Abstract
The catalogue of genes expressed at different levels in the two sexes is growing, and the mechanisms underlying sex differences in regulation of the mammalian transcriptomes are being explored. Here we report that the expression of the imprinted non-protein-coding maternally expressed gene H19 was female-biased specifically in the female mouse eye (1.9-fold, p = 3.0E−6) while not being sex-biased in other somatic tissues. The female-to-male expression fold-change of H19 fell in the range expected from an effect of biallelic versus monoallelic expression. Recently, the possibility of sex-specific parent-of-origin allelic expression has been debated. This led us to hypothesize that H19 might express biallelically in the female mouse eye, thus escape its silencing imprint on the paternal allele specifically in this tissue. We therefore performed a sex-specific imprinting assay of H19 in female and male eye derived from a cross between Mus musculus and Mus spretus. However, this analysis demonstrated that H19 was exclusively expressed from the maternal gene copy, disproving the escape hypothesis. Instead, this supports that the female-biased expression of H19 is the result of upregulation of the single maternal. Furthermore, if H19 would have been expressed from both gene copies in the female eye, an associated downregulation of Insulin-like growth factor 2 (Igf2) was expected, since H19 and Igf2 compete for a common enhancer element located in the H19/Igf2 imprinted domain. On the contrary we found that also Igf2 was significantly upregulated in its expression in the female eye (1.2-fold, p = 6.1E−3), in further agreement with the conclusion that H19 is monoallelically elevated in females. The female-biased expression of H19 and Igf2 specifically in the eye may contribute to our understanding of sex differences in normal as well as abnormal eye physiology and processes.
Background
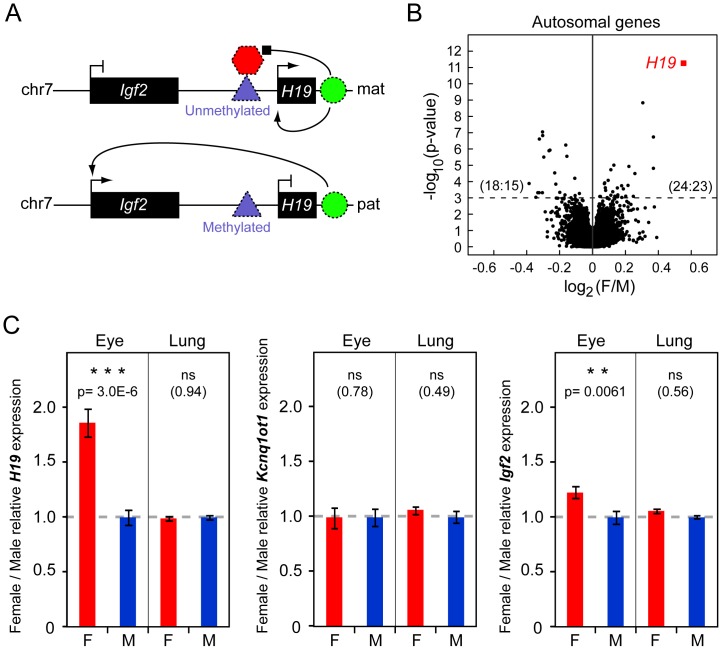
Mounting evidence has cemented our awareness of the importance of taking molecular sexual dimorphism into account for reaching a fuller understanding of normal physiology [1], [2], [3], [4], [5] as well as pathological conditions with sex-biased characteristics [6], [7], [8]. Sex differences in autosomal gene expression can vary extensively between tissues [9], [10] and are known to be regulated not only by sex hormones, but also directly by genes located on the X and Y chromosomes [11], and even via interaction with the X-inactive chromatin in female cells [12]. Recently a new mode of sexual gene expression bias was reported in the mouse. An RNA-sequencing analysis showed that sex-specific effects on the expression of the paternal versus the maternal allele (i.e. sex-specific imprinting effects) were wide-spread [13], although the validity of some of these results has later been questioned [14]. Moreover, a quantitative trait locus analysis reported sex-dependent imprinting effects on complex traits in mice [15]. However, the existence of sex-specific imprinting effects is not yet widely accepted, and possible underlying mechanisms remain to be described. Imprinted genes are of particular interest for understanding the evolution of sexual dimorphism, particularly in the context of competition between the sexes. The parental conflict hypothesis [16] states that the skew in maternal versus paternal optimal investment during the generation of offspring has led to the evolution of imprinted genes. The father’s fitness can be increased by delivering genes to the offspring that promote increased energy consumption at the expense of the mother, while the mother responds by activating growth inhibiting genes. The first imprinted locus to be identified in mouse, and likely the most studied imprinted locus, is the H19/Igf2 domain [17], [18], [19]. This locus encodes the paternally expressed Insulin growth factor 2 (Igf2), which is a major fetal growth factor, thus fitting well within the parental conflict hypothesis. The other gene in this locus, H19, is a long non-coding RNA expressed from the maternal allele. The specific expression of paternal Igf2 and maternal H19 is achieved by a mutually exclusive interaction with an enhancer element located in the H19/Igf2 imprinting domain ( Figure 1A ) [20], [21], [22].
Figure 1. Gene expression analysis. A.
Schematic model of allelic regulation in the H19/Igf2 imprinted domain, and premise for the experimental approach. The imprinting control region (ICR, triangle) is unmethylated on the maternal allele (mat), allowing for the expression of H19mat and the binding of the CCCTC-binding factor (hexagon) which insulates Igf2mat from interaction with an enhancer element (circle) located downstream of H19. Thus Igf2mat is normally silenced when H19mat is expressed. In contrast, on the paternal allele (pat), the ICR is methylated, preventing expression of H19pat and blocking CCCTC-binding to the ICR which allows Igf2pat to interact with the enhancer element and to be expressed. B. Volcano plot, separating female-biased (upper right quadrant) and male-biased (upper left quadrant) autosomal genes in the mouse eye in our microarray screen. H19 (red square) is identified as a candidate female-biased gene (p = 1.3E−12, female/male fold-change = 1.5). y-axis: –log10(p-value, two-sided t-test), x-axis: log2(female/male) expression ratio. The dotted line represents the significance threshold p = 0.001, and numbers within parenthesis denote the number of significant probes : unique genes in each sex. nfemales = 88, nmales = 88. C. RT-qPCR assays of female (F) and male (M) eye and lung tissues. Expression is normalized to the geometric mean of Gapdh and Actb and shown relative to the mean male expression in each tissue. P-values are given according to a two-sided t-test and error bars denote standard error of the mean. nfemales, eye = 19, nmales, eye = 19, nfemales, lung = 16, nmales, lung = 18.
Recently, we performed a large-scale microarray analysis of sexually dimorphic gene expression in somatic mouse tissues incorporating more than 700 microarray hybridizations [23]. In this analysis, H19 was identified as the top female-biased gene candidate among expressed autosomal transcripts in the mouse eye (1.5 fold, p = 1.3E−12, Figure 1B ), while H19 was not significantly sex-biased in other tissues analyzed (lung, liver, kidney, striatum, and hippocampus). H19 is expressed in several compartments of the mouse eye, including the retina, iris, ciliary bodies, eyecup and the cornea (Figure S1). Given that the female-bias of H19 fell in the range expected by an effect of biallelic versus monoallelic expression [10], [24], and the recent reports of a large number of imprinted genes showing sex-specific imprinting effects [13], we here investigated the possibility that H19 might “escape” its silencing imprint on the paternal allele specifically in the female eye. If so, this would open new avenues for the exploration of the mechanisms behind H19/Igf2 imprinting, since the molecular characteristics of different epigenetic states of the H19/Igf2 imprinted domain could be compared both within a tissue (by comparing male and female eye) and between tissues (by comparing female eye with other tissues).
Results and Discussion
Gene Expression Analysis
To validate the eye-specific female-bias of H19 expression we performed an RT-qPCR analysis of female and male eye, and included lung as a control tissue, using samples that were biologically independent from the specimens employed in the microarray experiments. As expected, this analysis confirmed a female-elevated expression of H19 in the eye (1.9 fold, p = 3.0E−6, Figure 1C ) while no difference was observed in the lung. To ensure that the sex-bias was specific to H19 transcription, and not a general effect in the eye tissues, we analyzed the expression of Kcnq1ot1, an imprinted long non-coding RNA located at mouse chromosome 7, 0.5 Mbp downstream of H19, and found no sex difference in expression of this control gene ( Figure 1C ).
Sex-specific Imprinting Assay of H19
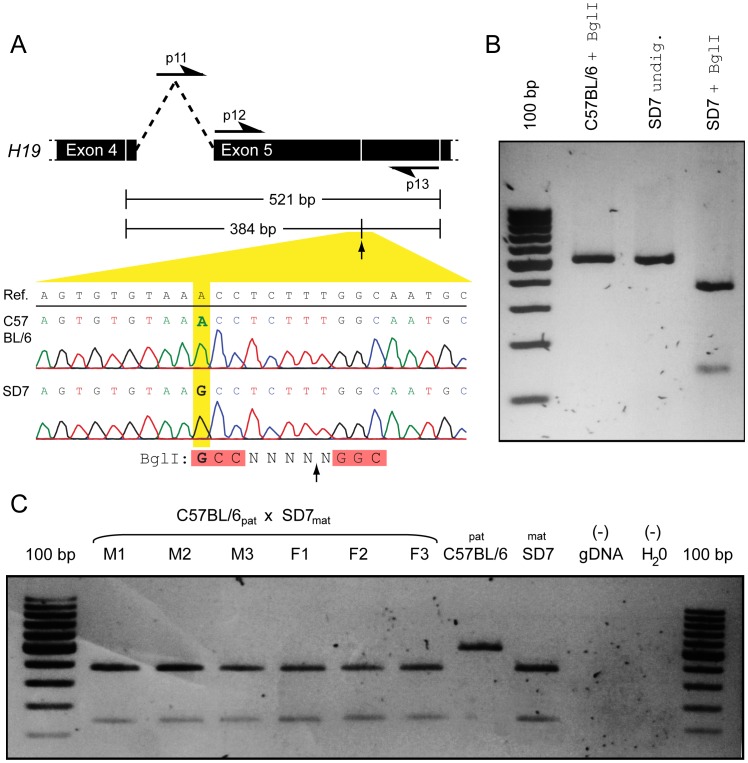
Next, to investigate whether the upregulation of H19 in the female eye was an effect of expression from both alleles (i.e. loss of imprinting) or an effect of upregulation of the maternal gene copy, we conducted a sex-specific imprinting assay. To distinguish between the paternal and maternal allele, we generated F1 offspring carrying polymorphisms within the H19 gene by crossing the mouse strains C57BL/6 (♂) and SD7 (♀). SD7 is a mouse strain maintained on a C57BL/6 background with the distal arm of chromosome 7 derived from the distant mouse subspecies Mus spretus [25]. Thus the F1s were heterozygous for the H19 allele. To identify single nucleotide polymorphisms (SNPs), we amplified and partially sequenced H19 in SD7 and C57BL/6. We identified one SNP in exon 4; five SNPs in exon 5; and 2 intronic polymorphisms (Figure S2. The SD7 sequence was deposited in GenBank: accession number JX869491). This, together with a Restriction Fragment Length Polymorphism (RFLP) analysis, confirmed the existence of an SD7-specific BglI restriction site within exon 5 ( Figure 2A and B ). We then carried out an RFLP analysis of H19 cDNA derived from the eye of three male and three female F1s. This analysis clearly showed that H19 was exclusively expressed from the maternal allele in males as well as in females ( Figure 2C ), disproving the hypothesis that H19 escape imprinting in the female eye. From this it follows that the female-elevated expression is derived from an eye-specific upregulation of maternal H19 expression relative to that in males. Furthermore, if H19 would have been biallelically expressed in the female eye we expected an associated downregulation of Igf2 (See the model in Figure 1A ), and we therefore performed an RT-qPCR analysis of Igf2. On the contrary, and in further refutation to the escape hypothesis, we found that also Igf2 was significantly female-biased in the mouse eye (1.2 fold, p = 0.0061, Figure 1C ). These results conclude that female-specific elevated expression of H19 is not due to loss of imprinting, rather due to hitherto unknown female eye-specific transcriptional regulatory mechanisms.
Figure 2. Sex-specific imprinting assay of H19. A.
RFLP experimental design. DNA sequencing of C57BL/6 and SD7 confirmed an SD7-specific BglI restriction site located within H19 exon 5. p11, p12 and p13 designate the locations of the RFLP primers listed in Table 1. B. Confirmation of SD7-specificity of the BglI restriction. PCR products of H19 amplified from BL6 and SD7 gDNA (using primers p12 and p13) digested with BglI, and an undigested SD7 sample. C. Imprinting assay of H19 in male and female mice. PCR products of H19 amplified from eye cDNA derived from three F1 males (M1-3) and three F1 females (F1-3) of the ♂C57BL/6×♀SD7 cross (using primers p11 and p13) digested with BglI. Controls for the paternal (C57BL/6) and maternal (SD7) allele, and negative controls for the PCR are shown to the right.
In the context of our results, it is of interest that epigenetic sex differences in the imprinted H19/Igf2 domain have been previously observed. For example, an analysis of embryonic germ cells showed that H19 is more strongly methylated in males than in females at 11.5 and 12.5 dpc, and likewise for Igf2 at 12.5 dpc [26]. This is an intrinsic cell-autonomous effect of the sex chromosome complement (XY versus XX) and not an effect of differences in systemic sex hormone levels [27]. These results correlate with our observation of robust female-biased expression of H19 and slight female-bias of Igf2. Sex differences in a range of aspects in eye anatomy and function are known in normal as well as in disease conditions (reviewed in [28]). For example, malignant glaucoma is about twice as common in females as in males [29]. Cataloguing sex-biased molecular characteristics will be crucial for approaching a more complete understanding sex differences in eye physiology. Of further interest is that H19 and Igf2 are both oncogenes known to be deregulated in various types of tumors [30], [31], [32], [33], [34], [35], H19 often being overexpressed in the tumorous tissues. The functional consequences and the evolutionary ground of the eye-specific female-elevated expression of H19 and Igf2 are now open for further in-depth investigation.
Conclusion
We showed that the expression of the imprinted genes H19 and Igf2 is female-biased in the mouse eye, while not sex-biased in other tissues analyzed. We speculated that the elevation of H19 might be a result of escape from its silencing genomic imprint on the paternal allele specifically in the female eye. Our imprinting assay showed that this was not the case, and therefore H19 upregulation emerges from the single maternal allele from which it is expressed. The awareness of sex differences in gene expression is important for understanding the molecular basis of sexual dimorphism in normal physiology and in disease, as well as for understanding of the evolution of imprinted and non imprinted traits with sex-biased characteristics.
Materials and Methods
Ethics Statement
The use of the experimental animals in this study was approved by the appropriate Swedish ethical committee, permits: c79/9 and 307-2011, Jordbruksverket.
Gene Expression Analysis
Microarray analysis
The eye microarray analysis included 176 RMA normalized microarrays (nfemale = 88, nmale = 88, MOE340 Affymetrix, Mus musculus BXD strains) and is described in detail elsewhere [23]. This data is available in the GeneNetwork depository: http://www.genenetwork.org, “Hamilton Eye Institute Mouse Eye M430v2 Data Set (Sept08) RMA”, Accession number: GN207. The significance criterion was p<0.001, two-sided t-test. We confirmed that the female-bias of H19 was not unique to BXD strains by similar analysis of a panel of female/male pairs from 20 different mouse subspecies and strains (p = 0.0006 two-sided paired t-test, mean fold-change: 1.2, nfemale = 20, nmale = 20, Table S1). Expression data for Figure S1 was obtained from BioGPS [36] (http://biogps.org, GeneAtlas MOE430 gcrma dataset; 1448194_a_at). RT-qPCR analysis: Total RNA was extracted from male and female eyes (nfemale = 19, nmale = 19, Mus musculus C57BL/6 strain) and lung (nfemale = 16, nmale = 18) after weaning (>P21, Swedish ethical committee permit: c79/9, Jordbruksverket) using Trizol (Invitrogen). RNA was reverse transcribed using a Dynamo cDNA synthesis kit F-470L (Finnzymes) and the following reagents: 0.95 µg total RNA, 15 ng/µl random hexamers, 10 U M-MuLV RNase H- reverse transcriptase, 1 × RT buffer, ddH2O, in a total reaction volume of 20 µl. Incubations were performed in a PTC-100 Peltier Thermal Cycler (MJ Research): 25°C; 10 min, 37°C; 45 min, 85°C; 5 min. cDNA samples were subsequently diluted 1∶10 in ddH2O. Reactions contained 0.3 µM of each primer ( Table 1A ), 1×Power SYBR Green Master Mix (Applied Biosystems), 4 µl diluted cDNA sample and ddH2O in a total reaction volume of 30 µl. Thermal cycles were: 50°C; 2 min, 95°C; 10 min, 40 cycles: 95°C; 15 s, 60°C; 1 min. To ensure that single PCR products of intended lengths were amplified, a melting program was executed subsequent to the quantifications and bands of expected sizes were inspected after gel electrophoresis. Copy numbers were determined relative to a cDNA dilution series. Expression was normalized to the geometric mean of Gapdh and Actb. The criterion for differential expression was p<0.05, two-tailed t-test. Information on overall expression of H19 and Igf2 in eye versus lung is included in Figure S3.
Table 1. Primers.
| Table 1a. qPCR primers. | ||
| H19 | forward | AGAGGACAGAAGGGCAGTCA |
| reverse | TGGGTGGACAATTAGGTGGT | |
| Kcnq1ot1 | forward | AGAGGACAGAAGGGCAGTCA |
| reverse | TGGGTGGACAATTAGGTGGT | |
| Igf2 | forward | CGCTTCAGTTTGTCTGTTCG |
| reverse | AAGCAGCACTCTTCCACGAT | |
| Gapdh | forward | GCCTTCCGTGTTCCTACC |
| reverse | GCCTGCTTCACCACCTTC | |
| Actb | forward | TGTTACCAACTGGGACGACA |
| reverse | GGGGTGTTGAAGGTCTCAAA | |
| Table 1b . H19 sequencing primers. | ||
| H19 seq | forward | CCAGGTCTCCAGCAGAGGT |
| reverse | TTTATTGATGGACCCAGGAC | |
| Table 1c . H19 RFLP primers. | ||
| H19 p11 | forward | TGCTCCAAGGTGAAGCTGAAAG |
| H19 p12 | forward | GTGAAGCTGAAAGAACAGATGGTG |
| H19 p13 | reverse | GTAGGGCATGTTGAACACTTTATG |
The primer sequences are given 5′ to 3′.
Imprinting Assay
DNA sequencing
PCR amplification of H19 gDNA for Sanger sequencing was performed: 94°C; 2 min, 35× (94°C; 10 s, 55°C; 40 s, 72°C; 90 s), 72°C; 10 min. Each reaction contained 100 ng gDNA, 0.75 U DreamTac DNA polymerase (Fermentas), 2.5 µl DreamTaq buffer including MgCl2 (Fermentas), 0.3 µM of each primer ( Table 1B ), 0.4 µM dNTPs, ddH2O to a total volume of 25 µl. DNA sequencing was performed at GATC-Biotech, Stockholm, using the primers in Table 1B . RFLP analysis: Eye and lung tissues were dissected from F1 mice (♂C57BL/6×♀SD7) after weaning. Total RNA was extracted using Trizol (Invitrogen) and treated with DNase I (Fermentas). Reverse transcription and PCR was performed as described above using the primers listed in Table 1C . 10 µl PCR product was incubated with 20 U BglI restriction enzyme (Fermentas), 3 µl Buffer O (Fermentas), ddH2O to a total volume of 30 µl, 37°C; 28 h. Fragments were separated on a 2% agarose gel and stained with ethidium bromide. This protocol was based on an imprinting assay described by Sasaki et al. [37]. The Swedish ethical committee permit for the C57BL/6 and SD7 animals used in the imprinting assay is 307-2011, Jordbruksverket.
Supporting Information
The expression level of H19 in a panel of mouse tissues, including sub compartments of the eye (1448194_a_at, MOE430 Affymetrix, BioGPS: http://biogps.org ).
(PDF)
A. Chromatogram showing eight C57BL/6:SD7 polymorphisms (pm) within the H19 gene, as identified by Sanger sequencing. B. Alignment between a H19 mouse reference sequence and the SD7 sequence.
(PDF)
The overall expression level of H19 (nfemales = 19, nmales = 19) and Igf2 (nfemales, lung = 16, nmales, lung = 16) in eye and lung. P-values are given according to a two-sided t-test and error bars denote standard error of the mean.
(PDF)
Eye H19 intensities from analysis of 1 female and 1 male microarray hybridization in 20 mouse strains/subspecies (1448194_a_at, MOE340 Affymetrix, GeneNetwork depository: http://www.genenetwork.org, Accession number: GN207). RNA pools of 4–8 eyes from 2–4 individuals are included in each hybridization.
(PDF)
Acknowledgments
We are thankful to Gaurav K. Pandey for help with the breeding of F1 mice.
Funding Statement
This work was supported by the grants from the Swedish Cancer Research foundation (Cancerfonden: Kontrakt no. 100422), Swedish Research Council (VR-M: K2011-66X-20781-04-3; VR-NT: 621-2011-4996), and Barncancerfonden (PROJ11/067) to C.K. The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jazin E, Cahill L (2010) Sex differences in molecular neuroscience: from fruit flies to humans. Nat Rev Neurosci 11: 9–17. [DOI] [PubMed] [Google Scholar]
- 2. Arnold AP, Lusis AJ (2012) Understanding the sexome: measuring and reporting sex differences in gene systems. Endocrinology 153: 2551–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aubin-Horth N, Renn SC (2009) Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol Ecol 18: 3763–3780. [DOI] [PubMed] [Google Scholar]
- 4. Hines M (2011) Gender development and the human brain. Annu Rev Neurosci 34: 69–88. [DOI] [PubMed] [Google Scholar]
- 5. Reinius B, Saetre P, Leonard JA, Blekhman R, Merino-Martinez R, et al. (2008) An evolutionarily conserved sexual signature in the primate brain. PLoS Genet 4: e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ober C, Loisel DA, Gilad Y (2008) Sex-specific genetic architecture of human disease. Nat Rev Genet 9: 911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cook MB, Dawsey SM, Freedman ND, Inskip PD, Wichner SM, et al. (2009) Sex disparities in cancer incidence by period and age. Cancer Epidemiol Biomarkers Prev 18: 1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ober C, Vercelli D (2011) Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet 27: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang X, Schadt EE, Wang S, Wang H, Arnold AP, et al. (2006) Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res 16: 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reinius B, Shi C, Hengshuo L, Sandhu KS, Radomska KJ, et al. (2010) Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics 11: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCarthy MM, Arnold AP (2011) Reframing sexual differentiation of the brain. Nat Neurosci 14: 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wijchers PJ, Yandim C, Panousopoulou E, Ahmad M, Harker N, et al. (2010) Sexual dimorphism in mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Dev Cell 19: 477–484. [DOI] [PubMed] [Google Scholar]
- 13. Gregg C, Zhang J, Butler JE, Haig D, Dulac C (2010) Sex-specific parent-of-origin allelic expression in the mouse brain. Science 329: 682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeVeale B, van der Kooy D, Babak T (2012) Critical evaluation of imprinted gene expression by RNA-Seq: a new perspective. PLoS Genet 8: e1002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hager R, Cheverud JM, Leamy LJ, Wolf JB (2008) Sex dependent imprinting effects on complex traits in mice. BMC Evol Biol 8: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moore T, Haig D (1991) Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 7: 45–49. [DOI] [PubMed] [Google Scholar]
- 17. Bartolomei MS, Zemel S, Tilghman SM (1991) Parental imprinting of the mouse H19 gene. Nature 351: 153–155. [DOI] [PubMed] [Google Scholar]
- 18. Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N (1991) The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349: 84–87. [DOI] [PubMed] [Google Scholar]
- 19. DeChiara TM, Robertson EJ, Efstratiadis A (1991) Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64: 849–859. [DOI] [PubMed] [Google Scholar]
- 20. Weth O, Renkawitz R (2011) CTCF function is modulated by neighboring DNA binding factors. Biochem Cell Biol 89: 459–468. [DOI] [PubMed] [Google Scholar]
- 21. Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, et al. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489. [DOI] [PubMed] [Google Scholar]
- 22. Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, et al. (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10: 853–856. [DOI] [PubMed] [Google Scholar]
- 23. Reinius B, Johansson MM, Radomska KJ, Morrow EH, Pandey GK, et al. (2012) Abundance of female-biased and paucity of male-biased somatically expressed genes on the mouse X-chromosome. BMC Genomics 13: 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Talebizadeh Z, Simon SD, Butler MG (2006) X chromosome gene expression in human tissues: male and female comparisons. Genomics 88: 675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dean W, Bowden L, Aitchison A, Klose J, Moore T, et al. (1998) Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development 125: 2273–2282. [DOI] [PubMed] [Google Scholar]
- 26. Tada T, Tada M, Hilton K, Barton SC, Sado T, et al. (1998) Epigenotype switching of imprintable loci in embryonic germ cells. Dev Genes Evol 207: 551–561. [DOI] [PubMed] [Google Scholar]
- 27. Durcova-Hills G, Burgoyne P, McLaren A (2004) Analysis of sex differences in EGC imprinting. Dev Biol 268: 105–110. [DOI] [PubMed] [Google Scholar]
- 28. Wagner H, Fink BA, Zadnik K (2008) Sex- and gender-based differences in healthy and diseased eyes. Optometry 79: 636–652. [DOI] [PubMed] [Google Scholar]
- 29. Razeghinejad MR, Amini H, Esfandiari H (2005) Lesser anterior chamber dimensions in women may be a predisposing factor for malignant glaucoma. Med Hypotheses 64: 572–574. [DOI] [PubMed] [Google Scholar]
- 30. Hibi K, Nakamura H, Hirai A, Fujikake Y, Kasai Y, et al. (1996) Loss of H19 imprinting in esophageal cancer. Cancer Res 56: 480–482. [PubMed] [Google Scholar]
- 31. Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, et al. (2002) Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 23: 1885–1895. [DOI] [PubMed] [Google Scholar]
- 32. Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, et al. (2006) The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res 66: 5330–5337. [DOI] [PubMed] [Google Scholar]
- 33. Tanos V, Ariel I, Prus D, De-Groot N, Hochberg A (2004) H19 and IGF2 gene expression in human normal, hyperplastic, and malignant endometrium. Int J Gynecol Cancer 14: 521–525. [DOI] [PubMed] [Google Scholar]
- 34. Ariel I, Lustig O, Schneider T, Pizov G, Sappir M, et al. (1995) The imprinted H19 gene as a tumor marker in bladder carcinoma. Urology 45: 335–338. [DOI] [PubMed] [Google Scholar]
- 35. Ariel I, Sughayer M, Fellig Y, Pizov G, Ayesh S, et al. (2000) The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol Pathol 53: 320–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, et al. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sasaki H, Ferguson-Smith AC, Shum AS, Barton SC, Surani MA (1995) Temporal and spatial regulation of H19 imprinting in normal and uniparental mouse embryos. Development 121: 4195–4202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression level of H19 in a panel of mouse tissues, including sub compartments of the eye (1448194_a_at, MOE430 Affymetrix, BioGPS: http://biogps.org ).
(PDF)
A. Chromatogram showing eight C57BL/6:SD7 polymorphisms (pm) within the H19 gene, as identified by Sanger sequencing. B. Alignment between a H19 mouse reference sequence and the SD7 sequence.
(PDF)
The overall expression level of H19 (nfemales = 19, nmales = 19) and Igf2 (nfemales, lung = 16, nmales, lung = 16) in eye and lung. P-values are given according to a two-sided t-test and error bars denote standard error of the mean.
(PDF)
Eye H19 intensities from analysis of 1 female and 1 male microarray hybridization in 20 mouse strains/subspecies (1448194_a_at, MOE340 Affymetrix, GeneNetwork depository: http://www.genenetwork.org, Accession number: GN207). RNA pools of 4–8 eyes from 2–4 individuals are included in each hybridization.
(PDF)