Abstract
Epigenetic mechanisms, including DNA methylation, that underlie neuropsychiatric conditions have become a promising area of research. Most commonly used DNA sources in such studies are peripheral (whole) blood (WB), saliva (SL), and lymphoblastoid cell lines (LCLs); thus, the question of the consistency of DNA methylation patterns in those cells is of particular interest. To investigate this question we performed comparative analyses of methylation patterns in WB, SL, and LCLs derived from the same individuals, using Illumina HumanMethylation27 BeadChip arrays. Our results showed that DNA methylation patterns in SL are relatively consistent with those in WB, whereas the patterns in LCLs are similarly distinct from both WB and SL. The results indicated that due to multiple random and directed changes in DNA methylation throughout cell culturing, LCLs are not a reliable source of DNA for epigenetic studies and should be used with caution when investigating epigenetic mechanisms underlying biological processes.
Keywords: Lymphoblastoid cell lines, Saliva, Whole blood, DNA methylation, Methylation pattern
Introduction
DNA methylation is a well-known mechanism of genes activity regulation that provides valuable information on development, typical functioning and disorders (Laird 2003), including neuropsychiatric conditions (Abdolmaleky et al. 2004, 2008; Feng and Fan 2009; Hsieh and Elisch 2010; Mill et al. 2008; Nohesara et al. 2011). Studies have provided evidence that individuals with neuropsychiatric disorders can be distinguished by epigenetic encoding disturbances found in both the brain and secondary tissues (Abdolmaleky et al. 2008; Feng and Fan 2009; Hsieh and Elisch 2010; Iwamoto et al. 2004; Mill et al. 2008). Among those “secondary tissues,” peripheral blood (whole blood, WB), saliva (SL), and lymphoblastoid cell lines (LCLs) are most commonly used as sources of genomic DNA in epigenetic research (Hu et al. 2006; Kaminsky et al. 2009; Nguyen et al. 2010; Nohesara et al. 2011; Sapienza et al. 2011; Tierling et al. 2011). Consequently, the question of the consistency of the methylation profiles derived from these cells and tissues is of particular interest. One study has reported that, at least in the context of investigating genomic sex effects on DNA methylation, SL DNA has shown a pattern of methylation consistent with WB DNA (Liu et al. 2010). Yet, the degree of convergence between patterns of methylation for LCLs and WB or SL might be different as random DNA methylation pattern changes may be observed in LCLs when compared to the original B-lymphocytes from which they originated (Brennan et al. 2009; Grafodatskaya et al. 2010).
Here, we present the results of a comparison between whole genome methylation profiles of WB, LCLs, and SL samples derived from the same individuals. The aims of this study were twofold: (1) to establish the degree of differences/similarities in the DNA methylation patterns in the tissues most commonly used in epigenetic studies; and (2) to explore the scope of epigenetic alterations occurring in LCLs.
Materials and methods
Sample collection and processing
WB and SL samples were derived from 14 healthy individuals (ten females and four males ranging in age from 7 to 61 years old) from an indigenous Slavic population of Northern Russia. Informed consent was obtained from all participants who donated biological samples. Two vials of saliva were collected from each individual using BD vacutainers containing ACD (BD, Franklin Lakes, NJ) and Oragene-DNA collection kits (DNAgenotek, Ontario, Canada), respectively.
LCLs were established by transforming lymphocytes with the Epstein-Barr Virus (EBV) according to standard procedures (Neitzel 1986); these were cultured for 4 weeks, then cryopreserved prior to DNA extraction. Genomic DNA was extracted from WB samples using the FlexiGene DNA Kit, according to the manufacturer’s protocol (Qiagen, Mississauga, ON); from LCLs using phenol–chloroform and ethanol precipitation; and from SL following the Oragene Laboratory protocol.
DNA methylation analysis
Analysis of DNA methylation profiles in the three sets of tissues from the same individuals was performed using Infinium HumanMethylation27 BeadChip assay (Illumina, San Diego, CA), which contains 27,578 CpG targets covering 14,495 genes. Bisulfite treatment, whole genome amplification, labeling, hybridization and scanning were performed at the Yale Center for Genomic Analysis http://medicine.yale.edu/keck/ycga/index.aspx).
The methylation data generated by the array were analyzed using the Illumina GenomeStudio software package. The methylation status of each CpG site was measured as the ratio of signal from methylated probe to the sum of both methylated and unmethylated signals (β value, ranges from 0, unmethylated, to 1, fully methylated). All CpG sites with a detection p value ≥0.001 were removed from later analyses; the p values were obtained using a background model in the Genome Studio. Three technical replicates were run across different BeadChips; pairwise comparison of these replicates showed consistent and highly reproducible methylation level measurements (r2 varied between 0.98 and 0.99); on average 27,566 ± 12 probes showed no significant (p ≤ 0.01) differences in β values.
Differential methylation analysis
The Illumina methylation data were processed and analyzed using the Methylation Module v1.8 of the Illumina GenomeStudio. For comparison of whole-genome methylation profiles across the samples, correlations and hierarchical clustering were used; the results of the analysis are provided in the form of a dendrogram. To prevent variability among samples within a tissue set related to gender, CpG sites localized on sex chromosomes were excluded, leaving 26,273 sites to be analyzed.
Differential methylation analysis between sets of samples representing three tissues was performed based on differences in the mean beta value (Avgβ) of each CpG site, or Delta Avgβ (Δβ). To account for multiple testing, the Illumina Custom Error Model with the False Discovery Rate (FDR) corrections was applied; we ran 1000 permutations and included FDR up to 20 %. Targets showing significant intergroup differences in methylation levels (the methylation Difference Score, DiffScore >|30|, p <0.001) more than 0.2 were considered to be differentially methylated (Grafodatskaya et al. 2010).
Functional annotation of differentially methylated genes (DMEGs)
To identify common biological processes and pathways, molecular functions, and cellular components for genes that showed differential methylation in the tissues analyzed, we applied the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics software (Dennis et al. 2003; Huang et al. 2008). For this analysis the default (medium stringency) setting of the DAVID analysis tool was used, which compares the enrichment of gene ontology (GO) with the list of differentially methylated genes (DMEGs) using Fisher’s exact test. Annotation clusters with enrichment scores>1.3 (p <0.05) were included in the analysis. GO terms with FDRs <20 % were used to avoid reporting false positives and to reduce the large number of associations identified by DAVID in the functional annotation charts. The p values and the Benjamini corrections of the scores from the tool were used as inclusion criteria in the trimming of the clusters to overrepresented term lists.
Results
We examined whole genome methylation patterns in three types of biological samples—whole blood (WB), saliva (SL) and lymphoblastoid cell lines (LCLs), which are widely used as a source of genomic DNA in epigenetic studies, and performed a comparative DNA methylation analysis across these three tissues derived from the same 14 individuals. Global methylation levels of DNA from three sets of samples (SLs, LCLs and WBs) were carried out using the Illumina Infinium HumanMethylation27 BeadChip.
The resulting methylation data were analyzed by means of pairwise comparison of genome-wide DNA-methylation profiles across WB, SL and LCLs samples, and differential methylation analysis between groups of samples corresponding to different tissues. The analysis was performed based on 26,273 Illumina targets, which had passed detection quality filtering for differential methylation analysis and were localized on autosomes. To assess the DNA methylation differences between different tissues in GO terms we performed functional annotation of genes, which showed differential methylation in pairwise comparisons across the three sets of samples, WB, SL and LCLs.
Comparison of global DNA methylation profiles
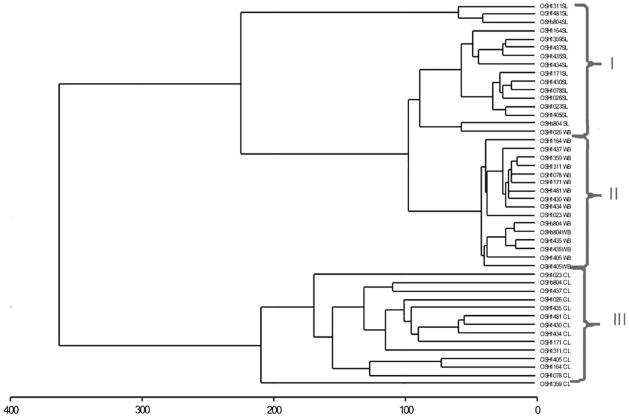
To assess the main differences in the genome-wide DNA-methylation of the three studied sets of samples, WB, SL and LCLs, we performed Euclidean hierarchical clustering analysis of the Illumina methylation data. The results of the analysis are represented in a dendrogram (Fig. 1), which leads to the following observations. First, the source of DNA is the main factor in sample differentiation: the dendrogram shows a clear separation of WB, LCLs, and SL into three distinct clusters, marked in the figure by Roman numerals. Second, according to the distances between samples within a cluster, WB samples (Fig. 1; cluster II) show minimal interindividual variability with the highest correlations (r2 = 0.992 ± 0.002) between individual methylation profiles compared to SL and LCLs. Whereas SL and LCLs (Fig. 1; clusters I and III) are characterized by average and distinctively high interindividual variability of their methylation profiles; r2 = 0.980 ± 0.019 and 0.955 ± 0.018, respectively. Third, the methylation profiles of DNA from SL and WB show a greater similarity in the comparison across all three groups, whereas LCLs cluster maximally remotely from the two others. The last observation on the distance of LCLs from SL and WB was confirmed by correlation analysis between methylation levels across these three tissues. The correlation coefficient for SL compared to WB (r2 = 0.967) was much higher than those for LCLs compared to WB, and LCLs compared to SL (r2 = 0.880 and 0.844, respectively). These correlations across WB, SL, and LCLs as groups of samples are consistent with the data on the correlations between the cell/tissue types within individuals (Online Resource, Table S1).
Fig. 1.
Euclidean hierarchical clustering of 14 individuals based on the pair-wise comparison of the whole genome methylation profiles in WB, SL and LCLs. The dendrogram shows a clear separation of the samples from the different tissues into three distinct clusters, those containing samples from SL, whole blood (WB), and LCLs, marked as clusters I, II, and III, respectively
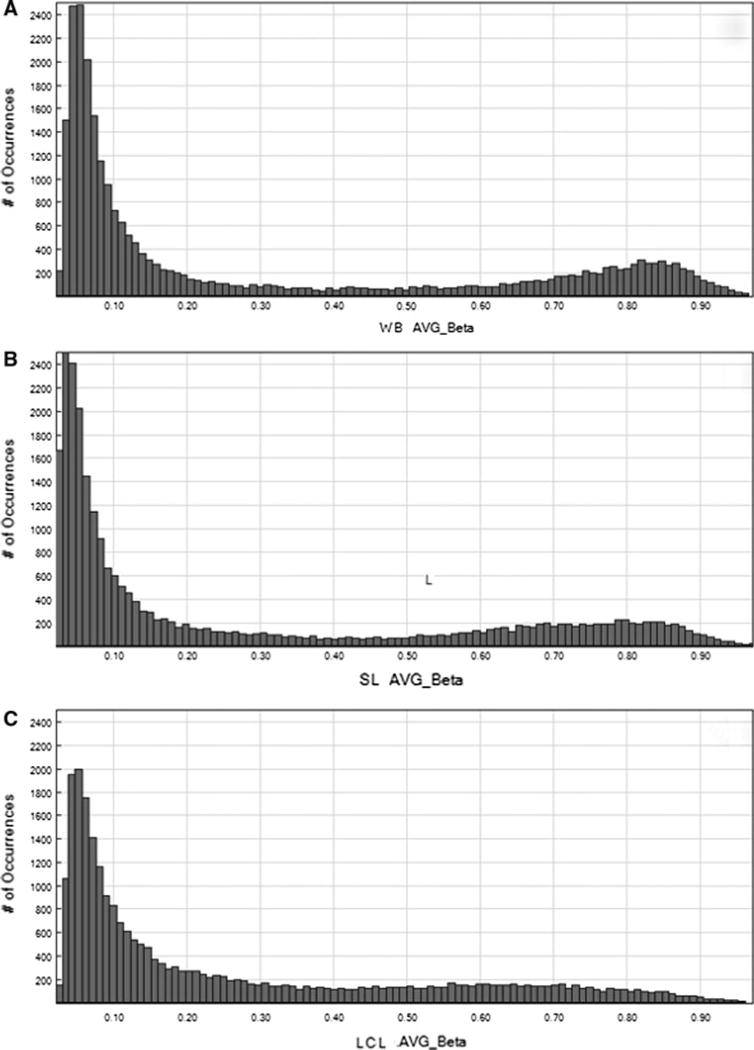
Comparison of the distributions of the methylation levels across the three sets, SL, WB and LCLs, showed that hypo and hypermethylation patterns are especially different between these three cell types. Specifically, the methylation profiles of LCLs are characterized by a lower frequency of CpG sites with high methylation levels, as well as unmethylated CpG sites compared to WB and SL (Fig. 2). This analysis shows that LCLs differ predominantly in areas of up and down regulated genes.
Fig. 2.
The distributions of the methylation levels (Avgβ) of 26,273 CpG sites contained in the Illumina Infinium27 array that had passed detection quality filtering and were localized on autosomes, in three groups of DNA samples derived from blood (a), saliva (b) and lymphoblastoid cell lines (c)
Differential methylation analyses and functional annotation of DMEGs
Data was then analyzed to detect differential methylation patterns of DNA obtained from three groups of samples corresponding to the three cell types. CpG sites showing significant (p <0.001) intergroup differences of at least 1.2-fold change in methylation levels were considered to be differentially methylated. The results of the pair-wise comparison between sets of samples are presented in supplementary materials (Online Resource, Table S2–S4). The distribution of DMEGs across pairwise comparisons is shown in Fig. 3.
Fig. 3.
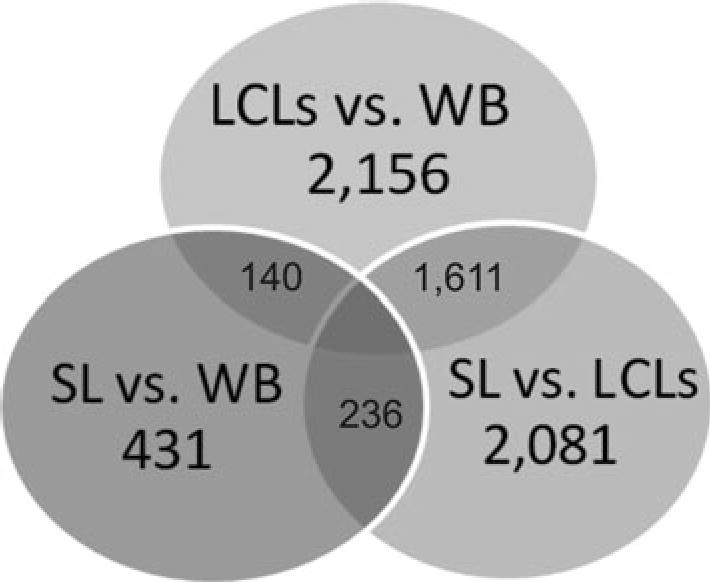
Venn diagram representing the results of differential methylation analysis across WB, SL and LCLs. The numbers in the circles represent both the number of differentially methylated genes detected in each pair-wise comparison and the number of overlapping genes across different comparisons
The minimal number of differentially methylated sites was found in the comparison of DNA from WB and SL: of the 27,578 CpG sites initially analyzed, 488 sites showed a significant difference in methylation level measurements (Online Resource, Table S2; Fig. 3). These 488 CpG sites are located on 431 genes, which represents ~3 % of the total number of genes (14,495) contained in the Illumina’s array (Online Resource, Table S5). The majority of sites (n = 370; ~76 %) are downregulated in SL compared to WB. The remaining sites were characterized by upregulation (n = 118; ~ 24 %) in SL. This corresponds to the results in Fig. 2, where blood showed more occurrences of hypermethylated sites compared to SL.
Whereas several thousand differentially methylated CpG sites were found in the comparisons of these two cell types with LCLs, differential methylation analysis between WB and LCLs groups revealed that 2,661 sites exhibited a methylation level difference at the same level of significance (Online Resource, Table S3; Fig. 3). These 2,661 CpG sites are located on 2,156 genes, which represent ~15 % of the total number of genes analyzed (Online Resource, Table S5). As in the WB and SL analysis, a great majority of the methylated sites (n = 2,545; ~96 %) were downregulated in LCLs compared to WB. Also evident in Fig. 2 is that LCLs exhibited lower levels of hypermethylated and hypomethylated sites when compared to WB and SL. The remaining sites were characterized by upregulation in LCLs (n = 116; ~4 %). Similar differences in methylation levels were found in the comparison between DNA from SL and LCLs: 2,513 sites (localized on 2,081 different genes) were found to be differentially methylated between these tissues, and most of them (~79 %) were hypomethylated in DNA from LCLs (Online Resource, Table S4, S5; Fig. 3).
Functional annotation of differentially methylated genes
The functional annotation of genes showed significant enrichment of the lists of DMEGs in some of the GO terms. Most of the genes differentially methylated in WB and SL are involved in the regulation of the immune response and body fluid levels, as well as genes belonging to the plasma membrane functional group (Table 1). There was a high concordance in GO terms overrepresented in the lists of DMEGs in LCLs compared to WB and SL. These genes are involved predominantly in the immune response, and the control of cellular activity and signaling systems, specifically genes coding glycoproteins, cytokines, and their receptors (Table 2).
Table 1.
Gene Ontology (GO) terms overrepresented in the list of genes, which showed differential methylation between DNA samples from SL and WB
| GO term | Genes, count | Genes (%) | p value | Fold enrichment | Benjamini | FDR |
|---|---|---|---|---|---|---|
| GO: 0005887—integral to plasma membrane | 61 | 16.05 | 5.53E–10 | 2.33 | 0.0000 | 0.0000 |
| GO: 0031226—intrinsic to plasma membrane | 62 | 16.32 | 4.84E–10 | 2.31 | 0.0000 | 0.0000 |
| GO: 0044459—plasma membrane part | 87 | 22.89 | 1.92E–08 | 1.79 | 0.0000 | 0.0000 |
| GO: 0005886—plasma membrane | 121 | 31.84 | 1.49E–06 | 1.45 | 0.0001 | 0.0020 |
| GO: 0006955—immune response | 35 | 9.21 | 1.40E–05 | 2.25 | 0.0060 | 0.0237 |
| GO: 0009611—response to wounding | 30 | 7.89 | 8.91E–06 | 2.51 | 0.0077 | 0.0151 |
| GO: 0044421—extracellular region part | 40 | 10.53 | 1.37E–04 | 1.89 | 0.0084 | 0.1832 |
| GO: 0004867—serine-type endopeptidase inhibitor activity | 11 | 2.89 | 3.79E–05 | 5.33 | 0.0203 | 0.0552 |
| GO: 0050878—regulation of body fluid levels | 13 | 3.42 | 8.02E–05 | 4.09 | 0.0273 | 0.1354 |
There were annotated 381 of 431 DMEGs. The GO terms, for which the Benjamini corrections <0.05 and FDR <0.20, are listed
Table 2.
Functional annotation and clusters of genes that showed significant differences of their methylation levels in lymphoblastoid cell lines (LCLs) compared to WB and SL
| Term | LCLs versus WB
|
LCLs versus SL
|
||||||
|---|---|---|---|---|---|---|---|---|
| Cluster# (EScore) | Fold enrichment | Benjamini | FDR | Cluster# (EScore) | Fold enrichment | Benjamini | FDR | |
| Disulfide bond | 1 (75.8) | 2.11 | 9.96E–79 | 2.01E–78 | 1 (76.8) | 2.14 | 1.26E–80 | 2.65E–80 |
| Signal | 2.02 | 3.54E–77 | 2.14E–76 | 2.05 | 3.08E–79 | 1.29E–78 | ||
| Signal peptide | 2.01 | 4.79E–75 | 2.14E–75 | 2.04 | 3.58E–77 | 1.66E–77 | ||
| Glycoprotein | 1.76 | 2.33E–64 | 1.89E–63 | 1.82 | 2.18E–71 | 1.38E–70 | ||
| Glycosylation site: N-linked (GlcNAc…) | – | – | – | 1.80 | 6.99E–64 | 9.76E–64 | ||
| Secreted | 2 (59.0) | 2.59 | 5.07E–79 | 2.05E–78 | 2 (53.4) | 2.41 | 5.20E–62 | 4.37E–61 |
| Signal | 2.02 | 3.54E–77 | 2.14E–76 | 2.05 | 3.08E–79 | 1.29E–78 | ||
| Signal peptide | 2.01 | 4.79E–75 | 2.14E–75 | 2.04 | 3.58E–77 | 1.66E–77 | ||
| GO: 0005576—extracellular region | 1.97 | 8.99E–52 | 2.56E–51 | 1.89 | 1.13E–42 | 3.17E–42 | ||
| GO: 0005615—extracellular space | 2.46 | 1.05E–31 | 5.97E–31 | 2.32 | 1.12E–25 | 6.28E–25 | ||
| GO: 0044421—extracellular region part | 2.10 | 8.15E–28 | 6.96E–27 | 1.99 | 3.55E–22 | 4.99E–21 | ||
| Glycoprotein | 3 (48.8) | 1.76 | 2.33E–64 | 1.89E–63 | 3 (27.9) | 1.82 | 2.18E–71 | 1.38E–70 |
| Glycosylation site: N-linked (GlcNAc…) | 1.73 | 3.44E–56 | 4.61E–56 | 1.80 | 6.99E–64 | 9.76E–64 | ||
| Topological domain: cytoplasmic | 1.49 | 1.30E–19 | 3.48E–19 | 1.78 | 1.14E–35 | 2.12E–35 | ||
| GO: 0006952—defense response | 4 (20.8) | 2.64 | 3.05E–31 | 1.49E–31 | 5 (20.1) | 2.64 | 1.88E–31 | 1.81E–31 |
| GO: 0006954—inflammatory response | 2.48 | 1.82E–12 | 2.66E–12 | 2.41 | 3.26E–11 | 4.71E–11 | ||
| GO: 0009611—response to wounding | 2.05 | 3.20E–11 | 6.25E–11 | 2.02 | 1.74E–10 | 5.02E–10 | ||
| Cytokine | 5 (15.8) | 3.69 | 2.24E–18 | 3.17E–17 | 7 (9.9) | 3.49 | 1.18E–15 | 2.73E–14 |
| GO: 0005125—cytokine activity | 3.36 | 1.72E–16 | 2.35E–16 | 3.07 | 1.50E–12 | 1.98E–12 | ||
| hsa04060: cytokine–cytokine receptor interaction | 2.11 | 1.01E–07 | 1.38E–06 | 2.35 | 1.68E–10 | 1.17E–09 | ||
| GO: 0031226—intrinsic to plasma membrane | 6 (15.1) | 1.71 | 7.98E–16 | 9.09E–15 | 4 (22.6) | 1.90 | 4.05E–24 | 3.42E–23 |
| GO: 0005887—integral to plasma membrane | 1.71 | 6.87E–16 | 9.78E–15 | 1.89 | 4.04E–23 | 4.54E–22 | ||
| GO: 0044459—plasma membrane part | 1.46 | 4.69E–14 | 7.99E–13 | 1.57 | 7.11E–21 | 1.20E–19 | ||
| GO: 0005886—plasma membrane | 1.26 | 7.70E–10 | 1.53E–08 | 1.35 | 6.24E–17 | 1.23E–15 | ||
| Topological domain: cytoplasmic | 7 (12.2) | 1.49 | 1.30E–19 | 3.48E–19 | 3 (27.9) | 1.63 | 8.09E–31 | 1.88E–30 |
| Membrane | 1.25 | 1.13E–12 | 2.28E–11 | 1.30 | 6.76E–18 | 9.95E–17 | ||
| GO: 0005886—plasma membrane | 1.26 | 7.70E–10 | 1.53E–08 | 1.35 | 6.24E–17 | 1.23E–15 | ||
| Transmembrane region | 1.25 | 3.14E–08 | 9.85E–08 | 1.34 | 1.19E–15 | 3.87E–15 | ||
| Transmembrane | 1.25 | 2.49E–09 | 1.01E–07 | 1.34 | 5.12E–16 | 9.68E–15 | ||
| Cell membrane | 1.47 | 3.92E–11 | 1.11E–09 | 1.57 | 9.79E–16 | 2.06E–14 | ||
| Topological domain: extracellular | 1.65 | 3.06E–26 | 6.85E–26 | – | – | – | ||
| GO: 0008544—epidermis development | 8 (9.9) | 2.85 | 3.04E–10 | 8.92E–10 | 9 (6.6) | 2.41 | 4.80E–06 | 3.23E–05 |
| GO: 0030216—keratinocyte differentiation | 4.40 | 3.32E–10 | 1.14E–09 | 3.58 | 5.81E–06 | 4.47E–05 | ||
| GO: 0007398—ectoderm development | 2.73 | 6.19E–10 | 2.42E–09 | 2.42 | 1.12E–06 | 6.49E–06 | ||
| GO: 0009913—epidermal cell differentiation | 4.17 | 5.74E–10 | 2.81E–09 | 3.41 | 8.19E–06 | 6.70E–05 | ||
| Keratinization | 5.84 | 2.68E–10 | 8.67E–09 | 4.90 | 6.57E–07 | 3.73E–05 | ||
| GO: 0030855—epithelial cell differentiation | 3.08 | 2.26E–09 | 1.33E–08 | 2.42 | 2.05E–04 | 2.75E–03 | ||
| GO: 0031424—keratinization | 4.80 | 6.87E–08 | 6.38E–07 | 3.96 | 1.08E–04 | 1.24E–03 | ||
| IPR003267: small proline-rich | 6.51 | 5.77E–07 | 1.57E–06 | 4.97 | 8.95E–04 | 1.38E–03 | ||
| GO: 0009617—response to bacterium | 9 (8.9) | 2.67 | 4.08E–09 | 2.59E–08 | 6 (10.2) | 2.89 | 4.59E–11 | 1.11E––10 |
| Antimicrobial | 4.19 | 2.82E–09 | 1.20E–07 | 4.60 | 5.17E–11 | 1.52E–09 | ||
| GO: 0042742—defense response to bacterium | 3.18 | 3.16E–08 | 2.48E–07 | 3.46 | 5.94E–10 | 2.00E–09 | ||
| Antibiotic | 4.21 | 4.99E–09 | 2.32E–07 | 4.32 | 3.15E–09 | 1.32E–07 | ||
| GO: 0042330—taxis | 10 (8.4) | 3.05 | 1.62E–10 | 3.96E–10 | 7 (9.9) | 3.13 | 3.54E–11 | 6.82E–11 |
| GO: 0006935—chemotaxis | 3.05 | 1.62E–10 | 3.96E–10 | 3.13 | 3.54E–11 | 6.82E–11 | ||
| Chemotaxis | 4.28 | 8.28E–10 | 2.85E–08 | 4.38 | 4.10E–10 | 1.55E–08 | ||
| IPR000827: small chemokine, C–C group, conserved site | 6.91 | 1.16E–07 | 1.05E–07 | 7.04 | 4.35E–08 | 7.92E–08 | ||
| IPR001811: small chemokine, interleukin-8-like | 5.21 | 2.12E–07 | 3.85E–07 | 5.56 | 3.17E–08 | 2.88E–08 | ||
| SM00199: SCY | 5.07 | 1.07E–07 | 4.59E–07 | 5.24 | 1.54E–08 | 6.44E–08 | ||
| GO: 0008009—chemokine activity | 4.68 | 1.48E–07 | 1.22E–06 | 4.90 | 5.57E–08 | 1.47E–07 | ||
| GO: 0042379—chemokine receptor binding | 4.39 | 3.70E–07 | 5.06E–06 | 4.60 | 1.73E–07 | 6.86E–07 | ||
| PIRSF001950: small inducible chemokine, C/CC types | 5.37 | 5.78E–06 | 1.17E–05 | 5.60 | 3.24E–06 | 6.44E–06 | ||
| GO: 0007610—behavior | 1.64 | 6.06E–04 | 1.29E–02 | 1.81 | 5.89E–06 | 4.25E–05 | ||
| GO: 0007626—locomotory behavior | 1.92 | 2.57E–04 | 4.65E–03 | 2.07 | 1.35E–05 | 1.24E–04 | ||
The top list of DAVID annotation clusters is presented
The differences in methylation levels observed may be a result of tissue specific regulation. We performed a functional annotation of DMEGs, using the DAVID tissue expression annotation tools. For the WB versus SL comparison, this assumption was confirmed; namely, the list of DMEGs was overrepresented by genes known to be expressed in white blood cells and the salivary gland (Table 3). In contrast, the spectrum of GO terms for genes differentially methylated in WB versus LCLs was wider and included a number of tissues—from the cerebellum to the uterus, as well as cultivated cells—bone marrow CD105+ endothelial cells and Burkitt’s lymphoma cell lines (Table 3).
Table 3.
The annotation of genes that have shown differential methylation in whole blood compared to saliva (WB vs. SL) and lymphoblastoid cell lines (WB vs. LCLs), in terms of their expression in different tissues
| Term | Gene count | Gene (%) | p value | Fold enrichment | Benjamini |
|---|---|---|---|---|---|
| WB versus SL | |||||
| WBCs, plaque macrophage | 38 | 10.00 | 1.47E–04 | 1.93 | 1.68E–02 |
| WBCs, monocyte-depleted mononuclear cells | 39 | 10.26 | 1.32E–03 | 1.70 | 3.21E–02 |
| WBCs, monocyte | 48 | 12.63 | 2.28E–03 | 1.55 | 4.83E–02 |
| Salivary gland | 247 | 65.00 | 1.85E–02 | 1.07 | 8.20E–02 |
| WB versus LCLs | |||||
| Cerebellum | 1110 | 60.72 | 5.88E–23 | 1.15 | 4.58E–21 |
| Salivary gland | 1140 | 62.36 | 4.45E–22 | 1.14 | 1.74E–20 |
| Burkitt lymphoma (Raji) | 1018 | 55.69 | 3.21E–09 | 1.10 | 3.13E–08 |
| Tonsil | 939 | 51.37 | 7.55E–05 | 1.07 | 3.68E–04 |
| Tongue | 386 | 21.12 | 5.35E–05 | 1.18 | 2.98E–04 |
| Whole blood | 479 | 26.20 | 5.37E–05 | 1.15 | 2.79E–04 |
| BM-CD105+ endothelial cells | 427 | 23.36 | 4.08E–04 | 1.14 | 1.77E–03 |
| Uterus | 293 | 16.03 | 1.74E–03 | 1.17 | 6.17E–03 |
Discussion
We compared DNA methylation profiles between WB, SL and LCLs from the same individuals using cluster and linear-regression analyses of the methylation profiles, and analysis of DMEGs. The results showed that LCLs have the most distinct methylation patterns compared to those in WB and SL. These results are not surprising and are consistent with published studies that warn researchers about the methylation changes in LCLs due to cell culturing (Brennan et al. 2009; Calıskan et al. 2011; Grafodatskaya et al. 2010; Sun et al. 2010; Sugawara et al. 2011). In contrast, WB and SL showed a relatively similar methylation pattern that is in line with a previous study (Liu et al. 2010). The DNA methylation differences found between WB and SL might be explained in terms of tissue specific methylation; thus most of the DMEGs are genes coding membrane complexes and are involved in immune response, as well as genes known to be expressed specifically in white blood cells and the salivary gland.
The distinctiveness of methylation patterns in LCLs might be caused by a complex of factors, including the mono-cellular nature of LCLs, which are composed only of B-lymphocytes, and their modifications throughout the culturing procedure. The compositional differences might be a result in the differential regulation of genes involved in the immune response, cellular activity and signaling systems, as was found in LCLs. At the same time, the EBV transformation of B-lymphocytes, preceding the cell culturing, caused uncontrolled growth, proliferation and abnormal cell signaling. DMEGs in LCLs were found to be predominantly involved in the control of cellular activity and signaling systems, specifically genes coding signal peptides, such as cytokines and chemokines. This is consistent with a study reporting that EBV-mediated transformations rely extensively on interference with cytokine signaling networks (Mosialos 2001). Additionally it was found that the list of DMEGs in LCLs was enriched in genes found to be expressed in other cultivated cells—bone marrow CD105+ endothelial cells and Burkitt’s lymphoma Raji cell line; the latter, Burkitt’s lymphoma, is known to be associated with EBV infection (Fujita et al. 2004; Maeda et al. 2009). This finding provides further evidence that cell culturing procedures are responsible for the specificity of the methylation pattern in LCLs and its distance from those in WB and SL cells.
Taken together, the results of the study suggest that due to multiple random and directed changes of methylation patterns, LCLs are not a reliable source of DNA for epigenetic studies, as opposed to peripheral blood and saliva. As a result, LCLs should be used with particular caution to identify the epigenetic mechanisms underlying biological processes and their violations, due to disorders associated with DNA methylation variants.
Supplementary Material
Acknowledgments
This work was supported by Awards DC007665 as administered by the National Institute of Deafness and Communication Disorders, P50 HD052120 as administered by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and Grant R25HL088730 (BioSTEP) from NIH-National Heart, Lung, and Blood Institute. Grantees undertaking such projects are encouraged to freely express their professional judgment. Therefore, this article does not necessarily reflect the position or policies of the National Institutes of Health, and no official endorsement should be inferred. The authors alone are responsible for the content and writing of the article.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10519-012-9579-1) contains supplementary material, which is available to authorized users.
Contributor Information
Tara M. Thompson, Yale University, New Haven, CT, USA
Duaa Sharfi, Yale University, New Haven, CT, USA. University of Illinois, Chicago, IL, USA.
Maria Lee, Yale University, New Haven, CT, USA.
Carolyn M. Yrigollen, Yale University, New Haven, CT, USA. University of California, Davis, CA, USA
Oksana Yu Naumova, Yale University, New Haven, CT, USA. Vavilov Institute of General Genetics of the Russian Academy of Sciences, Moscow, Russian Federation.
Elena L. Grigorenko, Email: elena.grigorenko@yale.edu, Yale University, New Haven, CT, USA. Moscow State University for Psychology and Education, Moscow, Russian Federation. Columbia University, New York, NY, USA. Child Study Center, Department of Psychology, Department of Epidemiology and Public Health, Yale University, 230 South Frontage Road, New Haven, CT 06519-1124, USA
References
- Abdolmaleky HM, Smith CL, Faraone SV, Shafa R, Stone W, Glatt SJ, Tsuang MT. Methylomics in psychiatry: modulation of gene–environment interactions may be through DNA methylation. Am J Med Genet. 2004;127B:51–59. doi: 10.1002/ajmg.b.20142. [DOI] [PubMed] [Google Scholar]
- Abdolmaleky HM, Zhou JR, Thiagalingam S, Smith CL. Epigenetic and pharmacoepigenomic studies of major psychoses and potentials for therapeutics. Pharmacogenomics. 2008;9:1809–1823. doi: 10.2217/14622416.9.12.1809. [DOI] [PubMed] [Google Scholar]
- Brennan EP, Ehrich M, Brazil DP, Crean JK, Murphy M, Sadlier DM, et al. Comparative analysis of DNA methylation profiles in peripheral blood leukocytes versus lymphoblastoid cell lines. Epigenetics. 2009;4:159–164. doi: 10.4161/epi.4.3.8793. [DOI] [PubMed] [Google Scholar]
- Calıskan M, Cusanovich DA, Ober C, Gilad Y. The effects of EBV transformation on gene expression levels and methylation profiles. Hum Mol Genet. 2011;20:1643–1652. doi: 10.1093/hmg/ddr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis GJ, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- Fujita S, Buziba N, Kumatori A, Senba M, Yamaguchi A, Toriyama K. Early stage of Epstein-Barr virus lytic infection leading to the “starry sky” pattern formation in endemic Burkitt lymphoma. Arch Pathol Lab Med. 2004;128:549–552. doi: 10.5858/2004-128-549-ESOEVL. [DOI] [PubMed] [Google Scholar]
- Grafodatskaya D, Choufani S, Ferreira JC, Butcher DT, Lou Y, Zhao C, et al. EBV transformation and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics. 2010;95:73–83. doi: 10.1016/j.ygeno.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Elisch AJ. Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: unraveling the genome to understand the mind. Neurobiol Dis. 2010;39:73–84. doi: 10.1016/j.nbd.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VW, Frank BC, Heine S, Lee NH, Quakenbush J. Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics. 2006;7:118. doi: 10.1186/1471-2164-7-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004;9:406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- Laird PW. The power and promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Liu J, Morgan M, Hutchison K, Calhoun VD. A study of the influence of sex on genome wide methylation. PLoS ONE. 2010;5(4):e10028. doi: 10.1371/journal.pone.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda E, Akahane M, Kiryu S. Spectrum of Epstein-Barr virus-related diseases: a pictorial review. Jpn J Radiol. 2009;27:4–19. doi: 10.1007/s11604-008-0291-2. [DOI] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosialos G. Cytokine signaling and EBV cell transformation. Cytokine Growth Factor Rev. 2001;12:259–270. doi: 10.1016/s1359-6101(00)00035-6. [DOI] [PubMed] [Google Scholar]
- Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet. 1986;73:320–326. doi: 10.1007/BF00279094. [DOI] [PubMed] [Google Scholar]
- Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohesara S, Ghadirivasfi M, Mostafavi S, Eskandari M-R, Ahmadkhaniha H, Thiagalingam S, Abdolmaleky HM. DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. J Psychiatr Res. 2011;45:1432–1438. doi: 10.1016/j.jpsychires.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Sapienza C, Lee J, Powell J, Erinle O, Yafai F, Reichert J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6:20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- Sugawara H, Iwamoto K, Bundo M, Ueda J, Ishigooka J, Kato T. Comprehensive DNA methylation analysis of human peripheral blood leukocytes and lymphoblastoid cell lines. Epigenetics. 2011;6:508–515. doi: 10.4161/epi.6.4.14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YV, Turner ST, Smith JA, Hammond PI, Lazarus A, Van De Rostyne JL, et al. Comparison of the DNA methylation profiles of human peripheral blood cells and transformed B-lymphocytes. Hum Genet. 2010;127:651–658. doi: 10.1007/s00439-010-0810-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierling S, Souren NY, Reither S, Zang KD, Meng-Hentschel J, Leitner D, et al. DNA methylation studies on imprinted loci in a male monozygotic twin pair discordant for Beckwith-Wiedemann syndrome. Clin Genet. 2011;79:546–553. doi: 10.1111/j.1399-0004.2010.01482.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.