Abstract
Molecular imaging, the visualization of molecular and cellular markers, is a promising method for detection of dysplasia and early cancer in the esophagus and can potentially be used to identify regions of interest for biopsy or tumor margins for resection. EGFR is a previously-reported cell surface receptor with stepwise increases in expression during the progression from Barrett’s metaplasia to adenocarcinoma. In this work, a 200-nm fluorescent nanoparticle contrast agent was synthesized for targeted imaging of EGFR through a series of surface modifications to dye-encapsulated polystyrene particles. Amino-functionalized polystyrene particles were PEGylated using a heterobifunctional PEG linker. Subsequently, thiolated M225 antibodies were conjugated to maleimide functional groups on attached PEGs for EGFR targeting. In vitro binding studies using flow cytometry demonstrated specific binding of M225-PEG-NP to EGFR-expressing cells with minimal non-specific binding in EGFR− cells. Binding was shown to increase proportionally with the number of conjugated M225 antibodies. Adsorbed formulations with unmodified M225 antibodies, M225 + PEG-NP, were synthesized using the same antibody feeds used in M225-PEG-NP synthesis to determine the contribution of adsorbed antibodies to EGFR targeting. Adsorbed antibodies were less efficient at mediated nanoparticle targeting to EGFR than conjugated antibodies. Finally, M225-PEG-NP demonstrated binding to EGFR-expressing regions in human esophageal tissue sections.
Introduction
Esophageal adenocarcinoma (EAC) is an epithelial cancer arising from the columnar glandular epithelium, and it has dramatically increased in incidence in recent decades.1 Patients with Barrett’s esophagus (BE) have an increased risk of developing EAC.2 Barrett’s esophagus is a metaplastic condition caused by gastroesophageal reflux disease (GERD) in which oxidative stress from chronic exposure to gastric acids leads squamous epithelium in the esophagus to change into columnar epithelium.3,4 From Barrett’s metaplasia, tissue may progress into premalignant dysplastic tissue and, finally, to adenocarcinoma, a phenomenon referred to as the metaplasia-dyplasia-adenocarcinoma sequence.5,6 High-grade dysplasia (HGD) is a validated predictive marker for adenocarcinoma and approximately 59% of patients with BE with HGD progress to EAC within 5 years of diagnosis.7-10 Pre-emptive measures such as esophageal mucosal resections, ablative therapies, or esophagectomies may be prescribed when high-grade dysplastic lesions or intramucosal adenocarcinoma are found in Barrett’s tissue. Current clinical practice for diagnosis involves endoscopic imaging and four-quadrant biopsy of Barrett’s tissue, which is easily visible due to its characteristic salmon pink coloration. However, dysplastic BE is not easily distinguishable from non-dysplastic BE using conventional white light endoscopy and the former can present in small patches and be multifocal. Thus, biopsy samples are prone to sampling error potentially resulting in false negative findings for dysplasia as well as cancer.11,12 Contrast agents labeling areas with dysplasia in Barrett’s esophagus are potentially useful tools to direct biopsy of areas with the greatest probability of dysplasia or cancer.
Imaging of aberrant molecular expression in cells and tissues is a promising method for early detection of dysplasia or cancer since molecular changes occur at earlier time points than visible physical changes.13 Targeted fluorescent contrast agents have been developed for the purpose of molecular imaging and have recently been shown to be effective in labeling dysplastic and cancerous tissue for endoscopic imaging.14-17 These contrast agents were synthesized through bioconjugation of proteins or peptides to fluorophores. Li et al reported the isolation of a peptide against dysplastic mucosa using phage display and further demonstrated preferential binding of the peptide-flourescein isothiocyanate (FITC) conjugate to Barrett’s esophagus with dysplasia.15 More recently, Bird-Lieberman et al. demonstrated the use of Alexa Fluor 680-conjugated wheat germ agglutinin (WGA) as a negative contrast agent for sialic acid which has decreased expression in EAC compared to non-dysplastic Barrett’s esophagus.17
The aforementioned peptide and protein constructs demonstrate the potential of molecular imaging for early detection of dysplasia in the esophagus. However, there is still a need for improvement of the current contrast agents. Peptide ligands have relatively low binding affinity to their targets (typical KD ~10-100 μM compared to nanomolar affinities for antibodies). In addition, fluorophores directly conjugated to ligands are susceptible to photobleaching when exposed to light and oxygen.18 In contrast, fluorophore-loaded polystyrene nanoparticles (NP) exhibit high photostability due to partitioning of dyes from the oxygenated environment.18,19 These fluorescent NP are commercially-available in well-characterized formulations in a range of sizes and colors. In addition, these particles are highly fluorescent (e.g. over 100,000 fluorophore equivalents in a 200-nm particle). Amino- and carboxylic acid-modified NP can be readily functionalized with ligands for molecular recognition. Thus, NP-based formulations of targeted contrast agents have the advantage of better photostability in vivo as well as greater equivalent fluorophore signal per binding event with fluorescence intensities much greater than that of tissue autofluorescence. Localization of these NPs on tissue would thus be easily visualized with a fluorescent endoscope.
This work reports the synthesis, characterization, and evaluation of a fluorescent NP-based imaging probe for targeted optical imaging of EGFR as a proof-of-concept for antibody-functionalized nanoparticle contrast agents for endoscopy. EGFR is a transmembrane receptor tyrosine kinase in the ErbB family of receptors. Ligand binding to the extracellular portion of EGFR induces phosphorylation of intracellular tyrosine kinase domains leading to downstream cell signaling for proliferation, migration, invasion, and inhibition of apoptosis in cancer cells.20 Thus, its upregulated expression has been commonly cited in the pathogenesis for many epithelial cancers and NP targeted to EGFR have been developed for both drug delivery and molecular imaging applications.21-27 A majority of these particles were designed and evaluated for systemic delivery while very few have been synthesized with the intention of topical application in the gastrointestinal tract. Recently, EGFR expression was shown to increase in a stepwise manner during the progression from Barrett’s metaplasia to low-grade dysplasia to high-grade dysplasia and finally to esophageal adenocarcinoma.28 Therefore, differential EGFR expression during the metaplasia-dysplasia-adenocarcinoma sequence of the esophagus provides an opportunity to evaluate the potential of antibody-functionalized nanoparticles for optical molecular imaging of the esophagus.
In the present work, a fluorescence-based probe against the EGFR protein was synthesized by conjugation of M225, a murine monoclonal antibody against human EGFR, to fluorescent polystyrene NP via a polyethylene glycol (PEG) linker. Specific binding of these NP-based contrast agents to EGFR-expressing cells was tested in vitro in A431 and NCI-H520 cultured cells using flow cytometry. A431 is a commonly used model cell line for evaluation of EGFR targeting due to its high EGFR expression29, and NCI-H520 has been previously shown to express no EGFR30. EGFR-targeted fluorescent NP were then used to label human esophageal tissue ex vivo to determine areas of NP localization. The heterogeneous composition and differential EGFR expression of tissues offered a more robust evaluation of EGFR-targeting NP and its potential for in vivo translatability.
Experimental Procedures
M225-PEG-NP synthesis
200-nm amine-modified red fluorescent (580/605) polystyrene Fluospheres® (Invitrogen, F8763) were sonicated for 30 minutes. The Fluospheres® were then reacted with 5 molar excess of 5000 MW maleimide-PEG-SCM (Laysan Bio Inc) to surface amine groups for 2.5 hours in sodium phosphate buffer (50 mM sodium phosphate, pH 7.5) at a reaction concentration of 5 mg Fluospheres®/ml. PEGylated NP were then pelleted at 10,000 rcf for 15 min. The supernatant was discarded to remove unreacted PEG. Mouse anti-human EGFR monoclonal antibody (M225) (Emory Vaccine Center) was thiolated using 10 molar excess of Traut’s reagent in phosphate buffered saline (50 mM sodium phosphate, 150 mM sodium chloride, pH 8) for 1 hour. Thiolated M225 antibody was purified using a PD-10 desalting column (GE Healthcare Life Sciences, 17-0851-01) Pre-adsorbed with bovine serum albumin (1ml, 1% BSA in PBS) and collected fractions were concentrated with an Amicon Ultra-4 centrifugal filter (Millipore) at 4000 rcf for 10 min. Final antibody concentration was measured using a Nanodrop 2000c spectrophotometer. Thiolated M225 antibodies were added to PEGylated NPs at antibody to NP molar ratios of 100, 500, and 900 and reacted overnight under stirring conditions. M225-PEG-NP formulations were then dialyzed against deionized water in 300kDa MWCO Spectra/Por CE dialysis membranes (Spectrum, 131450) for 2 days at 4°C to remove unreacted M225 and dialyzed for 1 day against 150 mM sodium chloride for buffer exchange.
M225 + PEG-NP synthesis
NP with adsorbed antibodies were prepared using the same PEGylation process outlined in the previous section. Unmodified M225 antibodies were added to PEG-NP at antibody to NP molar ratios of 100, 500, and 900 and adsorbed overnight under stirring conditions. M225 + PEG-NP formulations were then dialyzed against deionized water in 300kDa MWCO Spectra/Por CE dialysis membranes for 2 days at 4°C to remove unreacted M225 and dialyzed for 1 day against 150 mM sodium chloride for buffer exchange.
Characterization of M225-PEG-NP and M225 + PEG-NP
NP were sized using dynamic light scattering (DLS; Brookhaven Instruments Zeta PALS analyzer). For antibodyquantification, M225 antibodies were radiolabeled as previously described with slight modification.31,32 N-succinimidyl [2,3-3H] propionate in DMF (American Radiolabeled Chemicals, ART0135A) was reacted with M225 antibodies at a molar ratio of 1 radiolabel for every 5 antibodiesin pH 8.5 100 mM borate buffer with a final antibody concentration of 1 mg/ml. Radiolabeled antibodies were thiolated, purified using a PD-10 column, and reacted with PEGylated NPs as previously described to synthesize M225-PEG-NP nanoparticles. Unmodified radiolabeled antibodies were adsorbed to PEGylated NPs to yield M225 + PEG-NP. Absorbance at 280 nm and scintillation counting of an aliquot of radiolabeled antibody were used to determine counts per labeled antibody. Tritium radioactivity of radiolabeled antibodies and finished nanoparticles was measured using a Beckman LS 6500 scintillation counter. 100 μl of sample was dissolved in 5mL Ultima Gold scintillation liquid and read on the counter for 10 minutes.
Cell culture
Human A431 (epidermoid carcinoma) cells and NCI-H520 (lung squamous cell carcinoma) cells purchased from ATCC were maintained in DMEM and RPMI 1640 media, respectively. Media was supplemented with 10% fetal bovine serum and 1% antibiotic antimycotic solution. EGFR expression in A431 and NCI-H520 cells was evaluated using flow cytometry to confirm that A431 cells are EGFR+ and NCI-H520 cells are EGFR−. Cells were incubated with 300 μl of 5 μg/ml M225 antibody in FACS buffer (1% bovine serum albumin in PBS) for 15 minutes at 4°C, washed twice, and incubated in 300 ul of a 1:100 dilution of FITC-conjugated sheep anti-mouse IgG in FACS buffer. Cells were washed twice and then analyzed by flow cytometry (Supporting Information Figure S1).
In vitro binding experiments
Preferential and specific binding of M225-PEG-NP nanoparticles to EGFR-expressing cells was confirmed using flow cytometry. 2×105 A431 (EGFR+) cells and NCI-H520 (EGFR−) cells were incubated with 300 μL of 10 pM M225-PEG-NP formulations for 15 minutes at 37°C and washed twice using FACS buffer solution. 10 pM nanoparticle concentrations were used because it was determined to be the concentration at which bimodal populations in M225-PEG-NP-treated cells did not exist (data not shown), and at which fluorescence intensities would not saturate the detector for flow cytometry. For competitive conditions, both cell types were Pre-blocked with 300 μL of free M225 antibody (13.3 μM in FACS buffer) for 1 hour at room temperature. Following Pre-blocking, cells were incubated with 300 μL of 10 pM M225-PEG-NP formulations for 15 minutes at 37°C and washed twice with FACS buffer. Flow cytometry was then completed using a Miltenyi MACSQuant Analyzer. The extent of binding due to adsorbed antibodies versus chemically conjugated antibodies was determined by comparing binding observed in formulations with unmodified M225 antibody versus formulations with thiolated M225 antibody. To further show specificity of M225-PEG-NP to EGFR, H520 and A431 cells were grown on glass coverslips and treated with PEG-NP or M225-PEG-NP for 30 minutes at room temperature, counterstained with 2 μg/mL DAPI for 10 minutes, mounted onto slides with Fluoromount, and imaged using a Zeiss LSM 510 META confocal microscope.
Evaluation of cytotoxicity
5×105 H520 and A431 cells were treated with 200 μL unmodified NP, PEG NP, and M225-PEG-NP (10 pM-76 nM, 0.026-2.0 mg/mL)in phosphate buffered saline (PBS) for 1 hr at 37°C. Cells were washed twice and incubated with 300 μl of 5 μg/ml DAPI in PBSfor 5 minutes at room temperature. DAPI was used as a viability exclusion dye to stain dead or dying cells as previously described33 since other available viability exclusion dyes had significant spectral overlap with NPs. Cells were washed twice more, and flow cytometry was completed. H520 and A431 cells treated with DAPI only were used as negative controls for cell death while cells treated with 0.2 mg/ml poly(ethylenimine) (PEI) and DAPI were used as positive controls for cell death. Cells on DAPI versus forward scatter dot plots were gated to determine the percentage of DAPI− cells which correlate to the percentage of live cells.
Ex vivo M225-PEG-NP labeling of EGFR in human esophageal tissue
Approval from the University of Washington institutional review board (IRB) was obtained to acquire tissue from patients with Barrett’s esophagus and EAC. Informed consent was obtained from patients undergoing endoscopy procedures for collection of esophageal biopsy specimen for research purposes. Endoscopic biopsies were fresh frozen in OCT and sectioned into 7 μm thick sections. Sections were stained with hematoxylin and eosin and classified by a pathologist as normal esophagus squamous (SQ), Barrett’s metaplasia (BE), low-grade (LGD), high-grade dysplasia (HGD), and esophageal adenocarcinoma (EAC). In total, 13 biopsy samples (3 SQ, 5 BE, 1 BE and EAC, 3 EAC, and 1 EAC and HGD) from 6 patients were used for histology. Tissue sections were stained for EGFR using M225 antibody and the secondary Alexa Fluor® 488 goat anti-mouse IgG (Invitrogen). Adjacent tissue sections were stained with M225-PEG-NP for 30 minutes at 37°C and counterstained with DAPI for 10 minutes at room temperature to demonstrate localization of M225-PEG-NP in areas of EGFR-expression. Adjacent tissue sections were incubated with PEG-NP for 30 minutes at 37°C as negative controls. Sections were then imaged using a Zeiss LSM 510 META confocal microscope.
Statistical analysis
Student t-tests were completed to determine statistical significance of the binding data. P-values less than 0.05 were considered significant.
Results & Discussion
Preparation and characterization of M225-PEG-NPs
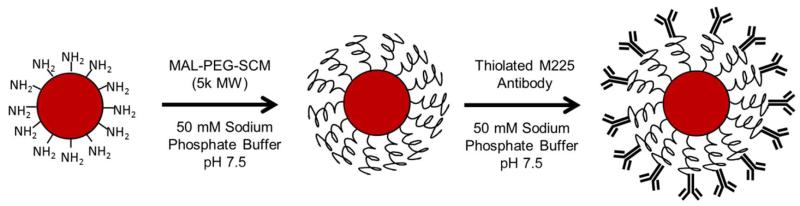
Anti-EGFR fluorescent NPs were synthesized by conjugation of the M225 murine antibody to polystyrene particles via a PEG linker. Unmodified polystyrene NPs are prone to flocculation in the presence of proteins and in physiologic salt concentrations.34 Particles were therefore PEGylated to increase the hydrophilicity of NP surfaces and prevent NP aggregation, to neutralize charge from surface amine groups thereby minimizing nonspecific binding to cells, and to provide a point of attachment for the targeting ligand. Intervening linkers have been previously shown to be more effective for antibody conjugation to nanoparticle surfaces than direct conjugation possibly due to reduced steric hindrance from bulky nanoparticle surfaces.35 PEG linkers with 5000 Da were selected because this size has been previously demonstrated to provide sufficient steric stabilization to keep nanoparticles disperse in physiologic conditions.36
Initially, varying ratios of shorter (2k) monofunctional mPEG-SCM to longer (5k) heterobifunctional maleimide-PEG-SCM (10:0, 7:3, 5:5, 3:7, 0:10) were reacted to amino-functionalized fluorescent polystyrene NPs for more protruded display of M225 antibodies from NP surfaces. The use of bidispersed protective polymers has been reported to provide improved receptor binding by displayed targeting ligands.37,38 The shorter polymer is proposed to increase accessibility of ligands conjugated to the longer polymer chain while providing a protective brush layer to the particle surface. However, introduction of shorter 2000 MW mPEG-SCM increased the amount of non-specific cell interaction of NP as assessed by flow cytometry (data not shown). This may be attributed to less effective charge shielding of residual protonated amine groups thus allowing electrostatic interaction between negative cell plasma membranes and positive NP surfaces. Surface-conjugated PEG has been shown to exist in two conformations: the mushroom conformation or brush conformation of which the former has poorer shielding capabilities. In general, larger PEG favor the brush conformation.39 In light of this, only 5000 MW maleimide-PEG-SCM was used to PEGylate NPs. Following PEGylation, thiolated M225 antibodies were reacted to maleimide groups on NP-attached PEG linkers for targeting to EGFR (Scheme 1).
Scheme 1.
Synthesis of M225-PEG-NP nanoparticles. Note: not drawn to scale.
The stability of M225-PEG-NP in suspension could be observed macroscopically. Unmodified NP controls sedimented during storage at 4°C while PEGylated formulations stayed in suspension. Particle size measurements from DLS indicate minor aggregation in nanoparticle formulations in 150 mM sodium chloride solution (Table 1). The effective diameter for unmodified and PEGylated nanoparticles were 640 ± 9 and 500 ± 4 nm, respectively. Thus, PEGylation was shown to reduce aggregation. Nanoparticles with conjugated or adsorbed M225 had effective diameters ranging from 505-586 nm and 523-735 nm, respectively, with diameter increasing proportionally with antibody feed.
Table 1.
Properties of M225-PEG-NP and M225 + PEG-NP formulations.
| Sample | Molar Feed Ratio of Thiolated Antibody to NP |
Molar Feed Ratio of Unmodified Antibody to NP |
Number of Conjugated or Absorbed Antibody Per NP |
Effective Diameter (nm) |
Standard Deviation (nm) |
Polydispersity |
|---|---|---|---|---|---|---|
| Unmodified NP | --- | --- | --- | 639.5 | 8.9 | 0.068 |
| PEG-NP | --- | --- | --- | 500.2 | 3.6 | 0.137 |
| M225-PEG-NP (100) | 100 | --- | 12 | 505.3 | 7.1 | 0.181 |
| M225 + PEG-NP (100) | --- | 100 | 7 | 520.3 | 4.0 | 0.217 |
| M225-PEG-NP (500) | 500 | --- | 60 | 537.4 | 3.9 | 0.209 |
| M225 + PEG-NP (500) | --- | 500 | 36 | 678.2 | 1 1.2 | 0.274 |
| M225-PEG-NP (900) | 900 | --- | 89 | 585.6 | 10.3 | 0.208 |
| M225 + PEG-NP (900) | --- | 900 | 62 | 735.3 | 20.3 | 0.289 |
The average number of M225 associated per particle was quantified by antibody radiolabeling. Addition of thiolated M225 to PEGylated NP at molar ratios of 100, 500, and 900 yielded M225-PEG-NP with an average of 12, 60, and 89 antibodies per nanoparticle, respectively, corresponding to ~10-12% conjugation efficiency. These efficiencies are comparable to those previously observed in conjugation of M225 antibodies to gold surfaces using dithiol-PEG-hydrazide linkers, which is a highly favorable reaction due to the presence of dense conjugation sites provided by surface Au atoms.40
To account for the effect of non-specific antibody adsorption to NP surfaces, PEGylated NP were also incubated with non-thiolated, unreactive M225. These formulations are termed “M225 + PEG-NP.” Incubation of unreactive M225 with PEG-NP at molar ratios of 100, 500, and 900 yielded NP with on average 7, 36, and 62 associated antibodies per particle. Thus, antibody loading by non-specific NP adsorption is ~30-40% lower.
Specific and M225-dose dependent binding of M225-PEG-NP to EGFR+ cells
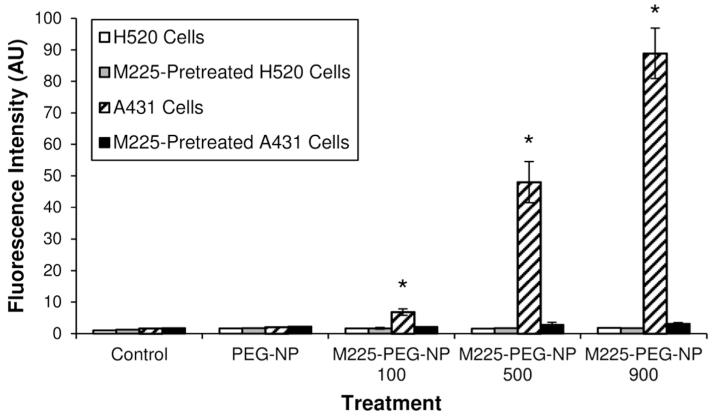
The antibody-modified NPs were tested as EGFR-specific probes using EGFR+ A431 epithelial carcinoma cells and EGFR− H520 lung carcinoma cells. The EGFR expression of these cell lines were confirmed by flow cytometry (Supporting Information Figure 1). Cultured cells were treated with PEG-NP and M225-PEG-NP formulations for 15 min at 37°C before particle association with cells was determined by flow cytometry. A431 (EGFR+) cells treated with M225-PEG-NP 100, 500, 900 nanoparticles had mean fluorescence intensities (MFI) of 6.8 ± 1.0, 48.0 ± 6.5, and 88.9 ± 8.0, respectively, which were significantly greater (p < 0.05) than fluorescence intensities of untreated controls and A431 cells treated with PEGylated NPs (1.7 ± 0.0 and 2.1 ± 0.0, respectively) (Figure 1). M225-PEG-NP exhibited no significant binding in EGFR− H520 cells (p > 0.05). Thus, conjugation of M225 to the surface of PEGylated NP resulted in efficient labeling of EGFR+ cells. In addition, M225-PEG-NP nanoparticles exhibited antibody dose-dependent binding to EGFR-expressing cells with binding increasing proportionally with number of conjugated antibodies.
Figure 1.
Specific and M225 dose-dependent binding of M225-PEG-NP to EGFR+ cells. M225-PEG-NP nanoparticles show significantly greater binding to A431 (EGFR+) cells compared to untreated controls, A431 cells treated with PEGylated beads, and M225-preblocked A431 cells treated with M225-PEG-NP nanoparticles *(p < 0.05) thus demonstrating binding specificity of nanoparticles for EGFR. Nanoparticle binding to A431 cells was directly proportional to the number of surface-conjugated M225 antibodies. PEGylated beads and M225-PEG-NP nanoparticles showed no significant binding in H520 (EGFR-) cells (p > 0.05).
To confirm binding specificity of M225-PEG-NP to EGFR, both cell lines were Pre-treated with free M225 antibody to block EGFR and subsequently treated with M225-PEG-NP (Figure 1). Resulting fluorescence intensities (2.2 ± 0.1, 2.8 ± 0.7, 3.1 ± 0.4 for M225-PEG-NP 100, 500, and 900, respectively) were comparable to baseline fluorescence intensities of untreated cells (1.8 ± 0.0) and of A431 cells treated with PEGylated NPs (2.2 ± 0.1). This indicates that the majority of binding observed in A431 cells without M225 Pre-treatment was due to specific binding of NP-bound M225 antibodies to EGFR rather than non-specific differences, such as differences in phagocytic activity of the cells. No changes in binding behavior were observed in H520 cells.
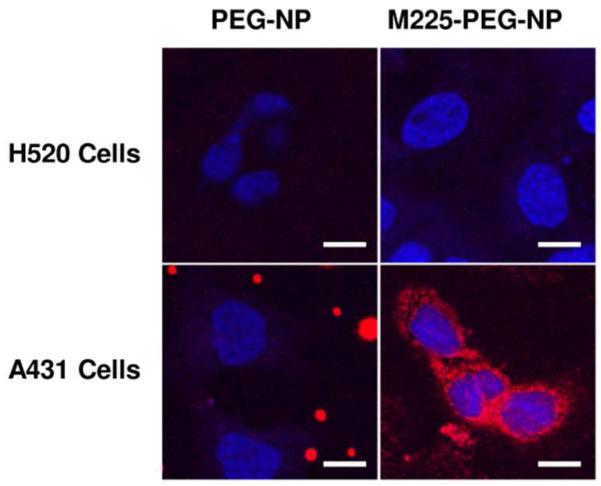
Confocal images of H520 and A431 cells treated with PEG-NP and M225-PEG-NP support binding specificity of M225-PEG-NP to EGFR (Figure 2). PEG-NP-treated cells demonstrated no background signal, and M225-PEG-NP-treated A431 cells showed significantly greater fluorescence signal compared to H520 cells.
Figure 2.
Confocal images of in vitro staining of H520 (EGFR−) cells and A431 (EGFR+) cells with PEG-NP and M225-PEG-NP and DAPI.
Immunoreactivity of adsorbed versus conjugated M225 antibodies
Passive antibody adsorption to particle surfaces has been frequently used in applications such as immunoassays. However, immunoreactivity of adsorbed antibodies on solid supports is highly dependent on Pre-activation of adsorbing surfaces, buffer pH and ionic strength, and temperature as these factors will affect antibody conformation and orientation.41 In general, physical adsorption of monoclonal antibodies has been shown to reduce or eliminate antigen binding capabilities due to steric hindrance from the surface of the solid support, antibody conformational changes, as well as limited intramolecular flexibility due to multi-site attachment to the support.41,42 Thus, there is strong motivation to conjugate M225 antibodies to NP surfaces instead of relying on passive adsorption.
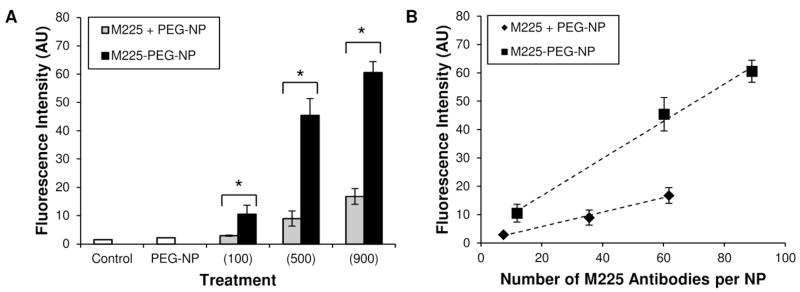
To compare the effect of M225 immobilization by conjugation versus adsorption on nanoparticle binding to EGFR-expressing cells, in vitro binding studies using flow cytometry were completed using M225-PEG-NP and M225 + PEG-NP each prepared using 100, 500, and 900 M225 per NP. Conjugated nanoparticle formulations showed 3.58, 5.07, and 3.61 fold greater binding than adsorbed nanoparticle formulations (Figure 3a). Thus, even assuming similar levels of M225 adsorption on M225-PEG-NP as M225 + PEG-NP, this 60-70% of Ab (for M225-PEG-NP 100, ) would be responsible for only 8-25% of binding (for M225-PEG-NP 100, ) of M225-PEG-NP to EGFR+ cells. Reduced immunoreactivity of adsorbed antibodies could account for this discrepancy as well as masking of antigen-binding domains.37 When lines are fitted to plots of binding versus number of surface-associated antibodies, the slope for conjugated formulations is approximately 2.6 times greater than the slope for adsorbed formulations indicating that addition of subsequent antibodies through conjugation results in greater increases in binding activity than addition of subsequent antibodies through adsorption (Figure 3b). Thus, bioconjugation of antibodies to nanoparticle surfaces was shown to be efficient in terms of antibody loading as well as retained immunoreactivity.
Figure 3.
Comparison of EGFR-targeting of M225-adsorbed formulations (M225 + PEG-NP) versus M225-conjugated formulations (M225-PEG-NP). (A) In vitro binding studies showed significantly greater binding of M225-PEG-NP than M225+PEG-NP to A431 (EGFR+) cells *(p < 0.05). M225-PEG-NP 100, 500, and 900 demonstrated 3.58, 5.07, and 3.61 fold greater binding than M225 + PEG-NP, respectively. (B) Fitted lines to plots of fluorescence intensity (from in vitro binding studies) versus number of antibodies per nanoparticle indicate lower immunoreactivity of antibodies in M225+PEG-NP compared to surface-associated antibodies in M225-PEG-NP. Slopes of the fitted lines for M225+PEG-NP and M225-PEG-NP are 0.254 and 0.657, respectively.
Cytotoxicity of M225-PEG-NP
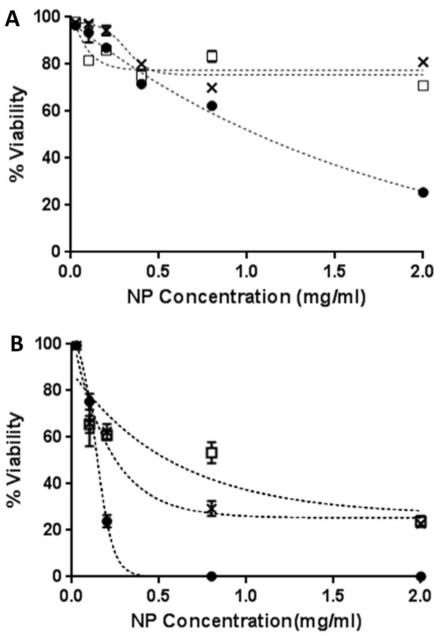
Clinical contrast agents should not have deleterious effects on normal tissue. Therefore, DAPI was used as a viability exclusion dye to quantify cell viability after treatment of cells with unmodified NP, PEG-NP, and M225-PEG-NP 900. Like propidium iodide (PI), DAPI is a DNA intercalating dye which enters dead or dying cells through compromised cell membranes. H520 and A431 cells were treated with 0.026-2.0 mg/ml unmodified NP, PEG-NP, and M225-PEG-NP 900 for 1 hr at body temperature and stained with DAPI. Unmodified NP demonstrated considerably higher cytotoxicity than PEG-NP and M225-PEG-NP 900 in both cell lines. For example, survival of A431 cells treated with 0.2 mg/mL Np was only 24% but was 62% and 61% for PEG-NP and M225-PEG-NP, respectively (Figure 4). Reduction of surface charge density through PEGylation has previously been shown to reduce cytotoxicity in amine-terminated poly(amidoamine) dendrimers and may possibly contribute to the reduction of cytotoxicity in this instance.43,44 Alternatively, particle stabilization by PEG may contribute toward improved biocompatibility of materials. Cytotoxic effects were more pronounced in the A431 cell line compared to the H520 cell line. The IC50 values of PEG-NP and M225-PEG-NP in H520 cells was > 2 mg/mL and in A431 cells was ~ 0.25 mg/mL for PEG-NP and ~ 0.8 mg/mL for M225-PEG-NP. These values are ~10 and 40-fold higher than expected concentrations necessary for in vivo applications. In addition, the incubation time of 1hr is significantly longer than any practical application time in patients undergoing endoscopy and thus toxicities should be less than that observed in this study.
Figure 4.
Cytotoxicity of unmodified NP (•), PEG-NP (x), and M225-PEG-NP (□) in H520 cells (A) and A431 cells (B) determined by flow cytometry using DAPI as a cell viability exclusion dye.
M225-PEG-NP labeling of EGFR in human esophageal tissue
Standard immunohistochemistry was completed on transverse tissue sections from normal esophageal squamous (3 samples from 3 patients), Barrett’s metaplasia (6 samples from 3 patients), and adenocarcinoma tissue (5 samples from 3 patients) using free M225 antibody and Alexa Fluor® 488 goat anti-mouse IgG for evaluation of EGFR expression in the different tissue types. Tissues with low-grade and high-grade dysplasia were scarce, illustrating the difficulty in identifying dysplastic lesions using conventional white light endoscopy. Contrary to previous reports on EGFR expression28,45, we found that Barrett’s metaplasia tissue sections exhibited much stronger EGFR staining than adenocarcinoma tissue sections (Figure 5 and Supporting Information Figure S2, second row). As expected, EGFR staining was weakest in normal esophagus squamous tissue sections. However, it is important to note that the tissue sample size is significantly smaller in this study than in previous studies that report the stepwise increase in EGFR staining during the histological progression from BE to EAC.28,45
Figure 5.
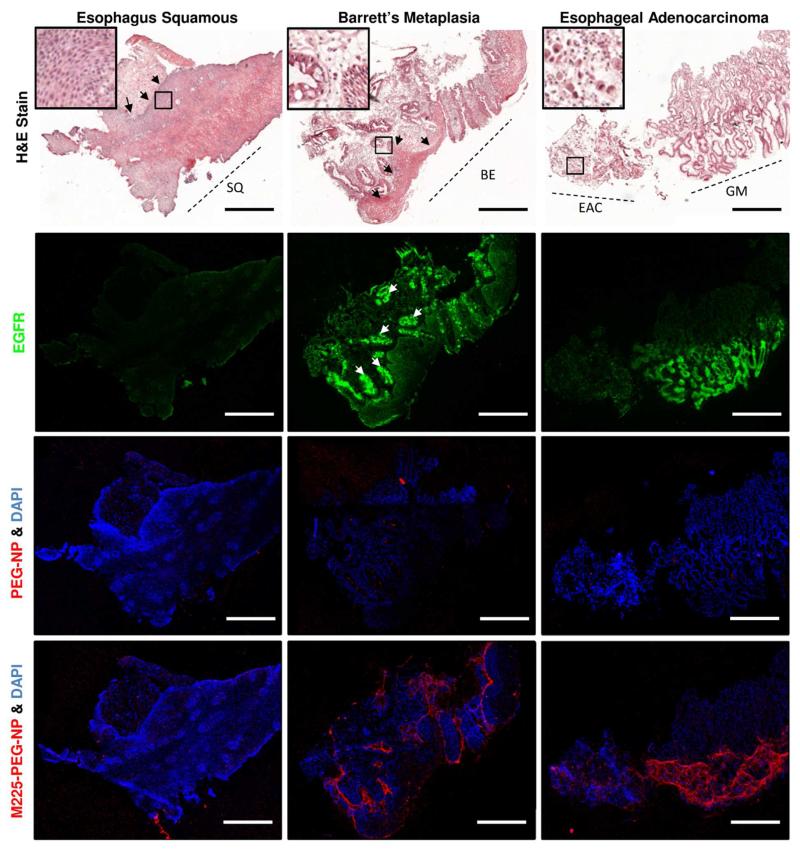
Ex vivo staining of esophagus squamous (SQ), Barrett’s metaplasia (BE), and esophageal adenocarcinoma (EAC) and gastric mucosa (GM) biopsy sections with hematoxylin and eosin (top row), M225 antibody and Alexa Fluor 488 goat anti-mouse IgG (second row), PEG-NP with DAPI counterstain (third row), and M225-PEG-NP 900 with DAPI counterstain (bottom row). Standard immunohistochemistry (middle row) showed high EGFR expression in the glands (white arrows) and basal epithelium (black arrows) in BE tissue sections. M225-PEG-NP bound to the same corresponding areas in adjacent tissue sections. Scale bar, 500 μm.
Different staining patterns were observed in each type of tissue representing the histological progression from BE to EAC. EGFR stains were diffuse in esophageal adenocarcinoma tissue sections. In contrast, EGFR stains were localized in Barrett’s metaplasia to the glands and columnar epithelium. In addition, stronger EGFR staining was seen in basal epithelium adjacent to Barrett’s metaplasia and adenocarcinoma compared to basal epithelium from normal squamous epithelium which is consistent with observations made in cervical cancer, another example of epithelial cancer.46 These observations suggest the potential use of EGFR as a biomarker of risk for metaplastic and neoplastic progression into adjacent tissues. For the purposes of this study, differential EGFR expression patterns in transverse esophageal tissue sections provided an opportunity to evaluate the binding of M225-PEG-NP for targeted imaging of EGFR under ex vivo conditions.
M225-PEG-NP were applied to tissue sections adjacent to EGFR-stained sections to demonstrate co-localization of EGFR and M225-PEG-NP (Figure 5 and Supporting Information Figure S2 and S3, second and third row). M225-PEG-NP demonstrated minimal binding in esophagus squamous epithelium and esophageal adenocarcinoma. The greatest amount of binding was observed in Barrett’s metaplasia tissue with localization of nanoparticles in areas of concentrated EGFR expression. Slight differences in staining can be attributed to the two stains being on adjacent sections rather than on the same tissue sections. The latter was not possible due to possible artifacts from interaction of NP-bound M225 antibodies and fluorescent secondary antibodies used for IHC. Differences in staining can also be attributed to tissue contour. Nanoparticles are more likely to be physically entrapped by tissues on slides during the staining process especially in the case of transverse tissues. Adjacent sections incubated with PEG-NP demonstrated low background fluorescence (Figure 5, bottom row). Correlation between tissue pathology and extent of M225-PEG-NP binding will be further investigated ex vivo in larger esophageal mucosal resection (EMR) specimen in future studies. En face imaging of M225-PEG-NP-labeled EMR specimen will be a more suitable model for the evaluation of molecular imaging of EGFR for the detection of dysplastic Barrett’s esophagus.
Conclusions
In summary, fluorescent nanoparticles for targeted optical imaging of EGFR were synthesized through a series of surface modifications to amino-functionalized dye-filled polystyrene NPs including PEGylation and bioconjugation of M225 antibodies. M225-PEG-NP demonstrated specific binding to EGFR and conjugation of M225 to NP surfaces via a PEG linker was shown to display higher immunoreactivity than direct adsorption of M225 to PEGylated NP surfaces. M225-PEG-NP were shown to bind to human esophageal tissue sections in areas of EGFR expression, demonstrating the potential of targeted fluorescent nanoparticles for in vivo molecular imaging using endoscopy. Contingent on biomarker specificity, such molecular contrast agents could potentially be used for targeted biopsies during endoscopic surveillance of the gastrointestinal tract in addition to delineating tumor margins for endoscopic mucosal resectioning.
Supplementary Material
Acknowledgements
This work is supported by NIH EB007636. Confocal microscopy images were acquired at the University of Washington Keck Microscopy Facility and TEM images at the University of Washington Nanotechnology User Facility (NTUF).
Abbreviations
- BE
Barrett’s esophagus/metaplasia
- LGD
low-grade dysplasia
- HGD
high-grade dysplasia
- EAC
esophageal adenocarcinoma
Footnotes
Supporting Information Available: Supplementary data demonstrating EGFR expression or lack thereof in A431 and H520 cell lines and additional confocal images of ex vivo esophageal tissue stains with M225 antibody and M225-PEG-NP. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–6. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- (2).Cameron AJ, Ott BJ, Payne WS. THE INCIDENCE OF ADENOCARCINOMA IN COLUMNAR-LINED (BARRETTS) ESOPHAGUS. New England Journal of Medicine. 1985;313:857–859. doi: 10.1056/NEJM198510033131404. [DOI] [PubMed] [Google Scholar]
- (3).Wetscher GJ, Hinder RA, Bagchi D, Hinder PR, Bagchi M, Perdikis G, McGinn T. Reflux esophagitis in humans is mediated by oxygen-derived free radicals. Am J Surg. 1995;170:552–6. doi: 10.1016/s0002-9610(99)80014-2. discussion 556-7. [DOI] [PubMed] [Google Scholar]
- (4).Olyaee M, Sontag S, Salman W, Schnell T, Mobarhan S, Eiznhamer D, Keshavarzian A. Mucosal reactive oxygen species production in oesophagitis and Barrett’s oesophagus. Gut. 1995;37:168–73. doi: 10.1136/gut.37.2.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Hameeteman W, Tytgat GN, Houthoff HJ, van den Tweel JG. Barrett’s esophagus: development of dysplasia and adenocarcinoma. Gastroenterology. 1989;96:1249–56. doi: 10.1016/s0016-5085(89)80011-3. [DOI] [PubMed] [Google Scholar]
- (6).Jankowski JA, Wright NA, Meltzer SJ, Triadafilopoulos G, Geboes K, Casson AG, Kerr D, Young LS. Molecular evolution of the metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. American Journal of Pathology. 1999;154:965–973. doi: 10.1016/S0002-9440(10)65346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Heitmiller RF, Redmond M, Hamilton SR. Barrett’s esophagus with high-grade dysplasia. An indication for prophylactic esophagectomy. Ann Surg. 1996;224:66–71. doi: 10.1097/00000658-199607000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hamilton SR, Smith RR. The relationship between columnar epithelial dysplasia and invasive adenocarcinoma arising in Barrett’s esophagus. Am J Clin Pathol. 1987;87:301–12. doi: 10.1093/ajcp/87.3.301. [DOI] [PubMed] [Google Scholar]
- (9).Montgomery E, Goldblum JR, Greenson JK, Haber MM, Lamps LW, Lauwers GY, Lazenby AJ, Lewin DN, Robert ME, Washington K, Zahurak ML, Hart J. Dysplasia as a predictive marker for invasive carcinoma in Barrett esophagus: A follow-up study based on 138 cases from a diagnostic variability study. Human Pathology. 2001;32:379–388. doi: 10.1053/hupa.2001.23511. [DOI] [PubMed] [Google Scholar]
- (10).Reid BJ, Levine DS, Longton G, Blount PL, Rabinovitch PS. Predictors of progression to cancer in Barrett’s esophagus: baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95:1669–76. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kariv R, Plesec TP, Goldblum JR, Bronner M, Oldenburgh M, Rice TW, Falk GW. The Seattle protocol does not more reliably predict the detection of cancer at the time of esophagectomy than a less intensive surveillance protocol. Clin Gastroenterol Hepatol. 2009;7:653–8. doi: 10.1016/j.cgh.2008.11.024. quiz 606. [DOI] [PubMed] [Google Scholar]
- (12).Goldblum JR. Barrett’s esophagus and Barrett’s-related dysplasia. Mod Pathol. 2003;16:316–24. doi: 10.1097/01.MP.0000062996.66432.12. [DOI] [PubMed] [Google Scholar]
- (13).Kelloff GJ, Krohn KA, Larson SM, Weissleder R, Mankoff DA, Hoffman JM, Link JM, Guyton KZ, Eckelman WC, Scher HI, O’Shaughnessy J, Cheson BD, Sigman CC, Tatum JL, Mills GQ, Sullivan DC, Woodcock J. The progress and promise of molecular imaging probes in oncologic drug development. Clinical Cancer Research. 2005;11:7967–7985. doi: 10.1158/1078-0432.CCR-05-1302. [DOI] [PubMed] [Google Scholar]
- (14).Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH, Wang TD. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat Med. 2008;14:454–8. doi: 10.1038/nm1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Li M, Anastassiades CP, Joshi B, Kormack CM, Piraka C, Elmunzer BJ, Turgeon DK, Johnson TD, Appelman H, Beer DG, Wang TD. Affinity peptide for targeted detection of dysplasia in Barrett’s esophagus. Gastroenterology. 2010;139:1472–1480. doi: 10.1053/j.gastro.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Miller SJ, Joshi BP, Feng Y, Gaustad A, Fearon ER, Wang TD. In vivo fluorescence-based endoscopic detection of colon dysplasia in the mouse using a novel peptide probe. PLoS One. 2011;6:e17384. doi: 10.1371/journal.pone.0017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Bird-Lieberman EL, Neves AA, Lao-Sirieix P, O’Donovan M, Novelli M, Lovat LB, Eng WS, Mahal LK, Brindle KM, Fitzgerald RC. Molecualr imaging using fluorescent lectins permits rapid endoscopic identification of dysplasia in Barrett’s esophagus. Nature Medicine. 2012;18:315–321. doi: 10.1038/nm.2616. [DOI] [PubMed] [Google Scholar]
- (18).Murcia MJ, Naumann CA. Biofunctionalization of fluorescent nanoparticles. In: Kumar C, editor. Nanotechnologies for the life sciences. Wiley-VCH Verlag GmbH; Weinheim: 2005. [Google Scholar]
- (19).Hahn MA, Singh AK, Sharma P, Brown SC, Moudgil BM. Nanoparticles as contrast agents for in-vivo bioimaging: current status and future perspectives. Anal Bioanal Chem. 2011;399:3–27. doi: 10.1007/s00216-010-4207-5. [DOI] [PubMed] [Google Scholar]
- (20).Khan JA, Kudgus RA, Szabolcs A, Dutta S, Wang E, Cao S, Curran GL, Shah V, Curley S, Mukhopadhyay D, Robertson JD, Bhattacharya R, Mukherjee P. Designing nanoconjugates to effectively target pancreatic cancer cells in vitro and in vivo. PLoS One. 2011;6:e20347. doi: 10.1371/journal.pone.0020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).El-Sayed IH, Huang X, El-Sayed MA. Surface plasmon resonance scattering and absorption of anti-EGFR antibody conjugated gold nanoparticles in cancer diagnostics: applications in oral cancer. Nano Lett. 2005;5:829–34. doi: 10.1021/nl050074e. [DOI] [PubMed] [Google Scholar]
- (22).Diagaradjane P, Orenstein-Cardona JM, Colon-Casasnovas NE, Deorukhkar A, Shentu S, Kuno N, Schwartz DL, Gelovani JG, Krishnan S. Imaging epidermal growth factor receptor expression in vivo: Pharmacokinetic and biodistribution characterization of a bioconjugated quantum dot nanoprobe. Clinical Cancer Research. 2008;14:731–741. doi: 10.1158/1078-0432.CCR-07-1958. [DOI] [PubMed] [Google Scholar]
- (23).Yang LL, Mao H, Wang YA, Cao ZH, Peng XH, Wang XX, Duan HW, Ni CC, Yuan QG, Adams G, Smith MQ, Wood WC, Gao XH, Nie SM. Single Chain Epidermal Growth Factor Receptor Antibody Conjugated Nanoparticles for in vivo Tumor Targeting and Imaging. Small. 2009;5:235–243. doi: 10.1002/smll.200800714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Acharya S, Dilnawaz F, Sahoo SK. Targeted epidermal growth factor receptor nanoparticle bioconjugates for breast cancer therapy. Biomaterials. 2009;30:5737–50. doi: 10.1016/j.biomaterials.2009.07.008. [DOI] [PubMed] [Google Scholar]
- (25).Glazer ES, Curley SA. Radiofrequency Field-Induced Thermal Cytotoxicity in Cancer Cells Treated With Fluorescent Nanoparticles. Cancer. 2010;116:3285–3293. doi: 10.1002/cncr.25135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lee H, Fonge H, Hoang B, Reilly RM, Allen C. The effects of particle size and molecular targeting on the intratumoral and subcellular distribution of polymeric nanoparticles. Mol Pharm. 2010;7:1195–208. doi: 10.1021/mp100038h. [DOI] [PubMed] [Google Scholar]
- (27).Yang J, Lim EK, Lee HJ, Park J, Lee SC, Lee K, Yoon HG, Suh JS, Huh YM, Haam S. Fluorescent magnetic nanohybrids as multimodal imaging agents for human epithelial cancer detection. Biomaterials. 2008;29:2548–55. doi: 10.1016/j.biomaterials.2007.12.036. [DOI] [PubMed] [Google Scholar]
- (28).Cronin J, McAdam E, Danikas A, Tselepis C, Griffiths P, Baxter J, Thomas L, Manson J, Jenkins G. Epidermal growth factor receptor (EGFR) is overexpressed in high-grade dysplasia and adenocarcinoma of the esophagus and may represent a biomarker of hisotlogical progression in Barrett’s esophagus (BE) Am J Gastroenterol. 2011;106:46–56. doi: 10.1038/ajg.2010.433. [DOI] [PubMed] [Google Scholar]
- (29).Masui H, Castro L, Mendelsohn J. Consumption of EGF by A431 cells: evidence for receptor recycling. J Cell Biol. 1993;120:85–93. doi: 10.1083/jcb.120.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Raben D, Helfrich B, Chan DC, Ciardiello F, Zhao L, Franklin W, Baron AE, Zeng C, Johnson TK, Bunn PA., Jr. The effects of cetuximab alone and in combination with radiation and/or chemotherapy in lung cancer. Clin Cancer Res. 2005;11:795–805. [PubMed] [Google Scholar]
- (31).Kummer U. Tritium radiolabeling of antibodies to high specific activity with N-succinimidyl [2,3-3H]propionate: use in detecting and analyzing monoclonal antibodies. Methods Enzymol. 1986;121:670–8. doi: 10.1016/0076-6879(86)21066-6. [DOI] [PubMed] [Google Scholar]
- (32).Tang YS, Davis A-M, Kitcher JP. N-succinimidyl propionate: Characterisation and optimum conditions for use as a tritium labelling reagent for proteins. Journal of Labelled Compounds. 1983;20:277–284. [Google Scholar]
- (33).Gordon KM, Duckett L, Daul B, Petrie HT. A simple method for detecting up to five immunofluorescent parameters together with DNA staining for cell cycle or viability on a benchtop flow cytometer. J Immunol Methods. 2003;275:113–21. doi: 10.1016/s0022-1759(03)00009-7. [DOI] [PubMed] [Google Scholar]
- (34).Popielarski SR, Pun SH, Davis ME. A nanoparticle-based model delivery system to guide the rational design of gene delivery to the liver. 1 Synthesis and characterization. Bioconjug Chem. 2005;16:1063–70. doi: 10.1021/bc050113d. [DOI] [PubMed] [Google Scholar]
- (35).Lee J, Choi Y, Kim K, Hong S, Park H-Y, Lee T, Park GJ, Song R. Characterization and cancer cell specific binding properties of anti-EGFR antibody conjugated quantum dots. Bioconjugate Chem. 2010;21:940–946. doi: 10.1021/bc9004975. [DOI] [PubMed] [Google Scholar]
- (36).Gref R, Luck M, Quellec P, Marchand M, Dellacherie E, Harnisch S, Blunk T, Muller RH. ‘Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (PEG): influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids and Surfaces B-Biointerfaces. 2000;18:301–313. doi: 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- (37).Chen SF, Liu LY, Zhou J, Jiang SY. Controlling antibody orientation on charged self-assembled monolayers. Langmuir. 2003;19:2859–2864. [Google Scholar]
- (38).Lin JJ, Silas JA, Bermudez H, Milam VT, Bates FS, Hammer DA. The effect of polymer chain length and surface density on the adhesiveness of functionalized polymersomes. Langmuir. 2004;20:5493–5500. doi: 10.1021/la036417a. [DOI] [PubMed] [Google Scholar]
- (39).Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanoparticle PEGylation for imaging and therapy. Nanomedicine (Lond) 2011;6:715–28. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ma LL, Tam JO, Willsey BW, Rigdon D, Ramesh R, Sokolov K, Johnston KP. Selective targeting of antibody conjugated multifunctional nanoclusters (nanoroses) to epidermal growth factor receptors in cancer cells. Langmuir. 2011;27:7681–7690. doi: 10.1021/la200659z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).van Erp R, Linders YE, van Sommeren AP, Gribnau TC. Characterization of monoclonal antibodies physically adsorbed onto polystyrene latex particles. J Immunol Methods. 1992;152:191–9. doi: 10.1016/0022-1759(92)90140-o. [DOI] [PubMed] [Google Scholar]
- (42).Wood WG, Gadow A. Immobilisation of antibodies and antigens on macro solid phases--a comparison between adsorptive and covalent binding. A critical study of macro solid phases for use in immunoassay systems. Part I. J Clin Chem Clin Biochem. 1983;21:789–97. doi: 10.1515/cclm.1983.21.12.789. [DOI] [PubMed] [Google Scholar]
- (43).Wang W, Xiong W, Wan J, Sun X, Xu H, Yang X. The decrease of PAMAM dendrimer-induced cytotoxicity by PEGylation via attenuation of oxidative stress. Nanotechnology. 2009;20:105103. doi: 10.1088/0957-4484/20/10/105103. [DOI] [PubMed] [Google Scholar]
- (44).Fant K, Esbjorner EK, Jenkins A, Grossel MC, Lincoln P, Norden B. Effects of PEGylation and Acetylation of PAMAM Dendrimers on DNA Binding, Cytotoxicity and in Vitro Transfection Efficiency. Mol Pharm. 2010;7:1734–46. doi: 10.1021/mp1001312. [DOI] [PubMed] [Google Scholar]
- (45).Li Y, Wo JM, Ray MB, Jones W, Su RR, Ellis S, Martin RC. Cyclooxygenase-2 and epithelial growth factor receptor up-regulation during progression of Barrett’s esophagus to adenocarcinoma. World J Gastroenterol. 2006;12:928–34. doi: 10.3748/wjg.v12.i6.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Boiko IV, Mitchell MF, Hu W, Pandey DK, Mathevet P, Malpica A, Hittelman WN. Epidermal growth factor receptor expression in cervical intraepithelial neoplasia and its modulation during an alpha-difluoromethylornithine chemoprevention trial. Clin Cancer Res. 1998;4:1383–91. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.