Abstract
Myotonic dystrophy type 1 is a complex multisystemic inherited disorder, which displays multiple debilitating neurological manifestations. Despite recent progress in the understanding of the molecular pathogenesis of myotonic dystrophy type 1 in skeletal muscle and heart, the pathways affected in the central nervous system are largely unknown. To address this question, we studied the only transgenic mouse line expressing CTG trinucleotide repeats in the central nervous system. These mice recreate molecular features of RNA toxicity, such as RNA foci accumulation and missplicing. They exhibit relevant behavioural and cognitive phenotypes, deficits in short-term synaptic plasticity, as well as changes in neurochemical levels. In the search for disease intermediates affected by disease mutation, a global proteomics approach revealed RAB3A upregulation and synapsin I hyperphosphorylation in the central nervous system of transgenic mice, transfected cells and post-mortem brains of patients with myotonic dystrophy type 1. These protein defects were associated with electrophysiological and behavioural deficits in mice and altered spontaneous neurosecretion in cell culture. Taking advantage of a relevant transgenic mouse of a complex human disease, we found a novel connection between physiological phenotypes and synaptic protein dysregulation, indicative of synaptic dysfunction in myotonic dystrophy type 1 brain pathology.
Keywords: myotonic dystrophy, transgenic mice, synaptic transmission, RAB3A, synapsin I
Introduction
Myotonic dystrophy is the most common form of adult muscular dystrophy (Harper, 2001). Myotonic dystrophy type 1 is caused by the expansion of a CTG trinucleotide repeat in the 3′-UTR of the dystrophia myotonica-protein kinase (DMPK) gene (Brook et al., 1992; Fu et al., 1992; Mahadevan et al., 1992). CTG trinucleotide repeat number correlates directly with disease severity and inversely with age of onset. The myotonic dystrophy type 1 repeat shows a marked tendency towards further expansion in intergenerational transmissions and somatic tissues (Gomes-Pereira and Monckton, 2006). The prevailing model of disease pathogenesis points to a trans-dominant effect of expanded DMPK transcripts, which accumulate in nuclear foci, interfering with at least two families of alternative splicing regulators: the muscleblind-like (MBNL) and CUGBP/Elav-like (CELF) proteins. MBNL1 loss of function, through sequestration into ribonuclear foci (Miller et al., 2000), and CELF1 upregulation (Timchenko et al., 2001) disturb a developmentally regulated splicing programme, resulting in aberrant expression of embryonic isoforms in adult skeletal muscle and heart (Ranum and Cooper, 2006). Missplicing explains important myotonic dystrophy type 1 symptoms, such as myotonia (Charlet et al., 2002; Mankodi et al., 2002; Lueck et al., 2007) and insulin resistance (Savkur et al., 2001).
Although traditionally considered a muscle disease, myotonic dystrophy type 1 presents many debilitating neurological manifestations. Patients with adult-onset myotonic dystrophy type 1 show prevalent hypersomnia and fatigue, as well as visuoconstructive impairment, attention deficits, reduced initiative and apathy (Harper, 2001; Meola and Sansone, 2007), suggesting executive dysfunction and the involvement of the frontal lobes (Meola et al., 2003; Gaul et al., 2006; Meola and Sansone, 2007; Sistiaga et al., 2010). Reduced IQs were measured in one-third of adult and in the majority of childhood-onset patients (Angeard et al., 2007; Meola and Sansone, 2007), whereas moderate to severe mental retardation is a feature of congenital myotonic dystrophy type 1 (Harper, 2001). Additional signs of personality disorder, such as increased anxiety, depression and anhedonia, were reported in a proportion of patients with myotonic dystrophy type 1 (Bungener et al., 1998; Delaporte, 1998; Meola et al., 2003; Winblad et al., 2005). Region-specific structural abnormalities (including white and grey matter affection) and metabolic changes, revealed by imaging studies, may contribute to myotonic dystrophy type 1 neuropsychological manifestations (Romeo et al., 2010; Weber et al., 2010; Minnerop et al., 2011).
RNA foci accumulate in post-mortem myotonic dystrophy type 1 brain and co-localize with MBNL1 and MBNL2 (Jiang et al., 2004). Nevertheless, the mechanistic links between the genetic mutation and neuropsychological abnormalities remain elusive (Meola and Sansone, 2007). We previously generated transgenic mice expressing DMPK transcripts in multiple tissues under the control of the human DMPK gene promoter, within the environment of the human myotonic dystrophy type 1 locus. In contrast with control DM20 lines (mice overexpressing short 20 CTG tracts), homozygous mice from two independent DM300 expansion lines (carrying 500–600 CTG), expressing sufficient toxic DMPK transcripts, showed wide RNA foci accumulation in a variety of tissues and developed a multisystemic phenotype (Seznec et al., 2001; Guiraud-Dogan et al., 2007; Panaite et al., 2008). Dramatic intergenerational instability in DM300 mice generated DMSXL animals carrying >1000 CTG, who develop a more severe phenotype (Gomes-Pereira et al., 2007; Huguet et al., 2012). We now used DMSXL mice to characterize RNA toxicity in the brain and to identify disease intermediates and pathways affected by myotonic dystrophy type 1 in the CNS. We gathered electrophysiological, neurochemical and molecular evidence of synaptic protein dysregulation and synaptic dysfunction, which translates into myotonic dystrophy type 1-associated behaviour deficits and, most certainly, mediates neurological symptoms.
Materials and methods
Transgenic mice
The myotonic dystrophy transgenic mice used in this study carried 45 kb of human genomic DNA cloned from a patient with myotonic dystrophy type 1 as described previously (Seznec et al., 2000; Gomes-Pereira et al., 2007). Transgenic mice were raised and kept at Centre d’Exploration et de Recherche Fonctionnelle Expérimentale (CERFE, Genopole, Evry, France). Animal housing, care and handling were carried out according to the French and European legislations and the ethical guidelines of the host institution. Genotyping procedures are described in the Supplementary material. The generation, characterization and genotyping of Mbnl1 and Mbnl2 knockout mice are described elsewhere (Kanadia et al., 2003; Charizanis et al., 2012).
Quantitative real-time reverse transcription polymerase chain reaction quantification of DMPK and Rab3A transcripts
Total RNA extraction and complementary DNA synthesis were performed as previously described (Gomes-Pereira et al., 2007). DMPK, Dmpk and Rab3A transcripts were amplified in a 7300 Real Time PCR System (Applied Biosystems) using SYBR® Green detection and oligonucleotide primer sequences described in Supplementary Table 1. DMPK and Rab3A messenger RNA levels were expressed relative to 18S transcripts. The quantification of DMPK transcript levels was performed on a complementary DNA sample synthesized from equal RNA quantities extracted from three individual DMSXL homozygous mice. Rab3A transcript levels were quantified in six individual DMSXL homozygotes and wild-type control mice. Two independent replicate experiments were performed.
Fluorescent in situ hybridization
Ribonuclear inclusions were detected with a 5′-FITC-labelled 2′-O-methyl-(CAG)10 oligonucleotide probe or a 5′-Cy3-labelled (CAG)5 peptide nucleic acid probe, as previously described (Seznec et al., 2001). RNA foci quantification was performed using Discovery Automaton (Roche) (Bassez et al., 2008). Mouse brain regions of interest were anatomically identified, and the percentage of cells containing nuclear foci was calculated based on the observation of >100 cells in each brain region, in each mouse.
Immunofluorescence combined with fluorescent in situ hybridization
After the SSC post-hybridization wash of the fluorescent in situ hybridization procedure, sections were incubated in primary antibodies overnight at 4°C (antibody references and working dilutions are listed in Supplementary Table 2), washed five times with PBS for 2 min and then incubated in secondary antibody and 0.001% (m/v) diamidino-2-phenylindole for 15 min at room temperature. Sections were washed five times in PBS before mounting.
Reverse transcription polymerase chain reaction analysis of alternative splicing
Total RNA extraction, complementary DNA synthesis and semi-quantitative reverse transcriptase PCR analysis were performed as previously described (Gomes-Pereira et al., 2007), using oligonucleotide primers described in Supplementary Table 1. The inclusion ratio of alternative exons in each individual animal was determined in two replicate PCR amplifications to minimize experimental variation. The percentage of exon inclusion was calculated as [exon inclusion band/(exon inclusion band + exon exclusion band)] × 100.
Behavioural analysis
Because DMSXL mice display muscle deficits (Huguet et al., 2012), behavioural tests were carefully selected, designed and interpreted to minimize the confounding effects of muscular impairment on brain functional deficits. All the experiments were performed according to the guidelines of the French Ministry of Agriculture for laboratory animal experimentation (law 87-848; Agreement N: A-45-234-8), and the mouse genotype was unknown to the researcher. Mouse behavioural testing was conducted to reduce animal stress to the minimum. Three groups of animals were studied, to limit the number of tests performed on each animal. Group A: open-field, Y maze, passive avoidance and sucrose intake; Group B: marble-burying and Morris water maze; and Group C: saccharine intake. Further details on behavioural phenotyping are described in the Supplementary material.
Electrophysiological profiling
Input/output properties, paired-pulse facilitation long-term depression and long-term potentiation were assessed on male DMSXL homozygotes and wild-type control mice, aged 7 months (input/output, paired-pulse facilitation and long-term depression) or 4 months (long-term potentiation). Mice were sacrificed by fast decapitation, without previous anaesthesia. Brains were processed in oxygenated buffer and artificial CSF as previously described (Steidl et al., 2006). Extracellular field excitatory post-synaptic potentials were measured with Multi-Electrode Arrays technology (100-µm distant electrodes) on 350-µm-thick hippocampal slices. One of the electrodes stimulated Schaeffer collaterals at the CA3/CA1 interface. The stimulus, consisting of a monopolar biphasic current pulse (negative for 60 μs and then positive for 60 μs), evoked responses (field excitatory post-synaptic potentials) in the CA1 region. Input/output properties: input/output curves were plotted by measuring the response evoked by stimulation intensities varying between 100 and 800 µA. Details on the electrophysiological procedures are included in the Supplementary material.
Two-dimensional protein electrophoresis and mass spectrometry
Ten-month-old female mice were used to compare the global proteomic profile in frontal cortex and brainstem between six homozygotes carrying expanded repeats (∼500–600 CTG) and six females carrying short 20-CTG control tracts. Isoelectric focusing and mass spectrometry were performed as previously described (Sergeant et al., 2003). CELF1 and CELF2 phosphorylation was assessed by 2D gel electrophoresis and immunodetection, using the antibodies shown in Supplementary Table 2.
Western blot analysis
Primary antibody references and working dilutions are shown in Supplementary Table 2.
Neurosecretion assays
Neurosecretion assays in cell culture were performed as described previously (Lee et al., 2007).
Human tissue samples
Autopsy materials were obtained from nine patients with myotonic dystrophy type 1 (Patients G–O; mean age: 60.9 years, range: 32–73 years) and six non-myotonic dystrophy type 1 control subjects (Patients A–F; three with no neurological disease, one with Charcot–Marie–Tooth disease, one with a brain tumour and one with limb-girdle muscular dystrophy; mean age: 68.2 years, range: 53–79 years). Six patients with myotonic dystrophy type 1 (Patients I, J, M, N and O) had signs of classical adult-onset myotonic dystrophy type 1, and they all died of complications related to the disease (respiratory failure or heart disease). Patient G developed late-onset myotonic dystrophy type 1. Patient H carried a large CTG repeat expansion in blood and brain tissue; her clinical form of myotonic dystrophy type 1 was not fully described by the clinicians, but she presented gait problems and general brain atrophy evidenced by neuroimaging. The clinical form of Patient L was not fully described; she carried large CTG repeats, presented gait problems and mild cognitive impairment, and her imaging assessment revealed general brain atrophy. Patient K exhibited mental retardation and was diagnosed with congenital myotonic dystrophy type 1. He died at the age of 32 years from pneumonia. Neuropshychological profiling and neuroimaging were not systematically conducted on the majority of these patients. Further patient details are given in Supplementary Table 3. All experiments using human samples were approved by the Ethics Committee of Asahikawa Medical Centre and Okayama University. Written informed consent specimen use for research was obtained from all patients.
Statistical analysis
Statistical analyses were performed with JMP 5, Prism 5 and Excel software. When two groups were compared, a two-tailed Student’s t-test for statistical significance was performed, unless otherwise stated. The significance level was set at P < 0.05 for all statistical analyses. All data are expressed as mean ± standard error of the mean (SEM), unless otherwise stated.
Results
Expression of myotonic dystrophy type 1 CTG expansions results in RNA foci accumulation and missplicing in the central nervous system of DMSXL mice
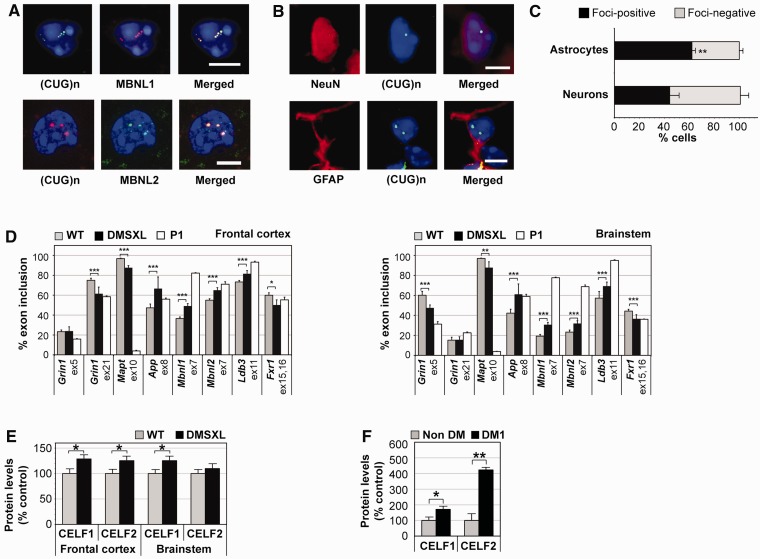
The toxic RNA hypothesis proposes that accumulation of CUG-containing transcripts is the initiating pathogenic event in myotonic dystrophy type 1. To assess to what extent DMSXL mice recreate central molecular aspects of myotonic dystrophy type 1, we measured DMPK transgene expression in CNS regions dissected from 1-month-old DMSXL homozygous mice and found expanded transcripts in all CNS regions investigated, with some regional differences (Supplementary Fig. 1A); DMPK transcript levels were higher in the hippocampus, thalamus/hypothalamus, cerebellum and brainstem than in frontal and temporal cortex and in striatum. Overall, the expression levels of the DMPK transgene in frontal cortex and cerebellum were approximately three times higher than those of the endogenous Dmpk gene (Huguet et al., 2012). Expanded DMPK transcripts accumulated in 1 to ∼20 ribonuclear inclusions per cell nucleus in both neurons and astrocytes (Fig. 1A) and co-localized with MBNL1 and MBNL2 (Fig. 1B). Astrocytes showed a significantly higher frequency of foci than neurons overall (62 versus 44% foci-containing nuclei) (Fig. 1C). Interestingly, foci distribution was not homogeneous throughout DMSXL brains: they were present in 20% nuclei of striatum, up to 70% in brain cortex and ∼80% in the dorsal raphe and raphe magnus nuclei of brainstem (Supplementary Fig. 1B). In addition to the accumulation of RNA foci in neurons previously reported in myotonic dystrophy type 1 human brains (Jiang et al., 2004), we found nuclear foci in human astrocytes (Supplementary Fig. 1C).
Figure 1.
Expression of an expanded DMPK transgene induces foci accumulation and splicing dysregulation in the CNS. (A) Fluorescent in situ hybridization and immunofluorescence revealed nuclear foci of transgenic DMPK messenger RNA in both NeuN-positive neurons and GFAP-positive astrocytes, in multiple regions of the brain of 1-month-old DMSXL homozygotes. (B) MBNL1 and MBNL2 co-localized with nuclear RNA foci. No RNA foci were observed in DM20 and wild-type control animals (data not shown). Punctuated MBNL2 staining was occasionally detected and did not correspond to RNA aggregates. Diamidino-2-phenylindole was used for nuclear staining. Scale bar = 5 µm. (C) Quantification of the percentage of astrocytes and neurons exhibiting nuclear RNA aggregates (±SEM) throughout the brain of 1-month-old DMSXL homozygotes (n = 2) (**P < 0.01, χ2 test). (D) Percentage of inclusion of alternative exons in messenger RNA transcripts encoding: GRIN1/NMDAR1 glutamate receptor, ATP2A1/SERCA1 endoplasmic calcium ATPase, microtubule-associated protein tau (MAPT/TAU), amyloid beta precursor protein (APP), insulin receptor (INSR), MBNL1 and MBNL2 splicing regulators, LDB3/CYPHER cytoskeleton-interacting protein and FXR1 RNA-binding protein. The analysis was performed in frontal cortex and brainstem of homozygous DMSXL mice (n = 9) and wild-type control mice (n = 9) at age 1 month. Splicing profiles were compared with those of wild-type newborns collected as post-natal Day 1 (n = 3). The graphs show the average fractional inclusion of the specified exon in triplicate assays (±SEM). (E) Quantification of CELF proteins in homozygous DMSXL (n = 3) and wild-type mice (n = 3) at age 1 month. (F) Quantification of CELF proteins in frontal cortex of adult myotonic dystrophy type 1 individuals (n = 9) and non-myotonic dystrophy control subjects (n = 3). The graphs in E and F show the average steady-state levels relative to normalized control subjects. *P < 0.05, **P < 0.01, ***P < 0.001. WT = wild-type.
To investigate the trans-dominant effect of DMPK transcripts in the CNS, we studied the alternative splicing of candidate genes. We focused primarily on foci-rich brain regions relevant to disease manifestations. Whereas frontal cortex is involved in executive tasks (Robbins and Arnsten, 2009), brainstem is involved in the control of sleep cycles, respiratory and cardiac function (Izac and Eeg, 2006). Mild region-specific missplicing events were detected in frontal cortex and brainstem of 1-month-old DMSXL homozygotes (Fig. 1D and Supplementary Fig. 2A).
The missplicing of Mbnl1 exon 7 and Ldb3 exon 11, two alternative exons specifically regulated by MBNL1 activity (Kalsotra et al., 2008), confirmed the impact of the sequestration of MBNL proteins (Fig. 1B) on alternative splicing deregulation. Similarly, mild missplicing of CELF1-dependent exons 15 and 16 of Fxr1 predicted a role of CELF1 in CNS spliceopathy. To confirm this hypothesis, we quantified the steady-state levels of CELF proteins and found significant CELF1 and CELF2 upregulation (of ∼30%) in DMSXL frontal cortex (Fig. 1E and Supplementary Fig. 1D). In DMSXL brainstem, only CELF1 was significantly upregulated. CELF1 upregulation was associated with mild protein hyperphosphorylation in DMSXL frontal cortex and brainstem (Supplementary Fig. 1E). CELF2 phosphorylation levels did not differ between genotypes in frontal cortex and seemed to decrease in DMSXL brainstem. The upregulation of CELF proteins was more pronounced in human myotonic dystrophy type 1 frontal cortex: CELF1 levels were 70% higher and CELF2 showed a 4-fold increase relative to non-myotonic dystrophy individuals (Fig. 1F and Supplementary Fig. 1F).
Interestingly, wild-type splicing profiles were region-specific (e.g. Grin1/Nmdar1; Fig. 1D and Supplementary Fig. 2A). Regional differences were associated with varying steady-state levels of MBNL and CELF proteins between frontal cortex and brainstem in adult mice (Supplementary Fig. 1G), suggesting a determining role of the ratio between antagonistic splicing regulators in the establishment of adult splicing profiles.
We studied the alternative splicing of alternative genes throughout wild-type mouse brain development (Supplementary Fig. 2B), and found that the DMSXL spliceopathy in the CNS modified certain transcripts towards embryonic/newborn splicing profiles. For instance, the splicing patterns of Grin1 exon 21 and Mbnl2 exon 7 in DMSXL frontal cortex, as well as App exon 8 and Frx1 exons 15/16 in DMSXL brainstem, resembled, to a limited extent, the splicing patterns of newborn wild-type mice. The effect was less pronounced in other alternative exons that displayed intermediate inclusion ratios in DMSXL mice of between those characteristic of the embryonic and adult developmental stages (e.g. Ldb3 exon 11 in frontal cortex and brainstem; Grin1 exon 5 in brainstem).
CTG repeat expansions induce mouse behavioural abnormalities
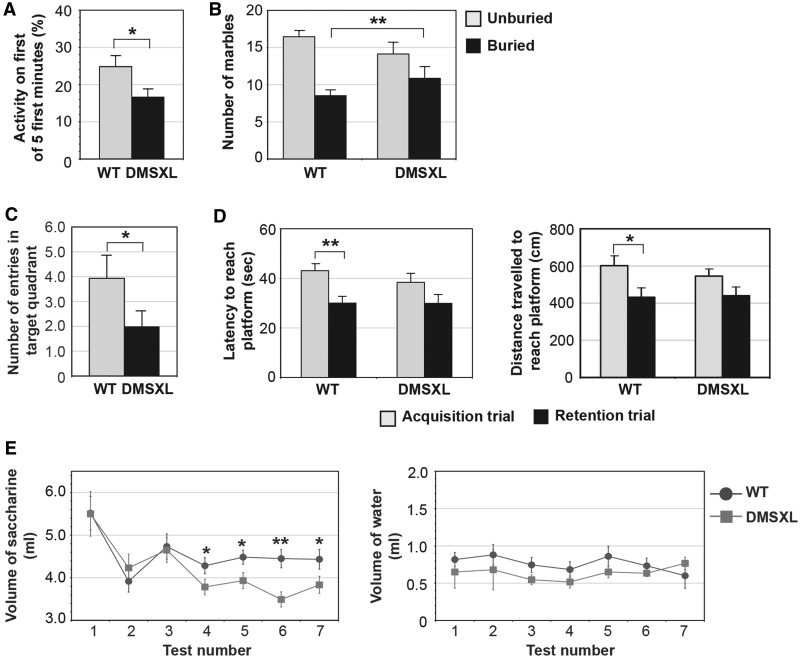
Following the validation of toxic RNA expression, foci accumulation and missplicing in DMSXL brains, we then investigated the impact of expanded DMPK transcripts on mouse behaviour and cognition through blinded phenotyping of adult DMSXL homozygotes. We first assessed mouse activity in the open-field test. DMSXL mice displayed overall levels of horizontal (P = 0.402, Student’s t-test) and vertical activity (P = 0.355, Student’s t-test) similar with those of wild-type control mice, excluding a major effect of muscular deficits on mouse performance in this test (Supplementary Fig. 3A). However, DMSXL mice showed a significant decrease in exploratory activity, shortly after transfer into the open-field arena. Although the total number of rearings did not differ between the two genotypes, the percentage of rearings over the first minute out of the first 5 min spent in the arena was significantly lower in DMSXL mice (Fig. 2A). This result reveals freezing behaviour in response to a new and unfamiliar environment and indicates novelty-induced inhibition, suggestive of increased anxiety. This was confirmed by the assessment of obsessive–compulsive behaviour using a marble-burying test. DMSXL mice displayed a statistically significant shift towards a higher number of fully buried marbles (Fig. 2B), indicative of increased anxiety.
Figure 2.
DMSXL exhibit novelty-induced inhibition, anxiety, spatial and working memory impairment and anhedonia. (A) Assessment of novelty-induced inhibition in DMSXL homozygotes (n = 16) and age-matched control mice (n = 16). The graph represents the percentage of the number of rearings (±SEM) in the first minute out of the first 5 min spent in the first open-field session. (B) Assessment of mouse anxiety levels by the marble-burying test. The graph represents the average number of marbles (±SEM) unburied and buried by DMSXL (n = 15) and wild-type mice (n = 15). The DMSXL line shows a significant shift towards a higher number of buried marbles (P = 0.0087, Fisher’s exact test). (C) Spatial memory assessment by the Morris water maze test. The graph represents the number of entries in the target quadrant (±SEM) during the probe trial, for DMSXL (n = 15) and wild-type (n = 15) mice. (D) Working memory assessment by the Morris water maze test. The graphs represent the average time and distance travelled (±SEM) to reach the platform in acquisition and retention trials for both genotypes (n = 15 per genotype). (E) Saccharine consumption test for anhedonia. The graph on the left represents the average volume of saccharine solution (±SEM) drank by DMSXL homozygotes (n = 12) and wild-type control mice (n = 12). The volume of water consumed once saccharine became available did not differ between the two genotypes (graph on the right). *P < 0.05; **P < 0.01. WT = wild-type.
We then examined whether the expression of expanded DMPK transcripts resulted in memory impairment as observed in patients with myotonic dystrophy type 1. Mouse spatial memory was assessed by the Morris water maze. The platform was located in the same target quadrant during the training period, and it was removed during the probe trial. DMSXL mice showed impaired spatial memory, illustrated by a significantly lower number of entries in the target quadrant during the probe trial (Fig. 2C). DMSXL swimming speed was not significantly different between genotypes (Supplementary Fig. 3B), excluding biases introduced by impaired motor performance. In the assessment of working memory, a component of the executive function (Robbins and Arnsten, 2009), both genotypes were capable of finding the platform in the acquisition trial. The apparent lower latency (P = 0.3066, Student's t-test) and shorter distance (P = 0.4490, Student’s t-test) travelled by DMSXL mice were not statistically different from those of wild-type control mice. Both genotypes showed similar performances in the acquisition trial. As expected, wild-type mice performed significantly better in the retention than in acquisition trial (latency: P = 0.0019; distance travelled: P = 0.0148; Student’s t-test), indicating that working memory was present. In contrast, DMSXL mouse performance did not improve significantly between the two trials (latency: P = 0.1916; distance travelled: P = 0.2726; Student’s t-test), suggesting possible mild working memory impairment (Fig. 2D). Non-spatial long-term memory, assessed by the passive avoidance conditioning test, was not affected in DMSXL mice (Supplementary Fig. 3C).
Anhedonia, previously reported in patients with myotonic dystrophy type 1, was assessed by the consumption of a highly palatable solution of saccharine (Schweizer et al., 2009). The similar volume of water drank during the habituation period by DMSXL and wild-type mice (Day 1: P = 0.136; Day 2: P = 0.538; Student’s t-test) shows similar satiety between the two genotypes (Supplementary Fig. 3D). Once saccharine became available, both animal groups showed a pronounced initial interest in the saccharine solution, showing again similar satiety over the first 3 days of testing (Test 1: P = 0.980; Test 2: P = 0.450; Test 3: P = 0.843; Student’s t-test). However, as the test progressed, DMSXL interest in the saccharine solution was significantly lower, as compared with wild-type control mice (Test 4: P = 0.043; Test 5: P = 0.037; Test 6: P = 0.003; Test 7: P = 0.043; Student’s t-test), suggesting an anhedonic-like behaviour (Fig. 2E).
DMSXL mice exhibit deficits in short-term synaptic plasticity
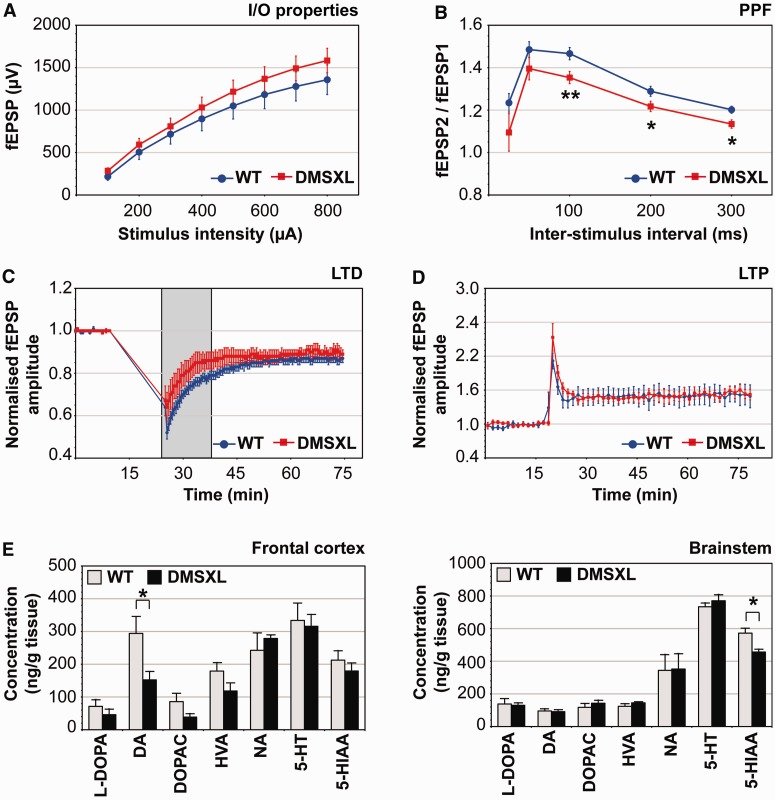
In parallel with the behavioural and cognitive phenotyping of DMSXL mice, we have assessed the physiological impact of toxic DMPK transcripts on synaptic function. To this end, we performed the electrophysiological profiling of DMSXL hippocampus, a brain region showing foci accumulation and missplicing (Supplementary Fig. 4). We first examined basal synaptic transmission by stimulating hippocampal Schaffer collaterals at increasing intensities and generating input/output curves from measures of field excitatory post-synaptic potentials. DMSXL homozygotes exhibited slightly higher field excitatory post-synaptic potentials relative to wild-type control mice, but the overall input/output curves were similar between both genotypes (Fig. 3A), indicating no major deficits in basal transmission. We next investigated some short-term plasticity properties through the quantification of paired-pulse facilitation. DMSXL slices displayed significantly reduced paired-pulse facilitation ratios, indicative of presynaptic dysfunction (Fig. 3B). Finally, we measured long-term depression and potentiation, which support some forms of learning and memory. Standard long-term depression and long-term potentiation protocols did not reveal overt abnormalities in DMSXL mice (Fig. 3C and D). However, early after low-frequency stimulation, long-term depression amplitude was slightly lower in DMSXL than in wild-type slices (36 ± 7% versus 47 ± 3%), which might suggest mild impairment of the post-synaptic response. The difference between genotypes, however, did not reach statistical significance (P = 0.142, repeated measures two-way ANOVA). Depression stabilized after 20 min, being similar for both genotypes at the endpoint of the experiment. In summary, the number of animals studied did not show marked abnormalities in long-term plasticity, but revealed significant deficits in short-term synaptic plasticity.
Figure 3.
DMSXL mice exhibit deficits in short-term synaptic plasticity and changes in neurochemical levels. (A) Electrophysiological profiling of DMSXL mice and age-matched control mice. Input/output (I/O) characteristics in the CA1 region. Mean value of field excitatory post-synaptic potentials (fEPSP) (±SEM) is expressed as a function of stimulation intensity. (B) Paired-pulse facilitation (PPF) in the CA1 region. The mean ratio of the second peak compared with the first one (fEPSP2/fEPSP1, ±SEM) is expressed as a function of the interstimulus interval. Paired-pulse facilitation values were significantly lower in DMSXL hippocampal slices (repeated measures two-way ANOVA). The difference between genotypes was more pronounced for interstimulus intervals of 100, 200 and 300 ms. (C) Long-term depression (LTD) in the CA1 region. Mean value of normalized field excitatory post-synaptic potentials amplitude (±SEM) is expressed as a function of time. Long-term depression amplitude was slightly lower in DMSXL slices, shortly after low frequency stimulation (grey box), but overall long-term depression amplitude was not significantly different between DMSXL and wild-type mice. (D) Long-term potentiation (LTP) in the CA1 region. Mean value of normalized field excitatory postsynaptic potentials amplitude (±SEM) is expressed as a function of time. Long-term potentiation did not significantly differ between DMSXL and wild-type mice. Input/output, paired-pulse facilitation and long-term depression data correspond to values averaged from 16 independent slices prepared from DMSXL (n = 5) and wild-type mice (n = 5). Long-term potentiation data correspond to values averaged from 10 independent slices prepared from four DMSXL (n = 4) and five independent slices from three wild-type mice (n = 3). (E) Quantification of neurochemicals in the brain of DMSXL (n = 5) and wild-type control mice (n = 5) at age 4 months. Average concentration (±SEM) of l-3,4-dihydroxyphenylalanine (L-DOPA), dopamine (DA), 3,4-dihydroxyphenylacetic acid (DOPAC), homovanillic acid (HVA), noradrenaline (NA), serotonin (5-HT) and 5-hydroxyindoleacetic acid (5-HIAA) are plotted for frontal cortex and brainstem. *P < 0.05, **P < 0.01.
DMSXL brains show abnormal levels of dopamine and serotonin metabolites
Neurotransmitter dysregulation can cause behavioural and electrophysiological dysfunction. Little is known about neurochemical signalling and metabolism in myotonic dystrophy type 1 brains. To further dissect the neurological phenotype of DMSXL mice and assess whether behavioural and electrophysiological phenotypes were associated with changes in neurotransmitter levels, we measured key neurosignalling molecules in DMSXL brains. Adult DMSXL mice revealed a significant reduction of dopamine in the frontal cortex, as well as a tendency to decreased levels of its precursor (l-DOPA) and metabolites (3,4-dihydroxyphenylalanine, homovanillic acid). A significant decrease of 5-hydroxyindoleacetic acid (5-HIAA, the main serotonin metabolite) was detected in the brainstem of DMSXL mice (Fig. 3E).
Synaptic protein abnormalities in DMSXL mice and patients with myotonic dystrophy type 1
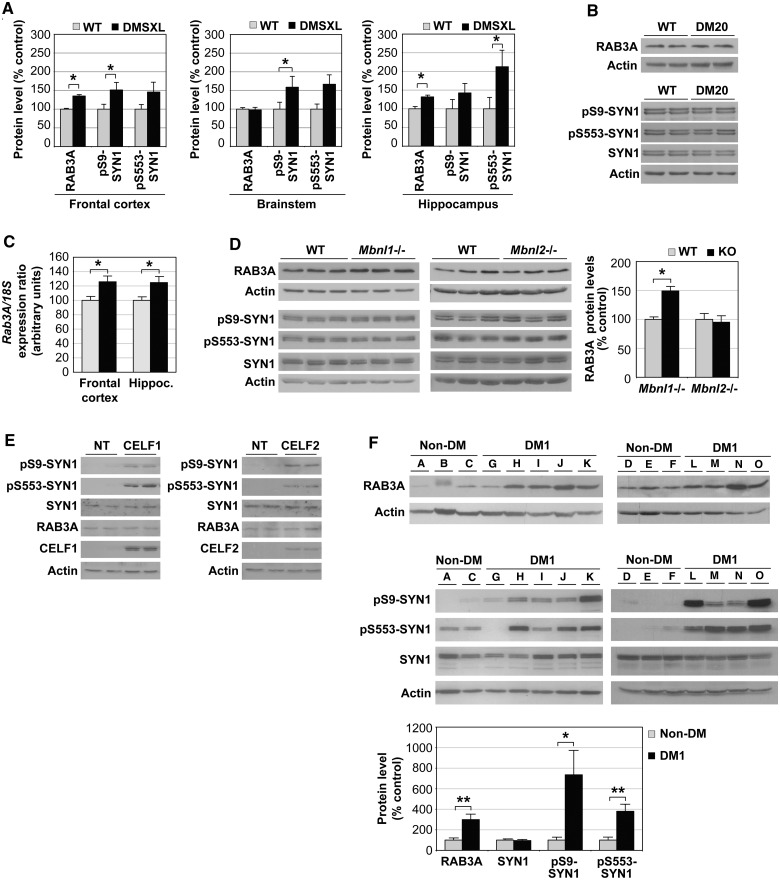
To identify pathways affected in the CNS that might contribute to the behavioural and electrophysiological phenotypes, we compared the proteomic profiles of adult homozygous mice carrying CTG expansions with those of DM20 control mice overexpressing short DMPK transcripts. The use of DM20 control mice excluded the identification of disease intermediates possibly affected by overexpression of DMPK protein. The proteomics analysis suggested altered expression of RAB3A and post-translational modifications of synapsin I (SYN1). Western blot quantification confirmed a statistically significant upregulation of RAB3A and hyperphosphorylation of serine residues of SYN1 in DMSXL frontal cortex and hippocampus at age 4 months (Fig. 4A and Supplementary Fig. 5A and B). To further test if synaptic protein dysregulation could be mediated by overexpression of short DMPK transcripts and protein, we analysed DM20 mice carrying 20 CTG repeats by western blot. This DMPK overexpressing control line did not show RAB3A upregulation or SYN1 hyperphosphorylation (Fig. 4B), indicating that the defects observed are mediated by the expression of expanded CUG RNA repeats. We extended the analysis to other synaptic proteins to assess the extent of synaptic dysfunction, but found no additional abnormalities (Supplementary Fig. 5C and D). Our data suggest that, rather than interfering with synaptic proteins in general, the repeat expansion may affect (directly or indirectly) a limited number of synaptic targets.
Figure 4.
Abnormal metabolism of RAB3A and SYN1 in the CNS of DMSXL mice and patients with myotonic dystrophy type 1. (A) Average steady-state levels (±SEM) of RAB3A and phosphorylated SYN1 in the frontal cortex, brainstem and hippocampus of homozygous DMSXL mice (n = 9), relative to normalized age-matched wild-type (WT) control mice (n = 9) at age 4 months. (B) Western blot analysis of RAB3A protein levels and SYN1 phosphorylation in 4-month-old DM20 and wild-type mice (n = 2 per genotype). (C) RAB3A protein upregulation in DMSXL brains is associated with increased transcript levels. Real-time quantitative PCR of Rab3A messenger RNA in frontal cortex and hippocampus of DMSXL (n = 6) and wild-type mice (n = 6). The graph shows the average Rab3A relative expression (±SEM) at age 4 months. *P < 0.05. (D) Western blot analysis of RAB3A protein levels and SYN1 phosphorylation in frontal cortex of knockout (KO) mice inactivated for Mbnl1- and Mbnl2-deficient mice. The graph on the right represents the average RAB3A steady state-levels (±SEM) in knockout mice (n = 3), relative to normalized age-matched wild-type control mice (n = 3). (E) Western blot analysis of RAB3A protein levels and SYN1 phosphorylation in PC12 cells overexpressing CELF1 or CELF2. NT = non-transfected cells. (F) RAB3A upregulation and SYN1 hyperphosphorylation in frontal cortex of adult patients with myotonic dystrophy type 1 (DM1; n = 9), relative to non-myotonic dystrophy control subjects (non-DM; n = 6). One non-myotonic dystrophy control subjects (Patient B) did not show a suitable SYN1 signal (possibly because of protein degradation) and was excluded from the analysis. *P < 0.05, **P < 0.01.
To address the mechanisms of synaptic protein dysregulation, we first tested if these abnormalities were associated with missplicing of known alternative exons of Rab3A and Syn1 transcripts. Reverse transcriptase PCR analysis revealed that Rab3A and Syn1 alternative splicing was not affected in DMSXL mice at age 4 months (Supplementary Fig. 5E), an age when synaptic protein abnormalities are detected. RAB3A and SYN1 protein changes are not mediated by spliceopathy of the candidate alternative exons studied. Interestingly, RAB3A upregulation was associated with a significant increase of messenger RNA transcript levels in DMSXL frontal cortex and hippocampus (Fig. 4C).
We then tested if Mbnl1 or Mbnl2 inactivation was sufficient to dysregulate synaptic proteins, through the analysis of knockout mice (Kanadia et al., 2003; Charizanis et al., 2012). Western blot analysis revealed that Mbnl1 inactivation (but not Mbnl2) resulted in a significant increase of RAB3A protein levels in mouse frontal cortex (Fig. 4D). Neither Mbnl1 nor Mbnl2 loss of function affected SYN1 phosphorylation levels. To test whether SYN1 hyperphosphorylation is mediated by upregulation of CELF proteins, we transfected PC12 cells with expression vectors encoding CELF1 or CELF2. We studied SYN1 phosphorylation by western blot and found protein hyperphosphorylation in PC12 cells overexpressing CELF1 or CELF2 (Fig. 4E). This analysis indicated that upregulation of CELF proteins is sufficient to dysregulate SYN1 phosphorylation levels.
To ascertain whether RAB3A and SYN1 dysregulation in mice reflected a pathophysiological event of the human condition, we investigated post-mortem myotonic dystrophy type 1 frontal cortex (Fig. 4F). Western blot quantification confirmed statistically significant RAB3A upregulation in patients with myotonic dystrophy type 1 when compared with subjects with non-myotonic dystrophy type 1 (P = 0.0054, Student’s t-test). SYN1 was hyperphosphorylated at Ser9 (P = 0.0282, Student’s t-test) and Ser553 (P = 0.0032, Student’s t-test) amino acid residues in individuals with myotonic dystrophy type 1. Abnormal phosphorylation was not accompanied by changes in protein steady-state levels (P = 0.7658, Student’s t-test). These results show that proteins playing important roles in synaptic function are dysregulated in myotonic dystrophy type 1.
Expanded CUG-containing DMPK transcripts affect neuronal exocytosis in culture
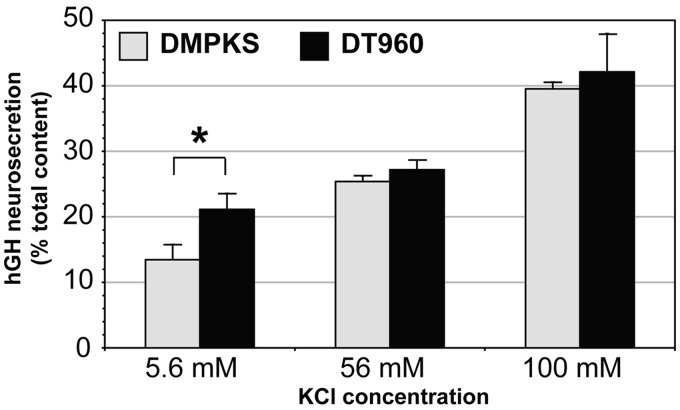
To investigate the functional consequences of myotonic dystrophy type 1 repeat expansions, and in particular, altered expression of synaptic proteins on vesicle trafficking, we studied exocytosis in an established transfected cell culture model of neurosecretion. Regulated exocytosis is triggered by extracellular stimulus and exhibits low-basal spontaneous secretion in the absence of stimulation (Sudhof, 2004). Basal and regulated neurosecretion can be measured in cultured PC12 cells transiently transfected with human growth hormone (Sugita, 2004; Lee et al., 2007). The percentage of human growth hormone secreted before and after osmotic stimulation serves as an estimate of basal and regulated neurosecretion, respectively. To assess the effect of CTG repeats on vesicle trafficking, PC12 cells were co-transfected with a human growth hormone-producing plasmid and with DMPK constructs carrying a CTG expansion or no CTG repeats. Expanded CUG-containing RNA accumulated in the nucleus of PC12 cells, co-localized with MBNL1 and MBNL2 and induced mild missplicing. More importantly, transfected PC12 cells exhibited RAB3A upregulation and SYN1 hyperphosphorylation (Supplementary Fig. 6). The neurosecretion assay demonstrated that the expression of toxic RNA repeats enhanced basal neurosecretion in the absence of stimulation, relative to no-repeat control constructs (Fig. 5). The effect was observed in four independent experiments with a significant average basal secretion enhancement of 60 ± 16%, during 15 min of incubation. Interestingly, CUG-containing transcripts did not disturb regulated neurosecretion after osmotic stimulation.
Figure 5.
Toxic CUG repeats dysregulate neurosecretion in culture. Quantification of the effect of CTG repeat expansion on neurosecretion. The average secretion from human growth hormone-expressing PC12 cells co-transfected with expanded DT960 or no-expansion DMPKS plasmids is plotted as a per cent of total human growth hormone content (±SEM). Basal secretion was measured in control medium containing 5.6 mM KCl. Stimulus-dependent secretion was measured in media containing 56 or 100 mM KCl. hCG = human growth hormone.
Discussion
To explore myotonic dystrophy type 1 neuropathology, we studied myotonic dystrophy type 1 transgenic mice that reproduce key molecular aspects of RNA toxicity and exhibit relevant behavioural phenotypes. DMSXL mice display neurochemical and electrophysiological signs of synaptic dysfunction, which are associated with molecular abnormalities in synaptic proteins, observed not only in mice but also in transfected cells and post-mortem myotonic dystrophy type 1 brain samples. The association between physiological and molecular phenotypes of the synapse indicates synaptic dysfunction in myotonic dystrophy type 1 neuropathophysiology.
The expression of expanded DMPK transcripts in the CNS of transgenic mice results in the accumulation of RNA foci in neurons and astrocytes. RNA toxicity in both cell types indicates that myotonic dystrophy type 1 may be associated not only with neuronal dysfunction but also with glial abnormalities. Interestingly, brain areas showing the highest foci content (e.g. frontal cortex) did not necessarily express the highest DMPK levels, suggesting that RNA aggregation into foci may depend on region- and/or cell-type–specific factors. As in skeletal (Lin et al., 2006; Orengo et al., 2008) and cardiac (Wang et al., 2007) muscles, CUG toxicity interferes with developmental alternative splicing in the CNS, increasing to a limited extent the abnormal expression of embryonic isoforms of some transcripts in adult DMSXL brains. Splicing abnormalities in the CNS may contribute to DMSXL phenotype. In particular, Grin1/Nmdar1 and Mapt/Tau splicing defects likely participate in synaptic dysfunction detected in DMSXL mice. Further experiments are required to investigate how Grin1/Nmdar1 missplicing influences intracellular localization and function of this receptor, as well as the impact of Mapt/Tau missplicing on myotonic dystrophy type 1 neurofibrillary degeneration.
The missplicing of alternative exons specifically regulated by MBNL or CELF proteins (Kalsotra et al., 2008) indicated that MBNL loss of function by protein sequestration and CELF upregulation induce missplicing in the CNS. In support of this view, Mbnl1- or Mbnl2-deficient mice show abnormal splicing in brain (Charizanis et al., 2012; Suenaga et al., 2012).
CELF1 upregulation in the CNS appears to be mediated by mild protein hyperphosphorylation, as in myotonic dystrophy type 1 heart (Kuyumcu-Martinez et al., 2007). It remains to be investigated whether PKC activity is increased in the myotonic dystrophy type 1 brains. In contrast, CELF2 upregulation in DMSXL frontal cortex was not associated with protein hyperphosphorylation, suggesting alternative mechanisms of protein regulation. The marked upregulation of CELF2 in human myotonic dystrophy type 1 frontal cortex predicts a pathogenic role for this protein in myotonic dystrophy type 1 neuropathogenesis. The modulation of CELF2 expression by miRNA species, recently reported in a transgenic mouse model of spinal-bulbar muscular atrophy (SBMA), suggests new possibilities for miRNA-mediated therapies in myotonic dystrophy type 1 (Miyazaki et al., 2012).
The behavioural phenotyping of DMSXL mice revealed reduced exploratory activity, increased anxiety, spatial memory impairment and anhedonia, which resemble myotonic dystrophy type 1 neurological manifestations. Previous neuropsychological assessment of patients with myotonic dystrophy type 1 revealed low scores in the exploratory scale (Winblad et al., 2005), a higher prevalence of anxiety-related behaviours (Delaporte, 1998; Meola et al., 2003), visual–spatial impairment (Modoni et al., 2004) and anhedonia associated with emotional blunting and depressive symptomatology (Bungener et al., 1998). The phenotypic parallel between patients and transgenic mice illustrates the impact of toxic DMPK transcripts on CNS physiology and corroborates the use of the DMSXL mouse line to recreate myotonic dystrophy type 1 brain pathology. Mild deficits in working memory were also found in DMSXL mice. Although not statistically significant, the initial difference between the two genotypes in the acquisition trial, during the working memory assessment, might confound the analysis, and the results should be interpreted with some caution. Alternative behavioural tests (e.g. elevated or multiple arm radial mazes) might prove useful to provide definitive evidence of working memory deficits in DMSXL mice.
The behavioural abnormalities of DMSXL mice are associated with deficits in short-term plasticity, as well as changes in neurochemicals, suggesting altered synaptic function and neurotransmission in response to the CTG repeat expansion. The neurochemical data, in particular, provide insight into the neuronal circuits affected by myotonic dystrophy type 1. Decreased dopamine in frontal cortex may account for motivation and reward deficits (Arias-Carrion and Poppel, 2007), while the reduced serotonin metabolism in brainstem may increase susceptibility to depressive-like behaviours (Werner and Covenas, 2010), thereby contributing to anhedonia. The high foci content in dopaminergic (substantia nigra) and serotonergic (raphe nucleus) brain centres may contribute, at least partially, to the neurochemical deficits of DMSXL mice. In humans, loss of catecholaminergic neurons (dopamine is an abundant catecholamine) and serotonin-containing neurons were previously reported (Ono et al., 1998a, b). The involvement of dopaminergic and serotonergic pathways in myotonic dystrophy type 1 neuropathology requires further investigation, and may provide insight into future means of therapeutic intervention, such as the modulation of the dopaminergic and serotinergic circuits.
In the search for pathways affected by the myotonic dystrophy type 1 repeat mutation in the CNS, we found RAB3A upregulation and SYN1 hyperphosphorylation, not only in transgenic mice expressing large CTG expansions but also in post-mortem myotonic dystrophy type 1 brains. Transgenic mice overexpressing short DMPK transcripts did not show abnormal synaptic protein dysregulation, indicating that RAB3A and SYN1 misregulation is specifically associated with expanded transcripts.
Decreased glucose metabolism (Fiorelli et al., 1992) and blood perfusion (Meola et al., 1999) have been previously described in the frontal lobe of patients with myotonic dystrophy type 1. Since abnormal glucose metabolism can alter the protein content of synaptic vesicles (Gaspar et al., 2010), it is reasonable to speculate that changes in the exocytocic machinery in frontal cortex might result (at least partially) from brain hypoperfusion and/or hypometabolism. Despite the possible contribution of altered glucose metabolism to synaptic dysfunction, cells transfected with myotonic dystrophy type 1 repeat expansions displayed RAB3A upregulation and SYN1 hyperphosphorylation in culture, supporting the view that synaptic protein dysregulation is also a direct consequence of CUG RNA toxicity, rather than a simple indirect effect, secondary to general brain dysfunction. To further support this hypothesis, the analysis of complementary animal and cell models of myotonic dystrophy type 1 revealed that RAB3A upregulation is mediated by MBNL1 inactivation, and SYN1 hyperphosphorylation is mediated by upregulation of CELF proteins. We propose that transcriptional dysregulation (Osborne et al., 2009) or altered post-transcriptional regulation of messenger RNA decay (Masuda et al., 2012) through loss of function of MBNL proteins could mediate RAB3A upregulation. Hyperphosphorylation of SYN1 may result from dysregulated kinase and/or phosphatase activities, as a result of altered CELF levels. Dysregulation of micro-RNAs, also reported in myotonic dystrophy type 1 skeletal muscle and heart (Gambardella et al., 2010; Perbellini et al., 2011; Rau et al., 2011), may extend to the CNS, opening new avenues for future research.
RAB3A is an abundant synaptic vesicle protein that regulates neurotransmission (Sudhof, 2004). Mouse Rab3a inactivation results in increased hippocampal paired-pulse facilitation (Geppert et al., 1997), while RAB3A overexpression in cell culture activates spontaneous exocytosis (Schluter et al., 2002), similar to PC12 cells transfected with expanded DMPK constructs. Given the involvement of RAB3A in short-term synaptic plasticity and neurotransmitter release, RAB3A upregulation in DMSXL mice likely contributes to altered paired-pulse facilitation and mediates the increase in basal neurosecretion in transfected PC12 cells. In addition, RAB3A has been implicated in visual–spatial learning (D’Adamo et al., 2004). Therefore, altered RAB3A expression may contribute to the cognitive deficits of DMSXL mice and patients with myotonic dystrophy type 1. Synapsins comprise the most abundant proteins in synaptic vesicles. SYN1 serves as a phosphorylation-dependent regulator of neurotransmitter release (Rosahl et al., 1993). Whereas non-phosphorylated SYN1 attaches synaptic vesicles to the actin cytoskeleton, stimulation-dependent phosphorylation decreases the affinity for synaptic vesicles and potentiates exocytosis (Fdez and Hilfiker, 2006). Consistent with a role for SYN1 in short-term plasticity (Fiumara et al., 2007), chemically induced hyperphosphorylation of SYN1 in vivo was previously associated with decreased paired-pulse facilitation (Tallent et al., 2009), like in DMSXL mice. The marked RAB3A upregulation and abnormal SYN1 hyperphosphorylation in post-mortem myotonic dystrophy type 1 brains corroborate their role in myotonic dystrophy type 1 neuropathology. RAB3A and SYN1 variability between patients may be associated with different degrees of disease severity, as previously reported for splicing abnormalities in skeletal muscle and heart (Wang et al., 2007; Orengo et al., 2008). In summary, we found RAB3A upregulation and SYN1 hyperphosphorylation in myotonic dystrophy type 1 transgenic mice, transfected cells and in human myotonic dystrophy type 1 brain samples. These protein defects were associated with electrophysiological and behavioural abnormalities in mice, as well as altered spontaneous neurosecretion in cell culture.
If changes in synaptic proteins reflect greater abnormalities in the dynamics and microstructure of the brain cell membrane, the molecular abnormalities identified might have wider implications and may correlate with the prevalent white matter lesions reported in myotonic dystrophy type 1 brains (Minnerop et al., 2011). Future MRI of DMSXL brain integrity will address this possibility through the characterization of tissue changes in response to the myotonic dystrophy type 1 mutation. Additional imaging measurements of regional brain blood flow and/or glucose metabolism in transgenic mice by PET and/or single-photon emission computed tomography must be performed to evaluate and localize the functional impact of myotonic dystrophy type 1 in the CNS and provide insight into the underlying mechanisms.
There is currently debate whether CNS dysfunction in myotonic dystrophy type 1 is neurodegenerative, neurodevelopmental or neurofunctional. Although clinical data are not enough to answer this difficult question, transgenic mice may provide significant insight. The molecular and electrophysiological abnormalities detected in DMSXL mice are consistent with functional deficits in adult brain. Future analyses of cell loss and the CNS investigation throughout mouse development will be required to address the contribution of neurodegeneration and impaired neurodevelopment, respectively, towards myotonic dystrophy type 1 CNS dysfunction. It remains possible that these processes are not mutually exclusive, and that they all participate in myotonic dystrophy type 1 brain pathophysiology.
In the context of the preclinical assessment of future therapies, it will be interesting to investigate whether therapeutic schemes targeting the CNS (and in particular RAB3A and SYN1) will be able to reverse the behavioural phenotypes (e.g. visual–spatial memory impairment, increased anxiety and anhedonia) and/or electrophysiological profiles (e.g. paired-pulse facilitation deficits) of DMSXL mice. In cell culture, RAB3A and SYN1 can modulate miniature endplate currents of neurons (Chiappalone et al., 2009; Wang et al., 2011). It is tempting to investigate whether RAB3A knockdown and/or SYN1 dephosphorylation in primary cultures can rescue the electrophysiological phenotype of DMSXL neurons.
Using molecular and physiological approaches to explore the molecular mechanisms of a complex human disease, we generated evidence of the impact of myotonic dystrophy type 1 on synaptic proteins, vesicle secretion, neurotransmission and synaptic plasticity. The validation of our molecular findings in post-mortem myotonic dystrophy type 1 brains substantiates a role of synaptic dysfunction in myotonic dystrophy type 1 through changes in proteins involved in the regulation of synaptic vesicle release.
Funding
ANR (Agence Nationale de Recherche, France; “DM1MICE” project to G.G.); AFM (Association Française contre les Myopathies, France; ‘DM Brain’ project to M.G.-P.); Prosensa (The Netherlands, to M.G.-P.); INSERM (Institute National de la Santé et Recherche Médicale, France); Université Paris Descartes (Paris, France); CONACyT (Consejo Nacional de Ciência y Tecnología, Mexico, to O.H.-H.); Ministère Français de la Recherche et Technologie (to G.S.); NIH (National Institutes of Health, AR046799 and NS058901 to M.S.S.).
Supplementary material
Supplementary material is available at Brain online.
Acknowledgements
The authors thank Dr Shuzo Sugita and Dr Thomas Cooper for providing the phGHCMV5, DMPKs, DT960, CELF1 and CELF2 plasmids and Dr Glenn Morris and Dr Manuel Hernández for providing the anti-MBNL2 and anti-ß-actin antibodies, respectively. They are grateful to Amine Bouallague and to the personnel of CERFE (Centre d’Exploration et de Recherche Fonctionelle Expérimentale, Genopole, Evry, France) for attentively caring for the mice. They thank their colleagues at Inserm U781 and the DM1 French Splicing Network for helpful discussions.
Glossary
Abbreviations
- CA
cornu ammonis
- CELF
CUGBP/Elav-like factor
- CUGBP
CUG RNA binding protein
- CYPHER
Z-band alternatively spliced PDZ-motif protein
- DM
myotonic dystrophy
- DMSXL
DM300 mice carrying a >1000 CTG expansion
- FITC
fluorescein isothiocyanate
- FXR
fragile X mental retardation
- GFAP
glial fibrillary acidic protein
- GRIN
glutamate receptor ionotropic, N-methyl D-aspartate
- LDB
LIM domain binding
- MBNL
muscleblind-like
- NMDAR
N-methyl D-aspartate receptor
- PC
pheochromocytoma
- PKC
protein kinase C
- RAB
RAS oncogene family
- SERCA
sarcoplasmic/endoplasmic reticulum calcium ATPase
- SSC
saline-sodium citrate
- UTR
untranslated region.
References
- Angeard N, Gargiulo M, Jacquette A, Radvanyi H, Eymard B, Heron D. Cognitive profile in childhood myotonic dystrophy type 1: is there a global impairment? Neuromuscul Disord. 2007;17:451–8. doi: 10.1016/j.nmd.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Arias-Carrion O, Poppel E. Dopamine, learning, and reward-seeking behavior. Acta Neurobiol Exp (Wars) 2007;67:481–8. doi: 10.55782/ane-2007-1664. [DOI] [PubMed] [Google Scholar]
- Bassez G, Chapoy E, Bastuji-Garin S, Radvanyi-Hoffman H, Authier FJ, Pellissier JF, et al. Type 2 myotonic dystrophy can be predicted by the combination of Type 2 muscle fiber central nucleation and scattered atrophy. J Neuropathol Exp Neurol. 2008;67:319–25. doi: 10.1097/NEN.0b013e31816b4acc. [DOI] [PubMed] [Google Scholar]
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;69:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Bungener C, Jouvent R, Delaporte C. Psychopathological and emotional deficits in myotonic dystrophy. J Neurol Neurosurg Psychiatry. 1998;65:353–6. doi: 10.1136/jnnp.65.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–50. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- Chiappalone M, Casagrande S, Tedesco M, Valtorta F, Baldelli P, Martinoia S, et al. Opposite changes in glutamatergic and GABAergic transmission underlie the diffuse hyperexcitability of synapsin I-deficient cortical networks. Cereb Cortex. 2009;19:1422–39. doi: 10.1093/cercor/bhn182. [DOI] [PubMed] [Google Scholar]
- D'Adamo P, Wolfer DP, Kopp C, Tobler I, Toniolo D, Lipp HP. Mice deficient for the synaptic vesicle protein Rab3a show impaired spatial reversal learning and increased explorative activity but none of the behavioral changes shown by mice deficient for the Rab3a regulator Gdi1. Eur J Neurosci. 2004;19:1895–905. doi: 10.1111/j.1460-9568.2004.03270.x. [DOI] [PubMed] [Google Scholar]
- Delaporte C. Personality patterns in patients with myotonic dystrophy. Arch Neurol. 1998;55:635–40. doi: 10.1001/archneur.55.5.635. [DOI] [PubMed] [Google Scholar]
- Fdez E, Hilfiker S. Vesicle pools and synapsins: new insights into old enigmas. Brain Cell Biol. 2006;35:107–15. doi: 10.1007/s11068-007-9013-4. [DOI] [PubMed] [Google Scholar]
- Fiorelli M, Duboc D, Mazoyer BM, Blin J, Eymard B, Fardeau M, et al. Decreased cerebral glucose utilization in myotonic dystrophy. Neurology. 1992;42:91–4. doi: 10.1212/wnl.42.1.91. [DOI] [PubMed] [Google Scholar]
- Fiumara F, Milanese C, Corradi A, Giovedi S, Leitinger G, Menegon A, et al. Phosphorylation of synapsin domain A is required for post-tetanic potentiation. J Cell Sci. 2007;120:3228–37. doi: 10.1242/jcs.012005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YH, Pizzuti A, Fenwick RG, Jr, King J, Rajnarayan S, Dunne PW, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–8. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- Gambardella S, Rinaldi F, Lepore SM, Viola A, Loro E, Angelini C, et al. Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J Transl Med. 2010;8:48. doi: 10.1186/1479-5876-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar JM, Baptista FI, Galvao J, Castilho AF, Cunha RA, Ambrosio AF. Diabetes differentially affects the content of exocytotic proteins in hippocampal and retinal nerve terminals. Neuroscience. 2010;169:1589–600. doi: 10.1016/j.neuroscience.2010.06.021. [DOI] [PubMed] [Google Scholar]
- Gaul C, Schmidt T, Windisch G, Wieser T, Muller T, Vielhaber S, et al. Subtle cognitive dysfunction in adult onset myotonic dystrophy type 1 (DM1) and Type 2 (DM2) Neurology. 2006;67:350–2. doi: 10.1212/01.wnl.0000225180.27833.c1. [DOI] [PubMed] [Google Scholar]
- Geppert M, Goda Y, Stevens CF, Sudhof TC. The small GTP-binding protein Rab3A regulates a late step in synaptic vesicle fusion. Nature. 1997;387:810–4. doi: 10.1038/42954. [DOI] [PubMed] [Google Scholar]
- Gomes-Pereira M, Foiry L, Nicole A, Huguet A, Junien C, Munnich A, et al. CTG trinucleotide repeat “big jumps”: large expansions, small mice. PLoS Genet. 2007;3:e52. doi: 10.1371/journal.pgen.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Pereira M, Monckton DG. Chemical modifiers of unstable expanded simple sequence repeats: what goes up, could come down. Mutat Res. 2006;598:15–34. doi: 10.1016/j.mrfmmm.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Guiraud-Dogan C, Huguet A, Gomes-Pereira M, Brisson E, Bassez G, Junien C, et al. DM1 CTG expansions affect insulin receptor isoforms expression in various tissues of transgenic mice. Biochim Biophys Acta. 2007;1772:1183–91. doi: 10.1016/j.bbadis.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Harper PS. Myotonic dystrophy. 3rd edn. London: WB Saunders; 2001. [Google Scholar]
- Huguet A, Medja F, Nicole A, Vignaud A, Ferry A, Guiraud-Dogan C, et al. Molecular, physiological and motor performance defects in DMSXL mice carrying >1000 CTG repeat from the human DM1 locus. PLoS Genet London. 2012;8:e1003043. doi: 10.1371/journal.pgen.1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izac SM, Eeg TR. Basic anatomy and physiology of sleep. Am J Electroneurodiagnostic Technol. 2006;46:18–38. [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–88. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105:20333–8. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–80. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Seo HS, Ha I, Chung SH. Overexpression of BACE1 stimulates spontaneous basal secretion in PC12 cells. Neurosci Lett. 2007;421:178–83. doi: 10.1016/j.neulet.2007.01.082. [DOI] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–97. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Lueck JD, Lungu C, Mankodi A, Osborne RJ, Welle SL, Dirksen RT, et al. Chloride channelopathy in myotonic dystrophy resulting from loss of posttranscriptional regulation for CLCN1. Am J Physiol Cell Physiol. 2007;292:C1291–7. doi: 10.1152/ajpcell.00336.2006. [DOI] [PubMed] [Google Scholar]
- Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992;255:1253–5. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- Masuda A, Andersen HS, Doktor TK, Okamoto T, Ito M, Andresen BS, et al. CUGBP1 and MBNL1 preferentially bind to 3' UTRs and facilitate mRNA decay. Sci Rep. 2012;2:209. doi: 10.1038/srep00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola G, Sansone V. Cerebral involvement in myotonic dystrophies. Muscle Nerve. 2007;36:294–306. doi: 10.1002/mus.20800. [DOI] [PubMed] [Google Scholar]
- Meola G, Sansone V, Perani D, Colleluori A, Cappa S, Cotelli M, et al. Reduced cerebral blood flow and impaired visual-spatial function in proximal myotonic myopathy. Neurology. 1999;53:1042–50. doi: 10.1212/wnl.53.5.1042. [DOI] [PubMed] [Google Scholar]
- Meola G, Sansone V, Perani D, Scarone S, Cappa S, Dragoni C, et al. Executive dysfunction and avoidant personality trait in myotonic dystrophy type 1 (DM-1) and in proximal myotonic myopathy (PROMM/DM-2) Neuromuscul Disord. 2003;13:813–21. doi: 10.1016/s0960-8966(03)00137-8. [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–48. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnerop M, Weber B, Schoene-Bake JC, Roeske S, Mirbach S, Anspach C, et al. The brain in myotonic dystrophy 1 and 2: evidence for a predominant white matter disease. Brain. 2011;134(Pt 12):3530–46. doi: 10.1093/brain/awr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y, Adachi H, Katsuno M, Minamiyama M, Jiang YM, Huang Z, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012;18:1136–41. doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]
- Modoni A, Silvestri G, Pomponi MG, Mangiola F, Tonali PA, Marra C. Characterization of the pattern of cognitive impairment in myotonic dystrophy type 1. Arch Neurol. 2004;61:1943–7. doi: 10.1001/archneur.61.12.1943. [DOI] [PubMed] [Google Scholar]
- Ono S, Takahashi K, Jinnai K, Kanda F, Fukuoka Y, Kurisaki H, et al. Loss of serotonin-containing neurons in the raphe of patients with myotonic dystrophy: a quantitative immunohistochemical study and relation to hypersomnia. Neurology. 1998a;50:535–8. doi: 10.1212/wnl.50.2.535. [DOI] [PubMed] [Google Scholar]
- Ono S, Takahashi K, Jinnai K, Kanda F, Fukuoka Y, Kurisaki H, et al. Loss of catecholaminergic neurons in the medullary reticular formation in myotonic dystrophy. Neurology. 1998b;51:1121–4. doi: 10.1212/wnl.51.4.1121. [DOI] [PubMed] [Google Scholar]
- Orengo JP, Chambon P, Metzger D, Mosier DR, Snipes GJ, Cooper TA. Expanded CTG repeats within the DMPK 3' UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc Natl Acad Sci USA. 2008;105:2646–51. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne RJ, Lin X, Welle S, Sobczak K, O'Rourke JR, Swanson MS, et al. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet. 2009;18:1471–81. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaite PA, Gantelet E, Kraftsik R, Gourdon G, Kuntzer T, Barakat-Walter I. Myotonic dystrophy transgenic mice exhibit pathologic abnormalities in diaphragm neuromuscular junctions and phrenic nerves. J Neuropathol Exp Neurol. 2008;67:763–72. doi: 10.1097/NEN.0b013e318180ec64. [DOI] [PubMed] [Google Scholar]
- Perbellini R, Greco S, Sarra-Ferraris G, Cardani R, Capogrossi MC, Meola G, et al. Dysregulation and cellular mislocalization of specific miRNAs in myotonic dystrophy type 1. Neuromuscul Disord. 2011;21:81–8. doi: 10.1016/j.nmd.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–77. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol. 2011;18:840–5. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- Robbins TW, Arnsten AF. The neuropsychopharmacology of fronto-executive function: monoaminergic modulation. Annu Rev Neurosci. 2009;32:267–87. doi: 10.1146/annurev.neuro.051508.135535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo V, Pegoraro E, Squarzanti F, Soraru G, Ferrati C, Ermani M, et al. Retrospective study on PET-SPECT imaging in a large cohort of myotonic dystrophy type 1 patients. Neurol Sci. 2010;31:757–63. doi: 10.1007/s10072-010-0406-2. [DOI] [PubMed] [Google Scholar]
- Rosahl TW, Geppert M, Spillane D, Herz J, Hammer RE, Malenka RC, et al. Short-term synaptic plasticity is altered in mice lacking synapsin I. Cell. 1993;75:661–70. doi: 10.1016/0092-8674(93)90487-b. [DOI] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–7. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- Schluter OM, Khvotchev M, Jahn R, Sudhof TC. Localization versus function of Rab3 proteins. Evidence for a common regulatory role in controlling fusion. J Biol Chem. 2002;277:40919–29. doi: 10.1074/jbc.M203704200. [DOI] [PubMed] [Google Scholar]
- Schweizer MC, Henniger MS, Sillaber I. Chronic mild stress (CMS) in mice: of anhedonia, 'anomalous anxiolysis' and activity. PLoS One. 2009;4:e4326. doi: 10.1371/journal.pone.0004326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant N, Bombois S, Ghestem A, Drobecq H, Kostanjevecki V, Missiaen C, et al. Truncated beta-amyloid peptide species in pre-clinical Alzheimer's disease as new targets for the vaccination approach. J Neurochem. 2003;85:1581–91. doi: 10.1046/j.1471-4159.2003.01818.x. [DOI] [PubMed] [Google Scholar]
- Seznec H, Agbulut O, Sergeant N, Savouret C, Ghestem A, Tabti N, et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum Mol Genet. 2001;10:2717–26. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- Seznec H, Lia-Baldini AS, Duros C, Fouquet C, Lacroix C, Hofmann-Radvanyi H, et al. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum Mol Genet. 2000;9:1185–94. doi: 10.1093/hmg/9.8.1185. [DOI] [PubMed] [Google Scholar]
- Sistiaga A, Urreta I, Jodar M, Cobo AM, Emparanza J, Otaegui D, et al. Cognitive/personality pattern and triplet expansion size in adult myotonic dystrophy type 1 (DM1): CTG repeats, cognition and personality in DM1. Psychol Med. 2010;40:487–95. doi: 10.1017/S0033291709990602. [DOI] [PubMed] [Google Scholar]
- Steidl EM, Neveu E, Bertrand D, Buisson B. The adult rat hippocampal slice revisited with multi-electrode arrays. Brain Res. 2006;1096:70–84. doi: 10.1016/j.brainres.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–47. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Suenaga K, Lee KY, Nakamori M, Tatsumi Y, Takahashi MP, Fujimura H, et al. Muscleblind-like 1 knockout mice reveal novel splicing defects in the myotonic dystrophy brain. PLoS One. 2012;7:e33218. doi: 10.1371/journal.pone.0033218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S. Human growth hormone co-transfection assay to study molecular mechanisms of neurosecretion in PC12 cells. Methods. 2004;33:267–72. doi: 10.1016/j.ymeth.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Tallent MK, Varghis N, Skorobogatko Y, Hernandez-Cuebas L, Whelan K, Vocadlo DJ, et al. In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation. J Biol Chem. 2009;284:174–81. doi: 10.1074/jbc.M807431200. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820–6. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007;117:2802–11. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Q, Yang S, Bucan M, Rich MM, Engisch KL. Impaired activity-dependent plasticity of quantal amplitude at the neuromuscular junction of Rab3A deletion and Rab3A earlybird mutant mice. J Neurosci. 2011;31:3580–8. doi: 10.1523/JNEUROSCI.5278-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber YG, Roebling R, Kassubek J, Hoffmann S, Rosenbohm A, Wolf M, et al. Comparative analysis of brain structure, metabolism, and cognition in myotonic dystrophy 1 and 2. Neurology. 2010;74:1108–17. doi: 10.1212/WNL.0b013e3181d8c35f. [DOI] [PubMed] [Google Scholar]
- Werner FM, Covenas R. Classical neurotransmitters and neuropeptides involved in major depression: a review. Int J Neurosci. 2010;120:455–70. doi: 10.3109/00207454.2010.483651. [DOI] [PubMed] [Google Scholar]
- Winblad S, Lindberg C, Hansen S. Temperament and character in patients with classical myotonic dystrophy type 1 (DM-1) Neuromuscul Disord. 2005;15:287–92. doi: 10.1016/j.nmd.2004.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.