Abstract
Chlamydia trachomatis is the most common bacterial sexually transmitted pathogen in the world. To identify new vaccine candidates a protein microarray was constructed by expressing the open reading frames (ORFs) from Chlamydia mouse pneumonitis (MoPn). C57BL/6, C3H/HeN and BALB/c mice were immunized either intranasally or intravaginally with live MoPn elementary bodies (EB). Two additional groups were immunized by the intramuscular plus subcutaneous routes with UV-treated EB, using CpG and Montanide as adjuvants to favor a Th1 response, or Alum, to elicit a Th2 response. Serum samples collected from the three strains of mice were tested in the microarray. The array included the expression of 909 proteins from the 921 ORFs of the MoPn genome and plasmid. A total of 530 ORFs were recognized by at least one serum sample. Of these, 36 reacted with sera from the three strains of mice immunized with live EB. These antigens included proteins that were previously described as immunogenic such as MOMP and HSP60. In addition, we uncovered new immunogens, including 11 hypothetical proteins. In summary, we have identified new immunodominant chlamydial proteins that can be tested for their ability to induce protection in animal models and subsequently in humans.
Keywords: Chlamydia, Antigens, Microarrays, Vaccine, Mouse model
1. Introduction
Chlamydia trachomatis is the most common sexually transmitted bacterial pathogen and the leading cause of preventable blindness in the world [1–3]. In the U.S.A. 1.2 million chlamydial infections were reported to CDC in 2009 [1]. In addition, the majority of the cases were not reported since most of these genital infections are asymptomatic [2,4,5]. Chlamydial infections can be treated with antibiotics however, due to its asymptomatic nature, most of them go untreated [4,6]. Untreated chlamydial infections can progress to serious reproductive and other health problems with both short-term and long-term consequences [7,8]. Furthermore, delayed or inadequate treatments fail to protect against long-term sequelae [7,8]. Therefore, a vaccine is the most effective way to control this disease [9–12].
Chlamydial vaccine development, to protect against trachoma, started in the 1960s using whole, inactivated, organisms [2,13,14]. Even though some of the vaccine formulations generated protection, the protection was short-lived and serovar, or serogroup specific [2,15]. Even in some instances, particularly if a low strength vaccine preparation was used, it appeared that vaccination might have enhanced the severity of ocular disease when individuals became re-infected, a problem that may also occur using live-attenuated vaccines[2,3,16]. Therefore, the need for a subunit vaccine is urgent.
Following the discovery of the major outer membrane protein (MOMP) of C. trachomatis, renewed attempts to develop a vaccine against genital infections were made in the 1980s[15,17]. Even though mice immunized with MOMP showed significant protection against chlamydial infection, some limitations were discovered [18–20]. For example, some of the protection generated by MOMP is dependent on the native 3-dimensional structure of the protein. Protection with recombinant MOMP was not as robust as that resulting from vaccination with the native MOMP [21,22]. Extraction of the native form of MOMP cannot be scaled up at a reasonable cost to manufacture for a human vaccine and therefore, alternative antigens need to be identified to formulate a vaccine.
Recent advances in generating whole proteome chips have led to a fast way to screen proteins that can generate an immune response [23,24]. Molina et al. [25] used sera from immunized mice and a chip containing approximately 25% of the C. trachomatis MoPn genome and identified seven immunodominant antigens that were recognized by immunized mouse sera. Cruz-Fisher et al. [26] generated a proteome chip of the C. trachomatis MoPn genome and identified 185 proteins that were recognized by sera of BALB/c female mice immunized by this bacterium. The great amount of variability in the human population will require that a Chlamydia vaccine includes antigens that can be recognized by individuals with multiple immunogenetic backgrounds. To address this issue, using three different strains of mice, we identified dominant novel antigens that can be further tested for their ability to induce a protective response against chlamydial infections in animal models and eventually in humans.
2. Materials and methods
2.1. Preparation and titration of stocks of C. trachomatis MoPn
The mouse C. trachomatis biovar (MoPn strain Nigg II), also called Chlamydia muridarum, was purchased from the American Type Culture Collection (Manassas, VA) and was grown in McCoy cells [27]. Eagle’s minimal essential medium was supplemented with 5% fetal bovine serum (EMEM-FBS) and 1 μg/mL of cycloheximide. Purification of elementary bodies (EB) was done as described by Caldwell et al. [28]. The EB stock was stored at −70 °C in SPG buffer (0.2 M sucrose, 0.02 M sodium phosphate, pH 7.2, and 5 mM glutamic acid). The number of inclusion forming units (IFU) of the stock was determined by titration on HeLa 229 cells.
2.2. Immunization of mice
Three weeks old female BALB/c (H-2d), C3H/HeN (H-2k), and C57BL/6 (H-2b) mice were purchased from Charles River Laboratories (Wilmington, MA). Groups of 12 mice were immunized as follows. For live intranasal (i.n.) immunization, BALB/c and C57BL/6 mice received 104 IFU of MoPn and C3H/HeN mice were inoculated with 101 IFU and for intravaginal (i.vag.) delivery mice received 105 IFU/mouse [29–31]. The animals immunized intravaginally were treated with 2.0 mg of medroxy-progesterone acetate (Greenstone, Peapack, NJ) subcutaneously (s.c.) 7 days before inoculation [32,33]. EB were inactivated by exposure to a UV transilluminator box (UV-EB) emitting at a wavelength of 302 nm (Spectroline, Westbury, NY) for 10 min as previously described [34]. For the combined intramuscular and subcutaneous (i.m.+s.c.) routes, the mice were vaccinated with 106 IFU of UV-EB per mouse three times 2 weeks apart [34]. To elicit a Th1 response, one of the groups immunized by the i.m.+s.c. routes was vaccinated using UV-EB with CpG oligodeoxynucleotide (ODN) 1826 (10 μg/mouse/immunization; Coley Pharmaceutical, Ottawa, Canada) and Montanide ISA 720 (Seppic, Inc., Fairfield, NJ) as adjuvants [18]. The Montanide was mixed at a 70:30 (vol/vol) ratio of the final preparation. To induce a Th2 response, a second group was immunized i.m.+s.c. using alum (250 μg/ mouse/immunization; 0.3% aluminum hydroxide solution; Alhydrogel 85; Superfos, Denmark) as the adjuvant [35]. As a negative-control group, mice were immunized with ovalbumin and the three combined adjuvants. Another control group was not immunized. Serum samples were collected before immunization and then at two-weeks intervals up to 180 days post immunization. All the samples were tested with the microarray. In order to be able to perform all serological tests with the same sample, the sera from each group of mice were pooled. To identify antigens based on their ability to induce antibodies that are long term persistent after immunization, we selected those that gave a positive signal for at least five or more time points or were positive for three consecutive data points. The experiment was repeated once. All animal protocols were approved by the University of California, Irvine (Irvine, CA), IACUC.
2.3. Microarray probing and data collection
Mouse serum samples were probed on the full proteome Chlamydia MoPn protein chip [26,36,37]. Briefly, samples were diluted 1:100 with 1× protein array blocking buffer (Whatman, Piscataway, NJ) containing 10% Escherichia coli lysate (McLab, San Francisco, CA) and incubated at room temperature for 30 min with constant agitation. The microarrays were rehydrated in 1× protein array blocking buffer for 30 min and probed with the diluted serum samples for 2 h at room temperature with constant agitation [23]. The slides were then washed three times with Tris-buffered saline (TBS) containing 0.05% Tween 20 (TTBS) and incubated with biotin-conjugated goat anti-mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). After three washes with TTBS, the bound secondary antibodies were detected using streptavidin-conjugated Sensilight P3 (Columbia Biosciences, Columbia, MD), diluted according to the manufacturer’s recommendations. The slides were washed three times with TTBS and three times with TBS, followed by a final wash with ultrapure water. The slides were air dried by centrifugation and scanned in a ScanArray Express HT microarray scanner (PerkinElmer, Waltham, MA), and the fluorescence signal was quantified using QuantArray software (PerkinElmer, Waltham, MA). All samples were tested in triplicate.
All antigen-specific signal intensities were first corrected for background noise by using QuantArray software (PerkinElmer, Waltham, MA). The data was transformed using the log variant asinh, normalized using the variance stabilization and normalization (VSN) package in the statistical programming language known as R, and transformed back for positive selection and creation of graph [23,26,38]. The antigen signal intensity data were then averaged, no DNA control plus 2 standard deviations was removed. Next, the signal of each antigen at the day before immunization was subtracted from that specific antigen signal post-vaccination. The antigen signal from the ovalbumin-immunized group was then subtracted from the antigen signal of groups immunized with UV-EB while the signal from the control non immunized group was removed from the groups immunized with live EB for the same time point [23,38].
2.4. Western blot
The Western blot was performed as previously described with MoPn EB as the antigen [18]. Briefly, 40 μg of purified EB was loaded on a 7.5-cm-wide slab polyacrylamide gel. Following transfer to a nitrocellulose membrane, the nonspecific binding was blocked with BLOTTO (bovine lacto transfer technique optimizer; 5% [wt/vol] nonfat dry milk, 2 mM CaCl2, 50 mM Tris–HCl [pH 8.0]) overnight at cold room, and then serum samples were added to the membrane strips and incubated overnight at 4 °C. The membrane was washed and incubated with horseradish peroxidase-conjugated goat anti-mouse antibody for 1 h, followed by visualization of the bands by developing with 0.01% hydrogen peroxide and the substrate 4-chloro-1-naphthol.
2.5. Bioinformatics analysis
Computational prediction of protein cellular role, enzyme class, and gene ontology utilized ProtFun 2.2 (http://www.cbs.dtu. dk/services/ProtFun). Signal peptide prediction and cellular location prediction used PSORTb, version 3.02, software [39] (http://www.psort.org/psortb). Enrichment statistical analysis was using Fisher’s exact test to calculate P-value (http://www.graphpad.com/quickcalcs/contingency1.cfm).
3. Results
3.1. Validation of microarray
Antibodies that recognize the N-terminal poly-His tag and the C-terminal HA tag were used to assess protein expression as previously described [23]. Poly-His and HA staining were done in technical quadruplicates, i.e. 4 microarrays for poly-His and 4 microarrays for HA. The microarrays were scanned with the PerkinElmer ProscanArray HT dual laser microarray scanner, and the intensity for each spot was quantified using the ProscanArray software package. Antigens with mean signal intensities greater than the average control value plus two standard deviations, were considered positive for the detection of the tag. Tag detection was used as a measure of protein expression.
From a total of 921 ORFs, of the C. trachomatis MoPn genome and plasmid, the protein microarray included the expression products of 909 ORFs, as well as the appropriate positive and negative controls (mouse IgG and no-DNA RTS reaction). ORFs TC0437, TC0438 and TC0439, due to their size (>3000 bp) were expressed as three fragments each. We were not able to clone 12 genes [26]. Each of the 909 expressed proteins and controls were printed three times in each microarray. Of the 909 ORFs arrayed, 908 stained positive for the N-terminal poly-His and 888 stained positive for the C-terminal HA tag [26]. A total of 887 ORFs, representing 96.3% (887 out of 921) of the MoPn genome and plasmid, were fully expressed as demonstrated by a positive signal for both His and HA. All 909-cloned ORFs were probed with mouse sera.
3.2. Identification of immunodominant antigens
Groups of BALB/c, C3H/HeN, and C57BL/6 female mice were immunized one time with live Chlamydia by the i.n. or the i.vag. routes. Two additional groups were immunized three times, two weeks apart with UV-EB by the i.m.+s.c. routes. One of the UV-EB preparations was mixed with CpG plus Montanide, adjuvants that favor a Th1 response. A second group of mice was immunized with UV-EB mixed with Alum, an adjuvant that elicits a Th2 response. Two negative control groups were immunized with ovalbumin as the antigen mixed with all the adjuvants, and the second control was not immunized. Serum samples were collected at two-weeks intervals up to 180 days post immunization. All serum samples from the three different strains of mice were used to probe the microarray chip. In order to select antigens based on their ability to induce antibodies that are persistent over long term after immunizations, we chose those that gave a positive signal for at least five or more time points or were positive for three consecutive time points.
Our analysis identified a total of 530 ORFs that gave at least one positive signal from all three strains of mice and all four routes of immunization, which represents 57.1% of the Chlamydia MoPn genome (Table 1; Supplemental). Since immunization with live Chlamydia elicits a more robust protection than that resulting from vaccination with non-viable organisms, and in addition some antigens may be present only when Chlamydia is replicating, we decided to select our positive antigens based on the result from both groups vaccinated with live EB.
Table 1.
Immunodominant antigens recognized by the sera of the three strains of mice immunized with live EB and their predicted function.
| Protein ID | Live EB, i.n. |
Live EB, i.vag. |
UV-EB, CpG/Monta. |
UV-EB, alum | Predicted functional category |
CT ortholog | CT ortholog seq identity |
|---|---|---|---|---|---|---|---|
| TC0035 | + | + | Hypothetical proteins | CT664 | 82 | ||
| TC0045 | + | + | Cellular processes | CT674 | 95 | ||
| TC0050 | + | + | Protein synthesis | CT679 | 89 | ||
| TC0052 | + | + | + | + | Transport and binding proteins | CT681 | 85 |
| TC0117 | + | + | Hypothetical proteins | CT741 | 93 | ||
| TC0137 | + | + | Cell envelope | CT756 | 86 | ||
| TC0140 | + | + | + | + | Hypothetical proteins | CT759 | 83 |
| TC0177 | + | + | Hypothetical proteins | CT795 | 65 | ||
| TC0189 | + | + | Hypothetical proteins | CT805 | 89 | ||
| TC0229 | + | + | Cellular processes | CT841 | 94 | ||
| TC0268 | + | + | No data | CT875 | 48 | ||
| TC0328 | + | + | Hypothetical proteins | CT058 | 55 | ||
| TC0344 | + | + | Hypothetical proteins | CT072 | 86 | ||
| TC0386 | + | + | Protein fate | CT110 | 98 | ||
| TC0387 | + | + | Protein fate | CT111 | 99 | ||
| TC0392 | + | + | No data | CT116 | 49 | ||
| TC0396 | + | + | Cellular processes | CT119 | 52 | ||
| TC0399 | + | + | Fatty acid and phospholipid metabolism | CT123 | 87 | ||
| TC0437 | + | + | Cellular processes | CT166 | 46 | ||
| TC0438 | + | + | Cellular processes | CT166 | 67 | ||
| TC0439 | + | + | Cellular processes | CT166 | 47 | ||
| TC0512 | + | + | Cell envelope | CT241 | 93 | ||
| TC0582 | + | + | Energy metabolism | CT308 | 97 | ||
| TC0589 | + | + | Transcription | CT315 | 98 | ||
| TC0590 | + | + | Protein synthesis | CT316 | 98 | ||
| TC0660 | + | + | Transport and binding proteins | CT381 | 82 | ||
| TC0680 | + | + | Energy metabolism | CT400 | 87 | ||
| TC0721 | + | + | + | + | Protein synthesis | CT437 | 96 |
| TC0726 | + | + | Cell envelope | CT442 | 59 | ||
| TC0727 | + | + | + | + | Cell envelope | CT443 | 92 |
| TC0773 | + | + | Transport and binding proteins | CT486 | 91 | ||
| TC0794 | + | + | Transcription | CT507 | 97 | ||
| TC0816 | + | + | Hypothetical proteins | CT529 | 61 | ||
| TC0828 | + | + | Cellular processes | CT541 | 88 | ||
| TC0848 | + | + | Cellular processes | CT559 | 89 | ||
| TC0854 | + | + | Hypothetical proteins | CT565 | 90 |
A total of 406 positive antigens were identified in the live EB immunization groups (Table 1; Supplemental). Out of the 406 positive signals, sera from BALB/c and C3H/HeN mice recognized 282 and 236 proteins respectively, while the sera from C57BL/6 mice only reacted with 139 antigens. Of these, 36 were dominant antigens among all the live EB immunization routes in all three strains of mice (Table 1). The 36 dominant antigens included the following known immunogens: TC0052 (MOMP), TC0386 (60 kDa chaperonin HSP), TC0387 (10 kDa chaperonin HSP), TC0396 (inclusion membrane localized protein, IncA), TC0660 (amino acid ABC transporter, periplasmic amino acid-binding protein), TC0726 (sulfur rich protein), TC0727 (60 kDa cysteine-rich outer membrane protein), TC0828 (peptidyl-proply cis-trans isomerase, Mip), and TC0848 (type III secretion protein, SctJ). Four of the 36 antigens (TC0052, TC0140, TC0721, and TC0727) were also positive in the three strains of mice immunized with UV-EB and either CpG/Montanide or Alum as adjuvants. As shown in Table 1, all these Chlamydia MoPn proteins have orthologs, with high degree of sequence identity, in the human C. trachomatis serovars.
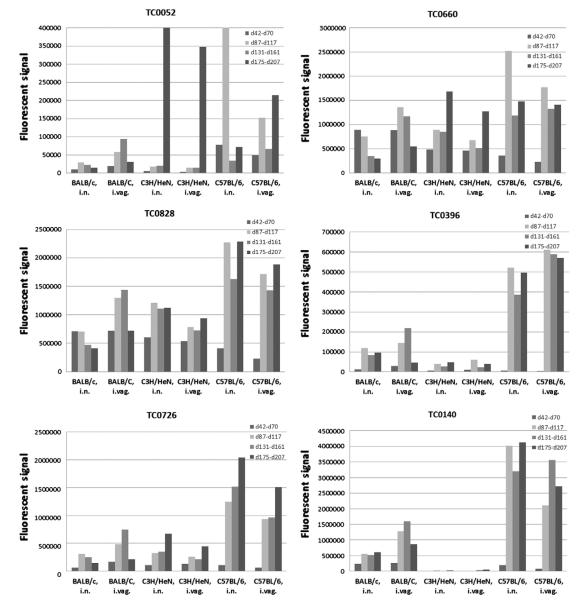
Fig. 1 shows the signal intensity of the top 6 immunodominant antigens, as determined by the lowest P-value, over time in the three strains of mice using different routes of immunization. The majority of the antibody response profile can be grouped into three different kinds. In one of them, for example TC0726 in C57BL/6, mice immunized by the i.n. and i.vag. routes, the antibody response shows a continuous increase in signal intensity throughout the study. The second kind shows an initial increase in antibody response and, after the antibody level reaches a high point, the signal then gradually decreases. Examples are, TC0396 in BALB/c mice immunized i.vag. and TC0660 in BALB/c mice vaccinated by the i.n. and i.vag. routes. The third kind of expression profile shows an initial increase in the antibody response and after reaching a maximum, the signal remains at that high point for the rest of the study. TC0828 in C3H/HeN mice immunized by the i.n. and i.vag. routes and TC0140 in C57BL/6 mice immunized i.n., showed this kind of expression pattern. These results may help identify the stage of the infection and to select antigens for vaccine formulation.
Fig. 1.
Signal intensity of the top six immunodominant Chlamydia MoPn antigens. These antigens were selected based on their low P-value.
3.3. Western blot
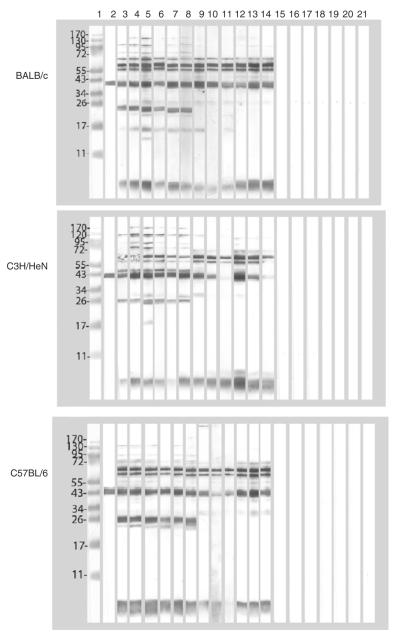
Western blots were performed using EB as the antigen and sera collected at 60, 120 and 180 days post-immunization (Fig. 2). Like with the microarray, a broader antibody response was observed with the sera from the BALB/c and C3H/HeN mice in comparison to the C57BL/6. The sera from all the three strains of mice, those immunized with live and also those vaccinated with UV-EB, reacted with the 70 kDa (TC0721), the 60 kDa crp (TC0727), MOMP (TC0052) and LPS. Animals immunized i.n. with live MoPn, and in particular C3H/HeN and BALB/c mice, produced antibodies against several components with MW higher than 100 kDa. Weaker high MW bands were observed with the serum samples from the mice immunized i.vag. The 28 kDa (TC0396; Inc A) and the 24 kDa (TC0140; hypothetical protein) reacted strongly only with the sera from mice vaccinated with live EB. Sera collected from the groups of mice immunized with MEM-0 or ova, as well as the pre-immunization sera, showed no bands.
Fig. 2.
Western blots of serum samples from control and immunized BALB/c, C3H/HeN and C57BL/6 mice reacted with MoPn EB. Lane 1) MW standards; lane 2) control monoclonal antibody to MOMP. Serum samples from mice immunized with: lane 3) live EB, i.n. at 60 days post-immunization (d.p.i.); lane 4) live EB, i.n. at 120 d.p.i.; lane 5) live EB, i.n. at 180 d.p.i.; lane 6) live EB, i.vag. at 60 d.p.i.; lane 7) live EB, i.vag. at 120 d.p.i.; lane 8) live EB, i.vag. at 180 d.p.i.; lane 9) UV-EB+CpG+Montanide at 60 d.p.i.; lane 10) UV-EB+CpG+Montanide at 120 d.p.i.; lane 11) UV-EB+CpG+Montanide at 180 d.p.i.; lane 12) UV-EB+Alum at 60 d.p.i.; lane 13) UV-EB+Alumat 120 d.p.i.; lane 14) UV-EB+Alumat 180 d.p.i.; lane 15) Ova+CpG+Montanide+Alumat 60 d.p.i.; lane 16) Ova+CpG+Montanide+Alumat 120 d.p.i.; lane 17) Ova+CpG+Montanide+Alumat 180 d.p.i.; lane 18)MEM-0 at 60 d.p.i.; lane 19) MEM-0 at 120 d.p.i.; lane 20) MEM-0 at 180 d.p.i.; lane 21) pre-immunization serum.
3.4. Assignment of cell function
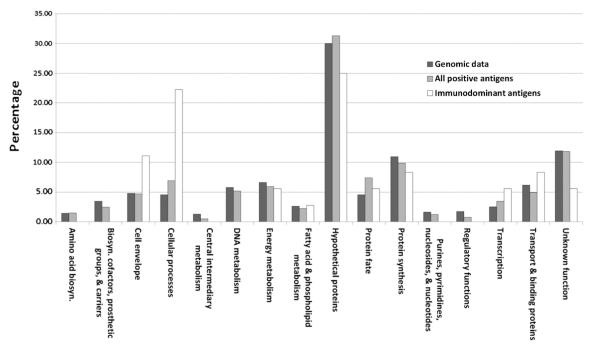
Using the Comprehensive Microbial Resource (CMR) from the J. Craig Venter Institute (JCVI; http://www.jcvi.org), we assigned the predicted cellular roles to the 406 antigens selected using the protein microarray. Each protein is assigned to one main cellular role category. The greatest number of the total positive antigens is categorized as hypothetical proteins (31.28%; 127/406). This is in similar proportion to the percentage represented in the Chlamydia MoPn genome (29.97%). Fig. 3 shows that most of the categorized cellular roles of all the identified positive antigens are in close proportion to the percentage represented in the Chlamydia MoPn genome. In contrast, for the immunodominant antigens we see a significant difference in the proportion of predicted cellular function roles when compared with either the whole genome or all the identified positive antigens (Table 1, Supplemental Table 1, Fig. 3). For example, the proportion of hypothetical proteins decreased in the dominant antigens when compared with either the whole genome or the positive antigens (25.00% vs. 29.97% and 31.28%). In contrast, proteins from the cell envelope (11.11% vs. 4.78%), cellular processes (22.22% vs. 4.56%), and transcription (5.56% vs. 2.50%) categories have a significantly higher representation in the immunodominant antigen selection when compared with the other two selections. Interestingly, the dominant antigen selection did not yield any protein in the “amino acid biosynthesis”, “biosynthesis of cofactors, prosthetic groups, and carriers”, “central intermediary metabolism”, “DNA metabolism”, “purines, pyrimidines, nucleosides, and nucleotides”, and the “regulatory function” categories.
Fig. 3.
Functional roles of the antigens of the whole Chlamydia MoPn genome, all the positive antigens identified following immunization with live EB and the immunodominant antigens for the three strains of mice tested.
In addition to the assigned protein functional categories, we also analyzed enrichment based on computationally predicted features (Table 2). Signal peptide and cellular localization were predicted from ORF sequence with pSORTb software (www.psort.org). Proteins predicted to contain signal peptide, and therefore membrane proteins, were significantly enriched for all three strains of mice immunized with live EB (1.28-fold enrichment for BALB/c, 1.18-fold enrichment for C3H/HeN, and 1.51-fold enrichment for C57BL/6). Further enrichment was observed with proteins that are dominant in all strains of mice (2.33-fold). The four proteins identified in this group were: TC0052, TC0117, TC0512 and TC0590.
Table 2.
Computational predicted localization enrichment of the Chlamydia MoPn proteins.
| Computational value | Total no. of ORF |
BALB/c |
C3H/HeN |
C57BL/6 |
Common |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hits | Fold change | P value | Hits | Fold change | P value | Hits | Fold change | P value | Hits | Fold change | P value | ||
| Signal peptide | |||||||||||||
| No | 878 | 269 | 0.99 | 0.8844 | 225 | 0.99 | 0.9585 | 133 | 0.98 | 0.8466 | 32 | 0.94 | 0.8085 |
| Yes | 43 | 13 | 1.28 | 0.4371 | 11 | 1.18 | 0.6124 | 6 | 1.51 | 0.3016 | 4 | 2.33 | 0.0371 |
| Cellular localization | |||||||||||||
| Cytoplasmic | 435 | 126 | 0.94 | 0.5865 | 127 | 1.14 | 0.3144 | 56 | 0.83 | 0.2524 | 11 | 0.63 | 0.2091 |
| Cytoplasmic membrane | 201 | 62 | 1.00 | 1.0000 | 39 | 0.76 | 0.1540 | 24 | 0.77 | 0.2749 | 7 | 0.87 | 0.6866 |
| Extracellular | 9 | 5 | 1.80 | 0.3403 | 6 | 2.60 | 0.0992 | 4 | 2.88 | 0.0871 | 1 | 2.78 | 0.3296 |
| Outer membrane | 15 | 5 | 1.08 | 0.5944 | 10 | 2.60 | 0.0243 | 9 | 3.89 | 0.0031 | 5 | 8.35 | 0.0011 |
| Periplasmic | 10 | 5 | 1.62 | 0.3669 | 4 | 1.56 | 0.5029 | 3 | 1.94 | 0.4011 | 1 | 2.51 | 0.3557 |
| Unknown | 251 | 79 | 1.05 | 0.7731 | 50 | 0.79 | 0.1714 | 43 | 1.19 | 0.3464 | 11 | 1.17 | 0.6005 |
| Total | 921 | 282 | 236 | 139 | 36 | ||||||||
For cellular localization, the three different strains of mice that were immunized with live EB showed different patterns of enrichment. In BALB/c mice, extracellular and periplasmic localizations showed the biggest enrichment (1.8-fold and 1.62-fold enrichment, respectively), while cytoplasmic membrane, outer membrane, and unknown localizations showed no significant enrichment. In the case of C3H/HeN mice, four cellular localizations showed enrichment (cytoplasmic, extracellular, outer membrane, and periplasmic) with extracellular and outer membrane showing the biggest enrichment (2.6-fold enrichment). For C57BL/6 mice, three cell localizations also showed enrichment (extracellular, outer membrane, and periplasmic) with outer membrane localization showing the most significant enrichment (3.89-fold). The biggest difference in enrichment was observed in the predicted localization of the immunodominant antigens of the three strains of mice. For outer membrane localization, pSORTb predicted 15 proteins from the complete Chlamydia MoPn genome and 5 of them were among the positive antigens, representing an 8.35-fold enrichment. These five proteins were: TC0045, TC0052, TC0512, TC0727 and TC0828. Finally, in the case of extracellular and periplasmic localizations, the program predicted 9 and 10 proteins respectively, and our positive antigen selection yielded one protein from each category, representing 2.78-fold and 2.51-fold enrichment.
4. Discussion
In this study, we have used a proteomic approach to search for novel antigens that can be used to formulate a subunit vaccine against chlamydial infections. Sera from three strains of mice immunized with live and nonviable EB were screened in a protein microarray expressing more than 99% of the ORFome of the Chlamydia MoPn genomic and plasmid DNA. Using this approach, we have identified 36 chlamydial immunodominant antigens that are reactive in all three strains of mice.
In the past, most of the vaccines had been generated using whole inactivated or attenuated organisms [40,41]. Recently, due to safety concerns, vaccines have been developed using limited number of recombinant proteins [42,43]. However, in order to formulate a subunit vaccine, multiple antigens that can generate an immune response need to be identified and tested for their ability to protect animal models. The large genome size of the infectious organism had been a big hurdle that researchers had to overcome in order to identify reactive antigens. The recent development of high-throughput methods for generating whole proteome microarrays has facilitated the discovery of reactive antigens from the entire proteome of pathogenic organisms.
Experience with several types of vaccine against infectious pathogens has shown that, in addition to the specific immunization protocol, induction of a protective immune response is dependent on the genetic background of the individual [40,41]. As a result of the widespread presence of chlamydial infections throughout the world this consideration is particularly important in the case of a vaccine for C. trachomatis. To identify antigens that will be recognized by individuals with various genetic backgrounds here, we tested three genetically diverse strains of mice that have been shown to have different susceptibilities to a chlamydial infection [31,44,45]. For example, when C3H/HeN, BALB/c and C57BL/6 mice were infected intravaginally with Chlamydia MoPn the C3H/HeN were found to be the most susceptible while the C57BL/6 were the most resistant [31]. In addition, the course and outcome of the infection differed markedly between these strains of mice [31,44–47]. Furthermore, significant differences in the innate and acquired immune responses have been reported between these three strains of mice following genital and pulmonary infections [45,46,48,49].
To identify dominant chlamydial antigens that can elicit an antibody response, we immunized C3H/HeN, BALB/c and C57BL/6 mice with live and inactivated EB and screened the sera with the Chlamydia whole proteome microarray [23]. We used several criteria to select the immunodominant antigens. Specifically we identified proteins that showed a prolonged immune response (i.e., 3 consecutive or 5 non-consecutive positive time points) and are positive in all three strains of mice tested. Previous studies have shown that immunization with live EB using the i.n. route provides the best protection in mice against a genital challenge [29,50]. Therefore, to prioritize the antigen selection, we used the results obtained with animals immunized with live Chlamydia EB. With these selection criteria, we identified 36 immunodominant antigens.
Several of our selected antigens have previously been identified as potential vaccine candidates. Some of them have been shown to elicit an immune response and even, in some cases, protection in animal models against a chlamydial challenge. These antigens include TC0052 (MOMP), TC0268 (hypothetical protein), TC0512 (outer membrane protein, putative), TC0727 (60-kDa outer membrane protein), and TC0816 (hypothetical protein) [18,25,26,28,51–58]. Of these, TC0052 (MOMP) and TC0727 (60-kDa outer membrane protein) were also recognized by sera from mice immunized with UV-EB. Members of the putative type III secretion system of Chlamydia have also been studied for their potential as vaccine candidates since several of these proteins are shared among Gram-negative pathogenic bacteria [10]. In this study, our immunodominant antigens include two type III secretion proteins, TC0045 (SctC) and TC0848 (SctJ) [51,59].
As expected, the reactive antigens are not found in all the functional categories throughout the proteome and no single category is completely dominant in reactivity. For example, proteins with predicted function of central intermediary metabolism, DNA metabolism, nucleotides, and regulatory functions are not selected. However, proteins with predicted function of cell envelope, cellular processes, and transcription are enriched in our selected reactive antigens. As expected, our signal peptide and cellular localization enrichment analysis also showed that antigens with surface membrane localization, i.e. extracellular, outer membrane, and periplasmic, and antigens with signal peptide are enriched in our reactive antigen selection. Four proteins with a signal peptide: TC0052, TC0117, TC0512 and TC0590 and five outer membrane proteins: TC0045, TC0052, TC0512, TC0727 and TC0828 were identified as immunodominant in these two categories. The two proteins dominant to both groups, MOMP (TC0052) and the Omp85 analog (TC0512), are well-known outer membrane chlamydial antigens [37,60,61].
The largest group of reactive antigens identified in this study is classified as hypothetical proteins. Among the 36 dominant antigens, 11 are hypothetical proteins and out of these 9 are classified as conserved hypothetical proteins. Two of these 11 hypothetical proteins have already been shown to be able to induce an immune response, TC0268 and TC0816 [62,63]. Additionally, the C. trachomatis homolog of TC0177 and TC0392 (CT0795 and CT0116, respectively) were also shown to be able to induce antibodies in patient sera [62,63]. The discovery of additional hypothetical proteins unique to Chlamydia supports the possibility that there will be more vaccine candidates among them.
In addition, from the list of the antigens that we have identified as reactive, homologs of several of them were also found in patients infected with Chlamydia. These antigens include TC0396 (inclusion membrane localized protein, IncA), TC0660 (amino acid ABC transporter, periplasmic amino acid-binding protein), TC0726 (sulfur rich protein), TC0229 (cell division protein FtsH), TC0590 (ribosomal protein L7/L12), and TC0386 (60 kDa chaperonin) [62–64]. These antigens have been shown to be able to elicit a CD4+ T-cell and/or antibody response, indicating that these proteins may be good candidates for vaccine development.
There are some limitations with the approach we have used for this study. For example, due to the fact that this is a high throughput method of generating clones and proteins, we cannot quantitate the amount of each protein being spotted on the array [23]. By testing for the expression of the poly(His) and the HA tags of each ORF using mAbs, we can confirm that the ORF is expressed partially or in its entirety. However, the binding of antibodies to the epitope of poly(His) and HA tags also depends on several other factors such as the folding and the conformation of the expressed ORF [23]. As a result, we cannot make a direct quantitative comparison of the antibody response between different antigens. However, we can follow the antibody response of a particular antigen since the amount of protein per spot for each protein is constant from array to array.
Another limitation of a bacterial-based protein expression system like ours is that the modification of the expressed proteins, such as glycosylation, phosphorylation, or lipidation, cannot be determined. If these modifications affect the immunogenicity of a specific epitope, we may or may not be able to detect a particular antigen. However, using the same type of microarray as the one utilized in this study, it was shown that sera from vaccinated humans and animals recognized all known glycosylated proteins of vaccinia virus [38]. Therefore, we can conclude that, at least some antibodies to these proteins are directed to domains that are not affected by post-transcriptional modifications. Similarly, conformational epitopes and epitopes dependent on disulfide bridges may or may not form correctly in the protein array used in this study. Here, we have shown that MOMP, and the 60 kDa cysteine-rich proteins, were recognized by sera of animals immunized with live EB in all three strains of mice. This finding supports the possibility that most antigens elicit antibodies against a wide array of epitopes. On the other hand, certain antigens that have been found to elicit a strong antibody response and protection, such as TCA04 (Pgp3), failed to meet our criteria for selection [65]. In the case of Pgp3, a possibility is that the protein expressed in the microarray lacked the correct conformation. The antibody response to Pgp3 is preferentially directed to conformational epitopes, and this may explain our failure to identify it as an immunodominant antigen [66]. However, this is unlikely since only sera from BALB/c mice immunized i.n. failed to meet our criteria for positivity. Interestingly, TC0248 (chlamydial protease-like factor, CPAF), a known chlamydial immunogen, was recognized by sera from the BALB/c and C57BL/6 mice but not by the samples from C3H/HeN mice most likely reflecting the ability of some epitopes to be only recognized by mouse strains with specific genetic backgrounds [62,67].
In conclusion, we screened the whole Chlamydia ORFome using serum samples from three different strains of mice and were able to identify 36 antigens that elicited an antibody response in all three strains of mice following immunization by two different routes with live EB. All these proteins have orthologs in the C. trachomatis human serovars with a high degree of sequence identity. Several of these proteins have not been previously identified as immunogenic and can be further studied for their ability to protect mice against a Chlamydia infection. Antigens found to be protective in mice and other animal models, could be considered for clinical trials in humans.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grants R41 AI072847 and RO1 AI067888 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jprot.2012.08.017.
REFERENCES
- [1].Chlamydia screening among sexually active young female enrollees of health plans—United States, 2000–2007. MMWR Morb Mortal Wkly Rep. 2009;58:362–5. [PubMed] [Google Scholar]
- [2].Schachter J, Dawson C. Human chlamydial infections: littleton. PSG Publishing Co; 1978. [Google Scholar]
- [3].Taylor HR. Trachoma: a blinding scourge from the Bronze Age to the twenty-first century. 1st ed Victoria; East Melbourne, Vic: 2008. [Google Scholar]
- [4].Miller WC, Ford CA, Morris M, Handcock MS, Schmitz JL, Hobbs MM, et al. Prevalence of chlamydial and gonococcal infections among young adults in the United States. JAMA. 2004;291:2229–36. doi: 10.1001/jama.291.18.2229. [DOI] [PubMed] [Google Scholar]
- [5].Stamm W. Chlamydia trachomatis infections of the adult. In: KK Holmes PS, Stamm WE, Piot P, Wasserheit JW, Corey L, Cohen MS, Watts DH, editors. Sexually transmitted diseases. McGrawHill Book Co; New York: 2008. pp. 575–93. [Google Scholar]
- [6].Ness RB, Soper DE, Holley RL, Peipert J, Randall H, Sweet RL, et al. Effectiveness of inpatient and outpatient treatment strategies for women with pelvic inflammatory disease: results from the Pelvic Inflammatory Disease Evaluation and Clinical Health (PEACH) Randomized Trial. Am J Obstet Gynecol. 2002;186:929–37. doi: 10.1067/mob.2002.121625. [DOI] [PubMed] [Google Scholar]
- [7].Ness RB, Smith KJ, Chang CC, Schisterman EF, Bass DC. Prediction of pelvic inflammatory disease among young, single, sexually active women. Sex Transm Dis. 2006;33:137–42. doi: 10.1097/01.olq.0000187205.67390.d1. [DOI] [PubMed] [Google Scholar]
- [8].Westrom L, Joesoef R, Reynolds G, Hagdu A, Thompson SE. Pelvic inflammatory disease and fertility. A cohort study of 1844 women with laparoscopically verified disease and 657 control women with normal laparoscopic results. Sex Transm Dis. 1992;19:185–92. [PubMed] [Google Scholar]
- [9].de la Maza LM, Peterson EM. Vaccines for Chlamydia trachomatis infections. Curr Opin Investig Drugs. 2002;3:980–6. [PubMed] [Google Scholar]
- [10].Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–61. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- [11].Rockey DD, Wang J, Lei L, Zhong G. Chlamydia vaccine candidates and tools for chlamydial antigen discovery. Expert Rev Vaccines. 2009;8:1365–77. doi: 10.1586/erv.09.98. [DOI] [PubMed] [Google Scholar]
- [12].de la Maza MA, de la Maza LM. A new computer model for estimating the impact of vaccination protocols and its application to the study of Chlamydia trachomatis genital infections. Vaccine. 1995;13:119–27. doi: 10.1016/0264-410x(95)80022-6. [DOI] [PubMed] [Google Scholar]
- [13].Taylor HR, Johnson SL, Prendergast RA, Schachter J, Dawson CR, Silverstein AM. An animal model of trachoma II. The importance of repeated reinfection. Invest Ophthalmol Vis Sci. 1982;23:507–15. [PubMed] [Google Scholar]
- [14].Wang SP, Grayston JT, Alexander ER. Trachoma vaccine studies in monkeys. Am J Ophthalmol. 1967;63(Suppl.):1615–30. doi: 10.1016/0002-9394(67)94155-4. [DOI] [PubMed] [Google Scholar]
- [15].Grayston JT, Wang SP. The potential for vaccine against infection of the genital tract with Chlamydia trachomatis. Sex Transm Dis. 1978;5:73–7. doi: 10.1097/00007435-197804000-00011. [DOI] [PubMed] [Google Scholar]
- [16].Kari L, Whitmire WM, Olivares-Zavaleta N, Goheen MM, Taylor LD, Carlson JH, et al. A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J Exp Med. 2011;208:2217–23. doi: 10.1084/jem.20111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Grayston JT, Wang S. New knowledge of chlamydiae and the diseases they cause. J Infect Dis. 1975;132:87–105. doi: 10.1093/infdis/132.1.87. [DOI] [PubMed] [Google Scholar]
- [18].Pal S, Peterson EM, de la Maza LM. Vaccination with the Chlamydia trachomatis major outer membrane protein can elicit an immune response as protective as that resulting from inoculation with live bacteria. Infect Immun. 2005;73:8153–60. doi: 10.1128/IAI.73.12.8153-8160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Farris CM, Morrison SG, Morrison RP. CD4+ T cells and antibody are required for optimal major outer membrane protein vaccine-induced immunity to Chlamydia muridarum genital infection. Infect Immun. 2010;78:4374–83. doi: 10.1128/IAI.00622-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hansen J, Jensen KT, Follmann F, Agger EM, Theisen M, Andersen P. Liposome delivery of Chlamydia muridarum major outer membrane protein primes a Th1 response that protects against genital chlamydial infection in a mouse model. J Infect Dis. 2008;198:758–67. doi: 10.1086/590670. [DOI] [PubMed] [Google Scholar]
- [21].Sun G, Pal S, Weiland J, Peterson EM, de la Maza LM. Protection against an intranasal challenge by vaccines formulated with native and recombinant preparations of the Chlamydia trachomatis major outer membrane protein. Vaccine. 2009;27:5020–5. doi: 10.1016/j.vaccine.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tifrea DF, Sun G, Pal S, Zardeneta G, Cocco MJ, Popot JL, et al. Amphipols stabilize the Chlamydia major outer membrane protein and enhance its protective ability as a vaccine. Vaccine. 2011;29:4623–31. doi: 10.1016/j.vaccine.2011.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A. 2005;102:547–52. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Davies DH, Wyatt LS, Newman FK, Earl PL, Chun S, Hernandez JE, et al. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J Virol. 2008;82:652–63. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Molina DM, Pal S, Kayala MA, Teng A, Kim PJ, Baldi P, et al. Identification of immunodominant antigens of Chlamydia trachomatis using proteome microarrays. Vaccine. 2010;28:3014–24. doi: 10.1016/j.vaccine.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cruz-Fisher MI, Cheng C, Sun G, Pal S, Teng A, Molina DM, et al. Identification of immunodominant antigens by probing a whole Chlamydia trachomatis open reading frame proteome microarray using sera from immunized mice. Infect Immun. 2011;79:246–57. doi: 10.1128/IAI.00626-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nigg C. An unidentified virus which produces pneumonia and systemic infection in mice. Science. 1942;95:49–50. doi: 10.1126/science.95.2454.49-a. [DOI] [PubMed] [Google Scholar]
- [28].Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–76. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pal S, Fielder TJ, Peterson EM, de la Maza LM. Protection against infertility in a BALB/c mouse salpingitis model by intranasal immunization with the mouse pneumonitis biovar of Chlamydia trachomatis. Infect Immun. 1994;62:3354–62. doi: 10.1128/iai.62.8.3354-3362.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pal S, Hui W, Peterson EM, de la Maza LM. Factors influencing the induction of infertility in a mouse model of Chlamydia trachomatis ascending genital tract infection. J Med Microbiol. 1998;47:599–605. doi: 10.1099/00222615-47-7-599. [DOI] [PubMed] [Google Scholar]
- [31].de la Maza LM, Pal S, Khamesipour A, Peterson EM. Intravaginal inoculation of mice with the Chlamydia trachomatis mouse pneumonitis biovar results in infertility. Infect Immun. 1994;62:2094–7. doi: 10.1128/iai.62.5.2094-2097.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Morrison SG, Morrison RP. Resolution of secondary Chlamydia trachomatis genital tract infection in immune mice with depletion of both CD4+ and CD8+ T cells. Infect Immun. 2001;69:2643–9. doi: 10.1128/IAI.69.4.2643-2649.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tuffrey M, Taylor-Robinson D. Progesterone as a key factor in the development of a mouse model for genital-tract infection with Chlamydia trachomatis. FEMS Microbiol Lett. 1981;12:111–5. [Google Scholar]
- [34].Pal S, Rangel J, Peterson EM, de la Maza LM. Immunogenic and protective ability of the two developmental forms of Chlamydiae in a mouse model of infertility. Vaccine. 1999;18:752–61. doi: 10.1016/s0264-410x(99)00032-8. [DOI] [PubMed] [Google Scholar]
- [35].Hui GS, Hashimoto CN. Adjuvant formulations possess differing efficacy in the potentiation of antibody and cell mediated responses to a human malaria vaccine under selective immune genes knockout environment. Int Immunopharmacol. 2008;8:1012–22. doi: 10.1016/j.intimp.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–9. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- [38].Davies DH, Molina DM, Wrammert J, Miller J, Hirst S, Mu Y, et al. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007;7:1678–86. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- [39].Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, et al. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–23. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- [40].Levine MM. New generation vaccines. 4th ed Informa Healthcare USA; New York: 2010. [Google Scholar]
- [41].Plotkin SA, Orenstein WA, Offit PA. Vaccines. 5th ed Saunders/Elsevier; Philadelphia, Pa: 2008. [Google Scholar]
- [42].Suzich JA, Ghim SJ, Palmer-Hill FJ, White WI, Tamura JK, Bell JA, et al. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. Proc Natl Acad Sci U S A. 1995;92:11553–7. doi: 10.1073/pnas.92.25.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ionescu-Matiu I, Kennedy RC, Sparrow JT, Culwell AR, Sanchez Y, Melnick JL, et al. Epitopes associated with a synthetic hepatitis B surface antigen peptide. J Immunol. 1983;130:1947–52. [PubMed] [Google Scholar]
- [44].Tuffrey M, Falder P, Gale J, Taylor-Robinson D. Salpingitis in mice induced by human strains of Chlamydia trachomatis. Br J Exp Pathol. 1986;67:605–16. [PMC free article] [PubMed] [Google Scholar]
- [45].Darville T, Andrews CW, Jr, Laffoon KK, Shymasani W, Kishen LR, Rank RG. Mouse strain-dependent variation in the course and outcome of chlamydial genital tract infection is associated with differences in host response. Infect Immun. 1997;65:3065–73. doi: 10.1128/iai.65.8.3065-3073.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yang X, HayGlass KT, Brunham RC. Genetically determined differences in IL-10 and IFN-gamma responses correlate with clearance of Chlamydia trachomatis mouse pneumonitis infection. J Immunol. 1996;156:4338–44. [PubMed] [Google Scholar]
- [47].Pal S, Peterson EM, de la Maza LM. Susceptibility of mice to vaginal infection with Chlamydia trachomatis mouse pneumonitis is dependent on the age of the animal. Infect Immun. 2001;69:5203–6. doi: 10.1128/IAI.69.8.5203-5206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Darville T, Andrews CW, Jr, Sikes JD, Fraley PL, Rank RG. Early local cytokine profiles in strains of mice with different outcomes from chlamydial genital tract infection. Infect Immun. 2001;69:3556–61. doi: 10.1128/IAI.69.6.3556-3561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Stagg AJ, Tuffrey M, Woods C, Wunderink E, Knight SC. Protection against ascending infection of the genital tract by Chlamydia trachomatis is associated with recruitment of major histocompatibility complex class II antigen-presenting cells into uterine tissue. Infect Immun. 1998;66:3535–44. doi: 10.1128/iai.66.8.3535-3544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kelly KA, Robinson EA, Rank RG. Initial route of antigen administration alters the T-cell cytokine profile produced in response to the mouse pneumonitis biovar of Chlamydia trachomatis following genital infection. Infect Immun. 1996;64:4976–83. doi: 10.1128/iai.64.12.4976-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Coler RN, Bhatia A, Maisonneuve JF, Probst P, Barth B, Ovendale P, et al. Identification and characterization of novel recombinant vaccine antigens for immunization against genital Chlamydia trachomatis. FEMS Immunol Med Microbiol. 2009;55:258–70. doi: 10.1111/j.1574-695X.2008.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Karunakaran KP, Yu H, Foster LJ, Brunham RC. Development of a Chlamydia trachomatis T cell Vaccine. Hum Vaccin. 2011;6:676–80. doi: 10.4161/hv.6.8.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].McNeilly CL, Beagley KW, Moore RJ, Haring V, Timms P, Hafner LM. Expression library immunization confers partial protection against Chlamydia muridarum genital infection. Vaccine. 2007;25:2643–55. doi: 10.1016/j.vaccine.2006.12.019. [DOI] [PubMed] [Google Scholar]
- [54].Murthy AK, Chambers JP, Meier PA, Zhong G, Arulanandam BP. Intranasal vaccination with a secreted chlamydial protein enhances resolution of genital Chlamydia muridarum infection, protects against oviduct pathology, and is highly dependent upon endogenous gamma interferon production. Infect Immun. 2007;75:666–76. doi: 10.1128/IAI.01280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Starnbach MN, Loomis WP, Ovendale P, Regan D, Hess B, Alderson MR, et al. An inclusion membrane protein from Chlamydia trachomatis enters the MHC class I pathway and stimulates a CD8+ T cell response. J Immunol. 2003;171:4742–9. doi: 10.4049/jimmunol.171.9.4742. [DOI] [PubMed] [Google Scholar]
- [56].Yu H, Jiang X, Shen C, Karunakaran KP, Brunham RC. Novel Chlamydia muridarum T cell antigens induce protective immunity against lung and genital tract infection in murine models. J Immunol. 2009;182:1602–8. doi: 10.4049/jimmunol.182.3.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wang J, Zhang Y, Yu P, Zhong G. Immunodominant regions of a Chlamydia trachomatis type III secretion effector protein. Tarp. Clin Vaccine Immunol. 2010;17:1371–6. doi: 10.1128/CVI.00218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Fling SP, Sutherland RA, Steele LN, Hess B, D’Orazio SE, Maisonneuve J, et al. CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc Natl Acad Sci U S A. 2001;98:1160–5. doi: 10.1073/pnas.98.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yu H, Jiang X, Shen C, Karunakaran KP, Jiang J, Rosin NL, et al. Chlamydia muridarum T-cell antigens formulated with the adjuvant DDA/TDB induce immunity against infection that correlates with a high frequency of gamma interferon (IFN-gamma)/tumor necrosis factor alpha and IFN-gamma/interleukin-17 double-positive CD4+ T cells. Infect Immun. 2010;78:2272–82. doi: 10.1128/IAI.01374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Caldwell HD, Schachter J. Antigenic analysis of the major outer membrane protein of Chlamydia spp. Infect Immun. 1982;35:1024–31. doi: 10.1128/iai.35.3.1024-1031.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Carey AJ, Timms P, Rawlinson G, Brumm J, Nilsson K, Harris JM, et al. A multi-subunit chlamydial vaccine induces antibody and cell-mediated immunity in immunized koalas (Phascolarctos cinereus): comparison of three different adjuvants. Am J Reprod Immunol. 2010;63:161–72. doi: 10.1111/j.1600-0897.2009.00776.x. [DOI] [PubMed] [Google Scholar]
- [62].Wang J, Zhang Y, Lu C, Lei L, Yu P, Zhong G. A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J Immunol. 2010;185:1670–80. doi: 10.4049/jimmunol.1001240. [DOI] [PubMed] [Google Scholar]
- [63].Rodgers AK, Budrys NM, Gong S, Wang J, Holden A, Schenken RS, et al. Genome-wide identification of Chlamydia trachomatis antigens associated with tubal factor infertility. Fertil Steril. 2011;96:715–21. doi: 10.1016/j.fertnstert.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Finco O, Frigimelica E, Buricchi F, Petracca R, Galli G, Faenzi E, et al. Approach to discover T- and B-cell antigens of intracellular pathogens applied to the design of Chlamydia trachomatis vaccines. Proc Natl Acad Sci U S A. 2011;108:9969–74. doi: 10.1073/pnas.1101756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Donati M, Sambri V, Comanducci M, Di Leo K, Storni E, Giacani L, et al. DNA immunization with pgp3 gene of Chlamydia trachomatis inhibits the spread of chlamydial infection from the lower to the upper genital tract in C3H/HeN mice. Vaccine. 2003;21:1089–93. doi: 10.1016/s0264-410x(02)00631-x. [DOI] [PubMed] [Google Scholar]
- [66].Li Z, Zhong Y, Lei L, Wu Y, Wang S, Zhong G. Antibodies from women urogenitally infected with C. trachomatis predominantly recognized the plasmid protein pgp3 in a conformation-dependent manner. BMC Microbiol. 2008;8:90. doi: 10.1186/1471-2180-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sharma J, Zhong Y, Dong F, Piper JM, Wang G, Zhong G. Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins. Infect Immun. 2006;74:1490–9. doi: 10.1128/IAI.74.3.1490-1499.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.