEADSouth gene
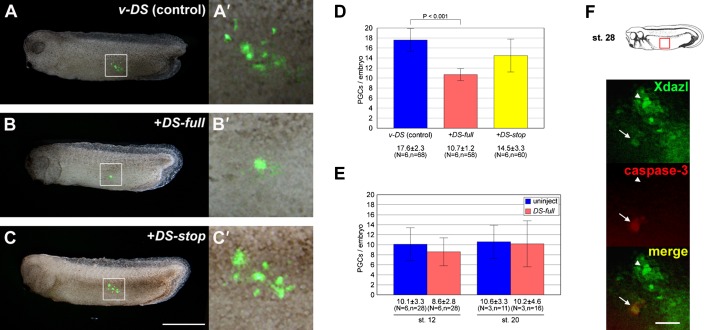
To elucidate the function of DEADSouth in PGC development, we synthesized full-length DEADSouth mRNA (DS-full) in vitro and injected it into the vegetal pole of fertilized eggs, together with Venus-DEADSouth 3′UTR mRNA (v-DS), to monitor PGCs in a living embryo (Kataoka et al., 2006). At stage 32, we externally observed that PGCs were significantly reduced in number (Fig. 2A–D). The number of PGCs per embryo injected with water (control) or DS-full were 17.6±2.3 and 10.7±1.2 (average ± s.d., P<0.001), respectively. No effect was observed in embryos injected with mRNA encoding nonsense DEADSouth (DS-stop), in which a stop codon was inserted downstream of the initial methionine of the DEADSouth ORF. This result indicated that the loss of PGCs was due to overexpression of DEADSouth protein.
Fig. 2. DEADSouth gene overexpression decreases the number of PGCs.
(A–C) Representative examples of stage 32 embryos injected with mRNA for v-DS only, v-DS plus DEADSouth-DS 3′UTR (DS-full) and v-DS plus nonsense DEADSouth-DS 3′UTR (DS-stop), respectively. Anterior is toward the left. Scale bar: 1 mm. (A′–C′) High magnification of the areas indicated in (A–C), respectively. (D) The number of PGCs per embryo at stage 32 injected with the indicated mRNAs. PGC numbers were determined by externally counting from both sides. (E) The number of PGCs per embryo at the indicated stage without/with DS-full mRNA. Embryos were dissociated, and EGFP-positive and large cells were counted as PGCs. ‘N’ and ‘n’ indicate the number of experiments and total embryos examined, respectively. The P-value was calculated by the Student's t-test. Error bars indicate s.d. (F) Apoptotic PGCs in DEADSouth-overexpressing embryos at stage 28. PGCs were observed after dissociating the embryonic region (red box) immunostained for Xdazl and caspase-3. Arrowheads and arrows indicate Xdazl-positive/caspase-3-negative cells (non-apoptotic PGCs) and Xdazl positive/caspase-3-positive cells (apoptotic PGCs), respectively. Scale bar: 100 µm.
To determine when the effect of DEADSouth gene overexpression appeared, we used mito-EGFP transgenic Xenopus. Embryos are more suitable for monitoring PGCs at an early stage, compared with the use of v-DS as a PGC tracer, because of no delayed EGFP expression. We dissociated embryos injected with DS-full and checked the PGCs in detail. At stages 12 and 20, no difference in PGC numbers between DS-full and the control (uninjected) was detected (Fig. 2E). DEADSouth-overexpressing embryos also showed no difference in PGC size and intracellular localization of germ plasm, compared with that of the control at stages 12, 20 and 32 (supplementary material Figs S1, S2). To determine whether the loss of PGCs resulted from apoptosis, we performed immunostaining for Xdazl and caspase-3 to detect apoptotic PGCs in DEADSouth-overexpressing embryos at stage 28 (Fig. 2F). As expected, 17% of PGCs (10 caspase-3-positives out of 60 Xdazl-positives) were apoptotic, compared with 4.8% of PGCs in control embryos (3 out of 63). This result indicated that the loss of PGCs in DEADSouth-overexpressing embryos, at least partially, resulted from apoptosis.
Knockdown of DEADSouth with an antisense LNA gapmer
To reveal the function of DEADSouth, we performed knockdown experiments using an antisense LNA gapmer. Because an antibody against DEADSouth is unavailable, it is difficult to evaluate the extent of knockdown after translational inhibition using morpholino oligos. Therefore, we chose an LNA gapmer as a knockdown reagent because it caused degradation of the target mRNA that was quantified easily by PCR. As shown in Fig. 3A, we designed an antisense LNA gapmer targeting around the start codon of DEADSouth mRNA and a mismatch gapmer as a control that was not expected to act on the target.
Fig. 3. An LNA gapmer causes degradation of targeted DEADSouth mRNA.
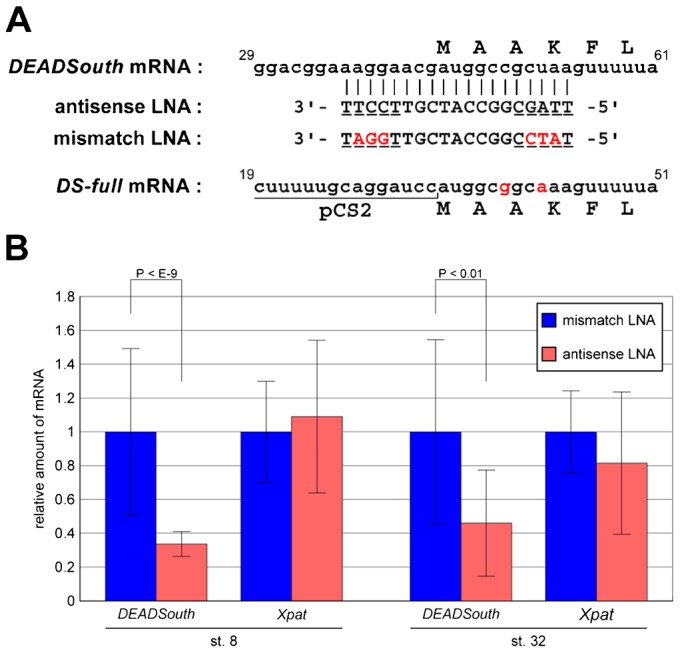
(A) Sequence alignment of DEADSouth mRNA (nucleotide position 29–61, accession no. AF190623), antisense and mismatch LNA gapmers and synthesized DEADSouth mRNA (DS-full). Upper letters indicate the amino acid sequence. LNA modifications and mismatch bases are underlined and in red, respectively. The sequence of DS-full mRNA was predicted from that of the template DNA. (B) Quantification of DEADSouth and Xpat mRNAs in stage 8 and 32 embryos injected with LNA gapmers. The amounts of these mRNAs were determined by real-time RT-PCR, normalized to EF1α mRNA levels as an internal control, and shown as relative amounts to the mRNA level in embryos injected with mismatch LNA gapmer. P-values and s.d. are indicated.
First, we evaluated the extent of DEADSouth mRNA degradation in embryos injected with the LNA gapmer by quantitative RT-PCR. The mRNA level of Xpat, which is expressed in a PGC-specific manner (Hudson and Woodland, 1998), was also examined as a control. Because PGC number and the amount of germ plasm depended on the embryos and stages, the amount of DEADSouth mRNA was normalized to that of EF1α mRNA and compared between embryos injected with antisense or mismatch LNA gapmers (Fig. 3B). At stages 8 and 32, DEADSouth mRNA levels in antisense LNA-injected embryos were reduced significantly compared with those in mismatch LNA gapmer-injected embryos (P<10−9 at stage 8). In contrast, Xpat mRNA levels in both embryos were similar at stages 8 and 32. These mRNA levels in mismatch LNA gapmer-injected embryos were similar to those in uninjected embryos (data not shown). These data indicated that injection of the LNA gapmer resulted in degradation of DEADSouth mRNA in a sequence-specific manner. Unfortunately, stage-dependent changes of the mRNA level could not be examined because the total amount of mRNA in the embryo changed during development.
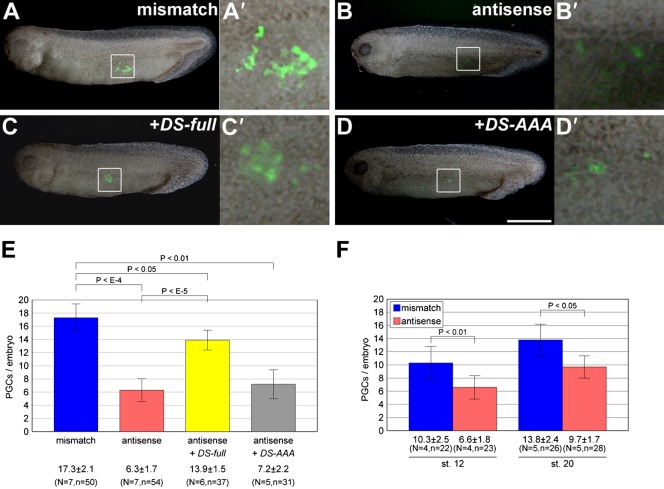
Second, we examined the effects of the LNA gapmer on PGC development by co-injection with v-DS as a PGC tracer. External observation at stage 32 showed that PGCs in antisense gapmer-injected embryos decreased remarkably in number compared with those in mismatch gapmer-injected embryos (antisense, 6.3±1.7; mismatch, 17.3±2.1). In addition, the reduction of PGC number in antisense gapmer-injected embryos was rescued by co-injection with DS-full mRNA that was ineffective for antisense gapmer targeting because of codon substitutions in the target region (Fig. 3). Although this rescue was significant (P<10−5 between antisense and antisense+DS-full), it appeared to be partial (P<0.05 between mismatch and antisense+DS-full). In contrast, this reduction was not rescued by co-injection with DS-AAA mRNA encoding a putatively inactive helicase of DEADSouth, in which the amino acid sequence SAT was substituted with AAA in motif III (Pause and Sonenberg, 1992) (Fig. 4). These results clearly show that DEADSouth RNA helicase is required for proper PGC development.
Fig. 4. The number of PGCs is decreased by knockdown of the DEADSouth gene.
(A–D) Representative examples of stage 32 embryos injected with the mismatch LNA gapmer, antisense LNA gapmer, antisense LNA gapmer plus DS-full and antisense LNA gapmer plus mutant DEADSouth-DS 3′UTR (DS-AAA), in addition to v-DS as a PGC-tracer, respectively. Scale bar: 1 mm. (A′–D′) High magnification of the areas indicated in (A–D), respectively. (E) The number of PGCs per embryo at stage 32 injected with the indicated LNA gapmer(s) and mRNAs. PGC numbers were determined by externally counting from both sides. (F) The number of PGCs per embryo at the indicated stages with mismatch or antisense LNA gapmers. The injected mito-EGFP embryos were dissociated, and EGFP-positive and large cells were counted as PGCs. N and n indicate the number of experiments and total embryos examined, respectively. P-values were calculated by the one-tailed t test. Error bars indicate s.d.
We also dissociated DEADSouth-knockdown embryos at stages 12, 20 and 32, and examined PGC number per embryo and size. For stage 12 and 20 embryos, mito-EGFP transgenic Xenopus were used because of easier PGC tracing. PGCs from knockdown embryos at stages 12, 20 and 32 showed less increasing numbers (6.6±1.8/embryo at stage 12 to 9.3±1.7/embryo at stage 32) in contrast to control embryos (mismatch LNA injected) (10.3±2.5/embryo at stage 12 to 21.9±3.4/embryo at stage 32). At stage 12, knockdown embryos already showed that the number of PGCs was significantly decreased compared with that of the control (P<0.01 between mismatch and antisense). The size distribution of DEADSouth-knockdown PGCs was also broader than that of the control (Fig. 5A). The average size of DEADSouth-knockdown PGCs was clearly larger than that of control at stages 12, 20 and 32, although PGCs in both DEADSouth-knockdown and control embryos became smaller and the difference between them appeared to be smaller as development proceeded.
Fig. 5. Knockdown of the DEADSouth gene affects PGC division and translocation to germ plasm.
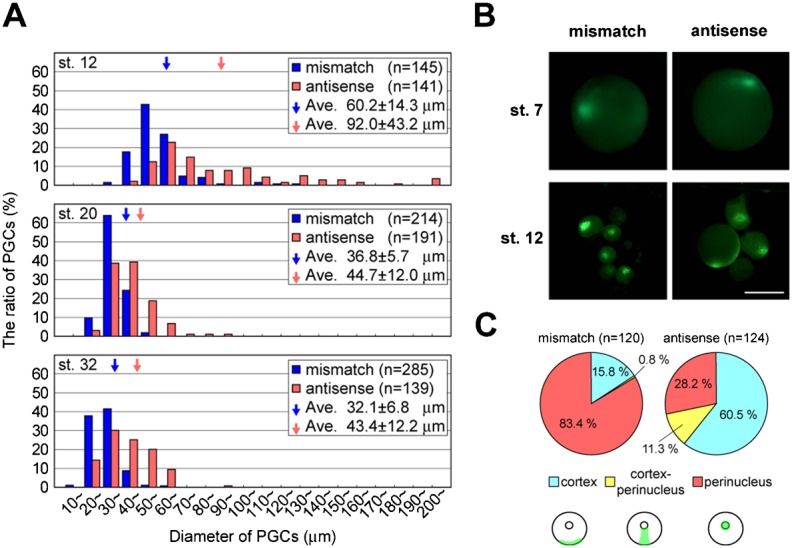
(A) Size distribution of PGCs at stages 12, 20 and 32 from embryos injected with mismatch or antisense LNA gapmers. All mito-EGFP- (at stages 12 and 20) and v-DS-labeled (at stage 32) PGCs from the injected embryos were isolated to measure their diameter. Total PGC numbers after each injection are shown as 100%. ‘n’ indicates total PGC numbers. Arrows indicate average diameters from the indicated experiments. (B) Localization of the germ plasm in PGCs isolated from mito-EGFP embryos at stages 7 and 12, which were injected with mismatch or antisense LNA gapmers. Scale bar: 100 µm. Note that at stage 12, PGCs injected with the antisense LNA gapmer are relatively large and contain the germ plasm beneath the cell membrane, compared with perinuclear localization of the germ plasm in PGCs with the mismatch LNA gapmer. (C) Ratio of PGCs with three localization patterns of germ plasm from stage 12 embryos injected with mismatch or antisense LNA gapmers. According to the localization patterns, 120 and 124 PGCs with mismatch or antisense gapmers, respectively, were classified into three groups; cortex, perinucleus and intermediate (cortex-perinucleus) shown at the bottom of the panel.
The decrease in number and increase in size of PGCs in DEADSouth-knockdown embryos appeared to be caused by inhibition of PGC division. In addition, we found a defect in the subcellular localization of germ plasm in DEADSouth-knockdown PGCs (Fig. 5B). In normal development, the germ plasm is present in a particular region of the cortex in PGCs before MBT. Then, it moves to a perinuclear region and is divided equally into two daughter PGCs after MBT. The germ plasm in PGCs from embryos injected with the mismatch gapmer appeared to behave similarly to those from uninjected embryos. Quantitative analysis demonstrated that 83.4% of PGCs had germ plasm in the perinuclear region at stage 12 (Fig. 5C). In contrast, only 28.2% of PGCs from DEADSouth-knockdown embryos showed a normal distribution of germ plasm, but the germ plasm remained in the cortex of most PGCs (60.5%). The germ plasm in DEADSouth knockdown-PGCs also appeared to be expanded in the cortex. These findings indicate that DEADSouth is involved in translocation of germ plasm in PGCs at MBT.
Functional substitution of vasa for DEADSouth
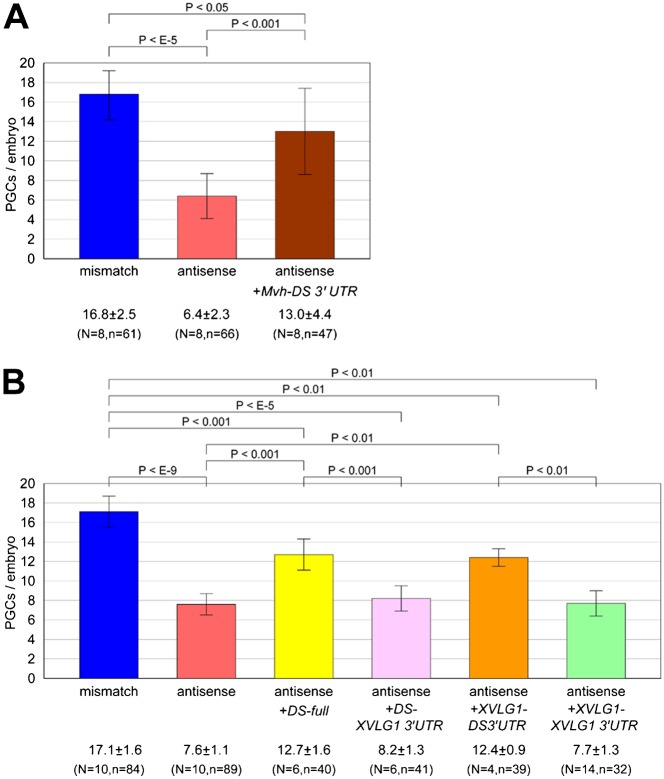
In Drosophila, the vasa gene encodes a DEAD-box RNA helicase that is required for germ cell development (Styhler et al., 1998). vasa homologs have been identified in various animals and are expressed specifically in their germline. Also, in Xenopus, vasa-like gene 1 (XVLG1) was identified as a vasa homolog (Komiya et al., 1994). However, XVLG1 is not expressed in a germline-specific manner (Ikenishi and Tanaka, 2000). Instead, DEADSouth was identified as another germline-specific DEAD-box RNA helicase (MacArthur et al., 2000). Therefore, we examined whether vasa genes could functionally substitute for the DEADSouth gene in Xenopus. We prepared two constructs, in which the ORF of Mvh or XVLG1 was fused with DEADSouth 3′UTR for PGC-specific expression. Fertilized eggs were injected with the mRNA of Mvh-DS 3′UTR or XVLG1-DS 3′UTR in addition to the antisense LNA gapmer and v-DS as a PGC tracer, and allowed to develop until stage 32 to externally observe PGCs (Fig. 6). Compared with mismatch gapmer-injected embryos (PGC number, 16.8±2.5/embryo), antisense gapmer-injected embryos showed significantly reduced PGC numbers (6.4±2.3/embryo) (Fig. 6A). Surprisingly, this reduction was restored by co-injection with Mvh-DS 3′UTR (13.0±4.4/embryo, P<0.001). Co-injection with XVLG1-DS3′UTR showed similar results. PGC numbers in embryos injected with the mismatch gapmer, antisense gapmer, and antisense gapmer plus XVLG1-DS 3′UTR were 17.1±1.6, 7.6±1.1 and 12.4±0.9, respectively (Fig. 6B). Although the rescue by XVLG1-DS 3′UTR was partial, it appeared to be similar to that by DS-full (12.7±1.6/embryo). These results indicated that the function of DEADSouth in PGC development can be substituted, at least in part, by another RNA helicase such as VASA.
Fig. 6. Knockdown of the DEADSouth gene is rescued by another type of DEAD-box RNA helicase.
(A) Rescue by injection of Mvh-DS 3′UTR mRNA. (B) Rescue by injection of XVLG1 and its dependency on the 3′UTR. At stage 32, PGCs were externally counted on both sides of embryos injected with the indicated mRNAs and v-DS mRNA as a PGC tracer, in addition to mismatch or antisense LNA gapmers. ‘N’ and ‘n’ indicate the number of experiments and total embryos examined, respectively. P-values and s.d. are indicated.
These constructs contained the 3′UTR of DEADSouth mRNA to ensure PGC-specific expression. Therefore, we evaluated the effect of the 3′UTR. Two constructs, in which DEADSouth or XVLG1 ORFs were followed by the XVLG1 3′UTR, were generated and subjected to assays to assess their rescue from DEADSouth knockdown. Surprisingly, both DEADSouth and XVLG1 ORFs fused with the DEADSouth 3′UTR rescued from DEADSouth knockdown, but could not when they were fused with the XVLG1 3′UTR (DS-XVLG1 3′UTR, 8.2±1.3/embryo; XVLG1-XVLG1 3′UTR, 7.7±1.3/embryo). This observation indicates that the rescue depends on the DEADSouth 3′UTR, because there is no distinct difference between the translational activities of the two mRNAs (data not shown).
Localization of DEADSouth mRNA and protein to germ plasm via the 3′UTR
To elucidate whether DEADSouth mRNA is localized to germ plasm via its 3′UTR, we examined the behavior of fluorescent-labeled mRNA with the DEADSouth 3′UTR in PGCs. Nonsense mRNA encoding a partially deleted XVLG1 ORF followed by the DEADSouth 3′UTR (XVLG1Δ-DS 3′UTR) or XVLG1 3′UTR (XVLG1Δ-XVLG1 3′UTR) was labeled with Cy3 fluorescent dye in vitro and injected at the vegetal pole of fertilized eggs from mito-EGFP Xenopus. At stage 7, we dissociated the embryos, isolated PGCs and observed Cy3-labeled mRNA in relation to the germ plasm (Fig. 7). The Cy3 signal was weak and dispersed throughout PGCs containing XVLG1Δ-XVLG1 3′UTR. In contrast, Cy3-labeled mRNA of XVLG1Δ-DS 3′UTR was accumulated in the germ plasm of PGCs, as demonstrated by co-localization with the mito-EGFP signal, indicating that the 3′UTR of DEADSouth mRNA includes a signal to localize or anchor mRNA to the germ plasm. In addition, we examined whether localization of DEADSouth protein was dependent on localization of the mRNA. mRNA encoding a DEADSouth-mCherry fusion protein fused with the DEADSouth 3′UTR (DS-mCherry-DS 3′UTR) or XVLG1 3′UTR (DS-mCherry-XVLG1 3′UTR) was injected as described above. As shown in Fig. 1, DEADSouth-mCherry fusion protein from DS-mCherry-DS 3′UTR mRNA was partially co-localized to the germ plasm in PGCs. DEADSouth-mCherry fusion protein from DS-mCherry-XVLG1 3′UTR mRNA was not localized to any region, but was distributed throughout the cells (supplementary material Fig. S3). This observation indicates that localization of the mRNA to the germ plasm is controlled by the 3′UTR of the mRNA, which localizes DEADSouth protein to the germ plasm.
Fig. 7. DEADSouth 3′UTR localizes the mRNA to the germ plasm.
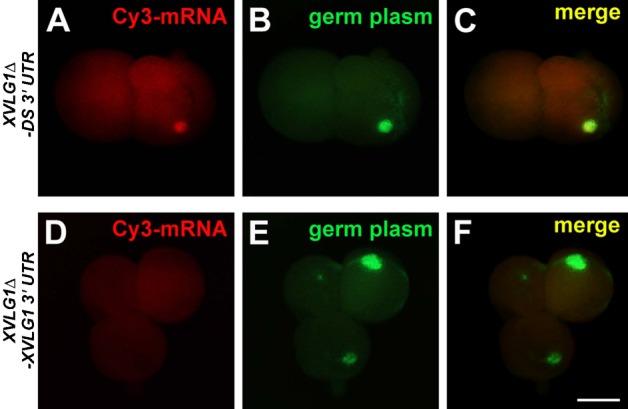
Localization of Cy3-labeled XVLG1Δ-DS 3′UTR mRNA (A–C) and Cy3-labeled XVLG1Δ-XVLG1 3′UTR mRNA (D–F) in PGCs at stage 7. PGCs were isolated from mito-EGFP embryos injected with these mRNAs and observed for Cy3 signals (mRNA, in red) and germ plasm (mitochondria, in green). Note that the Cy3 signal is superimposed on the germ plasm in (A–C). Scale bar: 10 µm.
Discussion
Germ plasm has been observed in oocytes of various animals including Drosophila and Xenopus (Ikenishi, 1998), and is required and sufficient for germline development (Illmensee and Mahowald, 1974; Okada et al., 1974; Tada et al., 2012). Many transcripts have been identified as RNA components of germ plasm (Cuykendall and Houston, 2010; King et al., 2005), and analyzed for their functions in Xenopus to understand the mechanisms underlying germline development (Houston and King, 2000; Horvay et al., 2006; Berekelya et al., 2007). DEADSouth is one of the RNA components of Xenopus germ plasm (MacArthur et al., 2000). It is interesting that DEADSouth is expressed in a PGC-specific manner, instead of XVLG1 (vasa homolog) that is expressed in somatic cells and PGCs during early Xenopus development (Ikenishi and Tanaka, 2000). We speculated that DEADSouth has a function similar to that of vasa and focused on DEADSouth in Xenopus germline development.
Recently, we have generated a transgenic Xenopus carrying a gene encoding EGFP fused with a mitochondrial targeting signal. Early embryos from the female have fluorescent germ plasm that is clearly visible until stage 20, because germ plasm is enriched with mitochondria supplied maternally (Taguchi et al., 2012). Together with a PGC visualization technique suitable for observation from about stage 12 onward (Kataoka et al., 2006), it was possible to trace PGCs in a living embryo during early development. Thus, we investigated the function of DEADSouth in the context of PGC development.
First, we investigated the intracellular localization of DEADSouth protein by expression of DEADSouth tagged with mCherry fluorescent protein. DEADSouth-mCherry fusion protein was detected in the germ plasm of PGCs at stage 12 (Fig. 1A–E). Unfortunately, we could not detect the localization at earlier stages because of low expression levels of the fusion protein. In zebrafish, vasa mRNA, but not its protein, is localized to the germ plasm before MBT and the protein is localized to the germ plasm at later stages (Knaut et al., 2000). As a control, mCherry protein was not localized to the germ plasm and distributed throughout PGCs, although mRNA was injected into the cortex of the vegetal pole where the germ plasm was present (Fig. 1F–J). These results suggest that DEADSouth protein includes a signal for localization or anchorage to the germ plasm. DEADSouth-mCherry fusion protein was also detected in the nuclei of PGCs. Considering that germ plasm contains determinants for PGC differentiation in Drosophila (Illmensee and Mahowald, 1974; Okada et al., 1974) and Xenopus (Buehr and Blackler, 1970; Tada et al., 2012), germ plasm migration to the nucleus at this stage suggests that some transport from the germ plasm to the nucleus triggers PGC differentiation. Because DEADSouth is an RNA helicase, it is feasible that DEADSouth is involved in the transport of RNAs from the germ plasm into the nucleus.
In mammals, GRTH/Ddx25 is a homolog of DEADSouth, which is expressed exclusively in testes (Sheng et al., 2003) and essential for completion of spermatogenesis (Tsai-Morris et al., 2004). DDX25 protein is a shuttle protein associated with RNA and localized to chromatoid bodies in spermatogonia, which have a morphology similar to that of germ plasm (Sheng et al., 2006). Amino acids regions 61–74 and 101–114 of mouse DDX25 protein have been identified as a nuclear export signal via CRM1 protein and a nuclear localization signal, respectively. We found some similarity in these regions between GRTH/DDX25 and DEADSouth, suggesting that DEADSouth functions as a shuttle protein between the germ plasm and nucleus. However, detailed studies using mutated constructs are required to evaluate whether these regions of DEADSouth play important roles in intracellular transport in PGCs. Translocation of DEADSouth from the germ plasm to the nucleus may be also regulated by phosphorylation (Sheng et al., 2006). Localization of endogenous DEADSouth protein also remains unknown.
To determine the possible function of DEADSouth in PGC development, we performed overexpression experiments in which in vitro synthesized DEADSouth mRNA including the 3′UTR was injected into fertilized eggs. Injected embryos showed normal PGC numbers and sizes at stages 12 and 20 (Fig. 2E; supplementary material Fig. S1). Translocation of germ plasm from the cortex to the perinuclear region appeared to occur normally before MBT (supplementary material Fig. S2). At stage 32, DEADSouth-overexpressing embryos showed a loss of PGCs, but the PGCs were normal in size and localization of the germ plasm. No PGCs were detected within the endodermal mass of DEADSouth-overexpressing embryos, indicating that the loss of PGCs was not due to a failure to migrate. Abnormal PGC migration is often observed at the tailbud stage in knockdown embryos for germ plasm-specific genes such as Xdazl, XDead end and XGRIP2.1 (Houston and King, 2000; Horvay et al., 2006; Tarbashevich et al., 2007). Recently, it has been shown that loss of PGCs in Nanos1-depleted embryos is due to apoptosis (Lai et al., 2012). Our results also suggest that loss of PGCs in DEADSouth-overexpressing embryos is, at least partially, due to apoptosis (Fig. 2F). Early Xenopus embryos might have a mechanism by which aberrant PGCs are eliminated by apoptosis.
However, PGC abnormalities were observed at stage 12 after depleting DEADSouth mRNA by injection of an antisense LNA gapmer. DEADSouth knockdown resulted in a decrease in number and increase in size of PGCs (Fig. 4; Fig. 5A), but no significant decrease in the total amount of germ plasm as indicated by the Xpat mRNA level (Fig. 3B). The diameter of DEADSouth-knockdown PGCs at stage 12 was about 1.5-fold longer than that of control PGCs, indicating that the volume of DEADSouth-knockdown PGCs was about twice as large as that of the control (Fig. 5A). These results indicate that knockdown of DEADSouth causes inhibition of PGC division. In addition, the knockdown phenotype was rescued by expression of normal DEADSouth, but not mutated DEADSouth encoding a putative inactive RNA helicase, suggesting that helicase activity is involved in PGC division.
Because we did not observe an abnormal cleavage pattern around the vegetal pole or defects in the size and number of PGCs in DEADSouth-knockdown embryos until stage 7 (data not shown), we concluded that PGC division was inhibited between stages 7 and 12. Interestingly, at that time, germ plasm moves from the cortex to perinucleus in PGCs followed by PGC specification (Whitington and Dixon, 1975; Venkatarama et al., 2010). We also found that aberrant translocation of germ plasm occurred in DEADSouth-knockdown embryos at stage 12, but not at stage 7 (Fig. 5B). In particular, germ plasm appeared to be expanded in the cortex of DEADSouth-knockdown PGCs at stage 12. Such expanded germ plasm was observable more often in larger PGCs. The ratio of PGCs with germ plasm at an aberrant position was higher in DEADSouth-knockdown embryos than that in control embryos (Fig. 5C). In DEADSouth-knockdown embryos, PGCs with germ plasm at the intermediate position between the cortex and perinucleus were also observable. Because translocation of germ plasm from the cortex to perinuclear region occurs prior to symmetric PGC division, we concluded that DEADSouth knockdown causes aberrant translocation of germ plasm, resulting in inhibited or delayed PGC division, which suggests a link between germ plasm translocation and PGC division. At later stages (stages 20 and 32), the size of PGCs in DEADSouth-knockdown embryos appeared to be closer to that of the control. Their germ plasm was also present at the normal position (data not shown). Such a phenotype may have been due to incomplete knockdown. Taken together, DEADSouth RNA helicase is required for proper PGC development. In particular, it is essential for germ plasm translocation required for symmetric division of PGCs after MBT. Moreover, translocation of germ plasm may be controlled via translational regulation by DEADSouth RNA helicase, which is similar to eIF4A (MacArthur et al., 2000).
GRTH/DDX25 protein is present in the chromatoid bodies of spermatids. In mice, GRTH/Ddx25-null mutation causes complete arrest of spermiogenesis and remarkably diminishes chromatoid bodies in round spermatids at the steps before arrest (Tsai-Morris et al., 2004). In addition, protein components of chromatoid bodies, such as MVH and MIWI, are completely excluded from chromatoid bodies in the knockout mouse (Sato et al., 2010). Together with its function as a shuttle protein, these findings suggest that GRTH/DDX25 is essential to govern the structure of chromatoid bodies for storage and processing of mRNAs. Similarly, DEADSouth protein may be essential to maintain the structure of germ plasm in Xenopus, which is supported by the observation that expanded germ plasm was present in DEADSouth-depleted PGCs.
Defects in DEADSouth-knockdown embryos were rescued by expression of XVLG1 or MVH RNA helicase belonging to the VASA/DDX4 family, which is different from the DDX25 family, possibly because of functional redundancy among DEAD-box RNA helicases. In mice, although Mvh is expressed both in the ovary and testis, no defect is detected in the ovaries of Mvh-knockout mice, suggesting that another helicase, such as PL10/DDX3, expressed in the ovary has functional redundancy with MVH (Tanaka et al., 2000). Recently, it was reported that VASA/DDX4 plays a role in cell cycle progression and germline development in Drosophila (Pek and Kai, 2011a) and sea urchin (Yajima and Wessel, 2011). In Drosophila, VASA appears to be involved in regulation of mitotic chromosome condensation/segregation in germline cells in a translation-independent manner. Furthermore, in human and Drosophila somatic cells, mitotic chromosome segregation is regulated similarly by Belle/DDX3 RNA helicase, instead of VASA/DDX4 (Pek and Kai, 2011b), suggesting functional redundancy between DDX3 and DDX4 helicases in somatic and germline cells, respectively. In Xenopus, XVLG1, a VASA homolog, appears to be involved in general cell specification and proliferation, rather than only in germline development, based on the expression pattern and perturbation of protein function (Ikenishi and Tanaka, 1997; Ikenishi and Tanaka, 2000). Our results suggest that XVLG1 and DEADSouth share VASA functions, including general and germline-specific functions, respectively, in early Xenopus development. To clarify such a functional redundancy and sharing between DEADSouth and VASA/DDX4, rescue experiments from vasa deficiency by the DEADSouth gene would be required. In addition, it will be necessary to determine the function of Centroid that is another DEAD-box RNA helicase gene expressed in germ plasm (Kloc and Chan, 2007).
Surprisingly, rescue from defects in DEADSouth-knockdown embryos depended on the 3′UTR of DEADSouth (Fig. 6B). mRNA with the 3′UTR of DEADSouth was localized to the germ plasm, but mRNA with the XVLG1 3′UTR was not localized to the germ plasm (Fig. 7). This finding indicates that the 3′UTR of DEADSouth contains a signal to anchor and/or localize the mRNA. Because DEADSouth protein also appears to have signal for localization to the germ plasm, the DEADSouth 3′UTR supports effective localization of the DEADSouth protein. We have previously demonstrated that the DEADSouth 3′UTR contains information for PGC-specific protein expression, probably via microRNA (Kataoka et al., 2006). It is very interesting to elucidate the relationship between PGC-specific expression and localization of mRNA in the context of translational regulation.
Finally, we demonstrated the utility of an antisense LNA gapmer with mito-EGFP transgenic embryos to study the functions of germ plasm-specific genes. In addition to previous methods, such as the host-transfer technique (Heasman et al., 1991) and PGC labeling with EGFP (Kataoka et al., 2006), these tools enable us to analyze the development of PGCs in Xenopus.
Supplementary Material
Acknowledgments
We thank Drs K. Abe (RIKEN BioResource Center) and T. Noce (Keio University) for the Mvh clone, Drs K. Mita (Tokushima Bunri University) and M. Yamashita (Hokkaido University) for the anti-Xdazl antibody, and Dr Y. Watai (Summit Pharmaceuticals International Corporation) for her technical support with the Nuance Fx digital camera. We also thank the members of our laboratory, especially Drs M. Mochii, K. Kataoka (Institute of Molecular Biotechnology, Austrian Academy of Science) and N. Kogo (RIKEN Brain Research Institute) for their support.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Berekelya L. A., Mikryukov A. A., Luchinskaya N. N., Ponomarev M. B., Woodland H. R., Belyavsky A. V. (2007). The protein encoded by the germ plasm RNA Germes associates with dynein light chains and functions in Xenopus germline development. Differentiation 75, 546–558 10.1111/j.1432-0436.2006.00160.x [DOI] [PubMed] [Google Scholar]
- Braasch D. A., Liu Y., Corey D. R. (2002). Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 30, 5160–5167 10.1093/nar/gkf651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M. L., Blackler A. W. (1970). Sterility and partial sterility in the South African clawed toad following the pricking of the egg. J. Embryol. Exp. Morphol. 23, 375–384. [PubMed] [Google Scholar]
- Cuykendall T. N., Houston D. W. (2010). Identification of germ plasm-associated transcripts by microarray analysis of Xenopus vegetal cortex RNA. Dev. Dyn. 239, 1838–1848 10.1002/dvdy.22304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziadek M., Dixon K. E. (1977). An autoradiographic analysis of nucleic acid synthesis in the presumptive primordial germ cells of Xenopus laevis. J. Embryol. Exp. Morphol. 37, 13–31. [PubMed] [Google Scholar]
- Elinson R. P., Sabo M. C., Fisher C., Yamaguchi T., Orii H., Nath K. (2011). Germ plasm in Eleutherodactylus coqui, a direct developing frog with large eggs. EvoDevo 2, 20 10.1186/2041-9139-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson E. A., Wessel G. M. (2010). Vasa genes: emerging roles in the germ line and in multipotent cells. Bioessays 32, 626–637 10.1002/bies.201000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J., Holwill S., Wylie C. C. (1991). Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. Methods In Cell Biology Vol. 36 Kay B K, Peng H B, ed685–695New York: Academic Press. [DOI] [PubMed] [Google Scholar]
- Horvay K., Claussen M., Katzer M., Landgrebe J., Pieler T. (2006). Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev. Biol. 291, 1–11 10.1016/j.ydbio.2005.06.013 [DOI] [PubMed] [Google Scholar]
- Houston D. W., King M. L. (2000). A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development 127, 447–456. [DOI] [PubMed] [Google Scholar]
- Hudson C., Woodland H. R. (1998). Xpat, a gene expressed specifically in germ plasm and primordial germ cells of Xenopus laevis. Mech. Dev. 73, 159–168 10.1016/S0925-4773(98)00047-1 [DOI] [PubMed] [Google Scholar]
- Ikenishi K. (1998). Germ plasm in Caenorhabditis elegans, Drosophila and Xenopus. Dev. Growth Differ. 40, 1–10 10.1046/j.1440-169X.1998.t01-4-00001.x [DOI] [PubMed] [Google Scholar]
- Ikenishi K., Tanaka T. S. (1997). Involvement of the protein of Xenopus vasa homolog (Xenopus vasa-like gene 1, XVLG1) in the differentiation of primordial germ cells. Dev. Growth Differ. 39, 625–633 10.1046/j.1440-169X.1997.t01-4-00010.x [DOI] [PubMed] [Google Scholar]
- Ikenishi K., Tanaka T. S. (2000). Spatio-temporal expression of Xenopus vasa homolog, XVLG1, in oocytes and embryos: the presence of XVLG1 RNA in somatic cells as well as germline cells. Dev. Growth Differ. 42, 95–103 10.1046/j.1440-169x.2000.00493.x [DOI] [PubMed] [Google Scholar]
- Illmensee K., Mahowald A. P. (1974). Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc. Natl. Acad. Sci. USA 71, 1016–1020 10.1073/pnas.71.4.1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K., Yamaguchi T., Orii H., Tazaki A., Watanabe K., Mochii M. (2006). Visualization of the Xenopus primordial germ cells using a green fluorescent protein controlled by cis elements of the 3′ untranslated region of the DEADSouth gene. Mech. Dev. 123, 746–760 10.1016/j.mod.2006.07.006 [DOI] [PubMed] [Google Scholar]
- King M. L., Messitt T. J., Mowry K. L. (2005). Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell 97, 19–33 10.1042/BC20040067 [DOI] [PubMed] [Google Scholar]
- Kloc M., Chan A. P. (2007). Centroid, a novel putative DEAD-box RNA helicase maternal mRNA, is localized in the mitochondrial cloud in Xenopus laevis oocytes. Int. J. Dev. Biol. 51, 701–706 10.1387/ijdb.072293mk [DOI] [PubMed] [Google Scholar]
- Knaut H., Pelegri F., Bohmann K., Schwarz H., Nüsslein-Volhard C. (2000). Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J. Cell Biol. 149, 875–888 10.1083/jcb.149.4.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T., Itoh K., Ikenishi K., Furusawa M. (1994). Isolation and characterization of a novel gene of the DEAD box protein family which is specifically expressed in germ cells of Xenopus laevis. Dev. Biol. 162, 354–363 10.1006/dbio.1994.1093 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Takamatsu K., Chuma S., Kojima-Kita K., Shiromoto Y., Asada N., Toyoda A., Fujiyama A. et al. (2010). MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 24, 887–892 10.1101/gad.1902110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurreck J., Wyszko E., Gillen C., Erdmann V. A. (2002). Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 30, 1911–1918 10.1093/nar/30.9.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznicki K. A., Smith P. A., Leung-Chiu W. M. A., Estevez A. O., Scott H. C., Bennett K. L. (2000). Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development 127, 2907–2916. [DOI] [PubMed] [Google Scholar]
- Lai F., Singh A., King M. L. (2012). Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development 139, 1476–1486 10.1242/dev.079608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur H., Houston D. W., Bubunenko M., Mosquera L., King M. L. (2000). DEADSouth is a germ plasm specific DEAD-box RNA helicase in Xenopus related to eIF4A. Mech. Dev. 95, 291–295 10.1016/S0925-4773(00)00357-9 [DOI] [PubMed] [Google Scholar]
- Mita K., Yamashita M. (2000). Expression of Xenopus Daz-like protein during gametogenesis and embryogenesis. Mech. Dev. 94, 251–255 10.1016/S0925-4773(00)00295-1 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. D., Faber J. (1994). Normal Table of Xenopus Laevis (Daudin): a Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis New York: Garland Publishing. [Google Scholar]
- Okada M., Kleinman I. A., Schneiderman H. A. (1974). Restoration of fertility in sterilized Drosophila eggs by transplantation of polar cytoplasm. Dev. Biol. 37, 43–54 10.1016/0012-1606(74)90168-7 [DOI] [PubMed] [Google Scholar]
- Pause A., Sonenberg N. (1992). Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11, 2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek J. W., Kai T. (2011a). A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr. Biol. 21, 39–44 10.1016/j.cub.2010.11.051 [DOI] [PubMed] [Google Scholar]
- Pek J. W., Kai T. (2011b). DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc. Natl. Acad. Sci. USA 108, 12007–12012 10.1073/pnas.1106245108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Tsai-Morris C.-H., Dufau M. L. (2010). Relevance of gonadotropin-regulated testicular RNA helicase (GRTH/DDX25) in the structural integrity of the chromatoid body during spermatogenesis. Biochim. Biophys. Acta 1803, 534–543 10.1016/j.bbamcr.2010.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., Tsai-Morris C.-H., Dufau M. L. (2003). Cell-specific and hormone-regulated expression of gonadotropin-regulated testicular RNA helicase gene (GRTH/Ddx25) resulting from alternative utilization of translation initiation codons in the rat testis. J. Biol. Chem. 278, 27796–27803 10.1074/jbc.M302411200 [DOI] [PubMed] [Google Scholar]
- Sheng Y., Tsai-Morris C.-H., Gutti R., Maeda Y., Dufau M. L. (2006). Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is a transport protein involved in gene-specific mRNA export and protein translation during spermatogenesis. J. Biol. Chem. 281, 35048–35056 10.1074/jbc.M605086200 [DOI] [PubMed] [Google Scholar]
- Styhler S., Nakamura A., Swan A., Suter B., Lasko P. (1998). vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125, 1569–1578. [DOI] [PubMed] [Google Scholar]
- Tada H., Mochii M., Orii H., Watanabe K. (2012). Ectopic formation of primordial germ cells by transplantation of the germ plasm: Direct evidence for germ cell determinant in Xenopus. Dev. Biol. 371, 86–93 10.1016/j.ydbio.2012.08.014 [DOI] [PubMed] [Google Scholar]
- Taguchi A., Tak M., 2nd, Motoishi M., Or H., 2nd, Moch M., 2nd, Watanabe K. (2012). Analysis of localization and reorganization of germ plasm in Xenopus transgenic line with fluorescence-labeled mitochondria. Dev. Growth Differ. 54, 767–776 10.1111/dgd.12005 [DOI] [PubMed] [Google Scholar]
- Tanaka S. S., Toyooka Y., Akasu R., Katoh-Fukui Y., Nakahara Y., Suzuki R., Yokoyama M., Noce T. (2000). The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 14, 841–853 10.1101/gad.14.7.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang P. Z., Tsai-Morris C.-H., Dufau M. L. (1999). A novel gonadotropin-regulated testicular RNA helicase. A new member of the dead-box family. J. Biol. Chem. 274, 37932–37940 10.1074/jbc.274.53.37932 [DOI] [PubMed] [Google Scholar]
- Tarbashevich K., Koebernick K., Pieler T. (2007). XGRIP2.1 is encoded by a vegetally localizing, maternal mRNA and functions in germ cell development and anteroposterior PGC positioning in Xenopus laevis. Dev. Biol. 311, 554–565 10.1016/j.ydbio.2007.09.012 [DOI] [PubMed] [Google Scholar]
- Toyooka Y., Tsunekawa N., Takahashi Y., Matsui Y., Satoh M., Noce T. (2000). Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech. Dev. 93, 139–149 10.1016/S0925-4773(00)00283-5 [DOI] [PubMed] [Google Scholar]
- Tsai-Morris C.-H., Sheng Y., Lee E., Lei K.-J., Dufau M. L. (2004). Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc. Natl. Acad. Sci. USA 101, 6373–6378 10.1073/pnas.0401855101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatarama T., Lai F., Luo X., Zhou Y., Newman K., King M. L. (2010). Repression of zygotic gene expression in the Xenopus germline. Development 137, 651–660 10.1242/dev.038554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitington P. M. D., Dixon K. E. (1975). Quantitative studies of germ plasm and germ cells during early embryogenesis of Xenopus laevis. J. Embryol. Exp. Morphol. 33, 57–74. [PubMed] [Google Scholar]
- Yajima M., Wessel G. M. (2011). The DEAD-box RNA helicase Vasa functions in embryonic mitotic progression in the sea urchin. Development 138, 2217–2222 10.1242/dev.065052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.