Abstract
Channelrhodopsins serve as photoreceptors that control the motility behavior of green flagellate algae and act as light-gated ion channels when heterologously expressed in animal cells. Here, we report direct measurements of proton transfer from the retinylidene Schiff base in several channelrhodopsin variants expressed in HEK293 cells. A fast outward-directed current precedes the passive channel current that has the opposite direction at physiological holding potentials. This rapid charge movement occurs on the timescale of the M intermediate formation in microbial rhodopsins, including that for channelrhodopsin from Chlamydomonas augustae and its mutants, reported in this study. Mutant analysis showed that the glutamate residue corresponding to Asp85 in bacteriorhodopsin acts as the primary acceptor of the Schiff-base proton in low-efficiency channelrhodopsins. Another photoactive-site residue corresponding to Asp212 in bacteriorhodopsin serves as an alternative proton acceptor and plays a more important role in channel opening than the primary acceptor. In more efficient channelrhodopsins from Chlamydomonas reinhardtii, Mesostigma viride, and Platymonas (Tetraselmis) subcordiformis, the fast current was apparently absent. The inverse correlation of the outward proton transfer and channel activity is consistent with channel function evolving in channelrhodopsins at the expense of their capacity for active proton transport.
Introduction
Genomes of microorganisms from all three domains of life encode retinylidene proteins with shared sequence homology that fulfill the functions of solar energy conversion and photosensory transduction (1). Members of a specific subclass of sensory rhodopsins mediate phototaxis and the photophobic response in green flagellate algae (2–4). Their photoexcitation leads to generation of a cascade of electrical currents that depolarize the cell membrane (5). When expressed in animal cells, these proteins act as light-gated cation channels, which has earned them the name channelrhodopsins (6,7). Because of this unique property, channelrhodopsins are widely used as molecular tools for regulation of neuronal firing, muscle contraction, and other cellular processes by light (8–10). Thirteen native channelopsin (i.e., channelrhodopsin apoprotein) sequences have been reported to date, and this number is expected to increase significantly as more algal genomic information becomes available.
In all microbial rhodopsins, retinal is attached by a protonated Schiff-base linkage to a lysyl amino group in the middle of the seventh transmembrane helix. Its photoisomerization triggers a cycle of photochemical reactions and conformational changes of the protein that drive ion transport or initiate the signaling process. A transient deprotonation of the Schiff base is a common step in the photocycles of microbial rhodopsins that function as proton pumps or sensory receptors. Schiff-base deprotonation and reprotonation are spectroscopically manifested as the appearance and decay, respectively, of a near-UV-absorbing intermediate designated the M intermediate. In rhodopsin proton pumps, the Schiff-base proton is transferred to an outwardly located acceptor, as a first step in proton translocation (11–13). A corresponding outward proton transfer occurs in haloarchaeal photosensors HsSRII and NpSRII (14–16), whereas in the dual-signaling HsSRI, the direction of the Schiff-base proton movement is inward or outward, reflecting the conformational state of the molecule with an inwardly or outwardly connected Schiff base, which determines whether the photomotility response is attractive or repellent, respectively (17–19).
Channelrhodopsin sequences form a distinct branch on a phylogenetic tree of microbial opsins (10,20), and the direction of the movement of the Schiff-base proton in channelrhodopsins cannot be predicted a priori. A recent crystal structure revealed that the overall composition of the Schiff-base counterion complex of a chimeric channelrhodopsin is similar to that of the proton pump bacteriorhodopsin (BR) (21). In fact, the position of the side chain of the Arg82 homolog in the chimera is closer to that in SRII photosensors (22). Channelrhodopsin-2 from Chlamydomonas reinhardtii (CrChR2) has been reported to show a weak proton-pumping activity (23), so its Schiff-base proton is assumed to move to an outwardly located acceptor (24). However, to the best of our knowledge, such movement has not been directly demonstrated in any channelrhodopsin.
Intramolecular charge displacements have been best studied in BR (25–27). They were first characterized in purple membranes attached to a lipid-impregnated polymer film (28) or a planar lipid bilayer (29), or oriented by an electric field (30). All these measuring systems yielded similar signals comprised of the following main phases: 1), an initial negative (i.e., directed toward the cytoplasmic side of the membrane) phase in the submicrosecond to microsecond time domain that reflects the formation of early intermediates of the photocycle (K and L); 2), a positive second phase on the timescale of tens of microseconds associated with the formation of the M intermediate; and 3), positive millisecond phases corresponding to later steps of the photocycle. Similar signals could also be detected in proteorhodopsin proton pumps and transducer-free haloarchaeal sensory rhodopsins by the same methods, or by measuring photoelectric signals in suspensions of intact Escherichia coli cells (31–33). Unfortunately, functional expression of channelrhodopsins in E. coli proved to be difficult. Functional CrChR2 has been produced and purified from Pichia, but so far its photoelectric activity after reconstitution in artificial membranes has only been studied under continuous light with low time resolution (23).
Expression of microbial rhodopsins in Xenopus oocytes or cultured mammalian cells offers a possibility to study their responses in natural biological membranes under defined voltage conditions (34). Here, we examined laser-flash-induced photoelectric currents generated by channelrhodopsins expressed in human embryonic kidney (HEK293) cells. We show that in several channelrhodopsin variants, channel opening is preceded by fast current transients similar to those observed in BR and other microbial rhodopsins, including a fast positive component indicating proton transfer from the Schiff base to an outwardly located acceptor(s). Other native channelrhodopsin variants we tested, including CrChR2, most frequently used in optogenetic applications, and PsChR, a novel variant we have cloned from the marine prasinophyte alga Platymonas (Tetraselmis) subcordiformis, do not show such fast currents. This unexpected diversity may provide a better insight into mechanisms of ionic conductance and evolution of channelrhodopsins.
Materials and Methods
Platymonas (Tetraselmis) subcordiformis and Dunaliella salina were obtained from the UTEX Culture Collection of Algae (#71 and #LB 1644, respectively) and grown in modified artificial seawater medium A (35) and Johnson's medium (36), respectively, under a 16/8 light/dark cycle (light: 2000 and 3000 lux, respectively). Cloning of channelopsin sequences from P. subcordiformis and D. salina is described in the Supporting Material.
The archaeorhodopsin-3 (AR-3) coding sequence was obtained from Edward S. Boyden (Massachusetts Institute of Technology, Cambridge, MA) and cloned into the E. coli expression vector pET28b(+) under control of an IPTG-inducible promoter, and into the mammalian expression vector (see below). E. coli strain BL21(DE3) was transformed with the AR-3-carrying expression vector, grown till OD600 = 0.4 and induced by IPTG in the presence of 5 μM of all-trans retinal. The culture was harvested after 4 h, washed in distilled water and transferred to low-ionic-strength medium consisting of (in mM) 1.5 NaCl, 0.15 CaCl2, 0.15 MgCl2, and 5 Tris, pH 7.2. Photocurrents in E. coli cell suspensions were evoked by a Vibrant HE 355 II tunable laser (5 ns, 35 mJ; Opotek, Carlsbad, CA) with flashes set to the wavelengths of maximum absorption of AR-3 and its mutant applied along the direction between two platinum electrodes and recorded as described previously (33).
The mammalian expression vector pcDNA3.1/VcChR1-EYFP that contains a human-codon-optimized version of the 7TM domain of VcChR1 flanked with BamHI and NotI sites (37) was provided by Karl Deisseroth (Stanford University, Stanford, CA). For expression of other channelrhodopsins, the VcChR1 sequence was replaced with algal cDNA fragments encoding the 7TM domains of channelrhodopsins CaChR1 (residues 1–352), DsChR1 (residues 1–365), MvChR1 (residues 1–331), CrChR2 (residues 1–309), and PsChR (residues 1–326). Mutations were introduced using a QuikChange XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) and verified by DNA sequencing.
HEK293 cells were transfected using the TransPass COS/293 transfection reagent (New England Biolabs, Ipswich, MA). All-trans retinal (Sigma, St. Louis, MO) was added as a stock solution in ethanol at a final concentration of 5 μM. Measurements were performed 48–72 h after transfection with an Axopatch 200B amplifier (Molecular Devices, Union City, CA) with a 10-kHz filter. The signals were digitized with a Digidata 1440A using pClamp 10 software (both from Molecular Devices, Eugene, OR) at a sampling rate of 4 μs/point. Patch pipettes with resistances of 2–5 MΩ were fabricated from borosilicate glass and filled with a solution containing (in mM) 126 KCl, 2 MgCl2, 0.5 CaCl2, 5 EGTA, and 25 HEPES, pH 7.4, unless otherwise indicated. The bath solution contained (in mM) 150 NaCl, 1.8 CaCl2, 1 MgCl2, 5 glucose, and 10 HEPES, pH 7.4, unless otherwise indicated. Flash excitation (pulsewidth 6 ns, energy 12 mJ) was via a Minilite Nd:YAG laser (Continuum, Santa Clara, CA) at wavelength 532 nm and an interval of 2 s between flashes, unless otherwise indicated. A laser artifact measured with a blocked optical path was digitally subtracted from the recorded traces. For further analysis, the signals were logarithmically averaged with a custom-created computer algorithm. Numerical data in the text are presented as the mean ± SE.
A Pichia clone that expresses the 7TM domain of wild-type CaChR1 was obtained as described previously (38), and a similar procedure was used to select clones that express the CaChR1_E169Q and CaChR1_D299N mutants. Cells were grown in buffered minimal methanol yeast medium, and expression was induced by the addition of 0.5% methanol every 24 h in the presence of 5 μM all-trans retinal. Cells were grown for two days, harvested by low-speed centrifugation, and disrupted by a bead beater. Membrane fragments were collected by centrifugation for 1 h at 48,000 rpm. The proteins were partially purified on a Ni-NTA agarose column (Qiagen, Hilden, Germany) after solubilization by incubation with 2% dodecyl maltoside for 1 h.
Absorption changes of Pichia-expressed pigments were induced by a Surelight I Nd-YAG laser flash (532 nm, 6 ns, 40 mJ; Continuum, Santa Clara, CA) and measured with a laboratory-constructed cross-beam measuring system, as described earlier (39). For calculation of the relative yields of the M-like intermediate formation in wild-type CaChR1 and its mutants, flash-induced absorbance changes at the maximal absorption of the M intermediate were normalized to the pigment concentration (maximal absorption of the unphotolysed state) and corrected for the relative absorption at the excitation wavelength (532 nm) according to fluence-response curves.
Results
Testing the measuring system with a proton pump
The time resolution of whole-cell patch-clamp measurements is limited by the series resistance and the membrane capacitance of the cell. To estimate the degree of signal integration by our measuring system and facilitate interpretation of fast currents generated in HEK cells by channelrhodopsins, we first examined signals from the proton pump archaerhodopsin-3 (AR-3, aka Arch (40,41)). Unlike channelrhodopsins, this protein can also be produced in E. coli cells, in suspensions of which intramolecular charge movements associated with rhodopsin photocycling can be recorded with time resolution at least an order of magnitude better than that observed for whole-cell patch-clamp recording (33). The outward proton-transfer current recorded from wild-type AR-3 in E. coli suspensions (Fig. 1, upper dashed curve) could also be detected by whole-cell patch clamp in HEK cells (Fig. 1, upper solid curve). However, in the latter system, its peak was almost 10-fold later than when measured in E. coli suspensions due to integration by the measuring circuit.
Figure 1.

Photoelectric signals generated by the wild-type proton pump AR-3 (above zero line) and its D95N mutant (below zero line) in E. coli suspensions (dashed lines) and HEK293 cells (solid lines) in response to a 6-ns laser flash (532 nm). The signals recorded in E. coli suspensions were normalized to those measured in HEK cells. The arrows show a shift of the peak times and a decrease in the relative amplitude of the negative phase caused by a lower time resolution of measurements in HEK cells compared to those in E. coli suspensions.
Initial stages of the charge transfer in rhodopsins associated with the formation of K and L intermediates of the photocycle occur on a submicrosecond timescale (for review, see Kaulen (26) and Dér and Keszthelyi (27)) and are not resolved even when recorded in E. coli suspensions. The initial negative current overlaps to a great extent the subsequent fast positive current associated with the formation of the M intermediate, and their kinetics and amplitudes influence each other (Fig. 1, upper dashed curve). When the positive current was suppressed by neutralization of the primary proton acceptor Asp95 in the AR-3_D95N mutant, both techniques registered a large, unresolved negative current (Fig. 1, lower), which was not resolved in HEK cells expressing the wild-type. In E. coli, where this signal was also visible in the wild-type, its apparent peak shifted to longer times in the mutant, as expected due to suppression of the subsequent positive peak. These results demonstrated that both the initial negative and subsequent fast positive currents generated by microbial rhodopsins can be detected and semiquantitatively analyzed in HEK cells, although their kinetics cannot be directly resolved and their strong overlap should be taken into account when interpreting the results obtained.
Channelrhodopsins with outward proton transfer
CaChR1from Chlamydomonas augustae demonstrates typical channel activity when expressed in HEK cells (38). Electrical signals generated by CaChR1 in response to a laser flash showed complex kinetics (Fig. 2). At negative holding voltages (Vh), a small negative peak appeared at ∼25 μs after the flash, followed by a larger positive peak at 130–140 μs and a negative peak of channel current at 1–2 ms. The relative contributions of all currents to the net signal kinetics were independent of the flash intensity and the time interval between successive flashes (Fig. S1 in the Supporting Material), indicating that they were initiated by excitation of the unphotolyzed pigment rather than subsequent photocycle intermediates. At the reversal potential (Vr) for passive channel current (∼0 mV), the trace was dominated by the fast positive current. Net signal traces recorded at other holding voltages (Fig. 2, B and C, black lines) could be fit with superposition of a properly scaled fast positive current (Fig. 2, B and C, red lines) and a slow passive channel current (Fig. 2, B and C, blue lines).
Figure 2.
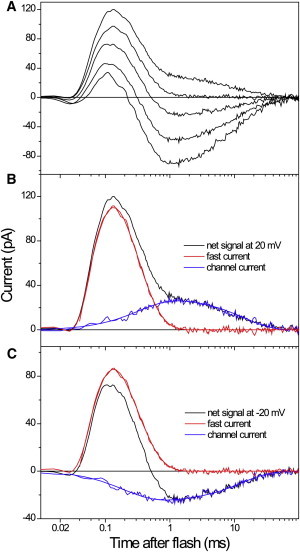
(A) Electrical currents generated by CaChR1 expressed in an HEK293 cell in response to a laser flash at the holding potential applied in 20-mV increments from −60 to 20 mV (bottom to top). (B and C) Decomposition of the net signals recorded at 20 mV (B) and −20 mV (C) in two components. Shown are the recorded traces (black lines), the properly scaled fast positive current, obtained by measuring the signal at 0 mV (red lines), and the channel current, revealed by subtraction of the fast current from the net signal (blue lines). The smooth lines are the results of fitting of each of the two components with two exponentials.
The voltage dependence of the fast positive current was evident from the presence of a distinct peak in the differential signals calculated by subtraction of traces recorded at different Vh (Fig. S2). The current-voltage relationship (I-V curve) of the current averaged over the time of the fast positive phase was linear and crossed the zero line at ∼−95 mV, i.e., it was dramatically shifted to negative voltages with respect to the reversal potential of channel current (Fig. 3, open squares and circles, respectively). When the amplitude of the fast positive current was corrected for the contribution of channel current, extrapolation of its voltage dependence predicted its complete inhibition at ∼−200 mV (Fig. 3, solid squares).
Figure 3.

Voltage dependence of the mean current measured between 50 and 150 μs (open squares) and between 0.75 and 1.55 ms (open circles) after the flash, and of the amplitude of the fast positive current (solid squares), corrected for contribution of channel current. Data are represented as the mean ± SE of experiments in 6 cells.
In a previous article, we reported functional expression and purification of CaChR1 from Pichia (38). Flash photolysis measurements of CaChR1 revealed a photocycle typical of microbial rhodopsins with K-, M-, and O-like intermediates (Fig. 4 A). Two major components of the M-like intermediate formation in CaChR1 had the time constants ∼20 and 110 μs and almost equal amplitudes (Fig. 4 B, black line). These values are only slightly slower than those reported earlier for the appearance of the M-like intermediate in the photocycle of ChR2 from C. reinhardtii (42). However, instead of a fast decay observed in CrChR2 on the timescale of several milliseconds, the concentration of the M-like intermediate of CaChR1 remained stable or even slightly increased with τ ∼ 3 ms (Fig. 4 B, black line) and decayed very slowly with time constants of 32 and 200 ms (data not shown). The fast components of the M formation in the photocycle of detergent-purified CaChR1, as probed by flash photolysis, roughly correlated with the time domain of the fast positive electrical current. An exact temporal correlation between the fast electrical and optical components could not be expected because on the one hand, the time resolution of patch-clamp current measurements was limited, and on the other, the photocycle rate of detergent-solubilized CaChR1 was around three times slower than that of the pigment in intact Pichia membranes (Fig. S3).
Figure 4.

(A) Spectral dependence of flash-induced absorbance changes in detergent-purified wild-type CaChR1. (B) Absorbance changes monitored at 390 nm in wild-type CaChR1 (black), CaChR1_E169Q (red), and CaChR1_D299N (green) in response to a 6-ns laser flash (532 nm).The traces in the main figure were arbitrarily scaled for better visualization of changes in the rates of the M intermediate accumulation. (Inset) Relative yields of the M formation.
Neither the amplitude nor voltage dependence of the fast positive current changed upon a 100-fold decrease in the Na+ concentration in the bath, whereas the amplitude of the channel current decreased by ∼28% and its Vr shifted ∼12 mV to negative values, indicating that predominantly proton-selective CaChR1 (38) is also permeable for Na+. The fast positive current also did not change when K+ in the pipette solution was substituted with Na+, which rules out a contribution of a passive K+ efflux to the current.
Weak dependence of the fast positive current on the membrane potential and cation composition of the media, as well as its semiquantitative correlation with the time course of the M-intermediate formation led us to the conclusion that it reflects proton transfer from the Schiff base to an outwardly located acceptor(s). This interpretation was confirmed in experiments with site-specific mutations. Neutralization of Glu169, which is homologous to the primary proton acceptor Asp85 in BR, led to a dramatic, almost 20-fold decrease in the amplitude of the fast positive current (Fig. 5 A, red line). Similar effects have been observed previously in the D85N mutant of BR (43,44) and the corresponding mutants of other microbial rhodopsins (33). The remaining positive current was slow, with a peak time of ∼220 μs. In the E169Q mutant, the unresolved fast negative signal related to early stages of the photocycle became visible, whereas in the wild-type CaChR1 it was apparently canceled by the fast outwardly directed proton movement. Remarkably, channel current in this mutant was suppressed to approximately the same degree as the fast positive current (Fig. 5 B), so that their ratio remained almost equal to that in the wild-type, 1.19 ± 0.46 (n = 5 cells) and 1.24 ± 0.32 (n = 7 cells), respectively.
Figure 5.
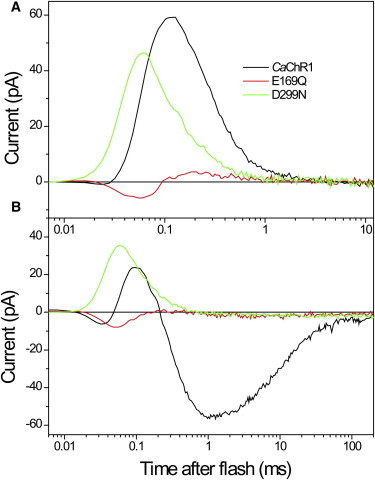
Electrical currents recorded from wild-type CaChR1 (black lines) and its E169Q (red lines) and D299N (green lines) mutants expressed in HEK293 cells at the reversal potential for channel currents to minimize their contribution to the signal kinetics (A), and at −60 mV holding potential (B).
Neutralization of Asp299, which is homologous to the second aspartate of the complex counterion of the Schiff base in BR (Asp212), produced different changes in photocurrents. The amplitude of the fast positive current measured at the reversal potential for channel current was only slightly reduced, and at −60 mV was even higher than that in the wild-type at the same voltage (Fig. 5, green lines). Moreover, the current in the D299N mutant accelerated relative to that in wild-type, so that its peak time decreased from ∼125 μs to ∼50 μs.
Despite only a slight reduction of the Schiff-base deprotonation current in the CaChR1_D299N mutant, its channel current was suppressed even more than that of the CaChR1_E169Q mutant. The ratio of the channel current measured at −60 mV to the fast positive current measured at the Vr for channel current was ∼25-fold smaller in the CaChR1_D299N mutant (0.048 ± 0.007, n = 8) than in the wild-type or in CaChR1_E169Q. The remaining small channel current in CaChR1_D299N was dramatically slower compared to that in the wild-type. In particular, the time constant of the fast component of channel closing increased from 10 ms in the wild-type to ∼400 ms in the mutant.
CaChR1_E169Q and CaChR1_D299N were expressed in Pichia, and their photocycles were analyzed by flash photolysis. Accumulation of an M intermediate was found in both mutants (Fig. 4 B). The yields of M formation in the mutants were even higher than in the wild-type (2.75- and 1.63-fold, respectively (Fig. 4 B, inset)) due to a much slower rate of its disappearance. In full agreement with the results of electrical measurements, the M intermediate formation was slowed down in the CaChR1_E169Q mutant and accelerated in the CaChR1_D299N mutant compared to its rate in the wild-type (Fig. 4 B). The half-rise time of the M intermediate formation in these mutants was around seven times larger and around six times smaller, respectively, relative to that observed in the wild-type. In all three proteins, the rise of the M intermediate formation could be fit with three exponentials and the decay with two exponentials (see the corresponding time constants in Table S1).
Summarizing the above results, our interpretation is that in CaChR1, the residue homologous to Asp85 in BR acts as a primary Schiff-base proton acceptor. The residue homologous to Asp212 appears to be an alternative proton acceptor, which nevertheless plays a key role in channel function. Neutralization of either of these residues changes the rate of the fast electric current and M formation. These changes can be explained by alteration of electrostatic interactions, hydrogen bonding, and elimination of some conformational changes in the complex counterion, as is known, for example, in the R82A/Q mutants of bacteriorhodopsin (43,45,46). Alternatively, two subpopulations of the pigment may exist, in each of which the proton transfers to one of the alternative acceptors and the rates of the two processes are different. In this case, neutralization of the acceptor for which the transfer is slower would result in acceleration of the signal, and vice versa. In deep-water blue-absorbing proteorhodopsin (39), neutralization of the Asp212 homolog selectively eliminated the second positive microsecond current component correlated with the second slower phase of the M formation (33).
A fast current, similar to that produced by CaChR1, was also generated by channelrhodopsin from Volvox carteri (VcChR1). This current, which we interpret by analogy as proton movement from the Schiff base to an outwardly located acceptor, was significantly faster in VcChR1 (peak time, ∼80 μs at Vr (Fig. 6)) than in CaChR1, which is consistent with a faster formation of the M photocycle intermediate by this pigment (47,48). It also demonstrated an even weaker dependence on the holding potential (Fig. S4). Faster generation of the positive current by VcChR1 could be one of the reasons for the apparent absence of the initial unresolved negative phase of the signal, in contrast to wild-type CaChR1 and the CaChR1_E169Q mutant. In a similar way, this initial component could not be resolved in the signals recorded from AR-3 expressed in HEK cells due to its cancellation by a subsequent fast positive current, although it was clearly visible when measured at a higher time resolution in E. coli suspensions (Fig. 1).
Figure 6.

(A) Electrical currents generated by VcChR1 expressed in an HEK293 cell in response to a laser flash at the holding potential applied in 20-mV increments from −60 to 40 mV. (B and C) Electrical currents recorded from wild-type VcChR1 (black lines) and its E118Q (red lines) and D248N (green lines) mutants expressed in HEK293 cells at the reversal potential for channel currents to minimize their contribution to the signal kinetics (B), and at −60 mV holding potential (C).
Under our measuring conditions, the Vr for channel current in VcChR1 was more positive (∼10 mV (Fig. S4)) than that in CaChR1, indicating a higher Na+/H+ permeability ratio. Signals generated by VcChR1 at Vr for channel currents differed from those generated by CaChR1 in that they showed a small additional negative wave at ∼0.5 ms, which could not be eliminated by variation of Vh (Fig. 6 B, black line). This current component may reflect either a slower inward transfer of the Schiff-base proton in molecules with the inwardly connected Schiff base, as has been found in SRI (17,18), or the existence of two open channel states with different ion selectivities, as has been suggested for CrChR2 (49).
Neutralization of either of the two residues homologous to those that form the Schiff-base counterion in BR had effects in VcChR1 qualitatively similar to those observed in CaChR1. Specifically, relative to the wild-type, the VcChR1_E118Q mutant (in which the residue corresponding to Asp85 in BR was mutated) demonstrated a significantly smaller and slower positive current, whereas its channel current was only slightly reduced (Fig. 6, B and C, red lines). In contrast, mutation of Asp248 (corresponding to Asp212 in BR) did not diminish the positive current but greatly suppressed channel current (Fig. 6, B and C, green lines). In the D248N mutant, the ratio of the channel current amplitude at −60 mV to that of the positive current at Vr was only 1.0 ± 0.3 (n = 3), as compared to 7.9 ± 2.0 (n = 5) in the wild-type and 19.2 ± 3.4 (n = 3) in the E118Q mutant. Remarkably, the reversal potential for the remaining channel current in the D248N mutant shifted to ∼0 mV, indicating that neutralization of this residue primarily suppresses cation conductance. These results indicate that in VcChR1, as in CaChR1, the residue corresponding to Asp85 in BR serves as the primary proton acceptor, whereas that corresponding to Asp212 is critical for channel opening.
Except for DsChR1 from D. salina, all channelrhodopsins as yet known contain a Glu residue in the position of the Schiff-base proton acceptor Asp85 in BR, whereas in DsChR1 this site is occupied by alanine (Ala178). Interestingly, the 7TM domain of DsChR1 cloned in our laboratory (accession no. JX983144) showed only 90% identity at the protein level with the previously reported sequence (10). Most differences were found in the N-terminal part of the protein, whereas Ala178 and all other residues so far identified as functionally important were conserved. DsChR1 showed much larger amplitude of the fast unresolved negative current (Fig. 7 A) compared to both wild-type pigments described above. Such large negative currents were also observed upon neutralization of the corresponding residues in the CaChR1_E169Q and VcChR1_E118Q mutants (Figs. 5 and 6, respectively, red lines), and can be explained by the reduction of the amplitude and rate of the subsequent positive proton transfer current. In DsChR1, the apparent peak time of this current was 406 ± 46 μs, n = 6.
Figure 7.
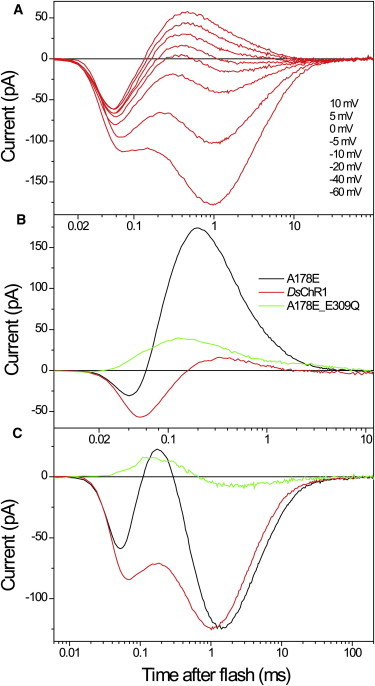
(A) Electrical currents generated by DsChR1 expressed in a HEK293 cell in response to a laser flash at the holding potentials indicated in the figure legend. (B and C) Electrical currents recorded from wild-type DsChR1 (red lines) and its A178E (black lines) and A178E_E309Q (green lines) mutants expressed in HEK293 cells at the reversal potential for channel currents to minimize their contribution to the signal kinetics (B), and at −60 mV holding potential (C).
We generated and tested the DsChR1 mutant in which Ala178 was replaced with Glu, the residue found in this position in all other known wild-type channelrhodopsins. In this mutant, the fast positive current dramatically increased and accelerated relative to that in the wild-type (Fig. 7 B, black line), so that the signal became similar to those recorded from wild-type CaChR1 and VcChR1, which naturally contain Glu in the corresponding position. The A178E mutation of DsChR1 did not significantly affect channel currents, as compared to the wild-type (Fig. 7 C), confirming the above conclusion that although the residue in this position can serve as the most efficient (i.e., primary) proton acceptor, it does not play a key role in channel function. We also generated and tested a double mutant in which only the homolog of Asp85 was present (DsChR1_A178E_E309Q). As in the case of the corresponding CaChR1 mutant, this protein showed accelerated and only moderately decreased outward proton transfer current (Fig. 7 B, green line), whereas the ratio of channel current to proton transfer current was significantly (∼15-fold) diminished (Fig. 7 C, green line). As in the case of VcChR1, neutralization of the homolog of Asp85 in BR did not change the reversal potential of channel current in DsChR1, whereas neutralization of the homolog of Asp212 shifted it to negative values by >20 mV.
Channelrhodopsins without detectable outward proton transfer
CrChR2 from C. reinhardtii is the best studied channelrhodopsin variant and the one most frequently used in optogenetic applications (50). Flash photolysis studies of detergent-purified CrChR2 have established that the formation of its blue-shifted M intermediate occurs with time constants in the 10–100 μs range (42,51), similar to that in CaChR1 (Fig. 4 B). However, no electrical currents corresponding to an outward proton transfer from the Schiff base could be resolved in this time domain upon laser excitation of wild-type CrChR2 at the reversal potential for channel current (Fig. 8). Similarly, no fast currents could be detected at the reversal potential in two other highly efficient channelrhodopsins: MvChR1 from Mesostigma viride (52), and PsChR, which we cloned from the marine alga Platymonas (Tetraselmis) subcordiformis (accession no. JX983143) (Fig. 8 A). The reversal potentials for channel currents in these rhodopsins were positive (9.5, 15, and 10 mV, respectively). That MvChR1 has the highest Vr value indicates that this channelrhodopsin has the highest relative cation permeability of the tested variants, which was confirmed by measuring the shifts of Vr upon a 100-fold decrease in the bath Na+ concentration (data not shown). Under our conditions, the mean amplitude of channel currents generated by the newly cloned PsChR was actually larger than that generated by CrChR2 (Fig. 8 B), which makes the former a promising candidate for optogenetic applications.
Figure 8.

Electrical currents recorded from CrChR2, MvChR1, and PsChR in HEK293 cells in response to a laser flash (solid lines). For comparison, traces from CaChR1 are shown (dashed line). Traces were recorded at the reversal potential for channel currents to minimize their contribution to the signal kinetics (A), and at −60 mV holding potential (B).
Discussion
In this study, we carried out comparative analysis of photoinduced electrical responses generated upon laser flash excitation by several channelrhodopsins. According to our results, all tested channelrhodopsins can be divided into two groups. Representatives of the first group (CaChR1, VcChR1, and DsChR1) exhibit the fast current that reflects transfer of the Schiff-base proton to an outwardly located acceptor(s), as in proton pumps, indicating that in these channelrhodopsins the Schiff-base accessibility switch is outwardly directed. The carboxylate residue in the position of Asp85 in BR may serve as the primary proton acceptor in these channelrhodopsins, because its neutralization resulted in a strong decrease in the outward proton transfer currents. Channel currents in the corresponding mutants decreased to the same extent, so that the amplitude ratio of channel to proton transfer currents remained close to that in the wild-type (Fig. 9 A). On the other hand, the residue in the position of Asp212 in BR is less important than the Asp85 homolog for proton transfer in channelrhodopsins but is critical for channel opening. Neutralization of this residue induced only minor changes in the outward proton transfer currents but dramatically decreased the amplitude of channel currents (Fig. 9 B). Moreover, the shifts in the reversal potentials of channel currents observed upon neutralization of the Asp212 homolog, but not the Asp85 homolog, in VcChR1 and DsChR1 indicate that it primarily controls cation, rather than proton, conductance. A similar effect of mutating the Asp212 homolog was reported for the CrChR1/CrChR2 chimera (21).
Figure 9.
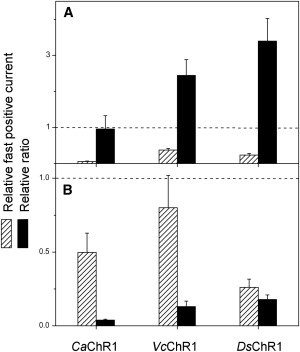
The effect of neutral residue substitution of the Asp85 (A) and Asp212 (B) homologs on peak proton transfer currents (hatched bars) and the ratio between peak channel currents and peak proton transfer currents (black bars) in CaChR1, VcChR1, and DsChR1. The channel currents were measured at −60 mV and the proton transfer currents at the Vr for channel currents. The data are mean values (n = 3–8 cells) measured upon neutralization of the corresponding carboxylate residues normalized to those in the wild-type for CaChR1 and VcChR1, and in the A178E mutant for DsChR1.
In the second group of channelrhodopsins (CrChR2, MvChR1, and PsChR), no fast currents that reflect proton transfer from the Schiff base have been detected. One possible explanation is that the Schiff base is not deprotonated during the photocycle. However, the formation of an M-like intermediate has been clearly demonstrated by flash photolysis experiments in detergent-purified CrChR2 ((42,51); our unpublished observations). We cannot exclude lateral transfer of the Schiff-base proton, which would not produce measurable current. However, the crystal structure of the chimeric channelrhodopsin and those of other microbial rhodopsins make this possibility very unlikely.
Calculations based on the CrChR1/CrChR2 chimera crystal structure suggest that the residue corresponding to Asp212 in BR is unprotonated and may therefore play the role of a proton acceptor (21), and time-resolved Fourier transfer infrared spectroscopy analysis in CrChR2 provides evidence for this suggestion (J. Heberle, Freie Universität Berlin, personal communication, 2012). These data imply that at least in CrChR2, an outward proton transfer current is expected, and its apparent absence in our recordings likely means that it is cancelled by an oppositely directed fast current. One possibility is that the outward proton transfer from the Schiff base is masked by a simultaneous inward proton transfer from a different residue. A possible candidate could be Asp90, which is unprotonated in the conducting state of the pigment (53), although it is not known whether its proton is indeed transferred in the cytoplasmic direction. Another possibility is that in these channelrhodopsins, outward proton transfer from the Schiff base coexists with the transfer to a cytoplasmic acceptor, which occurs at a similar rate, so that no net electrical signal can be detected. Molecules with oppositely directed Schiff bases may be in a fast dynamic equilibrium or may represent two subpopulations, as in SRI (17,18). This may mean that unlike in other microbial rhodopsins, Schiff-base connectivity changes in these channelrhodopsins may not be coupled to the canonical conformational change presumably responsible for channel opening, since the proton transfer precedes channel current. The amplitude of the recorded electric signal depends on the effective intramolecular dielectric constant (27). This constant can be different in different channelrhodopsins, which would result in a different degree of screening of the transient proton signal. However, it seems unlikely that this factor alone can account for the difference of almost two orders of magnitude, as observed in different channelrhodopsins. At present, the reasons for the apparent absence of the currents related to the Schiff-base deprotonation in the second group of channelrhodopsins remain a mystery.
Interestingly, the above-mentioned two groups of channelrhodopsins also greatly differ in the mean amplitude of their channel currents: the first group (CaChR1, VcChR1, and DsChR1) shows much lower efficiency compared to the second group (CrChR2, MvChR1, and PsChR). The difference in the mean channel currents between representatives of the two groups exceeds 10-fold, which cannot be attributed to different expression levels, as judged from tag fluorescence. It is therefore plausible that the difference in whole-cell currents reflects the difference in unitary conductance, which has so far been determined only for CrChR2 by noise analysis (23,54). The inverse relationship between the efficiency of channel function and detectable proton transfer from the Schiff base in different channelrhodopsins is visualized in Fig. 10. As no measurable fast current could be detected in highly efficient channelrhodopsins, we used maximal deviation from the baseline (i.e., noise of the current signal) as its upper limit. Fig. 10 shows that the relative amplitude of the fast current related to deprotonation of the Schiff base decreases 500-fold upon an ∼10-fold increase in channel efficiency. This suggests that the channel function evolved in channelrhodopsins at the expense of their ability for active proton transport.
Figure 10.

The relationship between the amplitudes of fast and channel currents generated by wild-type channelrhodopsins. The channel currents were measured at −60 mV and the fast currents at the Vr for channel currents. The data are mean values (n = 3–8 cells).
One should also bear in mind that light-gated channel activity exhibited by channelrhodopsins heterologously expressed in animal cells is only part of their molecular signaling function in native algal cells (55). Therefore, possible roles of proton transfer in channelrhodopsin signaling in natural systems needs to be examined separately.
Acknowledgments
We thank Elena N. Spudich for generation of the wild-type CrChR2 mammalian expression construct, Sing-Yi Hou for generation of the wild-type CaChR1 construct, and C. Elizabeth Lane for expert handling of cell cultures. We thank Jon K. Magnuson (Pacific Northwest National Laboratory, Richland, WA) for sequence information of DsChR1 mRNA. We also thank Karl Deisseroth (Stanford University, Stanford, CA) for the pcDNA3.1/VcChR1-EYFP mammalian expression vector, and Edward S. Boyden (Massachusetts Institute of Technology, Cambridge, MA) for the AR-3 gene, and both for valuable discussions.
This work was supported by grant R37GM027750 from the National Institute of General Medical Sciences, RC1AG035779 from the National Institute on Aging, the Hermann Eye Fund, and endowed chair AU-0009 from the Robert A. Welch Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes on Aging or General Medical Sciences, or the National Institutes of Health.
Supporting Material
References
- 1.Spudich J.L., Jung K.-H. Handbook of Photosensory Receptors. Wiley-VCH; Weinheim, Germany: 2005. Microbial rhodopsins: phylogenetic and functional diversity; pp. 1–23. [Google Scholar]
- 2.Sineshchekov O.A., Jung K.-H., Spudich J.L. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Govorunova E.G., Jung K.H., Spudich J.L. Chlamydomonas sensory rhodopsins A and B: cellular content and role in photophobic responses. Biophys. J. 2004;86:2342–2349. doi: 10.1016/S0006-3495(04)74291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthold P., Tsunoda S.P., Hegemann P. Channelrhodopsin-1 initiates phototaxis and photophobic responses in chlamydomonas by immediate light-induced depolarization. Plant Cell. 2008;20:1665–1677. doi: 10.1105/tpc.108.057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Litvin F.F., Sineshchekov O.A., Sineshchekov V.A. Photoreceptor electric potential in the phototaxis of the alga Haematococcus pluvialis. Nature. 1978;271:476–478. doi: 10.1038/271476a0. [DOI] [PubMed] [Google Scholar]
- 6.Nagel G., Ollig D., Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 7.Nagel G., Szellas T., Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow B.Y., Chuong A.S., Boyden E.S. Synthetic physiology strategies for adapting tools from nature for genetically targeted control of fast biological processes. Methods Enzymol. 2011;497:425–443. doi: 10.1016/B978-0-12-385075-1.00018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yizhar O., Fenno L., Zhang F., Hegemann P., Diesseroth K. Microbial opsins: a family of single-component tools for optical control of neural activity. Cold Spring Harb. Protoc. 2011;2011 doi: 10.1101/pdb.top102. top102. [DOI] [PubMed] [Google Scholar]
- 10.Zhang F., Vierock J., Deisseroth K. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mogi T., Stern L.J., Khorana H.G. Aspartic acid substitutions affect proton translocation by bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1988;85:4148–4152. doi: 10.1073/pnas.85.12.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butt H.J., Fendler K., Oesterhelt D. Aspartic acids 96 and 85 play a central role in the function of bacteriorhodopsin as a proton pump. EMBO J. 1989;8:1657–1663. doi: 10.1002/j.1460-2075.1989.tb03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramaniam S., Marti T., Khorana H.G. Protonation state of Asp (Glu)-85 regulates the purple-to-blue transition in bacteriorhodopsin mutants Arg-82⋯Ala and Asp-85⋯Glu: the blue form is inactive in proton translocation. Proc. Natl. Acad. Sci. USA. 1990;87:1013–1017. doi: 10.1073/pnas.87.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spudich E.N., Zhang W., Spudich J.L. Constitutive signaling by the phototaxis receptor sensory rhodopsin II from disruption of its protonated Schiff base-Asp-73 interhelical salt bridge. Proc. Natl. Acad. Sci. USA. 1997;94:4960–4965. doi: 10.1073/pnas.94.10.4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki J., Nara T., Spudich J.L. Constitutive activity in chimeras and deletions localize sensory rhodopsin II/HtrII signal relay to the membrane-inserted domain. Mol. Microbiol. 2007;66:1321–1330. doi: 10.1111/j.1365-2958.2007.05983.x. [DOI] [PubMed] [Google Scholar]
- 16.Holterhues J., Bordignon E., Steinhoff H.J. The signal transfer from the receptor NpSRII to the transducer NpHtrII is not hampered by the D75N mutation. Biophys. J. 2011;100:2275–2282. doi: 10.1016/j.bpj.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sineshchekov O.A., Sasaki J., Spudich J.L. A Schiff base connectivity switch in sensory rhodopsin signaling. Proc. Natl. Acad. Sci. USA. 2008;105:16159–16164. doi: 10.1073/pnas.0807486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sineshchekov O.A., Sasaki J., Spudich J.L. Attractant and repellent signaling conformers of sensory rhodopsin-transducer complexes. Biochemistry. 2010;49:6696–6704. doi: 10.1021/bi100798w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki J., Tsai A.L., Spudich J.L. Opposite displacement of helix F in attractant and repellent signaling by sensory rhodopsin-Htr complexes. J. Biol. Chem. 2011;286:18868–18877. doi: 10.1074/jbc.M110.200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma A.K., Spudich J.L., Doolittle W.F. Microbial rhodopsins: functional versatility and genetic mobility. Trends Microbiol. 2006;14:463–469. doi: 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Kato H.E., Zhang F., Nureki O. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luecke H., Schobert B., Spudich J.L. Crystal structure of sensory rhodopsin II at 2.4 Å: insights into color tuning and transducer interaction. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldbauer K., Zimmermann D., Bamberg E. Channelrhodopsin-2 is a leaky proton pump. Proc. Natl. Acad. Sci. USA. 2009;106:12317–12322. doi: 10.1073/pnas.0905852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nack M., Radu I., Heberle J. Kinetics of proton release and uptake by channelrhodopsin-2. FEBS Lett. 2012;586:1344–1348. doi: 10.1016/j.febslet.2012.03.047. [DOI] [PubMed] [Google Scholar]
- 25.Trissl H.-W. Photoelectric measurements of purple membranes. Photochem. Photobiol. 1990;51:793–818. [PubMed] [Google Scholar]
- 26.Kaulen A.D. Electrogenic processes and protein conformational changes accompanying the bacteriorhodopsin photocycle. Biochim. Biophys. Acta. 2000;1460:204–219. doi: 10.1016/s0005-2728(00)00140-7. [DOI] [PubMed] [Google Scholar]
- 27.Dér A., Keszthelyi L. Charge motion during the photocycle of bacteriorhodopsin. Biochemistry (Mosc.) 2001;66:1234–1248. doi: 10.1023/a:1013179101782. [DOI] [PubMed] [Google Scholar]
- 28.Drachev L.A., Kaulen A.D., Skulachev V.P. Time resolution of the intermediate steps in the bacteriorhodopsin-linked electrogenesis. FEBS Lett. 1978;87:161–167. doi: 10.1016/0014-5793(78)80157-4. [DOI] [PubMed] [Google Scholar]
- 29.Fahr A., Lauger P., Bamberg E. Photocurrent kinetics of purple-membrane sheets bound to planar bilayer membranes. J. Membr. Biol. 1981;60:51–62. [Google Scholar]
- 30.Keszthelyi L., Ormos P. Electric signals associated with the photocycle of bacteriorhodopsin. FEBS Lett. 1980;109:189–193. [Google Scholar]
- 31.Friedrich T., Geibel S., Bamberg E. Proteorhodopsin is a light-driven proton pump with variable vectoriality. J. Mol. Biol. 2002;321:821–838. doi: 10.1016/s0022-2836(02)00696-4. [DOI] [PubMed] [Google Scholar]
- 32.Váró G., Brown L.S., Lanyi J.K. Characterization of the photochemical reaction cycle of proteorhodopsin. Biophys. J. 2003;84:1202–1207. doi: 10.1016/S0006-3495(03)74934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sineshchekov O.A., Spudich J.L. Light-induced intramolecular charge movements in microbial rhodopsins in intact E. coli cells. Photochem. Photobiol. Sci. 2004;3:548–554. doi: 10.1039/b316207a. [DOI] [PubMed] [Google Scholar]
- 34.Nagel G., Möckel B., Bamberg E. Functional expression of bacteriorhodopsin in oocytes allows direct measurement of voltage dependence of light induced H+ pumping. FEBS Lett. 1995;377:263–266. doi: 10.1016/0014-5793(95)01356-3. [DOI] [PubMed] [Google Scholar]
- 35.McLachlan J. The culture of Dunaliella tertiolecta Butcher: a euryhaline organism. Can. J. Microbiol. 1960;6:367–379. [Google Scholar]
- 36.Haghjou M.M., Shariati M., Smirnoff N. The effect of acute high light and low temperature stresses on the ascorbate-glutathione cycle and superoxide dismutase activity in two Dunaliella salina strains. Physiol. Plant. 2009;135:272–280. doi: 10.1111/j.1399-3054.2008.01193.x. [DOI] [PubMed] [Google Scholar]
- 37.Optogenetics Resource Center. http://www.stanford.edu/group/dlab/optogenetics/.
- 38.Hou S.Y., Govorunova E.G., Spudich J.L. Diversity of Chlamydomonas channelrhodopsins. Photochem. Photobiol. 2012;88:119–128. doi: 10.1111/j.1751-1097.2011.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W.-W., Sineshchekov O.A., Spudich J.L. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J. Biol. Chem. 2003;278:33985–33991. doi: 10.1074/jbc.M305716200. [DOI] [PubMed] [Google Scholar]
- 40.Chow B.Y., Han X., Boyden E.S. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Husson S.J., Liewald J.F., Gottschalk A. Microbial light-activatable proton pumps as neuronal inhibitors to functionally dissect neuronal networks in C. elegans. PLoS ONE. 2012;7:e40937. doi: 10.1371/journal.pone.0040937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhoefen M.K., Bamann C., Wachtveitl J. The photocycle of channelrhodopsin-2: ultrafast reaction dynamics and subsequent reaction steps. ChemPhysChem. 2010;11:3113–3122. doi: 10.1002/cphc.201000181. [DOI] [PubMed] [Google Scholar]
- 43.Otto H., Marti T., Heyn M.P. Substitution of amino acids Asp-85, Asp-212, and Arg-82 in bacteriorhodopsin affects the proton release phase of the pump and the pK of the Schiff base. Proc. Natl. Acad. Sci. USA. 1990;87:1018–1022. doi: 10.1073/pnas.87.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gergely C., Varo G. Charge motions in the D85N and D212N mutants of bacteriorhodopsin. In: Rigaud J.L., editor. Structures and Functions of Retinal Proteins. John Libbey; Paris: 1992. pp. 193–196. [Google Scholar]
- 45.Balashov S.P., Govindjee R., Feng Y. Effect of the arginine-82 to alanine mutation in bacteriorhodopsin on dark adaptation, proton release, and the photochemical cycle. Biochemistry. 1993;32:10331–10343. doi: 10.1021/bi00090a008. [DOI] [PubMed] [Google Scholar]
- 46.Govindjee R., Misra S., Menick D.R. Arginine-82 regulates the pKa of the group responsible for the light-driven proton release in bacteriorhodopsin. Biophys. J. 1996;71:1011–1023. doi: 10.1016/S0006-3495(96)79302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ernst O.P., Sánchez Murcia P.A., Hegemann P. Photoactivation of channelrhodopsin. J. Biol. Chem. 2008;283:1637–1643. doi: 10.1074/jbc.M708039200. [DOI] [PubMed] [Google Scholar]
- 48.Kianianmomeni A., Stehfest K., Hallmann A. Channelrhodopsins of Volvox carteri are photochromic proteins that are specifically expressed in somatic cells under control of light, temperature, and the sex inducer. Plant Physiol. 2009;151:347–366. doi: 10.1104/pp.109.143297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berndt A., Prigge M., Hegemann P. Two open states with progressive proton selectivities in the branched channelrhodopsin-2 photocycle. Biophys. J. 2010;98:753–761. doi: 10.1016/j.bpj.2009.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang F., Wang L.P., Deisseroth K. Channelrhodopsin-2 and optical control of excitable cells. Nat. Methods. 2006;3:785–792. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- 51.Bamann C., Kirsch T., Bamberg E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J. Mol. Biol. 2008;375:686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 52.Govorunova E.G., Spudich E.N., Spudich J.L. New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. MBio. 2011;2 doi: 10.1128/mBio.00115-11. e00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eisenhauer K., Kuhne J., Gerwert K. In channelrhodopsin-2 Glu-90 is crucial for ion selectivity and is deprotonated during the photocycle. J. Biol. Chem. 2012;287:6904–6911. doi: 10.1074/jbc.M111.327700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin J.Y., Lin M.Z., Steinbach P., Tsien R.Y. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys. J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sineshchekov O.A., Govorunova E.G., Spudich J.L. Photosensory functions of channelrhodopsins in native algal cells. Photochem. Photobiol. 2009;85:556–563. doi: 10.1111/j.1751-1097.2008.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


