Summary
In moving cells dynamic microtubules (MTs) target and disassemble substrate adhesion sites (focal adhesions; FAs) in a process that enables the cell to detach from the substrate and propel itself forward. The short-range interactions between FAs and MT plus ends have been observed in several experimental systems, but the spatial overlap of these structures within the cell has precluded analysis of the putative long-range mechanisms by which MTs growing through the cell body reach FAs in the periphery of the cell. In the work described here cell geometry was controlled to remove the spatial overlap of cellular structures thus allowing for unambiguous observation of MT guidance. Specifically, micropatterning of living cells was combined with high-resolution in-cell imaging and gene product depletion by means of RNA interference to study the long-range MT guidance in quantitative detail. Cells were confined on adhesive triangular microislands that determined cell shape and ensured that FAs localized exclusively at the vertices of the triangular cells. It is shown that initial MT nucleation at the centrosome is random in direction, while the alignment of MT trajectories with the targets (i.e. FAs at vertices) increases with an increasing distance from the centrosome, indicating that MT growth is a non-random, guided process. The guided MT growth is dependent on the presence of FAs at the vertices. The depletion of either myosin IIA or myosin IIB results in depletion of F-actin bundles and spatially unguided MT growth. Taken together our findings provide quantitative evidence of a role for long-range MT guidance in MT targeting of FAs.
Key words: Cell micropatterning, Microtubules, Myosin II
Introduction
Systems that search targets with high precision, such as smart missiles, often use a two-step strategy whereby the long-range mechanism (e.g. global positioning system) guides the missile toward the approximate vicinity of the target while another, short-range mechanism (e.g. direct illumination with laser light) is used locally to determine the exact location of the target. The hypothesis underlying the present work is that in cells, a conceptually similar, two-tier mechanism may be used by dynamic microtubules (MTs) to target substrate adhesion sites (so-called focal adhesions; FAs). The MTs deliver cargoes and signals disassembling the FAs and enable the cell to detach from the substrate and to propel itself forward (Small et al., 2002). Typically, new MTs originate at the centrosome located in the central region of the cell and grow persistently through the cell body towards the periphery. At the periphery, MTs display frequent switches between growth and shrinking phases, so-called dynamic instability, which results in repetitive probing of peripheral target sites (Komarova et al., 2002). These short-range targeting interactions involving dynamic instability, binding and/or capture of MT plus ends by protein complexes at target sites (Kaverina et al., 1998; Fukata et al., 2002; Zaoui et al., 2010) have been well understood. However, the spatial overlap of cytoskeleton and adhesion structures within a cell has precluded quantification and adequate testing of putative long-range guidance mechanism. Here, we consider MT guidance as a process that results in adjustment(s) of a given MT trajectory so as to align it with the target FA site. To date, it has not been established whether throughout the cell body MTs grow in a random or in a non-random (guided) way (Kaverina et al., 1999; Kaverina et al., 1998; Kandere-Grzybowska et al., 2007; Small and Kaverina, 2003). If MT growth is non-random, it is not clear which elements – structural or chemical – provide the guidance. The possible guidance mechanisms include the structural elements such as filamentous actin, F-actin bundles (Small and Kaverina, 2003); diffusive chemical signals originating from the FAs [sometimes referred to as intracellular chemotaxis (Soh et al., 2010)]; or a transport mechanism whereby MTs are released from centrosome and transported toward the target sites (Salmon et al., 2002; Keating et al., 1997) (Gundersen and Bretscher, 2003; Hwang et al., 2003; Liakopoulos et al., 2003).
In a series of elegant papers, Kaverina and colleagues (Small and Kaverina, 2003; Kaverina et al., 1999; Kaverina et al., 1998; Krylyshkina et al., 2003) used live-cell, dual-wavelength fluorescence microscopy to visualize successive targeting of multiple FA sites, side-stepping or re-routing of the growing MTs, and repetitive probing/targeting of these sites (Kaverina et al., 1998) – these observations indicated that MT targeting to FAs could be a non-random process (Kaverina et al., 1998). The MT–FA interactions studied were confined to the cell periphery, while long-range guidance was not investigated. In subsequent papers, Kaverina and colleagues proposed that MT targeting to FAs could be due to MT guidance, but they did not quantify guidance and failed to find a role for F-actin bundles in the process (Kaverina et al., 1998). On the other hand, interdependence between FA and F-actin bundle formation (Geiger et al., 2009; Vicente-Manzanares et al., 2009) has provided a strong hint that the bundles might play a role in guiding the MT growth toward the FAs. In fact, MTs have been shown to grow along F-actin bundles in epithelial cells (Salmon et al., 2002) and skin keratinocytes (Kodama et al., 2003) and along filopodial actin bundles (no FAs at the end) in neurons (Schaefer et al., 2002; Kodama et al., 2003; Wu et al., 2008). In budding yeast, a slightly different non-random mechanism exists where during mitotic spindle assembly plus ends of selected MTs are captured by myosin V and transported along the actin cables towards the cortical anchoring site in the bud (Gundersen and Bretscher, 2003; Hwang et al., 2003; Liakopoulos et al., 2003). In summary, MT guidance has never been rigorously quantified and consequently it is not clear whether F-actin bundles are involved in long-range MT guidance to FAs. And if F-actin bundles are involved, then which type of bundles guide MTs?
The clarification of the long-range MT guidance mechanism requires observation and quantification of MT growth trajectories and their directions over the entire cell. One of the main challenges in obtaining quantitative measure of MT guidance is the spatial and temporal overlap of all involved structures – MTs, F-actin and FAs – within the cell. Specific practical difficulty with testing the potential role of F-actin bundles in MT guidance has been the inability to selectively depolymerize F-actin (for example, by using Latrunculin B) without causing cell retraction (Bliokh et al., 1980), thus making any quantification of the influence of F-actin on MT growth essentially meaningless. Here, we overcome both of these limitations by using micropatterned, triangular live cells (Fig. 1), in which the components of the cytoskeleton are spatially separated, and which do not change their shapes upon removal of the F-actin bundles. Using dual-wavelength, digital fluorescence live-cell imaging we quantify the directions of MT growth trajectories with respect to the FAs and the F-actin bundles. We show that when the bundles are present in the cell, MT growth is guided toward the FAs at the vertices; in contrast, when the bundles are removed, the MT growth becomes random/unguided. By depleting actin crosslinking proteins via RNA interference, we then show that MT guidance on F-actin requires crosslinking of F-actin by the common non-muscle myosin II isoforms IIA and IIB. Overall, our findings demonstrate that a significant, dynamic interaction between the two main cytoskeletal components, MTs and F-actin, is a cell-wide effect [i.e. not only local, as in some previous studies (Wu et al., 2008)] and is essential for guided MT growth towards FAs.
Fig. 1.
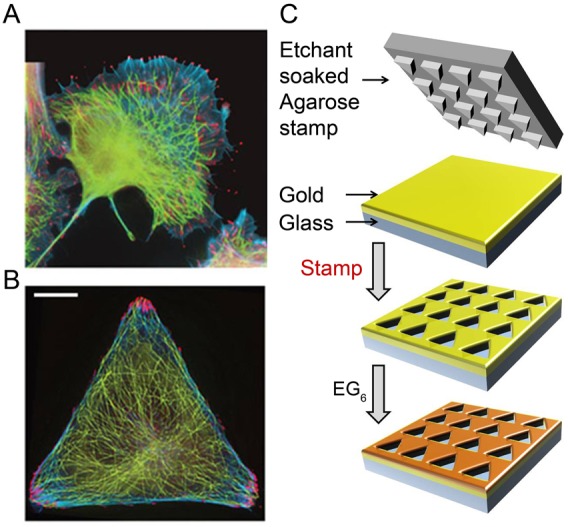
Control of cell geometry and cytoskeletal organization in micropatterned cells. (A) In unpatterned cells, different components of the cytoskeleton [here, FAs (red; phosphotyrosine antibody), MTs (green; tubulin antibody), F-actin bundles (blue; phalloidin)] overlap spatially throughout the cell thus complicate the analysis of their mutual orientations or dynamic processes such as MT guidance. (B) In contrast, in cells micropatterned on triangular islands, the cytoskeleton is spatially disentangled, with FAs located exclusively near the vertices of the triangle. Scale bars: 10 µm. (C) Substrates for cell patterning were prepared by wet etching (Kandere-Grzybowska et al., 2007; Kandere-Grzybowska et al., 2005; Kandere-Grzybowska et al., 2010; Mahmud et al., 2009; Klajn et al., 2004) using a hydrogel stamp to etch an array of microscopic islands in an e-beam evaporated, 35 nm gold layer supported by a 10 nm titanium adhesion layer on glass. The unetched gold was protected with a bioresistant SAM of HS(CH2)11(OCH2CH2)6OH, and the optically transparent islands were typically coated with cell-adhesion-promoting fibronectin. When the cells were plated, they spread on the islands and assumed their shapes.
Results
Non-random, guided microtubule growth revealed in micropatterned cells
Confinement of cells to polygonal microislands is known to cause spatial rearrangement of the cytoskeleton and localization of the FAs at the vertices of the islands (Kandere-Grzybowska et al., 2005; Brock et al., 2003). With well-defined locations of the FAs (as opposed to random distribution along the perimeter of an unpatterned cell, Fig. 1A), trajectories of the MTs emanating from the centrosome can be categorized with high accuracy as random/unguided or non-random/guided (see below). In the present study, we use mostly Rat2 fibroblasts because (1) they are highly motile cells that move using the MT/F–actin/FA system, (2) their cytoskeletal structures localize predictably in response to micropatterns and (3) they are readily transfectable (note: HeLa and B16F1 cells were also tested and give results qualitatively similar to Rat2s). The cells were plated onto arrays of triangular microislands (Fig. 1B) prepared by Wet Etching (Kandere-Grzybowska et al., 2007; Kandere-Grzybowska et al., 2005) method (Fig. 1C; also see Materials and Methods for further experimental details), whereby a micropatterned agarose stamp is soaked in a gold etchant and placed onto a thin layer (∼35 nm) of gold supported on glass. The etchant removes gold at the loci of the stamp/substrate contacts to give an array of transparent microislands surrounded by the opaque gold regions. These regions are subsequently rendered resistant to the adhesion of proteins or cells by forming a self-assembled monolayer (SAM) (Xia and Whitesides, 1998; Witt et al., 2004) of hexa(ethylene glycol)-terminated alkane thiols, EG6. The islands are then coated with fibronectin which promotes adhesion and spreading of the cells. When cells are plated (typically, at density ∼10,000 cm−2), they assume the triangular shape of the islands, reorganize spatially the components of their motility machinery (Kandere-Grzybowska et al., 2005), but otherwise operate this machinery with parameters similar to unpatterned cells [e.g. MT growth velocities were 17.3±2.2 µm/min for triangular cells vs 15.0±4.2 µm/min for unconstrained cells and similar to the values reported in previous works (Komarova et al., 2002; Kandere-Grzybowska et al., 2005)]. Importantly, the optically transparent, adhesive microislands are compatible with high-resolution imaging of fluorescently tagged fusion proteins (green fluorescent protein and related fusions) in live cells (Kandere-Grzybowska et al., 2005) allowing for simultaneous monitoring of either monomeric Cherry fusion with β-actin (mCherry–actin) and EB3 in fusion with GFP (EB3–GFP), or mCherry–myosinIIA and EB3–GFP.
The investigation of MT guidance requires quantification of the directions of MT growth trajectories. We visualized MT growth trajectories by live cell imaging of MT-end-binding protein EB3–GFP (Fig. 2A). By tracing comet-shaped EB3–GFP spots, we were then able to reconstruct the MT growth trajectories (Fig. 2B; typically ∼100 MT trajectories per cell imaged over a period of 5 min before photobleaching occurred), as described in detail in our previous works (Kandere-Grzybowska et al., 2005). Excluding MTs that grew along cell edges (see Materials and Methods for more details), we quantified the directionality of each MT growth trajectory with respect to the FAs (Fig. 2C,D: supplementary material Fig. S6) by an angle θ between the instantaneous MT growth velocity and the line joining current EB3–GFP position with the closest vertex of the triangular cell (Fig. 2E). This measure is sensitive to the nature of MT growth – if the growth is guided towards the FAs, the histogram of θ angles (over multiple trajectories) should feature a distinct maximum at θ = 0o; if, on the other hand, the MTs grow randomly, the histogram is expected to become more broadly distributed as illustrated for the computer-generated random/unguided growth trajectories in Fig. 2G,H (Random, black curve). In reality, the experimental MT growth trajectories observed on fibronectin-coated islands (on which cell adhesion is promoted by FA formation) correspond to the guided scenario as illustrated in Fig. 2H (fibronectin, red curve).
Fig. 2.
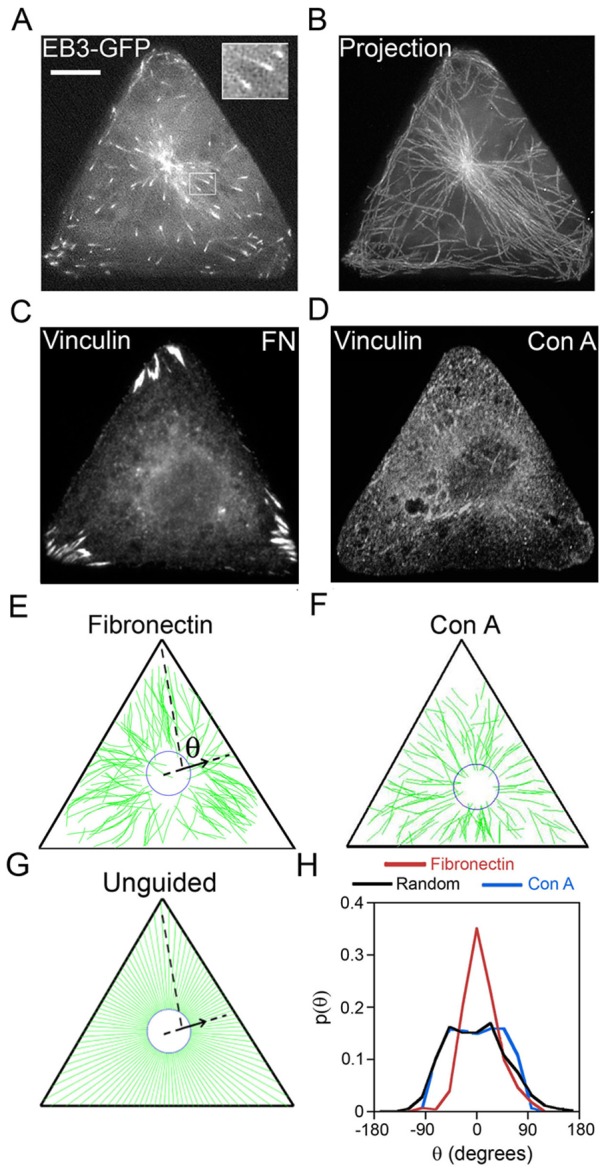
MT growth is guided towards focal adhesions located at the vertices of triangular cells. MT growth trajectories were reconstructed from time-lapse imaging of MT plus-end protein EB3–GFP in triangular Rat2 cells. (A) A single frame from a time-lapse movie of EB3–GFP dynamics (see also supplementary material Movie 1). Large, bright accumulation of EB3–GFP at the cell center marks the centrosome from which new MTs are nucleated. Small, comet-shaped EB3–GFP spots (see also enlarged inset in the top-right corner) mark growing plus ends of MTs. This comet shape makes it easy to identify the direction of movement and MT growth at any given time. (B) Reconstruction of representative MT growth trajectories by superposition of multiple (∼50) consecutive frames (total time, 150 sec). (C) FAs marked by vinculin are located exclusively at the vertices of the triangular cells plated on fibronectin. (D) Focal adhesions in cells plated on Con A are delocalized or cannot be discerned. Scale bar: 10 µm (A–D). (E) Example of reconstruction of all MT growth trajectories for a cell plated on fibronectin (shown in green) from one time-lapse movie of EB3–GFP dynamics (total time of observation for each cell was 5 min; image acquisition: one frame per 3 sec) by using house-written image recognition software. The arrow shows a vector parallel to the line joining EB3–GFP positions at two consecutive times; this vector corresponds to the direction of MT growth at a given time. The angle (θ) defined with respect to the nearest vertex of the triangle, is the angle by which the direction of growth of each MT at any given time deviates from its target (here, the vertex). (F) Reconstructed tracks for cells plated on Con A. (G) MT growth trajectories obtained computationally for the putative unguided case, whereby MTs grow radially outwards. (H) Normalized histograms/probability distributions of angles θ for experimental MT trajectories (red, on a fibronectin substrate; blue, on Con A, substrate; ∼500 MT growth trajectories in four cells for each substrate) and for the computationally generated, random/unguided (black) MT trajectories. On fibronectin, the spiked distribution indicates guided growth. On Con A, which promotes FA-independent adhesion, MT growth directions are unguided and very similar to computationally obtained random/unguided P(θ) distribution. A two-sample t-test shows that for cells on fibronectin substrate the probability (at θ = 0°) is greater than that for the random/unguided case at a 99% confidence level. In contrast, the probabilities for random/unguided case and for cells on Con A are statistically similar.
When analyzing such data in triangular cells, care must be taken to ascertain that the observed guidance is not due to the trivial fact that near the vertices there is simply less space for the MTs to grow, and it is the overall geometry of the cell that then constrains the MT trajectories toward the FAs. One way to eliminate this possibility is to analyze the distribution of the angles (θ) only within circles inscribed into triangular cells, where the ‘anisotropic’ cell shape is not directly felt. This is illustrated in Fig. 3, which shows regional analysis of MT growth directions over three regions/shells around the centrosome. Within the circular region closest to the contour of the centrosome, the initial growth directions of the MTs (indicative of MT nucleation) are essentially random/unguided (see Fig. 3, curves labeled ‘First EB3 spot’); in the subsequent shells, however, MT trajectories become increasingly aligned with the target FAs at vertices as the angles θ center around zero degrees. The observation that the more peripheral shell shows increased alignment of MT trajectories with the targets as compared to the inner shell is not indicative of the effects of cell geometry because all three regions are inscribed within triangular cell shape and the cell volume and thus molecular freedom is not yet reduced in these shells. Importantly, the regions near cell vertices – where molecular freedom is, indeed, reduced – are excluded from this analysis.
Fig. 3.
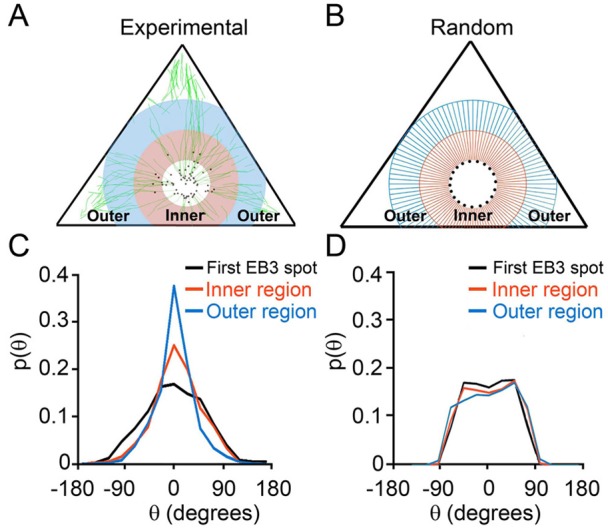
Regional analysis of the directions of MT growth trajectories. (A) An example in which three regions are analyzed: (1) the first region is closest to the centrosome and consists of the first EB3 spots (i.e. the first EB3 spot of each track) in the live-cell movie (shown as black dots in A and B); (2) the inner annular region (pink) of 6.5 µm width, containing the MT trajectories; and (3) an outer annular region (blue) of the same width, 6.5 µm (n = 8 cells; 619 trajectories). (B) The same regions but with unguided MT trajectories generated computationally. (C) Distributions based on the experimental data, such as those in A. (D) Distributions for the modeled unguided case. Comparison between C and D demonstrates that MTs are initially nucleated without preference in direction but their growth then becomes more guided (distributions peaking at θ = 0°) with an increasing distance from the centrosome. A two sample t-test shows that the probability (at θ = 0°) for the inner annular region (2) is greater than that for the random/unguided case, with 95% confidence. For the outer annular region (3), the analogous confidence level is 99%.
However, when cells are cultured on triangular islands coated with Concanavalin A (Fig. 2D,F,H; Con A, blue curve), a substrate that promotes FA-independent adhesion (Fig. 2D) (Rogers et al., 2003; Lin et al., 1996; Symons and Mitchison, 1991), MT growth directions are random/unguided over all regions of the cell (supplementary material Fig. S7A,B), as in the computationally generated unguided case (Fig. 2G, black curve). Taken together, these results demonstrate that MT growth in cells adherent via FAs is non-random, preferentially guided towards the vertices of the triangular cell, and is dependent on the presence of FAs at these vertices.
Cell-wide correlation of F-actin bundles and microtubule trajectories
One possibility is that MT growth toward FAs might be guided by internal F-actin bundles. To test the role of F-actin bundles, we performed dual-wavelength live cell imaging of cells simultaneously transfected with EB3–GFP and also with actin in fusion with mCherry (mCherry–actin) (Fig. 4A–C). Since large F-actin structures/bundles did not display changes during the short time interval of MT growth observation (3–5 min, actin images taken at the beginning and end of this time frame were nearly identical), we obtained time-lapse movies of GFP–EB3 dynamics and a single ‘static’ image of F-actin bundles for each cell. Visual inspection of the time-lapse movies color-combined with corresponding F-actin images revealed clear incidents of MTs growing along interior bundles towards the vertices/FAs (e.g. see regions indicated by arrows in Fig. 4B; supplementary material Movies 1, 2). To quantify the correlation between MT growth trajectories and F-actin bundles, we calculated (using a house-written image-processing code) the total length of the MT trajectories, LMT, and the total length of the F-actin bundles, LAB. Next, we overlaid the MT and F-actin bundle images (Fig. 4D), and calculated the total length LMT-AB of fragments where MT growth was spatially correlated with a proximal actin bundle (i.e. the MT growth trajectory was within ±250 nm of a bundle and aligned within 10° from that bundle). We then calculated the fraction of MT trajectories that correlate with the bundles, LMT-AB/LMT, and the fraction of bundles that correlate with MT growth trajectories, LAB-MT/LAB. This analysis, summarized in the histogram in Fig. 4E (Exp), revealed that ∼23% of all MT growth trajectories (LMT-AB/LMT = 0.23±0.12; ∼5400 µm of MT tracks in five cells were analyzed) were aligned with F-actin bundles and ∼40% of the bundles (LAB-MT/LAB = 0.41±0.17; ∼100 bundles of total length ∼1000 µm from five cells analyzed) were aligned with the MTs. These fractions were several tens of times higher than the colocalization measures that should be expected if the two types of structures were uncorrelated. To show this, we estimated the probabilities of colocalization in cells in which MTs and the F-actin bundles were assumed to be randomly distributed. The simulations in supplementary material Fig. S1 predict that for this uncorrelated case LMT-AB/LMT∼0.018±0.010 and LAB-MT/LAB∼0.056±0.032 (Fig. 4E, random); these values differ from their experimental counterparts with *P<0.01 in the Student's t-test. In other words, experimental MT growth trajectories are significantly correlated with F-actin bundles.
Fig. 4.
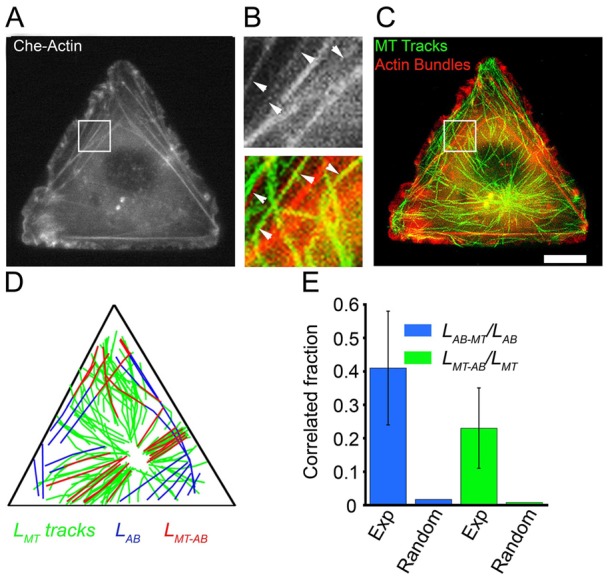
Cell-wide correlation of F-actin bundles and MT growth trajectories. (A–C) Dual-color time-lapse imaging of (A,C) mCherry–actin and (C) EB3–GFP (MT tracks from projection of ∼50 frames of EB3–GFP, shown in green, and merged with the F-actin image in red) in a triangular Rat2 cell. B shows enlarged images of the boxed regions in A and C (see also supplementary material Movies 1, 2). Scale bar: 10 µm (A,C). (D) Representative map of aligned, reconstructed F-actin bundles and MT growth trajectories (all actin bundles and MT growth trajectories in one cell are shown). Blue: F-actin bundles (ABs); green: MT growth trajectories/tracks (MT); red: F-actin bundles colocalized with MT growth trajectories (MT-AB). (E) Quantitative analysis of the colocalization of F-actin bundles and MT growth trajectories (Exp, experimental; Random, modeled random case; see main text for more details). A two-sample t-test shows that the experimental LMT-AB/LMT and LAB-MT/LAB values are greater than those of the simulated random case at the 99% confidence level.
Guided growth of microtubules to the vertices of the triangle requires internal F-actin bundles
The interior F-actin bundles of the cell can be distinguished from so-called edge bundles by differences in sensitivity to an inhibitor of Rho-associated kinase (ROCK) (Katoh et al., 2001a; Katoh et al., 2001b; Totsukawa et al., 2000; Totsukawa et al., 2004). We performed experiments aimed to determine whether the interior F-actin bundles are necessary for the observed MT guidance (Fig. 5). To this end, we selectively depleted interior bundles by treating the triangular cells with ROCK inhibitor Y27632. The assembly of interior/centrally located stress fibers requires ROCK and is sensitive to Y27632, whereas the edge bundles are less sensitive to ROCK inhibitors (Katoh et al., 2001a; Katoh et al., 2001b; Totsukawa et al., 2000; Totsukawa et al., 2004) allowing us to deplete interior bundles without affecting the triangular cell shape (Fig. 5A,C; supplementary material Fig. S4). MT growth velocities also were not affected by treatment of cells with Y27632 (supplementary material Fig. S9). In Rat2 cells on fibronectin treated with ROCK inhibitor (60 µM for 1 h), the number of internal bundles was reduced from 24.0±5.9 in untreated cells to 0.9±1.3 (Fig. 5A,C,F). Focal adhesions in these cells were delocalized from vertices (in control cells) to uniform distribution along the entire cell perimeter and are smaller in size (Fig. 5D; supplementary material Fig. S4). Most importantly, the distribution of MT growth trajectories in cells treated with the ROCK inhibitor was nearly identical to the computationally generated distributions for the random/unguided growth case (Fig. 5B; supplementary material Fig. S7C,D; Movie 3). The inhibition of stress fibers with myosin II inhibitor blebbistatin (Straight et al., 2003; Allingham et al., 2005) also rendered MT growth directions unguided and reduced the number of F-actin bundles (from 24.0±5.9 bundles in untreated cells to 1.5±0.8 in cells treated with 25 μM blebistatin for 2 h) while cells remained triangular (see supplementary material Fig. S2). Together, these results indicate that interior F-actin bundles are required for guided growth of MTs towards the vertices of the triangle/FAs.
Fig. 5.
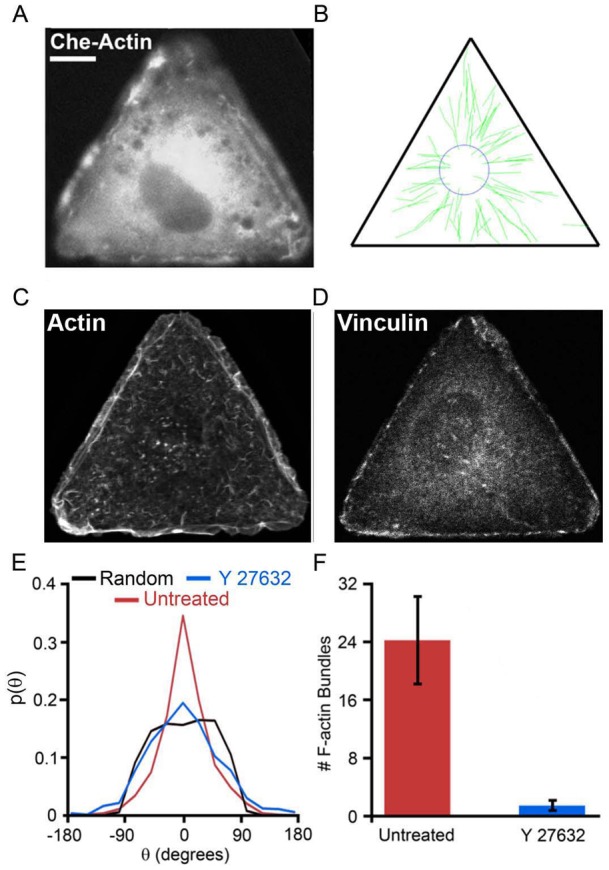
The effect of Rho-kinase inhibitor on F-actin bundle depletion and guided MT growth towards focal adhesions/vertices. Rat2 cells on triangular, fibronectin-coated islands were either untreated (see Fig. 3) or treated with ROCK inhibitor Y27632 (60 µM). (A) Cherry–actin distribution in a representative experimental cell shows depletion of F-actin bundles. (B) The reconstructed MT growth trajectories (green tracks) from the same cell; triangle indicates cell boundary. (C) F-actin and (D) focal adhesion (vinculin) distribution in cells treated with Y27632. Scale bar: 10 µm and is the same for all images. (E) The probability distributions of the MT growth directions; red, untreated; blue, Y27632; black, random/unguided. Data for Y27632-treated cells is based on 367 trajectories from five cells. (F) Quantification of the extent of F-actin bundle depletion. ROCK inhibitor depletes interior F-actin bundles and results in random/unguided MT growth. A two-sample t-test shows that the probability (at θ = 0°) for cells treated with Y27632 is statistically similar to the unguided case.
Guided growth of microtubules toward the vertices of the triangle requires myosin IIA
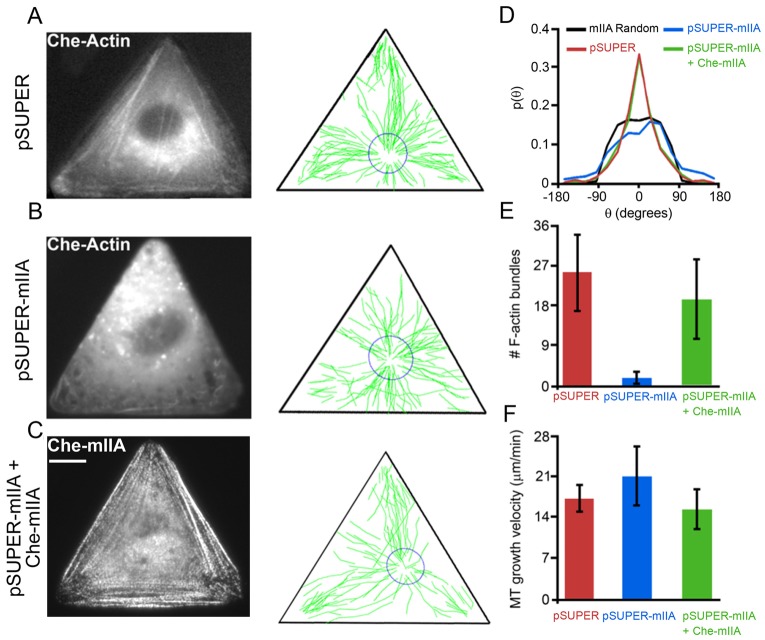
On a macromolecular level, F-actin bundles are formed by molecular crosslinking of multiple actin filaments (F-actin). One notable example is contractile motor protein non-muscle myosin II (Gupton and Waterman-Storer, 2006; Kolega, 1998; Vicente-Manzanares et al., 2007). In motile cells, myosin IIA initiates F-actin bundle formation while myosin IIB stabilizes these F-actin bundles (Wei and Adelstein, 2000; Vicente-Manzanares et al., 2011). To test the role of myosin IIA (MIIA) crosslinked bundles in MT guidance, we depleted MIIA from Rat2 cells by means of RNA interference. The knockdown vectors for depletion of MIIA and MIIB proteins with very high specificity were described previously (Vicente-Manzanares et al., 2007). Immunofluorescence staining revealed that in triangular Rat2 cells transfected with RNAi construct designed to deplete MIIA (pSUPER-MIIA), MIIA protein was depleted (see supplementary material Fig. S3 and S5) and the number of F-actin bundles was reduced from 24.0±5.9 bundles (n = 15 cells) in untreated control cells to 1.7±1.5 (n = 13) in MIIA-depleted cells. There were no discernible focal adhesion structures in MIIA-depleted cells (supplementary material Fig. S5). We simultaneously introduced the knockdown vector against MIIA (pSUPER-mIIA) with pmCherry-actin and pEB3-GFP DNAs in Rat2 cells and obtained time-series of EB3–GFP and a static image of mCherry–actin (Fig. 6). In live cells, mCherry–actin images were used to monitor the effectiveness of knockdown; the MIIA-depleted cells showed consistently very few large bundles (Fig. 6B,E). In cells transfected with control vector pSUPER, majority of MT trajectories were pointing toward the vertices (Fig. 6A), while in all MIIA-depleted cells MT trajectories were disorganized and many appeared to point towards the sides of the triangle (Fig. 6B). Probability distributions of MT growth directions in MIIA-depleted cells (Fig. 6D, blue curve) were almost identical to the computationally generated distributions for the unguided growth (Fig. 6D, black curve), and were significantly different from the trajectories guided toward the vertices in control cells spread on fibronectin (Fig. 6D, red curve; also see Fig. 2H). Based on these experiments, MT growth trajectories in MIIA-depleted cells can be classified as unguided.
Fig. 6.
Guided growth of MTs toward focal adhesions at the vertices of the triangles requires myosin IIA. (A–C) F-actin bundles [from mCherry–actin live images in A and B or from mCherry–myosin IIA (mChe–mIIA) image in C], and MT growth trajectories (reconstructed from EB3–GFP time-lapse movies) in triangular Rat2 cells transfected with (A) pSUPER (n = 4 cells; 470 trajectories), (B) pSUPER-mIIA (n = 7 cells; 699 trajectories), or (C) pSUPER-mIIA together with RNAi resistant mChe-mIIA DNAs (n = 6 cells; 597 trajectories). (D–F) Quantification of (D) the directions of MT growth trajectories, (E) depletion of F-actin bundles, and (F) MT growth velocities. The knockdown of MIIA with RNAi depletes F-actin bundles and renders MT trajectories disorganized and/or misdirected (note many tracks directed towards the sides of the triangle), an effect rescued by adding back exogenous RNAi-resistant mCherry–mIIA. Scale bar: 10 µm for all images. For the distributions in D, a two-sample t-test shows that the probabilities (at θ = 0°) for cells transfected with pSUPER or pSUPER-mIIA+Che-mIIA are greater than for the random/unguided case at the 99% confidence level. pSUPER-mIIA-transfected cells, however, are statistically similar to the unguided case.
It is worth noting that one previous study reported that myosin IIA deficiency results in MT stabilization (Even-Ram et al., 2007). In our triangular Rat2 cells, however, at least MT growth velocities were unaffected by MIIA knockdown (Fig. 6F). Also, the control vector (pSUPER) had no effect on the number of F-actin bundles, MT growth velocities, or MT trajectory directions (Fig. 6A,D–F; supplementary material Fig. S9). Furthermore, the introduction of exogenous RNAi resistant mCherry–MIIA into MIIA-depleted cells (pSUPER-MIIA + mChe-MIIA) resulted in re-forming of F-actin bundles and focal adhesions (supplementary material Fig. S5) and rescue of the guided growth of MTs (Fig. 6C–F; see also supplementary material Fig. S7 for regional analysis of MT trajectories). The last result indicates that unguided MT growth is indeed due to the depletion of MIIA protein, and not caused by non-specific, off-target effects due to RNAi. Depletion of myosin IIB similar to depletion of MIIA also results in depletion of all F-actin bundles and random/unguided MT growth trajectories (supplementary material Fig. S8). In sum, our results show that MIIA and MIIB are necessary for MT guidance towards FAs at the vertices of the triangle.
Discussion
We have combined digital fluorescence live cell microscopy with cell patterning to develop a material system in which the imposed shape removes the spatial overlap of cytoskeleton structures and translates into a well-defined organization of its cytoskeleton and thus allows for unprecedented quantitative analysis of MT guidance. The key findings are that (1) MTs start growing from the centrosome in random directions but, with increasing distance from the centrosome, MT trajectories become increasingly aligned with target FAs indicating that MT growth is non-random/guided process; (2) there are cell-wide correlation of MT trajectories with F-actin bundles; and (3) these internal F-actin bundles and their crosslinkers myosins IIA and IIB (MIIA and MIIB) are required for guided MT growth.
While cell micropatterning based on thiolate SAMs on gold has been around for almost two decades (Chen et al., 1997; Singhvi et al., 1994) the Wet Etched patterns – due to optical transparency of cell adhesive islands – offer distinct advantages over patterns micro-contact-printed on continuous, opaque gold substrates (Kandere-Grzybowska et al., 2007; Kandere-Grzybowska et al., 2005; Kandere-Grzybowska et al., 2010; Mahmud et al., 2009). In this respect, one of the distinguishing features of the present work is the demonstration that micropatterned cells are compatible not only with high resolution molecular dynamics imaging [which we demonstrated earlier (Kandere-Grzybowska et al., 2005)] but also with gene product depletion by way of RNA interference. It is the combination of these approaches that has proven essential in establishing the role of MIIA crosslinked F-actin bundles in MT guidance towards FAs. In this context, an interesting observation is that in the triangular cells all internal F-actin bundles (except those along the edges) are crucially dependent on MIIA as the depletion of MIIA results in almost complete removal of all large bundles. This is similar to unconstrained cells where bundling by other myosin isoform, MIIB, is dependent on MIIA and efficient MIIA knockdown results in complete depletion of all F-actin bundles (Vicente-Manzanares et al., 2011). Furthermore, the result that either depletion of Myosin IIA or Myosin IIB by themselves depletes all F-actin bundles are consistent with recent findings that in fibroblast cells MIIA initiates bundle formation whereas MIIB crosslinks and strengthens MIIA initiated bundles (Vicente-Manzanares et al., 2011). This property combined with the fact that depletion of MIIA or MIIB does not cause cell retraction from the microislands suggests a potential of our triangular cell system as a model platform with which to study MT interactions with F-actin bundles and FAs (Rodriguez et al., 2003; Waterman-Storer and Salmon, 1999; Wehrle-Haller and Imhof, 2003; Jordan and Wilson, 1998; Carragher and Frame, 2004) in quantitative detail.
While MT–FA interactions have been studied by other methods as well, these approaches were not suitable for studying MT guidance. In a recent notable example, MT targeting toward the FAs has been probed by simultaneous expression of GFP–EB1 to mark MT plus ends, DsRed–Zyxin to mark FAs, and CFP–actin to mark F-actin bundles, and by performing sophisticated three-wavelength confocal videomicroscopy (Wu et al., 2008). This method allowed for high resolution imaging of MT dynamics very close to the FAs and quantification of frequency of MT-plus-end contacts with FAs, but did not permit analysis of MT trajectory directions with respect to the FAs over the entire cell thus yielding no information about long-range guidance of MTs. By localizing the FAs predictably and reproducibly to the vertices of the micropatterned cells, our system offers an easier and likely a more versatile alternative, whereby long-range (and possibly also short-range) component of the complex process of MT/FA targeting could be analyzed by tracking just one structure (plus ends of the growing MTs marked by GFP–EB3 spots). MT growth is tracked over the entire cell and hence all (or, at least, the vast majority of) MT trajectories are accounted for, producing a detailed and complete description of the process. This description, with the help of automated image recognition and analysis software, is averaged over multiple cells of virtually identical shapes thus producing statistically significant datasets (e.g. conclusions of the present work are based on the analysis of >4000 trajectories from over 50 cells).
Another issue worth emphasizing is that the guidance of MTs on F-actin ‘rails’ is not necessary operative for all MTs at all times. This is evidenced by the fact that the colocalization fractions, LMT-AB and LAB-MT – while being much larger that for the random case – are still significantly smaller than unity, indicating that not all MTs are correlated with the actin bundles at all times. Perhaps, this finding is not surprising given that the characteristic widths of both types of fibers are submicrometer, and at this scale the effects of thermal noise and/or concentration fluctuations cannot be neglected. These stochastic effects can be especially important for the ‘dynamic’ MTs, which grow by the addition of nanoscopic tubulin monomers (and MT associated/regulating proteins), whose distribution within the cell fluctuates from one location to another (Niethammer et al., 2004). Thus, it can be expected that in response to the local concentration fluctuations, the MTs should be able to change their instantaneous growth direction before re-aligning with the guiding F-actin rail or, else, finding another rail to follow. In fact, both of these stochastic behaviors are observed experimentally and are illustrated in Fig. 7.
Fig. 7.
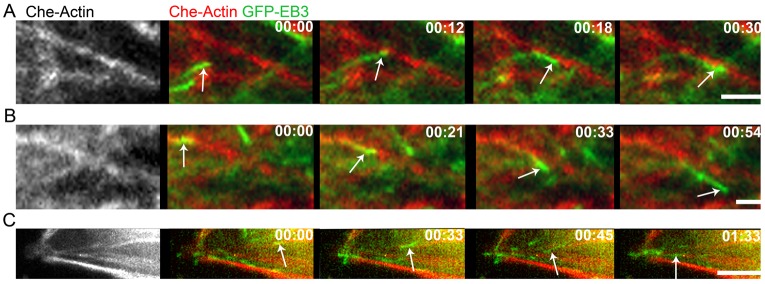
Dynamic effects accompanying MT interactions with F-actin bundles. (A) MT changes its growth direction upon encountering an F-actin bundle. (B) MT travels along and veers off an F-actin bundle. (C) MT switches from one F-actin bundle to another (at t = 45 sec). EB3–GFP (green); mCherry-actin (red). Time: min:sec. Scale bars: 2 µm.
Our work described here is distinct from the earlier work in unconstrained cells focusing on short-range interactions between MT and FAs (Kaverina et al., 1999; Kaverina et al., 1998; Wu et al., 2008) in that we demonstrate that long-range MT guidance plays a role in MT targeting to FAs. In particular, our key finding that MT targeting of FAs requires MIIA crosslinked F-actin bundles settles the previous disputes over the role of stress fibers in this process (Small and Kaverina, 2003; Kaverina et al., 1999; Kaverina et al., 1998; Ishizaki et al., 2001). The role for F-actin bundles in MT-FA targeting was previously called into question because fixed cell preparations of fibroblast cells (and also cancer cells in our unpublished observations) did not display obvious correlation of F-actin and MT cytoskeletons (Kaverina et al., 1998). Kaverina et al. suggested that ‘few short actin filaments’ splaying out of the FA sites may tether growing MTs and orient their growth (Kaverina et al., 1998). Such tethering mechanism may involve proteins or protein complexes that crosslink MTs and F-actin (Rodriguez et al., 2003) such as spectraplakin ACF7 (Wu et al., 2008), plectin (Svitkina et al., 1996), formin mDia1 (Ishizaki et al., 2001; Zaoui et al., 2008), CLASPs (Tsvetkov et al., 2007) or unconventional myosin in cooperation with plus end proteins (Small and Kaverina, 2003; Lantz and Miller, 1998). Such short-range mechanisms may play role locally once MT has been guided to the approximate location of its target. Our results indicate that in addition to these short-range mechanisms MT growth is guided towards FAs already in internal cytoplasm (but not yet at MT nucleation stage) and this guidance requires larger F-actin bundles (not just single actin filaments) crosslinked by MIIA. Lastly, our knockdown studies single out MIIA as an important mediator of the crosstalk between actomyosin and MT cytoskeletal systems (Even-Ram et al., 2007) and suggest that the previously hypothesized ‘at a distance’ effect of myosins (Vicente-Manzanares et al., 2007) on FA dynamics (and cell migration) is, in fact, due to the ability of myosin II (via bundle formation) to direct MT growth.
Admittedly, a notable drawback of our system is that a triangular cell exemplifies a stationary cell and cannot reproduce all the dynamic events (notably, cell polarization and coupling of front protrusion and rear retraction) occurring in motile cells. We believe, however, that the possibility to obtain high quality data sets – complete quantitative description of MT trajectories over entire cell – from cells with uniform shapes and with low cell-to-cell variability was crucial for the demonstration of MT guidance. In addition, it should be possible to extend our approach to examining MT growth trajectories in shape-controlled tear-drop cells, which have been shown to polarize (Jiang et al., 2005; Théry et al., 2006) and behave as if ‘running on a treadmill’ (Kandere-Grzybowska et al., 2010). Finally, while MIIA organizes F-actin to guide the MTs, we expect that other MT plus end/F-actin bundle interactions – mediated by (macro)molecules or their complexes – may be involved in the process. Elucidating the nature of these interactions can have implications for the design of small molecules inhibiting cell motility (by disrupting the MT guidance and thus reducing MT/FA targeting) – this goal remains a challenge for future research.
Materials and Methods
Cell micropatterning
Glass slides (2.2 ×2.2 cm, standard thickness; no. 1.5, Corning) were cleaned according to the following procedure: 15 min soaking in 1% (v/v) alcanox, 30 min sonication in acetone, and 30 min sonication in ethanol. Titanium (10 nm) and gold (35 nm) were deposited onto the cleaned glass coverslips by electron beam deposition (Edwards, Crawley, UK).
Polydimethylsiloxane, PDMS, masters presenting arrays of depressed, equilateral triangle features (54 µm on the side, area ∼1256 µm2) were fabricated by standard photolithography followed by molding, as described in detail before (Kandere-Grzybowska et al., 2007; Kandere-Grzybowska et al., 2005; Kandere-Grzybowska et al., 2010). Agarose stamps were prepared by casting hot, 8% (w/v) high gel strength agarose (EMD Biosciences) solution against the PDMS master. To remove air bubbles, the beaker with the agarose-covered PDMS master was placed in a desiccator under vacuum for 90 sec. After degassing, the agarose was cooled down to room temperature, peeled off the PDMS template, and cut into appropriately sized (∼1.5 ×1.5 ×1 cm) ‘stamps’ presenting an array of raised triangular microfeatures.
Subsequently, agarose stamps were soaked in 20% (v/v) gold etchant (type TFA, Transene Co. Inc., Danvers, MA) for 30 min, allowed to air-dry for 3 min, and placed feature-side down onto the titanium/gold-covered glass slides to microetch the desired patterns. Etched slides were washed with ethanol and dried under a stream of nitrogen. Unetched portions of the gold surface was derivatized with cell-adhesion-resistant self-assembled monolayer (SAM) of EG6 thiols [hexa(ethylene glycol)-terminated alkane thiols, HS-C11H22-(OCH2CH2)6OH, from ProChimia, Gdansk, Poland]. Each etched gold slide was placed in a Petri dish and covered with 1 ml of 4 mM EG6 thiol solution in ethanol for 12 h at 4°C. SAM-protected etched slides were washed with ethanol, dried under a stream of nitrogen, and attached to the bottom of plastic dishes with holes by using vacuum grease. SAM-protected slides were washed twice with phosphate-buffered saline (PBS), and were then incubated with fibronectin (Fn; 25 μg/ml, Sigma-Aldrich) or Concanavalin A (0.5 mg/ml, Sigma-Aldrich cat. no. C 5275) for 45 min at room temperature. Subsequently, slides were rinsed twice with PBS and used immediately for culturing cells. Rat2 cells were fully spread on the triangular microislands within 2 to 4 h from the time of plating.
Cell culture and drug treatments
Rat2 fibroblasts and HeLa cells were obtained from ATTC and cultured in DMEM culture medium supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) in atmosphere with 10% CO2. B16F1 cells were cultured as described previously (Kandere-Grzybowska et al., 2005; Kandere-Grzybowska et al., 2010). Rat2 fibroblasts cells were used in most experiments because they are highly motile and their motility employs the MT–FA system. Importantly, in these cells both MT plus-ends and F-actin could be adequately marked and imaged in the same cells by co-trasfection of EB3-GFP/mCherry-actin DNA constructs. For transfections, cells were cultured in 12-well plates in a standard culture medium (10% FBS–DMEM; 2 mL of medium/well) for 24 h. For F-actin/MT track imaging, cells were co-transfected with 1 µg of pEB3-GFP DNA and 1 µg of pmCherry-actin DNA using FuGENE transfection reagent (Roche) in a 4∶1 ratio of microliters of FuGENE to micrograms of plasmid DNA. For F-actin bundle depletion with pharmacological approach, cells were allowed to spread on triangular island for 12 h, then treated either with 60 µM Y27632 (Sigma-Aldrich, catalog no. Y0503) for 2 h, or with 25 µM blebbistatin (Sigma-Aldrich, catalog no. B0506) for 2 h. Imaging of F-actin/MT tracks was performed with drug (Y27632 or blebbistatin) present in the solution within 16 h from initial cell plating.
Plasmids and RNA interference
PmCherry-actin DNA was previously described by Applewhite and Borisy (Applewhite et al., 2007), and pEB3-GFP by Stepanova (Stepanova et al., 2003). pSUPER-mIIA and pmCherry-mIIA was described previously (Vicente-Manzanares et al., 2007). pSUPER was purchased from Oligoengine (Seattle, WA). For knockdown experiments, Rat2 cells were transfected with pSUPER (control) or pSUPER-mIIA (to deplete MIIA) vectors together with Cherry-actin (to directly monitor bundle depletion/knockdown effect in the same cells), cultured for 72–96 h, re-plated onto triangular islands, and imaged within 3 h of plating.
Live-cell digital fluorescence imaging of F-actin bundles and microtubule growth trajectories
Forty-eight hours after transfection, cells were re-plated on micropatterned fibronectin-coated slides attached to Petri dishes with holes and allowed to spread for 2–4 h before assuming the shapes of the triangular islands. For microscopy observation, cell culture medium was changed to Phenol-Red-free Leibowitz-15 medium supplemented with 10% FBS. During observation, the temperature of the medium covering the cells was kept constant at ∼37°C by a metallic heating ring surrounding the Petri dish and an objective heater. Time-lapse imaging of EB3–GFP dynamics and mCherry–actin was conducted using an inverted microscope (Diaphot 300; Nikon) equipped with a 63× objective and slow-scan CCD camera (model CH350; Photometrics, Tucson, AZ) driven by MetaMorph imaging software (Universal Imaging Corp., Worchester, PA). Time series of GFP–EB3 (total observation time per cell ∼5 min at 3 sec intervals) were obtained as previously described (Kandere-Grzybowska et al., 2005). The images of stationary (over 5 min time intervals) F-actin bundles marked by mCherry–actin were obtained at the beginning and at the end of EB3–GFP imaging. Since F-actin bundle reorganization occurs on time scales much slower than microtubule growth, the two actin images (∼5 min apart) showed almost identical actin bundle distribution justifying the use of single static actin bundle image. Metamorph software was used to combine the timelapse movie of EB3–GFP dynamics with F-actin bundle images for each cell.
Immunofluorescence staining
Microetched coverslips with triangular cells were washed with warm PBS, fixed with 3% formaldehyde (Ted Pella Inc.) solution for 10 min at room temperature, and permeabilized with 0.2% Triton X-100 (Thermo Scientific) for 10 min at this temperature (all solutions were in PBS). Myosin IIA was visualized by immunofluorescence staining using primary antibody specific to myosin IIA isoform (Covance, Emeryville, CA; cat. no. PRB-440P) and Rhodamine-conjugated anti-rabbit secondary antibody (Jackson ImmunoResearch, West Grove, PA; cat no. 111-025-144). F-actin was visualized in the same preparations by using Alexa-Fluor-647- or Alexa-Fluor-488-conjugated phalloidin (Molecular Probes/Invitrogen). The substrates were mounted using AquaPoly/Mount mounting medium (Polysciences Inc., cat. no. 18606) and imaged using an inverted fluorescence microscope (Nikon Diaphot 300) equipped with 63× magnification objective.
Data and statistical analysis
Microtubule growth trajectories were obtained by analyzing frame-to-frame displacements of EB3–GFP (EB) spots over time. ImageJ image analysis software was used to track the positions of each EB spot (new spot appearance marked the beginning of the track) in subsequent frames until the spot disappeared (indicating microtubule catastrophe and the end of the track) or reached cell edge (within ∼250 nm). MTs that grew along the edges of the triangle were excluded from the analysis. Next, x,y coordinates of all EB spots at each time point were extracted. A house-written Matlab® R2010b image processing algorithm was then used to reconstruct all MT growth trajectories in each cell from x,y coordinates of EB spots. From the xt, yt positions at time = t and the xt+Δt, yt+1 positions at time = t+Δt, the directions of growth of the MTs at each point of time were determined. To quantify the degree of MT/FA targeting, we defined θ as the angle between the instantaneous direction of MT growth and the line connecting the position xt, yt with the closest vertex of the triangle. After calculating θ for all time frames and all MT tracks, the frequency histograms for all angles (−180° ≤θ≤180°) were plotted and normalized into probability distributions. These distributions were compared with a unguided case of MT trajectories generated computationally. The case of unguided MT growth was represented by 100 radial tracks distributed evenly (every 3.6°) around the centrosome and growing towards the cell periphery. The values of θ for all tracks and times were converted into probability distributions and were used for comparisons with experimental data at various conditions.
F-actin bundles marked by mCherry–actin were also traced and reconstructed by the ImageJ software. To quantify the correlation between MT growth trajectories and F-actin bundles, the maps of reconstructed MT growth trajectories and F-actin bundles were overlaid for each cell. Using these aligned ‘maps’, the LAB, LMT, LMT-AB, and LAB-MT measures were calculated. MT and F-actin were classified as correlated/overlapping if the distance between them was less than the imaging resolution (∼250 nm), and the axes of the fibers were aligned to within less than 10°.
The experimentally observed fractions of correlated MT/F-actin lengths were LMT-AB/LMT = 0.23±0.12 and LAB-MT/LAB = 0.41±0.17. These values were compared with the case when MT and F-actin are distributed randomly across a triangular cell, independent of one another. This case was modeled using experimental values of average MT length ∼14 µm, average F-actin bundle length ∼17 µm, average number of MTs per cell ∼90, and average number of F-actin bundles per cell ∼18. Thus represented fibers were then distributed randomly over a triangular domain. Correlations between the two types of fibers were then calculated by the same criteria as for the experimental data (distance between the fibers <250 nm, alignment <10°). The results of this modeling were averaged over 100 realization of MT/F-actin random distributions and gave the values LMT-AB/LMT = 0.018±0.010 and LAB-MT/LAB = 0.056±0.032 (see supplementary material Fig. S2). Two sample t-test showed that the experimental LMT-AB/LMT and LAB-MT/LAB measures were greater than the simulated, random case at the 99% confidence level.
Supplementary Material
Acknowledgments
We thank Prof. Miguel Vicente-Manzanares (at the Universidad Autonoma, Madrid, Spain) for pSUPER-MIIA and mCherry-mIIA DNA constructs and for generous advice on their use.
Footnotes
Funding
This work was supported by National Institutes of Health [grant numbers 1R21CA137707-01 and U54CA119341 to B.A.G.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.110494/-/DC1
References
- Allingham J. S., Smith R., Rayment I. (2005). The structural basis of blebbistatin inhibition and specificity for myosin II. Nat. Struct. Mol. Biol. 12, 378–379 10.1038/nsmb908 [DOI] [PubMed] [Google Scholar]
- Applewhite D. A., Barzik M., Kojima S., Svitkina T. M., Gertler F. B., Borisy G. G. (2007). Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol. Biol. Cell 18, 2579–2591 10.1091/mbc.E06-11-0990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliokh Z. L., Domnina L. V., Ivanova O. Y., Pletjushkina O. Y., Svitkina T. M., Smolyaninov V. A., Vasiliev J. M., Gelfand I. M. (1980). Spreading of fibroblasts in medium containing cytochalasin B: formation of lamellar cytoplasm as a combination of several functional different processes. Proc. Natl. Acad. Sci. USA 77, 5919–5922 10.1073/pnas.77.10.5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock A., Chang E., Ho C-C., LeDuc P., Jiang X., Whitesides G. M., Ingber D. E. (2003). Geometric determinants of directional cell motility revealed using microcontact printing. Langmuir 19, 1611–1617 10.1021/la026394k [DOI] [PubMed] [Google Scholar]
- Carragher N. O., Frame M. C. (2004). Focal adhesion and actin dynamics: a place where kinases and proteases meet to promote invasion. Trends Cell Biol. 14, 241–249 10.1016/j.tcb.2004.03.011 [DOI] [PubMed] [Google Scholar]
- Chen C. S., Mrksich M., Huang S., Whitesides G. M., Ingber D. E. (1997). Geometric control of cell life and death. Science 276, 1425–1428 10.1126/science.276.5317.1425 [DOI] [PubMed] [Google Scholar]
- Even–Ram S., Doyle A. D., Conti M. A., Matsumoto K., Adelstein R. S., Yamada K. M. (2007). Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 9, 299–309 10.1038/ncb1540 [DOI] [PubMed] [Google Scholar]
- Fukata M., Watanabe T., Noritake J., Nakagawa M., Yamaga M., Kuroda S., Matsuura Y., Iwamatsu A., Perez F., Kaibuchi K. (2002). Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 109, 873–885 10.1016/S0092-8674(02)00800-0 [DOI] [PubMed] [Google Scholar]
- Geiger B., Spatz J. P., Bershadsky A. D. (2009). Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21–33 10.1038/nrm2593 [DOI] [PubMed] [Google Scholar]
- Gundersen G. G., Bretscher A. (2003). Cell biology. Microtubule asymmetry. Science 300, 2040–2041 10.1126/science.1084938 [DOI] [PubMed] [Google Scholar]
- Gupton S. L., Waterman–Storer C. M. (2006). Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell 125, 1361–1374 10.1016/j.cell.2006.05.029 [DOI] [PubMed] [Google Scholar]
- Hwang E., Kusch J., Barral Y., Huffaker T. C. (2003). Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 161, 483–488 10.1083/jcb.200302030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T., Morishima Y., Okamoto M., Furuyashiki T., Kato T., Narumiya S. (2001). Coordination of microtubules and the actin cytoskeleton by the Rho effector mDia1. Nat. Cell Biol. 3, 8–14 10.1038/35050598 [DOI] [PubMed] [Google Scholar]
- Jiang X., Bruzewicz D. A., Wong A. P., Piel M., Whitesides G. M. (2005). Directing cell migration with asymmetric micropatterns. Proc. Natl. Acad. Sci. USA 102, 975–978 10.1073/pnas.0408954102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M. A., Wilson L. (1998). Microtubules and actin filaments: dynamic targets for cancer chemotherapy. Curr. Opin. Cell Biol. 10, 123–130 10.1016/S0955-0674(98)80095-1 [DOI] [PubMed] [Google Scholar]
- Kandere–Grzybowska K., Campbell C., Komarova Y., Grzybowski B. A., Borisy G. G. (2005). Molecular dynamics imaging in micropatterned living cells. Nat. Methods 2, 739–741 10.1038/nmeth796 [DOI] [PubMed] [Google Scholar]
- Kandere–Grzybowska K., Campbell C. J., Mahmud G., Komarova Y., Soh S., Grzybowski B. A. (2007). Cell motility on micropatterned treadmills and tracks. Soft Matter 3, 672–679 10.1039/b617308j [DOI] [PubMed] [Google Scholar]
- Kandere–Grzybowska K., Soh S., Mahmud G., Komarova Y., Pilans D., Grzybowski B. A. (2010). Short-term molecular polarization of cells on symmetric and asymmetric micropatterns. Soft Matter 6, 3257–3268 10.1039/b922647h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kano Y., Amano M., Onishi H., Kaibuchi K., Fujiwara K. (2001a). Rho-kinase–mediated contraction of isolated stress fibers. J. Cell Biol. 153, 569–584 10.1083/jcb.153.3.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kano Y., Amano M., Kaibuchi K., Fujiwara K. (2001b). Stress fiber organization regulated by MLCK and Rho-kinase in cultured human fibroblasts. Am. J. Physiol., Cell Physiol. 280, C1669–C1679 [DOI] [PubMed] [Google Scholar]
- Kaverina I., Rottner K., Small J. V. (1998). Targeting, capture, and stabilization of microtubules at early focal adhesions. J. Cell Biol. 142, 181–190 10.1083/jcb.142.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Small J. V. (1999). Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J. Cell Biol. 146, 1033–1044 10.1083/jcb.146.5.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating T. J., Peloquin J. G., Rodionov V. I., Momcilovic D., Borisy G. G. (1997). Microtubule release from the centrosome. Proc. Natl. Acad. Sci. USA 94, 5078–5083 10.1073/pnas.94.10.5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klajn R., Fialkowski M., Bensemann I. T., Bitner A., Campbell C. J., Bishop K., Smoukov S., Grzybowski B. A. (2004). Multicolour micropatterning of thin films of dry gels. Nat. Mater. 3, 729–735 10.1038/nmat1231 [DOI] [PubMed] [Google Scholar]
- Kodama A., Karakesisoglou I., Wong E., Vaezi A., Fuchs E. (2003). ACF7: an essential integrator of microtubule dynamics. Cell 115, 343–354 10.1016/S0092-8674(03)00813-4 [DOI] [PubMed] [Google Scholar]
- Kolega J. (1998). Cytoplasmic dynamics of myosin IIA and IIB: spatial ‘sorting’ of isoforms in locomoting cells. J. Cell Sci. 111, 2085–2095 [DOI] [PubMed] [Google Scholar]
- Komarova Y. A., Vorobjev I. A., Borisy G. G. (2002). Life cycle of MTs: persistent growth in the cell interior, asymmetric transition frequencies and effects of the cell boundary. J. Cell Sci. 115, 3527–3539 [DOI] [PubMed] [Google Scholar]
- Krylyshkina O., Anderson K. I., Kaverina I., Upmann I., Manstein D. J., Small J. V., Toomre D. K. (2003). Nanometer targeting of microtubules to focal adhesions. J. Cell Biol. 161, 853–859 10.1083/jcb.200301102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz V. A., Miller K. G. (1998). A class VI unconventional myosin is associated with a homologue of a microtubule-binding protein, cytoplasmic linker protein-170, in neurons and at the posterior pole of Drosophila embryos. J. Cell Biol. 140, 897–910 10.1083/jcb.140.4.897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D., Kusch J., Grava S., Vogel J., Barral Y. (2003). Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112, 561–574 10.1016/S0092-8674(03)00119-3 [DOI] [PubMed] [Google Scholar]
- Lin C. H., Espreafico E. M., Mooseker M. S., Forscher P. (1996). Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron 16, 769–782 10.1016/S0896-6273(00)80097-5 [DOI] [PubMed] [Google Scholar]
- Mahmud G., Campbell C. J., Bishop K. J. M., Komarova Y. A., Chaga O., Soh S., Huda S., Kandere–Grzybowska K., Grzybowski B. A. (2009). Directing cell motions on micropatterned ratchets. Nat. Phys. 5, 606–612 10.1038/nphys1306 [DOI] [Google Scholar]
- Niethammer P., Bastiaens P., Karsenti E. (2004). Stathmin-tubulin interaction gradients in motile and mitotic cells. Science 303, 1862–1866 10.1126/science.1094108 [DOI] [PubMed] [Google Scholar]
- Rodriguez O. C., Schaefer A. W., Mandato C. A., Forscher P., Bement W. M., Waterman–Storer C. M. (2003). Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5, 599–609 10.1038/ncb0703-599 [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Wiedemann U., Stuurman N., Vale R. D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088 10.1083/jcb.200303023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon W. C., Adams M. C., Waterman–Storer C. M. (2002). Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 158, 31–37 10.1083/jcb.200203022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A. W., Kabir N., Forscher P. (2002). Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J. Cell Biol. 158, 139–152 10.1083/jcb.200203038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhvi R., Kumar A., Lopez G. P., Stephanopoulos G. N., Wang D. I., Whitesides G. M., Ingber D. E. (1994). Engineering cell shape and function. Science 264, 696–698 10.1126/science.8171320 [DOI] [PubMed] [Google Scholar]
- Small J. V., Kaverina I. (2003). Microtubules meet substrate adhesions to arrange cell polarity. Curr. Opin. Cell Biol. 15, 40–47 10.1016/S0955-0674(02)00008-X [DOI] [PubMed] [Google Scholar]
- Small J. V., Geiger B., Kaverina I., Bershadsky A. (2002). How do microtubules guide migrating cells? Nat. Rev. Mol. Cell Biol. 3, 957–964 10.1038/nrm971 [DOI] [PubMed] [Google Scholar]
- Soh S., Byrska M., Kandere–Grzybowska K., Grzybowski B. A. (2010). Reaction-diffusion systems in intracellular molecular transport and control. Angew. Chem. Int. Ed. Engl. 49, 4170–4198 10.1002/anie.200905513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T., Slemmer J., Hoogenraad C. C., Lansbergen G., Dortland B., De Zeeuw C. I., Grosveld F., van Cappellen G., Akhmanova A., Galjart N. (2003). Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23, 2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Cheung A., Limouze J., Chen I., Westwood N. J., Sellers J. R., Mitchison T. J. (2003). Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science 299, 1743–1747 10.1126/science.1081412 [DOI] [PubMed] [Google Scholar]
- Svitkina T. M., Verkhovsky A. B., Borisy G. G. (1996). Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J. Cell Biol. 135, 991–1007 10.1083/jcb.135.4.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M. H., Mitchison T. J. (1991). Control of actin polymerization in live and permeabilized fibroblasts. J. Cell Biol. 114, 503–513 10.1083/jcb.114.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M., Racine V., Piel M., Pépin A., Dimitrov A., Chen Y., Sibarita J-B., Bornens M. (2006). Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA 103, 19771–19776 10.1073/pnas.0609267103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G., Yamakita Y., Yamashiro S., Hartshorne D. J., Sasaki Y., Matsumura F. (2000). Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J. Cell Biol. 150, 797–806 10.1083/jcb.150.4.797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G., Wu Y., Sasaki Y., Hartshorne D. J., Yamakita Y., Yamashiro S., Matsumura F. (2004). Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J. Cell Biol. 164, 427–439 10.1083/jcb.200306172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov A. S., Samsonov A., Akhmanova A., Galjart N., Popov S. V. (2007). Microtubule-binding proteins CLASP1 and CLASP2 interact with actin filaments. Cell Motil. Cytoskeleton 64, 519–530 10.1002/cm.20201 [DOI] [PubMed] [Google Scholar]
- Vicente–Manzanares M., Zareno J., Whitmore L., Choi C. K., Horwitz A. F. (2007). Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 176, 573–580 10.1083/jcb.200612043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente–Manzanares M., Choi C. K., Horwitz A. R. (2009). Integrins in cell migration–the actin connection. J. Cell Sci. 122, 199–206 10.1242/jcs.018564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente–Manzanares M., Newell–Litwa K., Bachir A. I., Whitmore L. A., Horwitz A. R. (2011). Myosin IIA/IIB restrict adhesive and protrusive signaling to generate front-back polarity in migrating cells. J. Cell Biol. 193, 381–396 10.1083/jcb.201012159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman–Storer C. M., Salmon E. (1999). Positive feedback interactions between microtubule and actin dynamics during cell motility. Curr. Opin. Cell Biol. 11, 61–67 10.1016/S0955-0674(99)80008-8 [DOI] [PubMed] [Google Scholar]
- Wehrle–Haller B., Imhof B. A. (2003). Actin, microtubules and focal adhesion dynamics during cell migration. Int. J. Biochem. Cell Biol. 35, 39–50 10.1016/S1357-2725(02)00071-7 [DOI] [PubMed] [Google Scholar]
- Wei Q., Adelstein R. S. (2000). Conditional expression of a truncated fragment of nonmuscle myosin II-A alters cell shape but not cytokinesis in HeLa cells. Mol. Biol. Cell 11, 3617–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt D., Klajn R., Barski P., Grzybowski B. A. (2004). Applications, properties and synthesis of ω-functionalized n-alkanethiols and disulfides - the building blocks of self-assembled monolayers. Curr. Org. Chem. 8, 1763–1797 10.2174/1385272043369421 [DOI] [Google Scholar]
- Wu X., Kodama A., Fuchs E. (2008). ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell 135, 137–148 10.1016/j.cell.2008.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Whitesides G. M. (1998). Soft lithography. Angew. Chem. Int. Ed. 37, 550–575 [DOI] [PubMed] [Google Scholar]
- Zaoui K., Honoré S., Isnardon D., Braguer D., Badache A. (2008). Memo-RhoA-mDia1 signaling controls microtubules, the actin network, and adhesion site formation in migrating cells. J. Cell Biol. 183, 401–408 10.1083/jcb.200805107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaoui K., Benseddik K., Daou P., Salaün D., Badache A. (2010). ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc. Natl. Acad. Sci. USA 107, 18517–18522 10.1073/pnas.1000975107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.