Abstract
KCNK1, a member of the family of two-pore K+ ion channels, is specifically induced in the livers of male mice after phenobarbital treatment. Here, we have determined the molecular mechanism of this male-specific activation of the Kcnk1 gene and characterized KCNK1 as a phenobarbital-inducible antihyperplasia factor. Upon activation by phenobarbital, nuclear receptor CAR binds the 97-bp response element (−2441/−2345) within the Kcnk1 promoter. This binding is observed in the livers of male mice, but not in the livers of female mice and requires the pituitary gland, because hypophysectomy abrogates it. Hyperplasia further progressed in the livers of Kcnk1 −/− male mice compared with those of Kcnk1 +/+ males after phenobarbital treatment. Thus, KCNK1 suppresses phenobarbital-induced hyperplasia. These results indicate that phenobarbital treatment induces KCNK1 to elicit a male-specific and growth-suppressing signal. Thus, KCNK1 and Kcnk1 −/− mice provide an experimental tool for further investigation into the molecular mechanism of CAR-mediated promotion of the development of hepatocellular carcinoma in mice.
Key Words: phenobarbital, CAR, KCNK1, hepatic hyperplasia
Phenobarbital, a clinically used sedative, is a nongenotoxic carcinogen that promotes development of both spontaneous and chemically induced hepatocellular carcinoma (HCC) in rodents (Diwan et al., 1986; Preat et al., 1986). Phenobarbital promotion of HCC development is both sex dependent and strain dependent in mice; males predominantly develop HCC, and C3H/HeNCrlBR mice are tumor sensitive but C57BL/6 mice are tumor insensitive (Blanck et al., 1986; Diwan et al., 1986; Poole and Drinkwater, 1996). Phenobarbital promotes HCC development through activation of the constitutive active/androstane receptor (CAR; NR1I3), a member of the nuclear steroid/thyroid hormone receptor superfamily (Yamamoto et al., 2004). CAR represses its high constitutive activity by phosphorylating threonine 38 in the mouse liver. Phosphorylated CAR, which is sequestered in the cytoplasm, is dephosphorylated and activated to translocate into the nucleus after phenobarbital treatment (Mutoh et al., 2009). Activated CAR forms heterodimer with retinoid X receptor alpha (RXRα) and regulates expression of various genes, thereby altering hepatic functions such as drug and energy metabolism (Honkakoski et al., 1998; Kawamoto et al., 1999; Kodama and Negishi, 2006; Wada et al., 2009; Zelko and Negishi, 2000). Activated CAR also mediates induction of hepatic hypertrophy and hyperplasia in mice, which can result in the HCC development in mice (Huang et al., 2005; Yamamoto et al., 2004). Because CAR is equally activated in both sexes and strains after phenobarbital treatment, it has been speculated that phenobarbital-activated CAR regulates hepatic genes in male- and strain-specific manners. These genes may play critical roles in developing hepatic hypertrophy and/or hyperplasia as well as the subsequent HCC development. Various genes were previously suggested as candidates responsible for phenobarbital-promoted HCC development, such as Mmd2, Foxm1b, Gadd45β, and c-Myc (Blanco-Bose et al., 2008; Huang et al., 2005; Kalinichenko et al., 2004; Yamamoto et al., 2010). However, until now no such gene with this specificity and dependency has been known. Identifying a gene that is regulated in sex-specific and strain-dependent manner should help us to investigate the molecular mechanism of how CAR promotes HCC development. Now, for the first time, the two-pore K+ channel KCNK1 is identified to be specifically induced in the livers of male mice and in tumor-susceptible C3H/HeNCrlBR rather than in tumor-resistant C57BL/6 mice after phenobarbital treatment. Given these findings, we have investigated the molecular mechanism by which CAR regulates this male-specific activation of the Kcnk1 gene in mouse liver.
KCNK1 is a member of the family of two-pore K+ ion channels (Es-Salah-Lamoureux et al., 2010; Lotshaw, 2007). KCNK channel proteins function as regulator of cell volume, apoptosis, and proliferation and are involved with the development of various diseases. For example, estrogen induces KCNK5 to enhance proliferation of breast cancer cells (Alvarez-Baron et al., 2011). Reduced KCNK1 levels in cardiac atria have been associated with chronic atrial fibrillation in humans (Ellinghaus et al., 2005). KCNK1 has also been characterized as an essential suppressor of anchorage-independent growth in fibroblast (Beitzinger et al., 2008). These observations prompted us to investigate the role of KCNK1 in phenobarbital-induced hepatocyte growth.
In this study, utilizing real-time PCR, Western blots, and laser capture microdissection, we describe specific induction of KCNK1 in zone 3 of livers of male mice after phenobarbital treatment. Transient transfection and chromatin immunoprecipitation (ChIP) assays were performed to determine the molecular mechanism by which CAR specifically activates the Kcnk1 gene. Subsequently, Kcnk1 −/− mice were utilized to characterize KCNK1 as a phenobarbital-induced hepatocyte growth suppressor. Here, we present the hypothesis that KCNK1 is induced as a hepatocyte growth suppressor throughout the course of HCC development.
MATERIALS AND METHODS
Materials.
Phenobarbital was obtained from Sigma-Aldrich (St Louis, MO). Continuously releasing pellets of bromodeoxyuridine (BrdU; 10mg/pellet, 21 days release) were obtained from Innovative Research of America (Sarasota, FL). Antibodies against CAR and hepatocyte nuclear factor 4 alpha (HNF4α) were obtained from Perseus Proteomics (Tokyo, Japan). Antibodies against RXRα and laminB, normal IgG, and horseradish peroxidase–conjugated antimouse or antirabbit IgG were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody against caveolin was obtained from BD Biosciences (San Jose, CA). Antibody against KCNK1 peptide ASQSPPYEDGSADH (corresponding to residues 323–336 of mouse KCNK1) was produced by New England Peptide (Gardner, MA). Protease inhibitor cocktail (Complete Mini) was obtained from Roche Diagnostics (Mannheim, Germany). Expression plasmids for mouse CAR and human HNF4α were constructed as described previously (Inoue and Negishi, 2009). The reporter gene constructs of −2.6kb Kcnk1-Luc (2.6k), −2.1kb Kcnk1-Luc (2.1k), and −2.6kb Δ2.5/2.3kb Kcnk1-Luc (2.6kΔ2.5k/2.3k); −2.6kb ΔDR4 Kcnk1-Luc (ΔDR4), −2.6kb ΔDR1a Kcnk1-Luc (ΔDR1a), and −2.6kb ΔDR1b Kcnk1-Luc (ΔDR1b); and −2.6kb ΔDR1s Kcnk1-Luc (ΔDR1s) were produced as follows: a fragment of 2.6kb of Kcnk1 promoter was amplified from mouse genomic DNA and was cloned into pcDNA2.1-TOPO (Life Technologies, Carlsbad, CA). Subsequently, the fragment was subcloned into reporter plasmid pGL3-basic (Promega, Madison, WI), generating a reporter construct of −2.6kb Kcnk1-Luc. Other reporter constructs were produced by site-specific amplification using PCR.
Animals.
Adult (8–10 weeks) mice were used for all experiments. C3H/HeNCrlBR (Car +/+) male and female mice, and sham, hypophysectomized and gonadectomized C3H/HeNCrlBR male and female mice were purchased from Charles River Laboratories (Wilmington, MA). Car −/− C3H/HeNCrlBR (Car −/−) male and female mice were bred in our institute. Car −/− mice were established by cre-mediated deletion of exon 1 and then backcrossed to C3H/HeNCrlBR mice as described previously (Yamamoto et al., 2004). C57BL/6 (Kcnk1 +/+) male mice were purchased from Jackson Laboratory (Bar Harbor, ME). Kcnk1 −/− C57BL/6 (Kcnk1 −/−) male mice were kindly provided by Dr Jacques Barhanin (Nie et al., 2005) and were bred in our institute. All mice were maintained in a temperature- and light-controlled facility and had free to access to water and diet. All animal procedures were approved by the Animal Ethics Committee NIEHS, National Institutes of Health.
Treatments.
In all animal experiments, mice of each group were randomly divided into two groups and treated with vehicle control (PBS) or phenobarbital at a dose of 50 (only for BrdU assay) or 100mg/kg body weight (for all other experiments). Phenobarbital was given by ip injection. Because nuclear accumulation of CAR was reached plateau at 3h, we employed 3h treatment for Western blot of nuclear extract and ChIP assay. Twenty-four hour of phenobarbital treatment is well-established time frame for induction of mRNA and protein expression levels. To acquire clear staining, we employed 5 and 7 days phenobarbital treatment for immunohistochemistry and BrdU assay, respectively.
Real-time quantitative PCR.
Total liver RNAs were isolated from mouse livers treated with phenobarbital for 24h using Trizol Reagent (Life Technologies). Each zone of liver RNAs was collected by laser capture microdissection and isolated from mouse liver treated with phenobarbital for 24h using Picopure RNA Extraction Kit (Life Technologies). The areas of zones 1 and 3 were defined as a three-layer hepatocyte surrounding the portal vein and central vein, respectively. cDNAs were prepared using a High Capacity cDNA Archive Kit (Life Technologies). Real-time PCR was performed with the 7900HT Fast Real-Time PCR System (Life Technologies) with the following TaqMan probes or primers: CYP2B10 (Mm00456591_m1), KCNK1 (5′-TGCTCTCCACCACAGGCTATG-3′ and 5′-AGATGATGCAGAAGGCTTTGC-3′ or Mm00434624_m1). The reverse primer for detection of KCNK1 mRNA was designed within exon 2, resulting in no signal detection of KCNK1 mRNA in Kcnk1 −/− mice. TaqMan Rodent GAPDH Control Reagents (Life Technologies) was used as an internal control and to normalize expression levels of all other genes.
Western blot.
Cell membrane fractions were prepared from mouse livers treated with phenobarbital for 24h by the Mg2+ precipitation method as described previously (Konno et al., 2010). Briefly, mouse livers were homogenized with homogenization buffer (300mM mannitol, 5mM EGTA, 12mM Tris/HCl pH 7.1). MgCl2 was added to a final concentration of 10mM and the homogenate was centrifuged for 15min at 2300 × g. The supernatant was collected and centrifuged for 30min at 21,000 × g. The resulting pellet was cell membrane fraction. Nuclear extracts were prepared from mouse liver treated with phenobarbital for 3h as described previously (Honkakoski et al., 1998). Briefly, mouse livers were homogenized with homogenization buffer (0.3M sucrose, 10mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.6, 0.74mM spermidine, 0.15mM spermine, 0.1mM EDTA, 0.1mM EGTA, 10mM KCl, 1mM dithiothreitol (DTT), 0.5mM phenylmethanesulfonylfluoride (PMSF), and protease inhibitor cocktail). The nuclei were precipitated by centrifugation at 80,000 × g. The nuclear extracts were extracted from nuclei pellet with extraction buffer (12.5% glycerol, 2mM HEPES pH 7.6, 116mM KCl, 3mM MgCl2, 0.1mM EDTA, 1.0mM DTT, 0.1mM PMSF, and protease inhibitor cocktail). Proteins were separated on a 10% SDS-polyacrylamide gel and transferred to polyvinylidene fluoride membrane (GE Healthcare, Pittsburgh, PA). The membrane was incubated with given primary antibody. Horseradish peroxidase–conjugated antimouse or antirabbit antibody was used as the secondary antibody. Protein bands were visualized using ECL Plus Western blotting detection reagent (GE Healthcare).
Immunohistochemistry.
Paraffin-embedded liver sections (6 µm thick) were prepared from mice treated with phenobarbital for once a day for 5 days and subjected to immunohistochemistry with given primary antibody as described previously (Yamazaki et al., 2011). Briefly, the sections were deparaffinized, soaked into H2O2 for blocking endogenous peroxidase, and subjected to heat-induced epitope retrieval. Subsequently, the sections were stained with Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA) and anti-KCNK1 antibody. The slides were then counterstained with hematoxylin, dehydrated, and coverslipped.
Cell-based reporter gene assay.
HepG2 and COS1 cells were maintained in minimal essential medium and Dulbecco’s modified Eagle’s medium, respectively, supplemented with 10% (vol/vol) fetal bovine serum, antibiotics (100U/ml penicillin and 100 µg/ml streptomycin), and 2mM glutamine. Because HepG2 but not COS1 cells endogenously express HNF4α, HepG2 cells were used to examine CAR-mediated activation of the reporter gene and COS1 cells were used to examine HNF4α-mediated activation of reporter gene. Cells were placed on a 24-well plate at a density of 4 × 105 cells/well. Cells were transfected with reporter plasmids (200ng/well), expression plasmids (20ng/well), and phRL-TK plasmid (10ng/well, Promega) by Lipofectamine 2000 (Life Technologies). After 36h transfection, cells were collected and subjected to measurement of reporter activity as described previously (Inoue and Negishi, 2009). Briefly, the reporter activity was measured using Dual-Luciferase reporter assay system (Promega) by normalizing luciferase activity to Renilla luciferase activity.
ChIP assay.
Chromatin shearing was carried out using fresh mouse liver treated with phenobarbital for 3h as described previously (Saito et al., 2010). Briefly, the mouse liver tissue (1g) was collected, chopped on ice, and then crosslinked by 1% formaldehyde for 10min. Subsequently, crosslinking was quenched by glycine and crosslinked liver tissue was homogenized with PBS supplemented with PMSF. The crosslinked chromatin was then isolated from the homogenate by centrifugation and sonicated to produce sheared chromatin. The distribution of the length of sheared chromatin spanned a size of 200–600bp fragmented DNA. ChIP assay was performed using ChIP-IT Express (Active Motif, Carlsbad, CA) according to manufacturer’s protocol with 15 µg of sheared chromatin and 3 µg of antibody (anti-RXRα and -HNF4α), and normal IgG as negative control. The fragments of the 97-bp response element (RE) and a negative control region (−10kb upstream region from the transcription start site) of Kcnk1 promoter and the phenobarbital responsible enhancer module (PBREM) of Cyp2b10 promoter were amplified by PCR using following primers: 5′-GTAGTCCAGAGGACACACGCAG-3′; and 5′-AACCGAAGCTGCTGACCTTGA-3′ for the 97-bp RE, 5′-TGTGAG GGTCAGAGTCAGCC-3′, and 5′-AAATAAAGCAGCTCTTTGCTAGCC-3′ for a negative control region, and 5′-GCACTCCAGTGACTTAGGAGGA-3′ and 5′-GGAATACTGACCCAAGTTCAGTG-3′ for PBREM.
BrdU assay.
For BrdU assay, continuous releasing pellets of BrdU were implanted into mice 3h before the first injection of phenobarbital. Paraffin-embedded liver sections (6 µm thick) were prepared from mice treated with phenobarbital for once a day for 7 days (BrdU assay) as described in Immunohistochemistry section. BrdU assay was performed as described previously (Moser et al., 2009). Briefly, the sections were deparaffinized, soaked into H2O2 for blocking endogenous peroxidase, and subjected to heat-induced epitope retrieval. Subsequently, the sections were stained with Vectastain Elite ABC Kit and anti-BrdU antibody. The slides were then counterstained with hematoxylin, dehydrated, and coverslipped. The number of BrdU-positive and -negative hepatocytes nuclei was scored within at least five randomly selected fields of each zone (zones 1–3) at ×200 by light microscopy. The areas of zones 1 and 3 were defined as described above. The area of zone 2 was defined as midzone between zone 1 and 3. A total of at least 500 hepatocyte nuclei per each zone were scored from individual liver.
Statistical analysis.
All data were shown at the mean ± SD. The differences in data of real-time PCR and BrdU assay were determined by a one-way ANOVA for all groups followed by pairwise comparisons.
RESULTS
Male-Specific Induction of KCNK1 by Phenobarbital Treatment in Mouse Livers
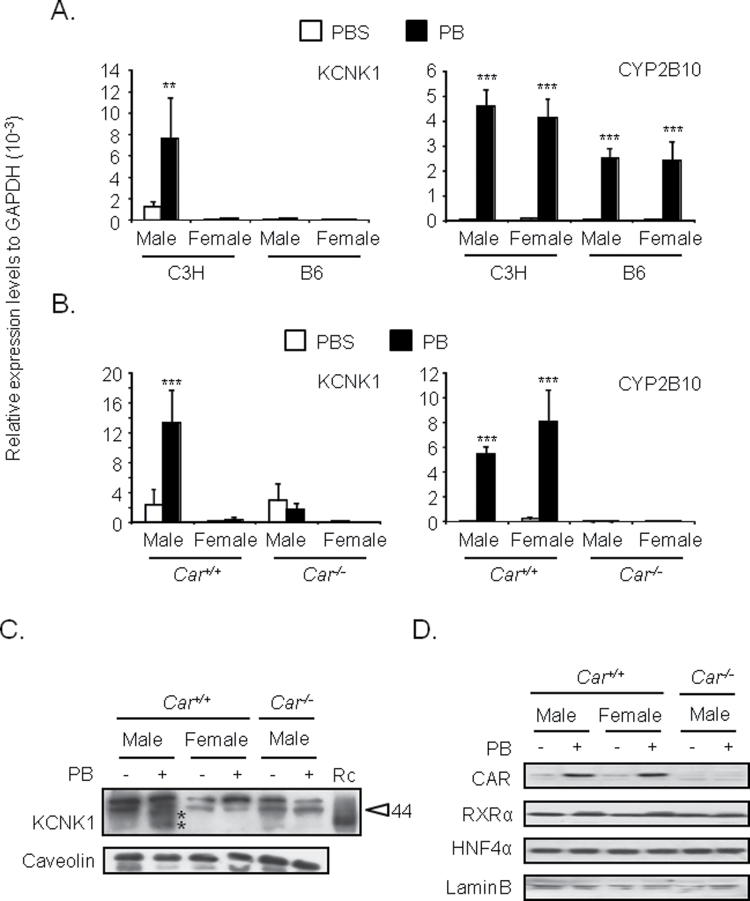
C3H/HeNCrlBR and C57BL/6 males and females were treated with PBS and phenobarbital, from which liver RNAs were prepared for subsequent real-time PCR analysis for KCNK1 mRNA. Phenobarbital treatment significantly increased KCNK1 mRNA in the livers of C3H/HeNCrlBR males but not in the livers of C3H/HeNCrlBR females or C57BL/6 males or females (Fig. 1A). No such sex and strain differences were observed with the classic CAR target CYP2B10 mRNA. The CAR and RXRα mRNA levels remain constant among sexes and strains (Supplementary fig. 1A). Car +/+ and Car −/− male and female mice in C3H/HeNCrlBR backgrounded were employed to confirm that CAR regulated this KCNK1 induction by phenobarbital. The mRNA levels of KCNK1 increased in the livers of Car +/+ male but not in the livers of either Car +/+ female or Car −/− male and female mice (Fig. 1B). The RXRα mRNA levels remain constant between Car +/+ and Car −/− males (Supplementary fig. 1B). The mRNA levels of CYP2B10 were equally increased in the livers of both Car +/+ male and female mice. Cell membrane fractions were also prepared from the livers of these mice and subjected to Western blot analysis. Western blot analysis confirmed that phenobarbital treatment increased protein levels of KCNK1 only in Car +/+ male mice (Fig. 1C). Although glycosylated KCNK1 appears broad band on a SDS gel protein, its estimated molecular size (37 and 40kDa) is consistent with the size reported previously (Lesage et al., 1996). These results indicated that CAR regulated male-specific induction of KCNK1 in mouse liver by phenobarbital treatment. Given the fact that phenobarbital treatment induced accumulation of CAR in the nucleus (Kawamoto et al., 1999), CAR protein levels were examined in the nuclear extracts prepared from the livers of these mice. Western blot analysis determined equal nuclear accumulations of CAR in the livers of both Car +/+ male and female mice (Fig. 1D). Thus, there were no sex-dependent differences in the mechanism by which phenobarbital triggers accumulation of CAR in the nucleus.
FIG. 1.
Male-specific induction of KCNK1 in mouse livers by phenobarbital.
Total RNAs, cell membrane fractions, and nuclear extracts were prepared from mouse livers as described in Materials and Methods section. (A and B) Expression levels of KCNK1 and CYP2B10 mRNA were determined. Values are expressed as the relative expression levels normalized to the expression levels of GAPDH mRNA. Data are mean ± SD (n = 3 or 4 in each group). **p < 0.01, ***p < 0.005 for PBS-treated mice vs. PB-treated mice, Tukey’s multiple comparison test. C3H, B6, and PB denote C3H/HeNCrlBR, C57BL/6, and phenobarbital, respectively. (C) Protein levels of KCNK1 in cell membrane fractions were determined. Protein levels of caveolin were determined as internal control. Data shown are representative of results from two to three individual experiments. The sizes of molecular markers (44kDa) are indicated as arrowhead. * represents putative molecular size of KCNK1 protein. Rc and PB denote bacterially expressed recombinant KCNK1 and phenobarbital. (D) Protein levels of CAR and RXRα in nuclear extracts were determined. Protein levels of laminB were determined as internal control. Data shown are representative of results from two to three individual experiments. PB = phenobarbital.
Zone 3-Specific Induction of KCNK1 in Mouse Livers
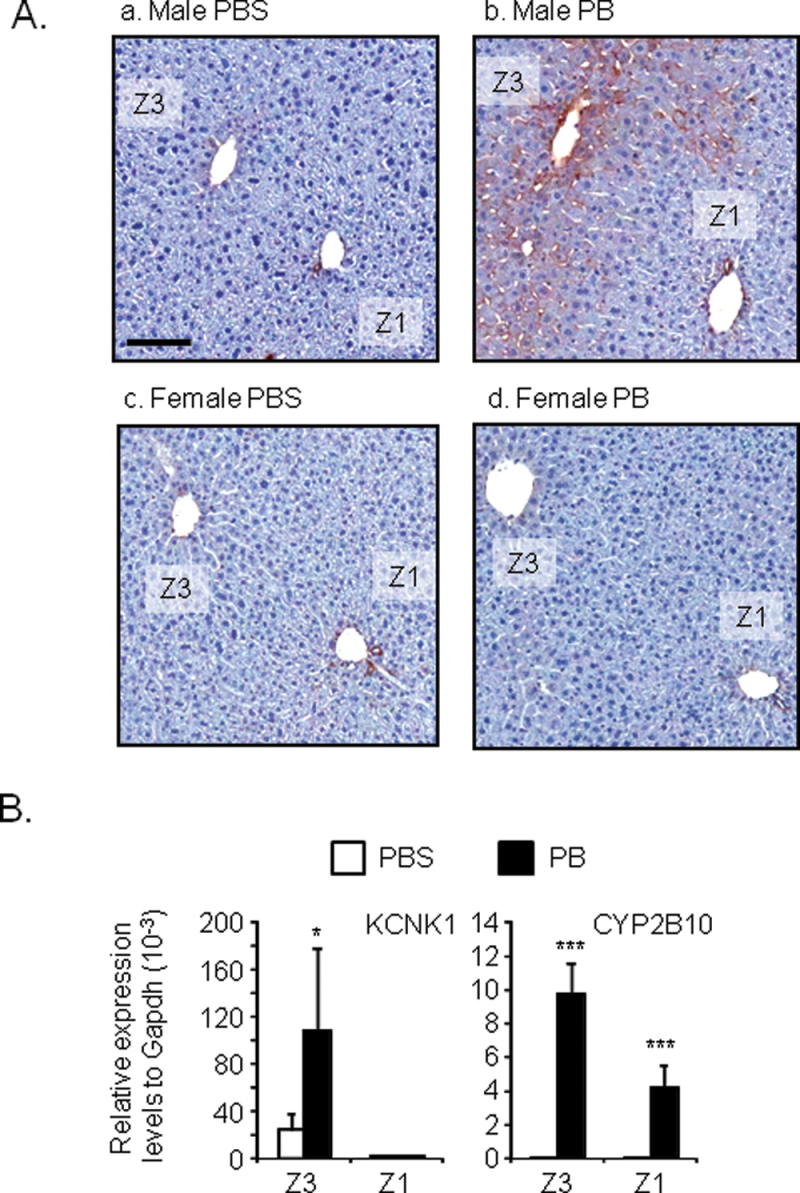
Immunohistochemistry was employed to visualize the localization of KCNK1 expression in mouse livers. Car +/+ male and female mice were treated with phenobarbital or PBS, from which liver sections were prepared and immunostained by an anti-KCNK1 antibody (Fig. 2).
FIG. 2.
Zone 3-specific induction of KCNK1 in mouse livers.
(A) Liver sections were prepared from mouse livers as described in Materials and Methods section. The localization of KCNK1 protein within the liver lobule was determined by immunohistochemistry. Data shown are representative photographs from section slides of three individual mice (original magnification ×200). Z1, Z3, and PB denote zone 1, zone 3, and phenobarbital, respectively. Scale bar = 100 µm. (B). Total RNAs were prepared from samples of laser capture microdissection as described in Materials and Methods section. Expression levels of KCNK1 and CYP2B10 mRNA were determined. Values are expressed as the relative expression levels normalized to the expression levels of GAPDH mRNA. Data are mean ± SD (n = 4 in each group). *p < 0.05, ***p < 0.005 for PBS-treated mice vs. PB-treated mice, Tukey’s multiple comparison test. PB = phenobarbital.
The specificity of this immune-staining was showed using the livers of Kcnk1 +/+ and Kcnk1 −/− mice (Supplementary fig. 2). This antibody lightly stained hepatocytes around central veins (zone 3) of livers of PBS-treated male mice (Fig. 2A). Phenobarbital treatment greatly intensified this staining in zone 3 and increased the area of positive staining toward zone 2 (the area between zone 1 and zone 3). The antibody did not stain hepatocytes in zone 1 of male livers. No staining was observed in hepatocytes in the livers of female mice before and after phenobarbital treatment. Thus, both basal expression and phenobarbital induction of KCNK1 were restricted in zone 3 of male livers. Bile duct epithelial cells in zone 1 were heavily stained in the livers of both male and female mice. Laser capture microdissection was employed to separately prepare hepatocytes from zone 1 and zone 3 of male mouse, from which RNAs were prepared and subjected to real-time PCR analysis. The qualities of zone preparations were evaluated by determination of the mRNA levels of glutamine-ammonia ligase (zone 3 marker) and serine dehydratase (zone 1 marker) (Supplementary fig. 3). Real-time PCR analysis confirmed the specificity of both basal mRNA levels of KCNK1 and phenobarbital induction of KCNK1 in hepatocytes in zone 3 than over hepatocytes in zone 1 of livers of male mice (Fig. 2B). Phenobarbital treatment increased the CYP2B10 mRNA levels in both zone 1 and zone 3.
Pituitary and Gonadal gland–dependent Regulation of Male-Specific KCNK1 Induction
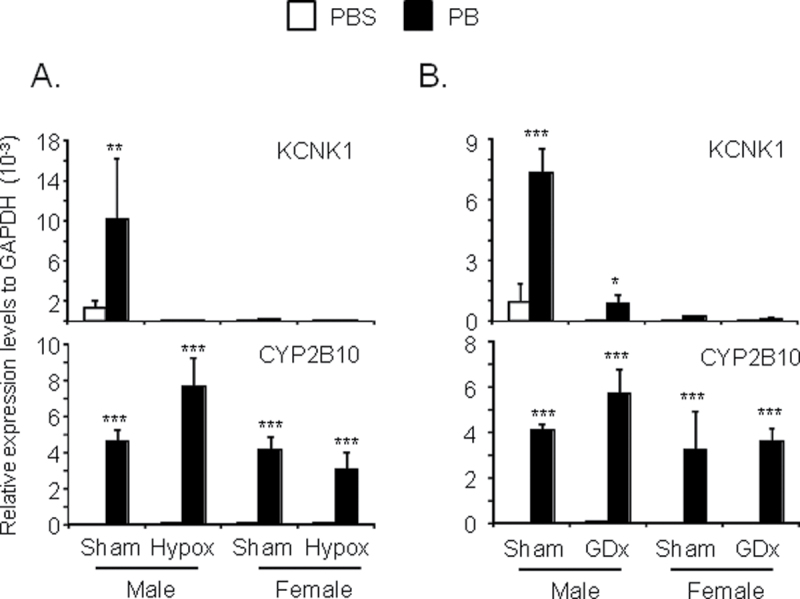
Sexual dimorphic expressions of hepatic genes are often regulated by growth hormone and/or sex hormone secreted from the pituitary and gonadal gland, respectively (Ahluwalia et al., 2004; Erdstein et al., 1984; Kobliakov et al., 1991). First, we utilized hypophysectomized mice to examine whether or not the male-specific induction of KCNK1 by phenobarbital treatment depended on the pituitary gland. Real-time PCR analysis demonstrated that hypophysectomy abolished phenobarbital induction of KCNK1 in the livers of male mice (Fig. 3A). Basal mRNA levels also decreased after hypophysectomy. On the other hand, neither basal nor induced levels of KCNK1 mRNA were affected in the livers of female mice by hypophysectomy. These results indicated that the pituitary gland regulated male-specific induction of KCNK1. However, growth hormone itself did not appear to be involved in this male-specific induction, because replacement of growth hormone was unable to restore induction in the livers of hypophysectomized male mice (data not shown). Second, we utilized gonadectomized mice to examine whether or not phenobarbital induction of KCNK1 was regulated by sex hormones. Real-time PCR analysis demonstrated that castrated male mice retained phenobarbital induction of KCNK1 in the livers (7.5-fold for sham male and 13.5-fold for castrated male) although basal mRNA levels were attenuated by castration (Fig. 3B). Neither basal nor induced levels of KCNK1 mRNA were changed in the livers of ovariectomized female mice. Thus, gonadal glands played no role in regulating male-specific induction of KCNK1. As expected from our observation that phenobarbital induction of CYP2B10 mRNA was not sex specific, neither hyperphysectomy nor gonadectomy affected their levels in livers.
FIG. 3.
KCNK1 induction in the livers of hypophysectomized and gonadectomized mice.
(A and B) Total RNAs, cell membrane fractions, and nuclear extracts were prepared from mouse livers as described in Materials and Methods section. Expression levels of KCNK1 and CYP2B10 mRNA were determined. Values are expressed as the relative expression levels normalized to the expression levels of GAPDH mRNA. Data are mean ± SD (n = 3 or 4 in each group). *p < 0.05, **p < 0.01, ***p < 0.005 for PBS-treated mice vs. PB-treated mice, Tukey’s multiple comparison test. PB, Hypox, and GDx denote phenobarbital, hypophysectomized, and gonadectomized, respectively.
RE of 97bp Within the Kcnk1 Promoter
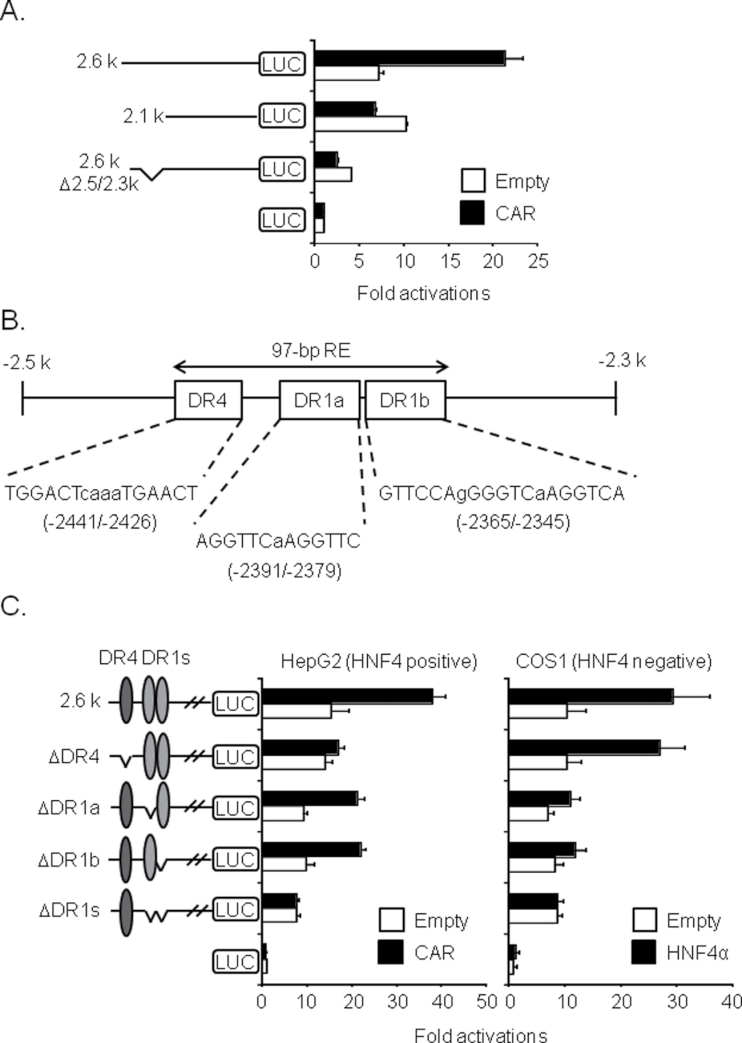
In efforts to determine the molecular mechanism by which CAR regulated the male-specific induction of KCNK1 by phenobarbital treatment, reporter gene assay delineated the CAR RE to the 97-bp DNA sequence (−2441/−2345) within the Kcnk1 promoter. First, CAR activated −2.6kb, but not −2.1kb Kcnk1-Luc reporter gene in HepG2 cells (Fig. 4A). Subsequent internal deletion indicated that the CAR response activity resided within the −2.5/−2.3kb region of the Kcnk1 promoter. This promoter region organizes one DR4 and two DR1 sites into the 97-bp DNA sequence starting with the position −2441 and ending by the −2345 (Fig. 4B). DR4 sites are canonical binding sites for CAR, whereas DR1 sites are canonical binding sites for HNF4α that plays essential role in the activation of the classical CAR target CYP2B6 promoter (Inoue and Negishi, 2009). Thus, these DR sites were internally deleted singularly or simultaneously within the context of the −2.6kb Kcnk1-Luc reporter gene. When DR4 was deleted, CAR no longer activated the −2.6kb Kcnk1-Luc reporter gene in HepG2 cells (Fig. 4C). When only one DR1 was deleted CAR was still able to activate the reporter gene, whereas CAR no longer activated it when both DR1s were simultaneously deleted. HNF4α activated the −2.6kb Kcnk1-Luc reporter gene in COS1 cells even when DR4 site was deleted. On the other hand, when both DR1s were deleted, HNF4α no longer activated the reporter gene. These results indicated that CAR primarily activated the −2.6kb Kcnk1-Luc reporter gene through DR4, while that activation required HNF4α on one of two DR1. Gel shift assay confirmed binding of CAR and HNF4α to DR4 and DR1s, respectively (data not shown). Hereafter, this 97-bp DNA sequence is simply called 97-bp RE.
FIG. 4.
CAR RE of 97-bp within Kcnk1 promoter.
(A and C) Reporter activities of Kcnk1 promoter were determined in HNF4-positive HepG2 and HNF4-negative COS1 cells transfected with or without CAR and HNF4α, respectively. Values are expressed as the relative intensity of Firefly luciferase activity (Kcnk1 promoter) normalized to the intensity of Renilla luciferase activity (internal control). Data are mean ± SD (n = 3 in each group). (B) The core sequences of DR4 site and DR1 sites within the 97-bp CAR RE (97-bp RE) are shown.
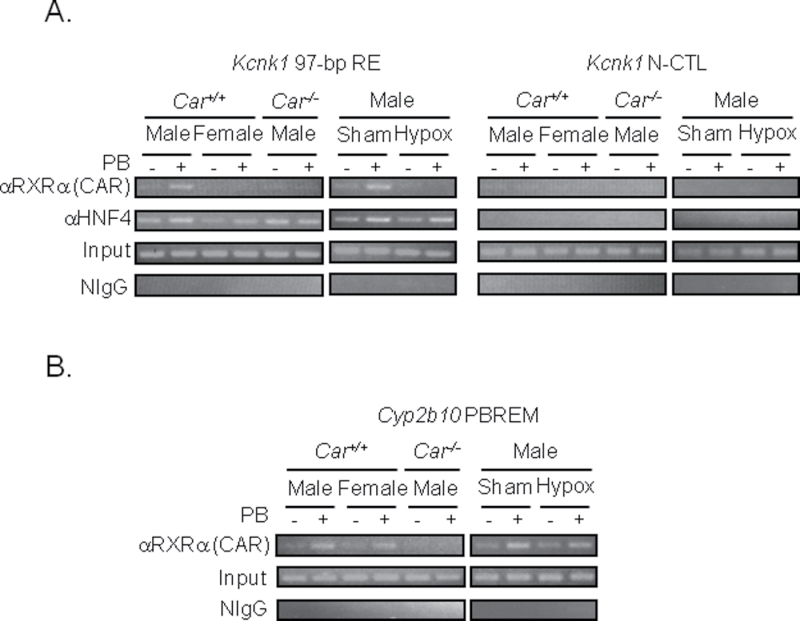
ChIP assay was employed to examine binding of CAR and HNF4α to 97-bp RE of the endogenous Kcnk1 gene in mouse liver. Because no CAR antibody suitable for ChIP assay was available, an anti-RXRα antibody was used to detect a CAR-RXRα complex, and the specificity of this complex was confirmed using the livers of Car −/− mice as negative control. Phenobarbital treatment increased binding of RXRα to 97-bp RE in the livers of male, but not in the livers of female, Car +/+ mice (Fig. 5A). No binding of RXRα was observed with Car −/− male mice, thereby indicating that RXRα binds to 97-bp RE represents as a CAR-RXRα complex. To examine the role of the pituitary gland in the regulation of CAR-RXRα binding to 97-bp RE, ChIP assay was also performed using the livers of hypophysectomized male mice. Hypophysectomy abrogated phenobarbital-induced binding of CAR-RXRα to 97-bp RE. Contrary to the binding of CAR-RXRα, binding of HNF4α to 97-bp RE remained constant throughout the experiments. This constant binding of HNF4α is consistent with constant expressions of HNF4α among sexes, strains and Car −/− (Supplementary fig. 1). No binding of CAR-RXRα and HNF4α to a −10-kb upstream region (negative control region; N-CTL) of the Kcnk1 gene was observed throughout the experiments. These results indicated that phenobarbital treatment triggered binding of CAR-RXRα to 97-bp RE, whereas HNF4α remained constantly bound to 97-bp RE. In addition, the pituitary gland regulated this binding of CAR-RXRα. As shown in Figures 1 and 3, CAR regulated the induction CYP2B10 in the livers of both male and female mice following phenobarbital treatment, and this induction was not regulated by pituitary grand. Consistent with these observations, ChIP assay demonstrated that CAR-RXRα bound to PBREM of the Cyp2b10 gene in the livers of Car +/+ male and female mice but not in the livers of Car −/− male mice, as well as in the livers of hypophysectomized mice (Fig. 5B).
FIG. 5.
Phenobarbital-induced CAR binding to the 97-bp RE in mouse livers.
(A and B) Precipitation of DNA fragments by RXRα and HNF4α was determined. Precipitation of DNA fragments by normal IgG was determined as negative control IgG (NIgG). DNA fragments without immunoprecipitation (Input) were used as positive control. Data shown are representative of results from two to three individual experiments. PB, Hypox, and N-CTL denote phenobarbital, hypophysectomized, and a negative control region, respectively.
Hepatic Hyperplasia in Kcnk1−/− Mice
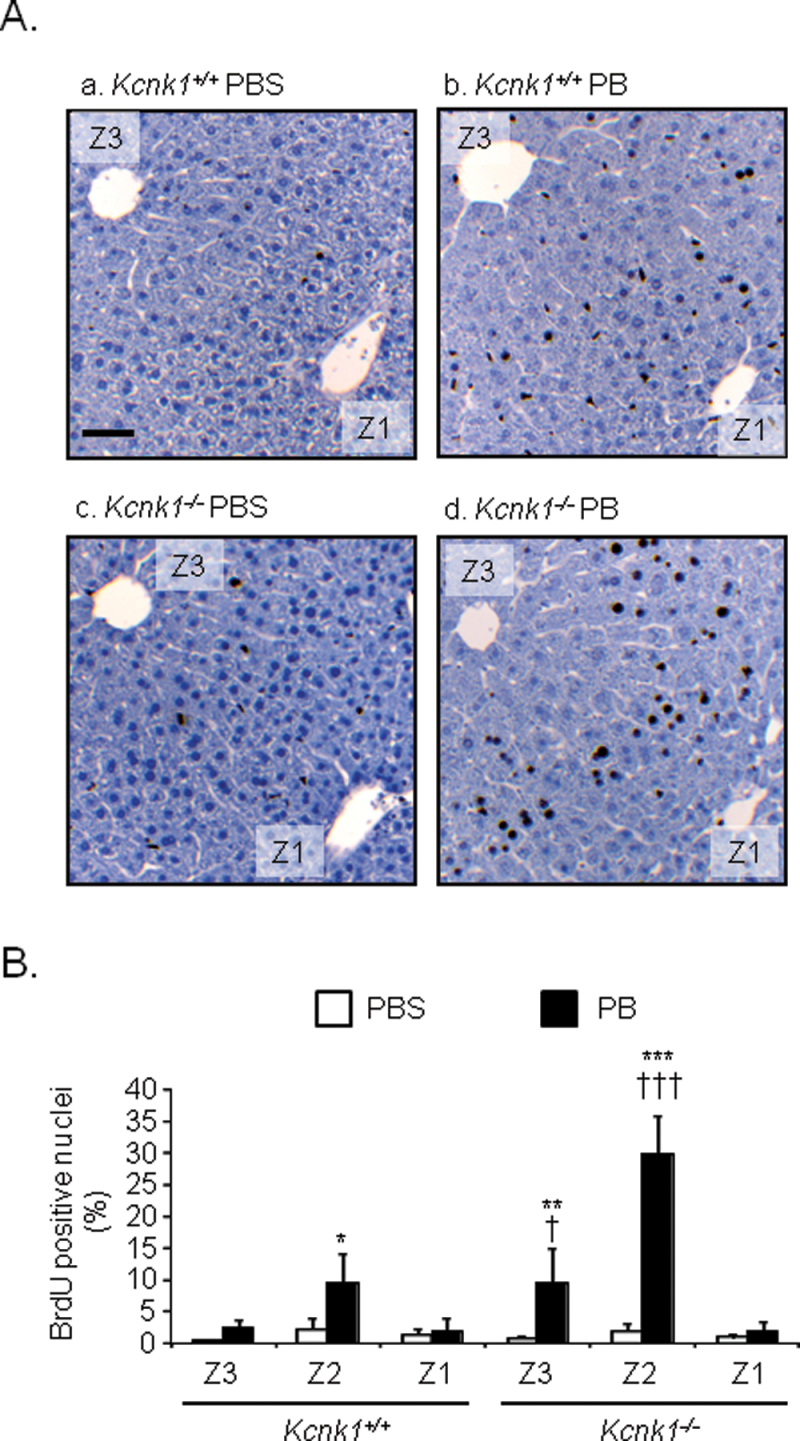
Phenobarbital treatment is known to induce hypertrophy and hyperplasia in the livers of male mice, which could develop into HCC (Blanck et al., 1986; El-Serag and Rudolph, 2007). Given report that KCNK1 suppressed anchorage-independent growth in fibroblast, we utilized Kcnk1 +/+ and Kcnk1 −/− male mice in C57BL/6 backgrounded to examine whether or not KCNK1 played any role in phenobarbital-induced hyperplasia in the liver. To examine the role of KCNK1 in hepatic hyperplasia, BrdU assays were employed. Liver sections, prepared form Kcnk1 +/+ and Kcnk1 −/− male mice implanted with BrdU pellets and subsequently treated with phenobarbital, were immunostained by an anti-BrdU antibody. A majority of BrdU-positive hepatocytes appeared in zone 2 of livers of both Kcnk1 +/+ and Kcnk1 −/− male mice after phenobarbital treatment (Fig. 6A). Actual counts of BrdU-positive hepatocytes revealed that their numbers were similar in the livers of both Kcnk1 +/+ and Kcnk1 −/− male mice before phenobarbital treatment. After phenobarbital treatment, these numbers increased 14 times in Kcnk1 −/− males but only 4 times in Kcnk1 +/+ males in BrdU-positive hepatocytes (Fig. 6B). Also, although to a lesser degree, the number of BrdU-positive hepatocytes was higher in zone 3 of livers of Kcnk1 −/− than of Kcnk1 +/+ males after phenobarbital treatment. No such differences were observed in zone 1. Increase of liver weight and hypertrophy was higher in Kcnk1 +/+ and Kcnk1 −/− male mice (data not shown). Thus, hepatic hyperplasia was augmented in the absence of KCNK1.
FIG. 6.
Hepatic hyperplasia in the livers of phenobarbital-treated Kcnk1 −/− mice.
(A and B) Liver sections were prepared from mouse livers as described in Materials and Methods section. The incorporation of BrdU within the liver lobule was determined by immunohistochemistry. (A) Data shown are representative photographs from section slides of five individual mice (original magnification ×200). Z1, Z3, and PB denote zone 1, zone 3, and phenobarbital, respectively. Scale bar = 100 µm. (B) BrdU-positive hepatocytes were counted. Values are shown as a percentage of BrdU-positive hepatocytes over total nuclei. Data are mean ± SD (n = 5 in each group). *p < 0.05, **p < 0.01, ***p < 0.005 for PBS-treated mice vs. PB-treated mice, † p < 0.05, ††† p < 0.005 for Kcnk1 +/+ vs. Kcnk1 −/− mice, Tukey’s multiple comparison test. PB = phenobarbital.
DISCUSSION
The K+ channel KCNK1 was induced in the livers of mice after phenobarbital treatment. This KCNK1 induction by phenobarbital was specific in livers of C3H/HeNCrlBR male mice and was under the control of the pituitary gland. In addition, the induction was also strain dependent as C3H/HeNCrlBR males induced KCNK1 to levels 45-fold higher than C57BL/6 males did. In response to phenobarbital, CAR specifically activated transcription of the Kcnk1 gene in the livers of C3H/HeNCrlBR males but not in the livers of C3H/HeNCrlBR females or hypophysectomized males. Thus, phenobarbital induction of the KCNK1 induction by CAR correlated with the male-, strain-, and pituitary gland–dependent susceptibility of HCC development in mice. KCNK1 appeared to function as a hepatocyte growth suppressor, because it attenuated hepatic hyperplasia induced by phenobarbital.
Drug induction of hepatic genes can be sex dependent in rodents. For example, it has long been known that the peroxisome proliferator clofibrate induces hepatic enzymes such as CYP4A in a sex-dependent manner (Sundseth and Waxman, 1992). In response to phenobarbital, CAR activated the Cyp3A44 gene in the livers of male mice but not in the livers of female mice (Anakk et al., 2007). CAR activators induced hepatic drug-metabolizing enzymes in a sexually dimorphic manner (Alnouti and Klaassen, 2008; Hernandez et al., 2009). Compared with the well-known growth hormone-STAT5b mechanism that regulates sexually dimorphic Cyp genes at their basal expression levels (Herrington and Carter-Su, 2001; Holloway et al., 2007), these studies were only descriptive in nature and not sufficient for understanding the molecular mechanisms by which sex-dependent inductions were regulated. On the other hand, this study delineates CAR-mediated male-specific activation of the Kcnk1 gene by phenobarbital to the 97-bp RE (−2441/−2345) within the promoter. CAR bound and activated 97-bp RE in only male livers after phenobarbital treatment. HNF4α and the pituitary gland were required for this activation. The pituitary gland regulated CAR binding to the 97-bp RE although growth hormone was not involved in this regulation. Binding of HNF4α was neither sex dependent nor regulated by the pituitary gland. Hormonal replacement experiments suggested that multiple pituitary hormones, such as prolactin and thyroid hormone in addition to growth hormone, were required to restore HCC development in hypophysectomized mice (Erdstein et al., 1984). Because the pituitary signal has now been found to regulate male-specific binding of CAR to the 97-bpRE and its molecular mechanism, determining the molecular mechanism of this specific binding remains key for future investigating into the mode of action of HCC development.
KCNK1 was induced in only zone 3 of livers, indicating that CAR activated the Kcnk1 gene in only this zone after phenobarbital treatment. The molecular mechanism of zone-specific gene activation is not well understood at the present time. However, the Wnt/β-catenin pathway has recently been suggested as a key regulator for zone-specific gene activation (Torre et al., 2011). The Cyp1a1 gene, which is expressed only in zone 3 of mouse livers, was reported to utilize this pathway to activate its transcription (Braeuning and Schwarz, 2010). The Cyp1a1 promoter contained an HNF4α-binding site, and this HNF4α interacted with LEF/TCF transcription factor to activate the promoter (Colletti et al., 2009). Because the LEF/TCF transcription factor is regulated by the Wnt/β-catenin pathway, HNF4α was suggested to play an essential role in the zone 3-specific activation of the Cyp1a1 gene. In CAR activation of the Kcnk1 gene, apparently HNF4α responds to neither the pituitary gland nor phenobarbital. However, the possibility remains that HNF4α is involved in zone 3-specific transcription of the Kcnk1 gene in mouse livers.
Phenobarbital-induced hyperplasia was augmented in the livers of Kcnk1 −/− males, which implicates KCNK1 as an antihyperplasia factor. This finding is somewhat unexpected, because KCNK1 induction is associated with HCC development following phenobarbital treatment. Whether or how does growth repression play a role in the phenobarbital promotion of HCC development is a critical question in the future investigations. Moreover, it has been suggested that both cell growth and death are involved in the tumor development. Therefore, this KCNK1 induction as an antihyperplasia factor may not conflict with susceptibility to HCC development. In addition to high male susceptibility, C3H/HeNCrlBR males are highly susceptible to HCC development compared with C57BL/6 males although both strains were utilized as experimental models in confirming an essential role of CAR in this promotion (Huang et al., 2005; Yamamoto et al., 2004). The degrees of phenobarbital-induced hyperplasia are different from these tumor susceptibilities; females exhibit a much severe hyperplasia than males do; C57BL/6 males more so than C3H/HeNCrlBR males as well (Ledda-Columbano et al., 2003; Lin et al., 1989). The induced levels of KCNK1 were 50 times higher in the livers of C3H/HeNCrlBR male than in the livers of female and 45 times higher in the livers of C3H/HeNCrlBR than in the livers of C57BL/6 males. Based on these KCNK1 levels, the antihyperplasia caused by KCNK1 induction should be higher in males than in females and is also higher in C3H/HeNCrlBR than in C57BL/6 males. Thus, KCNK1 induction, in fact, correlates with sex- and strain-predominant promotion of HCC development after phenobarbital treatment. KCNK1 is induced not only after a short treatment with phenobarbital as presented in this study but also continues to be induced after chronic treatment, as well as in tumor-bearing livers (data not shown). Another intriguing observation is the fact that, while KCNK1 is induced in zone 3, BrdU-stained hepatocytes accumulate in zone 2 and zone 3 regions. Where BrdU-stained hepatocytes arise and divide to cause hyperplasia after phenobarbital treatment is still an active area of investigation, this mechanism of which should be examined and how induced KCNK1 in zone 3 region represses hepatocyte division will be an important question for future investigations. Nevertheless, induction of KCNK1 appears to lead C3H/HeNCrlBR to be more susceptible for this promotion than C57BL/6 although its molecular mechanism remains inscrutable at the present time.
In conclusion, KCNK1 is an antihyperplasia factor (i.e., hepatocyte growth suppressor) and is specifically induced in the livers of C3H/HeNCrlBR males, consistent with phenobarbital promotion of HCC development. This specific induction is regulated by transcriptional activation of the Kcnk1 gene by CAR and the pituitary gland, both of which are essential factors for HCC development in mice. Thus, KCNK1 and Kcnk1 −/− mice provide us with an excellent experimental system to further investigate this molecular mechanism.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
Intramural Research Program of National Institute of Environmental Health Sciences at National Institutes of Health (Z01ES1005-01).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. A.F. Parlow and NHPP for providing recombinant human growth hormone; Dr. J. Barhanin for providing Kcnk1 −/− mice; Dr. R. Maronpot for suggestions and discussion to this work; J.A. Clark for implanting continuous releasing pellet of BrdU into mice; P. Stockton, J. Foley, and Dr. Y. Yamazaki for technical support of laser capture microdissection; the NIEHS immunohistochemistry core lab for performing immunohistochemistry and BrdU assay; and the NIEHS histology core lab for preparing liver section slides.
REFERENCES
- Ahluwalia A., Clodfelter K. H., Waxman D. J. (2004). Sexual dimorphism of rat liver gene expression: Regulatory role of growth hormone revealed by deoxyribonucleic Acid microarray analysis. Mol. Endocrinol. 18, 747–760 [DOI] [PubMed] [Google Scholar]
- Alnouti Y., Klaassen C. D. (2008). Regulation of sulfotransferase enzymes by prototypical microsomal enzyme inducers in mice. J. Pharmacol. Exp. Ther. 324, 612–621 [DOI] [PubMed] [Google Scholar]
- Alvarez-Baron C. P., Jonsson P., Thomas C., Dryer S. E., Williams C. (2011). The two-pore domain potassium channel KCNK5: Induction by estrogen receptor alpha and role in proliferation of breast cancer cells. Mol. Endocrinol. 25, 1326–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anakk S., Huang W., Staudinger J. L., Tan K., Cole T. J., Moore D. D., Strobel H. W. (2007). Gender dictates the nuclear receptor-mediated regulation of CYP3A44. Drug Metab. Dispos. 35, 36–42 [DOI] [PubMed] [Google Scholar]
- Beitzinger M., Hofmann L., Oswald C., Beinoraviciute-Kellner R., Sauer M., Griesmann H., Bretz A. C., Burek C., Rosenwald A., Stiewe T. (2008). p73 poses a barrier to malignant transformation by limiting anchorage-independent growth. EMBO J. 27, 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanck A., Hansson T., Gustafsson J. A., Eriksson L. C. (1986). Pituitary grafts modify sex differences in liver tumor formation in the rat following initiation with diethylnitrosamine and different promotion regimens. Carcinogenesis 7, 981–985 [DOI] [PubMed] [Google Scholar]
- Blanco-Bose W. E., Murphy M. J., Ehninger A., Offner S., Dubey C., Huang W., Moore D. D., Trumpp A. (2008). C-Myc and its target FoxM1 are critical downstream effectors of constitutive androstane receptor (CAR) mediated direct liver hyperplasia. Hepatology 48, 1302–1311 [DOI] [PubMed] [Google Scholar]
- Braeuning A., Schwarz M. (2010). beta-Catenin as a multilayer modulator of zonal cytochrome P450 expression in mouse liver. Biol. Chem. 391, 139–148 [DOI] [PubMed] [Google Scholar]
- Colletti M., Cicchini C., Conigliaro A., Santangelo L., Alonzi T., Pasquini E., Tripodi M., Amicone L. (2009). Convergence of Wnt signaling on the HNF4alpha-driven transcription in controlling liver zonation. Gastroenterology 137, 660–672 [DOI] [PubMed] [Google Scholar]
- Diwan B. A., Rice J. M., Ohshima M., Ward J. M. (1986). Interstrain differences in susceptibility to liver carcinogenesis initiated by N-nitrosodiethylamine and its promotion by phenobarbital in C57BL/6NCr, C3H/HeNCrMTV- and DBA/2NCr mice. Carcinogenesis 7, 215–220 [DOI] [PubMed] [Google Scholar]
- Ellinghaus P., Scheubel R. J., Dobrev D., Ravens U., Holtz J., Huetter J., Nielsch U., Morawietz H. (2005). Comparing the global mRNA expression profile of human atrial and ventricular myocardium with high-density oligonucleotide arrays. J. Thorac. Cardiovasc. Surg. 129, 1383–1390 [DOI] [PubMed] [Google Scholar]
- El-Serag H. B., Rudolph K. L. (2007). Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557–2576 [DOI] [PubMed] [Google Scholar]
- Erdstein J., Guyda H. J., Mishkin S. (1984). Effects of hypophysectomy and hormone replacement on the local and metastatic growth of Morris hepatoma 44. Cancer Res. 44, 2936–2941 [PubMed] [Google Scholar]
- Es-Salah-Lamoureux Z., Steele D. F., Fedida D. (2010). Research into the therapeutic roles of two-pore-domain potassium channels. Trends Pharmacol. Sci. 31, 587–595 [DOI] [PubMed] [Google Scholar]
- Hernandez J. P., Mota L. C., Huang W., Moore D. D., Baldwin W. S. (2009). Sexually dimorphic regulation and induction of P450s by the constitutive androstane receptor (CAR). Toxicology 256, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington J., Carter-Su C. (2001). Signaling pathways activated by the growth hormone receptor. Trends Endocrinol. Metab. 12, 252–257 [DOI] [PubMed] [Google Scholar]
- Holloway M. G., Cui Y., Laz E. V., Hosui A., Hennighausen L., Waxman D. J. (2007). Loss of sexually dimorphic liver gene expression upon hepatocyte-specific deletion of Stat5a-Stat5b locus. Endocrinology 148, 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P., Zelko I., Sueyoshi T., Negishi M. (1998). The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol. Cell. Biol. 18, 5652–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Zhang J., Washington M., Liu J., Parant J. M., Lozano G., Moore D. D. (2005). Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol. Endocrinol. 19, 1646–1653 [DOI] [PubMed] [Google Scholar]
- Inoue K., Negishi M. (2009). Early growth response 1 loops the CYP2B6 promoter for synergistic activation by the distal and proximal nuclear receptors CAR and HNF4alpha. FEBS Lett. 583, 2126–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko V. V., Major M. L., Wang X., Petrovic V., Kuechle J., Yoder H. M., Dennewitz M. B., Shin B., Datta A., Raychaudhuri P., et al. (2004). Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 18, 830–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T., Sueyoshi T., Zelko I., Moore R., Washburn K., Negishi M. (1999). Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol. Cell. Biol. 19, 6318–6322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobliakov V., Popova N., Rossi L. (1991). Regulation of the expression of the sex-specific isoforms of cytochrome P-450 in rat liver. Eur. J. Biochem. 195, 585–591 [DOI] [PubMed] [Google Scholar]
- Kodama S., Negishi M. (2006). Phenobarbital confers its diverse effects by activating the orphan nuclear receptor car. Drug Metab. Rev. 38, 75–87 [DOI] [PubMed] [Google Scholar]
- Konno Y., Moore R., Kamiya N., Negishi M. (2010). Nuclear xenobiotic receptor PXR-null mouse exhibits hypophosphatemia and represses the Na/Pi-cotransporter SLC34A2. Pharmacogenet. Genomics 20, 9–17 [DOI] [PubMed] [Google Scholar]
- Ledda-Columbano G. M., Pibiri M., Concas D., Molotzu F., Simbula G., Cossu C., Columbano A. (2003). Sex difference in the proliferative response of mouse hepatocytes to treatment with the CAR ligand, TCPOBOP. Carcinogenesis 24, 1059–1065 [DOI] [PubMed] [Google Scholar]
- Lesage F., Reyes R., Fink M., Duprat F., Guillemare E., Lazdunski M. (1996). Dimerization of TWIK-1 K+ channel subunits via a disulfide bridge. EMBO J. 15, 6400–6407 [PMC free article] [PubMed] [Google Scholar]
- Lin E. L., Klaunig J. E., Mattox J. K., Weghorst C. M., McFarland B. H., Pereira M. A. (1989). Comparison of the effects of acute and subacute treatment of phenobarbital in different strains of mice. Cancer Lett. 48, 43–51 [DOI] [PubMed] [Google Scholar]
- Lotshaw D. P. (2007). Biophysical, pharmacological, and functional characteristics of cloned and native mammalian two-pore domain K+ channels. Cell Biochem. Biophys. 47, 209–256 [DOI] [PubMed] [Google Scholar]
- Moser G. J., Foley J., Burnett M., Goldsworthy T. L., Maronpot R. (2009). Furan-induced dose-response relationships for liver cytotoxicity, cell proliferation, and tumorigenicity (furan-induced liver tumorigenicity). Exp. Toxicol. Pathol. 61, 101–111 [DOI] [PubMed] [Google Scholar]
- Mutoh S., Osabe M., Inoue K., Moore R., Pedersen L., Perera L., Rebolloso Y., Sueyoshi T., Negishi M. (2009). Dephosphorylation of threonine 38 is required for nuclear translocation and activation of human xenobiotic receptor CAR (NR1I3). J. Biol. Chem. 284, 34785–34792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X., Arrighi I., Kaissling B., Pfaff I., Mann J., Barhanin J., Vallon V. (2005). Expression and insights on function of potassium channel TWIK-1 in mouse kidney. Pflugers Arch. 451, 479–488 [DOI] [PubMed] [Google Scholar]
- Poole T. M., Drinkwater N. R. (1996). Strain dependent effects of sex hormones on hepatocarcinogenesis in mice. Carcinogenesis 17, 191–196 [DOI] [PubMed] [Google Scholar]
- Preat V., de Gerlache J., Lans M., Taper H., Roberfroid M. (1986). Comparative analysis of the effect of phenobarbital, dichlorodiphenyltrichloroethane, butylated hydroxytoluene and nafenopin on rat hepatocarcinogenesis. Carcinogenesis 7, 1025–1028 [DOI] [PubMed] [Google Scholar]
- Saito K., Kobayashi K., Mizuno Y., Furihata T., Chiba K. (2010). Constitutive androstane/active receptor is a target of retinoic acid receptor in humans. Biochem. Pharmacol. 80, 129–135 [DOI] [PubMed] [Google Scholar]
- Sundseth S. S., Waxman D. J. (1992). Sex-dependent expression and clofibrate inducibility of cytochrome P450 4A fatty acid omega-hydroxylases. Male specificity of liver and kidney CYP4A2 mRNA and tissue-specific regulation by growth hormone and testosterone. J. Biol. Chem. 267, 3915–3921 [PubMed] [Google Scholar]
- Torre C., Perret C., Colnot S. (2011). Transcription dynamics in a physiological process: β-catenin signaling directs liver metabolic zonation. Int. J. Biochem. Cell Biol. 43, 271–278 [DOI] [PubMed] [Google Scholar]
- Wada T., Gao J., Xie W. (2009). PXR and CAR in energy metabolism. Trends Endocrinol. Metab. 20, 273–279 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Moore R., Flavell R. A., Lu B., Negishi M. (2010). Nuclear receptor CAR represses TNFalpha-induced cell death by interacting with the anti-apoptotic GADD45B. PLoS One 5, e10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Moore R., Goldsworthy T. L., Negishi M., Maronpot R. R. (2004). The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 64, 7197–7200 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y., Moore R., Negishi M. (2011). Nuclear receptor CAR (NR1I3) is essential for DDC-induced liver injury and oval cell proliferation in mouse liver. Lab. Invest. 91, 1624–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelko I., Negishi M. (2000). Phenobarbital-elicited activation of nuclear receptor CAR in induction of cytochrome P450 genes. Biochem. Biophys. Res. Commun. 277, 1–6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.