Abstract
An RNA polymerase has been thought to transcribe by seeking out a promoter, initiating and then tracking down the template. We add tumor necrosis factor α to primary human cells, switch on transcription of a 221-kb gene and monitor promoter position during the ensuing transcription cycle (using RNA fluorescence in situ hybridization coupled to super-resolution localization, chromosome conformation capture and Monte Carlo simulations). Results are consistent with a polymerase immobilized in a ‘factory’ capturing a promoter and reeling in the template, as the transcript and promoter are extruded. Initially, the extruded promoter is tethered close to the factory and so likely to re-initiate; later, the tether becomes long enough to allow re-initiation in another factory. We suggest close tethering underlies enhancer function and transcriptional ‘bursting’.
INTRODUCTION
The traditional view of transcription involves an RNA polymerase that diffuses to a promoter wherever that promoter might be in the nucleus; then, after initiation, it transcribes by tracking like a locomotive down the template (1). However, an alternative sees polymerization occurring on the surface of a macro-molecular complex known as a ‘transcription factory’ (2–4). We define such a factory as a site containing at least two polymerases active on different templates. A factory contains ancillary machinery for capping, splicing and poly-adenylation, and the high local concentrations would promote efficient mRNA production (4). Support for this alternative has come, for example, from high-resolution imaging, which shows that nascent transcripts are found on the surface of ∼90-nm protein-rich structures (5,6); chromosome conformation capture [3C (7)], which indicates that genes often cluster together in nuclei when transcriptionally active (8–11) and biochemical purification of large complexes of >8 MDa, which contain the appropriate nascent transcripts and proteins (12). If factories are the active sites of transcription, the promoter would then have to diffuse to a factory, and—on initiation—transcription would occur as a transiently immobilized polymerase reels in the template as it extrudes the transcript (13). We wished to distinguish between the two alternatives by following the promoter of a gene during one transcription cycle.
For this analysis, we use human umbilical vein endothelial cells (HUVECs) arrested in the G0 phase of the cell cycle by serum starvation and then treated with tumor necrosis factor α (TNFα); this cytokine orchestrates the inflammatory response, and it signals through the transcription factor NFκB to activate a subset of genes within minutes (14). One responsive gene—SAMD4A—was chosen for analysis, as it has been studied previously in detail (13,15,16) and as its great length of 221 kb yields sufficient temporal and spatial resolution for the two main experimental techniques to be used—RNA fluorescence in situ hybridization (FISH) coupled to ‘super-resolution’ localization (17–19) and 3C coupled to quantitative polymerase chain reaction [qPCR (20)].
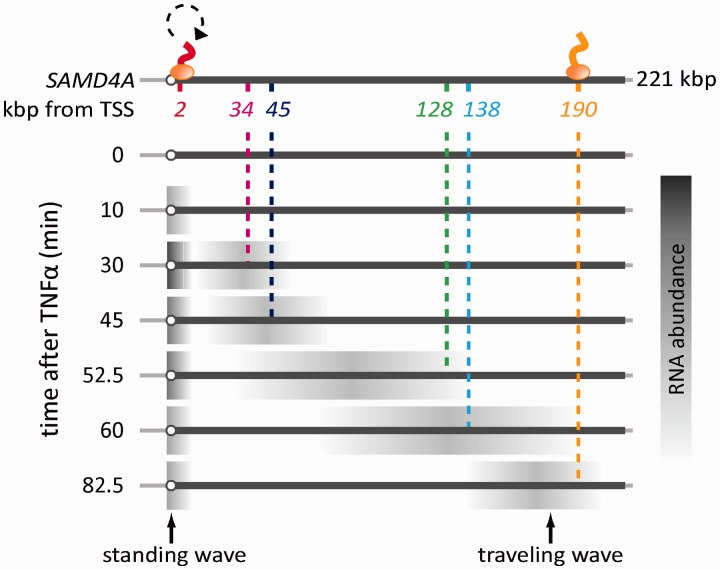
Data obtained using tiling microarrays (15) indicate that transcription of SAMD4A begins ∼10 min after adding TNFα. Then, a pioneering polymerase transcribes steadily at ∼3 kb/min to reach the terminus after ∼85 min; the resulting nascent transcripts made by a number of such polymerases are depicted in Figure 1 as a ‘traveling’ wave sweeping down the gene. Once pioneering polymerases leave the promoter, additional polymerases initiate; however, these soon abort within <10 kb. Successive cycles of initiation and abortion then generate additional transcripts (depicted as the ‘standing’ wave in Figure 1). As SAMD4A introns are so long relative to exons, both waves mainly contain intronic RNA. After ∼30 min, a ‘trough’ develops between the two waves; this can only result if intervening intronic RNA is removed and degraded co-transcriptionally and rapidly, and if the following polymerases soon abort (otherwise signal would fill the trough). The presence of polymerases and nascent (intronic) RNA at the points indicated has been confirmed using chromatin immunoprecipitation, chromatin immunoprecipitation coupled to next-generation sequencing, tiling microarrays, reverse transcriptase-PCR and RNA FISH (13,15,16). A similar ‘standing’ wave is seen after switching on transcription of four other long genes; however, we do not yet understand how the ‘following’ polymerase might sense the presence of a pioneer on the same gene (15).
Figure 1.
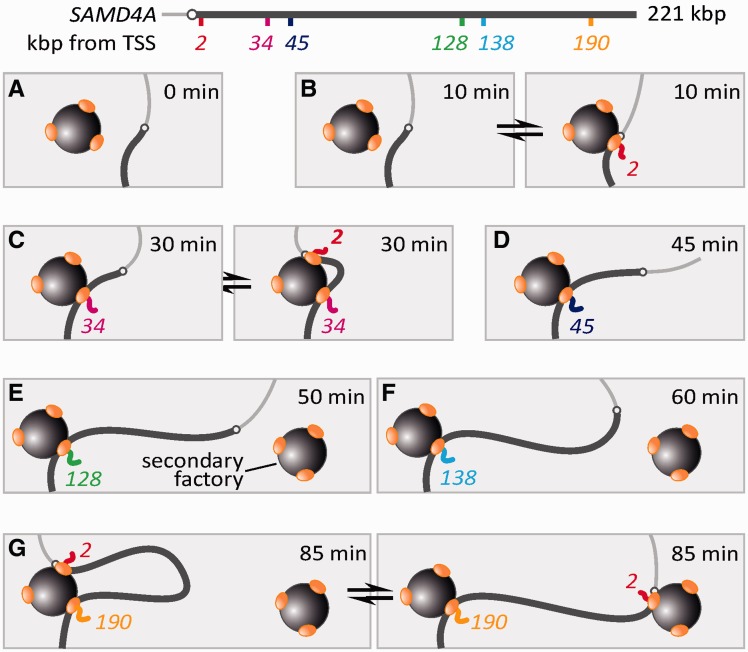
The first transcription cycle of SAMD4A after stimulation with TNFα. The cartoon illustrating results obtained previously using microarrays (15). HUVECs were treated with TNFα for different times, and total RNA applied to a tiling microarray spanning 221-kb SAMD4A; the map illustrates probes and their targets, which are named by distance in kb from the TSS (open circle). Before stimulation (0 min), no signal is detected. After 10 min, nascent RNA (gray, detected by Probe 2) appears at the promoter, indicative of rapid and synchronous initiation. The pioneering polymerase (oval) then transcribes steadily to reach Target 34 after 30 min, 45 after 45 min and eventually 190 after 82.5 min; this generates a ‘traveling’ wave that sweeps down the gene. After 10 min, an additional polymerase reinitiates but terminates prematurely (circular dotted arrow); a succession of such non-productive initiations create a ‘standing’ wave within 10 kb of the TSS. Thereafter, the gene is transcribed by two polymerases contributing respectively to the ‘traveling’ and ‘standing’ waves. The standing wave peaks at 30 min.
After switching on SAMD4A, we use (i) RNA FISH with intronic probes to localize a nascent transcript in the standing wave (which therefore marks the position of the promoter) relative to a transcript in the traveling wave (i.e. one copied from a segment of the gene that lies progressively further away as the time after stimulation increases); (ii) 3C to monitor the proximity between the two regions being transcribed and (iii) Monte Carlo simulations to model the various conformations of the locus at different stages during the transcription cycle. The results do not fit a model involving polymerases tracking along a randomly folded, uniformly packed, template. Rather, they are most simply explained if a promoter initiates in one factory, before that promoter is extruded from the factory as the pioneering polymerase transcribes. As a result, the promoter is now attached to the factory through a tether of increasing length. Initially, this tether is so short the promoter can only visit the same factory; if it then initiates again, nascent transcripts in the standing and traveling waves (and their templates) lie close together in 3D space on the surface of one factory. But later, when the tether becomes long enough to allow the promoter to visit either the same factory or a different one, the two nascent transcripts and their templates can either lie together or further apart. Although we studied two polymerases transcribing different parts of one long gene, our conclusions should be generally applicable to two polymerases transcribing any two adjacent transcription units on the chromosome. They also shed light on how enhancers might work and the process known as transcriptional ‘bursting’ (21–23). Neither of our two experimental techniques provides information on the other templates and transcripts that might be transcribed in the same factory by additional polymerases.
MATERIALS AND METHODS
Cell culture
HUVECs from pooled donors (Lonza) were grown to 80–90% confluence in Endothelial Basal Medium 2-MV with supplements (Lonza) and 5% foetal bovine serum (FBS), re-grown (‘starved’) for 16–18 h in Endothelial Basal Medium + 0.5% FBS, treated with TNFα (10 ng/ml; Peprotech) and harvested at different times post-stimulation; in some cases, 50 μΜ 5,6-dichloro-1-β-D-ribofuranosyl-benzimidazole (DRB; Sigma-Aldrich) was added 60 min before harvesting.
RNA FISH
RNA FISH was performed as described previously (13,15,24), using sets of five 50-mers (Gene Design, Japan) as probes targeting regions of <400 bp in SAMD4A, thus yielding diffraction-limited foci. In each 50-mer, roughly every 10th thymine residue was substituted by an amino-modifier C6-dT coupled to Alexa Fluor 488, 555 or 647 reactive dyes (Invitrogen). Probes were purified using G-50 columns (GE Healthcare), ethanol precipitated twice, concentrated using a Microcon-30 column (Millipore) and labeling efficiencies calculated using the Base:Dye ratio calculator (>8 fluors/100 nucleotides; Invitrogen; http://probes.invitrogen.com/resources/calc/basedyeratio.html). For each experiment, stimulated HUVECs grown on coverslips were fixed (17 min; room temperature) in 4% paraformaldehyde/0.05% acetic acid/0.15 M NaCl, washed 3× in phosphate buffered saline (PBS), permeabilized (5 min; 37°C) in 0.01% pepsin (pH 2.0), rinsed in water treated with diethyl-pyrocarbonate, post-fixed (5 min; 20°C) in 4% paraformaldehyde/PBS and stored (overnight; −20°C) in 70% ethanol. Coverslips were dehydrated in 70, 80, 90 and 100% ethanol and hybridized (overnight; 37°C in a moist chamber) with 25 ng labeled probes in 25% deionized formamide, 2× saline sodium citrate (SSC), 250 ng/ml sheared salmon sperm DNA, 5× Denhardt’s solution, 50 mM phosphate buffer (pH 7.0) and 1 mM ethylenediaminetetraacetic acid. Next, cells were washed once in 4× SSC (15 min; 37°C), three times in 2× SSC (10 min; 37°C) and mounted in Vectashield (Vector Laboratories) supplemented with 1 µg/ml 4,6-diamidino-2-phenylindole (Sigma).
Image analysis and separation measurements
Images were collected using an Axioplan 2 inverted microscope (Zeiss) with a CoolSNAP HQ camera (Photometrics) via MetaMorph 7.1 (Molecular Devices). Foci were initially selected manually and then checked by a computer algorithm to meet criteria for a diffraction-limited spot (i.e. Gaussian-like shape, signal-to-noise ratio and local contrast). Super-resolution localization was performed using the joint distribution algorithm (19), which is similar in effect to fitting a 2D Gaussian intensity profile to an image of a given focus using regression analysis (17,18). The translational and rotational misalignment between channels was measured using 0.1-μm TetraSpeck beads (Invitrogen). Misalignment was corrected using a 2D spatial transform (i.e. bi-linear interpolation following a local weighted mean of a minimum of 12 fiduciary points throughout the image). The relative distance between overlapping foci was determined after the position of each focus in a pair was identified to within 15 nm, but residual misalignment increased the error in distance measurements to 30 nm. All custom software routines were implemented in MATLAB and are available on request.
Monte Carlo simulations
For Model II in Figure 3B and Supplementary Figure S1B, we determine the distance expected between two polymerases—one contributing to the ‘standing’ wave and the second tracking down SAMD4A. The template is modeled as a self-avoiding polymer—a string of spherical subunits each of diameter σ (in this case, equal to 30 nm). Each subunit was packed with 3 kb of DNA, corresponding to 100 bp/nm—a packing commonly used in previous simulations (25–27). This corresponds to approximately six nucleosomes—each containing 185 bp DNA (28)—every 11-nm turn of the 30-nm fiber. The separation between two beads in the polymer was computed via Monte Carlo simulations of a linear self-avoiding walk off-lattice (29). In these simulations, the potential consists of two contributions. First, there is a steric repulsion between any two beads, which cannot approach closer together than the hard-core diameter of the bead (i.e. 30 nm). The second contribution to the potential is the bending-rigidity term (30),
 |
where kB is the Boltzmann constant, T is the temperature and ti is the ‘tangent’ vector, linking the i-th to the i+1-th bead in the chain of N beads. Here, the persistence length, ξ, varied between 40 and 150 nm, where the extremes represent typical values for eu- and hetero-chromatin, respectively (27,31–33). Note that setting ξ to σ leads to a slightly larger persistence length due to steric repulsion; we therefore refer to ξ = σ as a 40-nm persistence length. We used N = 158 to simulate a fragment of the chromosome containing SAMD4A. To plot the curves in Figure 3, we recorded the 2D projection of the 3D separation so that results can be compared with the data from microscopy (the average 2D projection can be shown, in our case, to be π/4 times the average 3D separation). We also sampled >10 000 statistically uncorrelated configurations. To generate statistically independent configurations, we used a dynamic Monte Carlo algorithm (29), consisting of local and non-local crankshaft moves, which rotate an internal portion of the loop while keeping the anchor points fixed.
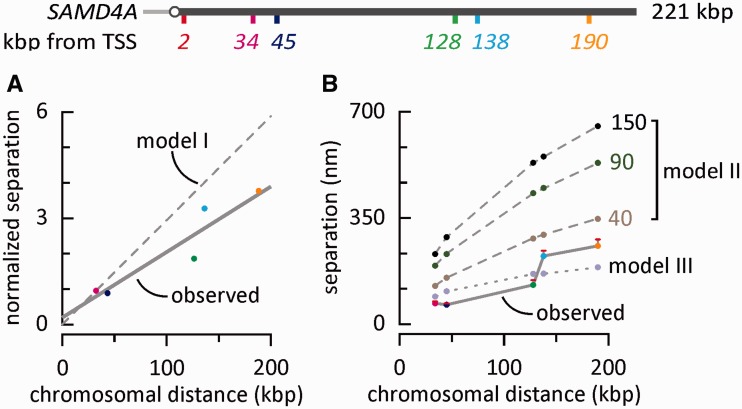
Figure 3.
Comparing expected and observed separations (with map of SAMD4A). Experimental data and color-coding are as in Figure 2. (A) Separations seen between probe pairs, normalized to the mean value obtained with 2 + 34. If polymerases track along a linear template, we would expect normalized separations to increase in proportion to the number of bases transcribed (dashed line—‘Model I’); however, observed separations have a different behavior (gray line; fitted by linear regression to the RNA FISH data from Figure 2D). (B) Separations between two points on a randomly folded (self-avoiding) polymer without (Model II) and with (Model III) an attraction between beads to provide a good fit to the experimental data (gray line, + SEM; from Figure 2G). In ‘Model II’, different persistence lengths found in eu- or hetero-chromatin (i.e. 40 and 150 nm), and an intermediate value, are included. In Model III, a persistence length of only 40 nm was used.
Model II leads to a spatial distance between any two points on the polymer that scales with the genetic distance according to a power law of 0.588 (34). Recent experiments (35,36) point to an exponent closer to 1/3 (although this evidence is mainly derived from larger genetic distances than analyzed here). Using the same general approach and conditions as for Model II, we also considered (as Model III in Figure 3B and Supplementary Figure S1C) a case including a non-specific self-attracting interaction between any two beads in the polymer (in the form of a square well, depth 0.7 kBT, attraction range 42 nm); this gives a scaling exponent of ∼1/3 (not shown). Here, we only consider a persistence length of 40 nm.
We also consider a further model (Model IV, Figures 6 and Supplementary Figure S1D) in which the chromosomal segment containing SAMD4A is a self-avoiding chromatin loop (again using the same general approach as for Model II). This loop is attached at each end to a primary factory (diameter 60 nm) at points located on opposite sides of the north pole at ±18° latitude; the loop is also attached to the factory at one other point (i.e. at 2, 34, 45, 128, 138 or 190). The primary factory is surrounded by six other similar factories, placed equally distant along the three coordinate axes (therefore, the primary factory is at the center of a cubic lattice). In most simulations, N = 158 (therefore, the contour length is 474 kb), the loop is tethered at each end through the promoters of CNIH and GCH1 (lying 141 kb upstream and 334 kb downstream of the transcription start site (TSS) of SAMD4A), and the center-to-center distance between primary and secondary factories is 450 nm. Here, the potential includes, in addition to the steric repulsion and bending-rigidity terms, a constraint—a strong harmonic spring (spring constant equal to 10 kBT/σ2; 34)—attaching the segment of DNA being transcribed (i.e. 2, 34, 45, 128, 138 or 190) to the primary factory. (Unlike Model III, Model IV does not assume any non-specific intrachromatin attraction.) To determine the probability that the TSS contacts either its own factory or a neighboring one, we generated >106 statistically independent configurations. We then recorded all ‘contacts’, defined as configurations where the surface of the bead at the TSS is within 30 nm of the surface of a factory, and classified these according to whether the contacting factory is the primary one or one of the six neighbors. For each set of parameters (persistence length, tether position and inter-factory distance), we record at least 1000 such contacts.
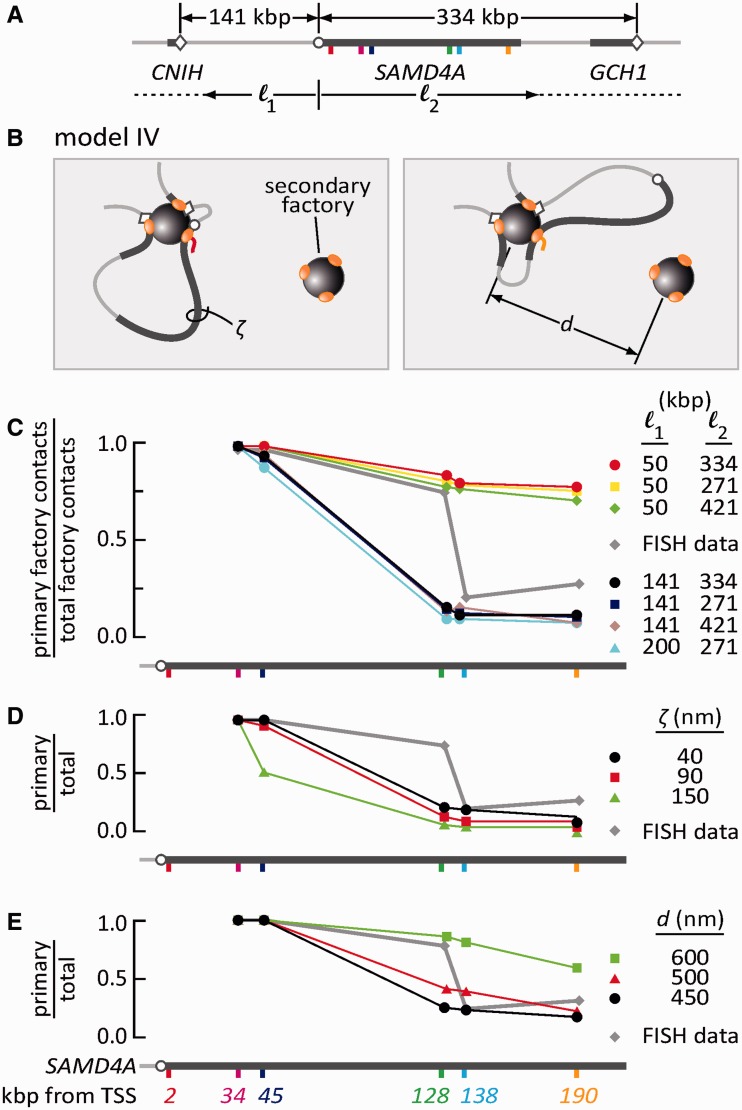
Figure 6.
Monte Carlo simulations analyzing how tethering affects association of the SAMD4A promoter with surrounding factories. (A) Map of the region around SAMD4A. 3C (not shown) shows the SAMD4A promoter contacts CNIH and GCH1 (promoters indicated as open diamonds). Distances l1 and l2 vary. (B) Model IV. The SAMD4A locus is modeled as a string of 30-nm beads (each representing 3 kb), tethered to a ‘primary’ factory (left) at three points (once within SAMD4A at one of the probe targets, and at l1 and l2 kb to the left and right, respectively); this factory is surrounded by six orthogonally positioned ‘secondary’ factories (only one shown; center-to-center distance, d). In the left-hand panel, the pioneering polymerase is transcribing region around Probe 2; the length of the tether connecting the promoter to the primary factory is so short that the promoter can never contact a secondary factory. In the right-hand panel, the pioneering polymerase is transcribing region around Probe 190; therefore, the tether is now long enough to allow the promoter to visit a secondary factory. A single simulation involves allowing such a structure to equilibrate, and—after >106 simulations involving >103 contacts for each condition analyzed—the fraction of contacts the promoter makes with the primary factory is expressed relative to the contacts made with all seven factories. Conditions analyzed included varying l1 and l2, persistence length (ξ) and inter-factory distance (d). In the panels below, values of l1, l2, ξ and d expected to be found in vivo provide the best fit to the RNA FISH data in Figure 2D (included for comparison, assuming that ‘occurrences’ seen between 0 and 160 nm and 0 and 500 nm reflect ‘primary factory contacts’ and ‘total factory contacts’, respectively). (C) Varying distance (l1 and l2 in kb) to flanking attachment points (when d = 450 nm and ξ = 40 nm). With the distances found in vivo (l1 = 141, l2 = 334; black), the SAMD4A promoter can only visit a secondary factory (indicated by a reduced fraction) when the body of the gene is attached at Position 128, 138 or 190. As intuition suggests, shortening l1 limits access to a secondary factory, whereas varying l2 has little effect. In (C) and (D), error bars are contained within the symbols. (D) Varying persistence length (when l1 = 141, l2 = 334 and d = 450 nm). Stiffening the fiber reduces the visits of the SAMD4A promoter to secondary factories. (E) Varying distance between factories (when l1 = 141, l2 = 334 and ξ = 40 nm). As distance between factories increases, the frequency that the fiber visits a secondary factory decreases.
3C coupled to qPCR
3C was performed as described (16). In brief, 107 cells were fixed (10 min; 20°C) in 1% paraformaldehyde (Electron Microscopy Sciences), aliquots of 106 cells in 0.125 M glycine in PBS spun, cells resuspended in the appropriate restriction enzyme buffer and lyzed (16 h; 37°C) in 0.3% SDS. After sequestering SDS by adding 1.8% Triton X-100 (1.5 h; 37°C), cells were treated overnight with HindIII (800 units; New England Biolabs), the enzyme heat-inactivated (25 min; 65°C) and digestion efficiency determined by qPCR; samples with digestion efficiencies >80% were ligated using T4 ligase (6000 units; New England Biolabs; DNA concentration <0.1 ng/μl; 3–5 days at 4°C to minimize unwanted ligations;37) and cross-links reversed (16 h; 65°C) in proteinase K (10 mg/ml; New England Biolabs) before DNA was purified using an EZNA MicroElute DNA clean-up kit (Omega BioTek). Non-digested/ligated, digested/non-ligated and amplification efficiency control templates (derived from bacterial artificial chromosomes, encoding the relevant genomic regions) were also prepared. qPCR was performed using a Rotor-Gene 3000 cycler (Corbett) and Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). Following incubation at 50°C for 2 min to activate the mix and 95°C for 5 min to denature templates, reactions were for 40 cycles at 95°C for 15 s and at 60°C for 50 s. The presence of single amplimers was confirmed by melting curve analysis and gel electrophoresis, and data were analyzed as described (20). Interaction frequencies were normalized using ‘loading’ (using a pair of convergent primers targeting one SAMD4A HindIII fragment), ‘inter-sample variation’ (using primers targeting adjacent HindIII fragments in the house-keeping gene, GAPDH) and ‘amplification efficiency’ controls (using templates encoded by bacterial artificial chromosomes); this should provide higher accuracy than obtained by Larkin et al. (16). Primers were designed using Primer 3.0 Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) using default ‘qPCR’ settings. Primer sequences are available on request.
Statistical analysis
P-values (two-tailed) from unpaired Student’s t-tests were calculated using GraphPad (http://www.graphpad.com); they were considered significant when <0.01. Errors in Monte Carlo simulations in Supplementary Figure S2 were calculated assuming Poisson statistics.
RESULTS
‘Super-resolution’ localization of nascent RNAs in the standing and traveling waves
We use RNA FISH with pairs of intronic probes—one red, the other green—to follow nascent transcripts in the standing and traveling waves. Probes (and their target sequences) are named by the distance of the complementary DNA sequence (in kb) from the SAMD4A TSS. In each case, Probe 2 is paired with a second probe targeting transcripts in the traveling wave (Figure 2A); the pairs are used at the times indicated in Figure 1 (i.e. when the traveling wave reaches the relevant segment). As a result, the targets of Probe 2 and its partner probe serve as proxies for the locations of the promoter and the segment being transcribed by the pioneering polymerase, respectively. The results of a typical experiment are illustrated in Figure 2B and C. Previous work (13,15,16) indicates that (i) essentially all HUVECs contain only two SAMD4A alleles (they are diploid and synchronized in the G0 phase of the cell cycle); (ii) the oligonucleotide probes used can detect single nascent RNAs efficiently, to yield subresolution spots (24); (iii) a yellow (colocalizing) spot results from targets copied from the same allele (as spot area is so small compared with nuclear area, a green focus can only overlap a red focus copied from a different allele in <1 nucleus in a thousand, assuming random distributions) and (iv) 30–35% alleles in the population are being transcribed by a pioneering polymerase at any one time after stimulation. The use of RNA FISH with intronic probes instead of DNA FISH has the great advantage that it allows us to focus on the minority of active alleles in the populatio, and not the majority of inactive ones. Our probe pairs are unlikely ever to target the same RNA molecule. First, tiling microarrays yield a trough between standing and traveling waves [Figure 1A (15)], indicative of rapid degradation of intervening intronic RNA. Second, the half-lives (assessed using quantitative reverse transcriptase-PCR and the transcriptional inhibitor DRB, 6-dichloro-1-β-D-ribofuranosyl-benzimidazole) of three different RNA segments complementary to positions 2, 34 and 128 kb from the TSS are all ∼5 min (not shown), and it takes the polymerase >4 such half-lives to transcribe from Target 2 to the closest target (i.e. 34)—and longer still to the others lying further downstream. Third, RNA FISH using probe pairs 34 + 45, 34 + 128, 34 + 190 and 45 + 128 all yield ≤1% nuclei with a yellow (colocalizing) spot; if targets lay within the same RNA molecule, whenever a 3′-target is seen, the 5′-target should also be seen—but the two are rarely (if ever) seen together because the 5′-target is degraded so rapidly (Figure 2C illustrates typical results).
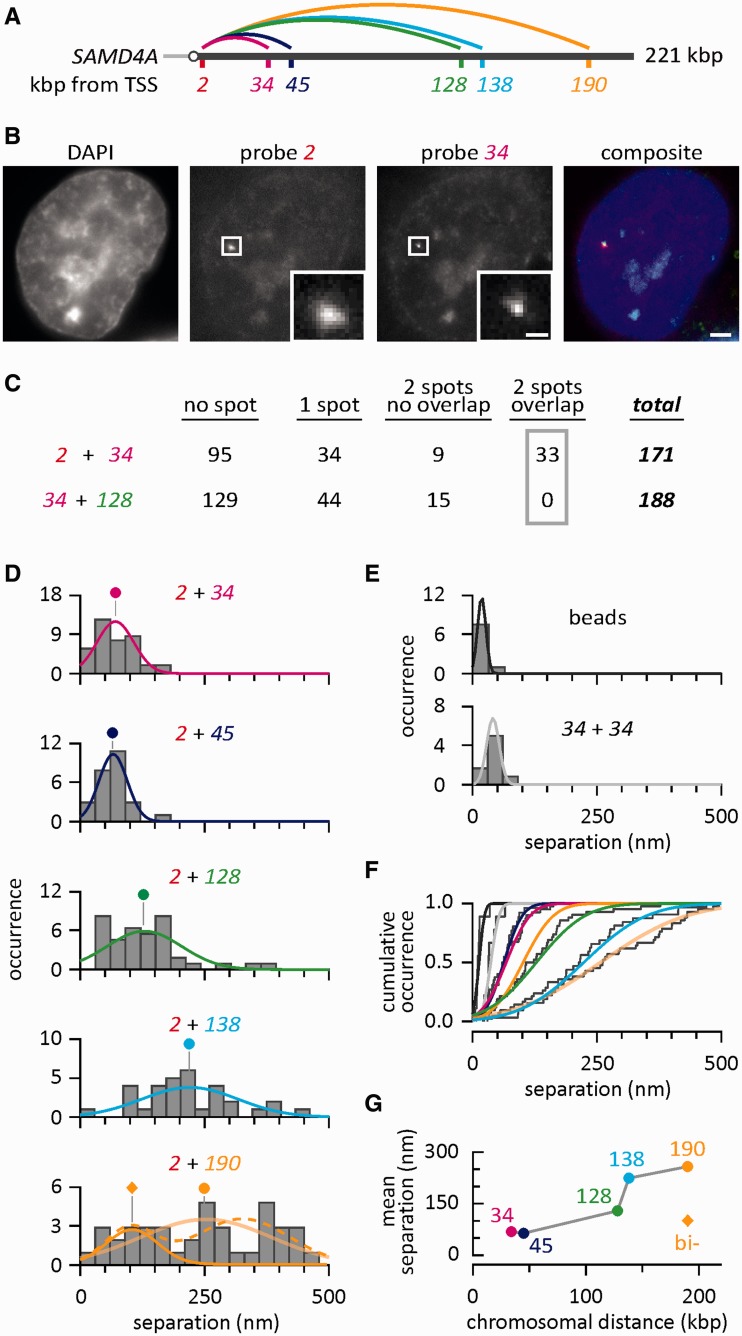
Figure 2.
Measured distances between pairs of nascent RNAs copied from one SAMD4A allele. HUVECs were treated with TNFα for 30–82.5 min, RNA FISH performed (using Probe 2 in a pairwise combination with Probe 34, 45, 128, 138 or 190 at 30, 45, 52.5, 60 or 82.5 min after stimulation—or Probes 34 + 128 at 45 min) and separations between probes determined with 30-nm precision. (A) Map of SAMD4A, indicating pairwise probe combinations (34 + 128 excluded). (B) Typical images of one nucleus obtained 30 min after stimulation. Bar: 2 µm (insets 500 nm, 90-nm pixels). (C) Some typical results. With probes 2 + 34 (30 min post-stimulation), 56% cells yield no signal, 20% cells give a single spot (either from Probe 2 or 34), 5% cells give non-overlapping spots (and thus mark transcripts copied from different alleles) and 19% cells yield overlapping signals; when Probe 2 is used in pairwise combinations with each of the other probes indicated in (A), essentially similar numbers are obtained. With Probe 34 + 128 (45 min post-stimulation), a similar percentage of cells with no signal (68%) is obtained, but no cells yield overlapping signals (confirming that these probes target two different RNA molecules). (D) The separation (nm) seen between Probe 2 and the second in the pair measured using images like those in (B). Histograms (30-nm bins; 40, 42, 26, 32 and 42 yellow foci analyzed for the examples given from top to bottom) illustrate the number of times a separation was seen (occurrence). Gaussian distributions are fitted to histograms and normalized by equalizing areas under curves to allow direct comparison of probabilities (circles indicate means). For 2 + 190, the histogram was also fitted with a bi-modal distribution (dotted line) and the left component plotted as a uni-modal distribution (diamond indicates mean). (E) Occurrences given by multispectral 100-nm beads (where the expected separation is zero if channel registration and localization are perfect), and a mixed red + green probe (34 + 34 used 30 min after stimulation) targeting the same region (which gives a slightly larger separation owing to the use of an odd number of oligomers in the probe set). (F) Cumulative occurrence (gray steps) with fitted curves color-coded as in panel (D). The curve for 2 + 138 (light blue) is closer to that for 2 + 190 (beige) than that for 2 + 128 (green), despite 138 being closer to 128. When 2 + 190 is fitted with a bi-modal distribution, the left component (orange) closely matches the curves given by 2 + 34 (red) and 2 + 45 (dark blue). (G) Mean separations, color- and shape-coded as in panel (D), plotted as a function of chromosomal distance (kb) between probe targets (diamond gives mean of left-hand component of bimodal distribution given by 2 + 190).
Thirty minutes after stimulation, RNA FISH with probe pair 2 + 34 yields yellow foci in 19% cells (Figure 2B and C); this indicates that two different transcripts copied from one allele lie close together. As the resolution afforded by conventional microscopy is too low to distinguish between nascent transcripts copied from DNA regions lying only 32 kb apart, we use a ‘super-resolution’ approach to measure (with 30-nm precision) the distance between the red and green foci underlying such yellow foci [like those in Figure 2B (16–19)]. Gaussian curves are fitted to the intensities of the underlying red and green foci, the positions of each peak measured with 15-nm precision and distances (separations) between peaks measured with 30-nm precision (the increased error results from residual misalignment between channels). Probes 2 + 34 yield a range of separations, with a mean of 68 nm; <5% separations are >150 nm (Figure 2D). Probes 2 + 45 give essentially the same pattern; however, the other pairs (2 + 128, 2 + 138 and 2 + 190) yield progressively wider separations (Figure 2D). These results point to a step change in separation; the first two pairs give comparable separations, and the last three progressively larger ones. The step change occurs at the time when the amplitude of the standing wave declines from its peak (Figure 1). Probes 34 + 128 yield no yellow foci (Figure 2C), indicating that their targets never lie on the same nascent RNA molecule (given earlier in the text).
Most separations given by red/green fluorescent beads are ≤30 nm (Figure 2E), and the distribution seen is that expected of a ‘perfectly’ colocalizing control measured with 30-nm precision. Overlapping red and green signals emitted by a mixture of red and green probes targeting just Segment 34 (i.e. red 34 + green 34; Figure 2E) also had a significantly smaller mean separation than those given by 2 + 34 or 2 + 45 (P < 0.05; two-tailed unpaired Student’s t-test).
We next analyzed the change in separation in detail. The data presented in Figure 2D are redrawn in Figure 2F as cumulative occurrences. Curves for Probes 2 + 34 (dark blue) and 2 + 45 (red) are similar, whereas that for 2 + 138 (light blue) lies closer to 2 + 190 (yellow) than 2 + 128 (green)—despite 138 being closer to 128 on the chromosome. The discontinuous behavior is confirmed by replotting the data in another way; 2 + 34 and 2 + 45 yield similar mean separations, and both are less than those obtained with 2 + 138 and 2 + 190 (Figure 2G).
Comparing models involving tracking and fixed polymerases
If we assume the simplest possible model involving a polymerase tracking along a linear template, the measured separation between a pioneer and a follower at the TSS should increase in direct proportion to the number of bases transcribed by the pioneer. Then, separations (normalizing to the one obtained with probes 2 + 34) should scale proportionately with the number of kb between targets (Figure 3A, dashed line given by ‘Model I’). This should hold true irrespective of levels of chromatin and/or transcript packing (if both remain invariant along the gene), and if nascent transcripts are randomly arranged around their templates. However, separations measured by FISH fall below this line (Figure 3A, gray line).
We next assume the template adopts a self-avoiding random walk and has a persistence length characteristic of hetero- or eu-chromatin, or an intermediate value (27,31–33). After generating >100 000 configurations using a Monte Carlo algorithm (29), we measure the spatial distance between two relevant points on the template; observed separations are again smaller (Figure 3B, compare the three dashed lines given by ‘Model II’ with the continuous line). (Supplementary Figure S1B also shows that the separations have different distributions from those observed experimentally.)
Recent experimental measurements suggest that the scaling of 3D separation with genetic distance can be fitted with an exponent of ∼1/3 (35,36); this contrasts with the exponent of 0.588 given by a self-avoiding walk (34). Although these experimental data were mainly obtained using longer genetic distances than analyzed here, we nevertheless considered the case in which the fiber folds more compactly than a self-avoiding walk by introducing a non-specific attraction that yielded an exponent of 1/3. The 3D separation is now in the observed range, although it shows a smoother variation with genetic distance than seen with our FISH data (compare dotted line given by Model III with the gray continuous line in Figure 3B). However, the distributions never show the multiple peaks (Supplementary Figure S1C) given by our FISH data (Figure 2D). Clearly, our experimental observations are inconsistent with all three models.
If active polymerases are clustered in factories, separations will change in a more complicated way (Figure 4). After initiating (Figure 4B, right), the pioneering polymerase reels in the template and extrudes the promoter, which will initially be tethered close to the factory so that it can only access a polymerase on the surface of the same factory; if it now re-initiates, its transcript will inevitably be found in the same factory as the pioneering transcript (Figure 4C, right). As a result, the mean separations seen by RNA FISH should lie in the range from zero to slightly more than the diameter of a factory. Separations should then remain within this range as the length of the tether connecting the promoter to the pioneer increases, and the promoter goes through repeated cycles of re-initiation and abortion [giving ‘bursty’ transcription (21–23)]. A promoter in a living cell is able to diffuse throughout the volume being discussed here every minute or so (38,39). However, this tether will eventually become long enough (Figure 4D–F) to allow the promoter to access a second factory in addition to the original one (Figure 4G). As inter-factory distances are between 450 and 600 nm in other cells (40), we would then expect the separations seen by FISH to increase up to these values. [Inter-factory distances used here are calculated using the densities given in Table 1 of Faro-Trindade and Cook (40), after correction for the larger factory diameter of 90 nm obtained more recently (5).]
Figure 4.
An interpretation of results obtained by RNA FISH (with map of SAMD4A). (A) Before stimulation (when NFκB is absent), the SAMD4A promoter is rarely active, even though it may diffuse to a polymerase (oval) in a factory (sphere). (B) After 10 min, NFκB has entered the nucleus and promotes transcriptional initiation when the promoter collides with the polymerase in the factory (yielding an RNA FISH signal with Probe 2). (C) By 30 min, the pioneering polymerase has reeled in the template, as promoter and transcript are extruded. The promoter, being tethered close to the factory, can now reinitiate at a second polymerase—but this polymerase will soon abort. The separations given by Probes 2 + 34 are consistent with the two targets being randomly distributed on the surface of one factory. (D) As the pioneer polymerase transcribes, the length of the tether connecting the promoter to the factory increases. (E) At 50 min, Probe 128 yields a signal, and the tether is not long enough to reach a second factory. (F) By 60 min, the tether has grown sufficiently to allow the promoter to reinitiate at a polymerase in a secondary factory. (G) By 85 min, the promoter is initiating in the first (left) or the second factory (right)—this produces a wider distribution of measured separations in the population.
The separations seen in Figure 2 fit this second model well. Thus, those given by Probes 2 + 34 range from 8 to 163 nm (Figure 2D), and the distribution is that expected after repeatedly and randomly scattering a pair of red and green points in a 35-nm shell around an 87-nm diameter sphere (as in 13; not shown). [The distribution is not the one expected after randomly scattering the red and green points within a sphere—as this would give a mean separation close to zero (as in the ‘perfectly-colocalizing’ control provided by the beads; Figure 2E). Therefore, this result is inconsistent with a model that requires direct contact between the two polymerases.] The separations given by 2 + 45 are similar to those given by 2 + 34 (Figure 2D)—consistent with the pioneering polymerase and the follower still being attached to the same factory (as in Figure 4C, right). Then, Probes 2 + 128, 2 + 138 and 2 + 190 yield the range of separations up to 600 nm expected if the pioneer and follower can now attach either to the same or a different factory in different cells in the population (as in Figure 4G). Moreover, a bimodal fit to the distribution seen with Probes 2 + 190 (Figure 2D, bottom) yields a left-hand component consistent with the two polymerases being bound to the same factory. The model in Figure 4 has one final consequence of interest here: once the promoter re-initiates at a secondary factory and then aborts, the long tether (compared with a short tether) ensures that the promoter will explore a much larger volume (41)—with the consequence that it will inevitably take longer to re-initiate at either the primary or the secondary factory. As a result, this model is consistent with the decline in the amplitude of the standing wave seen after 30 min (Figure 1). Taken together, these results support a model involving transcripts being generated on the surface of factories.
3C shows the templates of the standing and traveling waves lie close together
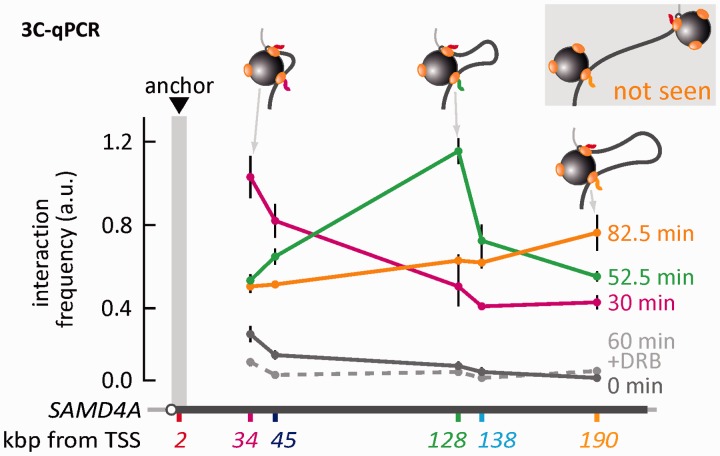
RNA FISH revealed that the two nascent transcripts are sometimes found close together. We next used an independent method—3C coupled to qPCR—to confirm that templates lie close together at the relevant times. In each case, one primer targets Position 2 (the ‘anchor’), whereas the second targets sites progressively further down the gene (Figure 5). Before stimulation, the anchor rarely contacts any other targets (gray line). But 30 min after stimulation, it interacts strongly with 34 and less so with sites further away (red line). After 52.5 min, it now interacts most strongly with 128 (green line); the transcriptional inhibitor, DRB, abolishes such interactions (gray dotted line). By 82.5 min, most contacts are with 190 (orange line), but this frequency is ∼1/3 lower than the maximum seen at 30 or 52.5 min. This is consistent with the tether now being long enough to allow the promoter to access a second factory (and thus to yield no 3C product with 190). All these results are difficult to reconcile with any model involving tracking polymerases; however, they are consistent with appearance of a subgene loop that enlarges with time (as in Figure 4).
Figure 5.
The development and growth of a subgene loop in SAMD4A. HUVECs were treated for 0–82.5 min with TNFα, and 3C coupled to qPCR performed using one primer targeting region 2 (anchor) and another targeting sites successively further 3′. In some cases, DRB was added 25 min before harvesting. The normalized interaction frequency (arbitrary units, a.u.) between the anchor and the sites indicated on the map is given. At 0 min (gray), the anchor rarely contacts other regions. By 30 min (red), it interacts strongly with Position 34 (a possible structure is indicated); however, it interacts less well with sites further away. After 52.5 min (green), it now interacts most strongly with Position 128 (a possible structure is indicated), and DRB abolishes all interactions (dotted line). By 82.5 min (orange), most contacts are with Position 190, but this frequency is ∼30% lower than the maximum seen at 30 or 52.5 min (two possible structures are indicated; the upper one does not yield a 3C product). Results are consistent with the appearance of a subgene loop that enlarges with time, and therefore with the model described in Figure 4.
Monte Carlo simulations of a loop tethered to a factory
3C shows that after stimulation, the major contacts made immediately upstream and downstream by the TSS of SAMD4A are with the CNIH and GCH1 promoters (42). Inspection of encyclopedia of DNA elements data on the University of California at Santa Cruz browser also indicates that this region of the genome in HUVECs bears histone marks—for example, H3K4me, H3K27Ac—characteristic of euchromatin (43,44). Therefore, we used standard techniques (25–31) to model the CNIH:SAMD4A:GCH1 segment (Figure 6A) as a semi-flexible polymer consisting of 30-nm beads, each representing ∼3 kb of DNA packaged into chromatin—a packing density typical of euchromatin (25,26,32,33). The polymer is looped by attachment at each end to a primary factory, as well as at an additional point within SAMD4A (corresponding to a target of a FISH probe); this primary factory is surrounded by six other factories along the orthogonal axes (Figure 6B, only one secondary factory is shown). We call this Model IV. No bead or factory is allowed to occupy the same volume as another, and—after generating >1 000 000 configurations using a Monte Carlo algorithm (29)—we record the frequency with which the SAMD4A TSS is seen within 30 nm of the surface of a factory and calculate the fraction of times that the TSS ‘visits’ the primary factory relative to all seven factories. We also model fibers with differing contour length (by varying the distances l1 and l2 to the left and right of the TSS of SAMD4A; Figure 6A), inter-factory distance and persistence length (ξ)—again covering the range used previously (27,31–33). We first consider a structure mimicking that found in vivo (Figure 6C, black line), which we will use as a reference. As expected, subdividing the CNIH:GCH1 loop by attaching Position 34 to the primary factory ensures that the SAMD4A promoter is now connected to that factory through a tether too short to allow the promoter to visit a secondary factory (Figure 6B, left); as a result, the fraction of visits to the primary factory is 1.0. At the other extreme, attaching 190 creates a tether long enough for the promoter to reach a secondary factory (Figure 6B, right); the fraction is now close to zero. As intuition suggests, decreasing l1 to 50 kb prevents this fall toward zero because the promoter is now tethered close to the primary factory from both the left and right (Figure 6C, compare the black with the topmost three curves). As intuition also suggests, increasing l1 (or varying l2) has little effect on the pattern (Figure 6C, compare the black with the bottom three curves). Clearly, the distance to the nearest tethering point is the major determinant of whether a promoter can visit a secondary factory (Supplementary Figure S2 provides additional data on the likelihood that other points in the locus will visit a primary or secondary factory). Using values of l1 and l2 mimicking those in vivo, we now vary the stiffness of the polymer between the extremes expected of eu- and hetero-chromatin (27,31–33). A short (flexible) persistence length expected of euchromatin (i.e. 40 nm) gives a much better fit to the RNA FISH data than that expected of (stiffer) heterochromatin (i.e. 150 nm; Figure 6D, compare black and green curves with gray curve). Similarly, an inter-factory distance of 500 nm—which is within the expected range (given earlier in the text)—provides the best fit (Figure 6E); moreover, the distribution of separations is also the one closest to the experimental data (Supplementary Figure S1D). We conclude that this simple model yields results that closely approximate experimental data.
DISCUSSION
We studied 221-kb SAMD4A after switching it on rapidly and synchronously with TNFα (15). Within 10 min, pioneering polymerases initiate and then transcribe steadily to terminate after ∼90 min; subsequently other polymerases initiate, but these soon abort. As a result, after 30 min, the long gene becomes transcribed by two polymerases—a pioneer and a succession of others that abort almost immediately—and these generate traveling and standing waves of nascent RNA throughout the course of our experiment (Figure 1). We analyzed the changing conformation of SAMD4A using RNA FISH with 30-nm precision to localize nascent transcripts (used as proxy markers for the pioneering plus following polymerases), 3C (to monitor contacts between the two segments of the gene that are being transcribed) and Monte Carlo modeling (to simulate the diffusion of the promoter). The application of these approaches to this system allowed us to gain insights into the transcription cycle that are difficult—if not impossible—to obtain using a continuously active, short, gene.
In particular, we focus on which of two views of transcription might apply (see ‘Introduction’ section). The traditional view assumes an active polymerase tracking down the template. A closely related variant involves a polymerase initiating at a hub or node in the chromatin network and tracking away. In both cases, the distance between transcripts made by a pioneer and a succession of (recursively aborting) followers at the TSS should increase in proportion to the number of bases transcribed by the pioneer. Here, we assume the template adopts a random walk and that both template and transcript packing remain unchanged from point to point along the gene—all natural assumptions associated with this model. However, RNA FISH coupled to ‘super-resolution’ localization shows that the distance between the two nascent transcripts does not increase in the expected way (Figures 2C and 3; Supplementary Figure S1); rather, observed separations are smaller and/or have different distributions. Clearly, one or other of our assumption must be wrong.
In contrast, all results are consistent with an alternative view involving active polymerases attached to the surface of a factory. According to this, once the pioneer initiates, the promoter is extruded away from the factory. Then, this promoter is initially tethered close to the factory, where it is likely to access another polymerase (Figure 4C). [Computer simulations confirm this intuition (41).] On re-initiation, the resulting nascent transcript (made by the following polymerase) will lie close to that made by the pioneer (Figure 4C, right)—and the same will be true of transcripts made by later followers during their cycles of initiation and abortion. As a result, the inter-transcript distance will remain constant (in Figure 2D, compare results for Probes 2 + 34 and 2 + 45). But as the pioneer continues to transcribe, the length of the tether connecting the promoter to the pioneer will eventually become long enough to allow the promoter to access a second factory in addition to the original one (Figure 4G, right). Then, the distance between transcripts should fill the range between zero and the inter-factory distance—and it does (Figure 2, Probes 2 + 138 and 2 + 190). 3C coupled to qPCR confirms that the templates for the two nascent transcripts lie close together at the relevant times (Figure 5). In addition, the results of the computer simulations support the intuition that the SAMD4A promoter, when embedded in a euchromatic loop mimicking the one found in vivo, is only able to visit a second factory once the tether becomes long enough (Figure 6C)—and then only if the chromatin fiber has the flexibility of euchromatin (Figure 6D), and if the inter-factory distance is in the expected range (Figure 6E).
We conclude that active polymerases are immobile molecular machines, and we imagine that the way the SAMD4A promoter explores nucleoplasmic space will be typical of all human promoters. Moreover, our conclusions should be generally applicable to any two polymerases transcribing any two adjacent transcription units on the chromosome. In addition, our results shed light on how enhancers might work and what might drive transcriptional ‘bursting’. Enhancers are classically defined as sequences that promote firing of flanking promoters; it turns out that many are transcribed into non-coding RNAs, and that ongoing transcription is required for their function (45–47). Just as the pioneering polymerase tethers the SAMD4A promoter close to a factory to enhance its firing frequency, we suggest a polymerase transcribing an enhancer will tether its target (genic) promoter close to a factory and do the same (48). Similarly, transcription at the level of a single gene occurs sporadically and cyclically, with successive initiations producing a ‘burst’ of transcripts followed by silence (21–23); such ‘bursting’ is usually explained by remodeling a permissive chromatin state into a restrictive one (49). Just as close tethering underlies repeated re-initiation by the SAMD4A promoter, we suggest close tethering also drives ‘bursting’ in other genes (50,51).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
Biotechnology and Biological Sciences Research Council via the ERASysBio+/FP7 initiative [BB/100467X/1 to P.R.C for funding for A.P.]; Wellcome Trust [085058 to P.R.C. for funding for A.P., 086017 to P.R.C. for funding for J.D.L.]. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Tatsuhiko Kodama for RNA FISH probes and Jon Bartlett for technical assistance.
REFERENCES
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science; 2002. [Google Scholar]
- 2.Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakalova L, Fraser P. Organization of transcription. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a000729. a000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook PR. A model for all genomes: the role of transcription factories. J. Mol. Biol. 2010;395:1–10. doi: 10.1016/j.jmb.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 5.Eskiw CH, Rapp A, Carter DR, Cook PR. RNA polymerase II activity is located on the surface of protein-rich transcription factories. J. Cell Sci. 2008;121:1999–2007. doi: 10.1242/jcs.027250. [DOI] [PubMed] [Google Scholar]
- 6.Eskiw CH, Fraser P. Ultrastructural study of transcription factories in mouse erythroblasts. J. Cell Sci. 2011;124:3676–3683. doi: 10.1242/jcs.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 8.Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 10.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh H-M, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melnik S, Deng B, Papantonis A, Baboo S, Carr IM, Cook PR. The proteomes of transcription factories containing RNA polymerases I, II, or III. Nat. Methods. 2011;8:962–968. doi: 10.1038/nmeth.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, Cook PR. Active RNA polymerases: mobile or immobile molecular machines? PLoS Biol. 2010;8:e1000419. doi: 10.1371/journal.pbio.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, Komura D, Kitakami J, Oshida N, Papantonis A, et al. A wave of nascent transcription on activated human genes. Proc. Natl Acad. Sci. USA. 2009;106:18357–18361. doi: 10.1073/pnas.0902573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larkin JD, Cook PR, Papantonis A. Dynamic reconfiguration of long human genes during one transcription cycle. Mol. Cell. Biol. 2012;32:2738–2747. doi: 10.1128/MCB.00179-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 19.Larkin JD, Cook PR. Maximum precision closed-form solution for localizing diffraction-limited spots in noisy images. Opt. Express. 2012;20:18478–18493. doi: 10.1364/OE.20.018478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 21.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr. Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 25.Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput. Biol. 2008;4:e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl Acad. Sci. USA. 2004;101:16495–16500. doi: 10.1073/pnas.0402766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook PR, Marenduzzo D. Entropic organization of interphase chromosomes. J. Cell Biol. 2009;186:825–834. doi: 10.1083/jcb.200903083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weidemann T, Wachsmuth M, Knoch TA, Müller G, Waldeck W, Langowski J. Counting nucleosomes in living cells with a combination of fluorescence correlation spectroscopy and confocal imaging. J. Mol. Biol. 2003;334:229–240. doi: 10.1016/j.jmb.2003.08.063. [DOI] [PubMed] [Google Scholar]
- 29.Landau DP, Binder K. A Guide to Monte Carlo Simulations in Statistical Physics. New York: Cambridge University Press; 2000. [Google Scholar]
- 30.Kratky O, Porod G. Diffuse small-angle scattering of X-rays in colloid systems. J. Colloid Sci. 1949;4:35–70. doi: 10.1016/0095-8522(49)90032-x. [DOI] [PubMed] [Google Scholar]
- 31.Langowski J. Polymer chain models of DNA and chromatin. Eur. Phys. J. E Soft Matter. 2006;19:241–249. doi: 10.1140/epje/i2005-10067-9. [DOI] [PubMed] [Google Scholar]
- 32.Bohn M, Heermann DW. Diffusion-driven looping provides a consistent framework for chromatin organization. PLoS One. 2010;5:e12218. doi: 10.1371/journal.pone.0012218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui Y, Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl Acad. Sci. USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doi M, Edwards SF. The Theory of Polymer Dynamics. Cambridge: Oxford Science Publications; 1986. [Google Scholar]
- 35.Mateos-Langerak J, Manfred Bohn M, de Leeuw W, Giromus O, Manders EMM, Verschure PJ, Indemans MHG, Gierman HJ, Heermann DW, van Driel R, et al. Spatially confined folding of chromatin in the interphase nucleus. Proc. Natl Acad. Sci. USA. 2009;106:3812–3817. doi: 10.1073/pnas.0809501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Göndör A, Rougier C, Ohlsson R. High-resolution circular chromosome conformation capture assay. Nat. Protoc. 2008;3:303–313. doi: 10.1038/nprot.2007.540. [DOI] [PubMed] [Google Scholar]
- 38.Levi V, Ruan Q, Plutz M, Belmont AS, Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 2005;89:4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wachsmuth M, Caudron-Herger M, Rippe K. Genome organization: balancing stability and plasticity. Biochim. Biophys. Acta. 2008;1783:2061–2079. doi: 10.1016/j.bbamcr.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 40.Faro-Trindade I, Cook PR. A conserved organization of transcription during embryonic stem cell differentiation and in cells with high C value. Mol. Biol. Cell. 2006;17:2910–2920. doi: 10.1091/mbc.E05-11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bon M, Marenduzzo D, Cook PR. Modeling a self-avoiding chromatin loop: relation to the packing problem, action-at-a-distance, and nuclear context. Structure. 2006;14:197–204. doi: 10.1016/j.str.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 42.Papantonis A, Kohro T, Baboo S, Larkin JD, Deng B, Short P, Tsutsumi S, Taylor S, Kanki Y, Kobayashi M, et al. TNFα signals through specialized factories where responsive coding and miRNA genes are transcribed. EMBO J. 2012;31:4404–4414. doi: 10.1038/emboj.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolovos P, Knoch TA, Grosveld FG, Cook PR, Papantonis A. Enhancers and silencers: an integrated and simple model for their function. Epigenetics Chromatin. 2012;5:1. doi: 10.1186/1756-8935-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raj A, van Oudenaarden A. Stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finan K, Cook PR. In: Transcriptional initiation: frequency, bursting, and transcription factories. Rippe K, editor. Weinheim, Germany: Genome Organization and Function in the Cell Nucleus. Wiley-VCH Verlag GmbH & Co, KGaA; 2012. pp. 235–254. [Google Scholar]
- 51.Lionnet T, Singer RH. Transcription goes digital. EMBO Rep. 2012;13:313–321. doi: 10.1038/embor.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.