Abstract
Energy transfer (ET) in phycobilisomes, a macrocomplex of phycobiliproteins and linker proteins, is a process that is difficult to understand completely. A model for a rod composed of two hexamers of Phycocyanin and two hexamers of Phycoerythrin was built using an in silico approach and the three-dimensional structures of both phycobiliproteins from Gracilaria chilensis. The model was characterized and showed 125 Å wide and 230 Å high, which agree with the dimensions of a piling of four hexamers as observed in the images of subcomplexes of phycobilisomes obtained by transmission electron microscopy. ET rates between every pair of chromophores in the model were calculated using the Förster approach, and the fastest rates were selected to draw preferential ET pathways along the rod. Every path indicates that the ET is funneled toward the chromophores located at Cysteines 82 in Phycoerythrin and 84 in Phycocyanin. The chromophores that face the exterior of the rod are phycoerythrobilins, and they also show a preferential ET toward the chromophores located at the center of the rod. The values calculated, in general, agree with the experimental data reported previously, which validates the use of this experimental approach.
Keywords: phycobilisome, antenna, Phycocyanin, Phycoerythrin, energy transfer
Introduction
Phycobilisomes (PBS) are light harvesting accessory protein complexes, present in red algae and cyanobacteria. The architecture of these protein complexes provides the framework in which the distribution of chromophores is in such geometry that allows the transfer of energy with the highest efficiency known in biological systems.1 Basically, the process of converting the energy from the sun in other forms of energy in plants and also in algae implies three steps, energy harvesting, energy transfer (ET), and energy dissipation. Phycobilisomes are involved in the first and second steps, because they are macromolecular systems organized to maximize energy harvesting and transfer, minimizing the dissipation of energy. The understanding of this process at molecular level is crucial to design artificial systems or biomimetics.2
PBS in red algae or cyanobacteria are composed of phycobiliproteins and linker proteins. In Gracilaria chilensis, PBS are organized in a core of allophycocyanin (APC) from which four to six rods radiate as shown in Figure 1(A). These rods are formed by R-Phycoerythrin (PE) and R-Phycocyanin (PC).3 Phycobiliproteins present a general organization of hexamers of (αβ) heterodimers. The hexamers present a ring-like shape that allows them to pile up to form the rod. The three-dimensional structures of PE (PDB code: 1eyx)4 and PC (PDB code: 2vb8)5 from this algae have been determined previously in our group, and the structure of both proteins is shown in Figure 1(B,C), respectively. Table I3, 6 shows their chromophore composition and the main spectroscopic absorption and emission wavelengths. The general chemical structure of the phycobilins is shown in Figure 1(D).
Figure 1.
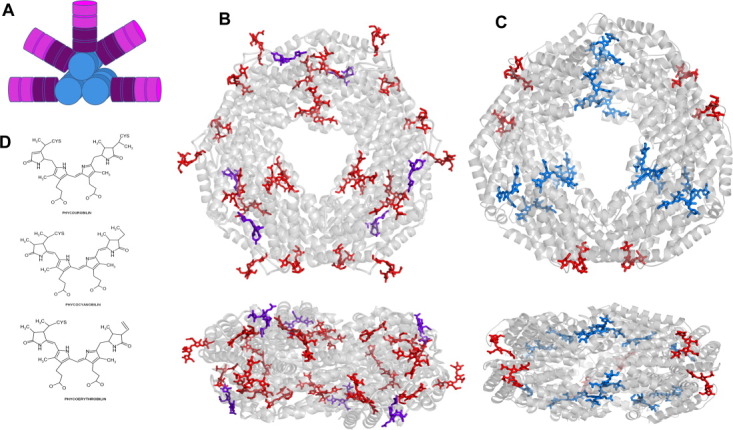
A: Schematic view of a phycobilisome. B: Three-dimensional structure of Phycoerythrin and C: Phycocyanin. Phycoerithrobilins are shown in red, phycocyanobilins in blue, and phycourobilins in violet. D: Structure of phycobilins.
Table I.
Characteristics of the Chromophores Present in R-Phycocyanin and R-Phycoerythrin from Gracilaria chilensis3, 6
| Composition | Chromophore | Cysteina | λA max | λE max |
|---|---|---|---|---|
| Phycoerythrin | 566 nm | 574 nm | ||
| α Subunit | Phycoerythrobilin | 82 | ||
| Phycoerythrobilin | 139 | |||
| β Subunit | Phycoerythrobilin | 82 | ||
| Phycoerythrobilin | 158 | |||
| Phycourobilin | 50–61b | |||
| Phycocyanin | 621 nm | 634 nm | ||
| α Subunit | Phycocyanobilin | 82 | ||
| β Subunit | Phycocyanobilin | 84 | ||
| Phycoerythrobilin | 153 | 540 nm |
The chromophores are covalently bound to cysteines at the position in the sequences shown in the table.
Phycourobilin is bound to Cysteines 50 and 61. Notice the erythrobilin bound to Cysteine 139 in the β subunit of Phycocyanin.
The conformation of each chromophore as well as their relative position in every hexamer and in a piling of hexamers is crucially important to propose the main ET pathways in a rod and explain the high efficiency. As discussed in the literature,7, 8 the transferred energy can be modeled by fluorescence resonance ET mechanism, where the transfer of the electronic excitation is produced by coulombic interactions represented by the weak donor–acceptor dipole–dipole coupling and strong dipole–dipole interaction9 involving the transfer of one electron from the lowest unoccupied molecular orbital (LUMO) of the donor chromophore to the LUMO of the acceptor chromophore and with the concomitant transfer of one electron from the highest occupied molecular orbital (HOMO) of the acceptor chromophore to the HOMO of the donor chromophore. It is noteworthy that at donor–acceptor distances lower than 20 Å, the interaction between the electronic systems is too strong to behave as localized systems, and they behave as a super chromophore (i.e., exciton coupling model).10 A good approach used to calculate ET constants for this system has been the Förster approach, which has been used since the first structural studies performed on Phycocyanin.11–14 Theoretical calculations have been performed for Phycocyanin using their three-dimensional structures,15–17 and Ref. 13 reported a comparison between theoretical and experimental results for the ET in monomers and trimers of Phycocyanin from Synechococcus sp. The results were very similar to the theoretical values supporting the use of this approach to this biological system.
Full modeling of PBS is a very hard task due the complexity of that kind of systems. So, in order to study the ET pathways in PBS, we have chosen a reduced model: an antenna (rod) formed by two molecules of Phycoerythrin and two molecules of Phycocyanin. As the number of molecules and composition of any rod are concerned, these depend on the light intensity and wavelength of the light.18 According to the architecture shown by the electron micrographs, we chose a 1:1 composition, which also has been observed in some phycobilisomes from red algae depending on the season, and we chose four molecules in the antenna, because in Gracilaria chilensis, most of the rods observed by electron microscopy have 4 or 5 U.19 Docking models were built and evaluated to simulate the rod by rotating and sliding one hexamer relative to the other, and the co-ordinates of each chromophore were extracted from the final model. Using that information, the ET constants were calculated in order to compare the theoretical transfer with the experimental data reported in the literature.
Results
Characterization of rods
The rods obtained as described in Materials and Methods section were characterized by electron microscopy. The micrograph [Fig. 2(B)] shows isolated hexamers and rods formed by three and four hexamers, showing the arrangement of the hexamers along the rod similar to those represented by the scheme included in the insert. The absorption and emission spectra [Fig. 2(A)] shows the presence of organized rods. The rod-enriched fraction contained some free PE as revealed by the fluorescence maximum at 574 nm, and rods composed of PE and PC as revealed by the emission at 634 nm upon irradiation at 566 nm. From our results, the rods present in the phycobilisomes of Gracilaria chilensis contain three to four hexamers, and they are highly enriched in PC and PE.1 The dimensions of the rod formed by four hexamers are 230 Å high and 125 Å wide, values that agree with the dimensions of the structural model for an antenna with similar components as shown in Figure 3.
Figure 2.
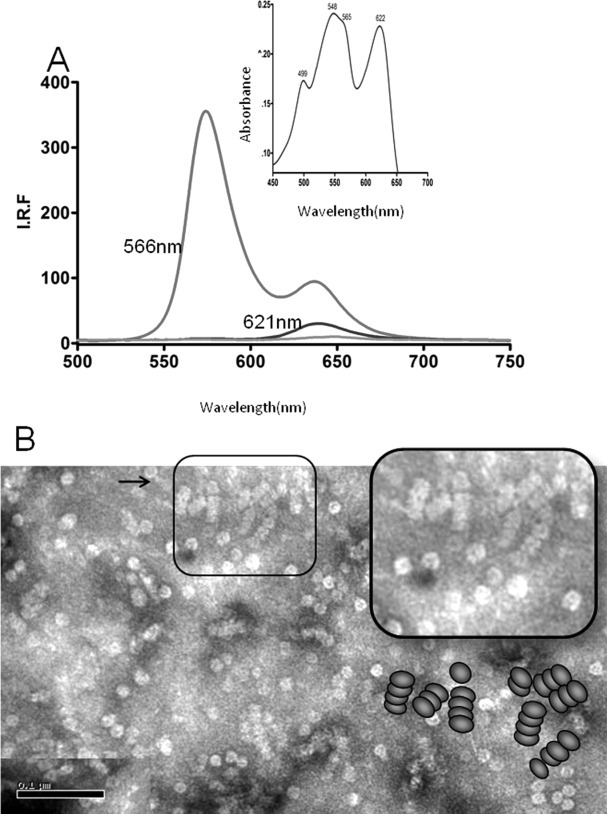
Characterization of rod-enriched fractions. A: Fluorescence spectrum upon excitation at 566 and 621 nm as indicated. B: Electron micrograph of the rod-enriched fraction. An enhancement of the image is shown in the insert, and the schematic representation on that region of the image.
Figure 3.
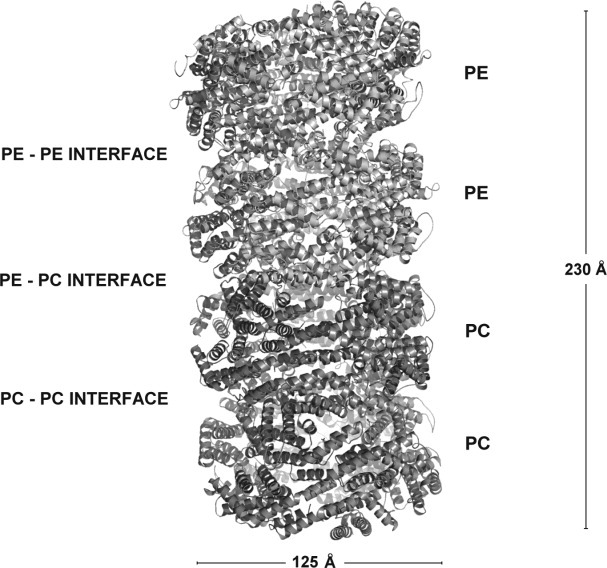
Ribbon representation of the model of the rod.
Molecular building of the antenna
The final model for the rod is shown in Figure 3 as a result of the molecular docking and dynamics.
The molecular dynamic procedure performed on the rigid model of the rod optimized the interaction area among phycobiliproteins and relaxed the structures coming from the rigid-docking process. Two hundred picoseconds of simulation showed to be enough to reach a structural convergence of the model, as the root mean square deviation (rmsd) of the backbone shows (Fig. 4). This procedure increased the number of salt bridges and hydrogen bonds that are summarized in Table II. The data show that after the MD, the PC–PC interaction surface shows an important hydrophobic component in comparison with the PE–PE or PC–PE interfaces in which besides the hydrophobic component, interfaces show higher number of salt bridges and hydrogen bonds.
Figure 4.
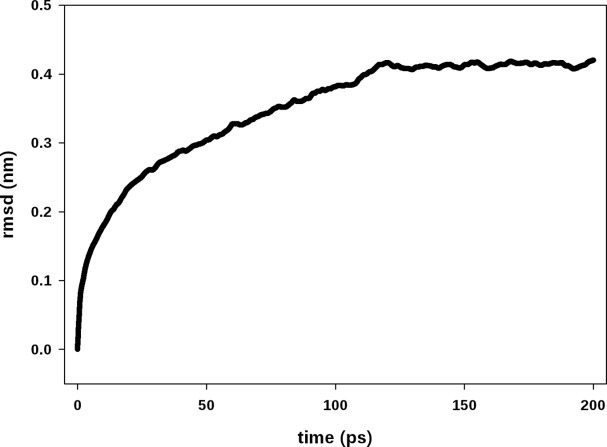
Root mean square deviation (rmsd) of the backbone along the molecular dynamic simulation. The structural convergence of the model was monitored following the rmsd of backbone, comparing the structure present at each frame with the starting coordinates. After 120 ps of simulation, there are no more changes, and the model was considered stable.
Table II.
| Characteristic/interface | PCII–PCI | PCII–PEI | PEI–PEII |
|---|---|---|---|
| Interaction surface (A2) | 2655 | 2447 | 2281 |
| aa residues (No.) | 287 | 252 | 254 |
| % Hydrophobic residues | 33 | 33 | 37 |
| % Hydrophobic area | 28.6 | 36.8 | 43.6 |
| Salt bridges (No.)b | 7 (3) | 9 (2) | 6 (1) |
| Hydrogen bonds (No.)b | 36 (31) | 30 (16) | 27 (8) |
Values in parentheses correspond to those observed before molecular dynamic simulation.
ET between chromophores in the antenna
The relaxed model of the rod shown in Figure 3 provided the co-ordinates for each of the 96 chromophores. The energy transfer (ET) constants were calculated for each pair of chromophores, some of the steps involved very low-transfer rates (>250 ps), so they were not considered while analyzing the transfer pathways. A summary of the fastest calculated transfer rates is shown in Table III and Figure 5.
Table III.
Main Transfer Rates in Pico Seconds (ps) Between Pairs of Chromophores in an Antenna of Two Hexameric Phycoerythrins and Two Hexameric Phycocyanins of a Phycobilisomea, Shown for One-Third of the Antennab
| 1 | Chromophore | PE (ps) intratrimer | PE (ps) intertrimer | PE/PE (ps) interface | PE/PC (ps) interface | PC (ps) intratrimer | PC (ps) intertrimer | PC/PC (ps) interface |
|---|---|---|---|---|---|---|---|---|
| K → P(a) | α82/β82 | 7 | ||||||
| K ← K(b) | α82/α139 | 35 | ||||||
| P → P(c) | β50/β82 | 104 | ||||||
| P → P(d) | β50/β158 | 43 | ||||||
| P → A(e) | β50/α 82 | 69 | ||||||
| P → B(f) | β82/β 82 | 85 | ||||||
| A → P(g) | α139/β158 | 66 | ||||||
| A → P(h) | α82/β50 | 89 | ||||||
| B → N(i) | β82/β50 | 65 | ||||||
| B → N(j) | β82/β82 | 142 | ||||||
| D → C(k) | β82/α82 | 27 | ||||||
| E → F(l) | α82/β84 | 63 | ||||||
| E → A(m) | α82/α82 | 47 | ||||||
| D → A(n) | β82/α82 | 97 | ||||||
| C → B(o) | β82/β84 | 24 | ||||||
| A ←→ F(p) | α84/β84 | 2 | ||||||
| F ←→ F(q) | β153/β84 | 240 | ||||||
| C → K(r) | α82/α82 | 32 | ||||||
| B → K(s) | β153/α82 | 135 | ||||||
| L → P(t) | α82/β84 | 177 | ||||||
| P → L(u) | β84/β84 | 5 | ||||||
| K → L(v) | α82/β84 | 65 | ||||||
| M → K(w) | α82/α82 | 230 |
On the left, the names of the subunits are indicated as well as a letter in parenthesis that identify each path in Figure 4. The chromophores involved in the transition are also indicated in the table.
The complete molecule can be rebuilt by symmetry and also a complete picture of the energy transfer possibilities.
Abbreviations: APC, allophycocyanin; PBS, phycobilisome; PC, phycocyanin; PE, phycoerythrin; PEB, phycoerythrobilin; PCB, phycocyanobilin; PUB, phycourobilin.
Figure 5.
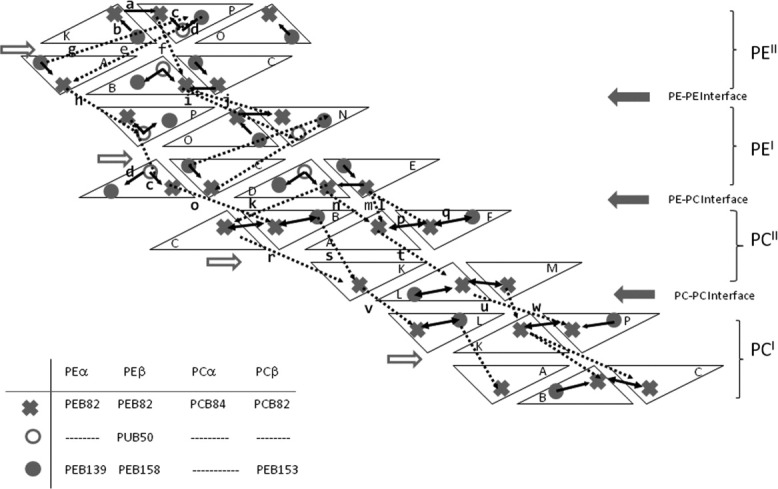
Representation of one-third of the antenna. Each subunit is represented as a triangle. The names of the subunits are indicated with letters: A, C, E, K, M, and O for α subunits and B, D, F, L, N, and P for β subunits. The solid arrows represent intratrimer paths. The intertrimer paths are represented by discontinuous arrows. An identification letter was added to the paths that are drawn in the figure and correlate them with the transfer rates indicated on Table III.
ET in PE
From the analysis of the transfer rates, it comes out that PEBβ82 from the upper trimer in one R-Phycoerythrin (PE) hexamer transfers to PEBβ82 in the lower trimer (path f), also PUBβ50 transfers to PEBα82 in the lower trimer (path e) and PEBα139 to PEBβ158 from the lower to the upper trimer (path g); the ET interhexamers can be observed from PEBα82 in PEII to PUBβ50 in PEI (path h) and from PEBβ82 in PEII to PEBβ82 and PUBβ50 in PEI (path j and path i, respectively). In Figure 5, the paths are represented only once on behalf of the clarity of the scheme.
ET rates from PE to PC
From PEBβ82 from R-Phycoerythrin (PE)I, the energy is transferred to PCBα82 (path k) and PCBβ84 in R-Phycocyanin (PC)II (path n) and from PEBα82 and PEI, the energy is transferred to PCBα82 (path m) and PCBβ84 in PCII (path l).
ET rates in PC
Once arrived to phycocyanin, the energy is transferred to the second trimer from PEBβ153 to PCBα82 (path s) and from PCBα82 to PCBα82 (path r) or PCBβ84 (path t). Between R-Phycocyanin (PC) hexamers, the energy is transferred from PEBα82 in PCII to PCBβ84 in PCI (path v) and to PCBα82 in PCI (path u), also from PCBβ84 in PCII to PCBβ84 in PCI (path w).
Also, some slower transfer rates are indicated in Figure 5 according to the values given in Table III, in order to show the possible flow of the energy along the rod. These ET paths always point to the transfer from the erythrobilins more exposed to the solvent to the chromophores facing the center of the rod, which are able to funnel the energy along the rod.
Discussion
In the present manuscript, we have built a minimum model of antenna of a phycobilisome. To achieve this, we used the combination of experimental and bioinformatic techniques. First, the isolation and spectroscopic characterization of the antennas from phycobilisomes showed a composition of three to four phycobiliproteins, highly enriched in PC and PE, without the presence of APC, the phycobiliprotein found only in the core of the phycobilisome.1, 21 The size of the subcomplexes is in the range of those reported for Porphyridium cruentum. A review published by Adir22 reported that, in general, the rods present a diameter of 110 Å and the high for each hexamer is 55 Å. Second, using a molecular-docking approach, we could ensemble in silico an antenna composed by 2 PEs and 2 PCs, model in agreement with electron microscopy results and with the spectroscopic characterization. The molecular-docking approach considers the protein as a rigid body because of the computational cost; however, it was necessary to perform a posterior relaxation of the system to achieve a more realistic model. In this way, the molecular dynamic simulation showed to be an excellent complementary approach for the rigid molecular docking, because interatomic distances in the rigid model shorter than the standard values were corrected by the procedure.23, 24 The final model shows an improvement at interaction surfaces level, specially an increase in the number of hydrogen bond and salt bridges.
The final model of the antenna allowed us to obtain the 3D co-ordinates of each chromophore. This information, in combination with the experimental parameters from the literature for Phycocyanin and the parameters recently determined in our laboratory for Phycoerythrin,25 was used to determine the transfer rate constants among the chromophores in the antenna, applying the Förster approach. The Förster approach used for the ET rates calculated (shown in Table III) is supported by experimental results for different subcomplexes of PBS as reported earlier, but also by the theoretical studies performed by Womick et al.,26, 27 who compared simulations for PE and C-Phycocyanin using a delocalized approach modified radiated transfer (MRT) and a localized approach (Förster). In both cases, the results were very similar, and the transfer rates were in the range of 100 fs–10 ps, toward the chromophores located at the center of the ring. The same group27 also reported an ET process in PE from PUB to PEB of 2.5–3 ps. Our results shown in Table III agree with the experimental results informed by Chen et al.28 for Phycoerythrin with ET rates from PEBα84 to PEBβ84 of 1–2 ps, and α84 to β84 from the next heterodimer of 30–40 ps, they are also consistent with the experimental values obtained for Phycocyanin from Anabaena variabilis, measured by time-resolved fluorescence and anisotropy spectra.29 These values are consistent with our results, which gave a ET rate of 2 ps, and in the trimer, our calculations also agree in time transfer rates in the vicinity of 50–100 ps.
ET rates in Phycocyanin and Phycocyanin complexes have been reported in literature previously. For example, a preliminary study of the ET pathways was reported,5 detecting differences, mainly due to the application of the Förster extended approach and because Gracilaria chilensis Phycocyanin contains erythrobilin at position 153 in the β subunit. In Ref. 6, we proposed two fast pathways in a complex formed by two hexamers of phycocyanin, one internal path involving chromophores in α82 and β84 and one external path involving chromophores at β153. The same pathways were also observed in reference for C-phycocyanin from Fremyella diplosyphon.30 The main differences specifically for PC from Gracilaria chilensis are in the external pathways proposed earlier, because the chromophore involved is now erythrobilin, which was not evidenced in the three-dimensional structure, and it was considered as cyanobilin. In this model, it is not possible to detect two clear pathways but a participation of most of the chromophores in the ET along the antenna toward PCBβ82. Experimental and theoretical studies12, 26, 31 on C-PC always point to the energy being funneled toward PCBβ82 from where the energy should be transferred to APC. Also, in PE, the energy is channeled toward PEBβ82; in both cases, these chromophores are facing the center of the hexamer. The fact that the γ subunits (one or two depending on the specie), associated to Phycoerythrin, are also chromophorylated and that its location is also proposed to be at the center of R-PE20 make us think the possibility that its presence may contribute to the transfer process. This idea is interesting, because, in R-PE, our results using the Förster approach show slow transfer rates interhexamers, and the presence of a chromophorylated molecule in the center of the ring could be the natural solution to preserve the efficiency of the system. Our results on the ET rates calculated for phycoerythrin were not comparable with other theoretical results, because only here the extended Förster approach was used, using the spectroscopic parameters reported by us.25 Nevertheless, our calculations are also in the order of magnitude of those reported by Chen et al.28
The analysis presented here combines theoretical and experimental tools and provides valuable information about the possible preferential pathways considering only the phycobiliprotein components of the phycobilisome.
Materials and Methods
Purification and characterization of rod-enriched fractions obtained from phycobilisomes from Gracilaria chilensis
Rods were obtained from purified phycobilisomes3, 19 obtained in 0.9M phosphate buffer pH 7. Phycobilisomes were dissociated by lowering the buffer concentration to 0.3M in the same buffer, while their fluorescence spectra were recorded. Emission at 634 nm (Phycocyanin) was observed after excitation at 566 nm, maximum absorption λ for phycoerythrin. The fractions were purified by fast protein liquid chromatography (FPLC) using gel filtration chromatography, using a Pharmacia Superdex 200 26/60 column; the fractions were characterized spectroscopically by analyzing the emission at 634 nm, corresponding to phycocyanin, after excitation at 566 nm, maximum absorption λ for phycoerythrin. The general characterization of the fractions was performed using a Shimadzu spectrofluorophotometer RF-5301PC. The rod-enriched fractions were selected by their spectroscopic characteristics (Fig. 2) and pooled to be used for observation by transmission electron microscopy.
Transmission electron microscopy
The carbon-coated Cu/Rh grids were irradiated by UV light for 5 min. Then a drop with the sample was deposited on the grid, fixed with glutaraldehyde 0.5% during 5 min, and washed with nanopure water; the staining was performed with 2% uranyl acetate for 2 min and observed in a JEOL/JEM1200 ExII electron microscope.
A model for a rod
The construction of the model was made in two steps: rigid molecular docking and molecular dynamic simulation to relax the system.
Rigid molecular docking
The co-ordinates of the three-dimensional structures of PE (PDB code: 1eyx) and PC (PDB code: 2bv8) were used to build a model for a rod formed by two hexamers of phycoerythrin and two hexamers of phycocyanin (PEII–PEI–PCII–PCI). The model was built as pairs; three different docking models were built: Phycoerythrin–Phycoerythrin (PEII–PEI); Phycoerythrin–Phycocyanin (PEI–PCII); and Phycocyanin–Phycocyanin (PCII–PCI), using a docking procedure performed with the software ZDOCK23, 24 with angular steps of 6°. The docking models were scored by the program, considering desolvation, and the electrostatic and hydrophobic contributions.32–34 For each pair, the first 500 models were evaluated. To evaluate the models, a visual inspection was accomplished to select those in agreement with the electron micrographs [Fig. 2(B)]; 15 complexes were selected from the PC–PC docking, 5 complexes from the PC–PE docking, and 8 complexes from the PE–PE docking with similar disposition. The selected complexes were evaluated analyzing its interaction surfaces, using the protein–protein interaction server ProtorP35 and by the number of H-bonds in the interface, using HBPLUS.36 The best complexes of each docking pair PE–PE, PE–PC, and PC–PC were selected and then assembled by fitting the redundant protein, using the software Swiss PDB Viewer37 to obtain a rigid rod model formed by two hexamers of PE and two hexamers of PC.
Molecular dynamics
A molecular dynamic protocol for the rigid rod model was performed in order to optimize the interaction area among phycobiliproteins by increasing the number of salt bridges and hydrogen bonds. This procedure was performed using the software GROMACS38, 39 and the OPLS/AA force field,40, 41 in which the topologies for the chromophores were added. The rigid rod was situated at the center of a box filled with water molecules (SPC model) and Na+ ions to equilibrate the system charge. An energy minimization through a steepest descent protocol implemented in GROMACS was performed as starting relaxation step, followed by a short molecular dynamic simulation of 20 ps with position restraint for the protein atoms in order to equilibrate the solvent. After the solvent equilibration, a full molecular dynamic simulation was performed. The total simulation time was 200 ps heating from 288 to 303 K in 10 ps, keeping this temperature for 140 ps, then cooling until 288 K in 40 ps, and maintaining at 288 K for 10 ps to produce the convergence of the system. To determine the structural convergence, the rmsd was monitored for the backbone of the proteins. After MD simulation, the system was subjected to a new energy minimization by steepest descent. The final model was considered as the rod model.
ET calculations
The co-ordinates of each of the 96 chromophores were obtained from the docking model, and using the protocol previously developed,29 applying the Förster theory for the resonance energy transfer (ET), it was possible to calculate the ET constants between every pair of chromophores and to propose preferential pathways through the antenna. In summary, the method consists in the calculation of transfer constants kDA = CGSI for every pair of chromophores, where C is a collection of constants, S are the spectroscopic properties of the interacting chromophores, I is the overlap integral between the emission and absorption spectra, and G is a geometric factor defined as κ2DA/R6DA. RDA is the distance between the donor–acceptor center of masses of the conjugate system of the chromophores, and κDA is the dipole orientation coefficient. The experimental values for phycocyanobilins were obtained from Ref. 42, and for phycourobilin and phycoerythrobilin, the experimental values were obtained from Ref. 24. ET steps with constants higher than 20 and 10 ns−1 (transfer times shorter than 50 and 100 ps) were used to propose molecular preferential ET pathways. The calculation was performed for every pair of chromophores.
Acknowledgments
We are grateful to FONDECYT and Universidad de Concepción in the context of the project FONDECYT: 108.0267 and project DIUC: 211.037.012-1.0
Glossary
Abbreviations
- APC
allophycocyanin
- PBS
phycobilisome
- PC
phycocyanin
- PE
phycoerythrin
- PEB
phycoerythrobilin
- PCB
phycocyanobilin
- PUB
phycourobilin
References
- 1.Glazer AN. Phycobilisomes: structure and dynamics. Annu Rev Microbiol. 1982;36:173–198. doi: 10.1146/annurev.mi.36.100182.001133. [DOI] [PubMed] [Google Scholar]
- 2.Mulder CL, Theogarajan L, Currie M, Mapel JK, Baldo MA, Vaughn M, Willard P, Bruce BD, Moss MW, McLain CE, Morseman JP. Luminescent solar concentrators employing phycobilisomes. Adv Mater. 2009;21:3181–3185. [Google Scholar]
- 3.Bunster M, Tellez J, Candia A. Characterization of phycobiliproteins present in Gracilaria chilensis. Bol Soc Chil Quím. 1997;42:449–455. [Google Scholar]
- 4.Contreras-Martel C, Martinez-Oyanedel J, Bunster M, Legrand P, Piras C, Vernede X, Fontecilla-Camps JC. Crystallization and 2.2 Å resolution structure of R-phycoerythrin from Gracilaria chilensis: a case of perfect hemihedral twinning. Acta Cryst. 2001;D57:52–60. doi: 10.1107/s0907444900015274. [DOI] [PubMed] [Google Scholar]
- 5.Contreras-Martel C, Matamala A, Bruna C, Poo-Caamano G, Almonacid D, Figueroa M, Martinez-Oyanedel J, Bunster M. The structure at 2Å resolution of Phycocyanin from Gracilaria chilensis and the energy transfer network in a PC-PC complex. Biophys Chem. 2007;125:388–396. doi: 10.1016/j.bpc.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Morales M. 2012. In vitro and in silico studies of the stability of Phycocyanin from Gracilaria chilensis, Dissertation for a Biochemistry degree, Universidad de Concepción, Chile.
- 7.Forster T. Mechanism of energy transfer. In: Florkin M, Stolz EH, editors. Comprehensive Biochemistry. Vol. 22. Amsterdam: Elsevier; 1967. pp. 61–80. [Google Scholar]
- 8.Förster T. Zwischenmolekulare energiewanderung und fluoreszenz. Ann Phys. 1948;437:55–75. [Google Scholar]
- 9.Dexter DL. A theory of sensitized luminescence in solids. J Chem Phys. 1953;21:836–850. [Google Scholar]
- 10.Forster T, Sinanoglu O. Modern Quantum Chemistry, Istanbul Lectures, Part 3: action of light and organic crystals. Vol. 3. New York: Academic Press; 1965. Delocalized excitation and excitation transfer; p. 93. [Google Scholar]
- 11.Beljonne D, Curutchet C, Scholes GD, Silbey RJ. Beyond Forster resonance energy transfer in biological and nanoscale systems. J Phys Chem B. 2009;113:6583–6599. doi: 10.1021/jp900708f. [DOI] [PubMed] [Google Scholar]
- 12.Debreczeny MP, Sauer K, Zhou J, Bryant DA. Comparison of calculated and experimentally resolved rate constants for excitation energy transfer in C-Phycocyanin. I. Monomers. J Phys Chem. 1995;99:8412–8519. [Google Scholar]
- 13.Debreczeny MP, Sauer K, Zhou J, Bryant DA. Comparison of calculated and experimentally resolved rate constants for excitation energy transfer in C-Phycocyanin. II. Trimers. J Phys Chem. 1995;99:8420–8431. [Google Scholar]
- 14.Demidov AA, Borisov AY. Computer simulation of energy migration in the C-phycocyanin of the blue-green algae Agmenellum quadruplicatum. Biophys J. 1993;64:1375–1384. doi: 10.1016/S0006-3495(93)81503-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi T, Degenkolb EO, Bersohn R, Rentzepis PM, MacColl R, Berns DS. Energy transfer among the chromophores in phycocyanins measured by picosecond kinetics. Biochemistry. 1979;18:5073–5078. doi: 10.1021/bi00590a008. [DOI] [PubMed] [Google Scholar]
- 16.Nield J, Rizkallah PJ, Barber J, Chayen NE. The 1.45Å three-dimensional structure of C-phycocyanin from the thermophilic cyanobacterium Synechococcus elongatus. J Struct Biol. 2003;141:149–155. doi: 10.1016/s1047-8477(02)00609-3. [DOI] [PubMed] [Google Scholar]
- 17.Padyana AK, Ramakumar S. Lateral energy transfer model for adjacent light-harvesting antennae rods of C-phycocyanins. Biochim Biophys Acta. 2006;1757:161–165. doi: 10.1016/j.bbabio.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Grossman AR, Schaefer MR, Chiang GG, Collier JL. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol Rev. 1993;57:725–749. doi: 10.1128/mr.57.3.725-749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mella C. 2011. Estudio de la arquitectura de un ficobilisoma de Gracilaria chilensis, Dissertation to obtain the Master in Biochemistry and Bioinformatics, Universidad de Concepción, Chile.
- 20.Apt KE, Metzner S, Grossman AR. The γ subunits of phycoerythryn from red alga: position in phycobilisomes and sequence characterization. J Phycol. 2001;37:64–70. [Google Scholar]
- 21.Arteni AA, Liu L, Aartsma TJ, Zhang Y, Zhou B, Boekema EJ. Structure and organization of phycobilisomes on membranes of the red alga Porphyridium cruentum. Photosynth Res. 2008;95:169–174. doi: 10.1007/s11120-007-9264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adir N. Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth Res. 2005;85:15–32. doi: 10.1007/s11120-004-2143-y. [DOI] [PubMed] [Google Scholar]
- 23.Chen R, Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins. 2002;47:281–294. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 24.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 25.Sepúlveda-Ugarte J, Brunet JE, Matamala AR, Martínez-Oyanedel J, Bunster M. Spectroscopic parameters of phycoerythrobilin and phycourobilin on phycoerythrin from Gracilaria chilensis. J Photochem Photobiol A: Chem. 2011;219:211–216. [Google Scholar]
- 26.Womick JM, Moran AM. Nature of excited states and relaxation mechanisms in C-phycocyanin. J Phys Chem B. 2009;113:15771–15782. doi: 10.1021/jp908093x. [DOI] [PubMed] [Google Scholar]
- 27.Womick JM, Liu H, Moran AM. Exciton delocalization and energy transport mechanisms in R-phycoerythrin. J Phys Chem A. 2011;115:2471–2482. doi: 10.1021/jp111720a. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Dang W, Xie J, Zhao J, Weng Y. Ultrafast energy transfer pathways in R-phycoerythrin from Polysiphonia urceolata. Photosynth Res. 2012;111:81–86. doi: 10.1007/s11120-011-9708-3. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Zhao J, Jiang L, Zheng X, Zhao F, Wang H. Studies on the energy transfer among the rod-core complex from phycobilisome of Anabaena variabilis by time resolved fluorescence emission and anisotropy spectra. Biochim Biophys Acta. 1997;1320:285–296. [Google Scholar]
- 30.Matamala AR, Almonacid DE, Figueroa MF, Martinez-Oyanedel J, Bunster MC. A semiempirical approach to the intra-phycocyanin and inter-phycocyanin fluorescence resonance energy-transfer pathways in phycobilisomes. J Comput Chem. 2007;28:1200–1207. doi: 10.1002/jcc.20628. [DOI] [PubMed] [Google Scholar]
- 31.Xie J, Zhao J, Peng C. Analysis of the disk-to-disk energy transfer processes in C-phycocyanin complexes by computer simulation technique. Photosynthetica. 2002;40:251. [Google Scholar]
- 32.Archakov AI, Govorun VM, Dubanov AV, Ivanov YD, Veselovsky AV, Lewi P, Janssen P. Protein-protein interactions as a target for drugs in proteomics. Proteomics. 2003;3:380–391. doi: 10.1002/pmic.200390053. [DOI] [PubMed] [Google Scholar]
- 33.Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci USA. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallone B, Miele AE, Vecchini P, Chiancone E, Brunori M. Free energy of burying hydrophobic residues in the interface between protein subunits. Proc Natl Acad Sci USA. 1998;95:6103–6107. doi: 10.1073/pnas.95.11.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds C, Damerell D, Jones S. ProtorP: a protein-protein interaction analysis server. Bioinformatics. 2009;25:413–414. doi: 10.1093/bioinformatics/btn584. [DOI] [PubMed] [Google Scholar]
- 36.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 37.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 38.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comp. 2008;4:435. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 39.Van Gunsteren WF, Hunenberger PH, Kovacs H, Mark AE, Schiffer CA. Investigation of protein unfolding and stability by computer simulation. Philos Trans R Soc Lond B Biol Sci. 1995;348:49–59. doi: 10.1098/rstb.1995.0045. [DOI] [PubMed] [Google Scholar]
- 40.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 41.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105:6474. [Google Scholar]
- 42.Apt KE, Collier JL, Grossman AR. Evolution of the phycobiliproteins. J Mol Biol. 1995;248:79–96. doi: 10.1006/jmbi.1995.0203. [DOI] [PubMed] [Google Scholar]


