Abstract
Water plays a fundamental role in the folding, structure, dynamics and function of proteins and peptides. The extracellular N-terminal domain of chemokine receptors is crucial in mediating binding affinity, receptor selectivity, and regulating function. The flexible N-terminal domain becomes ordered in membranes and membrane-mimetic assemblies, thereby indicating that the membrane could play an important role in regulating CXC chemokine receptor 1 (CXCR1) function. In view of the role of hydration in lipid-protein interactions in membranes, we explored the organization and dynamics of a 34-mer peptide of CXCR1 N-terminal domain in reverse micelles by utilizing a combination of fluorescence-based approaches and circular dichroism spectroscopy. Our results show that the secondary structure adopted by the CXCR1 N-domain is critically dependent on hydration. The tryptophan residues of the CXCR1 N-domain experience motional restriction and exhibit red edge excitation shift (REES) upon incorporation in reverse micelles. REES and fluorescence lifetime exhibit reduction with increasing reverse micellar hydration. Time-resolved fluorescence anisotropy measurements reveal the effect of hydration on peptide rotational dynamics. Taken together, these results constitute the first report demonstrating modulation in the organization and dynamics of the N-terminal domain of a chemokine receptor in membrane-like environment of varying hydration. We envisage that these results are relevant in the context of hydration in the function of G protein-coupled receptors.
Keywords: Chemokine receptor, REES, time-resolved anisotropy decay, reverse micelles, hydration, fluorescence lifetime
INTRODUCTION
The interaction of water with proteins is fundamental in the folding, structure, dynamics and function of proteins and peptides.1–5 Since life evolved in water, proteins are uniquely adapted to utilize the aqueous environment to facilitate their function. It is estimated that a threshold level of hydration is necessary to fully activate the dynamics and function of proteins.6 It is now well established that the dynamics of water molecules is slowed as a result of interaction with the protein surface7 and the presence of motionally restricted water molecules is essential for the organization and function of proteins. In addition, hydration plays a crucial role in lipid-protein interactions8 and function of membrane proteins.9
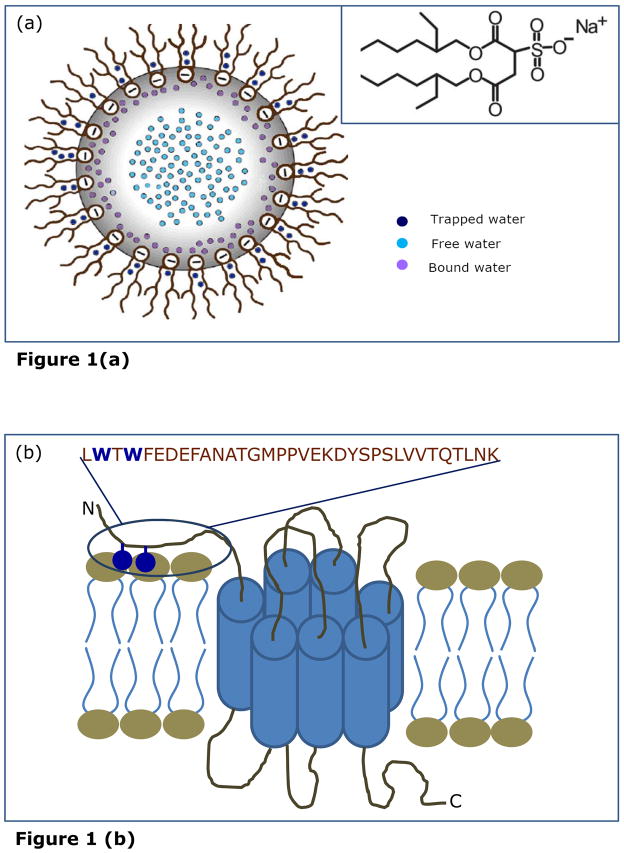
An emerging area of interest is the inherent complexity and confinement experienced by proteins in the cellular environment.10,11 Reverse micelles represent an appropriate molecular assembly for exploring constrained dynamics of proteins and peptides incorporated in them.12–18 Amphiphilic surfactants, such as sodium bis(2-ethylhexyl)sulfosuccinate (AOT), self assemble to form reverse micelles in non-polar solvents in which the polar head groups of the surfactant monomers cluster to form a micellar core and are directed toward the center of the assembly and the hydrophobic tails extend outward into the bulk organic phase (see Figure 1a).18,19 Reverse micelles are relatively simple yet versatile systems. They provide an attractive model system for biomembranes since they mimic a number of important and essential features of biological membranes although lacking much of the complexity associated with them. It is known that the dynamics of liquids in confined spaces is different than that of their bulk counterparts20,21 and this constitutes one of the main reasons for the popularity that reverse micelles enjoy as a model system in exploring water dynamics.22 The entrapped water in reverse micelles has properties that markedly differ from the properties of bulk water but similar in several aspects to those of biological interfacial water as found in membranes or protein interfaces.21,23–25 The interfacial water is crucial for the induction of secondary structure in peptides and proteins when bound to surfaces such as membranes or micelles as well as for variation of their local internal motion. Both experimental23–25 and theoretical26 approaches have shown that the crucial structural parameter of reverse micelles is [water]/[surfactant] molar ratio (w0) that determines the micellar size as well as the extent of deviation of the properties of the entrapped water from those of normal bulk water.27 Reverse micelles therefore represent a type of organized molecular assembly which offer the unique advantage of monitoring dynamics of molecules with varying degrees of hydration.
FIGURE 1.
(a) A schematic representation of reverse micellar organization showing highly structured yet heterogeneous water pools of graded dynamics, termed as trapped, bound and free water pools. The chemical structure of AOT is shown in the inset. Adapted and modified from ref. 18. (b) A schematic representation of the CXCR1 receptor. The sequence of the rabbit CXCR1 N-domain (34-mer peptide) is shown and the tryptophan residues are highlighted. The construct shown lacks 10 amino acids at the N-terminal end (this stretch of amino acids has earlier been shown to be not essential for ligand binding; see ref. 35 for details). The tryptophan residues could serve as anchors on the outer leaflet of the membrane.36 Adapted and modified from refs. 36 and 38.
Chemokines (chemotactic cytokines) represent a large family of small soluble proteins (typically 70–120 residues). They play an important modulatory role in innate immunity, inflammation, host cell defense against infection, embryogenesis and metastasis.28,29 Chemokines are classified either as CC, CXC, CX3C, or C based on the presence of conserved cysteine residues near the N-terminus. They induce transmembrane signaling by activation of a subclass of G protein-coupled receptors (GPCRs). Although the chemokine receptor family is the largest subfamily of peptide-binding GPCRs,30 the only chemokine receptor whose crystal structure is known is CXCR4.31 Very recently, the structure of membrane-bound CXCR1 has been reported using solid state NMR spectroscopy.32
From a variety of studies using chimeras and mutagenesis, it is well established that the extracellular N-terminal domain of chemokine receptors plays crucial roles in determining binding affinity, receptor selectivity, and in regulation of signaling activities.28,33,34 The chemokine receptor CXCR1 binds the chemokine interleukin-8 (IL-8) with high affinity.35–37 With the goal of addressing the role of the N-terminal domain in ligand specificity and affinity of CXCR1, the organization and dynamics of the CXCR1 N-domain were earlier studied in a micellar environment.35 More importantly, we recently showed that the CXCR1 N-domain preferentially interacts with membranes and experiences motional restriction upon binding to membranes.38 This motional restriction could result in loss of conformational entropy and therefore likely to influence ligand binding of the receptor. Interestingly, the CXCR1 N-terminal domain, although unstructurted in aqueous solution, assumes an ordered conformation in both micellar35 and membrane38 environments, thereby suggesting that the membrane bilayer could play an important role in regulating CXCR1 function.
As mentioned above, hydration is a crucial parameter in lipid-protein interactions in membranes8 and in the function of membrane proteins.9 Any alteration in the degree of hydration, particularly at the protein-lipid interface in membranes, could potentially lead to modifications of protein structure and function.39 Unfortunately, model membranes (liposomes) are not appropriate for exploring the effect of hydration on the organization and dynamics of peptides and proteins incorporated in them since controlled variation of water content is difficult to achieve in membranes. Reverse micelles represent a unique type of organized molecular assembly that offers the advantage of monitoring dynamics of molecules incorporated in them with varying states of hydration. The double chain anionic surfactant AOT (Figure 1a) has been extensively used to form reverse micelles in non-polar solvents. One of the advantages for using AOT is that reverse micelles formed by AOT can solubilize a large quantity of water in a non-polar solvent. In addition, reverse micelles formed by AOT retain a spherical shape over a wide range of w0. As a result of this, the radius of the entrapped water pool can be linearly related to w0.40 In order to explore the role of hydration in the membrane interaction of the CXCR1 N-domain (see Figure 1b), we monitored the organization and dynamics of the CXCR1 N-domain in AOT reverse micelles. Toward this goal, we applied a combination of steady state and time-resolved fluorescence approaches which include red edge excitation shift (REES), fluorescence anisotropy decay, along with circular dichroism (CD) spectroscopy. Our results show that the organization and dynamics of the CXCR1 N-domain in membrane-like environment is critically dependent on the level of hydration.
EXPERIMENTAL SECTION
Materials
The N-terminal domain peptide of CXCR1 (34-mer) was synthesized as described previously.35 The sequence of this peptide is LWTWFEDEFANATGMPPVEKDYSPSLVVTQTLNK (see Figure 1b). AOT was obtained from Serva (Heidelberg, Germany). The purity of AOT was confirmed by good agreement of its UV absorption spectrum with previously reported spectrum.19 Isooctane used was of spectroscopic grade. Water was purified through a Millipore (Bedford, MA) Milli-Q system and used throughout.
Sample Preparation
Reverse micelles of AOT containing the CXCR1 N-domain peptide were prepared without addition of any cosolvent as follows. CXCR1 N-domain peptide in methanol (48 nmol) was dried under a stream of nitrogen while being warmed gently (~35 °C). After further drying under a high vacuum for at least 4 h, 1.5 ml of 50 mM AOT in isooctane was added. Each sample was vortexed for 5 min followed by sonication in a bath sonicator (Laboratory Supplies, Hicksville, NY) for 20 min. Appropriate amounts of water were subsequently added to make reverse micellar dispersions of different [water]/[surfactant] molar ratios (w0). The optical density of the fluorescent samples at the excitation wavelength was low (generally < 0.2) in all cases. Background samples were prepared the same way except that the CXCR1 N-domain peptide was not added to them. Samples were kept in the dark for 12 h prior to fluorescence and CD measurements. All experiments were done at room temperature (~25 °C). The molar ratio of peptide to surfactant was carefully chosen to give optimum signal-to-noise ratio with minimal perturbation to the micellar organization and negligible interprobe interactions. The final peptide concentration in the reverse micelles was 32 μM while the concentration of AOT was 50 mM in all cases. This corresponds to a final molar ratio of peptide to surfactant of 1:1562 (mol/mol). At such a low peptide to surfactant molar ratio, not more than one peptide molecule would be present per reverse micelle on an average, which rules out any peptide aggregation effects, especially keeping in mind the aggregation number of AOT of ~ 40–300 in the range of wo used by us.41
Steady State Fluorescence Measurements
Steady state fluorescence measurements were performed with a Hitachi F-4010 spectrofluorometer (Tokyo, Japan) using 1 cm path length quartz cuvettes. Excitation and emission slits with a nominal bandpass of 5 nm were used for all measurements. All spectra were recorded using the correct spectrum mode. Background intensities of samples in which the peptide was omitted were subtracted from each sample spectrum to cancel out any contribution due to the solvent Raman peak and other scattering artifacts. The spectral shifts obtained with different sets of samples were identical in most cases. In other cases, the values were within ± 1 nm of the ones reported.
Time-resolved Fluorescence Measurements
Fluorescence lifetimes were calculated from time-resolved fluorescence intensity decays using IBH 5000F NanoLED equipment (Horiba Jobin Yvon, Edison, NJ) with DataStation software in the time-correlated single photon counting mode. A pulsed light emitting diode (LED) (NanoLED-17) was used as an excitation source. This LED generates optical pulse at 295 nm, of pulse duration less than 750 ps, and is run at 1 MHz repetition rate. The LED profile (instrument response function) was measured at the excitation wavelength using Ludox (colloidal silica) as the scatterer. In order to optimize the signal to noise ratio, 10,000 photon counts were collected in the peak channel. All experiments were performed using emission slits with a bandpass of 6 nm or less. The sample and the scatterer were alternated after every 5% acquisition to ensure compensation for shape and timing drifts occurring during the period of data collection. This arrangement also prevents any prolonged exposure of the sample to the excitation beam thereby avoiding any possible photodamage to the fluorophore. Data were stored and analyzed using DAS 6.2 software (Horiba Jobin Yvon, Edison, NJ). Fluorescence intensity decay curves so obtained were deconvoluted with the instrument response function and analyzed as a sum of exponential terms:
| (1) |
where F(t) is the fluorescence intensity at time t and αi is a pre-exponential factor representing the fractional contribution to the time-resolved decay of the component with a lifetime τi such that Σiαi = 1. The decay parameters were recovered using a nonlinear least squares iterative fitting procedure based on the Marquardt algorithm.42 The program also includes statistical and plotting subroutine packages.43 The goodness of the fit of a given set of observed data and the chosen function was evaluated by the χ2 ratio, the weighted residuals,44 and the autocorrelation function of the weighted residuals.45 A fit was considered acceptable when plots of the weighted residuals and the autocorrelation function showed random deviation about zero with a minimum χ2 value not more than 1.4. Intensity-averaged mean lifetimes <τ> for triexponential decays of fluorescence were calculated from the decay times and pre-exponential factors using the following equation: 46
| (2) |
Time-resolved Fluorescence Anisotropy Measurements and Data Analysis
Time-resolved fluorescence anisotropy decay measurements were carried out using a time correlated single photon counting (TCSPC) setup. A continuous-wave (CW) mode-locked frequency-doubled Nd:YAG (Vanguard, Spectra Physics) laser-driven dye (Rhodamine 6G) laser that generates 4–10 ps pulses was utilized for TCSPC measurements.47 The second harmonic output (295 nm) of an angle-tuned KDP crystal was used to excite the protein sample, and fluorescence emission was collected through a 320 nm cutoff filter followed by a monochromator. The filter prevents scattered light from reaching the detector. In fluorescence lifetime measurements used for the analysis of time-resolved anisotropy data, emission was monitored at the magic angle (54.7°) with respect to the excitation polarizer to eliminate the contribution from the decay of anisotropy. Fluorescence decay curves were obtained at the laser repetition rate of 4 MHz by a micro-channel plate photomultiplier (model R2809u, Hamamatsu Corp.) coupled to a TCSPC setup. The instrument response function (IRF) at 295 nm was obtained using a dilute colloidal suspension of dried non-dairy creamer. The full width at half maxima (FWHM) of the IRF was 40 ps and number of channels used was 1024. Fluorescence emission at magic angle (54.7°) was dispersed in a monochromator (spectral width 2.5 nm) and counted (3–4 × 103 s−1) by a microchannel plate photomultiplier, and processed through constant fraction discriminator, time-to-amplitude converter and multichannel analyzer. To optimize the signal to noise ratio, 20,000 photon counts were collected in the peak channel. All experiments were performed using excitation and emission slits with a nominal bandpass of 6 nm or less. The data stored in the multichannel analyzer was routinely transferred to an IBM PC for analysis. Fluorescence intensity decay curves so obtained were deconvoluted with the instrument response function and analyzed as a sum of exponential terms (see eq 1 above). The decay parameters were recovered using a nonlinear least squares iterative fitting procedure based on the Levenberg-Marquardt algorithm. A fit was considered acceptable when plots of the weighted residuals and the autocorrelation function showed random deviation about zero with a minimum χ2 value not more than 1.2. Intensity-averaged lifetimes (<τ>) for triexponential decays of fluorescence were calculated as mentioned above (see eq 2).
Time-resolved fluorescence anisotropy was analyzed as described previously.48–50 The fluorescence intensity decays were collected with the emission polarizer kept at parallel (I||) and perpendicular (I⊥) orientations with respect to the excitation polarizer. Anisotropy was calculated as:
| (3) |
where G(λ) is the grating factor (G-factor). The G-factor is defined as the ratio of the transmission efficiency of the grating for vertically polarized light to horizontally polarized light. The G-factor of the emission collection optics was determined in a separate experiment using a standard sample (NATA). The time-resolved anisotropy decay was analyzed based on the model:
| (4) |
| (5) |
where I||(t) and I⊥(t) are the decays of the parallel (||) and perpendicular (⊥) components of emission. The equation for time-resolved fluorescence anisotropy can be expressed as a biexponential decay:
| (6) |
where φi and βi represent the ith rotational correlation time and the corresponding pre-exponential factor in the exponential anisotropy decay such that Σβi = 1. r0 represents the anisotropy at zero time (initial anisotropy). The goodness of the fit of a given set of observed data and the chosen function was evaluated by the reduced χ2 ratio which is around 1.0 to 1.4.
Circular Dichroism Measurements
Circular Dichroism (CD) measurements were carried out at room temperature (~25 °C) on a JASCO J-815 spectropolarimeter (Tokyo, Japan) which was calibrated with (+)-10-camphorsulfonic acid. Spectra were scanned in a quartz optical cell with a path length of 0.1 cm, and recorded in 0.5 nm wavelength increments with a 2 sec response and a band width of 2 nm. For monitoring changes in secondary structure, spectra were scanned from 200 to 250 nm in the far-UV range at a scan rate of 50 nm/min. Each spectrum is the average of 6 scans with a full scale sensitivity of 100 mdeg. Spectra were corrected for background by subtraction of appropriate blanks. Data are represented as mean residue ellipticities and were calculated using the equation:
| (7) |
where θobs is the observed ellipticity in mdeg, l is the path length in cm, and C is the concentration of peptide bonds in mol/L.
RESULTS
CXCR1 N-domain Peptide Adopts β-sheet Conformation in Reverse Micelles
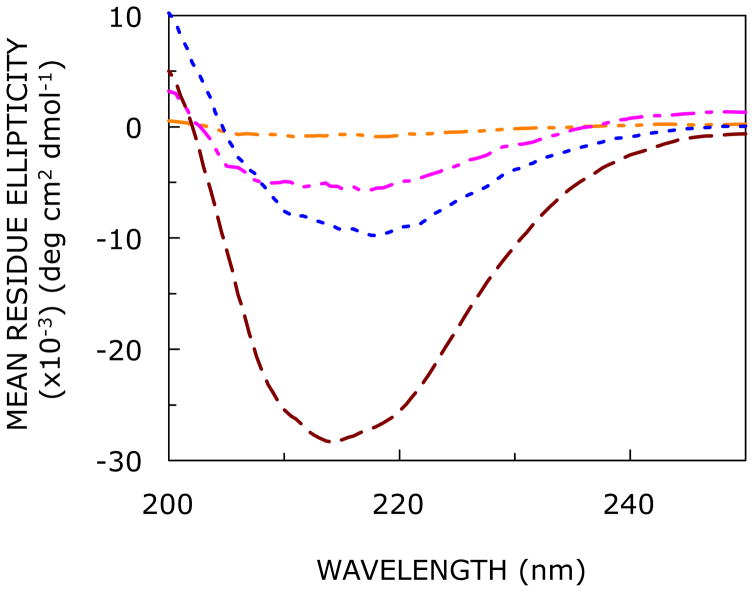
To investigate the effect of increasing hydration on the secondary structure of the N-terminal domain peptide of CXCR1 in the confined reverse micellar environment, we performed far-UV CD spectroscopy with varying w0. The CD spectra of the CXCR1 N-domain peptide in AOT reverse micelle with varying degrees of hydration are shown in Figure 2. At lower hydration (w0 = 5), the peptide does not exhibit appreciable optical activity, possibly due to suboptimal solubilization under these conditions. Interestingly, with increase in hydration (i.e., w0 = 10 and above) the peptide appears to adopt an ordered conformation. The extent of secondary structure shows an increase with increasing hydration (see Figure 2).
FIGURE 2.
Effect of increasing hydration on the far-UV CD spectra of the CXCR1 N-domain peptide in AOT reverse micelles corresponding to w0 = 5 (orange, — - - —), 10 (magenta, — - —-), 15 (blue, ····), and 20 (red, ----). The ratio of peptide to surfactant was 1:1562 (mol/mol) and peptide concentration was 32 μM in all cases. See Experimental Section for more details.
Fluorescence Characteristics of the CXCR1 N-domain in Reverse Micellar Environments
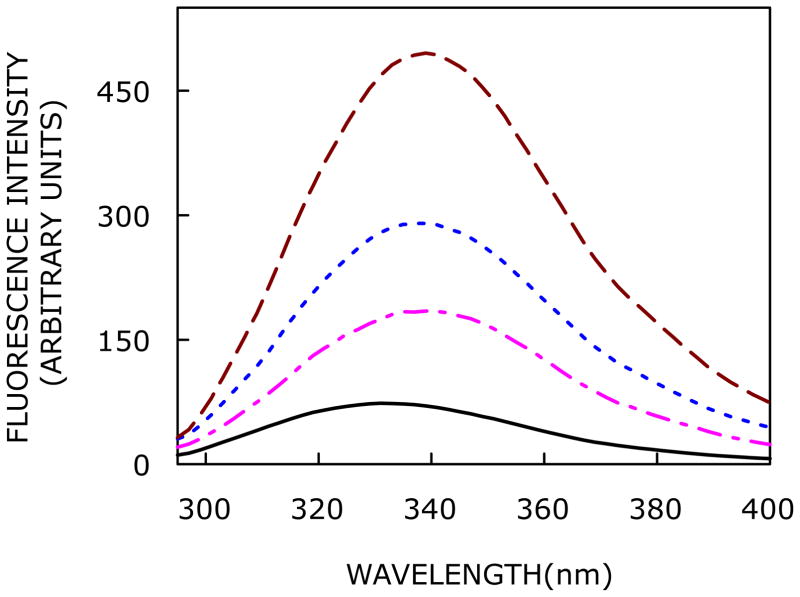
The N-terminal domain of CXCR1 has two closely positioned tryptophan residues (see Figure 1b). Tryptophan fluorescence is a useful indicator of protein microenvironment due to the presence of a polarity dependent change in the dipole moment (~5–6 D) associated with So → 1La transition (1La is the lowest singlet state in tryptophan and is the fluorescing state in most proteins).51,52 The fluorescence emission spectra of the CXCR1 N-domain peptide in AOT reverse micelles with increasing hydration (i.e., w0) are shown in Figure 3. The maximum of fluorescence emission (We have used the term maximum of fluorescence emission in a somewhat broader sense here. In every case, we have monitored the wavelength corresponding to maximum fluorescence intensity, as well as the center of mass of the fluorescence emission, in the symmetric part of the spectrum. In most cases, both these methods yielded the same wavelength. In cases where minor discrepancies were found, the center of mass of emission has been reported as the fluorescence maximum) of the CXCR1 N-domain peptide at the lowest hydration (w0 = 3) is 332 nm, which is significantly blue shifted relative to the emission maximum of the peptide in buffer (350 nm), and in the presence of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) vesicles (344 nm).38 The blue shift in emission maximum indicates the rather nonpolar nature of the site of localization of peptide tryptophans in the reverse micellar assembly. This could possibly be due to the reduced polarity associated with the water pool in reverse micelles.53 With increase in reverse micellar hydration (i.e., with increasing w0), the fluorescence emission maximum exhibits a progressive red shift due to exposure of peptide tryptophans to more polar environment (see also changes in fluorescence lifetime with increasing w0, Figure 6). The emission maximum is shifted from 332 to 339 nm (a red shift of 7 nm), corresponding to an increase in w0 from 3 to 20. The shift is more pronounced till w0 = 15 beyond which no significant change in the fluorescence emission maximum is observed. Interestingly, the red shift in emission maximum with increasing w0 is accompanied with a marked increase in fluorescence intensity when w0 is increased from 3 to 20 (see Figure 3). The relatively low intensity at low hydration (such as w0 = 3) could be due to self quenching of closely positioned tryptophan residues. With increase in hydration, the peptide adopts an ordered structure (Figure 2) resulting in increase in intensity.
FIGURE 3.
Effect of increasing hydration on fluorescence emission spectra of the CXCR1 N-domain peptide in AOT/isooctane reverse micelles. Fluorescence emission spectra are shown as a function of [water]/[surfactant] molar ratio (w0) in order of increasing intensity corresponding to w0 = 3 (black,—), 10 (magenta, — - —), 15 (blue, ····), and 20 (red, ----). The excitation wavelength used was 280 nm. All the other conditions are as in Figure 2. See Experimental Section for more details.
FIGURE 6.
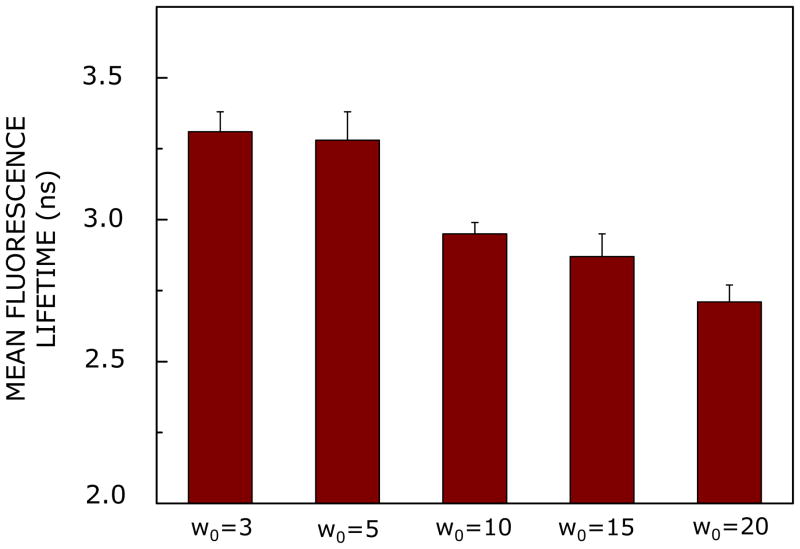
Effect of increasing hydration on mean fluorescence lifetime of the CXCR1 N-domain peptide in AOT reverse micelles. The excitation wavelength used was 295 nm and emission was monitored at 335 nm. Data shown are means ± SE of at least three independent measurements. Mean fluorescence lifetimes were calculated from Table 2 using eq 2. All other conditions are as in Figure 2. See Experimental Section for more details.
Red Edge Excitation Shift of the CXCR1 N-domain in Reverse Micellar Environments
A shift in the wavelength of maximum fluorescence emission toward higher wavelengths, caused by a shift in the excitation wavelength toward the red edge of the absorption band, is termed red edge excitation shift (REES).54–57 This effect is mostly observed with polar fluorophores in motionally restricted media such as viscous solutions or condensed phases where the dipolar relaxation time for the solvent shell around a fluorophore is comparable to or longer than its fluorescence lifetime. REES arises due to slow rates of solvent relaxation (reorientation) around an excited state fluorophore, which depends on the motional restriction imposed on the solvent molecules (or the dipolar environment, as in green fluorescent protein14) in the immediate vicinity of the fluorophore. While other fluorescence techniques yield information about the fluorophore itself, REES provides information about the relative rates of solvent relaxation that is not possible to obtain by other techniques. Since the dynamics of hydration is directly associated with the function of proteins, REES has proved to be a valuable tool to explore the organization and dynamics of soluble and membrane proteins under varying degrees of hydration.12,13,18 This makes the application of REES extremely useful since hydration plays a crucial modulatory role in a large number of important cellular events including protein folding.58 We have previously shown that REES can be used as a sensitive tool to monitor peptide conformations in membranes38,59 and membrane-mimetic environments.12–14,60
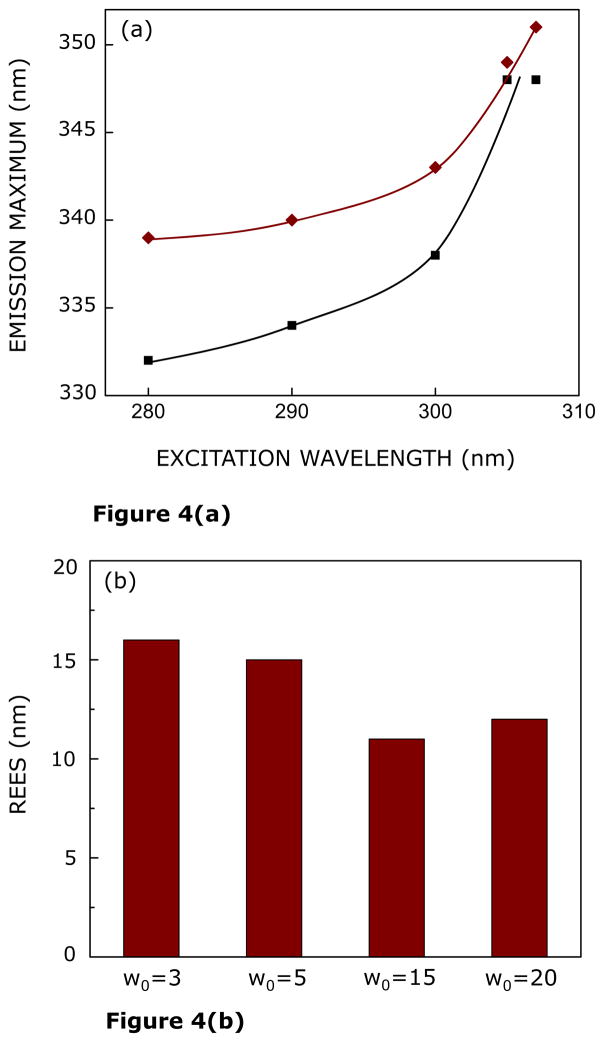
Figure 4a shows the shifts in the maxima of fluorescence emission of the CXCR1 N-domain peptide as a function of excitation wavelength in reverse micelles of varying hydration (w0 = 3 and 20). As the excitation wavelength is changed from 280 to 307 nm, the emission maximum is shifted from 332 to 348 nm at low hydration (w0 = 3). This shift corresponds to REES of 16 nm. It is possible that there could be further red shift if excitation is carried out beyond 307 nm. We found it difficult to work in this wavelength range due to low signal/noise ratio and artifacts due to the solvent Raman peak that sometimes remains even after background subtraction. Such dependence of the emission maximum on excitation wavelength is characteristic of REES and implies that the tryptophans in the CXCR1 N-domain peptide are localized in a motionally restricted region under these conditions. The interfacial region of reverse micelles is associated with bound water with characteristic dynamics.21,23–25 Since REES arises due to the dynamics of reorientational motion of solvent molecules, these results assume significance in the context of reports of slow (~ns) water relaxation in reverse micelles.61 At higher hydration (w0 = 20), the emission maximum displays a shift from 339 to 351 nm in response to change in excitation wavelength from 280 to 307 nm, corresponding to REES of 12 nm. The dependence of REES for the CXCR1 N-domain peptide on reverse micellar hydration is comprehensively shown in Figure 4b. The figure shows that there is an overall reduction in the magnitude of REES with increasing hydration. The reduction in REES is attributed to the sensitivity of the tryptophan residues to increase in free water content (Figure 1a) with increasing reverse micellar hydration.
FIGURE 4.
(a) Effect of changing excitation wavelength on the wavelength of maximum emission of the CXCR1 N-domain peptide in AOT reverse micelles corresponding to w0 = 3 (black, ■) and 20 (red, ◆). Lines joining the data points are provided merely as viewing guides. (b) Effect of increasing hydration (w0) on the magnitude of red edge excitation shift (REES) of the CXCR N-domain peptide in AOT reverse micelles. The magnitude of REES corresponds to the total shift in emission maximum when the excitation wavelength is changed from 280 to 307 nm. All other conditions are as in Figure 2. See Experimental Section for more details.
Time-resolved Anisotropy of the CXCR1 N-domain Peptide
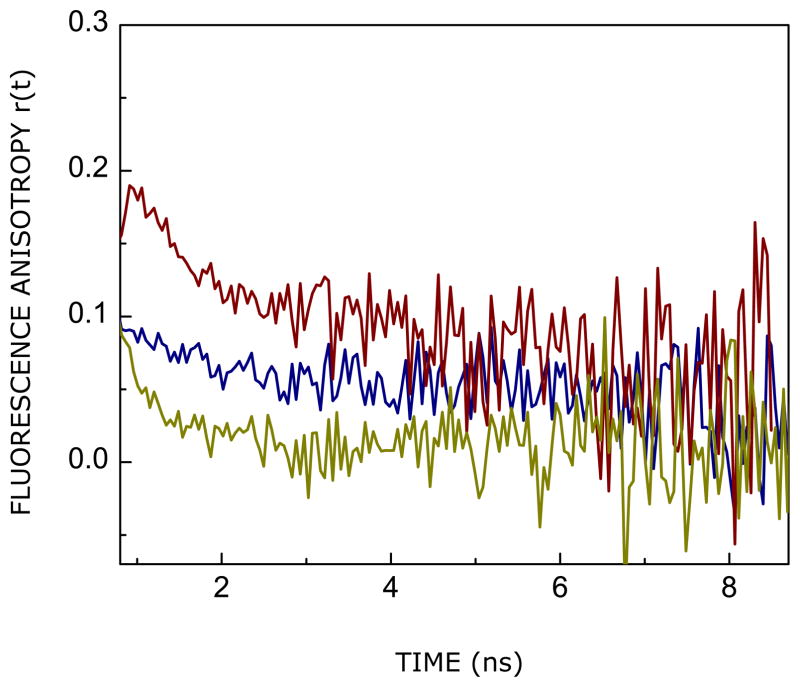
Picosecond time-resolved fluorescence anisotropy measurements of intrinsic fluorophores such as tryptophan provide insight into the local, segmental or global rotational dynamics of proteins.62 Figure 5 shows the time-resolved decay of fluorescence anisotropy of the tryptophan residues of the CXCR1 N-domain peptide in reverse micelles of varying hydration (w0 = 7 and w0 = 20). The anisotropy decay of the peptide in bulk water is shown as a control. As evident from the anisotropy decay, the tryptophan residues present in an environment of lower hydration in reverse micelles of w0 = 7 displays faster rotational dynamics relative to tryptophans present in higher hydration (w0 = 20). The anisotropy decay kinetics of the peptide in reverse micelles and bulk water could be fitted to a sum of two exponentials (see eq 6), and the values of the rotational correlation times (φ) from a representative fit is given in Table 1 (the biexponential fits and the statistical parameters used to check the goodness of the fit are shown in Supplementary Figure 1). The rotational correlation times exhibited by the peptide in reverse micelles are: φ1 in the range of 0.05–0.4 ns (local/segmental motion) and φ2 ~ 6–10 ns (global motion). This is consistent with previously reported values of the rotational correlation time of AOT reverse micelles.64 Interestingly, our results show that both rotational correlation times (φ1 and φ2) and their relative amplitude (β1 and β2) are influenced with increase in hydration (see Table 1). This difference in rotational dynamics could also be ascribed to motional constraint brought about by adoption of secondary structure. The φ2 value of ~6 ns (w0 = 20) is considerably higher than that observed in bulk water, probably indicating the motional restriction imposed as a result of conformational reorganization (see later) and enhanced viscosity in the reverse micellar environment. The magnitude of φ2 is longer in low hydration condition (w0 = 7) relative to the corresponding value in bulk water due to confinement in the reverse micellar assembly with slower tumbling rate. Table 1 shows that the amplitude (β1) of the sub-nanosecond rotational correlation time (local dynamics) shows a reduction upon increase in reverse micellar hydration, while the amplitude (β2) of the nanosecond rotational motion exhibits an increase under these conditions. The local dynamics is generally described by the “wobbling-in-cone” model65 where the tryptophan residue wobbles freely within a cone of semiangle related to the order of its environment. Our results indicate a small cone angle,65 corresponding to β1, associated with the local dynamics of tryptophans in higher hydration (w0 = 20). This could be due to motional constraint induced in local dynamics by adoption of secondary structure (see above). This is also reflected in a corresponding increase in steady state anisotropy at higher hydration.
FIGURE 5.
Time-resolved fluorescence anisotropy decay of tryptophan residues in the CXCR1 N-domain peptide in AOT reverse micelles corresponding to w0 = 7 (blue) and 20 (red). The fluorescence anisotropy decay of tryptophan residues of the same peptide in bulk water is shown for comparison (olive). The excitation wavelength was 295 nm and emission was monitored at 335 and 340 nm (for reverse micellar samples corresponding to w0 = 7 and 20, respectively), with a combination of a monochromator and a 320 nm cut-off filter, using a TCSPC setup. Emission was set at 350 nm for the peptide in water. All other conditions are as in Figure 2. See Experimental Section for more details.
Table 1.
Time-resolved Fluorescence Anisotropy Decay Parameters of the CXCR1 NDomain Peptide in AOT Reverse Micellesa
| Condition | β1 | φ1 (ns) | β2 | φ2 | r0b | rssc | χ2 |
|---|---|---|---|---|---|---|---|
| w0 = 7 | 0.68 | 0.06 | 0.32 | 8.88 | 0.23 | 0.07 | 1.01 |
| w0 = 20 | 0.37 | 0.36 | 0.63 | 6.25 | 0.23 | 0.14 | 1.22 |
| Bulk water | 0.86 | 0.10 | 0.14 | 1.15 | 0.22 | 0.03 | 1.24 |
The excitation wavelength was 295 nm. Emission was collected at 335, 340 and 350 nm for samples corresponding to w0 = 7, 20 and bulk water, respectively. All other conditions are as in Figure 2. The errors were 5–10% for all parameters. See Experimental Section for other details.
r0 is the fundamental anisotropy. The value of r0 was kept constant at 0.2563 in the analysis.
rss is the steady state anisotropy, obtained by integration of the area under the time-resolved anisotropy decay curve.
Hydration-dependent Lifetimes of the CXCR1 N-domain Peptide
Fluorescence lifetime serves as a faithful indicator of the local environment in which a given fluorophore is localized.66 A typical decay profile of the CXCR1 N-terminal domain peptide in AOT reverse micelles with its triexponential fitting and the statistical parameters used to check the goodness of the fit are shown in Supplementary Figure 2. The fluorescence lifetimes of the CXCR1 N-terminal peptide incorporated in reverse micelles are shown in Table 2. All fluorescence decays could be fitted well with a triexponential function. The intensity-averaged mean fluorescence lifetime was chosen as an important parameter since it is independent of the method of analysis and the number of exponentials used to fit the time-resolved fluorescence decay and were calculated using eq 2. Mean fluorescence lifetimes of the CXCR1 N-terminal peptide in AOT reverse micelle with increasing hydration are shown in Figure 6. The mean lifetime decrease from a value of ~3.3 ns at w0 = 3 to a value of ~2.7 ns at w0 = 20 (~18% reduction). In general, tryptophan lifetimes are known to be reduced when exposed to polar environments due to fast deactivating processes under such condition.67 The decrease of mean lifetime with increasing hydration could be due to an increase in reverse micellar polarity (as evident from the red shift with increasing hydration, see Figure 3).
Table 2.
Representative Fluorescence Lifetimes of the CXCR1 N-domain Peptide in AOT Reverse Micellesd
| w0 | α1 | τ1 (ns) | α2 | τ2 (ns) | α3 | τ3 (ns) | χ2 |
|---|---|---|---|---|---|---|---|
| 3 | 0.30 | 0.42 | 0.52 | 1.97 | 0.18 | 5.14 | 1.21 |
| 5 | 0.19 | 0.34 | 0.47 | 1.47 | 0.34 | 4.09 | 1.02 |
| 10 | 0.25 | 0.47 | 0.42 | 1.59 | 0.33 | 3.93 | 0.92 |
| 15 | 0.15 | 0.26 | 0.47 | 1.30 | 0.38 | 3.63 | 1.02 |
| 20 | 0.32 | 0.44 | 0.48 | 1.71 | 0.20 | 4.28 | 1.11 |
The excitation wavelength was 295 nm. Emission was monitored at 335 nm. All other conditions are as in Figure 2. See Experimental Section for other details.
DISCUSSION
The CXCR1 receptor and its natural ligand interleukin-8 (IL-8) play a crucial role in host immune response by regulating neutrophil recruitment and its activation in eliminating a wide variety of microbial infections.28,35,68 CXCR1 belongs to the GPCR superfamily and it is now well established that CXCR1 activation involves interactions between the N-terminal loop of IL-8 and N-terminal domain and the receptor extracellular loop residues of CXCR1.28,35,36,69 Recent NMR structure of CXCR1 reveals that its N-terminal domain is highly dynamic (and therefore the first 20 N-terminal amino acids could not be resolved),32 yet anchored to the membrane surface, and that membrane dissociation is critical upon IL-8 binding.36 Knowledge of the molecular mechanism and structural features that mediate membrane anchoring and ligand recognition is therefore essential to understand how these interactions mediate CXCR1 function.
Considering chemokine ligands are unusually large compared to conventional GPCR class A agonists, N-terminal domain of chemokine receptors are relatively short compared to the N-terminal domain of other GPCRs.36 The N-terminal domain represents the most variable region among chemokine receptors in terms of sequence and length. This built-in diversity is an important factor that provides the specificity for each receptor sub-type. Exploring the role of the N-terminal domain of CXCR1 using the whole receptor is challenging. Fortunately, the N-terminal domain peptide functions as an excellent surrogate for monitoring the structural and functional features of the N-terminal domain of the intact receptor. Interestingly, the recent NMR structure of membrane-bound CXCR1 lacks information regarding the first 20 amino acids at the N-terminal domain due to the dynamic nature of this region.32 We previously characterized the binding of IL-8 to the CXCR1 N-domain peptide, and demonstrated that chemokine binding is mediated by both packing and electrostatic interactions, and that these interactions play a major role in determining selectivity, affinity, and the activation process.35,69,70 We proposed that the N-terminal domain residues function as the initial docking site, and that this initial binding induces structural and dynamic changes both in the ligand and receptor, that are critical for receptor signaling mediated via the IL-8 N-terminal residues. We previously utilized the intrinsic tryptophan fluorescence of the CXCR1 N-domain to study membrane interactions and showed that this domain experiences motional restriction upon interacting with membranes.38 The motional restriction, in turn, could result in loss of conformational entropy which would influence ligand binding of the receptor. In the present work, we explored the role of water and hydration in organizing the N-terminal domain in membranes, keeping in mind the implications for ligand binding and activation.
We have previously shown that the N-terminal domain peptide interacts with micelles and membranes in a sequence-specific manner.35,38 The tryptophan residues exhibit REES of 19 nm in membranes while the magnitude of REES observed was 13 nm in micelles. This implies that solvent relaxation is faster and degree of motional restriction is less in micelles, due to more water penetration in the headgroup region relative to membranes and reverse micelles.35,38 The loss of conformational entropy is therefore critically dependent on the geometrical constraints and levels of hydration of the host assembly. This is further supported by comparison of fluorescence anisotropy of the CXCR1 N-domain peptide in micelles, reverse micelles and membranes. The steady state fluorescence anisotropy exhibited by the N-domain peptide in reverse micelles of higher hydration (w0 = 20, see Table 1) approaches the corresponding value in micelles and membranes.38 In addition, time-resolved anisotropy decay provide important insights into the rotational dynamics of the confined peptide with increasing hydration. The changes in the shorter and larger rotational correlation times with hydration clearly indicate that dynamics of the peptide in reverse micelles is influenced by water dynamics. As stated above, reverse micelles offer the unique advantage of monitoring organization and dynamics of embedded molecules with varying degrees of hydration. Various types of water pools in reverse micelles, characterized by graded dynamics, represent interesting models for water present in biological systems such as membranes. Three kinds of water populations (pools) have been shown to coexist in reverse micelles (Figure 1a). These are bound, trapped, and free water.23,24 Among these, free water pool has been reported to be available only at w0 values greater than ~10.71 In the absence of free water (w0 = 7), local motion predominantly governs the N-terminal domain dynamics, while with the presence of free water (w0 = 20), segmental or global motion of the peptide become predominant.
The reverse micellar water pool has been shown to substantially influence the conformational states and dynamics of peptides solubilized in it.72 The conformation of the N-terminal domain peptide in reverse micelles merits comment. Interestingly, the effect of constrained environment on the conformation of proteins and peptides appears to depend on the sequence and hydrophobicity. For example, while myelin basic protein73 and melittin12 fold into α-helical structures in reverse micellar environment, the peptide hormone adrenocorticotropin-(1–24) assumes a β-sheet structure.74 Other proteins such as cutinase75 become disordered in reverse micelles. In our case, the N-terminal domain of CXCR1 assumes ordered conformation with increased hydration.
These results assume relevance in the context of a recent report that the N-terminal domain of CXCR1 is anchored into the membrane through hydrophobic contacts mediated by tryptophan residue (see Figure 1b).36 We have previously shown that a scrambled peptide with identical amino acid composition and charge does not interact with either micelles and membranes. This suggests that it is the sequence that plays a predominant role (and not electrostatics) in the interaction between the N-terminal domain of CXCR1 and the interface of the host assembly. Moreover, IL-8 binds to CXCR1 N-domain in the free and micelle-bound forms,35 thereby indicating receptor residues that bind IL-8 are accessible both in the free and membrane-bound forms. Although the precise membrane-bound N-domain structure is not known, it is unlikely that the lysine side chain binds to host assembly headgroup, and most likely that backbone amide polar groups and tryptophan residues mediate the binding process. In addition, considering that the interactions are dynamic and interfacial, it is likely that the N-domain peptide lies on the membrane surface with minimal penetration to the membrane interior.
In this paper, we explored the organization and dynamics of the CXCR1 N-terminal domain in a membrane-mimetic reverse micellar environment with varying hydration. Our results show that the secondary structure adopted by the CXCR1 N-domain is critically dependent on hydration. This is supported by fluorescence spectroscopic studies of the CXCR1 N-domain with varying hydration. We show here that the tryptophan residues of the CXCR1 N-domain experience motional restriction and exhibit REES when incorporated in reverse micelles. Interestingly, REES and fluorescence lifetime display reduction with increasing hydration, indicating exposure of the tryptophan residues to more free water. The effect of hydration was reflected in the dynamics of the peptide tryptophan residues upon incorporation in reverse micelles. To the best of our knowledge, this is the first report demonstrating modulation in the organization and dynamics of the functionally important N-terminal domain of any chemokine receptor in membrane-mimetic environment of varying hydration. Our results assume significance in the context of the role of hydration in the function of chemokine receptors in particular and GPCRs in general.76,77
Supplementary Material
Acknowledgments
This work was supported by research grants from the Council of Scientific and Industrial Research (A.C.), and Department of Atomic Energy (G.K.), Govt. of India, and NIH grant R01-AI069152 to K.R. Ar.C. and S.H. thank the Council of Scientific and Industrial Research for the award of Senior Research Fellowships. P.B. was awarded a Summer Training Program Internship by the Centre for Cellular and Molecular Biology, Hyderabad. A.C. is an Adjunct Professor at the Special Centre for Molecular Medicine of Jawaharlal Nehru University (New Delhi, India) and Indian Institute of Science Education and Research (Mohali, India), and Honorary Professor at the Jawaharlal Nehru Centre for Advanced Scientific Research (Bangalore, India). A.C. and G.K. gratefully acknowledge J.C. Bose Fellowship (Department of Science and Technology, Govt. of India). We thank Saswata Sarkar for helpful discussions and members of A.C.’s research group for critically reading the manuscript.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Representative time-resolved anisotropy decay and time-resolved fluorescence intensity decay of CXCR1 N-terminal domain peptide in AOT reverse micelles corresponding to various hydration and statistical parameters used to check goodness of fit. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fenimore PW, Frauenfelder H, McMohan BH, Parak FG. Proc Natl Acad Sci USA. 2002;99:16047–16051. doi: 10.1073/pnas.212637899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattos C. Trends Biochem Sci. 2002;27:203–208. doi: 10.1016/s0968-0004(02)02067-4. [DOI] [PubMed] [Google Scholar]
- 3.Timasheff SN. Biochemistry. 2002;41:13473–13482. doi: 10.1021/bi020316e. [DOI] [PubMed] [Google Scholar]
- 4.Levy Y, Onuchic JN. Annu Rev Biophys Biomol Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 5.Hilser VJ. Nature. 2011;469:166–167. doi: 10.1038/469166a. [DOI] [PubMed] [Google Scholar]
- 6.Bizzarri AR, Cannistraro S. J Phys Chem B. 2002;106:6617–6633. [Google Scholar]
- 7.Jha A, Ishii K, Udgaonkar JB, Tahara T, Krishnamoorthy G. Biochemistry. 2011;50:397–408. doi: 10.1021/bi101440c. [DOI] [PubMed] [Google Scholar]
- 8.McAuley KE, Fyfe PK, Ridge JP, Isaacs NW, Cogdell RJ, Jones MR. Proc Natl Acad Sci USA. 1999;96:14706–14711. doi: 10.1073/pnas.96.26.14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kouyama T, Nishikawa T, Tokuhisa T, Okumura H. J Mol Biol. 2004;335:531–546. doi: 10.1016/j.jmb.2003.10.068. [DOI] [PubMed] [Google Scholar]
- 10.Mittal J, Best RB. Proc Natl Acad Sci USA. 2008;105:20233–20238. doi: 10.1073/pnas.0807742105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou H-X, Rivas G, Minton AP. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghuraman H, Chattopadhyay A. Langmuir. 2003;19:10332–10341. [Google Scholar]
- 13.Kelkar DA, Chattopadhyay A. Biophys J. 2005;88:1070–1080. doi: 10.1529/biophysj.104.051490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haldar S, Chattopadhyay A. J Phys Chem B. 2007;111:14436–14439. doi: 10.1021/jp076797z. [DOI] [PubMed] [Google Scholar]
- 15.Van Horn WD, Ogilvie ME, Flynn PF. J Am Chem Soc. 2009;131:8030–8039. doi: 10.1021/ja901871n. [DOI] [PubMed] [Google Scholar]
- 16.Tian J, Garcia AE. Biophys J. 2009;96:L57–L59. doi: 10.1016/j.bpj.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nucci NV, Pometun MS, Wand AJ. Nat Struct Mol Biol. 2011;18:245–250. doi: 10.1038/nsmb.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haldar S, Chattopadhyay A. In: Reviews in Fluorescence 2010. Geddes CD, editor. Springer; New York: 2012. pp. 155–172. [Google Scholar]
- 19.Luisi PL, Magid LJ. CRC Crit Rev Biochem. 1986;20:409–473. doi: 10.3109/10409238609081999. [DOI] [PubMed] [Google Scholar]
- 20.Granick S. Science. 1991;253:1374–1379. doi: 10.1126/science.253.5026.1374. [DOI] [PubMed] [Google Scholar]
- 21.Brubach J-B, Mermet A, Filabozzi A, Gerschel A, Lairez D, Krafft MP, Roy P. J Phys Chem B. 2001;105:430–435. [Google Scholar]
- 22.Levinger NE. Science. 2002;298:1722–1723. doi: 10.1126/science.1079322. [DOI] [PubMed] [Google Scholar]
- 23.Jain TK, Varshney M, Maitra A. J Phys Chem. 1989;93:7409–7416. [Google Scholar]
- 24.Ikushima Y, Saito N, Arai M. J Colloid Interface Sci. 1997;186:254–263. doi: 10.1006/jcis.1996.4641. [DOI] [PubMed] [Google Scholar]
- 25.Venables DS, Huang K, Schmuttenmaer CA. J Phys Chem B. 2001;105:9132–9138. [Google Scholar]
- 26.Faeder J, Ladanyi BM. J Phys Chem B. 2000;104:1033–1046. [Google Scholar]
- 27.Moilanen DE, Fenn EE, Wong D, Fayer MD. J Am Chem Soc. 2009;131:8318–8328. doi: 10.1021/ja901950b. [DOI] [PubMed] [Google Scholar]
- 28.Rajagopalan L, Rajarathnam K. Biosci Rep. 2006;26:325–329. doi: 10.1007/s10540-006-9025-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen SJ, Crown SE, Handel TM. Annu Rev Immunol. 2007;25:787–820. doi: 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- 30.Onuffer JJ, Horuk R. Trends Pharmacol Sci. 2002;23:459–467. doi: 10.1016/s0165-6147(02)02064-3. [DOI] [PubMed] [Google Scholar]
- 31.Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis AA, Marassi FM, et al. Nature. 2012;491:779–784. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prado GN, Suetomi K, Shumate D, Maxwell C, Ravindran A, Rajarathnam K, Navarro J. Biochemistry. 2007;46:8961–8968. doi: 10.1021/bi7004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szpakowska M, Fievez V, Arumugan K, van Nuland N, Schmit J-C, Chevigné A. Biochem Pharmacol. 2012;84:1366–1380. doi: 10.1016/j.bcp.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Rajagopalan L, Rajarathnam K. J Biol Chem. 2004;279:30000–30008. doi: 10.1074/jbc.M313883200. [DOI] [PubMed] [Google Scholar]
- 36.Park SH, Casagrande F, Cho L, Albrecht L, Opella SJ. J Mol Biol. 2011;414:194–203. doi: 10.1016/j.jmb.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernando H, Nagle GT, Rajarathnam K. FEBS J. 2007;274:41–51. doi: 10.1111/j.1742-4658.2006.05579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haldar S, Raghuraman H, Namani T, Rajarathnam K, Chattopadhyay A. Biochim Biophys Acta. 2010;1798:1056–1061. doi: 10.1016/j.bbamem.2010.02.029. [DOI] [PubMed] [Google Scholar]
- 39.Rand RP. Science. 1992;256:618. doi: 10.1126/science.256.5057.618. [DOI] [PubMed] [Google Scholar]
- 40.Eastoe J, Young WK, Robinson BH, Steytler DC. J Chem Soc Faraday Trans. 1990;86:2883–2889. [Google Scholar]
- 41.Amararene A, Gindre M, Le Huérou J-Y, Urbach W, Valdez D, Waks M. Phys Rev E. 2000;61:682–689. doi: 10.1103/physreve.61.682. [DOI] [PubMed] [Google Scholar]
- 42.Bevington PR. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill; New York: 1969. [Google Scholar]
- 43.O’Connor DV, Phillips D. Time-correlated Single Photon Counting. Academic Press; London: 1984. pp. 180–189. [Google Scholar]
- 44.Lampert RA, Chewter LA, Phillips D, O’Connor DV, Roberts AJ, Meech SR. Anal Chem. 1983;55:68–73. [Google Scholar]
- 45.Grinvald A, Steinberg IZ. Anal Biochem. 1974;59:583–598. doi: 10.1016/0003-2697(74)90312-1. [DOI] [PubMed] [Google Scholar]
- 46.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; New York: 2006. [Google Scholar]
- 47.Rami BR, Krishnamoorthy G, Udgaonkar JB. Biochemistry. 2003;42:7986–8000. doi: 10.1021/bi030006b. [DOI] [PubMed] [Google Scholar]
- 48.Mukherjee S, Kombrabail M, Krishnamoorthy G, Chattopadhyay A. Biochim Biophys Acta. 2007;1768:2130–2144. doi: 10.1016/j.bbamem.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 49.Paila YD, Kombrabail M, Krishnamoorthy G, Chattopadhyay A. J Phys Chem B. 2011;115:11439–11447. doi: 10.1021/jp201458h. [DOI] [PubMed] [Google Scholar]
- 50.Haldar S, Chaudhuri A, Gu H, Koeppe RE, Kombrabail M, Krishnamoorthy G, Chattopadhyay A. J Phys Chem B. 2012;116:11056–11064. doi: 10.1021/jp304846f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callis PR. Methods Enzymol. 1997;278:113–150. doi: 10.1016/s0076-6879(97)78009-1. [DOI] [PubMed] [Google Scholar]
- 52.Pierce DW, Boxer SG. Biophys J. 1995;68:1583–1591. doi: 10.1016/S0006-3495(95)80331-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen B, Huppert D, Solntsev KM, Tsfadia Y, Nachliel E, Gutman M. J Am Chem Soc. 2002;124:7539–7547. doi: 10.1021/ja012646c. [DOI] [PubMed] [Google Scholar]
- 54.Haldar S, Chaudhuri A, Chattopadhyay A. J Phys Chem B. 2011;115:5693–5706. doi: 10.1021/jp200255e. [DOI] [PubMed] [Google Scholar]
- 55.Raghuraman H, Kelkar DA, Chattopadhyay A. In: Reviews in Fluorescence 2005. Geddes CD, Lakowicz JR, editors. Springer; New York: 2005. pp. 199–222. [Google Scholar]
- 56.Chattopadhyay A. Chem Phys Lipids. 2003;122:3–17. doi: 10.1016/s0009-3084(02)00174-3. [DOI] [PubMed] [Google Scholar]
- 57.Demchenko AP. Methods Enzymol. 2008;450:59–78. doi: 10.1016/S0076-6879(08)03404-6. [DOI] [PubMed] [Google Scholar]
- 58.Wyttenbach T, Bowers MT. Chem Phys Lett. 2009;480:1–16. [Google Scholar]
- 59.Rawat SS, Kelkar DA, Chattopadhyay A. Biophys J. 2004;87:831–843. doi: 10.1529/biophysj.104.041715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rawat SS, Kelkar DA, Chattopadhyay A. Biophys J. 2005;89:3049–3058. doi: 10.1529/biophysj.105.060798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhattacharyya K. Acc Chem Res. 2003;36:95–101. doi: 10.1021/ar020067m. [DOI] [PubMed] [Google Scholar]
- 62.Krishnamoorthy G. Curr Sci. 2012;102:266–276. [Google Scholar]
- 63.Eftink MR, Selvidge LA, Callis PR, Rehms AA. J Phys Chem. 1990;94:3469–3479. [Google Scholar]
- 64.Vos K, Laane C, Weijers SR, van Hoek A, Veeger C, Visser AJWG. Eur J Biochem. 1987;169:259–268. doi: 10.1111/j.1432-1033.1987.tb13606.x. [DOI] [PubMed] [Google Scholar]
- 65.Kinosita KJ, Kawato S, Ikegami A. Biophys J. 1977;20:289–305. doi: 10.1016/S0006-3495(77)85550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prendergast FG. Curr Opin Struct Biol. 1991;1:1054–1059. [Google Scholar]
- 67.De Lauder WB, Wahl Ph. Biochim Biophys Acta. 1971;243:153–163. doi: 10.1016/0005-2795(71)90071-7. [DOI] [PubMed] [Google Scholar]
- 68.Mukaida N. Am J Physiol Lung Cell Mol Physiol. 2003;284:L566–L577. doi: 10.1152/ajplung.00233.2002. [DOI] [PubMed] [Google Scholar]
- 69.Ravindran A, Joseph PR, Rajarathnam K. Biochemistry. 2009;48:8795–8805. doi: 10.1021/bi901194p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rajarathnam K, Prado GN, Fernando H, Clark-Lewis I, Navarro J. Biochemistry. 2006;45:7882–7888. doi: 10.1021/bi0605944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goto A, Yoshioka H, Manabe M, Goto R. Langmuir. 1995;11:4873–4975. [Google Scholar]
- 72.Fiori S, Renner C, Cramer J, Pegoraro S, Moroder L. J Mol Biol. 1999;291:163–175. doi: 10.1006/jmbi.1999.2951. [DOI] [PubMed] [Google Scholar]
- 73.Nicot C, Vacher M, Vincent M, Gallay J, Waks M. Biochemistry. 1985;24:7024–7032. doi: 10.1021/bi00345a041. [DOI] [PubMed] [Google Scholar]
- 74.Gallay J, Vincent M, Nicot C, Waks M. Biochemistry. 1987;26:5738–5747. doi: 10.1021/bi00392a024. [DOI] [PubMed] [Google Scholar]
- 75.Melo EP, Costa SMB, Cabral JMS. Photochem Photobiol. 1996;63:169–175. [Google Scholar]
- 76.Mitchell DC, Litman BJ. Biochemistry. 1999;38:7617–7623. doi: 10.1021/bi990634m. [DOI] [PubMed] [Google Scholar]
- 77.Angel TE, Chance MR, Palczewski K. Proc Natl Acad Sci USA. 2009;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.