Abstract
Soda lakes are saline and alkaline ecosystems that are believed to have existed throughout the geological record of Earth. They are widely distributed across the globe, but are highly abundant in terrestrial biomes such as deserts and steppes and in geologically interesting regions such as the East African Rift valley. The unusual geochemistry of these lakes supports the growth of an impressive array of microorganisms that are of ecological and economic importance. Haloalkaliphilic Bacteria and Archaea belonging to all major trophic groups have been described from many soda lakes, including lakes with exceptionally high levels of heavy metals. Lonar Lake is a soda lake that is centered at an unusual meteorite impact structure in the Deccan basalts in India and its key physicochemical and microbiological characteristics are highlighted in this article. The occurrence of diverse functional groups of microbes, such as methanogens, methanotrophs, phototrophs, denitrifiers, sulfur oxidizers, sulfate reducers and syntrophs in soda lakes, suggests that these habitats harbor complex microbial food webs that (a) interconnect various biological cycles via redox coupling and (b) impact on the production and consumption of greenhouse gases. Soda lake microorganisms harbor several biotechnologically relevant enzymes and biomolecules (for example, cellulases, amylases, ectoine) and there is the need to augment bioprospecting efforts in soda lake environments with new integrated approaches. Importantly, some saline and alkaline lake ecosystems around the world need to be protected from anthropogenic pressures that threaten their long-term existence.
Keywords: Lonar Lake, soda lakes, alkaliphiles, stable-isotope probing, microbial prospecting
Introduction
Soda lakes are considered exceptional to all other aquatic ecosystems in simultaneously exhibiting high productivity rates (>10 g cm−2 per day), and high pH (9.0–12.0) and salinity (up to saturation concentrations) (Melack and Kilham, 1974; Grant, 2006). Although they are widely distributed across the globe (Grant and Sorokin, 2011), only a few have been studied (Supplementary Table S1). Many soda lakes experience massive seasonal or permanent microbial blooms often resulting in distinct coloration of the lake water (Grant et al., 1990). Soda lakes harbor considerably diverse microbial populations (Grant et al., 1990; Zavarzin et al., 1999). Cultivation-dependent and -independent surveys of soda lakes in the East African Rift valley resulted in the isolation of several hundred strains of aerobic, heterotrophic and (halo)alkaliphilic Bacteria and Archaea (Duckworth et al., 1996; reviewed in Grant and Sorokin, 2011) and the detection of several novel lineages of putative Bacteria and Archaea (Grant et al., 1999; Rees et al., 2004). The microbial diversity of Lonar Lake, a saline and alkaline ecosystem centered at a meteorite impact crater structure in the Deccan basalts in India, has been explored recently (Wani et al., 2006; Surakasi et al., 2007, 2010; Joshi et al., 2008; Antony et al., 2010, 2012a, 2012b). Microbes in soda lake ecosystems have attracted considerable attention as a source for biomolecules with biotechnological potential. Understanding life in soda lake environments may help answer key questions related to early life as fossilized alkaline playa lakes have been implied in the 2.3 billion-year-old Ventersdorp geological formation in South Africa (Karpeta, 1989; Jones et al., 1998). Here, we review the microbiology of Lonar Lake and other soda lake environments and implications for exploring and exploiting the functional diversity of microbial life at high pH.
Key physicochemical attributes of Lonar Lake and other soda lakes
Lonar crater (19°59′N, 76°31′E) is a simple, bowl-shaped, near-circular and remarkably well-preserved depression in the otherwise featureless Deccan Plateau in Buldhana district, Maharashtra state, India (Fredriksson et al., 1973; Maloof et al., 2010). The crater is 1.88±0.05 km wide and approximately 135 m deep, with the rim of the crater rising 30 m above the surrounding plains. Direct evidence of impact-associated shocked materials at the crater established meteorite impact origin as the leading hypothesis for its formation (Fredriksson et al., 1973). The age of the crater is between 12 000 years (Maloof et al., 2010) and 570 000 years (Jourdan et al., 2011). The crater cavity is filled with breccia (>225 m) and unconsolidated sediment (30–100 m) (Maloof et al., 2010). Most of the crater floor is covered by a saline (NaCl approximately 0.4%) and alkaline lake (pH approximately 10) that is approximately 6 m deep (Joshi et al., 2008; Maloof et al., 2010). Although several perennial freshwater springs and streams flow into the lake, there are no stream outlets from the lake (Maloof et al., 2010).
The physicochemical properties associated with Lonar Lake and other soda lakes (Table 1 and Supplementary Table S1) influence the structure and function of their microbial communities. Observations over the past several decades suggest declining levels of Lonar Lake water salinity and alkalinity (Joshi et al., 2008). Phosphate in surface waters of Lonar Lake was relatively lower (31.6 μℳ) than Mono Lake (Oremland et al., 2004; Table 1). Although Lonar Lake is not known to be particularly rich in heavy metals, the American soda lakes—Mono Lake and Searles Lake (Supplementary Table S1) —contain dissolved arsenic levels as high as 3000 and 200 μℳ, respectively (Oremland et al., 2004). Extremely high Uranium (U) concentrations have been reported in the soda lakes in Eastern Mongolia, with Shar Burdiin Lake potentially having the highest known concentrations of naturally occurring U (62.5 μℳ) in a surface water body (Linhoff et al., 2011). Sulfide concentration (∼200 mℳ) in the anaerobic waters of Soap Lake (USA) is probably the highest recorded so far in a natural water body (Sorokin et al., 2007). In comparison to Lonar Lake, the Rift valley soda lakes and Mono Lake have higher dissolved concentrations of chlorides, sulfates, sodium and potassium, whereas the reverse holds true in the case of dissolved organic nitrogen and nitrate levels (Table 1). Chemical analysis of the Lonar Lake sediments revealed high levels of iron (21.9 g kg−1), magnesium (10.9 g kg−1), phosphate (30.6 g kg−1), nitrate (7.9 g kg−1), total organic carbon (2.5 g kg−1) and total Kjeldhal nitrogen (1.9 g kg−1) (Antony et al., 2010).
Table 1. Comparison of Lonar Lake surface water chemistry with that of Rift valley soda lakes and Mono Lake [data compiled from multiple sources—Supplementary Table S1 and Table 1 References 1, 5, 8, 9 and 12 (see Supplementary Information)].
| Physicochemical characteristic | Lonar Lake | Lake Bogoria | Lake Nakuru | Lake Elmenteita | Mono Lake |
|---|---|---|---|---|---|
| pH | 9.8, 10a | 10 | 10 | 10 | 9.8 |
| Temp. (°C) | 26 | 27.9 | 24.1 | 23.1 | 20 |
| Salinity (%) | 1.1a | 4.9 | 1.8 | 2.6 | 8.4 |
| Dissolved oxygen (mg l−1) | 13.3 | 13 | 17 | 8 | 3.8 |
| Dissolved organic nitrogen (mg l−1) | 53.95 | 2.33 | 11.38 | 8.42 | ND |
| Total phosphorus (mg l−1) | 2.8, 31.6a,b | 6.2 | 2.4 | 1.2 | 400b |
| Nitrates (mg l−1) | 2.4a | 0.3 | 1.4 | 0.3 | ND |
| Cl− (meq l−1) | 71.9, 87.4a | 176 | 67.3 | 215 | 500 |
| SO42− (meq l−1) | 2.3, 1.0a | 8.2 | 5.0 | 14.1 | 260 |
| Na+ (meq l−1) | 136.1, 159.1a | 1348.7 | 413.1 | 718.8 | 1300 |
| K+ (meq l−1) | 0.4, 0.2a | 19.2 | 8.5 | 15.9 | 30.7 |
Abbreviations: ND, no data.Mean values are shown.
Data from analyses performed in May 2009 (Antony et al., unpublished).
Phosphate (μℳ).
Microbial diversity of Lonar Lake and other soda lakes
Overview of microbial communities in Lonar Lake
Abundances of cultivatable aerobic Bacteria in Lonar Lake water and sediment samples were approximately 102–104 CFU ml−1 and 102–106 CFU g−1, respectively (Joshi et al., 2008). Heterotrophic bacteria isolated from the lake water and sediment samples showed highest diversity and abundance within the Firmicutes phylum, followed by Gammaproteobacteria, Actinobacteria, Alphaproteobacteria and Betaproteobacteria phyla. Many isolates produce biotechnologically relevant enzymes such as lipase, amylase, cellulase and caseinase at alkaline pH (Joshi et al., 2008). Sediment-based enrichments supplemented with C1 substrates (methane, methanol, methylamine and dimethylsulfide) and C2 substrates (ethanol and acetate) yielded isolates related to Halomonas, Alkalimonas, Pseudomonas, Bacillus, Methylophaga, Paracoccus, Rhodobaca and Idiomarina spp. (Antony et al., unpublished; Antony et al., 2012a). Four novel heterotrophs (Nitritalea halakaliphila, Indibacter alkaliphilus, Cecembia lonarensis and Georgenia satyanarayanai) and one novel methylotroph (Methylophaga lonarensis) have been described from Lonar Lake (Kumar et al., 2010a, 2010b, 2012; Srinivas et al., 2012; Antony et al., 2012a).
Bacterial 16S rRNA genes retrieved from Lonar Lake sediments were mostly related to Firmicutes, Proteobacteria and Actinobacteria (Wani et al., 2006). Assessment of key structural genes (pmoA, mmoX, mxaF and mauA) revealed many methylotrophs (Supplementary Figure S1) (Antony et al., unpublished). Sequences related to Methylomicrobium-like methanotrophs, purple sulfur and non-sulfur photosynthetic bacteria were detected in the Arthrospira spp.-dominated blooms on the surface of Lonar Lake (Surakasi et al., 2010). Community DNA-based analysis of 18S rRNA genes in sediments revealed a high diversity of micro-eukaryotes related to alveolates, fungi, stramenopiles, choanoflagellates, amoebozoans and cercozoans, and many novel lineages (Antony et al., unpublished).
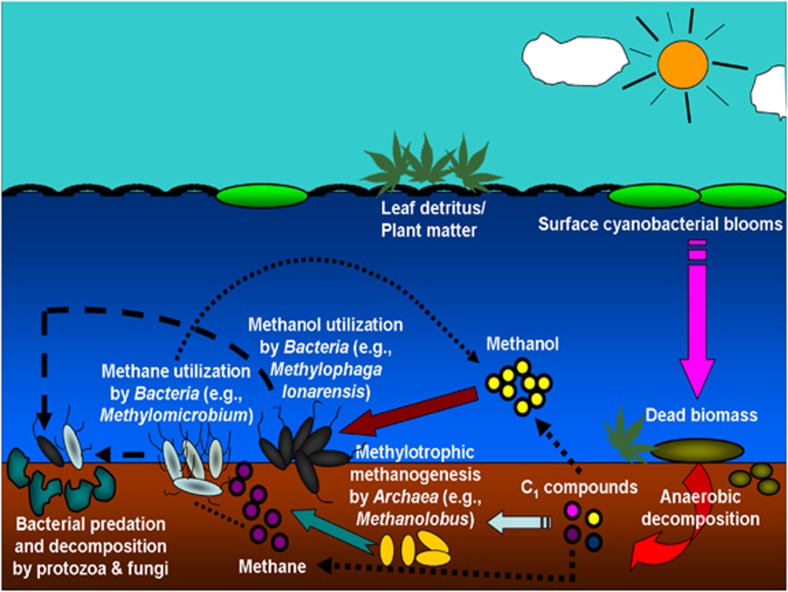
Methylomicrobium, Methylophaga and Bacillus spp. were the predominant active methylotrophs utilizing methane, methanol and methylamine, respectively, in DNA stable-isotope probing (DNA-SIP) experiments with Lonar Lake sediments (Antony et al., 2010). Many gene sequences recovered from the 13C-labeled DNA indicated the presence of novel methylotrophic Bacteria (Antony et al., 2010). Subsequently, the novel moderately haloalkaliphic methylotroph M. lonarensis was isolated from Lonar Lake sediments (Antony et al., 2012a). Interestingly, the partial 16S rRNA gene sequence of this isolate had high identity (99.8%) to some of the Methylophaga-like sequences retrieved by DNA-SIP experiment with methanol (Antony et al., 2012a). Methylophaga are known to utilize both methanol and methylamine, but methylamine-based DNA-SIP experiments did not identify any such members. This finding was corroborated by the ability of M. lonarensis to utilize methanol alone and not methylamine (Antony et al., 2012a). Since the genomic DNA of M. lonarensis contained mauA (encoding methylamine dehydrogenase) (GenBank accession number: JF906115) and since Methylophaga-like mauA phylotypes were obtained from sediments (Supplementary Figure S1), it is possible that there is niche differentiation among Lonar Lake methylotrophs in their preferences for specific C1 substrates due to the limited availability of nutrients in this extreme ecosystem. Eukaryotic small subunit rRNA genes retrieved from methane and methylamine DNA-SIP experiments were dominated by sequences related to Oxytricha longa (ciliated protozoan) and Candida spp. (fungi), respectively, suggesting either direct assimilation of C1 compounds by the active micro-eukaryotes present or flow of labeled carbon from prokaryotes to higher trophic levels of the lake food web through grazing or decomposition of (methylotrophic) biomass and metabolites. Results from these experiments revealed a high diversity of active microorganisms that might be directly or indirectly associated with the C1 food web in Lonar Lake sediments (Figure 1).
Figure 1.
Schematic outline of the hypothetical C1 cycle and associated microbes in Lonar Lake sediments.
Archaeal 16S rRNA genes retrieved from Lonar Lake sediment yielded five Crenarchaeotal phylotypes and eight Euryarchaeotal (mostly methanogen) phylotypes (Wani et al., 2006). Methanogenic enrichment cultures supplemented with H2:CO2, acetate and trimethylamine yielded sequences distantly related to Methanoculleus, Methanocalculus spp. and extant methanogens (Surakasi et al., 2007). Methanogenic species related to Methanosarcina, Methanocalculus and Methanoculleus have also been isolated from the Lonar Lake sediments (Thakker and Ranade, 2002; Surakasi et al., 2007). Primer sets targeting 16S rRNA genes and mcrA (encoding the α-subunit of methyl coenzyme M reductase) revealed the presence of several potentially novel methanogenic Archaea (Methanosarcinales and Methanomicrobiales) in Lonar Lake sediments (Antony et al., 2012b). Analysis of mcrA transcripts that were retrieved from methanol-consuming and methane-emitting sediment microcosms implicated Methanolobus-like organisms as the active Archaea involved in methylotrophic methanogenesis (Antony et al., 2012b). These methanogens may be linked to the ‘intermediary ecosystem metabolism' of Lonar Lake sediments, that is, the anaerobic network of trophically linked microbes that yield intermediates (for example, fatty acids and alcohols) that drive methanogenesis either directly or indirectly (McInerney and Bryant, 1981; Drake et al., 2009). Although much is known about methanogenic links to the intermediary ecosystem metabolism of methane-emitting ecosystems such as peatlands (Dedysh et al., 2006; Cadillo-Quiroz et al., 2008; Wüst et al., 2009; Hunger et al., 2011), the anaerobic trophic interactions that occur in Lonar Lake sediments are poorly understood. Resolving the primary and secondary syntrophic fermenters (McInerney et al., 2008) that might be integrated to the flow of carbon in sediments of Lonar Lake would be of value since little is known of these anaerobes in soda lake environments.
Overview of microbial communities in other soda lakes
Soda lakes in the East African Rift valley, Kulunda-Transbaikal steppes and the USA constitute some of the best characterized soda lakes in the world and several novel genera and species of autotrophic and heterotrophic microorganisms have been described from them (reviewed in Grant and Sorokin, 2011). Numbers of cultivatable aerobic and anaerobic Bacteria in the dilute East African soda lakes approximated 105–106 CFU ml−1 (Grant et al., 1990; Jones et al., 1998). Duckworth and co-workers (1996) isolated alkaliphilic Bacteria related to Halomonas, Bacillus and Arthrobacter and alkaliphilic Archaea related to Natronococcus and Natronobacterium from East African soda lakes. Photosynthetic primary productivity in these lakes has been attributed mainly to Arthrospira spp., although other cyanobacterial species related to Cyanospira, Synechococcus and Chroococcus have been observed seasonally (Jones et al., 1998). The possible contribution of extant anoxygenic phototrophs, such as Ectothiorhodospira, towards lake primary productivity has been noted (Grant 1992; Grant and Sorokin, 2011). Recently, an unusual cyanobacterium (‘Candidatus Gloeomargarita lithophora') capable of intracellular carbonate formation was detected in the modern-day microbialites of Lake Alchichica, Mexico (Couradeau et al., 2012). Chemoorganotrophic Halanaerobiales and Clostridiaceae constituted the predominant anaerobes in the sediments of East African soda lakes (Jones et al., 1998). The occurrence of acetogenic bacterial taxa, such as Natroniella acetigena, Thermosyntropha lipolytica and Tindallia magadiensis (Zavarzin et al., 1994, 1999), suggests that acetogens might be consumers of the products of other anaerobic microorganisms in these lake sediments (Grant and Sorokin, 2011). Several 16S rRNA gene sequences related to putative novel Archaea (Euryarchaeota) were retrieved from the alkaline saltern at Lake Magadi, Kenya (Grant et al., 1999). Haloalkaliphilic Archaea related to Natronomonas, Natrialba, Natronolimnobius and Halorubrum spp. have also been isolated from Lake Magadi and Inner Mongolian soda lakes (reviewed in Grant and Sorokin, 2011).
Depth-based molecular profiling of bacterial communities in the Mono Lake water column showed differences in diversity between surface and bottom layers. The former being abundant in Actinobacteria and the latter being abundant in Firmicutes (Humayoun et al., 2003). The diversity of microbial communities and their association with cycling of arsenic has been characterized in Mono Lake and Searles Lake (Oremland et al., 2004, 2005). Two novel arsenate-respiring heterotrophs, Bacillus selenitireducens and B. arsenicoselenatis, and three novel chemoautotrophs, Alkalilimnicola ehrlichii (strain MLHE-1), strain MLMS-1 and ‘Halarsenatibacter silvermanii' (strain SLAS-1), have been isolated from these lakes (Switzer Blum et al., 1998; Hoeft et al., 2004, 2007; Oremland et al., 2005). Structural gene-based diversity analysis of methanotrophic Bacteria in Mono Lake revealed changes in the methanotroph community structure with respect to methane oxidation activity and recovered many sequences that were affiliated to Methylobacter, Methylomicrobium, Methylothermus and Methylocystis spp. (Lin et al., 2005). Although anaerobic oxidation of methane is a predominant biological process in the lower waters of Mono Lake and Big Soda Lake (Iversen et al., 1987; Oremland et al., 1993), the microorganisms responsible have not been identified. Cultivation-dependent and -independent methods employed recently in Soap Lake (Central Washington) showed the purple phototrophic bacterial population to be comprised of novel members of the Chromatiaceae and Ectothiorhodospiraceae (Asao et al., 2011) and the sulfur-oxidizing bacterial community was dominated by Thioalkalimicrobium spp. (Sorokin et al., 2007). Several methylotrophs have been isolated from the Transbaikal soda lakes (Doronina et al., 2001; Kaluzhnaya et al., 2001) and DNA-SIP experiments showed that the active methanotrophs in Lake Suduntuiskii Torom and Lake Gorbunka were mainly Methylomicrobium, Methylobacter, Methylomonas and Methylothermus spp. (Lin et al., 2004). Since methanotrophs isolated from Transbaikal and East African soda lakes can also oxidize ammonia and carbon disulfide (Khmelenina et al., 2000; Sorokin et al., 2000a), it is likely that these Bacteria constitute an important functional link between C, N and S cycles in soda lake ecosystems (Trotsenko and Khmelenina, 2002). Haloalkaliphilic sulfur oxidizers of the genera Thioalkalivibrio and Thioalkalimicrobium were first isolated from Lake Hadyn in the Transbaikal region (Sorokin et al., 2000b). These strains were also detected in sediments of north-eastern Mongolian soda lakes (Sorokin et al., 2004). dsrAB analysis of Siberian soda lakes showed the sulfate-reducing bacterial population to be comprised of phylotypes related to the orders Desulfovibrionales and Desulfobacterales (Foti et al., 2007). Several lithotrophic sulfate-reducing bacteria of the genera Desulfonatronum, Desulfonatronovibrio, Desulfonatronospira and several heterotrophic sulfate-reducing bacteria affiliated to Desulfobotulus alkaliphilus, Desulfobacteraceae, ‘Desulfobulbus alkaliphilus' and Synthrophobacteraceae have been isolated from soda lakes in Kulunda Steppe (reviewed in Sorokin et al., 2011). The occurrence of both autotrophic sulfur-oxidizing microbes and heterotrophic sulfate-reducing microbes in soda lakes suggests that they might be functionally linked via ‘intercycle coupling' (Drake et al., 2008), that is, via redox junctions between C and S cycles. The occurrence of heterotrophic denitrifiers, such as Halomonas, Pseudomonas and Paracoccus spp. (Table 3), in soda lakes illustrate that the N and C cycles of these habitats are also interconnected when oxygen becomes limiting. Syntrophic associations between sulfur autotrophs related to Thioalkalivibrio in the presence/absence of other partial denitrifiers potentially mediate the denitrification process in the hypersaline soda lakes of the Wadi al Natrun (Sorokin et al., 2003).
Table 3. Some of the prominent Bacteria and Archaea (belonging to diverse metabolic groups) that are commonly detected in isolation-based and cultivation-independent microbial diversity surveys of soda lakes across Africa, North America and Eurasia.
| Heterotrophs | Phototrophs | Methanotrophs and methanogens |
|---|---|---|
| Halomonas | Ectothiorhodospira | Methylomicrobium |
| Spirochaeta | Synechococcus | Methylobacter |
| Bacillus | Synechocystis | Methanolobus |
| Alcaligenes | Rhodobaca | Methanosalsum |
| Arthrobacter | Thiocapsa | Acetogens |
| Nitrincola | Roseinatronobacter | Tindallia |
| Clostridium | ||
| Alkalilimnicola | Sulfur oxidizers and sulfate reducers | Nitrifiers and denitrifiers |
| Marinospirillum | Thioalkalivibrio | Nitrosomonas |
| Alkalimonas | Thioalkalimicrobium | Nitrobacter |
| Idiomarina | Desulfonatronovibrio | Pseudomonas |
| Halorubrum | Desulfonatronum | Paracoccus |
| Halobiforma | ||
| Natronococcus | ||
| Natronolimnobius |
Key steps in elucidating the microbial diversity of soda lake environments are summarized in Table 2. Although some differences have been observed in the microbial diversity profiles among the various soda lakes, as in the case of photosynthetic primary producers (Grant and Sorokin, 2011), the similarities are the most conspicuous (Table 3). This, of course, raises an interesting biogeography question as to whether soda lakes across Africa, North America and Eurasia share a core microbiota (Table 3) on account of dispersal over long geographical distances or whether microbiota evolved independently in each of these soda lakes under the influence of local environmental and geochemical factors.
Table 2. Key steps undertaken to elucidate the microbiology of soda lake environments.
| Major step | Soda lakes | Reference(s) |
|---|---|---|
| Cultivation of alkaliphilic Bacteria and Archaea | Rift valley soda lakes | Duckworth et al. (1996), Zavarzin et al. (1999) |
| Molecular survey of bacterial and/or archaeal diversity | Rift valley soda lakes, Mono Lake | Grant et al. (1999), Humayoun et al. (2003), Rees et al. (2004) |
| Investigations on methanogenesis and methanotrophy | Big Soda Lake, Mono Lake, Transbaikal soda lakes | Oremland et al. (1982, 1993), Iversen et al. (1987), Khmelenina et al. (2000) |
| Characterization of microbes associated with arsenic cycle | Mono Lake, Searles Lake | Reviewed in Oremland et al. (2004, 2005) |
| Characterization of microbes associated with sulfur cycle | Several African, North American and Eurasian soda lakes | Reviewed in Sorokin et al. (2011) |
| Characterization of microbes associated with C1 cycle | Tuva and Transbaikal soda lakes, Rift valley soda lakes, Lake Hotontyn, Mono Lake, Lonar Lake | Khmelenina et al. (1997), Sorokin et al. (2000a), Kaluzhnaya et al. (2001), Doronina et al. (2001, 2003); Lin et al. (2005), Antony et al. (2012a) |
| Identification of active microbes associated with C1 cycle | Transbaikal soda lakes, Lonar Lake | Lin et al. (2004), Antony et al. (2010, 2012b) |
| Investigations on nitrogen cycle and the characterization of microbes associated with this cycle | Big Soda Lake, Mono Lake, Siberian soda lakes, Mongolian soda lakes, Rift valley soda lakes | Oremland et al. (1988, Sorokin (1998), Sorokin et al. (1998, 2001), Ward et al. (2000), Carini and Joye (2008) |
| Characterization of diversity of chemo- or photoautotrophic Bacteria | Mono Lake, Kulunda steppe soda lakes, Wadi al Natrun soda lakes, Soap Lake | Giri et al. (2004), Kovaleva et al. (2011), Asao et al. (2011) |
Biotechnological potential of soda lake microbes
The global industrial enzyme market is estimated to be worth >US$1.6 billion and detergent enzymes account for approximately 40% of this market (Ito et al., 2005). Soda lakes are important sources of microbial enzymes that can function at high pH (Grant and Heaphy, 2010). Through classical recovery and screening methods, two industrial cellulases have been obtained from Gram-positive Kenyan soda lake isolates, which are marketed as IndiAge Neutra and Puradax by Genencor for use in textile and laundry processes (Sheridan, 2004; Grant and Heaphy, 2010). Alkaline proteases, lipases, amylases, chitinases and caseinases have also been reported in a wide range of Bacteria isolated from soda lake environments, such as Rift valley soda lakes, Lonar Lake and Mono Lake (Jones et al., 1998; LeCleir et al., 2007; Joshi et al., 2008). Halo(alkali)philic microorganisms use several different types of organic solutes as osmolytes (Ciulla et al., 1997). Ectoine is accumulated intracellularly by soda lake microorganisms such as methylotrophs (Trotsenko and Khmelenina, 2002; Doronina et al., 2003; Antony et al., 2012a) and sulfur-oxidizing bacteria (Sorokin et al., 2011). Ectoine has many biotechnologically relevant applications, especially in molecular biology, cosmetics and therapeutics (reviewed in Pastor et al., 2010). The search for novel genes, enzymes and other biomolecules from soda lake environments can be expedited through the application of ‘focused metagenomics' strategies such as DNA-SIP coupled with metagenomic library screening (reviewed in Moussard et al., 2011). If this approach can further be complemented with improved high-throughput screening methods and the latest advances in sequencing technology, it may lead to discovery of many commercially important enzymes and biomolecules from saline and alkaline lake ecosystems.
Future perspectives
The functional links between soda lake microorganisms and their ecophysiological roles in the cycling of major elements in soda lakes are poorly understood. The occurrence of diverse functional groups illustrates that soda lakes harbor complex microbial food webs that interconnect various biological cycles via redox coupling. Direct measurement of microbial processes and construction of elemental budgets are now needed to understand which processes are significant and how they interact in the soda lake ecosystem. To gain deeper insights into the functional guilds of microorganisms mediating the various biogeochemical processes, next-generation sequencing can be combined with advanced microbial ecology techniques such as SIP and Raman-FISH (Neufeld and Murrell, 2007). The use of microarrays, such as PhyloChip and GeoChip (Andersen et al., 2010), in conjunction with the measurement of specific biogeochemical parameters will aid in the rapid comparative profiling of the structure and activity of soda lake microbial communities on a spatiotemporal scale. Interestingly, microbial ecology studies conducted so far in soda lakes have been restricted to the bacterial and archaeal domains of life and little is known about the diversity, abundance and activity of micro-eukaryotes in such lakes. Similarly, information on the nature of trophic interactions between the various soda lake microbes and their gross contribution towards maintenance of overall ecosystem health is lacking. Future efforts in these areas will be important and productive.
Serious conservation and ecosystem management is needed to protect soda lakes, which may be perturbed by anthropogenic activity (Gebre-Mariam, 1998; Chakravarty, 2009). Microbial diversity of soda lake ecosystems is not only central to our better understanding of the limits of life at extreme pH but also to our search for novel useful biomolecules.
Acknowledgments
This work received financial support from the British Council UK-India Education and Research Initiative Grant SA07-061. CPA acknowledges a senior research fellowship from the Indian Council of Medical Research, Government of India. DK acknowledges an OCE postdoctoral fellowship from CSIRO, Australia.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andersen GL, He Z, Desantis TZ, Brodie EL, Zhou J.2010The use of microarrays in microbial ecologyIn: Liu WT, Jansson JK (eds).Environmental Molecular Microbiology Caister Academic Press: Norwich,87–110. [Google Scholar]
- Antony CP, Kumaresan D, Ferrando L, Boden R, Moussard H, Scavino AF, et al. Active methylotrophs in the sediments of Lonar Lake, a saline and alkaline ecosystem formed by meteor impact. ISME J. 2010;4:1470–1480. doi: 10.1038/ismej.2010.70. [DOI] [PubMed] [Google Scholar]
- Antony CP, Doronina NV, Boden R, Trotsenko YA, Shouche YS, Murrell JC. Methylophaga lonarensis, a novel moderately haloalkaliphilic methylotroph isolated from the soda lake sediments of a meteorite impact crater. Int J Syst Evol Microbiol. 2012a;62:1613–1618. doi: 10.1099/ijs.0.035089-0. [DOI] [PubMed] [Google Scholar]
- Antony CP, Murrell JC, Shouche YS. Molecular diversity of methanogens and identification of Methanolobus sp. as active methylotrophic Archaea in Lonar crater lake sediments. FEMS Microbiol Ecol. 2012b;81:43–51. doi: 10.1111/j.1574-6941.2011.01274.x. [DOI] [PubMed] [Google Scholar]
- Asao M, Pinkart HC, Madigan MT. Diversity of extremophilic purple phototrophic bacteria in Soap Lake, a Central Washington (USA) Soda Lake. Environ Microbiol. 2011;13:2146–2157. doi: 10.1111/j.1462-2920.2011.02449.x. [DOI] [PubMed] [Google Scholar]
- Cadillo-Quiroz H, Yashiro E, Yavitt JB, Zinder SH. Characterization of the archaeal community in a minerotrophic fen and terminal restriction fragment length polymorphism-directed isolation of a novel hydrogenotrophic methanogen. Appl Environ Microbiol. 2008;74:2059–2068. doi: 10.1128/AEM.02222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carini SA, Joye SB. Nitrification in Mono Lake, California: Activity and community composition during contrasting hydrological regimes. Limnol Oceanogr. 2008;53:2546–2557. [Google Scholar]
- Chakravarty I. Conservation and management for the Lonar crater-lake, Maharashtra, India. S Asian J Tour Herit. 2009;2:112–118. [Google Scholar]
- Ciulla RA, Diaz MR, Taylor BF, Roberts MF. Organic osmolytes in aerobic bacteria from Mono Lake, an alkaline, moderately hypersaline environment. Appl Environ Microbiol. 1997;63:220–226. doi: 10.1128/aem.63.1.220-226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couradeau E, Benzerara K, Gerard E, Moreira D, Bernard S, Brown GE, López-Garcia P. An early-branching microbialite cyanobacterium forms intracellular carbonates. Science. 2012;336:459–462. doi: 10.1126/science.1216171. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Pankratov TA, Belova SE, Kulichevskaya IS, Liesack W. Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl Environ Microbiol. 2006;72:2110–2117. doi: 10.1128/AEM.72.3.2110-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina NV, Darmaeva T, Trotsenko YA. New aerobic methylotrophic isolates from the Soda lakes of the southern Transbaikal. Mikrobiologiia. 2001;70:398–404. [PubMed] [Google Scholar]
- Doronina NV, Darmaeva TD, Trotsenko YA. Methylophaga alcalica sp. nov., a novel alkaliphilic and moderately halophilic, obligately methylotrophic bacterium from an East Mongolian saline soda lake. Int J Syst Evol Microbiol. 2003;53:223–229. doi: 10.1099/ijs.0.02267-0. [DOI] [PubMed] [Google Scholar]
- Drake HL, Gößner AS, Daniel SL.2008Old acetogens, new lightIn: Wiegel J, Maier RJ, Adams MWW (eds).Incredible Anaerobes: From Physiology to Genomics to Fuels New York Academy of Sciences: Boston, MA; 100–128. [DOI] [PubMed] [Google Scholar]
- Drake HL, Horn MA, Wüst PK. Intermediary ecosystem metabolism as a main driver of methanogenesis in acidic wetland soil. Environ Microbiol Rep. 2009;1:307–318. doi: 10.1111/j.1758-2229.2009.00050.x. [DOI] [PubMed] [Google Scholar]
- Duckworth AW, Grant WD, Jones BE, Van Steenbergen R. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol Ecol. 1996;19:181–191. [Google Scholar]
- Foti M, Sorokin DY, Lomans B, Mussman M, Zakharova EE, Pimenov NV, et al. Diversity, activity and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Appl Environ Microbiol. 2007;73:2093–2100. doi: 10.1128/AEM.02622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson K, Dube A, Milton DJ, Balasundaram MS. Lonar Lake, India: an impact crater in basalt. Science. 1973;180:862–864. doi: 10.1126/science.180.4088.862. [DOI] [PubMed] [Google Scholar]
- Gebre-Mariam Z.1998Human interactions and water quality in the Horn of AfricaIn: Schoneboom J (eds).Science in Africa- Emerging Water Management Problems A Publication of the Symposium at the American Association for the Advancement of Science (AAAS), Annual Meeting 1998: Philadelphia; 47–61. [Google Scholar]
- Giri BJ, Bano N, Hollibaugh JT. Distribution of RuBisco genotypes along a redox gradient in Mono Lake, California. Appl Environ Microbiol. 2004;70:3443–3448. doi: 10.1128/AEM.70.6.3443-3448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant WD, Jones BE, Mwatha WE. Alkaliphiles: ecology, diversity and applications. FEMS Microbiol Rev. 1990;75:255–270. [Google Scholar]
- Grant WD.1992Alkaline environmentsIn: Lederberg J (ed.)Encyclopaedia of Microbiology1st edn.Academic Publishers: London; 73–80. [Google Scholar]
- Grant S, Grant WD, Jones BE, Kato C, Li L. Novel archaeal phytotypes from an East African alkaline saltern. Extremophiles. 1999;3:139–145. doi: 10.1007/s007920050109. [DOI] [PubMed] [Google Scholar]
- Grant WD.2006Alkaline environments and biodiversity, in extremophiliesIn: Gerday Charles, Glansdorff Nicolas (eds).Encyclopedia of Life Support Systems (EOLSS) Developed under the Auspices of the UNESCO, Eolss Publishers: Oxford, UK; , http://www.eolss.net . [Google Scholar]
- Grant WD, Heaphy S. Metagenomics and recovery of enzyme genes from alkaline saline environments. Environ Technol. 2010;31:1135–1143. doi: 10.1080/09593331003646661. [DOI] [PubMed] [Google Scholar]
- Grant WD, Sorokin DY.2011Distribution and diversity of soda lake alkaliphilesIn: Horikoshi K, Antranikian G, Bull AT, Robb FT, Stetter KO (eds).Extremophiles Handbook vol. 1Springer: Tokyo; 27–54. [Google Scholar]
- Hoeft SE, Kulp TR, Stolz JF, Hollibaugh JT, Oremland RS. Dissimilatory arsenate reduction with sulfide as electron donor: Experiments with Mono Lake water and isolation of strain MLMS-1, a chemoautotrophic arsenate-respirer. Appl Env Microb. 2004;70:2741–2747. doi: 10.1128/AEM.70.5.2741-2747.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft SE, Blum JS, Stolz JF, Tabita FR, Witte B, King GM, et al. Alkalilimnicola ehrlichii sp. nov., a novel, arsenite-oxidizing haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int J Syst Evol Microbiol. 2007;57:504–512. doi: 10.1099/ijs.0.64576-0. [DOI] [PubMed] [Google Scholar]
- Humayoun SB, Bano N, Hollibaugh JT. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl Environ Microbiol. 2003;69:1030–1042. doi: 10.1128/AEM.69.2.1030-1042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger S, Schmidt O, Hilgarth M, Horn MA, Kolb S, Conrad R, et al. Competing formate- and carbon-dioxide utilizing prokaryotes in an anoxic methane-emitting fen soil. Appl Environ Microbiol. 2011;77:3773–3785. doi: 10.1128/AEM.00282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Kobayashi T, Hatada Y, Horikoshi K.2005Enzymes in modern detergentsIn: Barredo JL (eds).Microbial Enzymes and Biotransformations Humana Press: Totowa, NJ; 151–164. [Google Scholar]
- Iversen N, Oremland RS, Klug MJ. Big Soda Lake (Nevada). 3. Pelagic methanogenesis and anaerobic methane oxidation. Limnol Oceanogr. 1987;32:804–814. [Google Scholar]
- Jones BE, Grant WD, Duckworth AW, Owenson GG. Microbial diversity of soda lakes. Extremophiles. 1998;2:191–200. doi: 10.1007/s007920050060. [DOI] [PubMed] [Google Scholar]
- Joshi AA, Kanekar PP, Kelkar AS, Shouche YS, Vani AA, Borgave SB, Sarnaik SS. Cultivable bacterial diversity of alkaline Lonar Lake, India. Microb Ecol. 2008;55:163–172. doi: 10.1007/s00248-007-9264-8. [DOI] [PubMed] [Google Scholar]
- Jourdan FF, Moynier F, Koeberl C, Eroglu S. 40Ar/39Ar age of the Lonar crater and consequence for the geochronology of planetary impacts. Geology. 2011;39:671–674. [Google Scholar]
- Kaluzhnaya M, Khmelenina V, Eshinimaev B, Suzina N, Nikitin D, Solonin A, et al. Taxonomic characterization of new alkaliphilic and alkalitolerant methanotrophs from soda lakes of the Southeastern Transbaikal region and description of Methylomicrobium buryatense sp. nov. Syst Appl Microbiol. 2001;24:166–176. doi: 10.1078/0723-2020-00028. [DOI] [PubMed] [Google Scholar]
- Karpeta WP. Bedded cherts in the Rietgat Formation, Hartbeesfontain, South Africa: a late Archean to early Proterozoic magadiitic alkaline playa lake deposit. South Afr J Geol. 1989;92:29–36. [Google Scholar]
- Khmelenina VN, Kalyuzhnaya MG, Starostina NG, Suzina NE, Trotsenko YA. Isolation and characterization of halotolerant alkaliphilic methanotrophic bacteria from Tuva soda lakes. Curr Microbiol. 1997;35:257–261. [Google Scholar]
- Khmelenina VN, Eshinimaev BT, Kalyuzhnaya MG, Trotsenko I. Potential activity of methane and ammonia oxidation by methanotropic communities from soda lakes of the southern Transbaikal. Mikrobiologiia. 2000;69:553–558. [PubMed] [Google Scholar]
- Kovaleva OL, Tourova TP, Muyzer G, Kolganova TV, Sorokin DY. Diversity of RuBisco and ATP citrate lyase genes in soda lake sediments. FEMS Microbiol Ecol. 2011;75:37–47. doi: 10.1111/j.1574-6941.2010.00996.x. [DOI] [PubMed] [Google Scholar]
- Kumar PA, Srinivas TNR, Kumar PP, Madhu S, Shivaji S. Nitritalea halalkaliphila gen. nov., sp. nov., an alkaliphilic bacterium of the family ‘Cyclobacteriaceae', phylum Bacteroidetes. Int J Syst Evol Microbiol. 2010a;60:2320–2325. doi: 10.1099/ijs.0.020230-0. [DOI] [PubMed] [Google Scholar]
- Kumar PA, Srinivas TNR, Madhu S, Manorama R, Shivaji S. Indibacter alkaliphilus gen. nov., sp. nov., an alkaliphilic bacterium isolated from a haloalkaline lake. Int J Syst Evol Microbiol. 2010b;60:721–726. doi: 10.1099/ijs.0.014076. [DOI] [PubMed] [Google Scholar]
- Kumar PA, Srinivas TNR, Madhu S, Sravan R, Singh S, Naqvi SWA, et al. Cecembia lonarensis gen. nov., sp. nov., a novel haloalkalitolerant bacterium of the family ‘Cyclobacteriaceae', isolated from a haloalkaline lake and emended descriptions of the genera Indibacter, Nitritalea and Belliella. Int J Syst Evol Microbiol. 2012;62:2252–2258. doi: 10.1099/ijs.0.038604-0. [DOI] [PubMed] [Google Scholar]
- LeCleir GR, Buchan A, Maurer J, Moran MA, Hollibaugh JT. Comparison of chitinolytic enzymes from an alkaline, hypersaline lake and an estuary. Environ Microb. 2007;9:197–205. doi: 10.1111/j.1462-2920.2006.01128.x. [DOI] [PubMed] [Google Scholar]
- Lin JL, Radajewski S, Eshinimaev BT, Trotsenko YA, McDonald IR, Murrell JC. Molecular diversity of methanotrophs in Transbaikal soda lake sediments and identification of potentially active populations by stable isotope probing. Environ Microbiol. 2004;6:1049–1060. doi: 10.1111/j.1462-2920.2004.00635.x. [DOI] [PubMed] [Google Scholar]
- Lin JL, Joye SB, Scholten JC, Schafer H, McDonald IR, Murrell JC. Analysis of methane monooxygenase genes in Mono lake suggests that increased methane oxidation activity may correlate with a change in methanotroph community structure. Appl Environ Microbiol. 2005;71:6458–6462. doi: 10.1128/AEM.71.10.6458-6462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhoff BS, Bennett PC, Puntsag T, Gerel O. Geochemical evolution of uraniferous soda lakes in Eastern Mongolia. Environ Earth Sci. 2011;62:171–183. [Google Scholar]
- Maloof AC, Stewart ST, Weiss BP, Soule SA, Swanson-Hysell NL, Louzada KL, et al. Geology of Lonar Crater, India. Geol Soc Am Bull. 2010;122:109–126. [Google Scholar]
- McInerney MJ, Bryant MP.1981Basic principles of bioconversions in anaerobic digestion and methanogenesisIn: Sofer S, Zaborsky OR (eds).Biomass Conversion Processes for Energy and Fuels Plenum Press: New York, NY; 277–296. [Google Scholar]
- McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJM, Schink B, et al. 2008Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolismIn: Wiegel J, Maier RJ, Adams MWW (eds).Incredible Anaerobes: From Physiology to Genomics to Fuels New York Academy of Sciences: Boston, MA; 58–72. [DOI] [PubMed] [Google Scholar]
- Melack JM, Kilham P. Photosynthetic rates of phytoplankton in East African alkaline, saline lakes. Limnol Oceanogr. 1974;19:743–755. [Google Scholar]
- Moussard H, Smith TJ, Murrell JC.2011DNA-stable isotope probing and gene miningIn: Murrell JC, Whiteley A (eds).Stable Isotope Probing and Related Technologies ASM press: Washington, DC; 3–24. [Google Scholar]
- Neufeld JD, Murrell JC. Witnessing the last supper of uncultivated microbial cells with Raman-FISH. ISME J. 2007;4:269–270. doi: 10.1038/ismej.2007.55. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Marsh L, Des Marais DJ. Methanogenesis in Big Soda Lake, Nevada: an alkaline, moderately hypersaline desert lake. Appl Environ Microbiol. 1982;43:462–468. doi: 10.1128/aem.43.2.462-468.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland RS, Cloern JE, Sofer Z, Smith RL, Culbertson CW, Zehr J, et al. 1988Microbial and biogeochemical processes in Big Soda Lake, NevadaIn: Fleet AJ, Kelts K, Talbot MR (eds).Lacustrine Petroleum Source Rocks Geological Society Special Publication: London; No.4059–75. [Google Scholar]
- Oremland RS. Nitrogen fixation dynamics of two diazotrophic communities in Mono Lake, California. Appl Environ Microbiol. 1990;56:614–622. doi: 10.1128/aem.56.3.614-622.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland RS, Miller LG, Culbertson CW, Robinson S, Smith RL, Lovley DR, et al. 1993Aspects of the biogeochemistry of methane in Mono Lake and the Mono Basin of California, USAIn: Oremland RS (ed.)The Biogeochemistry of Global Change: Radiative Trace Gases Chapman & Hall: New York; 704–744. [Google Scholar]
- Oremland RS, Stolz JF, Hollibaugh JT. The microbial arsenic cycle in Mono Lake, California. FEMS Microb Ecol. 2004;48:15–27. doi: 10.1016/j.femsec.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Kulp TR, Switzer Blum J, Hoeft SE, Baesman S, Miller LG, Stolz JF. A microbial arsenic cycle in a salt saturated, βextreme environment. Science. 2005;308:1305–1308. doi: 10.1126/science.1110832. [DOI] [PubMed] [Google Scholar]
- Pastor JM, Salvador M, Argandona M, Bernal V, Reina-Bueno M, Csonka LN, et al. Ectoines in cell stress protection: uses and biotechnological production. Biotechnol Adv. 2010;28:782–801. doi: 10.1016/j.biotechadv.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Rees HC, Grant WD, Jones BE, Heaphy S. Diversity of Kenyan soda lake alkaliphiles assessed by molecular methods. Extremophiles. 2004;8:63–71. doi: 10.1007/s00792-003-0361-4. [DOI] [PubMed] [Google Scholar]
- Sheridan C. Kenyan dispute illuminates bioprospecting difficulties. Nat Biotechnol. 2004;22:1337. doi: 10.1038/nbt1104-1337. [DOI] [PubMed] [Google Scholar]
- Sorokin DY. Occurrence of nitrification in extremely alkaline environments. Microbiology (Moscow, English translation) 1998;67:335–339. [Google Scholar]
- Sorokin DY, Muyzer G, Brinkhoff T, Kuenen JG, Jetten M. Isolation and characterization of a novel facultatively alkaliphilic Nitrobacter species–Nb. alkalicus. Arch Microbiol. 1998;170:345–352. doi: 10.1007/s002030050652. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Jones BE, Kuenen JG. A novel obligately methylotrophic, methane-oxidizing Methylomicrobium species from a highly alkaline environment. Extremophiles. 2000a;4:145–155. doi: 10.1007/s007920070029. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Robertson LA, Kuenen JG. Isolation and characterization of obligately chemolithoautotrophic alkaliphilic sulfur-oxidizing bacteria. Ant V Leeuwenhoek. 2000b;77:251–260. doi: 10.1023/a:1002445704444. [DOI] [PubMed] [Google Scholar]
- Sorokin D, Tourova T, Schmid MC, Wagner M, Koops H, Kuenen JG, Jetten M. Isolation and properties of obligately chemolithoautotrophic and extremely alkali-tolerant ammonia-oxidizing bacteria from Mongolian soda lakes. Arch Microbiol. 2001;176:170–177. doi: 10.1007/s002030100310. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Antipov AN, Kuenen JG. Complete denitrification in coculture of obligately chemolithoautotrophic haloalkaliphilic sulfur-oxidizing bacteria from a hypersaline soda lake. Arch Microbiol. 2003;180:127–133. doi: 10.1007/s00203-003-0567-y. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Gorlenko VM, Namsaraev BB, Namsaraev ZB, Lysenko AM, Eshinimaev BT, et al. Prokaryotic communities of the north-eastern Mongolian soda lakes. Hydrobiologia. 2004;522:235–248. [Google Scholar]
- Sorokin DY, Foti M, Pinkart HC, Muyzer G. Sulfur-oxidizing bacteria in Soap Lake (Washington, USA), a meromictic, haloalkaline lake with an unprecedented high sulfide content. Appl Environ Microbiol. 2007;73:451–455. doi: 10.1128/AEM.02087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin DY, Kuenen JG, Muyzer G. The microbial sulfur cycle at extremely haloalkaline conditions of soda lakes. Front Microbiol. 2011;2:44. doi: 10.3389/fmicb.2011.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas A, Rahul K, Sasikala Ch, Subhash Y, Ramaprasad EV, Ramana ChV. Georgenia satyanarayanai sp. nov., an alkaliphilic and thermotolerant amylase producing actinobacterium isolated from a soda lake. Int J Syst Evol Microbiol. 2012;62:2405–2409. doi: 10.1099/ijs.0.036210-0. [DOI] [PubMed] [Google Scholar]
- Surakasi VP, Wani AA, Shouche YS, Ranade DR. Phylogenetic analysis of methanogenic enrichment cultures obtained from Lonar Lake in India: isolation of Methanocalculus sp. and Methanoculleus sp. Microb Ecol. 2007;54:697–704. doi: 10.1007/s00248-007-9228-z. [DOI] [PubMed] [Google Scholar]
- Surakasi VP, Antony CP, Sharma S, Patole MS, Shouche YS. Temporal bacterial diversity and detection of putative methanotrophs in surface mats of Lonar crater lake. J Basic Microbiol. 2010;50:465–474. doi: 10.1002/jobm.201000001. [DOI] [PubMed] [Google Scholar]
- Switzer Blum J, Burns Bindi A, Buzzelli J, Stolz JF, Oremland RS. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens sp. nov: two haloalkaliphiles from Mono Lake, California, that respire oxyanions of selenium and arsenic. Arch Microbiol. 1998;171:19–30. doi: 10.1007/s002030050673. [DOI] [PubMed] [Google Scholar]
- Thakker CD, Ranade DR. Alkalophilic Methanosarcina isolated from Lonar Lake. Curr Sci. 2002;82:455–458. [Google Scholar]
- Trotsenko YA, Khmelenina VN. Biology of extremophilic and extremotolerant methanotrophs. Arch Microbiol. 2002;177:123–131. doi: 10.1007/s00203-001-0368-0. [DOI] [PubMed] [Google Scholar]
- Wani AA, Surakasi VP, Siddharth J, Raghavan RG, Patole MS, Ranade D, Shouche YS. Molecular analyses of microbial diversity associated with the Lonar soda lake in India: an impact crater in a basalt area. Res Microbiol. 2006;157:928–937. doi: 10.1016/j.resmic.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Ward BB, Martino DP, Diaz MC, Joye SB. Analysis of ammonia-oxidizing bacteria from a hypersaline Mono Lake, California, on the basis of 16S rRNA sequences. Appl Environ Microbiol. 2000;66:2873–2881. doi: 10.1128/aem.66.7.2873-2881.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüst PK, Horn MA, Drake HL. Trophic links between fermenters and methanogens in a moderately acidic fen soil. Environ Microbiol. 2009;11:1395–1409. doi: 10.1111/j.1462-2920.2009.01867.x. [DOI] [PubMed] [Google Scholar]
- Zavarzin GA, Zhilina TN, Pusheva MA.1994Halophilic acetogenic bacteriaIn: Drake HL (eds).Acetogenesis Chapman & Hall: New York; 432–444. [Google Scholar]
- Zavarzin GA, Zhilina TN, Kevbrin VV. The alkaliphilic microbial community and its functional diversity. Microbiologiya. 1999;68:503–521. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.