Abstract
Warfarin is the most commonly prescribed oral anticoagulant worldwide despite its narrow therapeutic index and the notorious inter- and intra-individual variability in dose required for the target clinical effect. Pharmacogenetic polymorphisms are major determinants of warfarin pharmacokinetic and dynamics and included in several warfarin dosing algorithms. This review focuses on warfarin pharmacogenomics in sub-Saharan peoples, African Americans and admixed Brazilians. These ‘Black’ populations differ in several aspects, notably their extent of recent admixture with Europeans, a factor which impacts on the frequency distribution of pharmacogenomic polymorphisms relevant to warfarin dose requirement for the target clinical effect. Whereas a small number of polymorphisms in VKORC1 (3673G > A, rs9923231), CYP2C9 (alleles *2 and *3, rs1799853 and rs1057910, respectively) and arguably CYP4F2 (rs2108622), may capture most of the pharmacogenomic influence on warfarin dose variance in White populations, additional polymorphisms in these, and in other, genes (e.g. CALU rs339097) increase the predictive power of pharmacogenetic warfarin dosing algorithms in the Black populations examined. A personalized strategy for initiation of warfarin therapy, allowing for improved safety and cost-effectiveness for populations of African descent must take into account their pharmacogenomic diversity, as well as socio-economical, cultural and medical factors. Accounting for this heterogeneity in algorithms that are ‘friendly’ enough to be adopted by warfarin prescribers worldwide requires gathering information from trials at different population levels, but demands also a critical appraisal of racial/ethnic labels that are commonly used in the clinical pharmacology literature but do not accurately reflect genetic ancestry and population diversity.
Keywords: African ancestry, CYP2C9, global pharmacogenomics, population diversity, VKORC1, warfarin algorithms
Introduction
‘Pharmacogenetics deals with pharmacological responses and their modification by hereditary influences’. This definition, offered by Werner Kalow in the first book dedicated to pharmacogenetics [1], highlights the three pillars of this discipline: pharmacology, genetics and human diversity. Pharmacogenetics has evolved greatly over the 50 years elapsed since Kalow's book was published, was re-christened as pharmacogenomics in the fashion of the ‘omics’ revolution, but its conceptual development and praxis remain contingent upon a better understanding of human genomic diversity and its impact on drug pharmacokinetics and pharmacodynamics. The human evolutionary history is remarkably short. There is powerful evidence that the anatomically modern man emerged in Africa around 150 000–200 000 years ago and spread to other continents probably within the last 70 000 years. Thus, from a genomic perspective, we are all African-descendants, whether living in Africa or in a quite recent exile outside of Africa [2]. However, the present review on pharmacogenomics of warfarin in populations of African descent adopts a less inclusive approach, and examines sub-Saharan Africans, African-Americans and self-identified Black or Brown (‘pardo’ in Portuguese) Brazilians. These ‘Black’ populations differ in several aspects, notably their extent of recent admixture (within the last 500 years) with non-Africans, primarily Europeans. Estimates of European biogeographical ancestry based on autosomal genetic markers reveal that the average proportion of European ancestry ranges between 44–73% in Brown Brazilians, 29–54% in Black Brazilians and 4–35% in African Americans, depending on the geographical regions of Brazil [3, 4] and the United States [5] in which the studies were conducted. It is noteworthy that the individual proportions of African ancestry vary over wide ranges, and most importantly, in a continuous manner within each of these admixed populations from the Americas [3, 4, 6, 7], a pattern that impacts on the frequency distribution of pharmacogenomic polymorphisms in these peoples [8, 9]. Recent European admixture may also be extensive in some sub-Saharan peoples, such as ‘Cape Mixed Ancestry’ (CMA) from Cape Town, South Africa, and similarly to admixed American populations, the mean European proportion in CMA (19%) is a poor representation of the large inter-individual range of variation (0–86%; [10]). Beyond European gene flow, high levels of admixture of distinct ancestral African clusters prevail in most sub-Saharan African populations, reflecting historical migration events across the continent [10]. Collectively, the findings presented above indicate the diversity and heterogeneity of the populations of African descent considered in this review, and should caution against lumping them together as an ethnic, racial or continental group and extrapolating pharmacogenetic data across these peoples. Finally, we would like to refer to important studies on the pharmacogenomics of warfarin in populations from Africa, such as Ethiopians [11] and Egyptians [12], or having biogeographical African ancestry, such as Hispanics in the United States [13], which are not included in this review.
Warfarin, a ‘global’ anticoagulant
Warfarin remains the most commonly prescribed oral anticoagulant worldwide for the prevention and treatment of thromboembolic events, despite the notorious inter- and intra-individual variability in the dose required for the target clinical effect, which is commonly assessed by the international normalized ratio (INR). INR values between 2 and 3–3.5 are recommended for most warfarin therapeutic indications. Warfarin is a racemic mixture of R- and S-enantiomers, which differ in anticoagulant activity and are substrates of different cytochrome P-450 enzymes (CYPs). S-warfarin, the more potent enantiomer is metabolized by CYP2C9, whereas various CYPs contribute to the inactivation of R-warfarin. Both R- and S-warfarin affect the coagulation cascade by inhibiting the vitamin K epoxide reductase complex 1 (VKORC1) thereby preventing the carboxylation of clotting factors II, VII, IX and X (Figure 1). There is convincing evidence that genetic polymorphisms contribute to inter-individual differences in warfarin dose requirements, but the fraction of variability explained by pharmacogenetic polymorphisms varies considerably across populations. Figure 1 presents a schematic view of warfarin pharmacokinetics and pharmacodynamics and Tables 3 show the frequency distribution of genetic polymorphisms affecting these warfarin interactive pathways in the African populations examined in the present review.
Figure 1.
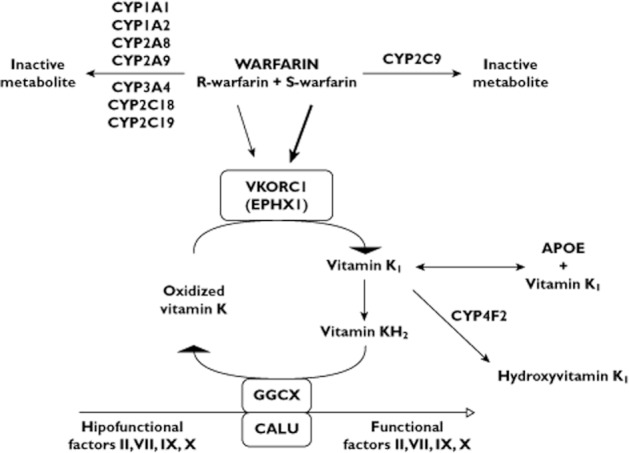
Warfarin interactive pathways. Warfarin R- and S- enantiomers are metabolized by different CYP enzymes and affect the coagulation cascade by inhibiting the vitamin K epoxide reductase complex 1 (VKORC1), preventing the carboxylation of clotting factors II, VII, IX and X. S-warfarin is the more potent enantiomer, as indicated by the thicker arrow. The influence of polymorphisms in the genes encoding CYP2C9, VKORC1, EPXH1, APOE, GGCX, CALU, CYP4F2 and factor VII on warfarin dose requirements is discussed in the text
Table 3.
CYP4F2, APOE, GGCX, FVII, CALU and EPHX1 polymorphisms in populations of African descent
| Polymorphisms | Populations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs# | Nucleotide change | Effect | African | African-American | Brazilian | ||||||
| Cohort (n) | Frequency (%) | Reference | n | Frequency (%) | Reference | Cohort (n) | Frequency (%) | Reference | |||
| CYP4F2 | |||||||||||
| rs2108622 | 1347C > T | V433M | Mozambican (206) | 8.7 | [58] | 717 | 7.2–11.7 | [24, 40, 58, 63] | Black (74) | 14.9 | [48] |
| YRI (113) | 5.8 | 61 | Brown (112) | 22.8 | [48] | ||||||
| APOE | |||||||||||
| rs429358 + | ε2 | 328 | 11.5, 12.6 | [15, 24] | |||||||
| rs7412 | ε3 | 328 | 66.2, 69.1 | [15, 24] | |||||||
| ε4 | 328 | 19.4, 21.2 | [15, 24] | ||||||||
| GGCX | |||||||||||
| rs10654848 | (CAA)8, (CAA)9 | intron 6 | 338 | 2.1, 0.1 | [27] | ||||||
| (CAA)10, (CAA)11 | 338 | 33.1, 13.9 | [27] | ||||||||
| (CAA)12, (CAA)13 | 338 | 17.6, 11.8 | [27] | ||||||||
| (CAA)14, (CAA)15 | 338 | 13.6, 5.0 | [27] | ||||||||
| (CAA)16, (CAA)17 | 338 | 2.5, 0.1 | [27] | ||||||||
| FVII | |||||||||||
| rs510335 | −401G > T | 5'UTR | 112 | 29.9 | 59 | Black (69) | 18.0 | [50] | |||
| Brown (108) | 21.0 | [50] | |||||||||
| rs510317 | −402G > A | 5'UTR | Black (69) | 21.0 | [50] | ||||||
| Brown (108) | 19.0 | [50] | |||||||||
| CALU | |||||||||||
| rs339097 | 10587T > C | intron 4 | YRI (113) | 18.6 | * | 435 | 10.4–16.3 | [18, 40, 61] | |||
| Mozambican (75) | 18.5 | * | |||||||||
| EPHX1 | |||||||||||
| rs2292566 | 26857G > A | K119K | YRI (113) | 17.7 | 61 | 216 | 6.8–15.1 | [32, 40, 61] | Black (73) | 12.3 | † |
| rs2740170 | 32001C > T | intron 3 | YRI (113) | 11.5 | 61 | 71 | 11.4, 19.4 | [32, 61] | |||
| rs2740171 | 32732C > A | intron 3 | YRI (113) | 38.5 | 61 | 71 | 31.6, 31.8 | [32, 61] | |||
| rs1051741 | 1071C > T | N357N | YRI (113) | 11.1 | 61 | 71 | 7.1, 9.1 | [32, 61] | |||
n, number of individuals.
Damasceno & Suarez-Kurtz, unpublished observations.
Suarez-Kurtz, unpublished observations
CYP2C9
The distribution of CYP2C9 polymorphisms among sub-Saharan populations is characterized by the absence or rarity (<1%) of the defective alleles *2 and *3, which are relatively common in Europeans. The frequency of these alleles in admixed African populations reflects their relative extent of European ancestry and increases progressively from African Americans and Brazilians with >80% African ancestry, to Black Brazilians and then to Brown Brazilians (Table 1). The low frequency of CYP2C9 *2 and *3 contributes to the poor performance of warfarin dosing algorithms which rely exclusively on these alleles as predictors of CYP2C9 phenotype in Africans [14–18]. Nevertheless, when present, CYP2C9 *2 and *3 associate with reduced warfarin requirements in populations of African descent: for example, Voora et al. [18] reported 20% and 34% reductions in warfarin dose per copy of alleles *2 and *3, respectively, in African American, whereas in Brown and Black Brazilians, the combined effect size of CYP2C9 *2 and *3 amounted to average reductions of 5.1 and 9.4 mg week–1 per variant allele, respectively [19]. A number of additional CYP2C9 variants, listed in Table 1, have been associated with warfarin dose requirements in different African cohorts. CYP2C9 *8, a 449G > A transition which leads to an arginine to histidine conversion [20] and lower S-warfarin clearance [21] accounts for significant reductions in warfarin dose in South-Africans [22], African Americans [23, 24] and Mozambicans (Damasceno & Suarez-Kurtz, unpublished observations). Notably, genotyping of CYP2C9 *8 alone prompted reclassification of the predicted metabolic phenotype in almost 10% of African-Americans, or when combined with CYP2C9 *5, *6 and *11, more than 15% [23]. CYP2C9 *5 and *11 are described in the CYP2C9 allele database nomenclature 25 as conferring decreased phenotypic activity, whereas CYP2C9 *6 has no description. However, CYP2C9 *6 was associated with decreased enzyme activity towards phenytoin, another CYP2C9 substrate [20]. A retrospective study of South African women associated warfarin dose requirement with CYP2C9 *11 and with a novel high frequency SNP, namely g.46028A > G, described as a possible splice site in intron 7. The combined effect of all CYP2C9 polymorphisms in this South African cohort was reported to explain 23.2% of the inter-individual variance in warfarin dose [22]. Resequencing of regions of evolutionary conservation and transcriptional binding prediction in CYP2C9 identified three novel, high frequency SNPs (rs2860905, rs7089580 and rs35511771) in African Americans, which on univariate analyses associated with warfarin requirements [26]. One of these SNPs, rs7089580, was retained in a multivariate model with other genetic and non-genetic covariates, accounting for a 3.7 mg week−1 increase in warfarin dose for each allele copy. This effect, which is in the opposite direction of the star CYP2C9 alleles, was not verified in another African American cohort [27].
Table 1.
CYP2C9 polymorphisms in populations of African descent
| Polymorphisms | Populations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs# | Nucleotide change | Effect | African | African-American | Brazilian | ||||||
| (allele) | Cohort (n) | Frequency (%) | Reference | n | Freqency (%) | Reference | Cohort (n) | Frequency (%) | Reference | ||
| rs1799853 | 430C > T (*2) | R144C | Beninese (103) | 0 | [20] | 1889 | 0–3.6 | [23, 26, 27, 29, 30, 40, 54–56] | >80% African ancestry (65) | 3.1 | [57] |
| Mozambican (103) | 0 | [58] | Black (756) | 6.6 | [19, 59] | ||||||
| South African (923) | 0 | [60] | Brown (816) | 8.2, 9.3 | [19, 59] | ||||||
| rs1057910 | 1075A > C (*3) | I359L | Beninese (103) | 0 | [20] | 1938 | 0.3–2.0 | [23, 26, 27, 29, 30, 40, 54–56, 61] | >80% African ancestry (65) | 2.3 | [57] |
| Mozambican (103) | 1.0 | [58] | Black (756) | 2.6, 3.3 | [19, 59] | ||||||
| South African (213) | 0.5 | [22] | Brown (816) | 5.1, 5.9 | [19, 59] | ||||||
| rs28371686 | 1080C > G (*5) | D360E | Beninese (103) | 1.8 | [20] | 1464 | 0.7–1.5 | [23, 26, 27, 29, 30, 55, 56] | >80% African ancestry (65) | 2.3 | [57] |
| Mozambican (103) | 1.9 | [58] | Black (756) | 0, 0.9 | [19, 59] | ||||||
| South African (213) | 0 | [22] | Brown (816) | 0.5, 0.8 | [19, 59] | ||||||
| rs9332131 | 818delA (*6) | 273 frameshift | Beninese (103) | 2.7 | [20] | 1051 | 0.4–1.7 | [23, 24, 29, 40, 55, 56] | |||
| Mozambican (103) | 0 | [58] | |||||||||
| South African (213) | 0.2 | [22] | |||||||||
| rs7900194 | 449G > A (*8) | R150H | Beninese (103) | 8.6 | [20] | 1111 | 4.7–7.2 | [23, 26, 27, 40] | Black (76) | 4.7 | [58] |
| Mozambican (103) | 10.8 | [58] | Brown (118) | 0.4 | [58] | ||||||
| South African (213) | 13.3 | [22] | |||||||||
| rs2256871 | 752A > G (*9) | H251R | Beninese (103) | 15.7 | [20] | ||||||
| South African (213) | 15.1 | [22] | |||||||||
| rs28371685 | 1003A > G (*11) | R335W | Beninese (103) | 2.7 | [20] | 1541 | 1.0–4.0 | [23, 26, 27, 29, 40, 55, 56, 61] | Black (756) | 0.7, 0.9 | [19, 59] |
| Mozambican (103) | 2.4 | [58] | Brown (816) | 0, 0.5 | [19, 59] | ||||||
| South African (213) | 3.9 | [22] | |||||||||
| rs2860905 | 3858G > A | intron 3 | YRI (111) | 26.6 | 61 | 378 | 22.0, 24.5 | [26, 61] | |||
| rs7089580 | 6787A > T | intron 3 | YRI (213) | 21.2 | 61 | 612 | 19.4–23.0 | [26, 27, 61] | |||
| 7880T > C | intron 3 | 329 | 2.0 | [26] | |||||||
| rs35511771 | 13706T > C | intron 5 | 329 | 24.0 | [26] | ||||||
| 46028A > G | intron 7 | South African (213) | 28.8 | [22] | |||||||
n, number of individuals.
VKORC1
VKORC1 encodes the vitamin K epoxide reductase subunit 1 (VKORC1), a key enzyme of the vitamin K cycle and molecular target of coumarin anticoagulants. The influence of VKORC1 polymorphisms on warfarin dose requirements provides a remarkable example of pharmacogenomic diversity worldwide. This is best documented by the original and the expanded International Warfarin Pharmacogenomic Consortium (IWPC) datasets, comprising 5700–6200 patients recruited from four continents and ascribed to three ‘racial’ groups, namely Asians, Blacks (predominantly African Americans) and Whites [17]. Univariate analyses showed that possession of the minor rs9923231 allele (3673A, also known as −1639A), which reduces hepatic VKORC1 expression, was associated with reduced warfarin dose requirements in the three groups. However, the percentage of inter-individual dose variation explained by this polymorphism was considerably lower in Blacks (4.2%) than in Whites (22.5%), a result which was largely explained by the lower frequency of rs9923231 in the Black study group (10.1%), compared with Whites (37.8%). A simulation exercise across the study populations showed that as the minor rs9923231 allele frequency increases, the percentage of variation in warfarin dose explained by this SNP also increases, with the highest variance explained at a frequency of 60–70% [17]. Accordingly, the low frequency of rs9923231 in sub-Saharan Africans (2.2–3.5%, Table 2) suggests that this SNP is a poor predictor of warfarin requirements in these populations. This inference may be extended to rs9934438, which is in extensive linkage disequilibrium (LD) with rs9923231 in sub-Saharan Africans [9]. European admixture coupled with the high frequency of these two SNPs in Europeans, account for rs9923231 and rs9934438 being considerably more common in African Americans (and consequently in the expanded IWPC Black cohort), Brown and Black Brazilians, than in sub-Saharan Africans. In Brazilians, the frequency distribution of rs9923231 is best described as a continuous function of the individual proportions of African ancestry, irrespective of self-reported colour or ‘race’ [9].
Table 2.
VKORC1 polymorphisms in populations of African descent
| Polymorphisms | Populations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs# | Nucleotide change | Effect | African | African-American | Brazilian | ||||||
| Cohort (n) | Frequency (%) | Reference | n | Frequency (%) | Reference | Cohort (n) | Frequency (%) | Reference | |||
| rs9923231 | 3673G > A | 5'UTR | Mozambican (143) | 3.5 | [9] | 1754 | 8.8–22.0 | [23, 26, 27, 31, 33, 40, 42, 56, 61] | Black (756) | 22.0, 23.8 | [19, 59] |
| Angolan (73) | 2.7 | [9] | Brown (816) | 30.6, 33.0 | [19, 59] | ||||||
| Black, 4 continents (368) | 10.1 | [17] | >80% African ancestry (68) | 21.1 | [9] | ||||||
| rs9934438 | 6484C > T | intron 1 | South African (993) | 4.0 | [60] | 1267 | 8.0–12.0 | [26, 29, 31, 40, 42, 49, 55, 61] | |||
| Beninese (51) | 2.0 | [62] | |||||||||
| Black, 4 continents (368) | 10.2 | [17] | |||||||||
| rs8050894 | 6853G > C | intron 2 | Mozambican (143) | 27.3 | [9] | 849 | 20.4–30.0 | [26, 29–32] | Black (756) | 35.7, 36.0 | [19, 59] |
| Black, 4 continents (368) | 26.4 | [17] | Brown (816) | 38.0, 38.6 | [19, 59] | ||||||
| Angolan (73) | 19.2 | [9] | >80% African ancestry (68) | 34.4 | [9] | ||||||
| rs2884737 | 5808T > G | intron 1 | Mozambican (143) | 1.1 | [9] | 698 | 3.5–7.1 | [24, 33, 55, 61] | Black (756) | 10.0, 12.8 | [19, 59] |
| Angolan (73) | 0 | [9] | Brown (816) | 13.5, 20.0 | [19, 59] | ||||||
| >80% African ancestry (68) | 10.7 | [9] | |||||||||
| rs7294 | 9041G > A | 3'UTR | South African (991) | 40.0 | [60] | 973 | 44.0–52.3 | [24, 26, 29, 32, 55, 61] | Black (756) | 37.9, 41.0 | [19, 59] |
| Mozambican (143) | 38.9 | [9] | Brown (816) | 37.2, 39.0 | [19, 59] | ||||||
| Angolan (73) | 40.4 | [9] | >80% African ancestry (68) | 46.9 | [9] | ||||||
| rs17886199 | 6915C > T | intron 2 | 258 | 3.7 | [33] | ||||||
| rs59502288 | 1989_1990insTGGATGA | 5'UTR | 329 | 42.0 | [26] | ||||||
| rs62057090 | 5374C > G | 5'UTR | 329 | 29.0 | [26] | ||||||
| rs61162043 | 6051G > A | 5'UTR | 570 | 47.0, 48.3 | [26, 27] | ||||||
| rs2359612 | 7566C > T | intron 2 | South African (974) | 24.0 | [60] | 804 | 15.9–23.3 | [26, 29, 32, 40, 61] | |||
| rs7200749 | 8773G > A | L120L | South African (113) | 25.7 | [22] | 353 | 18.2–26.5 | [32, 33, 61] | |||
| rs72547529 | 6557C > T | V66M | South African (110) | 0.4 | [22] | 509 | 0–1.6 | [23, 40, 55] | |||
n, number of individuals.
In addition to the tighly linked rs9923231 and rs9934438 SNPs, other VKORC1 polymorphisms have been associated with warfarin dose in patients of African descent. Lower warfarin requirements have been associated with rs8050894 and rs2884737 in an admixed Brazilian [19] and in some African American cohorts [24, 26], but these SNPs were not included in warfarin dosing algorithms derived for these groups. The rs8050894 was associated with warfarin dose in a multivariate model for Sudanese patients [28], but this was not verified in African Americans [29–31]. Two other VKORC1 polymorphisms, rs2359612 and rs61162043 were found to impact on warfarin dose in African American cohorts [24, 26, 27, 29, 32], whereas contradictory results were reported for rs17886199 [32, 33]. The rs61162043, located in the 5'UTR and not in LD with the other SNPs analyzed by Perera et al. [26] in an African American cohort, contributed 3.2% of inter-individual dose variation among patients.
VKORC1 variants associated with higher warfarin requirements include rs7200749 in South Africans [22] and rs7294 in an admixed Brazilian cohort [22], in African Americans [26] and in Sudanese [28]. These two SNPs were tightly linked in black South Africans, and explained 7.4% of warfarin dose variation in this group [22]. The association between rs7294 and warfarin dose was retained in multivariate modelling for Sudanese [28], but not African American patients [24, 26]. Two other variants, rs62057090 and rs59502288, were significantly associated with warfarin dose in African Americans in univariate, but not multivariate analyses [26]. A non-synonymous VKORC1 variant, V66M, conferred extreme warfarin resistance in one African-Caribbean [34], African Americans [23] and Black Brazilians [35], leading to the suggestion that inclusion of this SNP in genotyping panels might be considered for Black patients [23]. Another VKORC1 warfarin-resistant SNP, namely D36Y, prevalent in Ethiopians (MAF 15%; [36]) was not detected in Black South Africans [21] or African Americans [23].
Several reports assessed the extent of LD of VKORC1 polymorphisms in populations of African descent. Among rs9923231, rs2884737, rs8050894 and rs7294 no pairwise LD with a r2 > 0.5 was detected in Angolans and Mozambicans, whereas in Brown and Black Brazilians, rs9923231 and rs2884737 showed r2 values of 0.71 and 0.60, respectively [9]. Five haplotypes inferred from these four SNPs accounted for more 99% of the diversity in Angolans, Mozambicans and Brazilians with >80% African ancestry. Limdi et al. [33] reported that among 17 VKORC1 SNPs with minor allele frequency (MAF) >0.02 in African Americans, only two pairs (rs9923231 and rs9934438; rs17883591 and rs17886199) were in strong LD (r2 > 0.9). The strong LD between rs9923231 and rs9934438 was verified in another African American cohort [37]. The pairwise LD between all other SNPs identified by Limdi et al. [33] was weaker, with r2 values ranging from 0.2 to 0.6. These results include rs7294 and rs7200749 which are tightly linked in Black South Africans [22]. Twelve haplotypes comprising 11 SNPs with MAF > 0.08 accounted for 98.4% of the diversity in African Americans. Haplotypes inferred from common VKORC1 SNPs (MAF > 0.1) were equally informative as either rs9923231 and rs9934438 in capturing the genetic influence of VKORC1 in a cohort of Black individuals from four continents [17]. This, however, does not exclude the possibility that other SNPs, either per se or in haplotypes, might be more informative than rs9923231 and rs9934438 alone in distinct African-derived populations.
EPHX1
Microsomal epoxide hydrolase 1 (mEH), encoded by EPHX1, is another putative subunit of the vitamin K epoxide reductase complex (VKOR), which contains the vitamin K epoxide binding site. Two EPHX1 polymorphisms (rs2292566 and rs4653436) have been investigated in relation to warfarin dose requirements in Europeans, with controversial results and borderline associations at best (e.g. [38, 39]). No association between rs2292566 and warfarin dose was detected in African Americans [40] or Black Brazilians (Suarez-Kurtz, unpublished observations). Schelleman et al. [32] genotyped rs2292566 and 13 other SNPs in a small cohort of 22 African Americans and found that none associated with warfarin maintenance dose. The authors reported that two polymorphisms located in intron 3 (rs2740170 and rs2740171) and one exonic, synonymous SNP (rs1051741) were able to explain more than 10% of the dose variability (P > 0.05), and suggested that these SNPs should be retested in a study with larger sample sizes.
APOE
The main circulating form of vitamin K, phylloquinone, is bound to chylomicrons. The hepatic clearance of chylomicrons, and consequently vitamin K, is partly dependent on apolipoprotein E (APOE). Two variants in exon 4 of the APOE gene (rs429358 and rs7412), lead to formation of three alleles, ε2, ε3 and ε4, which are associated with low, intermediate and high vitamin K uptake by the liver, respectively. The frequency distribution of these APOE alleles varies across populations, ε3 being the most common in African populations. Cavallari et al. [41] observed that the time delay to achieve stable warfarin dose in African Americans was longer in patients with the APOE ε3/ε3 genotype, compared with other genotypes, and Kimmel et al. [15] reported that carriers of the ε4 allele required higher warfarin doses, but this was not replicated by others [24]. The APOE polymorphism has been included in some warfarin dosing algorithms for African Americans [42] (Table 4).
Table 4.
Warfarin pharmacogenomic algorithms for populations of African descent
| Study reference | VKORC1 | CYP2C9 alleles | Age | Body weight, BSA or BMI | Smoking status | INR | VTE/PE | Other factors | Cohort (n) | r2 or variation explained (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Momary et al. [30] | *2, *3, *5 | • | BSA | African American (115) | 33.0 | |||||
| Gage et al. [14] | 3673G > A | *2, *3 | • | BSA | • | INR | VTE | African American race | African American | |
| Co-medications | Derivation cohort (153) | 31.0 | ||||||||
| Validation cohort (45) | 40.0 | |||||||||
| Kimmel et al. [15] | 6484G > A | *2, *3 | • | BMI | Employment status | African American (111) | 54.6 | |||
| Co-medications | ||||||||||
| APOE (ε4) | ||||||||||
| Limdi et al. [33] | 3673G > A | *2, *3, *5, *6, *11 | • | BMI | • | Gender | African American (273) | 38.9 | ||
| Alcohol intake | ||||||||||
| Vitamin K intake | ||||||||||
| Co-morbidities | ||||||||||
| Co-medications | ||||||||||
| Perini et al. [19] | 3673G > A | *2, *3, *5, *11 | • | Body weight | VTE | Co-morbidities | Admixed Brazilians (390) | 50.4 | ||
| Co-medications | ||||||||||
| Schelleman et al. [49] | 6484G > A | • | BMI | Co-medications | African American (112) | 28.0 | ||||
| APOE (ε2, ε3, ε4) | ||||||||||
| FVII (−401G > T) | ||||||||||
| IWPC 16 | 3673G > A | *2, *3 | • | Body weight | Height | Black/African American (353) | 27.0 | |||
| Ethnicity | ||||||||||
| Suarez-Kurtz et al. [64] | 3673G > A | *2, *3, *5, *11 | • | Body weight | INR/dose | VTE | Co-morbidities | Admixed Brazilians (260) | 60.0 | |
| Co-medications | ||||||||||
| Cavallari et al. [24] | 3673G > A | *2, *3, *5, *6, *8, *11 | • | BSA | Co-morbidities | African American (226) | 36.4 | |||
| Voora et al. [18] | 3673G > A | *2, *3 | • | BSA | • | INR | VTE, PE | CALU (10587T > C) | African American (241) | – |
| Co-medications | ||||||||||
| Mitchell et al. [22] | 8773G > A, 9041G > A | *8, *9, *11 and 8 other alleles | • | Co-medications | South African (113) | 45.2 | ||||
| Perera et al. [26] | 6051G > A, 6484G > A | *2, *3, *5, *8, *11, 6787A > T | • | Body weight | VTE, PE | African American (330) | 40.0 | |||
| Shrif et al. [28] | 6853G > A, 9041G > A | *2, *5, *6, *11 | Body weight | INR | POL3S | Sudanese (203) | 36.7 | |||
| Co-medications | ||||||||||
| Ramirez et al. [40] | 3673G > A | *2, *3, *6, *8 | • | BSA | • | VTE | CALU (10587T > C) | African American (145) | 41.0 | |
| CYP4F2 (1347C > T) | ||||||||||
| Gender | ||||||||||
| Ethnicity | ||||||||||
| Co-medications | ||||||||||
| Co-morbidities |
BMI, body mass index; BSA, body surface area; PE, pulmonary embolism; VTE, venous thromboembolism.
GGCX
The protein encoded by GGCX, γ-glutamil carboxilase, is responsible for the post translational carboxylation of hypofunctional clotting factors II, VII, IX and X, with reduced vitamin K serving as a necessary cofactor. Three independent studies investigated the influence of GGCX polymorphisms in warfarin dose requirements in African-Americans [27, 32, 40]. The rs699664, a non-synonymous SNP in exon 8 was examined in all three studies and none verified the significant association between the minor allele rs699664 and higher warfarin dose requirements, previously reported for European and Asian patients [43, 44]. Of the other polymorphisms investigated in African Americans, only the rs10654848 microsatellite (CAA)n in intron 6 associated significantly with warfarin stable dose [27]. Carriers of either (CAA)16 or 17 repeat were over-represented among patients requiring warfarin doses higher than 7.5 mg day−1 vs. those who required lower doses. The GGCX rs10654848 genotype remained associated with warfarin dose in multivariable regression modelling, and was proposed as a predictor of higher than usual warfarin requirements in African Americans [27].
CALU
Calumenin is a chaperone protein found in the endoplasmic reticulum, where γ-glutamyl carboxylase and VKORC1 also localize. Calumenin exerts an inhibitory effect on the biosynthesis of the functional vitamin K-dependent clotting factors II, VII, IX and X [45]. A conserved intronic SNP (rs339097), rare in Europeans, but common in sub-Saharan Africans and African Americans (Table 3) was reported to associate with higher expression of CALU in human lymphoblastoid cells, but it is unclear whether this SNP directly affects calumenin expression or mRNA stability, or is in LD with causative SNP(s) within a large haplotype block [18]. Two studies have shown a significant association between the variant rs339097 allele and higher warfarin dose requirements in African American patients. In both studies, the association remained significant in multivariate regression modelling [18, 40].
CYP4F2
The enzyme encoded by the CYP4F2 gene catalyzes the conversion of vitamin K to hydroxyl vitamin K, acting as a counterpart to VKORC1 in limiting accumulation of vitamin K in hepatocytes. A genome-wide association in Europeans revealed an association of warfarin dose requirement with rs2108622 in CYP4F2, but only after adjusting for CYP2C9 and VKORC1 [46]. The frequency of rs2108622 differs markedly among populations, being considerably lower in sub-Saharan Africans and African Americans (0–9%) compared with Europeans and White North Americans (17–33%) 47. From a population perspective, it might be anticipated that rs2108622 will have a smaller influence, if any, on the warfarin dose requirements in Black individuals. The available data are consistent with this notion. No significant association between the CYP4F2 rs2108622 C > T genotype and warfarin dose was observed in African Americans [24, 40], Brown or Black Brazilians [48]. However, when rs2108622 was analyzed in a multivariate model in the overall Brazilian cohort, it was able to explain 1.1% of warfarin dose variability, an effect which the study authors considered too small to justify including this SNP in warfarin dosing algorithms for Brazilians [48].
FVII
Two common functional polymorphisms in the promoter region of the FVII gene, namely −401G > T and −402G > A have been investigated in relation to warfarin dose requirement in African-derived populations. In African Americans, the −401G > T genotype showed no statistically significant association with warfarin maintenance dose, but was nevertheless included in a dosing algorithm [49]. Neither −401G > T nor −402G > A associated with warfarin stable dose in an admixed Brazilian cohort, comprising 50% of self-identified Brown and Black individuals [50]. However, when the data were analyzed using the extreme discordant phenotype methodology [51] a significant difference emerged at the 5th percentile, such that the GG genotype was over-represented among patients at the low dose end of the warfarin dose distribution, compared with the high dose [50]. This result was not reproduced at higher cutoff points, suggesting that the impact of FVII −402G > A genotype on warfarin requirement is small, compared with VKORC1 and CYP2C9 polymorphisms.
Warfarin dosing algorithms
Several pharmacogenomic warfarin algorithms have been described and their performance, expressed by the correlation coefficient (r2) between predicted and prescribed doses ranges from 0.2 to 0.6, is consistently inferior in sub-Saharan Africans and African Americans, compared with European populations. This is best documented in studies enroling multiethnic cohorts, whether recruited worldwide (e.g. [16, 17]) or exclusively from the United States (e.g. [14, 49]). Indeed, Schelleman et al. [49] suggested that their pharmacogenetic dosing algorithms ‘performed only marginally better for African Americans when compared with giving 5 mg (warfarin) empirically’. This discrepant performance of warfarin algorithms in Caucasian vs. African Americans is not reversed by inclusion of a ‘race’ term in some algorithms [14, 16], which is not surprising, considering the complex interplay of genetic and non-genetic factors in modulating drug response, plus the fluctuation of racial definitions according to social context, geographic location, historical period and personal experience [52].
Table 4 compares the covariates and predictive performance of warfarin dosing algorithms tested in sub-Saharan Africans, African Americans and admixed Brazilians. All listed algorithms include polymorphisms in VKORC1 and CYP2C9, age and a measure for body mass. Most algorithms incorporate co-medication with CYP2C9 inhibitors (e.g. amiodarone) and inducers (e.g. carbamazepine), and some include African or Black ‘race’, gender, co-morbidities (notably thromboembolic conditions), smoking status, INR measurements and polymorphisms in other pharmacogenes (APOE, CALU, CYP4F2 and FVII). Whereas a small number of polymorphisms in CYP2C9 (alleles *2 and *3, rs1799853 and rs1057910, respectively), VKORC1 (3673G > A, rs9923231) and arguably CYP4F2 (rs2108622) may capture most of the pharmacogenomic influence on warfarin dose variance in White populations, additional pharmacogenetic variants increase the predictive power of warfarin dosing algorithms in sub-Saharan peoples and African Americans. This discordance is determined mainly by the absence or rarity of VKORC1 rs9923231, CYP2C9 *2 and *3 in Black populations (Tables 1 and 2). Other variants, notably CYP2C9 *8, replace CYP2C9 *2 and *3 as the major determinants of CYP2C9 metabolic phenotype in African-derived populations, and therefore must be incorporated in warfarin dosing algorithms for these peoples. Regarding VKORC1, no identified polymorphism(s) or inferred haplotype(s) account for as large a fraction of warfarin dose variance in Black populations as the tightly linked rs9923231 and rs9934438 do in European/Caucasian patients (see above). Although other pharmacogenetic variants have been found to associate with warfarin requirements in Blacks (e.g. CALU rs339097), their inclusion in dosing algorithms does not compensate for the reduced VKORC1 contribution, and consequently most algorithms explain only 27–41% of warfarin dose variation in sub-Saharan Africans and African Americans. Exceptions include one algorithm for Black South Africans (r2 = 45%; [22]) and another for African Americans (r2 = 54.6%; [15]). The latter algorithm was unique in comprising employment status as a covariate, and its higher predictive performance was not reproduced in subsequent studies in other African American cohorts.
Differently from North Americans, the performance of a warfarin dosing algorithm derived from an admixed Brazilian cohort, did not differ between White and Black patients, with r2 values of 51% and 52%, respectively [19]. Incorporation of an INR term to the algorithm increased its r2 to 60% in the overall cohort, irrespective of self-reported ‘race/colour’. The interplay of three factors may account for the distinct influence of ‘race’ or racial/ethnic labels on the performance of warfarin dosing algorithms in Brazilians vs. North Americans: (i) European gene flow is, on average, considerably larger in Black Brazilians (Introduction), (ii) self-reported race/colour correlates poorly with biogeographical genetic ancestry in Brazilians [3, 4], and (iii) distinct cultural semantic criteria impact the definition and self-adoption of racial/colour definitions across time and geographical space.
An alternative approach to the development of warfarin dosing algorithms, based on combining genome-wide association (GWA) studies with machine learning techniques, has been recently shown to improve the predictive performance in African Americans [53]. The peak performance (r2 = 66.4%) was obtained with a model that incorporated 200 SNPs identified by GWA in the study cohort. The adoption of this model in a natural clinical setting is uncertain.
A personalized strategy for initiation of warfarin therapy, allowing for improved safety and cost-effectiveness for the Black populations examined in this review must take into account their pharmacogenomic heterogeneity. Socio-economical, cultural and medical factors must also be considered, including access to and reliability of warfarin formulations, compliance to prescription, diet, concurrent diseases and co-medications, etc. Accounting for this diversity in algorithms that are ‘friendly’ enough to be adopted by warfarin prescribers worldwide requires gathering information from trials at different population levels, but demands also a critical appraisal of racial/ethnic labels that are commonly used in the clinical pharmacology literature but do not accurately reflect genetic ancestry and population diversity.
Competing Interests
There are no competing interests to declare.
G.S-K. is funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (Faperj) and Financiadora de Estudos e Projetos (Finep).
References
- 1.Kalow W. Pharmacogenetics, Heredity and the Response to Drugs. Philadelphia, PA: W.B Saunders Co; 1962. [Google Scholar]
- 2.Pääbo S. The mosaic that is our genome. Nature. 2003;421:409–412. doi: 10.1038/nature01400. [DOI] [PubMed] [Google Scholar]
- 3.Pena SD, Di Pietro G, Fuchshuber-Moraes M, Genro JP, Hutz MH, Kehdy FeS, Kohlrausch F, Magno LA, Montenegro RC, Moraes MO, de Moraes ME, de Moraes MR, Ojopi EB, Perini JA, Racciopi C, Ribeiro-Dos-Santos AK, Rios-Santos F, Romano-Silva MA, Sortica VA, Suarez-Kurtz G. The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS ONE. 2011;6:e17063. doi: 10.1371/journal.pone.0017063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suarez-Kurtz G. Pharmacogenetics in the Brazilian population. Front Pharmacogenet. 2010;1:1–10. doi: 10.3389/fphar.2010.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parra EJ, Suarez-Kurtz G. Admixture in North America. In: Suarez-Kurtz G Austin., editor. Pharmacogenomics in Admixed Populations. TX: Landes Bioscience; 2007. pp. 28–46. [Google Scholar]
- 6.Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo JM, Wambebe C, Tishkoff SA, Bustamante CD. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci U S A. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parra EJ, Kittles RA, Shriver MD. Implications of correlations between skin color and genetic ancestry for biomedical research. Nat Genet. 2004;36:S54–60. doi: 10.1038/ng1440. [DOI] [PubMed] [Google Scholar]
- 8.Suarez-Kurtz G, Perini JA, Bastos-Rodrigues L, Pena SD, Struchiner C. Impact of population admixture on the distribution of the CYP3A5*3 polymorphism. Pharmacogenomics. 2007;8:1299–1306. doi: 10.2217/14622416.8.10.1299. [DOI] [PubMed] [Google Scholar]
- 9.Suarez-Kurtz G, Amorim A, Damasceno A, Hutz MH, de Moraes MO, Ojopi EB, Pena SD, Perini JA, Prata MJ, Ribeiro-dos-Santos A, Romano-Silva MA, Teixeira D, Struchiner CJ. VKORC1 polymorphisms in Brazilians: comparison with the Portuguese and Portuguese-speaking Africans and pharmacogenetic implications. Pharmacogenomics. 2010;11:1257–1267. doi: 10.2217/pgs.10.89. [DOI] [PubMed] [Google Scholar]
- 10.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, Ibrahim M, Juma AT, Kotze MJ, Lema G, Moore JH, Mortensen H, Nyambo TB, Omar SA, Powell K, Pretorius GS, Smith MW, Thera MA, Wambebe C, Weber JL, Williams SM. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loebstein R, Dvoskin I, Halkin H, Vecsler M, Lubetsky A, Rechavi G, Amariglio N, Cohen Y, Ken-Dror G, Almog S, Gak E. A coding VKORC1 Asp36Tyr polymorphism predisposes to warfarin resistance. Blood. 2007;109:2477–2480. doi: 10.1182/blood-2006-08-038984. [DOI] [PubMed] [Google Scholar]
- 12.Shahin MH, Khalifa SI, Gong Y, Hammad LN, Sallam MT, El Shafey M, Ali SS, Mohamed ME, Langaee T, Johnson JA. Genetic and nongenetic factors associated with warfarin dose requirements in Egyptian patients. Pharmacogenet Genomics. 2011;21:130–135. doi: 10.1097/FPC.0b013e3283436b86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavallari LH, Momary KM, Patel SR, Shapiro NL, Nutescu E, Viana MA. Pharmacogenomics of warfarin dose requirements in Hispanics. Blood Cells Mol Dis. 2011;46:147–150. doi: 10.1016/j.bcmd.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, Milligan PE, Grice G, Lenzini P, Rettie AE, Aquilante CL, Grosso L, Marsh S, Langaee T, Farnett LE, Voora D, Veenstra DL, Glynn RJ, Barrett A, McLeod HL. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimmel SE, Christie J, Kealey C, Chen Z, Price M, Thorn CF, Brensinger CM, Newcomb CW, Whitehead AS. Apolipoprotein E genotype and warfarin dosing among Caucasians and African Americans. Pharmacogenomics J. 2008;8:53–60. doi: 10.1038/sj.tpj.6500445. [DOI] [PubMed] [Google Scholar]
- 16.Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MTM, Limdi NA, Page D, Roden DM, Wagner MJ, Caldwell MD, Johnson JA, Chen YT, Wen MS, Caraco Y, Achache I, Blotnick S, Muszkat M, Shin JG, Kim HS, Suarez-Kurtz G, Perini JA, Silva-Assuncao E, Andereson JL, Horne BD, Carlquist JF, Berg RL, Burmester JK, Goh BC, Lee SC, Kamali F, Sconce E, Daly AK, Wu AHB, Langaee TY, Feng H, Cavallari L, Momary K, Pirmohamed M, Jorgensen A, Toh CH, Williamson P, McLeod H, Evans JP, Weck KE, Brensinger C, Nakamura Y, Mushiroda T, Veenstra D, Meckley L, Rieder MJ, Rettie AE, Wadelius M, Melhus H, Stein CM, Schwartz U, Kurnik D, Deych E, Lenzini P, Eby C, Chen LY, Deloukas P, Motsinger-Reif A, Sagreiya H, Srinivasan BS, Lantz E, Chang T, Ritchie M, Lu LS. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limdi NA, Wadelius M, Cavallari L, Eriksson N, Crawford DC, Lee MT, Chen CH, Motsinger-Reif A, Sagreiya H, Liu N, Wu AH, Gage BF, Jorgensen A, Pirmohamed M, Shin JG, Suarez-Kurtz G, Kimmel SE, Johnson JA, Klein TE, Wagner MJ, Consortium IWP. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115:3827–3834. doi: 10.1182/blood-2009-12-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voora D, Koboldt DC, King CR, Lenzini PA, Eby CS, Porche-Sorbet R, Deych E, Crankshaw M, Milligan PE, McLeod HL, Patel SR, Cavallari LH, Ridker PM, Grice GR, Miller RD, Gage BF. A polymorphism in the VKORC1 regulator calumenin predicts higher warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:445–451. doi: 10.1038/clpt.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perini JA, Struchiner CJ, Silva-Assuncao E, Santana ISC, Rangel F, Ojopi EB, Dias-Neto E, Suarez-Kurtz G. Pharmacogenetics of warfarin: development of a dosing algorithm for Brazilian patients. Clin Pharmacol Ther. 2008;84:722–728. doi: 10.1038/clpt.2008.166. [DOI] [PubMed] [Google Scholar]
- 20.Allabi AC, Gala JL, Horsmans Y. CYP2C9, CYP2C19, ABCB1 (MDR1) genetic polymorphisms and phenytoin metabolism in a Black Beninese population. Pharmacogenet Genomics. 2005;15:779–786. doi: 10.1097/01.fpc.0000174787.92861.91. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Jeong H, Takahashi H, Drozda K, Patel SR, Shapiro NL, Nutescu EA, Cavallari LH. Decreased warfarin clearance associated with the CYP2C9 R150H (*8) polymorphism. Clin Pharmacol Ther. 2012;91:660–665. doi: 10.1038/clpt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell C, Gregersen N, Krause A. Novel CYP2C9 and VKORC1 gene variants associated with warfarin dosage variability in the South African black population. Pharmacogenomics. 2011;12:953–963. doi: 10.2217/pgs.11.36. [DOI] [PubMed] [Google Scholar]
- 23.Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. CYP2C9*8 is prevalent among African-Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10:1243–1255. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavallari LH, Langaee TY, Momary KM, Shapiro NL, Nutescu EA, Coty WA, Viana MA, Patel SR, Johnson JA. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:459–464. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 25.The Human Cytochrome P450 (CYP) Allele Nomenclature Database. CYP2C9 allele nomenclature. Available at http://www.cypalleles.ki.se/cyp2c9.htm (last accessed 18 May 2012)
- 26.Perera MA, Gamazon E, Cavallari LH, Patel SR, Poindexter S, Kittles RA, Nicolae D, Cox NJ. The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther. 2011;89:408–415. doi: 10.1038/clpt.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cavallari LH, Perera M, Wadelius M, Deloukas P, Taube G, Patel SR, Aquino-Michaels K, Viana MA, Shapiro NL, Nutescu EA. Association of the GGCX (CAA)16/17 repeat polymorphism with higher warfarin dose requirements in African Americans. Pharmacogenet Genomics. 2012;22:152–158. doi: 10.1097/FPC.0b013e32834f288f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shrif NE, Won HH, Lee ST, Park JH, Kim KK, Kim MJ, Kim S, Lee SY, Ki CS, Osman IM, Rhman EA, Ali IA, Idris MN, Kim JW. Evaluation of the effects of VKORC1 polymorphisms and haplotypes, CYP2C9 genotypes, and clinical factors on warfarin response in Sudanese patients. Eur J Clin Pharmacol. 2011;67:1119–1130. doi: 10.1007/s00228-011-1060-1. [DOI] [PubMed] [Google Scholar]
- 29.Limdi NA, Arnett DK, Goldstein JA, Beasley TM, McGwin G, Adler BK, Acton RT. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European-Americans and African-Americans. Pharmacogenomics. 2008;9:511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Momary KM, Shapiro NL, Viana MA, Nutescu EA, Helgason CM, Cavallari LH. Factors influencing warfarin dose requirements in African-Americans. Pharmacogenomics. 2007;8:1535–1544. doi: 10.2217/14622416.8.11.1535. [DOI] [PubMed] [Google Scholar]
- 31.Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadée W. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood. 2008;112:1013–1021. doi: 10.1182/blood-2008-03-144899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schelleman H, Brensinger CM, Chen J, Finkelman BS, Rieder MJ, Kimmel SE. New genetic variant that might improve warfarin dose prediction in African Americans. Br J Clin Pharmacol. 2010;70:393–399. doi: 10.1111/j.1365-2125.2010.03709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Limdi NA, Beasley TM, Crowley MR, Goldstein JA, Rieder MJ, Flockhart DA, Arnett DK, Acton RT, Liu N. VKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African-Americans and European-Americans. Pharmacogenomics. 2008;9:1445–1458. doi: 10.2217/14622416.9.10.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrington DJ, Underwood S, Morse C, Shearer MJ, Tuddenham EG, Mumford AD. Pharmacodynamic resistance to warfarin associated with a Val66Met substitution in vitamin K epoxide reductase complex subunit 1. Thromb Haemost. 2005;93:23–26. doi: 10.1160/TH04-08-0540. [DOI] [PubMed] [Google Scholar]
- 35.Orsi FA, Annichino Bizzacchi JM, de Paula EV, Ozelo MC, Langley MR, Weck KE. VKORC1 V66M mutation in African Brazilian patients resistant to oral anticoagulant therapy. Thromb Res. 2010;126:e206–210. doi: 10.1016/j.thromres.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Aklillu E, Leong C, Loebstein R, Halkin H, Gak E. VKORC1 Asp36Tyr warfarin resistance marker is common in Ethiopian individuals. Blood. 2008;111:3903–3904. doi: 10.1182/blood-2008-01-135863. [DOI] [PubMed] [Google Scholar]
- 37.Crawford DC, Brown-Gentry K, Rieder MJ. VKORC1 common variation and bone mineral density in the Third National Health and Nutrition Examination Survey. PLoS ONE. 2010;5:e15088. doi: 10.1371/journal.pone.0015088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pautas E, Moreau C, Gouin-Thibault I, Golmard JL, Mahe I, Legendre C, Taillandier-Heriche E, Durand-Gasselin B, Houllier AM, Verrier P, Beaune P, Loriot MA, Siguret V. Genetic factors (VKORC1, CYP2C9, EPHX1, and CYP4F2) are predictor variables for warfarin response in very elderly, frail inpatients. Clin Pharmacol Ther. 2010;87:57–64. doi: 10.1038/clpt.2009.178. [DOI] [PubMed] [Google Scholar]
- 39.Ciccacci C, Paolillo N, Di Fusco D, Novelli G, Borgiani P. EPHX1 polymorphisms are not associated with warfarin response in an Italian population. Clin Pharmacol Ther. 2011;89:791. doi: 10.1038/clpt.2011.31. [DOI] [PubMed] [Google Scholar]
- 40.Ramirez AH, Shi Y, Schildcrout JS, Delaney JT, Xu H, Oetjens MT, Zuvich RL, Basford MA, Bowton E, Jiang M, Speltz P, Zink R, Cowan J, Pulley JM, Ritchie MD, Masys DR, Roden DM, Crawford DC, Denny JC. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012;13:407–418. doi: 10.2217/pgs.11.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavallari LH, Butler C, Langaee TY, Wardak N, Patel SR, Viana MA, Shapiro NL, Nutescu EA. Association of apolipoprotein E genotype with duration of time to achieve a stable warfarin dose in African-American patients. Pharmacotherapy. 2011;31:785–792. doi: 10.1592/phco.31.8.785. [DOI] [PubMed] [Google Scholar]
- 42.Schelleman H, Chen Z, Kealey C, Whitehead AS, Christie J, Price M, Brensinger CM, Newcomb CW, Thorn CF, Samaha FF, Kimmel SE. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007;81:742–747. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- 43.Kimura R, Miyashita K, Kokubo Y, Akaiwa Y, Otsubo R, Nagatsuka K, Otsuki T, Okayama A, Minematsu K, Naritomi H, Honda S, Tomoike H, Miyata T. Genotypes of vitamin K epoxide reductase, gamma-glutamyl carboxylase, and cytochrome P4502C9 as determinants of daily warfarin dose in Japanese patients. Thromb Res. 2007;120:181–186. doi: 10.1016/j.thromres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Wadelius M, Chen LY, Downes K, Ghori J, Hunt S, Eriksson N, Wallerman O, Melhus H, Wadelius C, Bentley D, Deloukas P. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 45.Wajih N, Sane DC, Hutson SM, Wallin R. The inhibitory effect of calumenin on the vitamin K-dependent gamma-carboxylation system – characterization of the system in normal and warfarin-resistant rats. J Biol Chem. 2004;279:25276–25283. doi: 10.1074/jbc.M401645200. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.National Center for Biotechnology Information – dbSNP. Database of single nucleotide polymorphisms. Available at http://www.ncbi.nlm.nih.gov/projects/SNP/snp_refcgi?rs=2108622 (last accessed 18 May 2012)
- 48.Perini JA, Struchiner CJ, Silva-Assuncao E, Suarez-Kurtz G. Impact of CYP4F2 rs2108622 on the stable warfarin dose in an admixed patient cohort. Clin Pharmacol Ther. 2010;87:417–420. doi: 10.1038/clpt.2009.307. [DOI] [PubMed] [Google Scholar]
- 49.Schelleman H, Chen J, Chen Z, Christie J, Newcomb CW, Brensinger CM, Price M, Whitehead AS, Kealey C, Thorn CF, Samaha FF, Kimmel SE. Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin Pharmacol Ther. 2008;84:332–339. doi: 10.1038/clpt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuchshuber-Moraes M, Perini JA, Rosskopf D, Suarez-Kurtz G. Exploring warfarin pharmacogenomics with the extreme-discordant-phenotype methodology: impact of FVII polymorphisms on stable anticoagulation with warfarin. Eur J Clin Pharmacol. 2009;65:789–793. doi: 10.1007/s00228-009-0651-6. [DOI] [PubMed] [Google Scholar]
- 51.Nebert DW. Extreme discordant phenotype methodology: an intuitive approach to clinical pharmacogenetics. Eur J Pharmacol. 2000;410:107–120. doi: 10.1016/s0014-2999(00)00809-8. [DOI] [PubMed] [Google Scholar]
- 52.Suarez-Kurtz G. Population diversity and the performance of warfarin dosing algorithms. Br J Clin Pharmacol. 2011;72:451–453. doi: 10.1111/j.1365-2125.2011.04018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cosgun E, Limdi NA, Duarte CW. High-dimensional pharmacogenetic prediction of a continuous trait using machine learning techniques with application to warfarin dose prediction in African Americans. Bioinformatics. 2011;27:1384–1389. doi: 10.1093/bioinformatics/btr159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kealey C, Chen Z, Christie J, Thorn CF, Whitehead AS, Price M, Samaha FF, Kimmel SE. Warfarin and cytochrome P450 2C9 genotype: possible ethnic variation in warfarin sensitivity. Pharmacogenomics. 2007;8:217–225. doi: 10.2217/14622416.8.3.217. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi H, Wilkinson GR, Nutescu EA, Morita T, Ritchie MD, Scordo MG, Pengo V, Barban M, Padrini R, Ieiri I, Otsubo K, Kashima T, Kimura S, Kijima S, Echizen H. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-Americans. Pharmacogenet Genomics. 2006;16:101–110. doi: 10.1097/01.fpc.0000184955.08453.a8. [DOI] [PubMed] [Google Scholar]
- 56.Wu AH, Wang P, Smith A, Haller C, Drake K, Linder M, Valdes R. Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: comparison with other equations. Pharmacogenomics. 2008;9:169–178. doi: 10.2217/14622416.9.2.169. [DOI] [PubMed] [Google Scholar]
- 57.Suarez-Kurtz G, Genro JP, de Moraes MO, Ojopi EB, Pena SD, Perini JA, Ribeiro-Dos-Santos A, Romano-Silva MA, Santana I, Struchiner CJ. Global pharmacogenomics: impact of population diversity on the distribution of polymorphisms in the CYP2C cluster among Brazilians. Pharmacogenomics J. 2012;12:267–276. doi: 10.1038/tpj.2010.89. [DOI] [PubMed] [Google Scholar]
- 58.Vargens DD, Damasceno A, Petzl-Erler ML, Suarez-Kurtz G. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among Amerindians, Mozambicans and Brazilians. Pharmacogenomics. 2011;12:769–772. doi: 10.2217/pgs.11.35. [DOI] [PubMed] [Google Scholar]
- 59.Rede Nacional de Farmacogenética. The Brazilian Pharmacogenomic Network. Available at http://www.refargen.org.br (last accessed 18 May 2012)
- 60.Dandara C, Lombard Z, Du Plooy I, McLellan T, Norris SA, Ramsay M. Genetic variants in CYP (-1A2, -2C9, -2C19, -3A4 and -3A5), VKORC1 and ABCB1 genes in a black South African population: a window into diversity. Pharmacogenomics. 2011;12:1663–1670. doi: 10.2217/pgs.11.106. [DOI] [PubMed] [Google Scholar]
- 61.The International Project. Available at http://hapmap.ncb.inlm.nih.gov/ (last accessed 18 May 2012)
- 62.Allabi AC, Horsmans Y, Alvarez JC, Bigot A, Verbeeck RK, Yasar U, Gala JL. Acenocoumarol sensitivity and pharmacokinetic characterization of CYP2C9 *5/*8,*8/*11,*9/*11 and VKORC1*2 in black African healthy Beninese subjects. Eur J Drug Metab Pharmacokinet. 2012;37:125–132. doi: 10.1007/s13318-011-0056-7. [DOI] [PubMed] [Google Scholar]
- 63.Scott SA, Khasawneh R, Peter I, Kornreich R, Desnick RJ. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among racial and ethnic groups. Pharmacogenomics. 2010;11:781–791. doi: 10.2217/pgs.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suarez-Kurtz G, Perini JA, Silva-Assunção E, Struchiner CJ. Relative contribution of VKORC1, CYP2C9, and INR response to warfarin stable dose. Blood. 2009;113:4125–4126. doi: 10.1182/blood-2009-01-200600. [DOI] [PubMed] [Google Scholar]


