Abstract
Background
Genome-wide association studies (GWASs) of asthma have consistently implicated the ORM1-like 3 and gasdermin B (ORMDL3-GSDMB), IL33, IL-1 receptor–like 1 and IL-18 receptor 1 (IL1RL1-IL18R1), RAD50-IL13, thymic stromal lymphopoietin and WD repeat domain 36 region (TSLP-WDR36), and HLA-DR/DQ regions.
Objective
A GWAS of asthma was performed in a non-Hispanic white population.
Methods
A GWAS was performed in 813 Severe Asthma Research Program/Collaborative Studies on the Genetics of Asthma/Chicago Asthma Genetics Study cases and 1564 control subjects. The GWAS results were compared with those of the published GWASs of autoimmune diseases.
Result
Multiple single nucleotide polymorphisms in the TNFAIP3 interacting protein 1 (TNIP1) gene, which interacts with TNFAIP3 and inhibits the TNF-α–induced nuclear factor κB inflammation pathway, were associated with asthma: rs1422673 (P = 3.44 × 10−7) and rs10036748 (P = 1.41 × 10−6, r2 = 0.67). rs1422673 was also associated with asthma in the published GABRIEL (P = .018) and EVE (P = 1.31 × 10−5) studies. The minor allele T of rs20541 in IL13 is the risk allele for asthma but the protective allele for psoriasis. The minor allele T of rs2395185 in HLA-DRA is the risk allele for asthma but the protective allele for ulcerative colitis. The minor allele A of rs2872507 in GSDMB is the protective allele for asthma but the risk allele for rheumatoid arthritis, Crohn disease, and ulcerative colitis. The T allele of rs10036748 in the TNIP1 gene is the minor protective allele for asthma but the minor or major risk allele for systemic lupus erythematosus and systemic sclerosis in non-Hispanic white or Chinese subjects, respectively.
Conclusions
Our study suggests that single nucleotide polymorphisms associated with both asthma and autoimmune diseases might have opposite effects on immunopathogenesis. (J Allergy Clin Immunol 2012;130:861-8.)
Keywords: Asthma, genetics, genome-wide association study, TNFAIP3 interacting protein 1
Asthma is a common inflammatory airway disease that can be triggered in genetically susceptible subjects by various environmental exposures. Asthma is characterized by bronchial hyperresponsiveness, bronchodilator reversibility, and often by increased expression of TH2 cytokines, increased serum IgE levels, and atopy. In allergic asthmatic patients eosinophilic inflammation and TH2 cytokines dominate; in patients with severe/refractory asthma, neutrophilic inflammation and TNF-α/TH17 cytokines are involved.1 Genome-wide association studies (GWASs) of asthma and asthma-related traits have consistently identified 6 major regions: the ORM1-like 3 and gasdermin B (ORMDL3-GSDMB) region,2-4 interleukin 33 (IL33),3-5 the IL-1 receptor– like 1 and IL-18 receptor 1 (IL1RL1-IL18R1) region,3-5 the RAD50 homolog and IL-13 (RAD50-IL13) region,3,6 the thymic stromal lymphopoietin and WD repeat domain 36 region (TSLP-WDR36) region,3-5,7 and the major histocompatibility complex class II DR/DQ (HLA-DR/DQ) region.3,6,7
Autoimmune diseases arise through abnormal immune responses to self-antigens and are generally characterized by TH1-mediated inflammation. Autoimmune diseases cause extensive comorbidity among psoriasis, systemic lupus erythematosus (SLE), rheumatoid arthritis, Crohn disease, type I diabetes, multiple sclerosis, ulcerative colitis, and celiac disease, for example.8 Although both Crohn disease and ulcerative colitis are inflammatory bowel diseases, the TH1 process is dominant in patients with Crohn disease, whereas ulcerative colitis is likely to be a TH2 disease.9 Comparison of published GWAS results among patients with a variety of autoimmune diseases reveals that significant association at the gene or single nucleotide polymorphism (SNP) level is common among autoimmune diseases.8
Systematic comparison of genes or SNPs found to be significant in GWASs of both asthma and autoimmune diseases is very limited.8 Asthma and autoimmune diseases share extensive immunologic pathways but generally are believed to have different or opposite pathogenic T-cell mechanisms (oversimplified as the TH2 vs TH1 model). A counterregulatory model emphasizes the importance of regulatory T cells in patients with immune diseases, which inhibit both TH2-mediated allergic diseases and TH1-mediated autoimmune diseases.10 Here we report a GWAS of asthma in a non-Hispanic white population (813 cases and 1564 control subjects) from the Severe Asthma Research Program (SARP)/Collaborative Studies on the Genetics of Asthma (CSGA)/Chicago Asthma Genetics Study (CAG) to identify novel genes and to confirm previously identified genes involved in asthma. Several asthma candidate genes (HLA, IL13, and TNFAIP3 interacting protein 1 [TNIP1]) identified by us and others were associated with autoimmune diseases as well. Hence we compared SNPs in genes identified by using GWASs of asthma with those in autoimmune diseases to explore common genetic factors and disease causes.
Methods
Study subjects
Non-Hispanic white subjects were participants in the National Heart, Lung, and Blood Institute–funded SARP, the National Heart, Lung, and Blood Institute's CSGA, and the CAG (or CSGA enrolled in Chicago). Subjects with mild-to-severe asthma and nonasthmatic control subjects were recruited from SARP11,12 and CSGA13 centers with a similar protocol. CAG subjects with asthma and nonasthmatic control subjects were collected at the University of Chicago by using a similar protocol.4 Results from the GWAS in SARP/CSGA/CAG were included in the EVE consortium meta-analysis.4
Subjects with difficult-to-treat or severe asthma were recruited from the Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens (TENOR) multicenter study.6,14 SARP, CSGA, CAG, and TENOR studies were approved by the appropriate institutional review boards at the participating sites, including informed consent.
General population control subjects were obtained by using the Illumina iControlDB client (Illumina, Inc, San Diego, Calif) to download genotypes for 3294 white subjects with genotype data available from HumanHap 550 k products. General population control subjects (n 5 1892) were matched with 473 TENOR cases, as described previously.6 The remaining general population control subjects (n = 1011) and control subjects genotyped in SARP/CSGA/CAG (n = 5553) were merged and used as control subjects for SARP/CSGA/CAG cases (n = 5813).
DNA was isolated by using standard protocols, and SNP genotyping was performed with the Illumina HumanHap1M BeadChip or the Illumina HumanCNV370 BeadChip for SARP/CSGA/CAG4,15 and TENOR,6 respectively.
SARP/CSGA/CAG (813 cases vs 1564 control subjects) was used as the discovery dataset. The published TENOR,6 EVE,4 and GABRIEL3 studies were used to replicate top findings from SARP/CSGA/CAG. Meta-analysis P values from EVE and GABRIEL were reported by using a fixed-effects model.
Statistical analysis
Quality control was applied to SARP/CSGA/CAG cases and control subjects, TENOR cases, and Illumina control subjects separately because they were genotyped by using slightly different Illumina products, as described previously.6 In brief, subjects were removed if they (1) had genotyping call rates of less than 95%, (2) were discrepant or ambiguous for genetic sex (heterozygous haploid genotype percentage ≥0.01 for male subjects or X-chromosome homozygosity F ≥0.9 for female subjects), (3) failed the check for family relatedness (PI_HAT > 0.125), or (4) were detected as an outlier (>6 SDs for the first or second principal component). After subjects meeting these criteria were excluded, SNPs were removed if (1) call rates were 95% or less, (2) they were inconsistent with Hardy-Weinberg equilibrium (P < 10−4), or (3) they had a minor allele frequency of 0.05 or less. After quality control, SNPs shared between cases and control subjects were merged for analysis.
Logistic regression, assuming an additive disease model, was used for genome-wide association analysis of asthma susceptibility by using PLINK16 adjusted for age, sex, and significant principal components (n = 4) from EIGENSTRAT.17 Linkage disequilibrium (LD) was estimated with Haploview.18
Logistic regression of SNPs with well-replicated associations (rs2872507 in GSDMB, rs3939286 in IL33, rs13431828 in IL1RL1, rs20541 in IL13, rs1837253 in TSLP, and rs2395185 in HLA-DRA) were performed in SARP/CSGA/CAG and TENOR populations by using either 6 SNPs together or genetic scores with age and sex adjusted. Genetic scores were defined as follows: genotypes with 1 or 2 minor alleles were merged together and recoded to 0 as a protective category (if the minor allele was a protective allele) or to 1 as a risk category (if the minor allele was a risk allele). The percentage of deviance explained by an SNP was defined as deviance explained by an SNP/deviance of the null model with age and sex adjusted. The area under the receiver operating characteristic curve was calculated by using SAS software (SAS Institute, Inc, Cary, NC).
GWAS results from the published GABRIEL study3 were extracted from the European Genome-Phenome Archive (http://www.cng.fr/gabriel; accession no. EGAS00000000077 for the GABRIEL study). SNPs (P < 1.0 × 10−5) from GWASs of asthma or asthma-related traits2-7 were extracted from the NIH GWAS database19 (http://www.genome.gov/gwastudies/). SNPs (P < 1.0 × 10−5) from GWASs of autoimmune diseases were extracted from the NIH GWAS database19 (http://www.genome.gov/gwastudies/) if the same SNPs were significant (P<1.0 × 10−5) in a GWAS of asthma or asthma-related traits.2-7
Results
GWAS of asthma in SARP/CSGA/CAG
After quality control, a total of 474,271 SNPs in 813 SARP/CSGA/CAG cases, 553 SARP/CSGA/CAG control subjects, and 1,011 Illumina control subjects remained for the GWAS (see Table E1 in this article's Online Repository at www.jacionline.org). Population stratification was weak after adjustment for age, sex, and 4 significant principal components (genomic inflation factor, 1.018; see Fig E1 in this article's Online Repository at www.jacionline.org) and further adjusted by using genomic control.
TABLE E1.
Demographics (means ± SD) of subjects in SARP/CSGA/CAG and Illumina and phenotyped control subjects
| SARP/CSGA/CAG cases | SARP/CSGA/CAG control subjects | Illumina control subjects | |
|---|---|---|---|
| No. | 813 | 553 | 1011 |
| Age (y) | 32.2 ± 15.7 | 32.2 ± 10.5 | 30.4 ± 20.0 |
| Sex (% female) | 61.9 | 63.5 | 65.3 |
| Log total IgE(geometric mean) | 2.0 ± 0.7 (100.6) | 1.3 ± 0.7 (17.9) | NA |
| FEV1 (%) | 79.7 ± 21.5 | 98.1 ± 11.4 | NA |
| FVC (%) | 88.9 ± 17.6 | 99.8 ± 11.5 | NA |
| FEV1/FVC ratio | 0.73 ± 0.13 | 0.82 ± 0.08 | NA |
FVC, Forced vital capacity; NA, not applicable.
FIG E1.
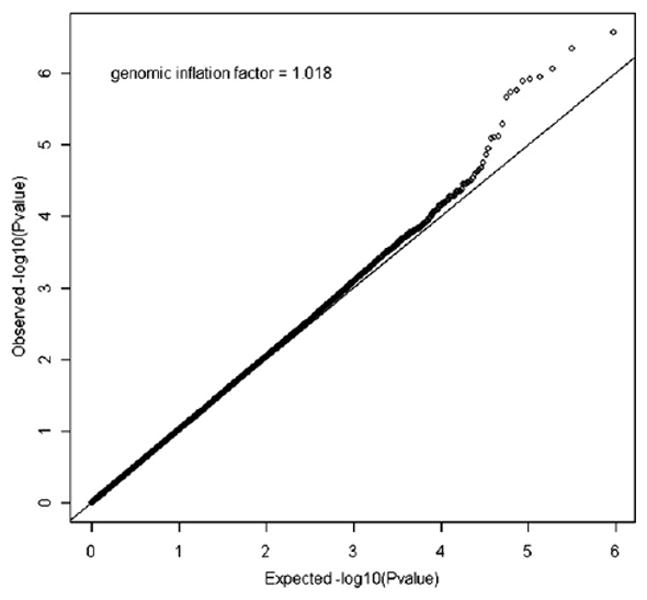
QQ-plot of the GWAS of asthma in SARP/CSGA/CAG. Negative log-transformed expected P values are shown on the x-axis. Negative log-transformed observed P values are shown on the y-axis.
A total of 59 SNPs had P values of 1.0 × 10−4 or less in all cases and control subjects (Fig 1 and see Table E2 in this article's Online Repository at www.jacionline.org). The TNIP1 region on 5q32-q33.1 had the strongest evidence for association with multiple SNPs strongly associated with asthma susceptibility: rs1422673 (P = 3.44 × 10−7; odds ratio [OR], 0.63) and rs10036748 (P =1.41 ×10−6; OR, 0.68; Table I and see Table E3 and Fig E2 in this article's Online Repository at www.jacionline.org). In the GABRIEL3 and EVE4 studies, rs1422673 was associated with asthma (P = .018 and 1.31 × 10−5 for meta-analysis, respectively), whereas in TENOR the SNP was not significant (P = .18), but the same trend was observed.6 The IL-1 receptor–like 2 (IL1RL2) and IL-1 receptor type I (IL1R1) region on 2q12 had SNPs that were strongly associated with asthma in the SARP/CSGA/CAG population (Table I) and replicated in both the TENOR6 and GABRIEL3 studies (Table I). SNPs in solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (SLC22A4); colony stimulating factor 2 (granulocyte-macrophage; CSF2); and IL-3 (colony-stimulating factor, multiple; IL3) region on 5q23-q31 was associated with asthma in SARP/CSGA/CAG (Table I). However, the significant SNPs in the SARP/CSGA/CAG population were different from those reported in the TENOR6 or GABRIEL3 studies. The ADAM metallopeptidase with thrombospondin type 1 motif, 14 (ADAMTS14), on 10q21 was associated with asthma (rs2791189: P = 5.73E-07; OR, 0.69) in SARP/CSGA/CAG but was not replicated in the GABRIEL3 or TENOR6 studies (Table I). To exclude the potential false-positive results caused by the difference between phenotyped control subjects and Illumina control subjects, P values of these SNPs were reported as well for the analysis in all cases and SARP/CSGA/CAG phenotyped control subjects only (see Table E2 in this article's Online Repository at www.jacionline.org). The results with or without Illumina control subjects were largely comparable. For example, rs1422673 was the highest ranked SNP in both analyses (P = 3.44 × 10−7 and P = 5.59 × 10−6, see Table E2).
FIG 1.
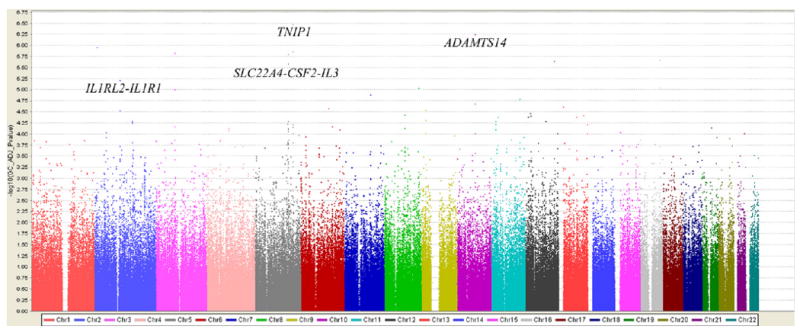
Genome-wide association of 474,271 SNPs in 813 SARP/CSGA/CAG cases, 553 SARP/CSGA/CAG control subjects, and 1,011 Illumina control subjects. The color scale of the x-axis represents chromosomes. Negative log-transformed genomic control–adjusted P values are shown on the y-axis.
TABLE E2.
Association results of 59 SNPs with genomic control–adjusted P values of 1.0E-04 or less in SARP/CSGA/CAG
| SNP | Chromosome | BP | Gene | Location | MAF | OR | L95 | U95 | Unadjusted P value* | GC-adjusted P valuey† | GC-adjusted P value(phenotypedcontrolsubjects) ‡ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1422673 | 5 | 150419181 | TNIP1 | Intron | T (0.1983) | 0.63 | 0.53 | 0.75 | 2.72E-07 | 3.44E-07 | 5.59E-06 |
| rs2791189 | 10 | 72187212 | ADAMTS14 | Intron | C (0.3613) | 0.69 | 0.60 | 0.80 | 4.56E-07 | 5.73E-07 | 4.17E-04 |
| rs11690527 | 2 | 12099276 | LPIN1 | Flanking_3UTR | T (0.3486) | 1.36 | 1.20 | 1.53 | 8.99E-07 | 1.12E-06 | 6.23E-05 |
| rs10036748 | 5 | 150438339 | TNIP1 | Intron | T (0.2603) | 0.68 | 0.58 | 0.79 | 1.14E-06 | 1.41E-06 | 6.50E-06 |
| rs7630534 | 3 | 74707813 | CNTN3 | Flanking_5UTR | A (0.2179) | 0.67 | 0.57 | 0.79 | 1.22E-06 | 1.50E-06 | 4.76E-04 |
| rs2073838 | 5 | 131677121 | SLC22A4 | Intron | A (0.08193) | 1.79 | 1.41 | 2.26 | 1.29E-06 | 1.59E-06 | 9.71E-04 |
| rs17656178 | 16 | 77758680 | WWOX | Intron | G (0.4623) | 0.73 | 0.64 | 0.83 | 1.79E-06 | 2.20E-06 | 1.58E-03 |
| rs10850603 | 12 | 115234120 | THRAP2 | Flanking_5UTR | C (0.06305) | 1.89 | 1.46 | 2.46 | 1.89E-06 | 2.31E-06 | 4.57E-04 |
| rs2306772 | 5 | 131703880 | SLC22A4 | Intron | A (0.08186) | 1.76 | 1.40 | 2.23 | 2.17E-06 | 2.65E-06 | 1.13E-03 |
| rs3755285 | 2 | 102210452 | IL1RL2 | Intron | G (0.267) | 1.41 | 1.21 | 1.63 | 5.13E-06 | 6.19E-06 | 2.48E-05 |
| rs11150140 | 16 | 77757915 | WWOX | Intron | A (0.4567) | 0.75 | 0.66 | 0.85 | 7.64E-06 | 9.15E-06 | 3.48E-03 |
| rs7465467 | 8 | 136056074 | ZNF406 | Flanking_5UTR | A (0.1134) | 1.56 | 1.28 | 1.90 | 7.96E-06 | 9.53E-06 | 1.42E-01 |
| rs11928037 | 3 | 74698943 | CNTN3 | Flanking_5UTR | G (0.2224) | 0.69 | 0.59 | 0.81 | 8.27E-06 | 9.89E-06 | 2.55E-03 |
| rs7779057 | 7 | 106382121 | PIK3CG | Flanking_3UTR | T (0.2034) | 1.42 | 1.22 | 1.66 | 1.11E-05 | 1.32E-05 | 3.15E-04 |
| rs7113799 | 11 | 112051515 | NCAM1 | Flanking_5UTR | T (0.06755) | 1.73 | 1.35 | 2.22 | 1.38E-05 | 1.64E-05 | 4.68E-03 |
| rs946742 | 10 | 72185697 | ADAMTS14 | Intron | C (0.4226) | 0.74 | 0.65 | 0.85 | 1.78E-05 | 2.10E-05 | 5.26E-04 |
| rs9581191 | 13 | 18556933 | TUBA2 | Flanking_3UTR | T (0.161) | 1.47 | 1.23 | 1.76 | 2.13E-05 | 2.51E-05 | 7.83E-03 |
| rs6935864 | 6 | 111497037 | SLC16A10 | Flanking_5UTR | C (0.2417) | 0.72 | 0.62 | 0.84 | 2.26E-05 | 2.66E-05 | 1.02E-02 |
| rs2129626 | 9 | 17061162 | C9orf39 | Flanking_5UTR | C (0.2769) | 0.73 | 0.63 | 0.85 | 2.46E-05 | 2.89E-05 | 2.17E-04 |
| rs12619383 | 2 | 102108150 | IL1R1 | Flanking_5UTR | G (0.1818) | 0.68 | 0.57 | 0.81 | 2.55E-05 | 2.99E-05 | 4.88E-05 |
| rs1427988 | 12 | 20204704 | PDE3A | Flanking_5UTR | G (0.2528) | 0.73 | 0.62 | 0.84 | 2.92E-05 | 3.42E-05 | 4.19E-04 |
| rs4415363 | 8 | 81282744 | TPD52 | Flanking_5UTR | T (0.342) | 1.33 | 1.16 | 1.53 | 3.21E-05 | 3.76E-05 | 8.97E-04 |
| rs4772201 | 13 | 98884260 | PHGDHL1 | Flanking_3UTR | G (0.1699) | 0.69 | 0.58 | 0.82 | 3.31E-05 | 3.87E-05 | 8.66E-06 |
| rs2955503 | 12 | 22090132 | CMAS | Flanking_5UTR | G (0.3733) | 0.75 | 0.65 | 0.86 | 3.34E-05 | 3.91E-05 | 8.80E-04 |
| rs4448642 | 11 | 27399727 | LGR4 | Intron | C (0.3378) | 1.33 | 1.16 | 1.52 | 3.56E-05 | 4.16E-05 | 2.31E-03 |
| rs10467606 | 13 | 70319802 | DACH1 | Flanking_3UTR | G (0.06247) | 1.71 | 1.33 | 2.21 | 3.59E-05 | 4.20E-05 | 1.27E-05 |
| rs12318822 | 12 | 12793539 | CDKN1B | Flanking_3UTR | G (0.3629) | 0.75 | 0.65 | 0.86 | 3.60E-05 | 4.20E-05 | 7.50E-04 |
| rs2774633 | 9 | 17060633 | C9orf39 | Flanking_5UTR | G (0.2789) | 0.74 | 0.64 | 0.85 | 4.30E-05 | 5.01E-05 | 3.61E-04 |
| rs4705893 | 5 | 131103820 | KIAA1961 | Intron | C (0.09512) | 1.57 | 1.26 | 1.94 | 4.49E-05 | 5.22E-05 | 7.42E-03 |
| rs1529757 | 12 | 80924312 | PPFIA2 | Flanking_5UTR | C (0.2726) | 1.35 | 1.17 | 1.56 | 4.51E-05 | 5.25E-05 | 4.66E-04 |
| rs11024979 | 11 | 19062621 | MRGPRX2 | Flanking_5UTR | T (0.1346) | 1.48 | 1.23 | 1.78 | 4.54E-05 | 5.28E-05 | 3.09E-04 |
| rs331125 | 2 | 150578696 | FLJ32955 | Flanking_5UTR | G (0.367) | 0.75 | 0.66 | 0.86 | 4.54E-05 | 5.28E-05 | 4.69E-03 |
| rs732227 | 2 | 150586238 | FLJ32955 | Flanking_5UTR | C (0.3885) | 1.32 | 1.15 | 1.51 | 4.90E-05 | 5.70E-05 | 6.76E-04 |
| rs17671387 | 5 | 130911895 | RAPGEF6 | Intron | G (0.07597) | 1.63 | 1.29 | 2.07 | 5.20E-05 | 6.04E-05 | 8.35E-03 |
| rs2702628 | 11 | 19503418 | NAV2 | Flanking_5UTR | A (0.387) | 1.31 | 1.15 | 1.50 | 5.24E-05 | 6.08E-05 | 3.95E-04 |
| rs2702646 | 11 | 19503096 | NAV2 | Flanking_5UTR | C (0.3868) | 1.31 | 1.15 | 1.50 | 5.25E-05 | 6.09E-05 | 3.95E-04 |
| rs3792783 | 5 | 150435925 | TNIP1 | Intron | C (0.1707) | 0.69 | 0.57 | 0.82 | 5.26E-05 | 6.11E-05 | 1.10E-04 |
| rs7330442 | 13 | 113656958 | FAM70B | Flanking_5UTR | T (0.1576) | 1.45 | 1.21 | 1.74 | 5.27E-05 | 6.12E-05 | 1.56E-03 |
| rs17012697 | 3 | 74755261 | CNTN3 | Flanking_5UTR | T (0.2066) | 0.71 | 0.60 | 0.84 | 5.97E-05 | 6.91E-05 | 1.78E-02 |
| rs1777220 | 6 | 126064295 | HEY2 | Flanking_5UTR | A (0.4218) | 1.31 | 1.15 | 1.50 | 5.98E-05 | 6.93E-05 | 1.41E-02 |
| rs3792785 | 5 | 150431843 | TNIP1 | Intron | G (0.1163) | 0.64 | 0.51 | 0.79 | 6.11E-05 | 7.07E-05 | 6.74E-05 |
| rs11672303 | 19 | 38418215 | SLC7A10 | Flanking_5UTR | C (0.1228) | 0.65 | 0.52 | 0.80 | 6.22E-05 | 7.20E-05 | 5.33E-04 |
| rs7814057 | 8 | 81289480 | TPD52 | Flanking_5UTR | C (0.3636) | 1.32 | 1.15 | 1.51 | 6.44E-05 | 7.45E-05 | 1.35E-03 |
| rs25879 | 5 | 131434943 | CSF2 | Flanking_5UTR | G (0.2217) | 1.37 | 1.17 | 1.60 | 6.45E-05 | 7.47E-05 | 2.89E-03 |
| rs12640395 | 4 | 87242418 | MAPK10 | Intron | A (0.1843) | 1.41 | 1.19 | 1.66 | 6.55E-05 | 7.58E-05 | 4.00E-04 |
| rs16936115 | 11 | 19065259 | MRGPRX2 | Flanking_5UTR | C (0.06542) | 1.68 | 1.30 | 2.17 | 6.79E-05 | 7.85E-05 | 3.90E-02 |
| rs2073506 | 5 | 131422637 | IL3 | Flanking_5UTR | A (0.09108) | 1.56 | 1.25 | 1.95 | 6.90E-05 | 7.97E-05 | 5.54E-03 |
| rs7768635 | 6 | 154869553 | CNKSR3 | Intron | T (0.1906) | 1.41 | 1.19 | 1.66 | 7.01E-05 | 8.09E-05 | 5.23E-02 |
| rs10766527 | 11 | 19057312 | MRGPRX2 | Flanking_5UTR | G (0.1374) | 1.46 | 1.21 | 1.76 | 7.10E-05 | 8.20E-05 | 5.62E-03 |
| rs11943376 | 4 | 87246843 | MAPK10 | Intron | T (0.1851) | 1.40 | 1.19 | 1.66 | 7.20E-05 | 8.31E-05 | 4.14E-04 |
| rs7721296 | 5 | 131643865 | PDLIM4 | Flanking_3UTR | C (0.09907) | 1.54 | 1.24 | 1.91 | 7.61E-05 | 8.78E-05 | 3.85E-03 |
| rs797700 | 2 | 47801605 | MSH6 | Flanking_5UTR | G (0.2472) | 1.36 | 1.17 | 1.58 | 8.17E-05 | 9.42E-05 | 9.10E-03 |
| rs11854309 | 15 | 22554134 | SNRPN | Flanking_5UTR | G (0.4144) | 0.77 | 0.67 | 0.88 | 8.19E-05 | 9.43E-05 | 1.70E-04 |
| rs7489746 | 13 | 113637032 | FAM70B | Intron | A (0.1676) | 1.42 | 1.19 | 1.70 | 8.58E-05 | 9.88E-05 | 4.85E-04 |
| rs2541232 | 10 | 72190936 | ADAMTS14 | Flanking_3UTR | C (0.4674) | 0.76 | 0.67 | 0.87 | 8.59E-05 | 9.89E-05 | 2.90E-03 |
| rs13285126 | 9 | 17367629 | C9orf39 | Intron | A (0.24) | 0.74 | 0.63 | 0.86 | 8.60E-05 | 9.90E-05 | 1.69E-03 |
| rs12422767 | 12 | 128241376 | KIAA1944 | Intron | T (0.1978) | 0.71 | 0.60 | 0.84 | 8.65E-05 | 9.95E-05 | 9.45E-04 |
| rs2233287 | 5 | 150420290 | TNIP1 | Intron | T (0.1011) | 0.62 | 0.49 | 0.79 | 8.65E-05 | 9.96E-05 | 1.28E-04 |
| rs13049896 | 21 | 42169257 | PRDM15 | Intron | A (0.09277) | 1.56 | 1.25 | 1.95 | 8.67E-05 | 9.98E-05 | 1.03E-02 |
BP, Base pair position based on hg18; L95, lower 95% CI; MAF, minor allele frequency; U95, upper 95% CI;UTR, untranslated region.
Unadjusted GWAS P values of 813 SARP/CSGA/CAG cases versus 1564 control subjects.
Genomic control–adjusted P values of 813 SARP/CSGA/CAG cases versus 1564 control subjects.
Genomic control–adjusted P values of 813 SARP/CSGA/CAG cases versus 553 phenotyped control subjects.
TABLE E3.
Association results of 27 SNPs in TNIP1 on chromosome 5
| No. | SNP | BP | Location | MAF | OR (95% CI) | P value* | ||
|---|---|---|---|---|---|---|---|---|
| SARP/CSGA/CAG | TENOR6 | GABRIEL3 | ||||||
| 1 | rs2277940 | 150389670 | Flanking_3 | C (0.072) | ||||
| 2 | rs3763009 | 150392333 | Intron | T (0.348) | ||||
| 3 | rs9324672 | 150400249 | Intron | A (0.124) | ||||
| 4 | rs2233299 | 150405660 | Coding | T (0.262) | 0.79 (0.68-0.92) | 2.67E-03 | .49 | .35 |
| 5 | rs11747787 | 150407061 | Intron | A (0.462) | ||||
| 6 | rs4292439 | 150408388 | Intron | C (0.184) | 1.22 (1.03-1.44) | .021 | .51 | .37 |
| 7 | rs2233297 | 150409433 | Intron | G (0.065) | 1.38 (1.07-1.78) | .016 | .23 | .78 |
| 8 | rs7713223 | 150410889 | Intron | C (0.135) | ||||
| 9 | rs7713567 | 150411148 | Intron | T (0.345) | 0.80 (0.70-0.92) | 1.93E-03 | .52 | .016 |
| 10 | rs888989 | 150411223 | Intron | C (0.199) | ||||
| 11 | rs2112635 | 150412346 | Intron | C (0.333) | 0.77 (0.67-0.89) | 4.25E-04 | .32 | .43 |
| 12 | rs871269 | 150412581 | Intron | T (0.318) | ||||
| 13 | rs17111695 | 150412639 | Intron | C (0.170) | ||||
| 14 | rs12516176 | 150415838 | Intron | C (0.268) | ||||
| 15 | rs1422673 | 150419181 | Intron | T (0.198) | 0.63 (0.53-0.75) | 3.44E-07 | .18 | .018 |
| 16 | rs2233287 | 150420290 | Intron | T (0.101) | 0.62 (0.49-0.79) | 9.96E-05 | .15 | .035 |
| 17 | rs17728260 | 150423198 | Intron | G (0.082) | ||||
| 18 | rs1422674 | 150425802 | Intron | G (0.089) | 0.78 (0.61-0.99) | .042 | .65 | .77 |
| 19 | rs3792789 | 150426161 | Intron | C (0.395) | ||||
| 20 | rs4958881 | 150430429 | Intron | C (0.135) | 0.67 (0.54-0.82) | 1.58E-04 | .38 | .11 |
| 21 | rs3792785 | 150431843 | Intron | G (0.116) | 0.64 (0.51-0.79) | 7.07E-05 | .27 | .095 |
| 22 | rs3792783 | 150435925 | Intron | C (0.171) | 0.69 (0.57-0.82) | 6.11E-05 | .69 | .13 |
| 23 | rs10036748 | 150438339 | Intron | T (0.260) | 0.68 (0.58-0.79) | 1.41E-06 | .41 | .11 |
| 24 | rs918498 | 150439981 | Intron | T (0.117) | 0.77 (0.63-0.96) | .020 | .97 | .68 |
| 25 | rs13168551 | 150442831 | Flanking_5 | C (0.424) | 0.79 (0.69-0.90) | 5.51E-04 | .37 | .12 |
| 26 | rs3792780 | 150446386 | Flanking_5 | G (0.386) | ||||
| 27 | rs3762999 | 150449619 | Flanking_5 | A (0.443) | ||||
Only entries with P values of less than .05 in SARP/CSGA/CAG are shown.
BP, Base pair position based on hg18; MAF, minor allele frequency.
Genomic control–adjusted P value for SARP/CSGA/CAG, TENOR, and GABRIEL.
FIG E2.
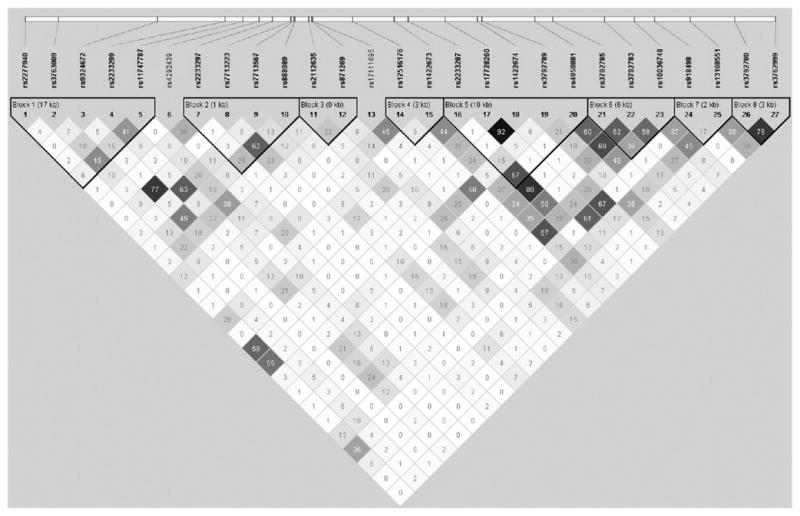
LD plot of 27 SNPs in TNIP1 regions. The r2 color scheme was used and labeled. 95% CIs on D′ were used to set up blocks.
Most consistently replicated SNPs in asthma
Six genomic loci have been identified consistently in a variety of GWASs of asthma and asthma-related traits: the ORMDL3-GSDMB region,2-4 IL33,3-5 the IL1RL1-IL18R1 region,3-5 the RAD50-IL13 region,3,6 the TSLP-WDR36 region,3-5,7 and the HLA-DR/DQ region.3,6,7 One SNP that was consistently associated with asthma in the SARP/CSGA/CAG, TENOR, and GABRIEL studies was selected from each of the regions: GSDMB (rs2872507), IL33 (rs3939286), IL1RL1 (rs13431828), IL13 (rs20541), TSLP (rs1837253), and HLA-DRA (rs2395185, Table II). Joint analyses of the 6 SNPs were performed in the SARP/CSGA/CAG and TENOR populations separately (see Table E4 in this article's Online Repository at www.jacionline.org). These SNPs explained the very limited variance of asthma risk either individually (percentage of deviance, 0.05% to 0.78%) or combined (percentage of deviance explained, 1.56% to 1.85%; area under the receiver operating characteristic curve, 0.581-0.592).
TABLE II.
Six SNPs in the 6 most consistently replicated genes/regions from the GWAS of asthma
| SNP | Gene | Chromosome | BP | Location | Risk/minorallele | P value | ||
|---|---|---|---|---|---|---|---|---|
| SARP/CSGA/CAG | TENOR6 | GABRIEL3 | ||||||
| rs2872507 | GSDMB | 17 | 35294289 | Flanking_3 | G/A | .006 | .054 | 5.70E-16 |
| rs3939286 | IL33 | 9 | 6200099 | Flanking_5 | A/A | .0018 | .094 | 4.53E-14 |
| rs13431828 | IL1RL1 | 2 | 102321085 | UTR | C/T | .0062 | .027 | 1.00E-10 |
| rs1837253 | TSLP | 5 | 110429771 | Flanking_5 | C/T | .00076 | .053 | 3.03E-10 |
| rs20541 | IL13 | 5 | 132023863 | Coding | T/T | .23 | .000417 | 1.95E-08 |
| rs2395185 | HLA-DRA | 6 | 32541145 | Flanking_3 | T/T | .00077 | .00015 | 1.84E-04 |
BP, Base pair position based on hg18; UTR, untranslated region.
TABLE E4.
Prediction of asthma susceptibility using 6 SNPs in GSDMB, IL33, IL1RL1, TSLP, IL13, and HLA-DRA
| SNP | Gene | SARP/CSGA/CAG | TENOR | AUC | |||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| P value | Deviance (%)* | AUC | P value | Deviance (%) | |||
| rs2872507 | GSDMB | .0063 | 0.25% | .11 | 0.12% | ||
|
| |||||||
| rs3939286 | IL33 | .033 | 0.15% | .061 | 0.17% | ||
|
| |||||||
| rs13431828 | IL1RL1 | 2.43E-04 | 0.46% | .03 | 0.23% | ||
|
| |||||||
| rs1837253 | TSLP | 4.78E-04 | 0.41% | .13 | 0.11% | ||
|
| |||||||
| rs20541 | IL13 | .22 | 0.05% | .0016 | 0.47% | ||
|
| |||||||
| rs2395185 | HLA-DRA | 3.84E-04 | 0.42% | 6.47E-05 | 0.78% | ||
|
| |||||||
| All 6 SNPs† | 1.12E-09 | 1.76% | 0.59 | 8.97E-07 | 1.85% | 0.592 | |
|
| |||||||
| Genetic Score‡ | 1.31E-11 | 1.56% | 0.581 | 7.65E-09 | 1.60% | 0.582 | |
AUC, Area under the receiver operating characteristic curve.
Percentage of deviance explained by SNPs in logistic regression model adjusted for age and sex.
All 6 SNPs were included in the logistic regression model.
A single score determined by counting the number of risk SNPs of 6 SNPs.
Comparison of asthma and autoimmune diseases
SNPs identified by means of GWASs of both asthma and autoimmune diseases were compared to elucidate common genetic factors for immune-mediated diseases (Table III3,6,20-30 and Fig 2). The candidate genes reported by GWASs of asthma based on the NIH GWAS database19 and the published literatures are ORMDL3-GSDMB,2-4 IL33,3-5 IL1RL1-IL18R1,3-5 RAD50-IL13,3,6 TSLP-WDR36 region,3-5,7 HLA-DR/DQ,3,6,7 PDE4D,31 TLE4,32 DENND1B,33 ADRA1B,34 PRNP,34 DPP10,34 SMAD3,3 IL2RB,3 RORA,3 SLC22A5,3 PYHIN1,4 NOTCH4,7 USP38-GAB,7 GATA3,7 IKZF4,7 IL6R,20 and C11orf30-LRRC32.20 The reported top SNPs in the above asthma candidate genes were searched on the NIH GWAS database19 and the published GWASs of autoimmune diseases. Data were included in Table III if the same SNPs were significant (P < 1 × 10−5) in both GWASs of asthma and autoimmune diseases.
TABLE III.
Comparison of IL13, TNIP1, GSDMB, HLA-DRA, SMAD3, LRRC32, and IKZF4 from the GWAS of asthma and autoimmune diseases
| Gene/SNP | Minor/major allele | GWASs | Function | |
|---|---|---|---|---|
| IL13 | Asthma (TENOR6 and GABRIEL3) | Psoriasis21 | TH2 | |
| rs20541 | T/C | T is minor risk allele | T is minor protective allele | |
| TNIP1 | Asthma (SARP/CSGA/CAG and GABRIEL3) | SLE25,26 and systemic sclerosis27 | Inflammation | |
| rs10036748 | T/C (white subjects) | T is minor protective allele | T is minor risk allele (white subjects)25,27 | |
| GSDMB | C/T (Chinese subjects) | NAAsthma (SARP/CSGA/CAG and TENOR6 and GABRIEL3) | T is major risk allele (Chinese subjects)26RA,22 CD,23 and UC24 | Inflammation? |
| rs2872507 | A/G | A is minor protective allele | A is minor risk allele | |
| HLA-DRA | Asthma (SARP/CSGA/CAG and TENOR6 and GABRIEL3) | UC28 | Antigenpresentation | |
| rs2395185 | T/G | T is minor risk allele | T is minor protective allele | |
| SMAD3 | Asthma (SARP/CSGA/CAG and GABRIEL3) | CD29 | Treg | |
| rs16950687 | G/A | G is minor risk allele | NA | |
| rs17293632 | T/C | NA | T is minor risk allele | |
| LRRC32 | Asthma20 | CD23 | ||
| rs7130588 | G/A | G is minor risk allele | NA | |
| rs7927894 | T/C | NA | T is minor risk allele | Treg? |
| IKZF4 | Asthma7 | T1D30 | ||
| rs1701704 | G/T | G is minor risk allele | G is minor risk allele | Treg? |
CD, Crohn disease; NA, not applicable; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; T1D, type I diabetes; Treg, regulatory T; UC, ulcerative colitis.
FIG 2.
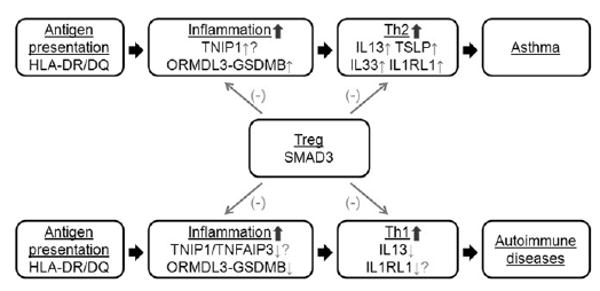
Comparison of GWASs of asthma and autoimmune diseases. Red upward-pointing arrows indicate higher gene expression levels or protein activities. Green downward-pointing arrows indicate the lower gene expression levels or protein activities. Question marks indicate the lack of available experimental evidence.
Minor allele T of rs20541 in IL13 is the risk allele for asthma in the TENOR and GABRIEL studies3,6 but the protective allele for psoriasis.21 The minor allele A of rs2872507 in GSDMB is the protective allele for asthma in the SARP/CSGA/CAG, TENOR, and GABRIEL studies3,6 but the risk allele for rheumatoid arthri-tis,22 Crohn disease,23 and ulcerative colitis.24 The T allele of rs10036748 in TNIP1 is the minor protective allele for asthma in the SARP/CSGA/CAG and GABRIEL studies3 but the minor or major risk allele for SLE in non-Hispanic white25 or Chinese26 populations, respectively. The T allele of rs2233287 (in weak LD with rs10036748: r2 = 0.29) is the minor protective allele for asthma in the SARP/CSGA/CAG study (see Table E3 and Fig E2) but the minor risk allele for systemic sclerosis.27 The minor allele T of rs2395185 in HLA-DRA is the risk allele for asthma in the SARP/CSGA/CAG, TENOR, and GABRIEL studies3,6 but the protective allele for ulcerative colitis.28
SNPs were not always in opposite effect directions for asthma and autoimmune diseases. For example, genetic association showed the same direction in asthma and autoimmune diseases for SMAD3 (Table III): the minor allele G of rs16950687 is the risk allele for asthma in the SARP/CSGA/CAG and GABRIEL studies3; the minor allele T of rs17293632 (in strong LD with rs16950687: r2 = 0.88) is the risk allele for Crohn disease.29 Similarly, the G allele of rs7130588in C11orf30-LRRC32 is the minor risk allele for asthma20; the T allele of rs7927894 (in perfect LD with rs7130588) is the minor risk allele for Crohn disease.23 The same risk allele has been shown to be associated with atopic dermatitis,35 allergic rhinitis,36 and increased total serum IgE levels.37 In addition, the G allele of rs1701704 in IKZF4 is the minor risk allele for asthma7 and type I diabetes.30SMAD3, LRRC32, and IKZF4 are all related to the regulatory T-cell pathway, which is the common negative regulatory process for the TH1 and TH2 pathways. Thus the same association direction of these genes is not contradictory with the opposite immunopathogenesis direction between asthma and autoimmune diseases.
In summary, genes involved in TH1/TH2 balance (IL13), the inflammation process (GSDMB and TNIP1), and antigen presentation (HLA-DRA) showed opposite genetic association direction between asthma and autoimmune diseases; however, genes involved in the regulatory T-cell pathway (SMAD3, C11orf30-LRRC32, and IKZF4) showed the same genetic association direction (Table III and Fig 2).
Discussion
In this study we identified a new candidate gene for asthma risk, TNIP1 (Table I and Table E3). The most significant SNP, rs1422673 (in intron 5 of TNIP1), was replicated in the published GABRIEL and EVE studies (P =.018 and 1.31 × 10−5 for meta-analysis, respectively) but not in the TENOR study (P = .18 but in the same direction). Because the results from the EVE consortium meta-analysis4 included the SARP/CSGA/CAG dataset, it should not be considered a totally independent replication. Because of the relatively weak replication of TNIP1, further confirmation studies are needed in independent populations.
Published GWASs of a variety of autoimmune diseases support a role of TNIP1 in immune-mediated diseases. GWASs of SLE in a Chinese Han population26 or North Americans of European descent25 identified rs10036748 (in intron 1 of TNIP1), which was confirmed in a replication study38; this SNP was also associated with asthma in our study and was in moderate LD with rs1422673 (r2 = 0.67, Table I and see Fig E2). GWASs of psoriasis in European21 and Chinese39 populations identified rs17728338 (12 kb upstream of TNIP1); this SNP is neither in LD with rs10036748 nor associated with asthma in our study (data not shown). Two distinct signals in the TNIP1 region are associated with SLE/asthma and psoriasis, respectively, suggesting that the regulatory mechanisms of TNIP1 expression might be transcription factor specific, tissue specific, or both. The presence of more than 1 signal in a genomic region is not uncommon. The most significant SNPs in the 2q12 region were located near IL1RL2 and IL1R1 (Table I and see Table E2) in our study but near IL1RL1 and IL18R1 in the GABRIEL study.3 Similarly, the most significant SNPs in the 5q23.3 region were located near SLC22A4 in our study (Table I and see Table E2) but near SLC22A5 in the GABRIEL study.3 Thus careful sequencing of these gene cluster regions is essential to identify all causal variants.
TABLE I.
Association results of the top SNPs in SARP/CSGA/CAG populations with asthma
| SNP | Gene | Chromosome | BP | Location | MAF | OR (95% CI) | P value* | ||
|---|---|---|---|---|---|---|---|---|---|
| SARP/CSGA/CAG | TENOR6 | GABRIEL3 | |||||||
| rs1422673 | TNIP1 | 5 | 150419181 | Intron | T (0.198) | 0.63 (0.53-0.75) | 3.44E-07 | .18 | .018 |
| rs10036748 | TNIP1 | 5 | 150438339 | Intron | T (0.260) | 0.68 (0.58-0.79) | 1.41E-06 | .41 | .11 |
| rs2791189 | ADAMTS14 | 10 | 72187212 | Intron | C (0.361) | 0.69 (0.60-0.80) | 5.73E-07 | .16 | .54 |
| rs946742 | ADAMTS14 | 10 | 72185697 | Intron | C (0.423) | 0.74 (0.65-0.85) | 2.10E-05 | .73 | .74 |
| rs3755285 | IL1RL2 | 2 | 102210452 | Intron | G (0.267) | 1.41 (1.21-1.63) | 6.19E-06 | .022 | .023 |
| rs12619383 | IL1R1 | 2 | 102108150 | Flanking_5 | G (0.182) | 0.68 (0.57-0.81) | 2.99E-05 | .2 | 2.16E-05 |
| rs2073838 | SLC22A4 | 5 | 131677121 | Intron | A (0.082) | 1.79 (1.41-2.26) | 1.59E-06 | .39 | .51 |
| rs2306772 | SLC22A4 | 5 | 131703880 | Intron | A (0.082) | 1.76 (1.40-2.23) | 2.65E-06 | .41 | .48 |
BP, Base pair position based on hg18; MAF, minor allele frequency.
Genomic control–adjusted P values for SARP/CSGA/CAG, TENOR, and GABRIEL.
At least 2 GWASs of asthma and related traits have identified the ORMDL3-GSDMB region,2-4IL33,3-5 the IL1RL1-IL18R1 re-gion,3-5 the RAD50-IL13 region,3,6 the TSLP-WDR36 region,3-5,7 and the HLA-DR/DQ region.3,6 In this study we confirmed that these 6 regions are associated with asthma susceptibility in both the SARP/CSGA/CAG and TENOR studies (Table II). We further analyzed the cumulative effect of the 6 most associated SNPs (in each of the 6 regions) on asthma risk in the SARP/CSGA/CAG and TENOR populations. Joint analyses indicated that the power to predict asthma susceptibility by using these 6 SNPs was limited (see Table E4), possibly because of the contributions of other small-effect common variants, large-effect rare variants, structural variants, epigenetic effects, gene-gene interaction, and gene-environment interaction. The consistent association and functional relevance of these 6 genes to asthma pathogenesis suggest that deep resequencing might be necessary to identify rare variants in addition to common variants and, more importantly, to determine the causal variants.
Asthma and autoimmune diseases share extensive immunologic pathways, but the comparison of shared GWAS-identified genes between asthma and autoimmune diseases is limited.8 To clarify common genetic factors for immune-mediated diseases, we compared risk and direction between shared SNPs identified by GWASs of asthma and autoimmune diseases (Table III and Fig 2).
The HLA region is the most consistent genomic locus identified for immune diseases and has an essential role in antigen presentation. rs2395185 in HLA-DRA was associated with asthma in the SARP/CSGA/CAG, TENOR, and GABRIEL studies,3,6 with the minor allele T associated with risk. In contrast, the rs2395185 T allele was protective for ulcerative colitis.28 A GWAS of pediatric asthma in Asian populations identified rs987870 (between HLA-DPA1 and HLA-DPB1) with the opposite direction of allelic effects from autoimmune diseases.40 One of the possible explanations of the opposite effect direction of HLA is that different antigen triggers might exist between asthma and autoimmune diseases (Table III and Fig 2).
The ORMDL3-GSDMB region is one of the most reproduced loci identified for immune diseases, with significant associations for asthma,3,6 rheumatoid arthritis,22 Crohn disease,23 and ulcerative colitis.24 The function of ORMDL3 or GSDMB is not totally determined, but a recent study indicated that ORMDL3 might regulate endoplasmic reticulum–mediated calcium signaling and the following unfolded-protein response and inflammation.41 Another functional study showed that variants in the ORMDL3-GSDMB region were associated with IL-17 secretion but not correlated with TH1/TH2 or regulatory T cytokines.42 An allele-specific chromatin remodeling study indicated that rs12936231 might be important for cis-regulation and common for asthma and autoimmune diseases.43 In addition, the same study observed that the risk alleles in the ORMDL3-GSDMB region were correlated with increased or decreased expression of ORMDL3 and GSDMB in patients with asthma or autoimmune diseases, respectively.43 In this study the minor allele A of rs2872507 (in moderate LD with rs12936231: r2 = 0.82) in GSDMB is the protective allele for asthma in the SARP/CSGA/CAG, TENOR, and GABRIEL studies3,6 but the risk allele for rheumatoid arthritis,22 Crohn disease,23 and ulcerative colitis24 (Table III and Fig 2).
TNIP1 is one of the most consistent genomic loci identified for autoimmune diseases.21,25,26,38,39 TNIP1 interacts with TNFAIP3 and inhibits the TNF-α–induced nuclear factor κB inflammatory pathway.44,45 The T allele of rs10036748 in TNIP1 is the protective allele for asthma in the SARP/CSGA/CAG and GABRIEL studies3 but the risk allele for SLE in non-Hispanic white25 and Chinese26 populations (Table III and Fig 2). The T allele of rs2233287 is the protective allele for asthma in SARP/CSGA/CAG (see Table E3 and Fig E2) but the risk allele for systemic sclerosis.27 Furthermore, reduced expression of the TNIP1 gene and protein was observed in lesional skin tissue from patients with systemic sclerosis.27 The opposite direction of effect for variants in TNIP1 and GSDMB indicates that the inflammation processes are disease specific for asthma and autoimmune diseases or gene-gene and gene-environment interaction might be involved.
The RAD50-IL13 region is one of the most consistent genomic loci identified for asthma3,6 and has an essential role in the TH2 pathway. rs20541 (Arg130Gln) in IL13 was associated with asthma in the TENOR and GABRIEL studies3,6 with the minor allele T as the risk allele. In contrast, the T allele of rs20541 was the protective allele for psoriasis in Europeans21 or Chinese subjects.39 IL-13 Arg130Gln has been shown to be functionally more active than wild-type IL-13 protein in inducing signal transducer and activator of transcription 6 phosphorylation, CD23 expression in monocytes, and IgE switching in B cells.46 The opposite effect direction for rs20541 supports the concept that asthma is TH2 driven but autoimmune diseases are TH1 driven (Table III and Fig 2).
SMAD3 is a transcription factor downstream of TGFB and important for regulatory T-cell and TH17 cell pathways.47,48 In the C11orf30-LRRC32 region C11orf30 might play a role in epithelial barrier function,35 and LRRC32 is a surface biomarker for regulatory T cells.49IKZF4 is a transcription factor essential for forkhead box protein 3–dependent gene silencing process in regulatory T cells.50 Genetic associations of SMAD3, C11orf30-LRRC32, and IKZF4 showed the same direction in asthma and autoimmune diseases, indicating that regulatory T cells might have similar effects by inhibiting either TH2 or TH1 pathways (Table III and Fig 2). Thus we can hypothesize that other genes involved in TH1/TH2 branching have opposite association direction between asthma and autoimmune diseases but genes in the regulatory T-cell pathway show the same genetic association direction.
The model presented here might be oversimplistic (Fig 2 and Table III). However, to avoid potential false-positive results, we only included SNPs significantly (P < 1.0 × 10−5) associated with both asthma and autoimmune diseases.19 Such comparison revealed that the risk alleles for genes involved in TH1/TH2 balance (IL13), inflammation (GSDMB and TNIP1), and antigen presentation (HLA-DRA) were reversed, whereas genes involved in the regulatory T-cell pathway (SMAD3, C11orf30-LRRC32, and IKZF4) showed the same effect direction. Similar genes were identified by using unbiased GWAS approaches for asthma and autoimmune diseases; however, different/opposite alleles appear to be the risk alleles. We hypothesize that the findings of a similar set of genes might help us understand the functional pathobiology involved in the development and progression of immune function disorders with very different disease characteristics. Further candidate gene studies and pathway analyses are warranted to extend these observations to study genes identified as important for autoimmune diseases and the effect directions for the risk alleles in subjects with asthma by using more relaxed criteria (P < .05).
There are exceptions to our model; for example, ulcerative colitis might be a TH2 disease instead of a TH1 disease.9 However, on the basis of the allele effect directions of HLA-DRA and GSDMB (Table III), ulcerative colitis is closer to other autoimmune diseases than asthma. Further cluster analysis, including asthma and autoimmune diseases based on shared SNPs, will help to tease out this relationship.
SNPs in TNIP1 were significantly associated with asthma in the SARP/CSGA/CAG population and was weakly associated in the GABRIEL and EVE studies but not in the TENOR study. TNIP1 is involved in the TNF-α–induced nuclear factor κB inflammation pathway. Inflammatory processes might differ between severe and mild-to-moderate asthma.51 For example, levels of the inflammatory cytokine TNF-α are increased in patients with severe/refractory corticosteroid-resistant asthma.52 Stratification by asthma severity might help to resolve any inconsistency with regard to the association of TNIP1 with asthma. Treatment with infliximab, an mAb against TNF-α, caused a decrease in the number of exacerbations in patients with moderate asthma.53 Treatment with etanercept, a soluble TNF-α receptor fusion protein, showed a reduction in sputum macrophage numbers and C-reactive protein levels in patients with corticosteroid-refractory asthma.54 Treatment with golimumab, a human mAb against TNF-α, did not demonstrate a favorable risk/benefit ratio in patients with severe persistent asthma.55 Stratification by SNPs in TNIP1, other TNF-a pathway genes, or both might be important to identify susceptible asthmatic patients in future anti–TNF-α drug clinical trials.
GWASs of asthma and asthma-related traits have consistently identified genes involved in antigen presentation (HLA-DR/DQ), inflammation (ORMDL3-GSDMB), and TH1/TH2 processes (IL33, IL1RL1-IL18R1, RAD50-IL13, and TSLP-WDR36). Pathway analysis using all genes in the biologically relevant pathways should be continued. Single SNPs in each of these 6 most consistent asthma genes can only explain limited variance, suggesting that the causal common/rare variants have not as yet been identified. Next-generation sequencing and study of the cumulative effects of all confirmed variants are necessary.
Key messages
SNPs in TNIP1 were associated with asthma.
Joint analyses on 6 SNPs in GSDMB, IL33, IL1RL1, IL13, TSLP, and HLA-DRA explained limited variance.
SNPs associated with both asthma and autoimmune diseases might have opposite effects on immunopathogenesis.
Acknowledgments
Severe Asthma Research Program (SARP) centers were supported by National Institutes of Health (NIH) grants HL69116, HL69130, HL69149, HL69155, HL69167, HL69170, HL69174, HL69349, UL1RR024992, M01RR018390, M01RR07122, M01RR03186, HL87665, and HL91762. Genetic studies for SARP and the Collaborative Studies on the Genetics of Asthma (CSGA) were funded by NIH grant HL87665. The STAMPEED study, including SARP/CSGA/Chicago Asthma Genetics Study data, was funded by NIH grant HL87665, HL72414, and HL101651. The clinical Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens study was supported by Genentech, Inc, and Novartis Pharmaceuticals Corp, and the genetic studies were funded by NIH grants HL76285 and HL87665.
We thank all investigators, staff, and participants in the SARP, TENOR, CSGA, and SARP/CSGA/CAG studies.
Abbreviations used
- CAG
Chicago Asthma Genetics Study
- CSGA
Collaborative Studies on the Genetics of Asthma
- GSDMB
Gasdermin B
- GWAS
Genome-wide association study
- IL1RL1
IL-1 receptor–like 1
- IL18R
IL-18 receptor 1
- LD
Linkage disequilibrium
- OR
Odds ratio
- ORMDL3
ORM1-like 3
- SARP
Severe Asthma Research Program
- SLE
Systemic lupus erythematosus
- SNP
Single nucleotide polymorphism
- TENOR
The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens
- TNIP1
TNFAIP3 interacting protein 1
- TSLP
Thymic stromal lymphopoietin
- WDR36
WD repeat domain 36
Footnotes
Disclosure of potential conflict of interest: W. C. Moore has been a subinvestigator on clinical trials supported by Aerovance, Amgen, AstraZeneca, Boehringer-Ingelheim, Centocor, Ception, Forest, Genentech, GlaxoSmithKline, MedImmune, Novartis, Pfizer, Sanofi Aventis, and Wyeth. W. W. Busse has received grants from the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) and the National Heart, Lung, and Blood Institute (NHLBI); is on a board for Merck; and has consulted for Amgen, AstraZeneca, Novartis, GlaxoSmithKline, MedImmune, and Genentech. D. L. Nicolae has received a grant from the NIH/NHLBI. C. Ober has received grants and travel support from the NIH and lecture honoraria from various entities. S. E. Wenzel has received grants from or has grants pending from Amgen, GlaxoSmithKline, MedImmune, Sanofi Aventis, and Genentech. The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.Bell GM, Reynolds G, Isaacs JD. Biologic therapies in non-rheumatic diseases: lessons for rheumatologists? Nat Rev Rheumatol. 2011;7:507–16. doi: 10.1038/nrrheum.2011.106. [DOI] [PubMed] [Google Scholar]
- 2.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 3.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–7. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010;125:328–35. e11. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–6. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 9.Fuss IJ. Is the Th1/Th2 paradigm of immune regulation applicable to IBD? Inflamm Bowel Dis. 2008;14(suppl 2):S110–2. doi: 10.1002/ibd.20683. [DOI] [PubMed] [Google Scholar]
- 10.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1:69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 11.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute's Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–13. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181:315–23. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Meyers DA, Ober C, Blumenthal MN, Mellen B, Barnes KC, et al. Genome-wide screen and identification of gene-gene interactions for asthma-susceptibility loci in three U.S. populations: collaborative study on the genetics of asthma. Am J Hum Genet. 2001;68:1437–46. doi: 10.1086/320589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolan CM, Fraher KE, Bleecker ER, Borish L, Chipps B, Hayden ML, et al. Design and baseline characteristics of the epidemiology and natural history of asthma: Outcomes and Treatment Regimens (TENOR) study: a large cohort of patients with severe or difficult-to-treat asthma. Ann Allergy Asthma Immunol. 2004;92:32–9. doi: 10.1016/S1081-1206(10)61707-3. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Howard TD, Moore WC, Ampleford EJ, Li H, Busse WW, et al. Importance of hedgehog interacting protein and other lung function genes in asthma. J Allergy Clin Immunol. 2011;127:1457–65. doi: 10.1016/j.jaci.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 18.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 19.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–14. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–9. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 26.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–7. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 27.Allanore Y, Saad M, Dieude P, Avouac J, Distler JH, Amouyel P, et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011;7:e1002091. doi: 10.1371/journal.pgen.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, et al. Ulcera-tive colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–20. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hakonarson H, Qu HQ, Bradfield JP, Marchand L, Kim CE, Glessner JT, et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008;57:1143–6. doi: 10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]
- 31.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–93. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY, et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in Mexican children. PLoS Genet. 2009;5:e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 34.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125:336–46. e4. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esparza-Gordillo J, Weidinger S, Folster-Holst R, Bauerfeind A, Ruschendorf F, Patone G, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet. 2009;41:596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- 36.Ramasamy A, Curjuric I, Coin LJ, Kumar A, McArdle WL, Imboden M, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Ampleford EJ, Howard TD, Moore WC, Li H, Busse WW, et al. The C11orf30-LRRC32 region is associated with total serum IgE levels in asthmatic patients. J Allergy Clin Immunol. 2012;129:575–8. e1–9. doi: 10.1016/j.jaci.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–33. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun LD, Cheng H, Wang ZX, Zhang AP, Wang PG, Xu JH, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42:1005–9. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noguchi E, Sakamoto H, Hirota T, Ochiai K, Imoto Y, Sakashita M, et al. Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genet. 2011;7:e1002170. doi: 10.1371/journal.pgen.1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19:111–21. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 42.Lluis A, Schedel M, Liu J, Illi S, Depner M, von Mutius E, et al. Asthma-associated polymorphisms in 17q21 influence cord blood ORMDL3 and GSDMA gene expression and IL-17 secretion. J Allergy Clin Immunol. 2011;127:1587–94. e6. doi: 10.1016/j.jaci.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 43.Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–93. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barnes PJ, Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–71. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 45.El Bakkouri K, Wullaert A, Haegman M, Heyninck K, Beyaert R. Adenoviral gene transfer of the NF-kappa B inhibitory protein ABIN-1 decreases allergic airway inflammation in a murine asthma model. J Biol Chem. 2005;280:17938–44. doi: 10.1074/jbc.M413588200. [DOI] [PubMed] [Google Scholar]
- 46.Vladich FD, Brazille SM, Stern D, Peck ML, Ghittoni R, Vercelli D. IL-13 R130Q, a common variant associated with allergy and asthma, enhances effector mechanisms essential for human allergic inflammation. J Clin Invest. 2005;115:747–54. doi: 10.1172/JCI22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, Gu H, et al. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 1999;18:1280–91. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez GJ, Zhang Z, Chung Y, Reynolds JM, Lin X, Jetten AM, et al. Smad3 differentially regulates the induction of regulatory and inflammatory T cell differentiation. J Biol Chem. 2009;284:35283–6. doi: 10.1074/jbc.C109.078238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM. GARP (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated FOXP3+ regulatory T cells. Proc Natl Acad Sci U S A. 2009;106:13445–50. doi: 10.1073/pnas.0901944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–6. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wenzel SE, Szefler SJ, Leung DY, Sloan SI, Rex MD, Martin RJ. Bronchoscopic evaluation of severe asthma. Persistent inflammation associated with high dose glucocorticoids. Am J Respir Crit Care Med. 1997;156:737–43. doi: 10.1164/ajrccm.156.3.9610046. [DOI] [PubMed] [Google Scholar]
- 52.Howarth PH, Babu KS, Arshad HS, Lau L, Buckley M, McConnell W, et al. Tumour necrosis factor (TNFalpha) as a novel therapeutic target in symptomatic corticosteroid dependent asthma. Thorax. 2005;60:1012–8. doi: 10.1136/thx.2005.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erin EM, Leaker BR, Nicholson GC, Tan AJ, Green LM, Neighbour H, et al. The effects of a monoclonal antibody directed against tumor necrosis factor-alpha in asthma. Am J Respir Crit Care Med. 2006;174:753–62. doi: 10.1164/rccm.200601-072OC. [DOI] [PubMed] [Google Scholar]
- 54.Morjaria JB, Chauhan AJ, Babu KS, Polosa R, Davies DE, Holgate ST. The role of a soluble TNFalpha receptor fusion protein (etanercept) in corticosteroid refractory asthma:a double blind, randomised, placebo controlled trial. Thorax. 2008;63:584–91. doi: 10.1136/thx.2007.086314. [DOI] [PubMed] [Google Scholar]
- 55.Wenzel SE, Barnes PJ, Bleecker ER, Bousquet J, Busse W, Dahlen SE, et al. A randomized, double-blind, placebo-controlled study of tumor necrosis factor-alpha blockade in severe persistent asthma. Am J Respir Crit Care Med. 2009;179:549–58. doi: 10.1164/rccm.200809-1512OC. [DOI] [PubMed] [Google Scholar]


