Abstract
Distinct regions of the eukaryotic genome are packaged into different types of chromatin, with euchromatin representing gene rich, transcriptionally active regions and heterochromatin more condensed and gene poor. The assembly and maintenance of heterochromatin is important for many aspects of genome control, including silencing of gene transcription, suppression of recombination, and to ensure proper chromosome segregation. The precise mechanisms underlying heterochromatin establishment and maintenance are still unclear, but much progress has been made towards understanding this process during the last few years, particularly from studies performed in fission yeast. In this review, we hope to provide a conceptual model of centromeric heterochromatin in fission yeast that integrates our current understanding of the competing forces of transcription, replication, and RNA decay that influence its assembly and propagation.
Keywords: Centromere, Heterochromatin, RNAi, Non-coding RNA
Heterochromatin
All eukaryotic genomes are packaged in chromatin, with the basic unit represented by the nucleosome consisting of a histone octamer wrapped by 147 bp of DNA. Histones are globular proteins, with a well-defined his-tone “core,” which mediates interactions between different histone partners, and less structured amino terminal tails, which extend out from the core nucleosome particle. Histones, and in particular histone tails, are subject to a variety of post-translational modifications, including acetylation, methylation, phosphorylation, and ubiquitination. These modifications play a crucial role in the assembly of higher order chromatin structures and in the recruitment of chromatin modifiers.
Heterochromatin is characterized by histone hypoacetylation and an enrichment of methyl marks on histones associated with repression. These marks along with the enzymes that produce these modifications and the proteins that recognize them are highly conserved from fission yeast to man (Kouzarides 2007). Methylation on lysine 9 (K9) of histone H3 is a key event in the assembly of heterochromatin (Rea et al. 2000; Bannister et al. 2001; Lachner et al. 2001). In fission yeast, a single histone H3 methyltransferase, Clr4, directs all methylation of K9 on histone H3 (H3K9me1, 2, or 3; Yamada et al. 2005), whereas in mammals, several proteins including Clr4’s homologs, the Suv39 proteins, can methylate K9. However, fission yeast lacks the enzymatic machinery for methylation of H3K27 and lacks DNA methylation; both of these marks correlate with heterochromatin in higher eukaryotes.
Heterochromatin has classically been perceived as being transcriptionally silent, but this view has been revolutionized with the perplexing observation from many organisms that transcription through a heterochromatic locus appears to be required for its silencing (reviewed in Huisinga et al. 2006). A frequently cited example of such transcription-coupled silencing occurs in female mammals during X chromosome inactivation. In this instance, there is increased expression of the long non-coding RNA, Xist, from one X chromosome, which then spreads in cis to initiate silencing of the entire chromosome.
A second common theme that has emerged from studies of heterochromatin assembly in many organisms is the importance of non-coding RNA and in many cases the cellular RNA interference (RNAi) pathway in silencing (Zaratiegui et al. 2007). For example, in many organisms, the RNAi pathway is required for germline silencing of transposable elements, which can constitute 50% of the genome (O’Donnell and Boeke 2007). In fission yeast and flies, the RNAi pathway is linked to the assembly of centromeric heterochromatin (Volpe et al. 2002; Pal-Bhadra et al. 2004; Deshpande et al. 2005), and studies in mammals have clearly demonstrated that gene silencing can be mediated via non-coding RNAs (Nagano and Fraser 2011).
Fission yeast centromeres
The fission yeast, Schizosaccharomyces pombe, has several constitutively heterochromatic loci, including telomeres, centromeres, and the mating type locus. The heterochromatin that assembles on these regions is similar, although the details of its assembly differ. Fission yeast centromeres range from 35 to 110 Kb in length and are composed of a central domain on which the kinetochore assembles flanked by outer-repeat sequences (otr, consisting of dg and dh repeats) coated in heterochromatin that resembles the pericentromeric heterochromatin of mammals (Takahashi et al. 2000; Partridge et al. 2000; Nakayama et al. 2001).
The central domain is the site on which the kineto-chore assembles, and as in all eukaryotes, the incorporation of a centromere specific histone H3 variant protein, Cnp1 (CENP-A in mammals) into nucleosomes of this domain directs assembly of a functional kinetochore. The central domain consists of a central core (cnt) of non-repetitive DNA flanked by inverted innermost repeats (imr). In the smallest centromere, centromere 1, there is approximately 4 Kb of cnt DNA, flanked by 5.6 Kb of imr DNA and 4.6 to 4.8 Kb of otr repeats. The larger centromeres (chromosomes 2 and 3) have similarly sized central domains, but more extensive repetition of the outer repeat sequences. The centromeric DNA is highly similar between the centromeres, with the exception of the imr repeats which are centromere specific. Homology ranges from 48% identity over a 1.4-Kb region of cnt to a striking 97% identity over 1.78 Kb of dg sequence (Wood et al. 2002). Fission yeast centromeres lack protein coding genes but contain clusters of tRNA genes within imr, flanking centromeres 2 and 3 and on the left side of centromere 1, that help insulate the distinct centromeric domains and prevent the spreading of centromeric heterochromatin into the chromosome arms (Noma et al. 2006; Scott et al. 2006).
Experiments performed with minichromosomes have demonstrated that DNA sequences from both the outer repeats and the central domain are required for centromere function (Takahashi et al. 1992; Baum et al. 1994; Folco et al. 2008). Interestingly, the role of the outer repeat sequences appears to be purely for heterochromatin assembly, since centromeres lacking outer repeat sequences gain function when an enzyme that drives heterochromatin assembly is tethered adjacent to central domain sequences (Kagansky et al. 2009).
Heterochromatin in fission yeast
The fission yeast has emerged as a very useful genetically tractable model in which to determine the mechanisms underlying the establishment and maintenance of heterochromatin. The reasons for this are conservation of both the key chromatin modifying enzymes required for assembly of heterochromatin and because fission yeast have a conserved RNAi machinery with single genes encoding each activity.
It was originally thought that fission yeast centromeres were transcriptionally inert, as a marker gene inserted within centromeric sequence exhibited classical position effect variegation (Allshire et al. 1994; Allshire et al. 1995). This silencing was thought to reflect spreading of heterochromatin over the gene blocking access of RNA polymerase II (pol II). However, more recent studies have demonstrated that centromeres are transcribed in both fission yeast and mammals, and that transcript abundance is regulated by the cellular RNAi apparatus (Volpe et al. 2002; Lehnertz et al. 2003; Kanellopoulou et al. 2005; Murchison et al. 2005). Centromeric transcription occurs during S phase, during a window of time in which the repressive histone marks at centromeres become diluted by DNA replication, permitting pol II access (Chen et al. 2008; Kloc et al. 2008).
H3K9 methylation and the Clr-C complex
In fission yeast, H3 K9 methylation is catalyzed by a single methyltransferase, Clr4, the homolog of human Suv39 and Drosophila Su(var)3–9 proteins (Rea et al. 2000). Clr4 is a component of a multi-subunit complex called Clr-C that also includes Rik1, Pcu4 (Cul4), Raf1 (Cmc1, Dos1, Clr8), and Raf2 (Cmc2, Dos2, Clr7; Li et al. 2005; Horn et al. 2005; Hong et al. 2005; Jia et al. 2005; Thon et al. 2005). These proteins, together with a ring box protein, Rbx1, are components of a cullin 4-dependent E3 ubiquitin ligase. Each component is necessary for heterochromatin assembly, and mutants deficient for these proteins completely lack H3K9me on centromeric repeats and at other normally heterochromatic loci, and lack centromeric siRNAs. However, their exact role in directing E3 ligase activity and promoting Clr4 function is not clear. What is known is that Pcu4, the cullin 4 protein that acts as a scaffold for assembly of ubiquitin ligases, associates with Raf2 (Thon et al. 2005), and Rik1 binds Raf1 (Li et al. 2005). Raf1 likely serves as the substrate acceptor (or DCAF) for the ubiquitin ligase activity, and interestingly, mutations that separate it from the rest of the complex show only loss of H3K9 methylation, with no impact on siRNA generation (Buscaino et al. 2012). Rik1 is likely to bind nucleic acids, since it shares homology with the UV DNA damage binding protein, and with CPSF-A, a subunit of the cleavage and poly-adenylation specificity complex (Neuwald and Poleksic 2000). Rik1 may recruit the complex to chromatin, since it remains localized even in mutant backgrounds such as pcu4, in which Clr4 is largely delocalized (Jia et al. 2005; Zhang et al. 2008). The target of Clr-C’s ubiquitin ligase activity is not known, although Clr-C can ubiquitinate histone H2B in vitro (Horn et al. 2005). Clr4 is thought to methylate targets other than just H3K9, since yeast bearing histone H3 mutants that block K9 methylation express higher levels of siRNAs from centromeric repeats than clr4 mutants that are deficient in methyltransferase activity (Gerace et al. 2010). One non-histone target that has been identified is Mlo3, a protein that is associated with the TRAMP complex and is involved in RNA surveillance and in the destruction of aberrant RNAs. Methylation by Clr4 is important for Mlo3 function since mutation of the methylation site on Mlo3 caused a reduction in centromeric siRNA production (Zhang et al. 2011).
The H3K9me mark in fission yeast is bound by the chromodomain-containing proteins Swi6 and Chp2 (homologs of HP1), by Chp1, and also by the Clr4 enzyme (Bannister et al. 2001; Nakayama et al. 2001; Partridge et al. 2002; Zhang et al. 2008; Sadaie et al. 2008). The ability of Clr4 to bind the product of its activity provides an example of an amplification switch where the protein that reads a chromatin mark can then write the mark on adjacent nucleosomes. Assembly of Swi6-containing heterochromatin is critical for centromere function in fission yeast and prevents the formation of aneuploid cells (Ekwall et al. 1995). Swi6 recruits cohesin to the outer repeats of the centromere (Bernard et al. 2001; Nonaka et al. 2002) which maintains sister chromatid cohesion until satisfaction of the spindle assembly checkpoints and ana-phase onset. Recruitment of cohesin appears to be the major role of Swi6 at centromeres, since artificially tethering cohesin to centromeres bypasses the requirement for Swi6 in centromere function during mitosis (Yamagishi et al. 2008).
In order to understand heterochromatin assembly, we need to understand how the Clr-C complex is recruited to chromatin. We still do not have a full understanding of this process, but what is clear is that different mechanisms can contribute to Clr-C recruitment at distinct loci. At centromeres, there is a high concentration of repetitive dg and dh sequences, which play a major role in centromeric heterochromatin assembly through the RNAi pathway (Volpe et al. 2002). Single copies of similar sequences are present at the mating type region and telomeres, but at these loci, heterochromatin is unaffected by loss of RNAi components, since alternative DNA sequences can recruit Clr-C independently of RNAi (Hall et al. 2002; Sadaie et al. 2004; Jia et al. 2004; Petrie et al. 2005). Centromeric heterochromatin is thus uniquely sensitive to the loss of the RNAi pathway, which leads to loss of silencing and decreased enrichment of H3K9me2 and Swi6 on the outer repeats of the centromere, causing aberrant chromosome segregation due to defects in centromere function (Volpe et al. 2002, 2003).
The RNAi pathway in fission yeast
Fission yeast has a scaled down RNAi machinery, with single genes encoding the key enzymes. Dicer (Dcr1) is the conserved ribonuclease that cleaves dsRNA, Argonaute (Ago1) is the conserved effector protein which binds siRNA and mediates sequence-specific destruction or inactivation of target (homologous) RNA (Buker et al. 2007; Irvine et al. 2006) and RNA-dependent RNA polymerase (Rdp1) is required for production of dsRNA to mediate the RNAi response (Motamedi et al. 2004; Sugiyama et al. 2005). Mutation of dcr1, ago1, or rdp1 leads to the accumulation of centromeric transcripts and disruption of centromeric heterochromatin (Volpe et al. 2002). This, coupled with the discovery of siRNAs derived from centromeric transcripts, seeded the idea that heterochromatin assembly was regulated by RNAi (Reinhart and Bartel 2002).
It has since been shown that RNA pol II preferentially transcribes one strand of centromeric sequence, and that this transcript (termed the pre-siRNA) is short-lived (Volpe et al. 2002; Djupedal et al. 2005). Rdp1 is thought to bind this transcript and use it as template to synthesize double stranded (ds) RNA which is then processed by dicer to form ds short interfering RNAs (siRNAs). These siRNAs are bound by an argonaute complex (ARC), in which the ribonuclease activity of Ago1 is blocked (Buker et al. 2007). siRNAs (and possibly Ago1 itself) are then passed to a second Ago1 containing complex, RITS, which is an RNAi effector complex in which argonaute is active (Irvine et al. 2006; Buker et al. 2007). In the RITS (RNA-induced initiation of transcriptional silencing) complex, Ago1 cleaves the “passenger strand” of the ds siRNA as the first target of its activity, leaving a single strand of siRNA bound to RITS, which can then program further sequence-dependent cleavage of RNA by Ago1. This cleavage activity of Ago1 may be important for co-transcriptional silencing of nascent centromeric transcripts, and for proper termination of centromeric transcripts.
Fission yeast strains that are defective for RNAi show an accumulation of centromeric transcripts, a loss of centromeric siRNAs, and a significant decrease in the levels of H3K9me2 at centromeres, resulting in a loss of centromeric heterochromatin. Similar phenotypes are seen for mutants in chp1, a chromodomain protein that is localized to centromeres (Partridge et al. 2000; Partridge et al. 2002). The similar phenotypes suggested that there would be a link between Chp1 and RNAi, and that link was made by purification of the RITS complex. Purification of Chp1 yielded a complex with Ago1, centromeric siRNAs, and an uncharacterized protein, Tas3 (Verdel et al. 2004). RITS weakly associates with a second RNAi effector complex, RDRC (RNA-dependent RNA polymerase complex), which includes Rdp1 required for dsRNA production (Motamedi et al. 2004). Localization of the two complexes is co-dependent and requires both Dcr1 and Clr4.
RITS
RITS and RDRC are key players in the assembly of centromeric heterochromatin; they localize to all sites of heterochromatin, and their chromatin association is dependent on Clr-C. Most intriguingly, centromeric recruitment of RITS/RDRC is necessary for the accumulation of high levels of H3K9me2 specifically at centromeres (Partridge et al. 2002; Verdel et al. 2004; Sadaie et al. 2004; Motamedi et al. 2004). One clue as to how Clr-C might be recruited to centromeres by RITS/RDRC is that the Rik1 component of Clr-C has been shown to physically associate with the RITS component Chp1 (Zhang et al. 2008). Also, the LIM domain containing protein Stc1 has been shown to physically bridge between Ago1 and Clr-C and to be required for heterochromatin assembly specifically at centromeres (Bayne et al. 2010). Interestingly, when Stc1 is tethered at a euchromatic site, assembly of heterochromatin occurs in a manner which is dependent on Clr-C, but independent of RNAi components (Bayne et al. 2010). A similar result is seen when the enzymatic domain of Clr4 is tethered to DNA, with heterochromatin assembly being independent of RNAi (Kagansky et al. 2009). Together these results would suggest that once Clr-C is physically recruited to the centromere by RITS/RDRC, that enrichment for H3K9me2 and heterochromatin assembly can occur independently of RNAi, through chromatin modifications leading to suppression of pol II recruitment to the locus (transcriptional gene silencing). However, recruitment of RITS and RDRC complexes to centromeres is also necessary for the turnover of centromeric transcripts both through Rdp1 mediated generation of dsRNA which is then cleaved by Dcr1 to generate siRNAs, and by the destruction of nascent transcripts via siRNA programmed RITS-mediated destruction. Thus, RITS/RDRC appear to play roles in both transcriptional gene silencing, by generating a chromatin environment refractory to RNA pol II access, and in co- or post-transcriptional silencing, by destruction of centromeric transcripts.
Targeting of RITS/RDRC to centromeres
The original model for the mechanism of RITS recruitment to centromeres put forward the idea that siRNAs bound by Ago1 in the RITS complex targeted RITS to either nascent transcripts or centromeric DNA. It was thought that once high levels of Clr4 activity were recruited to centromeres, H3K9Me2 stabilized the binding of RITS by providing a binding site for the chromo-domain of Chp1 (Verdel et al. 2004). However, mutation of the chromodomain of Chp1 revealed that chromatin association via Chp1 was critical for RITS function, and binding studies revealed that Chp1 has very high affinity for H3K9me2 peptides in vitro (Noma et al. 2004; Petrie et al. 2005; Schalch et al. 2009). Thus, we asked whether we could determine whether RITS recruitment and thus assembly of heterochromatin is driven by siRNA or by H3K9Me2. It may seem straightforward to ask whether siRNAs recruit RITS by knocking out dicer. However, dcr1 null cells not only lose siRNAs but have such reduced levels of H3K9Me2 at centromeres that heterochromatin cannot be maintained (Volpe et al. 2002; Sadaie et al. 2004). Similarly, knocking out the H3K9 methyltransferase Clr4 not only prevents methylation on K9 of histone H3, but also greatly impacts production of siRNAs (Nakayama et al. 2001; Buhler et al. 2006).
To attempt to dissect the mechanism of RITS recruitment, we designed specific mutations within the RITS complex to perturb either siRNA-mediated or H3K9Me2-mediated interactions of RITS. We blocked the association of Ago1 with the Tas3-Chp1 subcomplex by introducing mutations into the conserved GW domain of Tas3 that binds Ago1 (Partridge et al. 2007). We reasoned that removing Ago1 from RITS would abrogate the siRNA-dependent functions of the complex, yet leave other essential Ago1 functions intact in the cell. Indeed, this mutant (Tas3WG) yielded a very interesting phenotype, allowing for the maintenance of pre-assembled heterochromatin. This maintenance was dependent on efficient tethering of Ago1 to centromeres through siRNA mediated interactions, since introduction of a mutant Ago1 with reduced affinity for siRNA, while not affecting heterochromatin maintenance in isolation, caused complete loss of heterochromatin when introduced into TasWG cells (Partridge et al. 2007). In contrast, we found that mutation of Tas3 to remove Chp1 from RITS (Tas3Δ10–24) caused complete disruption of heterochromatin, with loss of H3K9me2 from centromeric repeats and a failure to generate centromeric siR-NAs (DeBeauchamp et al. 2008).
Transcriptional push me–pull you
These observations have led to a model in which there is an absolute requirement for the RNAi pathway for the maintenance of heterochromatin at fission yeast centromeres. In large part this is the case, but very interestingly, reducing the level of active transcription at centromeres in cells that are deficient for RNAi can allow for the maintenance of centromeric heterochromatin. Notably, this propagation of heterochromatin occurs in cells that completely lack centromeric siRNAs. Strains that maintain heterochromatin in the absence of RNAi include knockouts for Epe1 (Trewick et al. 2007), a protein that promotes transcription by pol II and which is somewhat surprisingly recruited to centromeres through interaction with the heterochromatic silencing protein Swi6 (Zofall and Grewal 2006; Isaac et al. 2007; Trewick et al. 2007), or knockouts for mst2+, a histone acetyltransferase which promotes acetylation of H3 on K14 (Reddy et al. 2011). Also, strains lacking another target of Clr4 methyltransferase activity, Mlo3, or its associated Cid14 protein, can assemble centromeric heterochromatin in the absence of the RNAi pathway (Reyes-Turcu et al. 2011). In all of these situations, compound mutants with RNAi components show decreased RNA pol II occupancy on centromeric repeats compared to that in RNAi defective backgrounds, decreased levels of centromeric transcripts, and higher levels of H3K9me2 and Swi6 on centromeric repeats than in RNAi deficient strains alone. Together, these results suggest that the major role for RNAi in the propagation of heterochromatin is to quash the generation of transcripts which otherwise disrupt the stability of heterochromatin.
Links between replication and transcription and redeposition of epigenetic marks
Why are centromeres transcribed if transcription is deleterious? The answer may rest in the fact that cells duplicate their DNA every cycle, and during the replication process, chromatin marks such as H3K9me are diluted by the deposition of newly synthesized histones onto the replicated DNA. Thus, replication effectively dilutes repressive marks, and temporarily alleviates the chromatin based suppression of pol II activity, leading to centromeric transcription during S phase. It is widely believed that the transcription of centromeres, although potentially deleterious, is critical for the inheritance of the heterochromatic state through cell division. Centromeric transcripts are converted into siRNAs that can help recruit RITS to centromeres and provide templating activity for Ago1-mediated suppression of transcripts. Ultimately, both of these pathways feed into the positive feedback loop for amplification of the RITS/RDRC signal, which can facilitate further recruitment of Clr-C leading to the accumulation of high levels of H3K9Me2 at centromeres (Partridge et al. 2002; Verdel et al. 2004; Motamedi et al. 2004). High levels of H3K9Me2 promote the assembly of heterochromatin through recruitment of chromodomain proteins such as Swi6, which are necessary for the spreading of heterochromatin (Partridge et al. 2000; Hall et al. 2002).
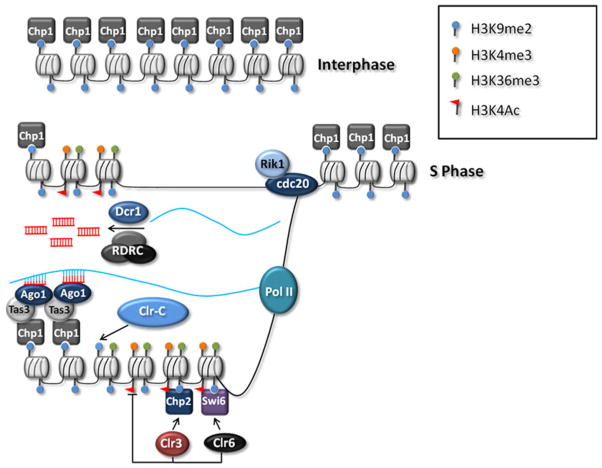
Here we present one model for how DNA replication ultimately leads to the propagation of the heterochromatin at centromeres (see Fig. 1). Following replication of DNA, there is a reduction in the levels of centromeric H3K9me2, and a wave of pol II transcription occurs, accompanied by the appearance of marks of transcriptional activity, such as H3K4me2, H3K36me3, and acetylation of histones (Kloc et al. 2008; Chen et al. 2008). One pertinent mark is the appearance of H3K4Ac. This mark reduces the affinity of Chp1 for binding chromatin, since the chromodomain of Chp1 which binds the H3K9me2/3 tail also makes a critical contact with the H3K4 moiety which is sensitive to acetylation (Schalch et al. 2009; Xhemalce and Kouzarides 2010). Transcription therefore effectively reduces the association of RITS with chromatin, and causes a “chromodomain switch,” allowing centromeric binding of the lower affinity chromodomain proteins, Chp2 and Swi6, whose binding is not sensitive to K4Ac status. Chp2 and Swi6 associate with large chromatin modifying complexes which contain deacetylase activities (Sadaie et al. 2008; Motamedi et al. 2008; Fischer et al. 2009). We suggest that the recruitment of Chp2 and Swi6 can help to reset (deacetylate) the chromatin, promoting the activity of Clr4 and allowing the wave of enrichment in H3K9me2 during late S phase. Notably, Rik1 enrichment at centromeres precedes that of H3K9me2 during S phase (Chen et al. 2008), and may correlate with the timing of centromeric replication by the elongating DNA polymerase Cdc20, which has recently been shown to bind Rik1 (Li et al. 2011). This Cdc20-Rik1 association may provide a mechanism for the replication-coupled recruitment of Clr-C to centromeric repeats. As seen for Chp1, Clr4’s ability to bind H3K9me2 is reduced by acetylation of K4 (Xhemalce and Kouzarides 2010). Hence, not only its ability to methylate K9, but also its ability to spread through chromatin may depend on the prior deacetylation of chromatin mediated by recruitment of Chp2 and Swi6-associated deacetylase activities.
Fig. 1.
Propagation of centromeric heterochromatin during DNA replication. Heterochromatin is characterized by H3K9me which serves to recruit RNAi components and Clr-C. During S-Phase, DNA replication dilutes the H3K9me mark and allows access to pol II, which leads to the appearance of marks of active transcription such as H3K4me3, H3K36me3, and H3K4Ac. The H3K4Ac reduces Chp1 binding affinity for H3K9me2/3, effectively reducing RITS levels associated with chromatin. This allows the lower affinity Swi6 and Chp2 to bind H3K9me and recruit chromatin modifying complexes containing deacetylases such as Clr3 and Clr6 which promote the removal of the H3K4Ac mark. It is likely that Clr4 is recruited though Rik1 associated with the elongating DNA polymerase Cdc20, allowing re-accumulation of the H3K9me mark. Concurrent with transcription, centromeric transcripts are converted into siRNA through the actions of RDRC and Dcr1. RITS is then recruited to the centromeric heterochromatin though Ago1’s binding of siRNA and interactions with Clr-C. RITS then recruits more Clr-C to further propagate the centromeric heterochromain
Recent work (Zaratiegui et al. 2011) has suggested further intriguing links between replication and transcription at centromeres, which may have profound implications for control of genome stability. Studies on pol II association with centromeres have shown that when replication is blocked with hydroxyurea, pol II localizes throughout the centromere. In contrast, in cycling cells, pol II is excluded from origins of DNA replication during S phase. Mapping of centromeric transcripts has demonstrated that centromeric transcripts in cells deficient in RNAi components extend beyond the predicted polyadenylation sites, suggesting that in RNAi defective cells, termination of pol II transcription is inefficient (Zaratiegui et al. 2011). A failure to terminate transcription during S phase can lead to stalling of pol II at replication forks and cause DNA damage. This may be in the form of excessive regions of association of RNA with single stranded DNA as in R-loops, which signal for DNA repair by homologous repair pathways (Bermejo et al. 2012). Consistent with this idea, synthetic interactions have been observed between mutants in RNAi and in DNA damage repair pathway proteins (Roguev et al. 2008; Zaratiegui et al. 2011).
Why does a defect in RNAi lead to inefficient termination of centromeric transcription? Presumably this reflects a requirement for Ago1 to cleave the nascent transcript. Ago1 uses siRNAs to target its cleavage activity, and the enrichment of centromeric siRNAs at sites just downstream of predicted polya-denylation sequences in wild type cells (Zaratiegui et al. 2011) would suggest that Ago1 is responsible for cleavage and hence termination of the centromeric transcripts.
Other major contributors to the silent state
Although RITS and the RNAi pathway are clearly major players in the assembly of centromeric heterochromatin, many other chromatin modifying complexes contribute to the heterochromatic environment. In particular, histone deacetylases play important roles in defining the hypoacetylated heterochromatic state. In fission yeast, the major histone deacetylases are Clr3, Clr6, and members of the sirtuin family. Clr3 exists in a large complex called SHREC which also includes a putative chromatin remodeling enzyme, Mit1 (Sugiyama et al. 2007; Motamedi et al. 2008). This complex is the fission yeast equivalent of NuRD/Mi2 complex, and the enzymatic activity of Clr3 is required for proper positioning of nucleosomes within the outer repeats of the centromere, which contributes to heterochromatin integrity (Sugiyama et al. 2007; Garcia et al. 2010). In the absence of SHREC components, there is a mild increase in the level of centromeric transcripts (Motamedi et al. 2008). This apparently minor effect is probably an underestimate of the real defect in transcriptional gene silencing in SHREC deficient cells since the RNAi pathway is operational and SHREC deficient cells have elevated levels of centromeric siRNAs (Sugiyama et al. 2007), suggestive of increased levels of the centromeric transcript that serves as template for siRNA production.
The target of Clr3 HDAC activity is thought to be predominantly acetylated lysine 14 on histone H3 (H3K14Ac; Bjerling et al. 2002). Interestingly, the Jia lab recently demonstrated that removal of the histone acetyltransferase responsible for H3K14Ac, Mst2, could override the requirement for the RNAi pathway in the maintenance of centromeric heterochromatin. Loss of Mst2 did not, however, compensate for loss of ClrC components or for loss of SHREC or other HDACs in heterochromatin maintenance (Reddy et al. 2011).
Clr6 is thought to be a more general HDAC, targeting multiple acetylated lysines in the H3 and H4 tail. Clr6 protein resides in two complexes, and attenuation of activity of either complex leads to defective TGS at centromeres and other heterochromatic loci (Nicolas et al. 2007). Clr6 can associate with a histone H3 and H4 chaperone complex containing Asf1 and HIRA, and the combined effect of these proteins is to promote nucleosomal occupancy and histone deacetylation to allow assembly of repressive chromatin (Blackwell et al. 2004; Yamane et al. 2011). Loss of Sir2 correlates with an increase in H3K9Ac, which is refractory to the activity of Clr4 in H3K9 methylation, and results in some attenuation of centromeric heterochromatin (Shankaranarayana et al. 2003).
De novo assembly of centromeric heterochromatin
RNAi-directed initiation
A popular model has been that RNAi drives heterochromatin initiation through processing of nascent transcripts by Rdp1 and Dcr1 to generate siRNAs for targeting of Ago1 to centromeres. Following Ago1 recruitment, Clr-C recruitment leads to deposition of H3K9me2 and eventual heterochromatin assembly (Verdel et al. 2004). In vitro, Rdp1 has primer independent RNA-dependent RNA polymerase activity, so this is a plausible mechanism for heterochromatin initiation (Motamedi et al. 2004; Sugiyama et al. 2005). However, low levels of H3K9 methylation are present in cells that are deficient for the RNAi pathway, indicating that Clr-C functions upstream of RNAi. Also, if this mechanism were correct, cells lacking Rdp1 would be expected to completely lack siRNAs. Instead, low levels of centromeric siRNAs are detectable in rdp1 mutants (Djupedal et al. 2009; Halic and Moazed 2010).
Several experiments have been performed to determine whether production of siRNAs against a target locus may suffice to elicit its silencing. Such experiments have met with varying success. However, at least two independent studies have confirmed that siRNAs derived from hairpin sequences can reduce transcription from homologous genomic loci in trans, by inducing heterochromatin formation in a manner that requires RDRC, Clr-C, Swi6, and Chp1 (Iida et al. 2008; Simmer et al. 2010). Furthermore, heterochromatin may form over endogenous loci with convergent transcripts, leading to transient silencing during G1-S phase (Gullerova and Proudfoot 2008). Importantly, convergent transcription may target the “autoregulation” of expression of various RNAi factors, thereby effecting an additional mechanism for control of gene expression throughout the cell cycle (Gullerova et al. 2011).
Other studies have suggested alternative methods to initiate heterochromatin assembly. One theory is that centromeric transcripts can form secondary structures that include regions of dsRNA that can be cleaved to generate siRNAs independent of Rdp1 activity (Djupedal et al. 2009). Another idea is that “primal” RNAs derived from random transcription can associate with Ago1 and be clipped by other nuclease activities to generate priRNAs that together with Ago1 can slice transcripts to recruit Rdp1 for dsRNA generation (Halic and Moazed 2010). Transcripts from genomic regions from which there is bidirectional transcription, such as the centromeric repeats would provide preferred targets for priRNA generation and slicing. This model suggests that Ago1 acts upstream of Clr-C and other components of RNAi, and to substantiate the model, Halic and Moazed show that levels of H3K9me2 in ago1 deficient cells are lower than in other RNAi mutants (Halic and Moazed 2010). This data is not supported by several other studies examining H3K9me2 in ago1 deficient backgrounds (Volpe et al. 2002; Shanker et al. 2010).
Clr-C directed initiation
Other work suggests that RNAi need not be the initiating signal for heterochromatin assembly. In studies using a crippled RITS complex, wherein Ago1 is dissociated from the complex by point mutation of GW sequences within Tas3, heterochromatin initiation is dependent on Clr-C, but not the RNAi pathway (Partridge et al. 2007; Shanker et al. 2010). Cells lacking Clr-C completely lack H3K9me2 on centromeric sequences, whereas cells deficient in RNAi retain some H3K9me2. This level of H3K9me2 is not sufficient to allow normal heterochromatin maintenance, but on reintroduction of the RNAi component into the Tas3WG background, it is sufficient to allow the re-establishment of heterochromatin. This did not occur on reintroduction of Clr-C components into the Tas3WG background (Partridge et al. 2007; Shanker et al. 2010). In addition, overexpression of Clr4 allowed deposition of H3K9me2 onto centromeric sequences in cells deficient for both Clr4 and RNAi, demonstrating that H3K9me2 deposition can be initiated independent of RNAi (Shanker et al. 2010). Consistent with this model of Clr-C acting upstream of RNAi for heterochromatin initiation, we found that heterochromatin initiation depends on the high affinity interaction of the Chp1 chromodomain with H3K9me2 (Schalch et al. 2009). Mutation of Chp1’s chromodomain to slightly weaken its affinity of interaction, dropping it from 200 nM to 1–2 uM (still higher affinity than seen for HP1), completely abolished heterochromatin initiation when Clr4 was transiently removed from these cells, in spite of these mutants showing normal levels of production of centromeric siRNAs (Schalch et al. 2009).
Additional recent studies suggest the existence of parallel pathways that can lead to heterochromatin assembly through degradation of antisense or convergent transcripts. Centromeric heterochromatin can assemble de novo independently of the RNAi pathway in cells bearing mutations in RNA quality control factors such as Mlo3 or Cid14 (Reyes-Turcu et al. 2011), a component of TRAMP, in which there is upregulation of antisense transcripts (Buhler et al. 2007; Buhler et al. 2008), or when Clr4 is overexpressed (Shanker et al. 2010). Interestingly, mutation of the exosome (rrp6 deletion), which causes similar upregulation of anti-sense transcription as seen in mlo3Δ cells (Zhang et al. 2011), does not lead to RNAi-independent heterochromatin assembly (Reyes-Turcu et al. 2011). Together, these data suggest that the exosome and RNAi pathways function in parallel, but that both are downstream of and dependent on RNA quality control mediated by TRAMP and Mlo3.
What then mediates the low levels of H3K9Me2 that are present at centromeres in cells that are defective in RNAi? Clearly, the enzyme is Clr4, but how is Clr4 recruited? Some clues as to this mechanism come from studies of other sites of heterochromatin in fission yeast, such as the telomeres and mating type locus. At these loci, RNAi-independent mechanisms play a redundant role in the recruitment of Clr4, thus abrogation of the RNAi pathway shows little impact on heterochromatin stability, whereas heterochromatin is lost at centromeres. At the mating type locus, it has been shown that transcription factors can recruit Clr4 to DNA sequences that lie outside the centromere homologous (cenH) sequences (Jia et al. 2004; Kim et al. 2004). Similar RNAi-independent mechanisms, involving recruitment of Clr4 via telomere specific factors, are thought to contribute to heterochromatin assembly at telomeres (Kanoh et al. 2005).
Given that transcription factors support the recruitment of Clr4 at Mat2/3, it is possible that recruitment of Clr4 to centromeres relies on pol II activity. RNA pol II generates the centromeric pre-siRNA transcript (Djupedal et al. 2005; Kato et al. 2005) and mutations have recently been identified within pol II subunits that disrupt assembly of centromeric heterochromatin. One mutant allows centromeric transcription but processing of transcripts is perturbed (Kato et al. 2005). A second mutant disrupts transcription of centromeric repeats (Djupedal et al. 2005). Models have therefore been proposed in which RNA pol II recruits chromatin-modifying enzymes to coordinate transcription with siRNA generation, chromatin modification, and heterochromatin assembly, in much the same way as it recruits RNA processing enzymes to the nascent transcript.
One factor that is thought to be critical for the nucleation of Clr4 binding at chromatin is Rik1. Rik1 has been proposed to be a key nucleator of heterochromatin, but its binding to chromatin was shown to be RNAi-dependent, in particular to be dependent on the catalytic activity of both Ago1 and Dcr1 (Zhang et al. 2008). This led many to believe that centromeric heterochromatin assembly was absolutely dependent on RNAi. More recent studies have demonstrated however, that Rik1 can bind chromatin independently of RNAi to nucleate heterochromatin assembly (Reyes-Turcu et al. 2011). Interestingly, Rik1 has recently been found in a complex with both DNA polymerase ε, and with Mms19 a transcriptional regulator which binds to pol II (Li et al. 2011). Transcription or replication could therefore provide routes for initial deposition of Rik1 at centromeres. However, somewhat inconsistent with this model, it was found that targeting of Rik1 to a euchromatic locus allowed generation of heterochromatin, but that the silencing that occurred was at the post-transcriptional level, with a requirement for RNAi components and no evidence of transcriptional gene silencing or involvement of Clr4 (Gerace et al. 2010). This could be an artifact of the targeting system, as mutants have been derived within Clr-C that can separate the siRNA generation role of the complex from its role in H3K9 methylation (Buscaino et al. 2012).
How conserved is the process of heterochromatin assembly at mammalian centromeres?
Dissection of the mechanism of action of the RITS/RDRC complexes has given us great insight into the workings of fission yeast centromeres, and highlighted the complex interplay between non-coding RNA and chromatin regulation. This model has provided a framework for conceptualizing interplay between other non-coding RNAs and chromatin modifiers which are implicated in widespread control of gene expression and genomic stability. This is especially important given the identification of thousands of non-coding RNAs in man, many of which bind important chromatin regulators such as the PRC2 complex, responsible for methylation of H3 on K27 and for regulation of key developmental and oncogenic switches (Guttman and Rinn 2012). However, it remains unclear as to the extent of involvement of the RNAi pathway in the maintenance and/or establishment of mammalian centromere function. Mammalian satellite repeats are clearly transcribed, and the level of those transcripts is accentuated in RNAi deficient backgrounds (Murchison et al. 2005; Kanellopoulou et al. 2005). However, there is scant further evidence for a direct role of RNAi in centromere regulation in mammals.
Exciting evidence has recently emerged for a role of centromeric non-coding RNAs (specifically for major satellite transcripts in mouse) for the de novo initiation of heterochromatin assembly (Maison et al. 2011). Major satellite transcripts associate specifically with Sumo modified HP1a. Mutation of HP1 to lose Sumo and RNA binding causes a specific defect not in the maintenance of pericentric heterochromatin, but in the de novo recruitment of HP1 to pericentric sequences in situations where normal heterochromatin has been perturbed, such as in cells deficient for Suv39 activity (and thus which lack H3K9me3- the canonical route for HP1 localization). This is a fascinating observation, as it implies that post translational modification of HP1 can facilitate its association with RNA to target it to a particular locus, and since mammalian HP1 physically associates with Suv39 (Melcher et al. 2000), the result suggests a mechanism for the initiation and subsequent spreading and propagation of mammalian centromeric heterochromatin.
Studies from numerous organisms with point, regional or holocentric centromeres now point to the central importance of balanced transcription for centromere regulation, whether it is implicated in regulation of kinetochore assembly or in the assembly of pericentric heterochromatin that influences chromosome segregation efficiency (O’Neill and Carone 2009, Chueh et al. 2009; Ohkuni and Kitagawa 2011; Gassmann et al. 2012; Chan et al. 2012). Recent work from the Verma lab has beautifully illustrated the importance of the tumor suppressor BRCA1 for assembly of heterochromatin in mammals (Zhu et al. 2011). BRCA1 binds to satellite DNAs at centromeres and monoubiquitinates H2A, a mark that correlates with transcriptional silencing. Brca1 deficiency causes loss of constitutive heterochromatin, with elevation of centromeric transcripts, loss of HP1 proteins, aneuploidy, and centrosome amplification. These phenotypes can be recapitulated by ectopic expression of satellite transcripts in otherwise wild type cells and are largely corrected in brca1 deficient cells by expression of a mimic of ubiquityl H2A. Since carriers of brca1 mutations are highly likely to develop breast and ovarian cancer, this data would suggest that the tumor suppressive role of BRCA1 is in maintenance of heterochromatin integrity. With the current explosion of interest in chromatin regulation, and identification of many mutants in chromatin regulators in the cancer genome sequencing projects, we look forward to achieving an even better understanding of the regulation of centromeric heterochromatin and how it is disrupted in disease in the next few years.
Acknowledgments
Our apologies to our colleagues whose work we have not cited due to space restraints. Thanks to Partridge lab members for the critical reading of this review. This work was supported by NIH grant R01-GM084045 (JFP), by Cancer Center support grant CCSG 2 P30 CA 21765, and by the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital. We dedicate this review to Jill Lahti for her courage, compassion, and determination.
Abbreviations
- ARC
Argonaute siRNA chaperone complex comprised of Ago1, Arb1, Arb2, and double-stranded siRNA
- Clr-C
Clr4 methyltransferase-containing complex mediating histone 3 lysine 9 methylation
- cnt
Non-repetitive central core DNA sequences of centromeres
- CPSF-A
Cleavage and polyadenylation specificity factor A component of multisubunit complex that plays central role in 3′ processing of mRNAs
- DCAF
DDB1/CUL4 associated factor substrate adaptor for ubiquitin ligase activity
- dg,dh
Repetitive DNA elements within otr
- HDAC
Histone deacetylase
- HIRA
Histone regulator A; ATP-dependent nucleosome remodeling complex
- imr
Innermost repeat DNA sequences internal to otr and distal to cnt
- NURD
Nucleosome remodeling deacetylase complex
- otr
Outer repeat DNA of centromeres
- RDRC
RNA-dependent RNA polymerase complex
- RITS
RNA-induced transcriptional silencing complex; comprised of fission yeast Chp1, Ago1, Tas3, and siRNA
- R-loops
Regions of RNA-associated single stranded DNA
- RNAi
RNA interference
- SHREC
Snf2, HDAC-containing repressor complex
- siRNA
Short interfering RNA
- TGS
Transcriptional gene silencing
- TRAMP
Trf4p/Air2p/Mtr4p polyadenylation complex
- Xist
Long, non-coding RNA with cis-acting silencing activity on mammalian female X chromosomes
Contributor Information
Benjamin J. Alper, Department of Biochemistry, St. Jude Children’s Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105, USA
Brandon R. Lowe, Department of Biochemistry, St. Jude Children’s Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105, USA. Integrated Program in Biomedical Sciences, University of Tennessee Health Science Center, 858 Madison Ave., Memphis, TN 38163, USA
Janet F. Partridge, Email: janet.partridge@stjude.org, Department of Biochemistry, St. Jude Children’s Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105, USA
References
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76(1):157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9(2):218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Baum M, Ngan VK, Clarke L. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell. 1994;5(7):747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140(5):666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell. 2012;45(6):710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294(5551):2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Bjerling P, Silverstein RA, Thon G, Caudy A, Grewal S, Ekwall K. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol Cell Biol. 2002;22(7):2170–2181. doi: 10.1128/MCB.22.7.2170-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell C, Martin KA, Greenall A, Pidoux A, Allshire RC, Whitehall SK. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol Cell Biol. 2004;24(10):4309–4320. doi: 10.1128/MCB.24.10.4309-4320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125(5):873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129(4):707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15(10):1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buker SM, Iida T, Buhler M, et al. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat Struct Mol Biol. 2007;14(3):200–207. doi: 10.1038/nsmb1211. [DOI] [PubMed] [Google Scholar]
- Buscaino A, White SA, Houston DR. Raf1 Is a DCAF for the Rik1 DDB1-like protein and has separable roles in siRNA generation and chromatin modification. PLoS Genet. 2012;8(2):e1002499. doi: 10.1371/journal.pgen.1002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FL, Marshall OJ, Saffery R, et al. Active transcription and essential role of RNA polymerase II at the centro-mere during mitosis. Proc Natl Acad Sci USA. 2012;109(6):1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451(7179):734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009;5(1):e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeauchamp JL, Moses A, Noffsinger VJ, et al. Chp1-Tas3 interaction is required to recruit RITS to fission yeast centromeres and for maintenance of centromeric heterochromatin. Mol Cell Biol. 2008;28(7):2154–2166. doi: 10.1128/MCB.01637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Schedl P. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 2005;19(14):1680–1685. doi: 10.1101/gad.1316805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19(19):2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Kos-Braun IC, Mosher RA, et al. Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA. EMBO J. 2009;28(24):3832–3844. doi: 10.1038/emboj.2009.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269(5229):1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA. 2009;106(22):8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319(5859):94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JF, Dumesic PA, Hartley PD, El-Samad H, Madhani HD. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 2010;24(16):1758–1771. doi: 10.1101/gad.1946410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Rechtsteiner A, Yuen KW, et al. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature. 2012;484(7395):534–537. doi: 10.1038/nature10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace EL, Halic M, Moazed D. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell. 2010;39(3):360–372. doi: 10.1016/j.molcel.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132(6):983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- Gullerova M, Moazed D, Proudfoot NJ. Autoregulation of convergent RNAi genes in fission yeast. Genes Dev. 2011;25(6):556–568. doi: 10.1101/gad.618611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140(4):504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297(5590):2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hong EJ, Villen J, Gerace EL, Gygi SP, Moazed D. A Cullin E3 Ubiquitin Ligase Complex Associates with Rik1 and the Clr4 Histone H3-K9 Methyltransferase and Is Required for RNAi-Mediated Heterochromatin Formation. RNABiol. 2005;2(3):106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19(14):1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Brower-Toland B, Elgin SC. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006;115(2):110–122. doi: 10.1007/s00412-006-0052-x. [DOI] [PubMed] [Google Scholar]
- Iida T, Nakayama J, Moazed D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol Cell. 2008;31(2):178–189. doi: 10.1016/j.molcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, et al. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313(5790):1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- Isaac S, Walfridsson J, Zohar T, et al. Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics. 2007;175(4):1549–1560. doi: 10.1534/genetics.106.068684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304(5679):1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7(10):1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, et al. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324(5935):1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol. 2005;15(20):1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309(5733):467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Kim HS, Choi ES, Shin JA, Jang YK, Park SD. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. J Biol Chem. 2004;279(41):42850–42859. doi: 10.1074/jbc.M407259200. [DOI] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18(7):490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13(14):1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ. Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol. 2005;15(16):1448–1457. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475(7355):244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, et al. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat Genet. 2011;43(3):220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol Cell Biol. 2000;20(10):3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119(6):789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32(6):778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292(5514): 110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Poleksic A. PSI-BLAST searches using hidden markov models of structural repeats: prediction of an unusual sliding DNA clamp and of beta-propellers in UV-damaged DNA-binding protein. Nucleic Acids Res. 2000;28(18):3570–3580. doi: 10.1093/nar/28.18.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Yamada T, Cam HP, FitzGerald PC, Kobayashi R, Grewal SI. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14(5):372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36(11):1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125(5):859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4(1):89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- O’Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129(1):37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K, Kitagawa K. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr Biol. 2011;21(20):1695–1703. doi: 10.1016/j.cub.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill RJ, Carone DM. The role of ncRNA in centromeres: a lesson from marsupials. Prog Mol Subcell Biol. 2009;48:77–101. doi: 10.1007/978-3-642-00182-6_4. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303(5658):669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14(7):783–791. [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol. 2002;12(19):1652–1660. doi: 10.1016/s0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Debeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell. 2007;26(4):593–602. doi: 10.1016/j.molcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Petrie VJ, Wuitschick JD, Givens CD, Kosinski AM, Partridge JF. RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol Cell Biol. 2005;25(6):2331–2346. doi: 10.1128/MCB.25.6.2331-2346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyl-transferases. Nature. 2000;406(6796):593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Reddy BD, Wang Y, Niu L, et al. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev. 2011;25(3):214–219. doi: 10.1101/gad.1993611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centro-mere heterochromatic repeats. Science. 2002;297(5588):1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol. 2011;18(10):1132–1138. doi: 10.1038/nsmb.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322(5900): 405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of hetero-chromatin in fission yeast. EMBO J. 2004;23(19):3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Kawaguchi R, Ohtani Y, et al. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol. 2008;28(23):6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Job G, Noffsinger VJ, et al. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34(1):36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol. 2006;16(2):119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol. 2003;13(14):1240–1246. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- Shanker S, Job G, George OL, Creamer KM, Shaban A, Partridge JF. Continuous requirement for the Clr4 complex but not RNAi for centromeric heterochromatin assembly in fission yeast harboring a disrupted RITS complex. PLoS Genet. 2010;6(10):e1001174. doi: 10.1371/journal.pgen.1001174. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Buscaino A, Kos-Braun IC, et al. Hairpin RNA induces secondary small interfering RNA synthesis and silencing in trans in fission yeast. EMBO Rep. 2010;11(2):112–118. doi: 10.1038/embor.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005;102(1):152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128(3):491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3(7):819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288(5474):2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Thon G, Hansen KR, Altes SP, et al. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics. 2005;171:1583–1595. doi: 10.1534/genetics.105.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 2007;26(22):4670–4682. doi: 10.1038/sj.emboj.7601892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303(5658):672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, et al. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11(2):137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415(6874):871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Kouzarides T. A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev. 2010;24(7):647–652. doi: 10.1101/gad.1881710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20(2):173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455(7210):251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SI. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell. 2011;41(1):56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128(4):763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Castel SE, Irvine DV, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479(7371):135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15(4):381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- Zhang K, Fischer T, Porter RL, et al. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science. 2011;331(6024):1624–1627. doi: 10.1126/science.1198712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM. BRCA1 tumor suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22(5):681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]