Abstract
Fungi are considered the primary decomposers of dead plant biomass in terrestrial ecosystems. However, current knowledge regarding the successive changes in fungal communities during litter decomposition is limited. Here we explored the development of the fungal community over 24 months of litter decomposition in a temperate forest with dominant Quercus petraea using 454-pyrosequencing of the fungal internal transcribed spacer (ITS) region and cellobiohydrolase I (cbhI) genes, which encode exocellulases, to specifically address cellulose decomposers. To quantify the involvement of phyllosphere fungi in litter decomposition, the fungal communities in live leaves and leaves immediately before abscission were also analysed. The results showed rapid succession of fungi with dramatic changes in the composition of the fungal community. Furthermore, most of the abundant taxa only temporarily dominated in the substrate. Fungal diversity was lowest at leaf senescence, increased until month 4 and did not significantly change during subsequent decomposition. Highly diverse community of phyllosphere fungi inhabits live oak leaves 2 months before abscission, and these phyllosphere taxa comprise a significant share of the fungal community during early decomposition up to the fourth month. Sequences assigned to the Ascomycota showed highest relative abundances in live leaves and during the early stages of decomposition. In contrast, the relative abundance of sequences assigned to the Basidiomycota phylum, particularly basidiomycetous yeasts, increased with time. Although cellulose was available in the litter during all stages of decomposition, the community of cellulolytic fungi changed substantially over time. The results indicate that litter decomposition is a highly complex process mediated by various fungal taxa.
Keywords: fungi, litter decomposition, cellulose, endophyte, temperate forests
Introduction
Plant litter represents a major source of organic carbon in forest soils. Its decomposition is a complex process that involves mineralisation and transformation of organic matter. Decomposition of plant litter is a key step in nutrient recycling (Berg et al., 2001). As most of the plant biomass-derived carbon in the temperate and boreal forests is mineralised in the litter layer, an understanding of this process and the microorganisms involved is essential for the identification of factors that affect global carbon fluxes.
Fungi are considered to be the key players in litter decomposition because of their ability to produce a wide range of extracellular enzymes, which allows them to efficiently attack the recalcitrant lignocellulose matrix that other organisms are unable to decompose (Kjoller and Struwe, 1982; de Boer et al., 2005). Biochemical decomposition of leaf litter is a sequential process that initially involves the loss of the less recalcitrant components (for example, oligosaccharides, organic acids, hemicellulose and cellulose) followed by the degradation of the remaining highly recalcitrant compounds (for example, lignin or suberin). Litter quality changes during the course of its transformation and so does the activity of litter-associated microorganisms (Dilly et al., 2001). These changes are accompanied by a succession of microbial litter decomposers that reflect the varied catabolic capabilities that are sequentially required to complete the process of litter decomposition (Frankland, 1998; Osono et al., 2006).
The ability of fungi to decompose leaf litter has been investigated many times under laboratory conditions (for example, Osono, 2007; Baldrian et al., 2011). Furthermore, many studies have combined litterbag techniques with cultivation-based methods followed by the isolation and identification of fungal decomposers (Koide et al., 2005; Osono, 2005; Zhang et al., 2008; Osono et al., 2009). Using these methods, fungi involved in the decomposition of litter have been divided into early, intermediate and late decomposers (Frankland, 1998; Tang et al., 2005). This observation was supported by a recent study performed by Šnajdr et al. (2011) as these three phases were distinguished during oak litter decomposition based on the differences in the activity of extracellular enzymes and the rates of decomposition of the individual litter constituents. In most previous studies, fungi from the Ascomycota phylum were found to dominate during the initial stages of litter decay along with a few basidiomycetous fungi. The abundance of fungi from the Ascomycota phylum decreases during the process of degradation as they are gradually replaced by fungi from the Basidiomycota phylum, especially the saprotrophic cord formers, during the later stages of decomposition (Frankland, 1998; Osono, 2007).
Plant organic matter transformation leads to the disappearance of easily utilisable compounds and to the formation of recalcitrant ones. As a consequence, the chemical and spatial heterogeneity of the substrate changes with time. This process can theoretically result in the formation of novel niches and a potential increase in fungal diversity or to the creation of more uniform environment with a potential decrease in diversity. Both scenarios have been reported from litter or wood (Melillo et al., 1989; Dickie et al., 2012) but the actual development of fungal community on decaying litter is so far unknown. Culture-dependent approaches are typically selective because only a small fraction of microbial taxa grow under the conditions used for strain isolation (Amann et al., 1995). Molecular methods, such as community fingerprinting or direct sequencing of cloned PCR sequences that have recently been applied to litter (Aneja et al., 2006; Kubartova et al., 2009) suffered from limited resolution. Therefore, next-generation sequencing approaches currently represent the only technique that can be used to sufficiently describe the development of fungal community composition during succession.
The degradation of plant leaves is not limited to the litter layer on the forest floor. Indeed, the decomposition process begins as soon as the leaf is formed (Stone, 1987). Phyllosphere fungi that are established in the interior or on the surfaces of live leaves have the advantage of gaining access to readily available nutrients in live leaves and later, after senescence, to the dead leave biomass. Recently, 454-pyrosequencing was used to assess fungal diversity in live oak leaves and demonstrated the presence of a diverse fungal community (Jumpponen and Jones, 2009a, 2009b). It is highly probable that at least some of these fungi participate in litter decomposition. There is some evidence that certain phyllosphere fungi are able to transform various components of litter because they produce the extracellular enzymes that are involved in decomposition in pure culture and their ability to decompose litter material has been described (Korkama-Rajala et al., 2008; Žifčáková et al., 2011). Although potential leaf endophytes have been isolated from litter in various stages of decomposition (Osono, 2002; Koide et al., 2005), their importance in the community of litter-associated fungi is currently unknown.
The aim of this study was to characterise the development of the fungal community composition over 24 months following litterfall in a temperate forest dominated by Quercus petraea. As some litter components, including cellulose, remain present in a considerable quantity during the entire 24-month period (Šnajdr et al., 2011), the fungi capable of cellulose decomposition may be present during all phases of decomposition. To address this possibility, the composition of the gene pool of the cbhI exocellulase gene, which is an enzyme that catalyses the rate-limiting step in the decomposition of cellulose (Baldrian and Valášková, 2008; Edwards et al., 2008), was monitored at various stages of litter decomposition. To evaluate the role of phyllosphere fungi in litter decomposition, fungal communities associated with live Q. petraea leaves and senescent leaves immediately before abscission were also analysed. The results of this study are discussed in light of previously published data derived from the same experiment where litter decomposition (mass loss), fungal and bacterial biomass content based on ergosterol and phospholipid fatty acid analysis, and the activity of the extracellular enzymes in the litterbags were explored by Šnajdr et al. (2011).
Materials and methods
Study site and sample collection
The study site was an oak (Q. petraea) forest in the Xaverovský Háj Natural Reserve, near Prague, Czech Republic (50°5′38"N, 14°36′48"E). The site was previously explored with respect to the activity of decomposition-related extracellular enzymes in the forest topsoil (Šnajdr et al., 2008) and during the successive transformation of Q. petraea litter (Šnajdr et al., 2011). In this study site, the saprotrophic fungi were characterised (Valášková et al., 2007; Baldrian et al., 2011). The soil was acidic cambisol with a litter thickness of 0.5–1.5 cm, a pH of 4.3, a C content of 46.2% and an N content of 1.76%. The mean annual temperature at the soil surface was 9.3 °C (winter mean 1.3 °C, summer mean 16.6 °C; Baldrian et al., in press).
The litterbag experiment was run as described previously (Šnajdr et al., 2011). Litter material (Q. petraea leaves, tree age 100–120 years) for litterbag construction was collected immediately after abscission and allowed to air dry at 20 °C. Litterbags containing 5 g of air-dried leaves (10 × 20 cm, 1 mm nylon mesh size) were placed on the top of the litter horizon at the study site at the end of the litterfall season. To prevent extensive desiccation, litterbags were overlaid with freshly fallen oak leaves. Litterbags were removed after 2, 4, 8, 12 and 24 months of incubation, three litterbags were collected at each sampling time for DNA extraction. For the analysis of the phyllosphere fungal community composition, live Q. petraea leaves were collected 2 months before abscission (August) by hand-picking (month −2), and senescent leaves were collected during the litterfall period by gently shaking oak twigs and collecting falling leaves before their contact with the soil (month 0). Collected material was transferred to the laboratory and processed immediately. Leaves or litter were cut into 0.25 cm2 pieces and used immediately for DNA extraction. The same material was also used for chemical analyses, measurement of enzyme activities and quantification of microbial biomass as described in Šnajdr et al. (2011).
454-Pyrosequencing of fungal internal transcribed spacer (ITS) and cellobiohydrolase I (cbhI) genes
The total genomic DNA was extracted from 300 mg of material using the Powersoil Kit (MoBio, Carlsbad, CA, USA). The primers ITS1/ITS4 (White et al., 1990) were used to amplify the ITS1 region, the 5.8S ribosomal DNA and the ITS2 region of the fungal ribosomal DNA. The primers cbhIF and cbhIR (Edwards et al., 2008) were used to amplify a partial sequence of the fungal cbhI gene. These primers amplify cbhI genes belonging to the GH7 family of fungi from the Basidiomycota, Ascomycota and Mucoromycotina unless the template contains intron in the primer sequence (Štursová et al., 2012).
A two-step PCR amplification using composite primers containing multiplex identifiers was performed to obtain amplicon libraries for 454-pyrosequencing following a previously described method (Baldrian et al., 2012). PCR amplicons were quantified using the Quant-iT PicoGreen Kit (Invitrogen, Grand Island, NY, USA). An equimolar mix of PCR products was prepared for each primer pair, and the pooled products were sequenced on a GS FLX Titanium platform (Roche, Basel, Switzerland). Fungal ITS sequences were analysed from all sampling times, and the cbhI gene diversity was analysed in the samples collected at −2, 0, 4 and 12 months.
Bioinformatic analysis
The pyrosequencing data were processed as described previously (Baldrian et al., 2012). Pyrosequencing noise reduction was performed using the Denoiser 0.851 (Reeder and Knight, 2010) and chimeric sequences were detected using UCHIME (Edgar, 2010) and deleted. Fungal sequences were shortened to 380 bases and clustered using cd-hit (Li and Godzik, 2006) at a 97% similarity level (O'Brien et al., 2005) to obtain the operational taxonomical units (OTUs). Consensus sequences were constructed for each cluster, and the closest hits were identified using the PlutoF pipeline (Tedersoo et al., 2010). For the cbhI gene, the sequences were truncated to 300 bp and clustered at a 96% similarity level (Baldrian et al., 2012) to obtain the OTUs. Consensus sequences were constructed, and the introns were removed. Data sets containing the cbhI sequences representing the OTUs and sequences retrieved from GenBank were aligned using SeaView 4 (http://pbil.univ-lyon1.fr/software/seaview.html) with MUSCLE (http://www.drive5.com/muscle/). Maximum likelihood phylogenetic trees were computed with the GTR substitutions model using GARLI (http://www.molecularevolution.org/sofware/phylogenetics/garli/garli_create_job). The OTUs of cbhI genes that clustered with sequences of known fungal taxa from the GenBank with bootstrap support >70% were taxonomically assigned to fungal phyla.
Sequence data have been deposited in the MG-RAST public database (http://metagenomics.anl.gov/, data set numbers 4497081.3 for fungal ITS region and 4497080.3 for cbhI genes).
Diversity and statistical analyses
Owing to the fact that the sampling depth achieved in this study did not allow to make realistic estimates of total diversity and since next-generation sequencing derived data were demonstrated to be affected by artefacts (Tedersoo et al., 2010), the only measure of diversity of OTUs used was the amount of the most abundant OTUs that represented 80% of all sequences. This metric in our opinion fairly represents the diversity of the quantitatively important part of the fungal or cbhI community. To avoid possible effects of variable sampling depth, these estimates were calculated for a data set containing 700 randomly chosen ITS sequences or 495 cbhI sequences from each litterbag. The sequences were clustered again as described above. The OTU richness and Chao1 were calculated using EstimateS 8.00 (http://viceroy.eeb.uconn.edu/estimates).
One-way analysis of variance with the Fisher's least significant difference post hoc test was used to analyse the significant differences in relative abundance of individual OTUs or fungal taxa among sampling times. Principal component analysis was performed with the relative abundance data of the 50 most abundant fungal genera. PC1 and PC2 loads were subjected to analysis of variance with the Fisher's least significant difference post hoc test. Differences with a P<0.05 were regarded as statistically significant.
Results
Fungal communities associated with oak leaves
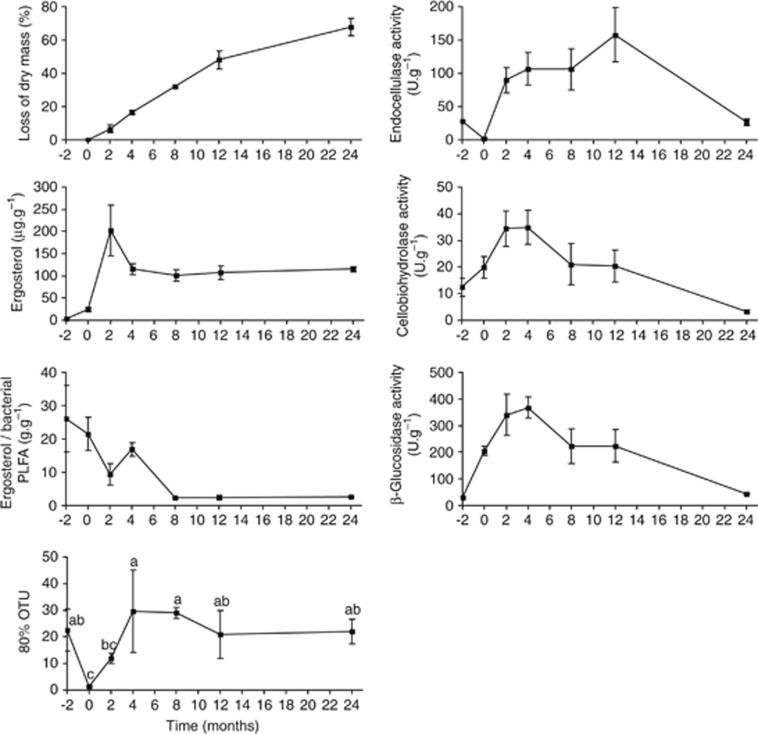
In total, 23 760 sequences of the fungal ITS region with>380 bp were used for analysis after denoising and removal of the chimeric sequences and sequences not belonging to fungi (<0.6%). These sequences clustered into 1874 OTUs (including 1193 singletons) at a 97% similarity level. Although 80% of all sequences at month −2 were represented by 23 dominant OTUs, at month 0 70–90% of sequences belonged to the single most abundant OTU (assigned to Mycosphaerella punctiformis). Within a relatively short time (month 4), the diversity peaked with 30 OTUs representing 80% of the total fungal community at month 8 and then levelled off (Figure 1).
Figure 1.
Loss of dry mass, activity of extracellular enzymes, development of fungal and bacterial biomass and estimates of fungal diversity in Q. petraea live leaves and leaves at different stages of decomposition. Fungal biomass is expressed as ergosterol content. The ratio of fungal and bacterial biomass is based on the ratio of ergosterol content and the content of bacteria-specific phospholipid fatty acids. Data on leaf chemistry and microbial biomass are derived from Šnajdr et al. (2011). 80% OTU represents the number of the most abundant OTUs, which represent 80% of all sequences. The data are shown as the means and s.e. from three litterbags.
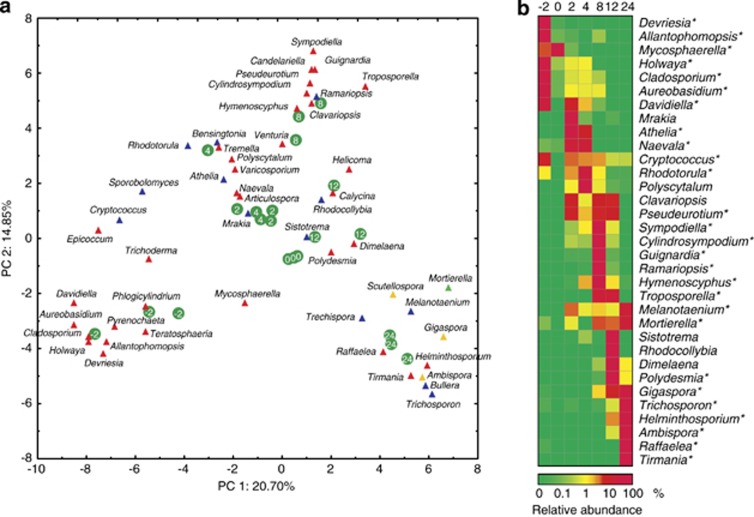
A total of 387 fungal genera were identified as the closest hits of individual OTUs. Mycosphaerella, Naevala, Troposporella and Trichosporon were the most abundant fungi in the amplicon pool. The 50 most abundant fungal OTUs with their closest identified hits and abundances are listed in Supplementary Table 1. In all, 40 of the 50 most abundant fungal OTUs and 27 of the top 33 genera demonstrated significant changes in abundance over time (Supplementary Table 1, Figure 2). Altogether, the ascomycetous OTU 0 (closest hit: Mycosphaerella punctiformis) and OTU 1 (Naevala minutissima) were the most abundant fungi. OTU 0 was predominant during the early stages of succession (month −2 and 0) but disappeared almost completely during later stages. OTU 1 and OTU 4 (Athelia) were highly abundant during the initial stages of litter decomposition (months 2 and 4). However, OTU 3 and OTU 11 (both Troposporella fumosa) dominated the later stages of decomposition (months 8 and 12) but were almost absent in other samples. The latest stages of litter decomposition were dominated by OTU 2 (Trichosporon porosum) and OTU 5 (Trichosporon miniliiforme) (Supplementary Table 1).
Figure 2.
(a) Principal component analysis of the relative abundance of the 50 most abundant fungal genera in Q. petraea live leaves and leaves at different stages of decomposition. Ascomycota—red, Basidiomycota—blue, Glomeromycota—yellow, Mucoromycotina—green. Green circles with numbers indicate the positions of individual samples (litterbags) with ages in months. (b) Time course of the relative abundance of dominant fungal genera in Q. petraea live leaves and leaves at different stages of decomposition. Mean abundances are shown for each time point. A statistically significant effect of time on abundance is indicated by an asterisk (P<0.05, analysis of variance (ANOVA) followed by Fisher's post hoc test).
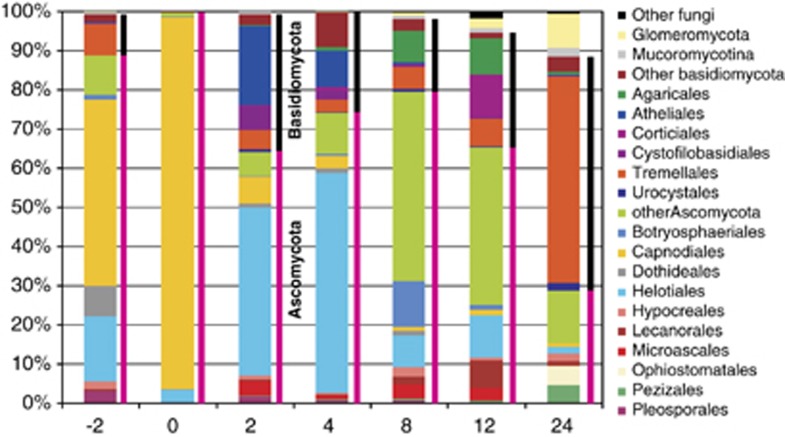
The majority of fungal sequences were assigned to the Ascomycota (71%) and Basidiomycota (26%) phylum. Glomeromycota were represented by 1.8% of all sequences, and fungi from Mucoromycotina comprised 0.76% of all sequences. Fungi from the Ascomycota phylum dominated in the live and senescent leaves, which is in contrast to month 24 when fungi from the Basidiomycota phylum represented 60% of the amplicons. Glomeromycota and Mucoromycotina sequences were rare until month 4 and then rapidly increased until month 8 and 24 when they represented 8.6% and 2.2% of the amplicon pool, respectively (Figure 3). The most abundant fungal orders were Capnodiales (22% of sequences), Helotiales (20%) and Tremellales (12%). Members of the Helotiales order were present during all phases of succession and peaked at month 2 and 4 together with fungi from the Atheliales family. Fungi belonging to the ascomycetous order Capnodiales dominated among amplicons from the live and senescent leaves, whereas the basidiomycetous order Tremellales predominated at later stages (Figure 3).
Figure 3.
Phylogenetic assignment of fungal sequences from Q. petraea live leaves and leaves at different stages of decomposition. The data are represented as the mean values from three litterbags.
Each sampling time was characterised by a specific fungal community, which was different from the community in the previous or the next stage. When the abundance of the top 50 fungal genera was analysed by principal component analysis, the first two canonical axes explained 20.70% and 14.85% of the total variability (Figure 2). The analysis of variance of the PC1 and PC2 loadings demonstrated that community changes over time were significant between any two dates except between months 2 and 4 (P<0.015). This is supported by the fact that most of the fungi that were highly abundant at a certain time only dominated for a short period. Among the 28 genera that represented >3% of the sequences at any particular sampling time, 20 did not exceed 1% at any other time (Figure 2).
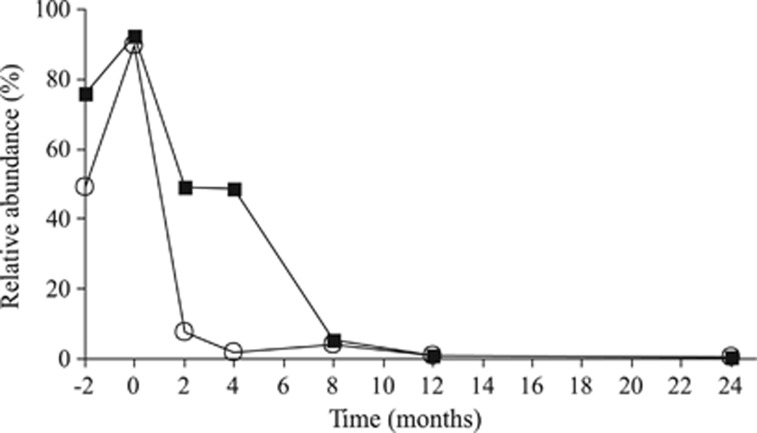
The amplicons from live Q. petraea leaves were dominated by the Ascomycota (88.5%) fungi, and Capnodiales, Helotiales, Dothideales and Pleosporales were the major orders. The Basidiomycota phylum (10.6%) was mainly represented by Tremellales (Figure 3). The fungal community on senescent leaves was dominated by the same OTUs that were dominant on live leaves, and the fungi that were highly abundant on live leaves comprised approximately 50% of all fungi until month 4 (Figure 4).
Figure 4.
The persistence of fungal OTUs recorded in live Q. petraea leaves (endophytes) during subsequent decomposition of litter. Data represent the sum of the relative abundances of dominant endophytes with abundances >5% in live leaves (open circles) and abundant endophytes (>1% black squares).
Cellulose-decomposing fungi associated with oak leaves
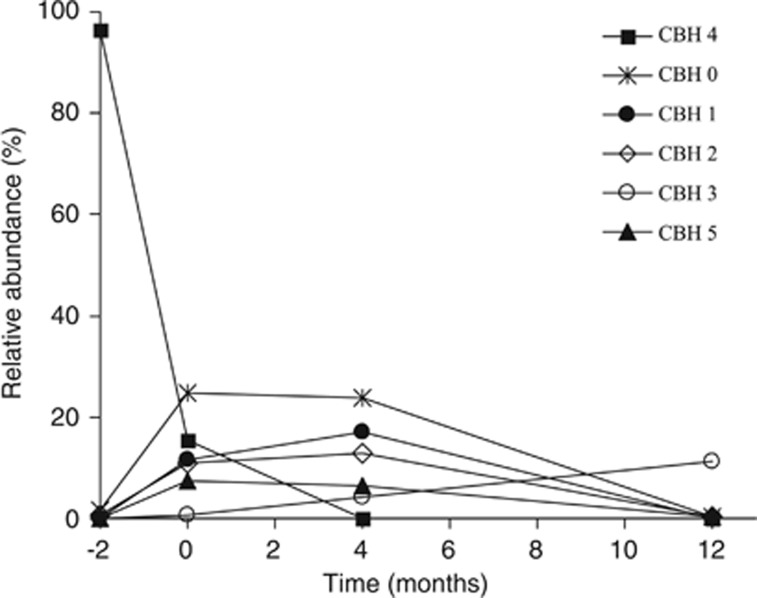
The gene cbhI encoding for cellobiohydrolase was used as a marker for the cellulolytic members of the fungal community. In total, 3351 denoised, non-chimeric sequences of cbhI were analysed. The sequences clustered into 235 OTUs (including 107 singletons). The diversity of the cellulolytic fungal community increased between months 4 and 12: 80% of the cbhI gene sequences were represented by only 7±1 and 8±1 dominant OTUs during months 0 and 4 while it was 26±2 at month 12.
The most abundant OTUs were OTU 4 and OTU 0, which represented 28% and 12.5% of all sequences, respectively. OTU 4 dominated during month −2 where it represented over 90% of all sequences. Among the other abundant OTUs, OTU 0, OTU 1 and OTU 2 predominated during months 0 and 4 but were almost absent at month 12. In contrast, OTU 3 was the most abundant at month 12 (Figure 5). This observation indicates that, despite the fact that cellulose was available during the entire decomposition process, specific cellulolytic fungi were present during different stages of decomposition. The abundance of one-half of the 30 most abundant OTUs significantly changed over time (Supplementary Table 2).
Figure 5.
Time course of the relative abundance of dominant cellulolytic fungi represented by the cbhI gene OTUs in Q. petraea live leaves and leaves at different stages of decomposition. The data are shown as the mean values from three litterbags.
Phylogenetic analysis of the cbhI gene sequences showed that 38% of all sequences clustered with sequences of known fungal taxa with >70% bootstrap support (Supplementary Figure 1). Of these sequences, the majority belonged to the Basidiomycota (29%) phylum, which was represented by two OTUs, and the other 9% belonged to seven ascomycetous OTUs. Sequence similarities >97% allowed us to determine the taxonomic affiliation of OTU 11 to the ectomycorrhizal basidiomycete Russula paludosa and OTU 46 to the ascomycete Aureobasidium pullulans.
Discussion
During the decomposition of Q. petraea leaves used in this study, approximately 70% of the total mass was lost within 24 months (Figure 1; Šnajdr et al., 2011). The C/N ratio decreased from 49 to 22 within 12 months and remained constant later. Fungal biomass increased rapidly from low values in the live and senescent leaves to a maximum at month 2 and remained lower but constant until the end of the experiment. The activity of cellulolytic enzymes was detected in live and senescent leaves, which indicates that decomposition started before leaf abscission (Figure 1; Šnajdr et al., 2011). Three distinct decomposition phases have been distinguished that are characterised by the sequential mass loss of extractables and hemicelluloses, cellulose, and lignin (Supplementary Figure 2; Šnajdr et al., 2011). This seems to be consistent with the culture-based observations that divided fungi into early, intermediate and late decomposers (Frankland, 1998; Osono and Takeda, 2001; Tang et al., 2005; Osono, 2007). The culture-dependent studies, however, tend to underestimate the total diversity of fungi and are biased towards rapidly growing species (Hering, 1967; Frankland, 1998). Thus, they do not provide reliable information about fungal communities associated with leaves/litter during its degradation. Despite several limitations (see for example Amend et al., 2010), next-generation sequencing seems to be better suitable to explore the fungal community because it can deliver information at higher quantitative resolution and is not biased towards easily culturable and fast growing taxa.
The live oak leaves used in this study harboured a relatively rich and even fungal community with its diversity comparable to previous reports from Quercus macrocarpa (Jumpponen and Jones, 2009a, 2009b). The low biomass of the fungi on live leaves is possibly a consequence of the action of the protective mechanisms of the plant. After leaf senescence, rapid proliferation of opportunistic Mycosphaerella spp. resulted in a sevenfold increase in fungal biomass but a rapid decrease in diversity. The rapid increase in fungal diversity after the litterfall was caused by the invasion of new colonisers and was detected during month 2 (Figure 1). Fungal diversity continued to increase until month 4, which indicates the arrival of new species on the substrate. However, the abundance of the most common fungal genera did not change significantly (Figure 2).
Fungi from the Ascomycota phylum prevailed in the live and senescent leaves on the trees (88.5% and 99.5% of amplicons, respectively). These data are in accordance with previous culture-based studies on various trees (Osono, 2002; Santamaría and Bayman, 2005) and the pyrosequencing analyses of live Q. macrocarpa leaves (Jumpponen and Jones, 2009a, 2009b). However, the most common genera recorded from a Q. macrocarpa phyllosphere in North America were quite distinct on the genus level. Of the major genera, Microsphaeropsis, Alternaria, Epicoccum, Aureobasidium, Phoma and Erysiphe were detected, and only Aureobasidium and Epicoccum were recovered from live leaves in this study with a >1% frequency, which demonstrates that either the tree species, geographic distance or different environmental conditions affect the composition of phyllosphere mycoflora.
Some fungi associated with living tree leaves are also found in association with decomposing leaf litter (Koide et al., 2005; Osono, 2006). The fact that some live leaf-associated fungi are able to produce extracellular enzymes or decompose sterile senescent leaves (Korkama-Rajala et al., 2008; Žifčáková et al., 2011) led to the hypothesis that certain taxa may change from endophytism to a saprotrophic strategy. In addition, molecular evidence indicates that fungi cultured from live leaves and decaying litter may indeed belong to the same taxa (Promputtha et al., 2007). This study shows that phyllosphere fungi are still quantitatively important during the subsequent stages of decomposition, at least until month 4 (Figures 2 and 4). Establishment in live, nonsenescent leaves created an opportunity for these fungi to readily exploit leaf-derived nutrients during decomposition after leaf senescence. Fungi belonging to the genera Holwaya, Cladosporium, Aureobasidium, Davidiella and Cryptococcus were predominant in living oak leaves, in senescent leaves nearly disappeared and their abundance rose again in early phases of litter decomposition. The genera Aureobasidium and Cladosporium contain well-known phyllosphere fungi that have been repeatedly isolated from various trees (Sadaka and Ponge, 2003; Slavikova et al., 2007; Unterseher and Schnittler, 2009), and their persistence until early decomposition has been reported (Sadaka and Ponge, 2003). OTUs belonging to the Mycosphaerella genus, which comprises pathogenic and saprotrophic species (Suto, 1999), were the most dominant in live leaves and made up a significant portion of the population at month 0 where they represented 90% of sequences, which indicates that they are both endophytes and efficient early saprotrophs.
The first year of our experiment was characterised by a relatively rapid loss of litter mass, a decrease in the C/N ratio and the cellulose content, and a relatively high activity of cellulolytic enzymes, which causes faster decomposition of cellulose (Supplementary Figure 2). These conditions were associated with the continuous dominance of fungi from the Ascomycota phylum, which are generally known to selectively decompose cellulose over lignin. Dominance of ascomycetous fungi in the early stages of beech litter decomposition was currently also demonstrated using metaproteomic approach (Schneider et al., 2012). Similar results were obtained when 8-week-old Fagus sylvatica litter was analysed, except that it also contained a significant proportion of fungi from the Mucoromycotina phylum (Aneja et al., 2006). Despite the fact that the Mucoromycotina fungi are often considered to be opportunistic microorganisms that are associated with nutrient-rich substrates, their abundance in Q. petraea litter was low until month 8. Among the fungal genera that were dominant during month 2, Naevala, Cryptococcus and Mycosphaerella were detectable in the live leaves, and the basidiomycetous genera Athelia and Mrakia appeared anew during month 2 and immediately became dominant, which demonstrates their ability to rapidly proliferate on fresh litter. Naevala, Athelia and Cryptococcus fungi maintained their prevalence until month 4, whereas Mrakia fungi nearly disappeared and were replaced by Polyscytalum and Rhodotorula fungi. Month 8 was characterised by an entirely different fungal community, which was most likely caused by a depletion of the majority of the readily available organic compounds and was associated with a sharp decrease in the phyllosphere fungi. Fungi belonging to the Glomeromycota and the Mucoromycotina phyla were detectable on litter beginning at month 8, and their abundances gradually increased until the end of the experiment. The fungal genera Troposporella, Guignardia, Ramariopsis, Sympodiella and Cryptococcus prevailed at month 8. Troposporella fungi have been recorded in seasonally flooded soil ecosystems (Carrino-Kyker and Swanson, 2008), where they were most likely involved in the decomposition of allochthonous carbon input. Troposporella remained frequent until month 12, whereas basidiomycetous Sistotrema and Rhodocollybia and lichenised ascomycetous Dimelaena fungi appeared for the first time at this stage. Rhodocollybia is a typical saprotroph (Valášková et al., 2007), and the polyphyletic genus Sistotrema contains both ectomycorrhizal and decomposer fungal species (Di Marino et al., 2008; Boberg et al., 2011).
During the second year, the rate of litter mass loss was relatively slow and the activity of cellulolytic enzymes decreased, which indicated that the easily accessible polysaccharides were depleted. Also, the substrate was richer in the recalcitrant lignin and nitrogen and characteristic with the increased activity of ligninolytic enzymes (Supplementary Figure 2). Fungi from the Basidiomycota phylum distinctively dominated over fungi from the Ascomycota phylum at month 24. In previous studies, basidiomycetous species, particularly the saprotrophic cord formers, have often been demonstrated to be late litter decomposers (Osono, 2007; Duong et al., 2008) because of their capability to synthesise enzymes required for the degradation of complex polymers (Baldrian, 2008). Interestingly, basidiomycetous cord formers were not among the most frequent taxa observed in our study. Instead, the basidiomycetous yeast genus Trichosporon comprised 50% of all sequences at month 24. This is consistent with a recent report that identified this species as the second most abundant cellulose decomposer in litter using stable isotope probing (Štursová et al., 2012). Although the cellulose content of the litter at this stage is relatively low (Šnajdr et al., 2011), it may be still sufficient to support the growth of yeasts and may not be sufficient to support the growth of the large mycelia of basidiomycetous cord formers. There is only a small overlap of the fungal community in litter and in the uppermost (organic) soil horizon at the site of study (unpublished data) with >50% of fungi in soil being ectomycorrhizal. Among the genera reported in this study, Mortierella, Cryptococcus, Trichosporon, Ambispora and Naevala are also frequent in soil, which can serve as a reservoir for their spread.
This study demonstrated that fungal succession during litter decomposition is much faster than so far expected from the culture-based studies (Figure 2). The fast appearance–disappearance of fungal taxa seems to contrast with the reported persistence of DNA from inactive fungi in decaying wood (Rajala et al., 2011) and to support the rapid turnover of early/intermediate/late saprotrophs (Lindahl and Finlay, 2006). The successional changes are likely governed not only by the relatively slow changes of the polysaccharide, lignin and nitrogen content in litter but possibly by other factors including more subtle changes in litter chemistry and interspecific fungal interactions.
Cellulose is the major polysaccharide in plant litter, and cellulose-degrading enzymes are thus an obvious target for the study of the decomposing microorganisms. Among these, cellobiohydrolases (exocellulases), which catalyses the rate-limiting step of cellulose decomposition (Baldrian and Valášková, 2008), and the cbhI gene represent suitable markers for the study of cellulolytic fungi (Edwards et al., 2008; Weber et al., 2011; Baldrian et al., 2012). Here, we show that several exocellulase-producing fungi are present in the live leaves of Q. petraea although the community is dominated by a single species (Figure 5). This OTU of cbhI genes was assigned to fungi from the Basidiomycota phylum, which is surprising if we consider the dominance of fungi from the Ascomycota phylum at this stage. The diversity of cellulolytic fungi is high in senescent leaves with 58 observed and >200 predicted OTUs. As the leaves are still attached to the trees, these fungi must have colonised the substrate before its contact with soil. Later in decomposition, estimates of cbhI richness were approximately 200 in number, which indicates that there are approximately 100 cellulolytic fungal species when multiple copies of the gene per fungal genome are considered (Edwards et al., 2008; Weber et al., 2011). The overall fungal diversity did not correlate with the diversity of fungi harbouring the cbhI gene, which indicates that the proportion of cellulolytic fungi changes during decomposition; no clear link was observed between the diversity of the cbhI genes and the activity of cellobiohydrolase. Although cellulose represents a substrate that is present in the litter during the entire decomposition process, the community of cellulolytic fungi also showed successive changes similar to those of the total fungal community with dominant OTUs appearing and disappearing (Figure 5). This may indicate that the individual cellulolytic fungi have specific additional nutritional requirements or competitive abilities. Interestingly, the sequences dominating in the cbhI gene pool at month 12 belonged to the ectomycorrhizal genus Russula. Our results are consistent with previous observations that identified cellulases and class II peroxidases in these fungi (Bodeker et al., 2009; Štursová et al., 2012) that may combine the mycorrhizal and saprotrophic lifestyle to some extent.
This study demonstrates that the composition of the fungal community changes with changing litter quality much faster than previously thought. Furthermore, similar changes during the decomposition process are observed among cellulolytic fungi, which indicates that succession is not only driven by the availability of the major nutrient sources but also by other factors, perhaps other nutritional requirements or the competitive abilities of individual taxa. The initial steps of decomposition where fungi dominate the decomposer community are characterised by a high involvement of fungi that occur on the live leaves of the tree. However, further research on both the structural and functional aspects of fungal community composition, for example, use of the metatranscriptomic or metaproteomic approaches, are needed to better understand the functional role of individual fungal taxa during decomposition.
Acknowledgments
This work was supported by the Ministry of Education, Youth and Sports of the Czech Republic (ME10152, LD12050) and by the Czech Science Foundation (P504/12/0709).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amann R, Ludwig W, Schleifer K. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend AS, Seifert KA, Bruns TD. Quantifying microbial communities with 454 pyrosequencing: does read abundance count. Mol Ecol. 2010;19:5555–5565. doi: 10.1111/j.1365-294X.2010.04898.x. [DOI] [PubMed] [Google Scholar]
- Aneja MK, Sharma S, Fleischmann F, Stich S, Heller W, Bahnweg G, et al. Microbial colonization of beech and spruce litter - influence of decomposition site and plant litter species on the diversity of microbial community. Microb Ecol. 2006;52:127–135. doi: 10.1007/s00248-006-9006-3. [DOI] [PubMed] [Google Scholar]
- Baldrian P.2008Enzymes of saprotrophic basidiomycetesIn: Boddy L, Frankland J, van West P (eds).Ecology of Saprotrophic Basidiomycetes Academic Press: New York; 19–41. [Google Scholar]
- Baldrian P, Valášková V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev. 2008;32:501–521. doi: 10.1111/j.1574-6976.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- Baldrian P, Voříšková J, Dobiášová P, Merhautová V, Lisá L, Valášková V. Production of extracellular enzymes and degradation of biopolymers by saprotrophic microfungi from the upper layers of forest soil. Plant Soil. 2011;338:111–125. [Google Scholar]
- Baldrian P, Kolařík M, Štursová M, Kopecký J, Valášková V, Větrovský T, et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012;6:248–258. doi: 10.1038/ismej.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrian P, Šnajdr J, Merhautová V, Dobiášová P, Cajthaml T, Valášková V.(in press). Responses of the extracellular enzyme activities in hardwood forest to soil temperature and seasonality and the potential effects of climate change Soil Biol Biochemdoi: 10.1016/j.soilbio.2012.01.020 [DOI]
- Berg B, McClaugherty C, Santo AVD, Johnson D. Humus buildup in boreal forests: effects of litter fall and its N concentration. Can J Forest Res. 2001;31:988–998. [Google Scholar]
- Boberg JB, Ihrmark K, Lindahl BD. Decomposing capacity of fungi commonly detected in Pinus sylvestris needle litter. Fungal Ecol. 2011;4:110–114. [Google Scholar]
- Bodeker ITM, Nygren CMR, Taylor AFS, Olson A, Lindahl BD. ClassII peroxidase-encoding genes are present in a phylogenetically wide range of ectomycorrhizal fungi. ISME J. 2009;3:1387–1395. doi: 10.1038/ismej.2009.77. [DOI] [PubMed] [Google Scholar]
- Carrino-Kyker SR, Swanson AK. Temporal and spatial patterns of eukaryotic and bacterial communities found in vernal pools. Appl Environ Microbiol. 2008;74:2554–2557. doi: 10.1128/AEM.01482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer W, Folman LB, Summerbell RC, Boddy L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Di Marino E, Scattolin L, Bodensteiner P, Agerer R. Sistotrema is a genus with ectomycorrhizal species - confirmation of what sequence studies already suggested. Mycol Prog. 2008;7:169–176. [Google Scholar]
- Dickie IA, Fukami T, Wilkie JP, Allen RB, Buchanan PK. Do assembly history effects attenuate from species to ecosystem properties? A field test with wood-inhabiting fungi. Ecol Lett. 2012;15:133–141. doi: 10.1111/j.1461-0248.2011.01722.x. [DOI] [PubMed] [Google Scholar]
- Dilly O, Bartsch S, Rosenbrock P, Buscot F, Munch JC. Shifts in physiological capabilities of the microbiota during the decomposition of leaf litter in a black alder (Alnus glutinosa (Gaertn.) L.) forest. Soil Biol Biochem. 2001;33:921–930. [Google Scholar]
- Duong LM, McKenzie EHC, Lumyong S, Hyde KD. Fungal succession on senescent leaves of Castanopsis diversifolia in Doi Suthep-Pui National Park, Thailand. Fungal Divers. 2008;30:23–36. [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edwards IP, Upchurch RA, Zak DR. Isolation of fungal cellobiohydrolase I genes from sporocarps and forest soils by PCR. Appl Environ Microbiol. 2008;74:3481–3489. doi: 10.1128/AEM.02893-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland JC. Fungal succession - unravelling the unpredictable. Mycol Res. 1998;102:1–15. [Google Scholar]
- Hering TF. Fungal decomposition of oak leaf litter. Trans Brit Mycol Soc. 1967;50:267–273. [Google Scholar]
- Jumpponen A, Jones KL. Seasonally dynamic fungal communities in the Quercus macrocarpa phyllosphere differ between urban and nonurban environments. New Phytol. 2009a;186:496–513. doi: 10.1111/j.1469-8137.2010.03197.x. [DOI] [PubMed] [Google Scholar]
- Jumpponen A, Jones KL. Massively parallel 454 sequencing indicates hyperdiverse fungal communities in temperate Quercus macrocarpa phyllosphere. New Phytol. 2009b;184:438–448. doi: 10.1111/j.1469-8137.2009.02990.x. [DOI] [PubMed] [Google Scholar]
- Kjoller A, Struwe S. Microfungi in ecosystems: fungal occurrence and activity in litter and soil. Oikos. 1982;39:289–422. [Google Scholar]
- Koide K, Osono T, Takeda H. Colonization and lignin decomposition of Camellia japonica leaf litter by endophytic fungi. Mycoscience. 2005;46:280–286. [Google Scholar]
- Korkama-Rajala T, Mueller MM, Pennanen T. Decomposition and fungi of needle litter from slow- and fast-growing norway spruce (Picea abies) clones. Microb Ecol. 2008;56:76–89. doi: 10.1007/s00248-007-9326-y. [DOI] [PubMed] [Google Scholar]
- Kubartova A, Ranger J, Berthelin J, Beguiristain T. Diversity and decomposing ability of Saprophytic fungi from temperate forest litter. Microb Ecol. 2009;58:98–107. doi: 10.1007/s00248-008-9458-8. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lindahl BD, Finlay RD. Activities of chitinolytic enzymes during primary and secondary colonization of wood by basidiomycetous fungi. New Phytol. 2006;169:389–397. doi: 10.1111/j.1469-8137.2005.01581.x. [DOI] [PubMed] [Google Scholar]
- Melillo J, Aber J, Linkins A, Ricca A, Fry B, Nadelhoffer K. Carbon and nitrogen dynamics along the decay continuum: plant litter to soil organic matter. Plant Soil. 1989;115:189–198. [Google Scholar]
- O‘Brien HE, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R. Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol. 2005;71:5544–5550. doi: 10.1128/AEM.71.9.5544-5550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osono T, Takeda H. Organic chemical and nutrient dynamics in decomposing beech leaf litter in relation to fungal ingrowth and succession during 3-year decomposition processes in a cool temperate deciduous forest in Japan. Ecol Res. 2001;16:649–670. [Google Scholar]
- Osono T. Phyllosphere fungi on leaf litter of Fagus crenata: occurrence, colonization, and succession. Can J Bot. 2002;80:460–469. [Google Scholar]
- Osono T. Colonization and succession of fungi during decomposition of Swida controversa leaf litter. Mycologia. 2005;97:589–597. doi: 10.3852/mycologia.97.3.589. [DOI] [PubMed] [Google Scholar]
- Osono T. Role of phyllosphere fungi of forest trees in the development of decomposer fungal communities and decomposition processes of leaf litter. Can J Microbiol. 2006;52:701–716. doi: 10.1139/w06-023. [DOI] [PubMed] [Google Scholar]
- Osono T. Ecology of ligninolytic fungi associated with leaf litter decomposition. Ecol Res. 2007;22:955–974. [Google Scholar]
- Osono T, Hirose D, Fujimaki R. Fungal colonization as affected by litter depth and decomposition stage of needle litter. Soil Biol Biochem. 2006;38:2743–2752. [Google Scholar]
- Osono T, Ishii Y, Takeda H, Seramethakun T, Khamyong S, To-Anun C, et al. Fungal succession and lignin decomposition on Shorea obutsa leaves in a tropical seasonal forest in northern Thailand. Fungal Divers. 2009;36:101–119. [Google Scholar]
- Promputtha I, Lumyong S, Dhanasekaran V, McKenzie E, Hyde K, Jeewon R. A phylogenetic evaluation of whether endophytes become saprotrophs at host senescence. Microb Ecol. 2007;53:579–590. doi: 10.1007/s00248-006-9117-x. [DOI] [PubMed] [Google Scholar]
- Rajala T, Peltoniemi M, Hantula J, Mäkipää R, Pennanen T. RNA reveals a succession of active fungi during the decay of Norway spruce logs. Fungal Ecol. 2011;4:437–448. [Google Scholar]
- Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaka N, Ponge JF. Fungal colonization of phyllosphere and litter of Quercus rotundifolia Lam. in a holm oak forest (High Atlas, Morocco) Biol Fertil Soils. 2003;39:30–36. [Google Scholar]
- Santamaría J, Bayman P. Fungal epiphytes and endophytes of coffee leaves (Coffea arabica) Microb Ecol. 2005;50:1–8. doi: 10.1007/s00248-004-0002-1. [DOI] [PubMed] [Google Scholar]
- Schneider T, Keiblinger KM, Schmid E, Sterflinger-Gleixner K, Ellersdorfer G, Roschitzki B, et al. Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions. ISME J. 2012;6:1749–1762. doi: 10.1038/ismej.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavikova E, Vadkertiova R, Vranova D. Yeasts colonizing the leaf surfaces. J Basic Microbiol. 2007;47:344–350. doi: 10.1002/jobm.200710310. [DOI] [PubMed] [Google Scholar]
- Stone JK. Initiation and development of latent infections by Rhabdocline parkeri on Douglas-fir. Can J Bot. 1987;65:2614–2621. [Google Scholar]
- Suto Y. Mycosphaerella chaenomelis sp. nov.: the teleomorph of Cercosporella sp., the causal fungus of frosty mildew in Chaenomeles sinensis, and its role as the primary infection source. Mycoscience. 1999;40:509–516. [Google Scholar]
- Šnajdr J, Valášková V, Merhautová V, Herinková J, Cajthaml T, Baldrian P. Spatial variability of enzyme activities and microbial biomass in the upper layers of Quercus petraea forest soil. Soil Biol Biochem. 2008;40:2068–2075. [Google Scholar]
- Šnajdr J, Cajthaml T, Valášková V, Merhautová V, Petránková M, Spetz P, et al. Transformation of Quercus petraea litter: successive changes in litter chemistry are reflected in differential enzyme activity and changes in the microbial community composition. FEMS Microbiol Ecol. 2011;75:291–303. doi: 10.1111/j.1574-6941.2010.00999.x. [DOI] [PubMed] [Google Scholar]
- Štursová M, Žifčáková L, Leigh MB, Burgess R, Baldrian P. Cellulose utilisation in forest litter and soil: identification of bacterial and fungal decomposers. FEMS Microbiol Ecol. 2012;80:735–746. doi: 10.1111/j.1574-6941.2012.01343.x. [DOI] [PubMed] [Google Scholar]
- Tang AMC, Jeewon R, Hyde KD. Succession of microfungal communities on decaying leaves of Castanopsis fissa. Can J Microbiol. 2005;51:967–974. doi: 10.1139/w05-086. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, Saar I, et al. 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol. 2010;188:291–301. doi: 10.1111/j.1469-8137.2010.03373.x. [DOI] [PubMed] [Google Scholar]
- Unterseher M, Schnittler M. Dilution-to-extinction cultivation of leaf-inhabiting endophytic fungi in beech (Fagus sylvatica L.) - different cultivation techniques influence fungal biodiversity assessment. Mycol Res. 2009;113:645–654. doi: 10.1016/j.mycres.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Valášková V, Šnajdr J, Bittner B, Cajthaml T, Merhautová V, Hoffichter M, et al. Production of lignocellulose-degrading enzymes and degradation of leaf litter by saprotrophic basidiomycetes isolated from a Quercus petraea forest. Soil Biol Biochem. 2007;39:2651–2660. [Google Scholar]
- Weber CF, Zak DR, Hungate BA, Jackson RB, Vilgalys R, Evans RD, et al. Responses of soil cellulolytic fungal communities to elevated atmospheric CO2 are complex and variable across five ecosystems. Environ Microbiol. 2011;13:2778–2793. doi: 10.1111/j.1462-2920.2011.02548.x. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW.1990Analysis of phylogenetic relationships by amplification and direct sequencing of ribosomal RNA genesIn: Innis MA, Gelfand DH, Sninsky JN, White TJ (eds).PCR Protocols: A Guide to Methods and Applications Academic Press: New York; 315–322. [Google Scholar]
- Zhang P, Tian X, He X, Song F, Ren L, Jiang P. Effect of litter quality on its decomposition in broadleaf and coniferous forest. Eur J Soil Biol. 2008;44:392–399. [Google Scholar]
- Žifčáková L, Dobiášová P, Kolářová Z, Koukol O, Baldrian P. Enzyme activities of fungi associated with Picea abies needles. Fungal Ecol. 2011;4:427–436. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.