Background: Phospholipid flippase mediates translocation of phospholipids between the bilayer leaflets.
Results: ATP8A1 and CDC50A form the flippase complex, and depletion of the complex causes a severe defect in cell migration.
Conclusion: The flippase-mediated translocation of phosphatidylethanolamine at the plasma membrane is involved in cell migration.
Significance: This study provides the first evidence that the phospholipid flippase plays a major role in cell migration.
Keywords: Cell Migration, Cytoskeleton, Lipid Transport, Membrane Bilayer, Membrane Lipids, Phospholipid, Type IV P-type ATPase, Phosphatidylethanolamine, Phosphatidylserine, Phospholipid Flippase
Abstract
Type IV P-type ATPases (P4-ATPases) and CDC50 family proteins form a putative phospholipid flippase complex that mediates the translocation of aminophospholipids such as phosphatidylserine (PS) and phosphatidylethanolamine (PE) from the outer to inner leaflets of the plasma membrane. In Chinese hamster ovary (CHO) cells, at least eight members of P4-ATPases were identified, but only a single CDC50 family protein, CDC50A, was expressed. We demonstrated that CDC50A associated with and recruited P4-ATPase ATP8A1 to the plasma membrane. Overexpression of CDC50A induced extensive cell spreading and greatly enhanced cell migration. Depletion of either CDC50A or ATP8A1 caused a severe defect in the formation of membrane ruffles, thereby inhibiting cell migration. Analyses of phospholipid translocation at the plasma membrane revealed that the depletion of CDC50A inhibited the inward translocation of both PS and PE, whereas the depletion of ATP8A1 inhibited the translocation of PE but not that of PS, suggesting that the inward translocation of cell-surface PE is involved in cell migration. This hypothesis was further examined by using a PE-binding peptide and a mutant cell line with defective PE synthesis; either cell-surface immobilization of PE by the PE-binding peptide or reduction in the cell-surface content of PE inhibited the formation of membrane ruffles, causing a severe defect in cell migration. These results indicate that the phospholipid flippase complex of ATP8A1 and CDC50A plays a major role in cell migration and suggest that the flippase-mediated translocation of PE at the plasma membrane is involved in the formation of membrane ruffles to promote cell migration.
Introduction
In eukaryotic plasma membranes, aminophospholipids such as PS2 and PE reside predominantly in the inner leaflet (1). This asymmetry is generated and maintained by an energy-dependent lipid-translocation machinery known as phospholipid flippase, which mediates the net transfer of PS and PE from the outer to inner leaflets of the plasma membrane bilayer (2–6). ATPase II (currently designated as ATP8A1), purified from bovine chromaffin granules, was identified as the first candidate for the phospholipid flippase (7), and a subsequent genome search revealed that ATP8A1 and its closest yeast homolog, Drs2p, were founding members of a novel subfamily of P-type ATPases known as type IV P-type ATPases (P4-ATPases) (8). Using the budding yeast Saccharomyces cerevisiae, we and other investigators have shown that the P4-ATPases translocate aminophospholipids and function together with Cdc50 family proteins (9–12). Among the five members of P4-ATPases expressed in the yeast, Drs2p, Dnf1p/Dnf2p, and Dnf3p have been shown to associate with the Cdc50 family proteins Cdc50p, Lem3p, and Crf1p, respectively (12, 13). The association between P4-ATPases and Cdc50 family proteins is required for their exit from the endoplasmic reticulum (ER) and for the proper cellular localization (12, 13). Cdc50p is also suggested to be a crucial component for the catalytic activity of P4-ATPases (14, 15), although a reconstitution study of Drs2p into proteoliposome and transplantation analysis of transmembrane segments between Drs2p and Dnf1p suggest that P4-ATPases play a dominant role in determining substrate specificity and in translocating phospholipids (16, 17).
In mammals, at least 14 members of P4-ATPases, designated ATP8A1 through ATP11C, and three CDC50 proteins (CDC50A, CDC50B, and CDC50C) have been identified (2, 3). The association between P4-ATPases and CDC50 proteins is required for the stable expression and proper subcellular localization of the complex (18–20), with the exception of ATP9A and ATP9B, which exit from the ER in the absence of CDC50 proteins (21). Among the P4-ATPases expressed in mammalian cells, ATP8A1, ATP8A2, ATP8B1, ATP8B3, ATP8B5, and ATP11C have been implicated in the translocation of phospholipids. ATP8A1 is the first and best characterized candidate for the phospholipid flippase in erythrocyte membranes (22, 23). The ATPase activity of ATP8A1 is strongly activated by PS and minimally activated by PE, but not by other negatively charged phospholipids such as phosphatidic acid and phosphatidylinositol (24). ATP8A1 is activated only by the naturally occurring sn-1,2-glycerol isomer of PS but not by the sn-2,3-glycerol isomer, although it does not show selectivity for the stereochemistry of the serine head group (24). In Caenorhabditis elegans deficient in the ATP8A1 ortholog tat-1, PS is abnormally exposed on the cell surface, suggesting that TAT-1 functions in maintaining the cell surface asymmetry of PS in vivo (25). ATP8A2 is expressed in the retina and throughout the brain, and its mutation has recently been shown to cause severe mental retardation and other neurological problems in humans (26). The purified ATP8A2·CDC50A complex exhibits PS-dependent ATPase activity and the ability to translocate fluorescence-labeled PS in proteoliposomes (20, 27, 28). ATP8B1 is expressed in the apical membrane of epithelial cells, and its mutations cause progressive familial intrahepatic cholestasis type 1 (PFIC1), a severe liver disease characterized primarily by impaired bile salt secretion from liver into bile (29–31). Induced expression of ATP8B1 in the mutant CHO cells with defective PS translocation stimulates the uptake of fluorescence-labeled PS, suggesting that ATP8B1 is involved in the PS translocation (18). Although the molecular mechanisms underlying the pathogenesis of PFIC1 remain elusive, defective function of ATP8B1 localized on the canalicular membrane of hepatocytes perturbs the membrane organization, which would sensitize the canalicular membrane to enhanced extraction of cholesterol by hydrophobic bile salts, leading to intrahepatic cholestasis (31). ATP8B3 and ATP8B5 (also known as FetA) are exclusively expressed in the testis and are involved in the translocation of PS and PE/PC, respectively (32, 33). The co-expression of ATP8B3 and ATP8B5 in acrosomes suggests that they play a role in the formation of acrosomes and in the acrosome reaction during fertilization (32, 33). Recent studies by two groups have indicated that ATP11C plays a crucial role in differentiation of B lymphocyte possibly through regulating the internalization of PS at the plasma membrane (34, 35). As described above, some of the P4-ATPases have been shown to exhibit the phospholipid flippase activity, but only fragmentary information is available about their substrate specificities and cellular functions.
During the migration of mammalian cells, the dynamic movements of the plasma membrane, which accompany the reorganization of cortical actin filaments at the leading edge, provide a driving force for cell motility (36, 37). It is likely that the rapid transbilayer movements of phospholipids are involved in the remodeling of the plasma membrane, but the role of phospholipid flippase in cell migration remains unknown. In this study, we provide the first evidence that the flippase complex of ATP8A1 and CDC50A plays a critical role in cell migration, and we discuss a possible role for the translocation of PE at the plasma membrane in cell migration.
EXPERIMENTAL PROCEDURES
Constructs and Reagents
Cdc50a cDNA and Atp8a1 cDNA were amplified by PCR using mouse EST clones AA238841 (Incyte Genomics) and AK220560 (Kazusa DNA Research Institute), respectively. cDNA fragments encoding full-length Cdc50a were subcloned into pCMV-Tag4A (Stratagene) for generating a C-terminal FLAG-tagged construct of Cdc50a. To generate the N-terminal GFP- and HA-tagged Atp8a1 constructs, cDNA fragments encoding full-length Atp8a1 were subcloned into pCAGGSneodelEcoRI. pEGFP-Rac1 was a gift from Dr. S. Narumiya (Kyoto University). For knockdown of CDC50A, cDNA fragments encoding full-length Cdc50a were cloned by reverse transcriptase (RT)-PCR from CHO cells and were sequenced. cDNA fragments encoding partial length Atp8a1, corresponding to nucleotides 1912–3495 of mouse Atp8a1, were also cloned from CHO cells and sequenced. For knockdown of Atp8a1 using siRNA, stealth siRNA, a 25-bp duplex oligoribonucleotide with a sense strand corresponding to nucleotides 1947–1971 (siRNA-1) or 2059–2083 (siRNA-2) of hamster Atp8a1 was designed and custom-synthesized (Invitrogen). Stealth RNAiTM siRNA negative control was used for control siRNA. Transient transfection of synthetic siRNA was achieved using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. For all experiments, the cells cultured for 3 days after transfection were used. The following reagents were obtained from commercial sources: annexin V, mouse monoclonal anti-FLAG, anti-α-tubulin, and anti-γ-tubulin antibodies (Sigma); rat monoclonal anti-HA antibody (Roche Applied Science); and mouse monoclonal anti-human transferrin receptor antibody (Zymed Laboratories Inc.). 1-Palmitoyl-2-(6-NBD-aminocaproyl) phosphatidylserine (NBD-PS), 1-palmitoyl-2-(6-NBD-aminocaproyl) phosphatidylethanolamine (NBD-PE), and 1-palmitoyl-2-(6-NBD-aminocaproyl) phosphatidylcholine (NBD-PC) were purchased from Avanti Polar Lipids.
Cell Lines
CHO cells were grown in Ham's F-12 medium containing 10% newborn calf serum. R-41, a CHO-derived mutant cell line, was established as a variant resistant to the PE-binding peptide (Ro09-0198) (38). Tetracycline-regulated cell lines expressing CDC50A were generated using the Tet-Off system. In brief, CHO cells were co-transfected with pT2-Cdc50a, which contains a tetracycline-repressible element and a Cdc50a gene, and pTA-Hyg vector, which contains the tetracycline-sensitive transactivator tTA and the hygromycin resistance gene. Hygromycin-resistant clones displaying enhanced CDC50A expression in the absence of tetracycline were selected. For establishing CDC50A- and ATP8A1-deficient cell lines using shRNA, two distinct target sequences corresponding to nucleotides 736–758 (cell line 7) and 1025–1057 (cell lines 11–2 and 11–3) of hamster Cdc50a and a target sequence corresponding to nucleotides 2673–2695 (cell lines 3 and 7) of hamster Atp8a1 were selected. DNA fragments encoding the shRNA sequences were subcloned into the H1 RNA polymerase III promoter vector, pTA-H1. The resulting vectors and the vector without insert were transected in CHO cells, and stable deficient or control clones were selected with 10 μg/ml puromycin. For establishing cell lines stably expressing GFP- and HA-tagged ATP8A1, FLAG-tagged CDC50A, and GFP-tagged Rac1, CHO cells were transfected with the corresponding vectors, and the cell lines were established by selecting with 500 μg/ml G418.
Generation of Anti-CDC50A and Anti-ATP8A1 Antibodies
Rabbit polyclonal antibodies against CDC50A were raised in New Zealand White female rabbits against the synthetic peptide (MAMNYSAKDEVDGGPAGC) that corresponds to 17 amino acids at the N-terminal end of CDC50A with the addition of an extra cysteine to the C-terminal end. Antibodies were isolated from the immune sera by affinity chromatography on a synthetic peptide-conjugated SulfoLink column (Pierce). Rabbit polyclonal antibodies against ATP8A1 were raised against the synthetic peptide (CSEVIRAYDTTKQRPDEW) at the C-terminal end of ATP8A1 with the addition of an extra cysteine to the N-terminal end as described above. The specificity of the antibodies was confirmed by immunoblotting and immunofluorescence staining where the expression of endogenous proteins was suppressed by introducing siRNA.
Expression Analysis of CDC50A and P-type ATPase
Primers for RT-PCR were synthesized according to the sequences of mouse CDC50 family proteins and mouse P-type ATPases. Amplified fragments corresponding to the nucleotides were as follows: Cdc50a, 90–968 bp; Cdc50b (accession number NM_178715), 217–1077 bp; Cdc50c (accession number NM_027651), 467–1101 bp; Atp8a1, 1880–2262 bp; Atp8a2, 2184–3023 bp; Atp8b1, 2440–3286 bp; Atp8b2, 2524–3067 bp; Atp8b3, 3147–3676 bp; Atp8b4, 165–699 bp; Atp9a, 2090–2607 bp; Atp9b, 2448–2996 bp; Atp10a, 3240–4214 bp; Atp10d, 2068–2892 bp; Atp11a, 1613–2274 bp; Atp11b, 2276–3095 bp; and Atp11c, 2221–2920 bp (for the accession number of each molecule, see Ref. 6). All of the amplified fragments shown in Fig. 1A were sequenced and identified as hamster homolog of CDC50A and ATPases.
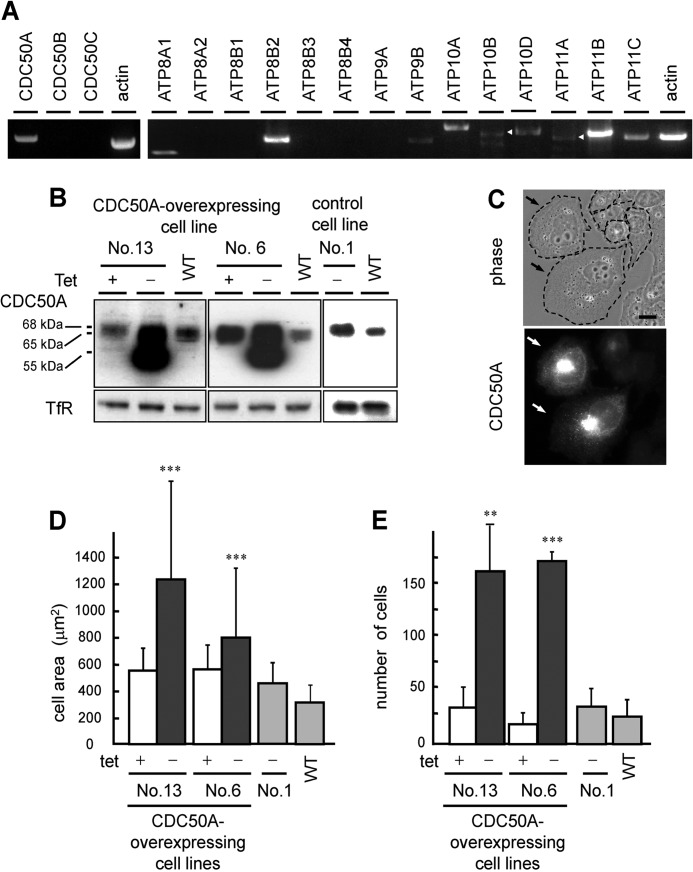
FIGURE 1.
Overexpression of CDC50A induces cell spreading and enhances cell migration. A, mRNA expression of CDC50 family proteins (left panel) and P4-ATPases (right panel), visualized by using RT-PCR. Arrowheads indicate the fragment corresponding to ATP10B and ATP11A, respectively. B, immunoblotting showing the expression of CDC50A in the wild-type CHO cells (WT), CDC50A-overexpressing cell lines (No. 6 and No. 13), and control cell line (No. 1). Anti-TfR was used as a loading control. C, phase-contrast image (top) and CDC50A expression visualized by anti-CDC50A antibody (bottom) in the CDC50A-overexpressing CHO cell line 13. D, cross-sectional areas of the wild-type (WT), CDC50A-overexpressing CHO cell lines 6 and 13, and control cell line 1 were measured and quantified by AxioVision LE 4.5 software. Values are means ± S.D. (n >100). E, migratory activity of the CDC50A-overexpressing cell lines was examined with the transwell assay. All experiments were performed in the presence (tet+) or absence (tet−) of tetracycline. Values are means ± S.D. from three independent experiments. Data were analyzed by using Student's t test. **, p < 0.01; ***, p < 0.0001.
Immunoblotting
Harvested cells were lysed in SET buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 250 mm sucrose) supplemented with protease inhibitors. The lysates were centrifuged at 100,000 × g for 1 h at 4 °C. The precipitated membrane fractions were washed and resuspended in SDS buffer (62.5 mm Tris-HCl, pH 7.5, 1% SDS, 10% glycerol). Protein concentration was determined by BCA protein assay reagent (Pierce). For treatment with peptide:N-glycosidase F (PNGase F) (Roche Applied Science), the membrane fractions were denatured by boiling in 0.75% 2-mercaptoethanol for 3 min, followed by incubation with PNGase F for 16 h at 37 °C according to the manufacturer's instructions. Immunoblotting analysis was performed by using anti-CDC50A antibody, anti-ATP8A1 antibody, and anti-human transferrin receptor (TfR) antibody as a loading control. Bound antibodies were detected with horseradish peroxidase-conjugated anti-rabbit IgG antibody, anti-mouse IgG antibody, and anti-rat IgG antibody using ECL Western blotting reagents (GE Healthcare).
Immunoprecipitation
HEK293 cells were transfected with the indicated plasmids and cultured for 2 days. Cells were lysed in a buffer consisting of 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, and protease inhibitors (Roche Applied Science) for 15 min on ice. Clarified cell lysates were immunoprecipitated by using anti-FLAG M2 affinity gel (Sigma) according to the manufacturer's instructions. Immunoblotting analysis was performed by using anti-HA antibody and anti-FLAG antibody.
Migration Assay
Transwell migration assays were performed using modified Boyden chambers containing Transwell filters (8-μm pore diameter; Corning Costar) according to the method described by Huttenlocher et al. (39). In brief, a total of 1 × 105 cells were seeded to the upper chamber, with the same medium located in the lower chamber. After migration for 3 h, cells on the lower side were fixed and counted. To assess cell migration using an in vitro wound healing assay, CHO cells grown in 24 wells as confluent monolayers were wounded by scraping with a P200 pipette tip. The migration of cells into the wound area was followed by time-lapse microscopy. The area of wound sealing was calculated using AxioVision LE 4.5 software (Carl Zeiss). Each experiment was performed a minimum of three times.
Phospholipid Translocation Assay
Cells grown on glass coverslips were washed with serum-free Ham's F-12 warmed to 37 °C and incubated in serum-free Ham's F-12 containing 2 μm NBD-labeled phospholipids for 30 min at 30 °C. The cells were then washed with DMEM/Ham's F-12 non-phenol red medium and photographed. To determine the amount of internalized fluorescent lipid analogs, the cells were incubated in 50 mm dithionite for 30 s at room temperature to quench the NBD lipids on the outer monolayer of the plasma membrane (40). After washing with DMEM/Ham's F-12, cells were photographed again. To quantify the internalization of NBD-labeled phospholipids, fluorescence images without the dithionite treatment were obtained by a laser-scanning confocal microscopy LSM 510. Signal intensities of the inside of the cells were measured using LSM 510-ConfoCor3 software. To examine the endocytosis, 5 μg/ml FM1-43, 10 μg/ml DiI-labeled low density lipoprotein (DiI-LDL), or 10 μg/ml cy3-labeled transferrin (cy3-Tf) were added to the serum-free Ham's F-12 medium instead of NBD lipids, and they were incubated for the indicated periods at 37 °C.
Peptide Sensitivity Assay
Cells were seeded at 1.5 × 104 cells on 48-well plates and cultured for 24 h. After washing with PBS, cells were incubated with the PE-binding peptide Ro09-0198 or PS-binding drug papuamide B in serum-free Ham's F-12 containing 0.1% fatty acid-free BSA for 1 h at 37 °C. Cell viability was determined by trypan blue exclusion assay.
Microscopy
Cells were visualized using a Zeiss Axiovert 200 M microscope equipped with ×63 Plan-Apochromat oil immersion objective. Optical sections were taken at 0.15-μm intervals throughout the depth of the cell, and cross-sectional area was measured using AxioVision LE 4.5 software (Carl Zeiss).
RESULTS
CDC50A Is Involved in Cell Migration
We identified at least eight members of P4-ATPases expressed in Chinese hamster ovary (CHO) cells (Fig. 1A). In contrast to the variation among the P4-ATPases, only a single CDC50 family protein, CDC50A, was expressed in CHO cells (Fig. 1A). To explore the cellular function of phospholipid flippase, we first focused on CDC50A and established CDC50A-overexpressing cell lines using a tetracycline-regulated expression system. The withdrawal of tetracycline from the culture medium resulted in a significant increase in the expression of CDC50A with apparent molecular masses of 68, 65, and 55 kDa (Fig. 1B). CDC50A contains five possible sites for N-linked glycosylation, and the treatment with PNGase F shifted the 68- and 65-kDa bands to the 55-kDa band, suggesting that the 55-kDa band observed with the CDC50A overexpression was due to the incomplete glycosylation. Cell line 1, which harbored the pT2-CDC50A vector but did not show any significant increase in the expression of CDC50A upon the withdrawal of tetracycline, is used as a control cell line (Fig. 1B).
Immunofluorescence staining showed that the amounts of CDC50A expressed in each cell varied significantly and that the CDC50A-overexpressing cells exhibited extensive cell spreading and membrane ruffling (Fig. 1C). Quantitative analysis showed that this overexpression resulted in an average 1.5–2-fold increase in cross-sectional area of the cells compared with the noninduced (tet+) and control cells (cell line 6 tet+ (556.4 ± 174.6 μm2) and tet− (800.1 ± 537.8 μm2); cell line 13 tet+ (549.6 ± 160.8 μm2) and tet− (1230.6 ± 806.2 μm2); WT (313.1 ± 125.7 μm2); cell line 1 (control) (473.9 ± 170.5 μm2), p < 0.0001; Fig. 1D). In a transwell migration assay, overexpression of CDC50A remarkably enhanced migratory activity compared with that of the noninduced (tet+) and control cells (cell line 6 tet+ (18.6 ± 8.8 cells) and tet− (169.0 ± 8.2 cells), p < 0.0001; cell line 13 tet+ (33.9 ± 18.6 cells) and tet− (159.5 ± 42.1 cells), p < 0.01; WT (25.5 ± 15.5 cells); cell line 1 (control) (40.4 ± 18 cells); Fig. 1E).
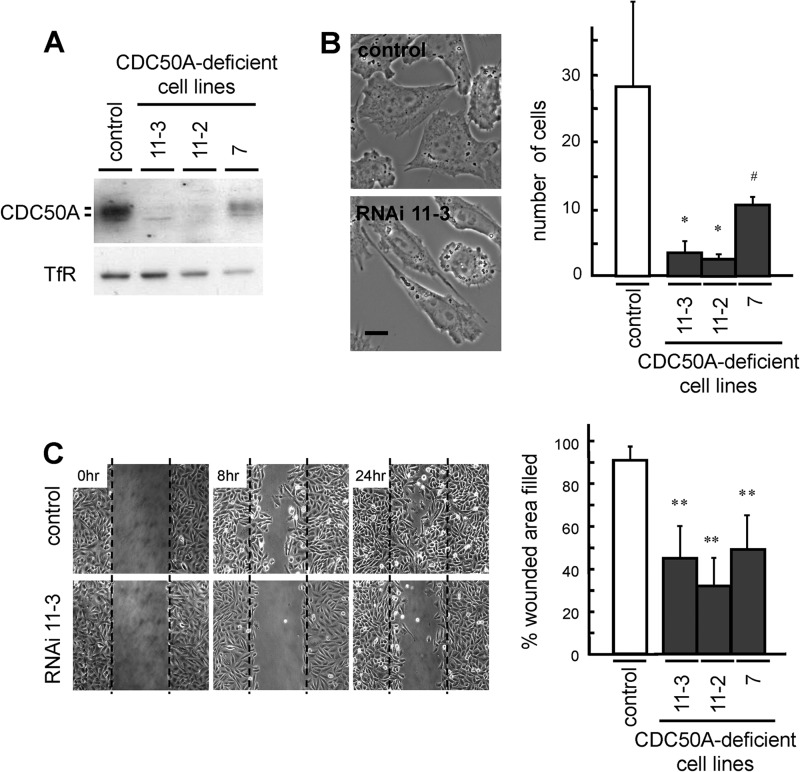
To further examine the involvement of CDC50A in cell migration, we established CDC50A-deficient cells (cell lines 7, 11-12, and 11-3) by stably expressing short hairpin RNA (shRNA). CDC50A expression was reduced to 7.6% (cell line 11-2), 9.1% (cell line 11-3), and 27.4% (cell line 7) of that expressed in the control cell line (Fig. 2A). The CDC50A-deficient cell lines exhibited an elongated spindle-like morphology, with reduced formation of membrane ruffles (Fig. 2B). The migratory activity of the CDC50A-deficient cells was significantly lower than that of the control cells (control (28.2 ± 12.5 cells); cell line 11-3 (3.6 ± 0.9 cells), p < 0.05; cell line 11-2 (3 ± 0.9 cells), p < 0.05; cell line 7 (10.5 ± 1.9 cells), p < 0.08; Fig. 2B). In an in vitro wound healing assay, CDC50A-deficient cells also exhibited markedly reduced migratory activity (control (90.1 ± 7.3%); cell line 11-3 (45.0 ± 14.9%), p < 0.01; cell line 11-2 (32.1 ± 6.8%), p < 0.01; cell line 7 (48.2 ± 6.5%), p < 0.01; Fig. 2C). These results indicate that CDC50A plays a critical role in cell migration.
FIGURE 2.
Suppressed expression of CDC50A inhibits cell migration. A, expression of CDC50A in the CDC50A-deficient cell lines and control vector-transfected cell line (control) was analyzed by immunoblotting using anti-CDC50A antibody. Anti-TfR antibody was used as a loading control. B, phase-contrast images of the control and CDC50A-deficient cell line 11-3 (RNAi 11-3) are shown (left). Migratory activities of the control and the CDC50A-deficient cell lines 7, 11-2, and 11-3 were examined by the transwell assay (right). Values are means ± S.D. from three independent experiments. Scale bar, 10 μm. C, migratory activities of the control and the CDC50A-deficient cell lines were examined by the wound healing assay. The control and CDC50A-deficient cell lines (RNAi 11–3) were wounded by tip scraping, and the same area of each dish was monitored microscopically at 0, 8, and 24 h (left). To quantify cellular migration, data are presented as the percent of wound closure after 18 h (right). At least three independent experiments were performed. Values are means ± S.D. from three independent experiments. At least three independent experiments were performed, and the typical results are shown. *, p < 0.05; **, p < 0.01; #, p < 0.08.
CDC50A Regulates Transbilayer Movement of Aminophospholipids at the Plasma Membrane
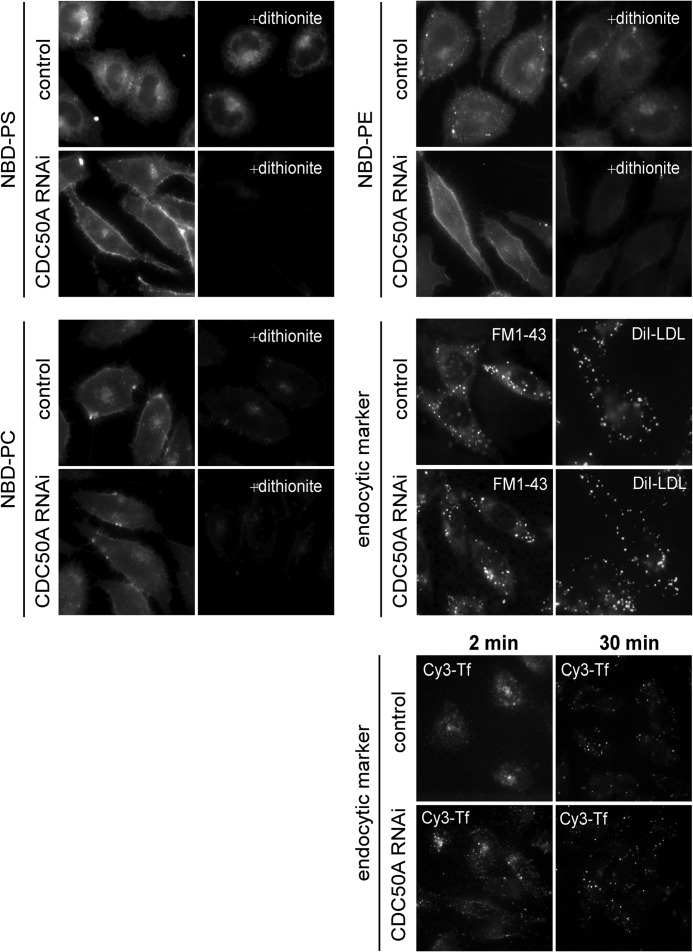
We next examined whether or not CDC50A is required for the translocation of phospholipids at the plasma membrane. We analyzed the uptake of fluorescence-labeled analogs of phospholipids using a dithionite quenching assay (40), in which phospholipids labeled with 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) in the outer layer of the plasma membrane were quenched due to chemical reduction of the nitro group by membrane-impermeable dithionite. In this assay, the translocation of NBD-labeled phospholipids was measured at 30 °C, at which temperature the uptake of a membrane marker FM1-43 via endocytosis was largely diminished, but the translocation of NBD-labeled phospholipids such as NBD-PS and NBD-PE was not significantly affected. As shown in Fig. 3, CDC50A-deficient cells showed significant defects in the internalization of NBD-PS and NBD-PE compared with control cells but no significant change in the internalization of NBD-labeled phosphatidylcholine (NBD-PC), suggesting that CDC50A plays a critical role in the translocation of aminophospholipid analogs (NBD-PS and NBD-PE) across the plasma membrane. We then examined the effect of CDC50A depletion on endocytosis using the endocytotic membrane markers such as FM1-43, DiI-labeled low density lipoprotein (DiI-LDL), and Cy3-labeled transferrin (Cy3-Tf). As shown in Fig. 3, CDC-50A-deficient cells exhibited no significant change in the uptake of the endocytotic membrane markers nor in the recycling of Cy3-Tf to the cell surface from the endosomal recycling compartments.
FIGURE 3.
CDC50A regulates transbilayer movement of aminophospholipids at the plasma membrane. The CDC50A-deficient cell line 11-3 (CDC50A RNAi) and the control cell line were incubated with 2 μm NBD-PS, NBD-PE, or NBD-PC for 30 min at 30 °C, and then they were washed and photographed. The cells incubated with the NBD-labeled phospholipids were further incubated with 50 mm dithionite for 30 s at room temperature, and photographed (+dithionite). FM1-43 (5 μg/ml), DiI-LDL (10 μg/ml), and Cy3-Tf (10 μg/ml) were added to the cells and incubated for indicated periods at 37 °C.
CDC50A Associates with and Recruits ATP8A1 to the Cell Surface in CHO Cells
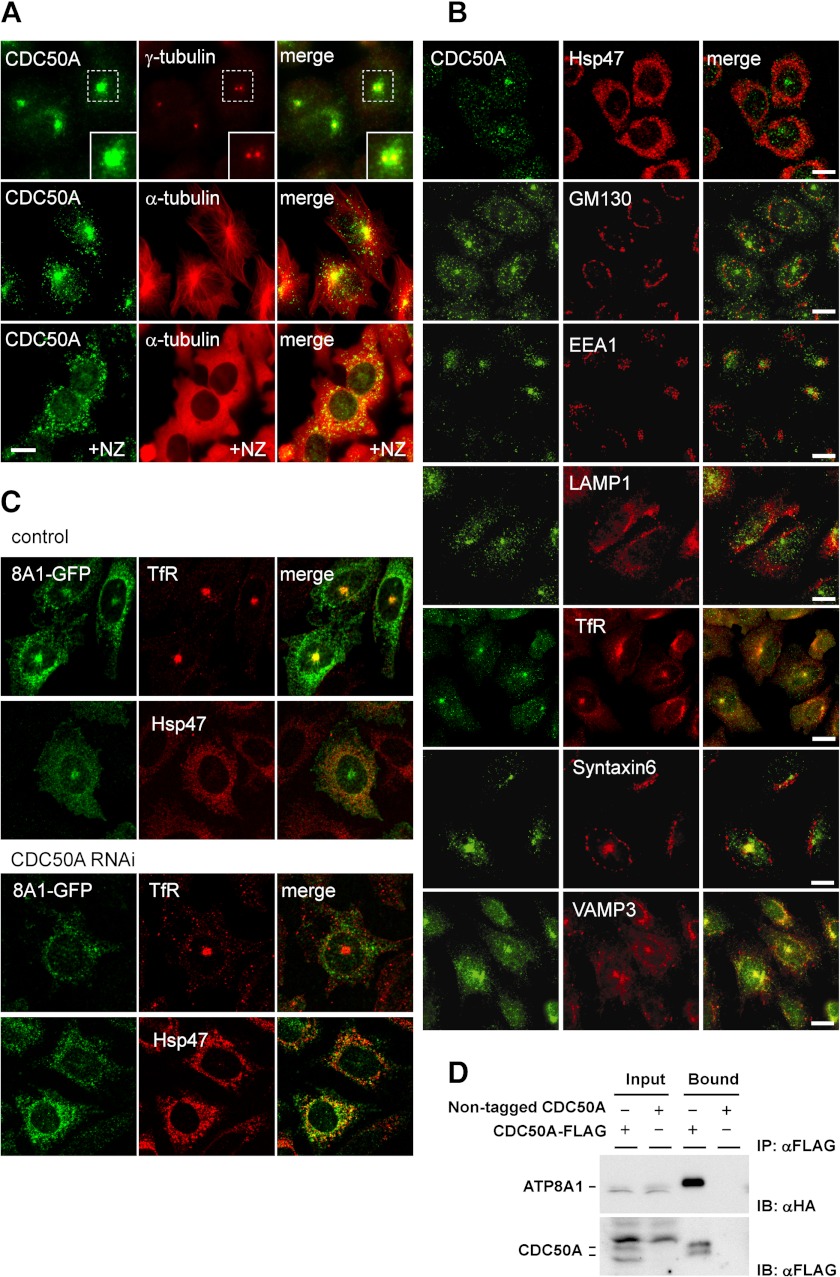
Among the P4-ATPases expressed in CHO cells, ATP8A1 is the only known candidate for phospholipid flippase and is shown to associate with CDC50A (19). In this study, we first studied the subcellular localization of CDC50A and ATP8A1 in CHO cells and confirmed the interaction of the two proteins by co-immunoprecipitation analysis. Immunofluorescence staining using the anti-CDC50A antibody exhibited a punctate distribution throughout the cytoplasm, with intense staining at the pericentriolar region; this staining was dispersed by treatment with nocodazole, a microtubule inhibitor (Fig. 4A). The localization of CDC50A partly overlapped with that of transferrin receptor, syntaxin 6, and VAMP3, suggesting that CDC50A mainly localized in the endosomal recycling compartments (Fig. 4B). In CHO cell lines that stably express low levels of green fluorescent protein (GFP)-tagged ATP8A1, ATP8A1 mainly localized at the recycling compartments and partially on the plasma membrane but not at the ER (Fig. 4C). In contrast, ATP8A1 accumulated in the ER in CDC50A-deficient cells (Fig. 4C). From the immunoprecipitation analysis, in which HA-tagged ATP8A1 and FLAG-tagged CDC50A were co-expressed in HEK293 cells, ATP8A1 was effectively precipitated by using the anti-FLAG antibody (Fig. 4D). These results demonstrate that CDC50A and ATP8A1 form a complex that is essential not only for the exit of ATP8A1 from the ER but also for its proper localization to the plasma membrane and recycling compartments. It is likely that the suppressed expression of CDC50A inhibited the recruitment of ATP8A1 to the plasma membrane, causing a defect in phospholipid translocation.
FIGURE 4.
CDC50A associates with and recruits ATP8A1 to recycling endosome and the plasma membrane. A, CHO cells were fixed and stained for CDC50A (CDC50A), and the same specimens were co-stained with the centrosomal marker γ-tubulin (γ-tubulin) and the cortical microtubule marker α-tubulin (α-tubulin). Right panels show merged images (merge). For nocodazole treatment, the cells were incubated with 33 μm nocodazole at 37 °C for 3 h (NZ). Insets in the top panels show the enlarged images of the regions framed with dashed lines. B, fluorescence micrographs showing the localization of CDC50A (CDC50A) and organelle markers; ER (Hsp47), cis-Golgi (GM130), early endosome (EEA1), lysosome (LAMP1), recycling endosome (human TfR), TGN and recycling endosome (Syntaxin6), early endosome, and recycling endosome (VAMP3). Right panels show merged images (merge). C, CDC50A-deficient cell line 11-3 (CDC50A RNAi) and the control cell line (control) were stably transfected with GFP-ATP8A1, and cell lines expressing GFP-ATP8A1 at low levels were cloned. The resulting cell lines were co-stained with TfR or Hsp47 and observed by fluorescence microscopy. D, HEK293 cells were transiently transfected with HA-ATP8A1 and CDC50A-FLAG or nontagged CDC50A constructs for 48 h. Cells were lysed and immunoprecipitated with anti-FLAG M2 affinity gel (IP: αFLAG). The eluates were analyzed by immunoblotting with anti-HA and anti-FLAG antibody (IB: αHA and IB: αFLAG, respectively). The scale bars in A–C indicate 10 μm.
ATP8A1 Plays a Role in Cell Migration
To investigate whether ATP8A1 is involved in cell migration, we next examined the migratory activity of ATP8A1-deficient CHO cells. To suppress ATP8A1 expression, we employed two different sequences (siRNA-1 and -2) for siRNA treatment (Fig. 5A). The ATP8A1-deficient cells had an elongated spindle-like shape with no membrane ruffles, exhibiting morphological features similar to those of the CDC50A-deficient cells (Fig. 2B). The ATP8A1-deficient cells were less motile than the control cells in the transwell migration assay (control (33.5 ± 9.6 cells); siRNA-1 (15.2 ± 3.7 cells) p < 0.05; siRNA-2 (8.9 ± 6.9 cells) p < 0.05; Fig. 5B) and in the in vitro wound healing assay (control (97.1 ± 2.2%); siRNA-1 (54.2 ± 16.1%) p < 0.05; siRNA-2 (55.3 ± 2.4%) p < 0.01; Fig. 5C). The role of ATP8A1 in cell migration was further confirmed using other motile cells such as NIH3T3 and mouse embryonic fibroblast (MEF). As shown in Fig. 5C, the migratory activities of both NIH3T3 and MEF cells were significantly suppressed by the depletion of ATP8A1. These results clearly demonstrate that ATP8A1 plays a critical role in the control of cell migration.
FIGURE 5.
Suppressed expression of ATP8A1 inhibits cell migration. A, immunoblotting showing the expression of ATP8A1 in the ATP8A1-deficient cells using siRNA. For knockdown by siRNA, CHO cells were transfected with siRNA against ATP8A1 (siRNA-1 or -2) and cultured for 3 days. Expression of ATP8A1 protein in the control siRNA-transfected cells (ctr) and ATP8A1-deficient cells were transfected with siRNA-1 (lane 1) or siRNA-2 (lane 2). Anti-TfR antibody was used as a loading control. B, CHO cells were transfected with siRNA against ATP8A1 (siRNA-1 or -2) and cultured for 3 days. Phase-contrast images of the control siRNA (ctr) and siRNA-1 (siRNA-1)-transfected cells are shown (left). The scale bar indicates 10 μm. Migratory activity of the control siRNA (ctr) and siRNA (siRNA-1 and -2)-transfected cells were examined by the transwell assay (right). Values are means ± S.D. from three independent experiments. C, migratory activities of the control siRNA-transfected cells (ctr) and the ATP8A1-deficient cells transfected with siRNA-1 or siRNA-2 were examined by the in vitro wound healing assay in CHO, NIH3T3, and MEF cells. Data are presented as the percent of wound closure after 18 h (left; CHO) or 8 h (middle; NIH3T3, right; MEF). At least three independent experiments were performed. Values are means ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.01.
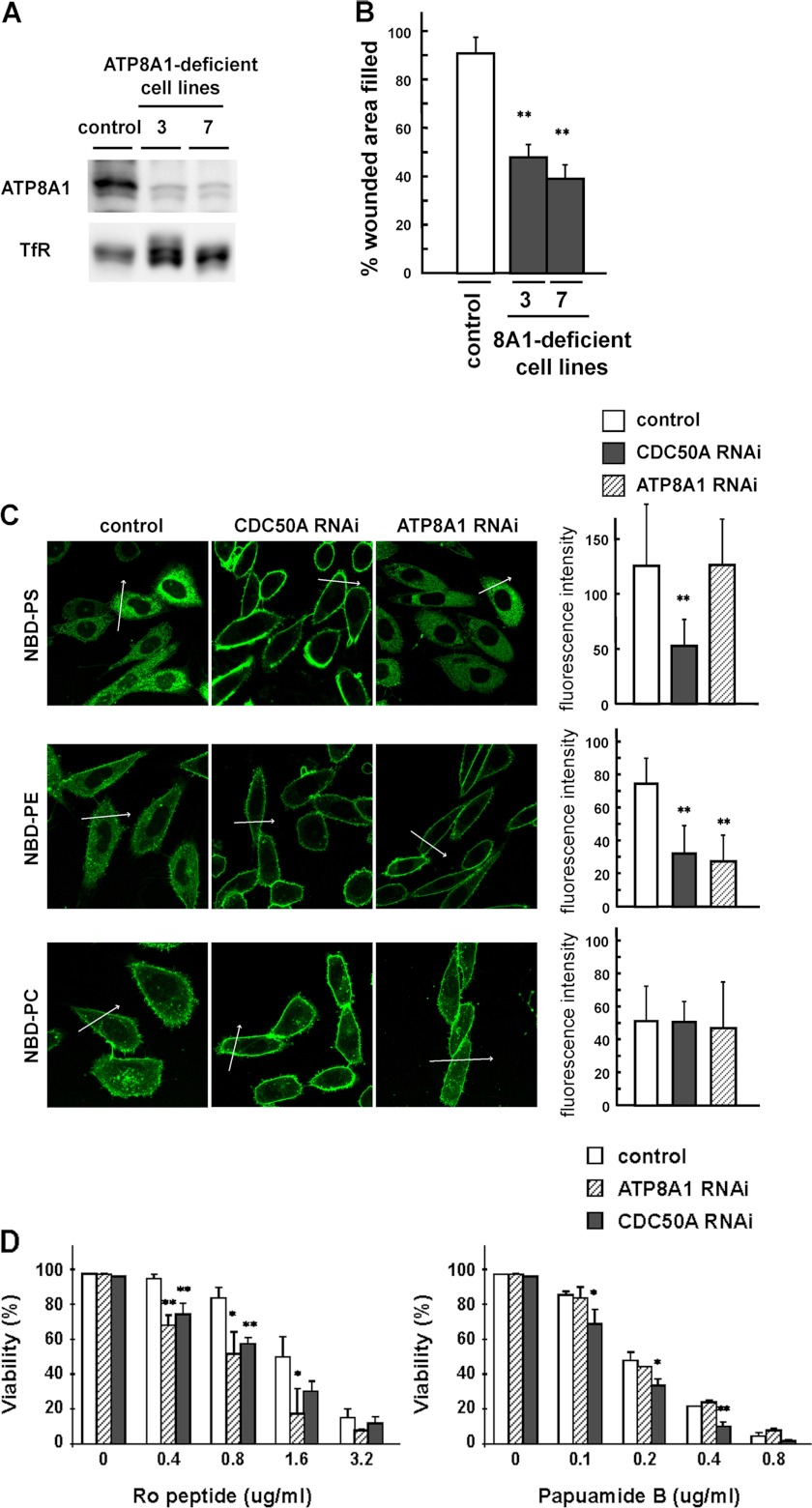
Inward Translocation of Cell-surface PE Is Involved in Cell Migration
To examine whether the translocation of particular phospholipids at the plasma membrane is involved in cell migration, we established the ATP8A1-deficient cell lines 3 and 7 by stably expressing shRNA (Fig. 6A) and performed the quantitative measurements of phospholipid translocation. The migratory activity of the ATP8A1-deficient cell lines was significantly suppressed in the in vitro wound healing assay (control (90.1 ± 7.3%); cell line 3 (48.0 ± 5.7%) p < 0.01; cell line 7 (38.9 ± 7.1%) p < 0.01; Fig. 6B). Quantitative measurements of the translocation of fluorescence-labeled phospholipids at the plasma membrane of the CDC50A-deficient cells (cell line 11-3) showed that the translocation of both NBD-PS and NBD-PE was significantly reduced (Fig. 6C), and these results showed good correlation with those observed with the dithionite quenching assay (Fig. 3). In contrast, in the ATP8A1-deficient cells (cell line 3), the translocation of NBD-PS was not affected but that of NBD-PE was significantly reduced (Fig. 6C). These observations indicate that the defective expression of ATP8A1 had no significant effect on the translocation of NBD-PS but effectively inhibited the translocation of NBD-PE at the plasma membrane. Furthermore, the ATP8A1-deficient cells exhibited a significantly increased sensitivity toward the PE-binding peptide (Ro09-0198) (9) compared with the control cells but not toward the PS-binding drug (papuamide B) (Fig. 6D) (41). The CDC50A-deficient cells showed an increased sensitivity toward both the PE- and PS-binding probes (Fig. 6D). These results indicate that the inward translocation of PE but not PS is affected in the ATP8A1-deficient cells. Because cell migration was significantly suppressed by the defective expression of ATP8A1, these observations raise the intriguing possibility that the ATP8A1-mediated translocation of PE at the plasma membrane is involved in cell migration.
FIGURE 6.
Suppressed expression of ATP8A1 affects the translocation of PE at the plasma membrane. A, immunoblotting showing the expression of ATP8A1 in the ATP8A1-deficient cell lines using stable expression of shRNA. Expression of ATP8A1 protein in the control vector-transfected cell line (control) and ATP8A1-deficient cell lines 3 and 7 were analyzed by immunoblotting using anti-ATP8A1 antibody. Anti-TfR antibody was used as a loading control. B, migratory activities of the control and the ATP8A1-deficient cell lines 3 and 7 were examined by the in vitro wound healing assay. Data are presented as the percent of wound closure after 18 h. At least three independent experiments were performed. Values are means ± S.D. from three independent experiments. C, NBD-labeled phospholipids were added to the control, the ATP8A1-deficient cell line 3 (ATP8A1 RNAi), and CDC50A-deficient cell line 11-3 (CDC50A RNAi). After incubation for 30 min at 30 °C, fluorescence images were obtained by confocal microscopy (left), and the signal intensities of inside of the cells were measured using LSM 510-ConfoCor3 software (right). Arrows show the position used for fluorescence intensity scans. Values are means ± S.D. (n >23). **, p < 0.01. D, control, the ATP8A1-deficient (ATP8A1 RNAi), and CDC50A-deficient (CDC50A RNAi) cell lines were seeded at 1.5 × 104 cells and were incubated with PE-binding peptide Ro09-0198 (left) or PS-binding drug papuamide B (right) at indicated concentrations. After 1 h of incubation at 37 °C, cell viability was determined by trypan blue exclusion assay. Student's t test was used to compare the control with ATP8A1- or CDC50A-deficient cells. p < 0.05 was considered statistically significant. *, p < 0.05; **, p < 0.01.
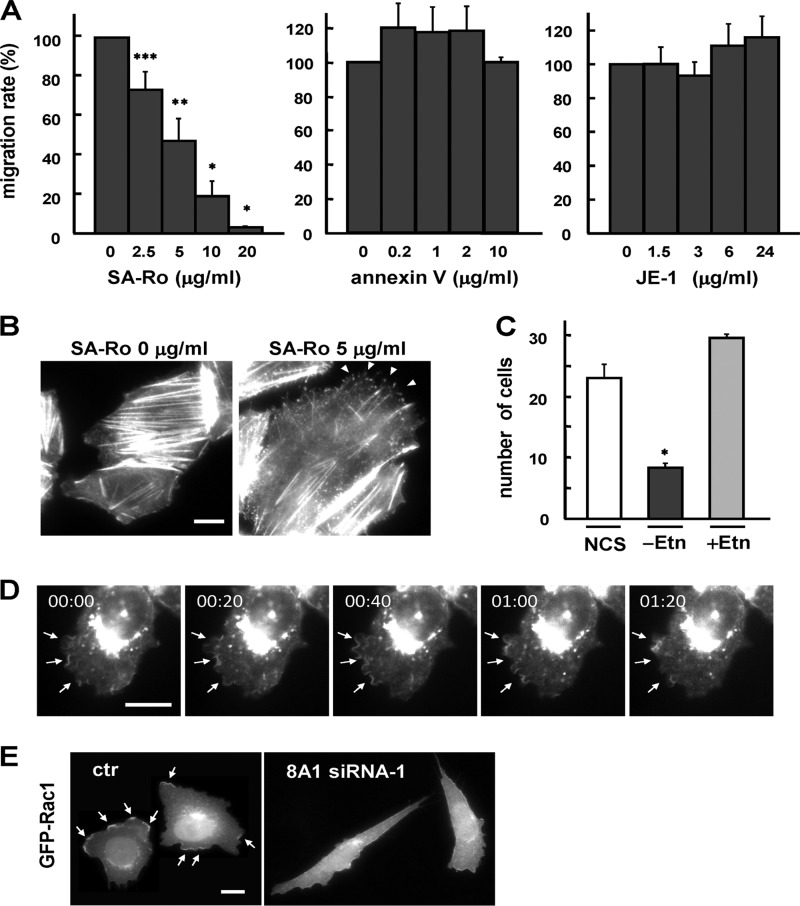
To examine the involvement of cell-surface PE in cell migration, we took two different approaches. We manipulated the cell-surface PE by the PE-binding peptide conjugated with streptavidin (SA-Ro) (42), and we used a CHO cell mutant that had a specific defect in PE biosynthesis (38). We previously showed that the cytolytic activity of the PE-binding peptide was abolished by the conjugation with streptavidin and that immobilization of cell-surface PE by SA-Ro at a concentration of 50–100 μg/ml effectively blocked disassembly of the contractile ring, resulting in the arrest of cytokinesis at the final stage of cell division (42, 43). As shown in Fig. 7A, SA-Ro effectively inhibited the enhanced cell motility of CDC50A-overexpressing cells in a dose-dependent manner, with 50% inhibition at 5 μg/ml, whereas other phospholipid-binding probes, including PS-binding protein annexin V and anti-PC monoclonal antibody JE-1 (44), had no significant effect on cell migration. F-actin staining of the CDC50A-overexpressing cells treated with SA-Ro showed that the assembly of cortical actin filaments beneath the membrane ruffles was collapsed by SA-Ro treatment, leading to the formation of spike-like membranes (Fig. 7B). To further investigate the role of PE in cell migration, we used a CHO mutant cell line, R-41, which is defective in the translocation of PS into the inner mitochondrial membrane, where PS is decarboxylated to PE (38). Because PE in mammalian cells is also produced by another pathway, the CDP-ethanolamine pathway, depletion of ethanolamine from culture medium caused a significant reduction in the cell-surface content as well as the overall content of PE in the R-41 mutant cells, but no significant change occurred in the contents of the other phospholipids, including PS (38). Thus, we examined the effect of ethanolamine depletion on the migratory activity of the R-41 mutant cells. The R-41 mutant cells showed normal migratory activity when cultured in normal medium, but the withdrawal of ethanolamine from the medium caused an elongated spindle-like shape (42) and a significant defect in cell migration, which was restored to the normal level by addition of ethanolamine (Fig. 7C). A time-lapse observation of localization of ATP8A1 in serum-induced actively migrating cells showed that ATP8A1 was localized on the highly motile ruffling membranes at the cell front (Fig. 7D). The translocation of Rac1, which activates the polymerization of actin filaments in the lamellipodial protrusions and promotes the formation of membrane ruffles, to the plasma membrane was also diminished in the ATP8A1-deficient cells (Fig. 7E). Taken together, these results suggest that the ATP8A1-mediated translocation of PE at the plasma membrane is involved in the remodeling of cortical actin filaments, which leads to the formation of membrane ruffles to promote cell migration.
FIGURE 7.
Inward translocation of cell surface PE is involved in cell migration. A, migratory activity of CDC50A-overexpressing cells was examined by the transwell assay, in the presence of PE-binding probe (SA-Ro), the PS-binding probe (annexin V), or anti-PC monoclonal antibody (JE-1) at the indicated concentrations. The number of migrating cells without addition of probes was normalized to a 100% migration rate. Values are means ± S.D. from three independent experiments. B, CDC50A-overexpressing cell line 13 was washed and incubated in serum-free Ham's F-12 containing 0.1% BSA and 700 μg/ml hygromycin B in the presence (left) or absence (right) of 5 μg/ml SA-Ro for 3 h at 37 °C. Cells were then fixed and stained for the distribution of F-actin using TRITC-labeled phalloidin. Arrowheads indicate the spike-like membrane protrusions. The scale bar indicates 10 μm. C, R-41 cells were cultured in Ham's F-12 medium containing 10% newborn calf serum (NCS). For ethanolamine depletion, R-41 cells were cultured in medium containing 10% dialyzed newborn calf serum (−Etn) for 48 h. For restoration of PE synthesis, R-41 cells were cultured in medium containing 10% dialyzed newborn calf serum and 20 μm ethanolamine-HCl (+Etn) for 48 h. The transwell assay was performed as described above. Values are means ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.01. D, localization of ATP8A1 in migrating cells. The cell line stably expressing GFP-ATP8A1 was serum-starved for 24 h and then incubated in the presence of 10% newborn calf serum. After serum stimulation for 50 min, images from GFP-positive cells were recorded every 20 s for 1.5 min. Representative cells are shown at 0, 20, 40, 60, and 80 s. Arrows indicate the highly motile ruffling membranes. E, CHO cell line stably expressing GFP-Rac1 was transiently transfected with control siRNA (ctr) and siRNA-1 (8A1 siRNA-1) and was observed after 3 days. Arrows indicate the distribution of Rac1 at the membrane ruffles.
DISCUSSION
ATP8A1 (ATPase II) was identified as the first candidate for the phospholipid flippase in mammals, but its cellular function remains unknown. We here showed that ATP8A1 was associated with CDC50A in CHO cells and depletion of either CDC50A or ATP8A1 strongly inhibited cell migration. This defective cell migration was confirmed by using two different migration assays as well as by using other motile cells such as NIH3T3 and MEF cells (Figs. 2, B and C, and 5, B and C). These results provide the first evidence that the phospholipid flippase complex of ATP8A1 and CDC50A plays a critical role in cell migration. As discussed below, we also suggest that the inward translocation of PE at the plasma membrane is pivotal for the formation of membrane ruffles to promote cell migration.
A previous study demonstrated that the ATPase activity of murine ATP8A1 that was purified to homogeneity was activated maximally by PS and minimally by PE in an in vitro assay using detergent micelles, suggesting that PS is the dominant substrate for ATP8A1 (24). We unexpectedly found that the translocation of NBD-PS at the plasma membrane was not affected in the ATP8A1-deficient cells, whereas that of NBD-PE was strongly inhibited (Fig. 6C). Because the translocation of both NBD-PS and NBD-PE was strongly inhibited in the CDC50A-deficient cells, it is likely that another P4-ATPase expressed in CHO cells partly replaces the PS-translocating activity in the ATP8A1-deficient cells. Analyses using the PE- and PS-binding peptides also showed that the transbilayer distribution of PE but not PS was altered in the ATP8A1-deficient cells (Fig. 6D). These observations prompted us to examine whether or not the ATP8A1-mediated translocation of PE at the plasma membrane is involved in cell migration. Studies using the PE-binding peptide (Ro09-0198) and mutant cells with defective PE-synthesis demonstrated that either blockage of PE translocation by the PE-binding peptide or reduction in the PE content strongly inhibited cell migration (Fig. 7). Although further detailed analyses of the substrate specificities of the P4-ATPases expressed in CHO cells will be needed to obtain deeper insight into the role of P4-ATPases in cell migration, this study raises the intriguing possibility that the ATP8A1-mediated translocation of PE at the plasma membrane is involved in cell migration.
A critical question here is how the translocation of PE at the plasma membrane affects the cell migration. In the budding yeast S. cerevisiae, the P4-ATPases Dnf1p and Dnf2p are associated with the Cdc50 family protein Lem3p and localize at the plasma membrane (12). In contrast to the Drs2p·Cdc50p flippase complex that dominantly translocates PS, the Dnf1p/Dnf2p·Lem3p complex has been shown to be responsible for the translocation of PE and PC at the plasma membrane (9–11, 45). Saito et al. (46) demonstrated that the Dnf1p/Dnf2p·Lem3p complex-mediated translocation of the phospholipids at the plasma membrane was required for the polarized cell growth of yeast. Based on the reconstitution study, they proposed that the increased concentration of either PE or PS in the inner leaflet of the plasma membrane stimulates the GTPase-activating protein activity of Rga1p and Rga2p toward Cdc42p, thereby controlling polarized cell growth by down-regulating Cdc42p signaling (46). A recent study by Das et al. (47) showed that the Dnf1p/Dnf2p·Lem3p complex-mediated translocation of PE at the plasma membrane was required for the fast Cdc42p recycling during the formation of cell polarity. They demonstrated that blockage of the inward translocation of PE by either depletion of the Dnf1p/Dnf2p·Lem3p complex or cell-surface immobilization of PE by the PE-binding peptide (Ro09-0198) disrupted the Cdc42p polarity maintained by guanine nucleotide dissociation inhibitor-mediated recycling. They proposed that the enrichment of PS in the inner leaflet of the plasma membrane, which is caused by the blockage of the PE translocation, strengthened the electrostatic interaction between the C-terminal polybasic domain of Cdc42p with the inner membrane leaflet, and thus it inhibited its capture by the cytosolic guanine nucleotide dissociation inhibitor protein Rdi1p for the fast recycling at the polar cortex. A further in vitro reconstitution study using supported lipid bilayers with different PE contents also supported a direct effect of the lipid composition on Cdc42p dissociation by Rdi1p. Based on these observations, it is plausible to speculate that the PE content in the inner leaflet of the plasma membrane, which may be maintained by the activities of the phospholipid flippase, may directly affect the recruitment and activation of molecules that play a crucial role in cell migration.
In mammalian cells, dynamic rearrangement of the actin cytoskeleton provides the driving force for cell migration, where Rho GTPases and their regulatory proteins play a dominant role in controlling signaling cascades (48, 49). Among Rho GTPases, Rac1 activates the polymerization of actin filaments in the lamellipodial protrusions and promotes the formation of membrane ruffles at the leading edge of migrating cells. F-actin staining of the cells treated with the PE-binding peptide exhibited an extensive disassembly of cortical actin filaments and the collapse of membrane ruffles, suggesting that the blockage of PE translocation at the plasma membrane directly affects the remodeling of cortical actin filaments and the formation of membrane ruffles (Fig. 7B). The translocation of Rac1 to the plasma membrane was diminished in the ATP8A1-deficient cells (Fig. 7E). Although the association of Rac1 to membranes was shown to be dependent on its C-terminal polybasic sequence (50, 51), it remains to be clarified whether or not the direct interaction between Rac1 and the membrane is affected by the membrane's lipid composition. Kooijman et al. (52) have shown that the ionization property of phosphatidic acid is strongly influenced by the interaction with other hydrogen donors such as PE present in the membrane; the hydrogen bond formation of phosphatidic acid with PE leads to a reduced pKa2 of the phosphomonoester head group, thereby resulting in a higher negative charge of the molecule in the physiological pH range. More importantly, the intermolecular hydrogen bond formation between PE and phosphoinositides such as phosphatidylinositol (4,5)-bisphosphate may also affect the ionization properties of the phosphoinositides in membranes (53). A time-lapse observation of localization of ATP8A1 in serum-induced actively migrating cells showed that ATP8A1 was localized on the highly motile ruffling membranes at the cell front, again suggesting the role of ATP8A1 in the formation of membrane ruffles to promote cell migration (Fig. 7D). It is intriguing to speculate that the local inward translocation of PE at the plasma membrane may influence the ionization property of phosphatidylinositol (4,5)-bisphosphate, which affects the membrane association and activation of various signaling molecules such as Rac1 and those with a pleckstrin homology domain, such as N-WASP and WAVE, that are involved in lamellipodium formation and extension (54, 55).
In addition to affecting the lipid composition and the electrostatic properties of the inner membrane leaflet, the phospholipid flippase complex has been shown to serve as a molecular scaffold to recruit the signaling molecules at the plasma membrane. In S. cerevisiae, Drs2p interacts directly with the Arf GTPase activator Gea2p, which is responsible for the formation of secretory granules/vesicles at the Golgi (56). In mammals, Arf6, one of the ARF GTPases, plays a crucial role in controlling cell migration through the activation of Rac1 as well as by regulating membrane recycling at the leading edge (57, 58). Although a 20-amino acid cytoplasmic tail of Drs2p, which was required for the interaction with Gea2p, is not conserved in the mammalian P4-ATPases, including ATP8A1 (56), this does not exclude the possibility that ATP8A1 may directly interact with signaling molecules such as Arf6 and its activator ARNO (58).
It is also noteworthy that the endocytic recycling of membranes contributes to cell migration by regulating the turnover of adhesion molecules such as integrins at the leading edge (59, 60). In C. elegans, the P4-ATPase TAT-1, the closest homolog of Drs2p and ATP8A1, localizes at the plasma membrane and is required for endocytosis and lysosome biogenesis as well as for the maintenance of PS asymmetry at the plasma membrane (25, 61, 62). TAT-1 forms a complex with the Cdc50 family protein CHAT-1, and the TAT-1·CHAT-1 complex is found in tubular membrane structures along the sorting and recycling pathway, where they regulate the multiple steps in the endocytic sorting and recycling processes (61, 62). As proposed for the functional role of Drs2p in vesicular trafficking (63, 64), the TAT-1·CHAT-1 complex-mediated translocation of aminophospholipids may create an imbalance in the numbers of phospholipids between the two leaflets, which deforms the membrane into a highly curved structure, thereby driving membrane tubulation and vesicle formation (62). Concerning the dominant localization of the ATP8A1·CDC50A complex in the recycling endosome and also in the membrane ruffles, it is plausible to speculate that the migration defects observed with the CDC50A- and ATP8A1-deficient cells are caused by the impaired endocytosis or recycling from endosome back to the plasma membrane. In the CDC50A-depleted cells, however, no significant change was observed in the uptake of the endocytotic membrane markers such as FM1-43 and DiI-LDL nor in the recycling of Cy3-Tf to the cell surface from the endosomal recycling compartments (Fig. 3). These results suggest that the depletion of CDC50A does not significantly affect clathrin-dependent endocytosis. Because recent studies have shown that clathrin-independent endocytosis pathways are involved in cell migration and signaling (65, 66), further detailed analyses of endocytic recycling of the molecules such as integrins will be needed to clarify the functional role of the ATP8A1·CDC50A complex in cell migration.
In summary, our study demonstrates that the phospholipid flippase complex of ATP8A1 and CDC50A plays a major role in cell migration and suggests that the flippase-mediated translocation of PE at the plasma membrane is involved in the formation of membrane ruffles. It remains to be defined which molecules in the migratory machinery coordinately function with the phospholipid flippase complex and how the localization and function of the effector molecules are controlled by the flippase-mediated changes in the transbilayer distribution of phospholipids.
Acknowledgments
We thank Dr. S. Narumiya (Kyoto University) and Dr. H. Niwa (RIKEN Center for Developmental Biology) for providing the pEGFP-Rac1 construct and the pCAGGSneodelEcoRI vector, respectively.
This work was supported by a grant-in-aid for scientific research, the 21st Century COE Program “Genome Science,” a grant-in-aid for the Global COE Program “International Center for Integrated Research and Advanced Education in Materials Science” from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by a research grant from Astellas Foundation for Research on Metabolic Disorders.
- PS
- phosphatidylserine
- PE
- phosphatidylethanolamine
- P4-ATPase
- type IV P-type ATPase
- PC
- phosphatidylcholine
- NBD
- 7-nitrobenz-2-oxa-1,3-diazol-4-yl
- PNGase F
- peptide:N-glycosidase F
- Tf
- transferrin
- TfR
- Tf receptor
- DiI
- 1,1′-dilinoleyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- ER
- endoplasmic reticulum
- MEF
- mouse embryonic fibroblast
- tet
- tetracycline.
REFERENCES
- 1. Zachowski A. (1993) Phospholipids in animal eukaryotic membranes. Transverse asymmetry and movement. Biochem. J. 294, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sebastian T. T., Baldridge R. D., Xu P., Graham T. R. (2012) Phospholipid flippases. Building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta 1821, 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tanaka K., Fujimura-Kamada K., Yamamoto T. (2011) Functions of phospholipid flippases. J. Biochem. 149, 131–143 [DOI] [PubMed] [Google Scholar]
- 4. Daleke D. L. (2007) Phospholipid flippases. J. Biol. Chem. 282, 821–825 [DOI] [PubMed] [Google Scholar]
- 5. Holthuis J. C., Levine T. P. (2005) Lipid traffic. Floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6, 209–220 [DOI] [PubMed] [Google Scholar]
- 6. Paulusma C. C., Oude Elferink R. P. (2005) The type 4 subfamily of P-type ATPases, putative aminophospholipid translocases with a role in human disease. Biochim. Biophys. Acta 1741, 11–24 [DOI] [PubMed] [Google Scholar]
- 7. Zachowski A., Henry J. P., Devaux P. F. (1989) Control of transmembrane lipid asymmetry in chromaffin granules by an ATP-dependent protein. Nature 340, 75–76 [DOI] [PubMed] [Google Scholar]
- 8. Tang X., Halleck M. S., Schlegel R. A., Williamson P. (1996) A subfamily of P-type ATPases with aminophospholipid transporting activity. Science 272, 1495–1497 [DOI] [PubMed] [Google Scholar]
- 9. Kato U., Emoto K., Fredriksson C., Nakamura H., Ohta A., Kobayashi T., Murakami-Murofushi K., Kobayashi T., Umeda M. (2002) A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J. Biol. Chem. 277, 37855–37862 [DOI] [PubMed] [Google Scholar]
- 10. Hanson P. K., Malone L., Birchmore J. L., Nichols J. W. (2003) Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J. Biol. Chem. 278, 36041–36050 [DOI] [PubMed] [Google Scholar]
- 11. Pomorski T., Lombardi R., Riezman H., Devaux P. F., van Meer G., Holthuis J. C. (2003) Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell 14, 1240–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. (2004) Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 3418–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Furuta N., Fujimura-Kamada K., Saito K., Yamamoto T., Tanaka K. (2007) Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell 18, 295–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lenoir G., Williamson P., Puts C. F., Holthuis J. C. (2009) Cdc50p plays a vital role in the ATPase reaction cycle of the putative aminophospholipid transporter Drs2p. J. Biol. Chem. 284, 17956–17967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takahashi Y., Fujimura-Kamada K., Kondo S., Tanaka K. (2011) Isolation and characterization of novel mutations in CDC50, the non-catalytic subunit of the Drs2p phospholipid flippase. J. Biochem. 149, 423–432 [DOI] [PubMed] [Google Scholar]
- 16. Zhou X., Graham T. R. (2009) Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc. Natl. Acad. Sci. U.S.A. 106, 16586–16591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baldridge R. D., Graham T. R. (2012) Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc. Natl. Acad. Sci. U.S.A. 109, E290–E298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paulusma C. C., Folmer D. E., Ho-Mok K. S., de Waart D. R., Hilarius P. M., Verhoeven A. J., Oude Elferink R. P. (2008) ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology 47, 268–278 [DOI] [PubMed] [Google Scholar]
- 19. van der Velden L. M., Wichers C. G., van Breevoort A. E., Coleman J. A., Molday R. S., Berger R., Klomp L. W., van de Graaf S. F. (2010) Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. J. Biol. Chem. 285, 40088–40096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coleman J. A., Molday R. S. (2011) Critical role of the β-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J. Biol. Chem. 286, 17205–17216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takatsu H., Baba K., Shima T., Umino H., Kato U., Umeda M., Nakayama K., Shin H. W. (2011) ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J. Biol. Chem. 286, 38159–38167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Daleke D. L., Lyles J. V. (2000) Identification and purification of aminophospholipid flippases. Biochim. Biophys. Acta 1486, 108–127 [DOI] [PubMed] [Google Scholar]
- 23. Smriti, Nemergut E. C., Daleke D. L. (2007) ATP-dependent transport of phosphatidylserine analogues in human erythrocytes. Biochemistry 46, 2249–2259 [DOI] [PubMed] [Google Scholar]
- 24. Paterson J. K., Renkema K., Burden L., Halleck M. S., Schlegel R. A., Williamson P., Daleke D. L. (2006) Lipid-specific activation of the murine P4-ATPase Atp8a1 (ATPase II). Biochemistry 45, 5367–5376 [DOI] [PubMed] [Google Scholar]
- 25. Darland-Ransom M., Wang X., Sun C. L., Mapes J., Gengyo-Ando K., Mitani S., Xue D. (2008) Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science 320, 528–531 [DOI] [PubMed] [Google Scholar]
- 26. Cacciagli P., Haddad M. R., Mignon-Ravix C., El-Waly B., Moncla A., Missirian C., Chabrol B., Villard L. (2010) Disruption of the ATP8A2 gene in a patient with a t(10;13) de novo balanced translocation and a severe neurological phenotype. Eur. J. Hum. Genet. 18, 1360–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coleman J. A., Kwok M. C., Molday R. S. (2009) Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J. Biol. Chem. 284, 32670–32679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coleman J. A., Vestergaard A. L., Molday R. S., Vilsen B., Andersen J. P. (2012) Critical role of a transmembrane lysine in aminophospholipid transport by mammalian photoreceptor P4-ATPase ATP8A2. Proc. Natl. Acad. Sci. U.S.A. 109, 1449–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bull L. N., van Eijk M. J., Pawlikowska L., DeYoung J. A., Juijn J. A., Liao M., Klomp L. W., Lomri N., Berger R., Scharschmidt B. F., Knisely A. S., Houwen R. H., Freimer N. B. (1998) A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat. Genet. 18, 219–224 [DOI] [PubMed] [Google Scholar]
- 30. Klomp L. W., Vargas J. C., van Mil S. W., Pawlikowska L., Strautnieks S. S., van Eijk M. J., Juijn J. A., Pabón-Peña C., Smith L. B., DeYoung J. A., Byrne J. A., Gombert J., van der Brugge G., Berger R., Jankowska I., Pawlowska J., Villa E., Knisely A. S., Thompson R. J., Freimer N. B., Houwen R. H., Bull L. N. (2004) Characterization of mutations in ATP8B1 associated with hereditary cholestasis. Hepatology 40, 27–38 [DOI] [PubMed] [Google Scholar]
- 31. Paulusma C. C., Groen A., Kunne C., Ho-Mok K. S., Spijkerboer A. L., Rudi de Waart D., Hoek F. J., Vreeling H., Hoeben K. A., van Marle J., Pawlikowska L., Bull L. N., Hofmann A. F., Knisely A. S., Oude Elferink R. P. (2006) Atp8b1 deficiency in mice reduces resistance of the canalicular membrane to hydrophobic bile salts and impairs bile salt transport. Hepatology 44, 195–204 [DOI] [PubMed] [Google Scholar]
- 32. Wang L., Beserra C., Garbers D. L. (2004) A novel aminophospholipid transporter exclusively expressed in spermatozoa is required for membrane lipid asymmetry and normal fertilization. Dev. Biol. 267, 203–215 [DOI] [PubMed] [Google Scholar]
- 33. Xu P., Okkeri J., Hanisch S., Hu R. Y., Xu Q., Pomorski T. G., Ding X. Y. (2009) Identification of a novel mouse P4-ATPase family member highly expressed during spermatogenesis. J. Cell Sci. 122, 2866–2876 [DOI] [PubMed] [Google Scholar]
- 34. Siggs O. M., Arnold C. N., Huber C., Pirie E., Xia Y., Lin P., Nemazee D., Beutler B. (2011) The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nat. Immunol. 12, 434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yabas M., Teh C. E., Frankenreiter S., Lal D., Roots C. M., Whittle B., Andrews D. T., Zhang Y., Teoh N. C., Sprent J., Tze L. E., Kucharska E. M., Kofler J., Farell G. C., Bröer S., Goodnow C. C., Enders A. (2011) ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat. Immunol. 12, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rottner K., Stradal T. E. (2011) Actin dynamics and turnover in cell motility. Curr. Opin. Cell Biol. 23, 569–578 [DOI] [PubMed] [Google Scholar]
- 37. Ridley A. J. (2011) Life at the leading edge. Cell 145, 1012–1022 [DOI] [PubMed] [Google Scholar]
- 38. Emoto K., Kuge O., Nishijima M., Umeda M. (1999) Isolation of a Chinese hamster ovary cell mutant defective in intramitochondrial transport of phosphatidylserine. Proc. Natl. Acad. Sci. U.S.A. 96, 12400–12405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huttenlocher A., Ginsberg M. H., Horwitz A. F. (1996) Modulation of cell migration by integrin-mediated cytoskeletal linkages and ligand-binding affinity. J. Cell Biol. 134, 1551–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McIntyre J. C., Sleight R. G. (1991) Fluorescence assay for phospholipid membrane asymmetry. Biochemistry 30, 11819–11827 [DOI] [PubMed] [Google Scholar]
- 41. Chen S., Wang J., Muthusamy B. P., Liu K., Zare S., Andersen R. J., Graham T. R. (2006) Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic 7, 1503–1517 [DOI] [PubMed] [Google Scholar]
- 42. Emoto K., Umeda M. (2000) An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J. Cell Biol. 149, 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emoto K., Inadome H., Kanaho Y., Narumiya S., Umeda M. (2005) Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J. Biol. Chem. 280, 37901–37907 [DOI] [PubMed] [Google Scholar]
- 44. Nam K. S., Igarashi K., Umeda M., Inoue K. (1990) Production and characterization of monoclonal antibodies that specifically bind to phosphatidylcholine. Biochim. Biophys. Acta 1046, 89–96 [DOI] [PubMed] [Google Scholar]
- 45. Stevens H. C., Malone L., Nichols J. W. (2008) The putative aminophospholipid translocases, DNF1 and DNF2, are not required for 7-nitrobenz-2-oxa-1,3-diazol-4-yl-phosphatidylserine flip across the plasma membrane of Saccharomyces cerevisiae. J. Biol. Chem. 283, 35060–35069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saito K., Fujimura-Kamada K., Hanamatsu H., Kato U., Umeda M., Kozminski K. G., Tanaka K. (2007) Transbilayer phospholipid flipping regulates Cdc42p signaling during polarized cell growth via Rga GTPase-activating proteins. Dev. Cell 13, 743–751 [DOI] [PubMed] [Google Scholar]
- 47. Das A., Slaughter B. D., Unruh J. R., Bradford W. D., Alexander R., Rubinstein B., Li R. (2012) Flippase-mediated phospholipid asymmetry promotes fast Cdc42 recycling in dynamic maintenance of cell polarity. Nat. Cell Biol. 14, 304–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fukata M., Nakagawa M., Kaibuchi K. (2003) Roles of Rho-family GTPases in cell polarisation and directional migration. Curr. Opin. Cell Biol. 15, 590–597 [DOI] [PubMed] [Google Scholar]
- 49. Raftopoulou M., Hall A. (2004) Cell migration. Rho GTPases lead the way. Dev. Biol. 265, 23–32 [DOI] [PubMed] [Google Scholar]
- 50. Kreck M. L., Freeman J. L., Abo A., Lambeth J. D. (1996) Membrane association of Rac is required for high activity of the respiratory burst oxidase. Biochemistry 35, 15683–15692 [DOI] [PubMed] [Google Scholar]
- 51. Gorzalczany Y., Alloul N., Sigal N., Weinbaum C., Pick E. (2002) A prenylated p67phox-Rac1 chimera elicits NADPH-dependent superoxide production by phagocyte membranes in the absence of an activator and of p47phox. Conversion of a pagan NADPH oxidase to monotheism. J. Biol. Chem. 277, 18605–18610 [DOI] [PubMed] [Google Scholar]
- 52. Kooijman E. E., Carter K. M., van Laar E. G., Chupin V., Burger K. N., de Kruijff B. (2005) What makes the bioactive lipids phosphatidic acid and lysophosphatidic acid so special? Biochemistry 44, 17007–17015 [DOI] [PubMed] [Google Scholar]
- 53. Kooijman E. E., King K. E., Gangoda M., Gericke A. (2009) Ionization properties of phosphatidylinositol polyphosphates in mixed model membranes. Biochemistry 48, 9360–9371 [DOI] [PubMed] [Google Scholar]
- 54. Ling K., Schill N. J., Wagoner M. P., Sun Y., Anderson R. A. (2006) Movin' on up. The role of PtdIns(4,5)P2 in cell migration. Trends Cell Biol. 16, 276–284 [DOI] [PubMed] [Google Scholar]
- 55. Takenawa T., Suetsugu S. (2007) The WASP-WAVE protein network. Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37–48 [DOI] [PubMed] [Google Scholar]
- 56. Chantalat S., Park S. K., Hua Z., Liu K., Gobin R., Peyroche A., Rambourg A., Graham T. R., Jackson C. L. (2004) The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J. Cell Sci. 117, 711–722 [DOI] [PubMed] [Google Scholar]
- 57. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins. Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 58. Myers K. R., Casanova J. E. (2008) Regulation of actin cytoskeleton dynamics by Arf family GTPases. Trends Cell Biol. 18, 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Caswell P., Norman J. (2008) Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 18, 257–263 [DOI] [PubMed] [Google Scholar]
- 60. Parsons J. T., Horwitz A. R., Schwartz M. A. (2010) Cell adhesion. Integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ruaud A. F., Nilsson L., Richard F., Larsen M. K., Bessereau J. L., Tuck S. (2009) The C. elegans P4-ATPase TAT-1 regulates lysosome biogenesis and endocytosis. Traffic 10, 88–100 [DOI] [PubMed] [Google Scholar]
- 62. Chen B., Jiang Y., Zeng S., Yan J., Li X., Zhang Y., Zou W., Wang X. (2010) Endocytic sorting and recycling require membrane phosphatidylserine asymmetry maintained by TAT-1/CHAT-1. PLoS Genet. 6, e1001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Graham T. R. (2004) Flippases and vesicle-mediated protein transport. Trends Cell Biol. 14, 670–677 [DOI] [PubMed] [Google Scholar]
- 64. Pomorski T., Menon A. K. (2006) Lipid flippases and their biological functions. Cell. Mol. Life Sci. 63, 2908–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ivaska J., Heino J. (2011) Cooperation between integrins and growth factor receptors in signaling and endocytosis. Annu. Rev. Cell Dev. Biol. 27, 291–320 [DOI] [PubMed] [Google Scholar]
- 66. Parachoniak C. A., Park M. (2012) Dynamics of receptor trafficking in tumorigenicity. Trends Cell Biol. 22, 231–240 [DOI] [PubMed] [Google Scholar]