Abstract
The ascidian (sea squirt) C. intestinalis has become an important model organism for the study of cis-regulation. This is largely due to the technology that has been developed for assessing cis-regulatory activity through the use of transient reporter transgenes introduced into fertilized eggs. This technique allows the rapid and inexpensive testing of endogenous or altered DNA for regulatory activity in vivo. This review examines evidence that C. intestinalis cis-regulatory elements are located more closely to coding regions than in other model organisms. I go on to compare the organization of cis-regulatory elements and conserved non-coding sequences in Ciona, mammals, and other deuterostomes for three representative C.intestinalis genes, Pax6, FoxAa, and the DlxA-B cluster, along with homologs in the other species. These comparisons point out some of the similarities and differences between cis-regulatory elements and their study in the various model organisms. Finally, I provide illustrations of how C. intestinalis lends itself to detailed study of the structure of cis-regulatory elements, which have led, and promise to continue to lead, to important insights into the fundamentals of transcriptional regulation.
Keywords: Chordate, tunicate, amphioxus, transgenics, reporter, transcription, promoters, enhancers.
CIONA AS A MODEL FOR CIS-REGULATORY STUDY
Ciona intestinalis has emerged as an important model organism for the study of genomic regulation in the last 15 years. C. intestinalis is an ascidian - a member of the class Urochordata, which along with the cephalochordates are considered basal chordates Fig. (1). They have a simple chordate body plan, with a dorsal nervous system, axial skeleton (notochord), ventral heart, and an elaborate pharyngial apparatus. The developmental biology of ascidians has been the subject of detailed study for over 100 years, in part because of their stereotyped cell lineage, which enables experimental study of the relative roles of cytoplasmic inheritance and cell-cell signaling in development [1, 2]. C. intestinalis has particular advantages as a laboratory model organism, since it is a common marine organism, and thousands of synchronously, and rapidly, developing fertilized embryos can be obtained easily by in vitro fertilization on a daily basis.
Fig. (1).
Phylogeny of selected bilaterian animals. Taxa with abundant experimental cis-regulatory data are denoted by boxes around the taxon name. Tree topology based on a consensus of recent phylogenetic work eg. [16, 17].
The advent of genomics has increased interest in Ciona for several reasons [3]. It has a very small genome - approximately 160 Mb. [4], around the size of Drosophila, which facilitated the release of whole genome sequences of the congeneric species C. intestinalis and C. savignyii, by 2003. The genome is compact, with only about 7.5 kb. per gene, making it relatively easy to capture significant portions of transcriptional units in readily cloneable pieces. Significantly, ascidians lack the genome duplications present in vertebrates, so they have only one paralog of many genes that have been duplicated in vertebrates. This lack of duplicate genes reduces the amount of functional overlap and redundancy, simplifying functional genomic study. These characteristics of Ciona, among others, have enabled the generation of a host of bioinformatic resources, as summarized in Table 1. This review concentrates on the progress in cis-regulatory study in C. intestinalis. It does not cover the large and growing literature on other aspects of genomic research in Ciona, such as the elicidation of gene regulatory networks (eg. [5-7]).
Table 1.
Bioinformatic Resources for Ascidian Research
| Database | Content | URL | Ref. |
|---|---|---|---|
| Ghost | EST sequences, ISH data, genome browser (C. intestinalis, v.1), EST clones | ghost.zool.kyoto-u.ac.jp/SearchGenomekh.html#CDNA | [53, 54] |
| JGI C. intestinalis genome | C. intestinalis v. 2 genome browser | genome.jgi-psf.org/Cioin2/Cioin2.home.html | [4] |
| Whitehead C. savignyi genome | C. savignyi release 1 genome browser | www.broadinstitute.org/annotation/ciona/index.html | n/a |
| ANISEED | ESTs, gene models, ISH data, genome browser (C. intestinalis, v.1), CRE database | www.aniseed.cnrs.fr/ | [55] |
| Tunicate Portal | Links to tunicate bioinformatic resources | www.tunicate-portal.org | n/a |
| CiAID | ISH data on adult stages, anatomical resources | ioinfo.s.chiba-u.jp/ciaid/atlas2/jv2.htm | [56] |
| DBTGR | CRE database | dbtgr.hgc.jp | [57] |
| CiPRO | Proteomics database | cipro.ibio.jp | [58] |
| FABA | Interactive anatomical body atlas - embryonic | chordate.bpni.bio.keio.ac.jp/faba/top.html | [59] |
| FABA2 | Interactive anatomical body atlas - after hatching | chordate.bpni.bio.keio.ac.jp/faba2/ | [59] |
| CiTRES | GFP reporter transgenic lines | http://marinebio.nbrp.jp/ciona/index.jsp | [60] |
Perhaps the most important reason for the interest in Ciona for cis-regulatory study is the development of electroporation as a technique for introducing reporter transgenes into eggs or zygotes [8-11]. This method enables the simple and rapid generation of many transgenic embryos, which develop to the larval stage in around 18 hours. The typical in vivo assays performed in C. intestinalis are transient - only observed in the generation electroporated with the transgene. For many successful studies, these transient assays have allowed rapid testing of large numbers of experimental cis-regulatory element (CRE) variants at low cost [12]. However, some workers have also successfully transmitted transgenes to the germline and reared subsequent generations of transgenic animals. In addition, transposon mediated enhancer trap lines have been reported recently (reviewed in [11]).
While C. intestinalis is a chordate, its genome has some significant differences with those of vertebrates. It is very AT rich, with only about 35% GC content. The proportion of repetitive sequence has not been determined precisely, but it is much lower than in many vertebrates, such as humans. It does however, harbor several transposon types [13]. Methylation patterns are also different from vertebrates. Coding sequences of genes are preferentially methylated, but intergenic regions have only half that level of methylation. In vertebrates, on the other hand, the genome is more uniformly methylated, except at promoters, and the overall level of methylation is twice as high as in C. intestinalis [14]. Like other invertebrates, C. intestinalis has a very high level of nucleotide polymorphism, around 1.2% SNPs and indels between alleles. The high allelic variation makes genome assembly a difficult task [4, 15].
Recent phylogenomic studies have placed the ascidian urochordates as the sister group to the Vertebrata [16, 17] (Fig. 1). Since cis-regulatory experiments are more difficult in other basal chordates, such as amphioxus and lamprey, Ciona has become a major model organism for the functional study of the evolution of chordate genomic regulation and the evolution of development [18-20].
BASIC CHARACTERISTICS OF CIONA CIS-REGULATORY ELEMENTS
Nearly all reported experimentally confirmed cis-regulatory elements (CREs) in Ciona are enhancers, which activate transcription in a more or less tissue-specific manner. This is probably due to the fact that other elements, such as insulators or silencers, are more difficult to identify. Table 2 is a compilation of most, if not all, transcription factor and signaling molecule enhancer elements reported to date for C. intestinalis. (The table was limited to these catagories in the interest of space, and because these genes have attracted much of the interest in the field.) A 2005 review of ascidian cis-regulatory elements listed 11 genes of these types with experimentally determined CREs [21]. As of this writing, the total is up to 63 genes as listed in Table 2, several with multiple CREs determined. Many other CREs have been reported in Ciona in other functional catagories.
Table 2.
List of Experimentally Verified Transcription Factor and Signaling Gene Enhancers in Ciona
| Ciona Gene | CRE | Expression Pattern | Ref. |
|---|---|---|---|
| Ci-achaete-scute-a-like2 | A | Anterior ant. sens. ves., mesenchyme, tail muscles | [55]+ |
| Ci-ADMP | A | CNS, epidermis, mesoderm | [55]+ |
| Ci-AP2-like2 | A | Tail epidermis | [55]+ |
| Ci-AP4 | A | Mesenchyme, muscle | [55] |
| B | Mesenchyme, muscle | ||
| Ci-Brachyury | A | Notochord | [8] |
| Ci-chordin | A | CNS, mesoderm | [55] |
| Ci-COE | A | Mesenchyme, muscle | [42] |
| Ci-Delta2 | A | Epidermis, siphon primordia, TLCs, TVCs | [55] |
| Ci-derriere-like | A | Epidermis | [55]+ |
| Ci-DllA | A1 | Anterior neuroectoderm at tailbud stage | [61] Irvine lab unpubl. |
| Ci-DllB | B1 | Pan-animal hemisphere at gastrula stage | [37] |
| Ci-DMRT1 | A | CNS, mesenchyme, tail muscles, palps | [55]+ |
| Ci-ELK1 | A | CNS, notochord, muscles, endoderm | [55]+ |
| B | Mesenchyme, b-epidermis, nerve cord and b-muscle | ||
| C | a6.5 & b6.5 at 110-cell stage | ||
| Ci-Emx | A | Silencer | [55]+ |
| Ci-EphrinA-c | A | Head endoderm, notochord | [55]+ |
| Ci-EphrinA-d | A | Ectoderm and mesenchyme | [55]+ |
| Ci-ERF-a | A | a6.5 & b6.5 | [55]+ |
| Ci-ets | A | Tail muscles, mesenchyme, part of brain | [55]+ |
| Ci-Ets97D | A | Mesenchyme, notochord | [55]+ |
| Ci-Eya | A | Epidermis, mesench., neurohyp., palps | [40] |
| Ci-fog | A | Pan-animal hemisphere at 32-cell stage | [62] |
| Ci-FoxAa | A | Lateral CNS | [48] |
| B | CNS | ||
| C | Autoregulation | ||
| D | Notochord & endoderm | ||
| E | Ectopic epidermis | ||
| Ci-FoxB | A | Notochord, epidermis | [55]+ |
| B | Mesenchyme, neck, muscle, visc. ganglion | ||
| Ci-FoxC | A | Palps, sensory vesicle | [55]+ |
| Ci-FoxD | A | A5.1, A5.2, B5.1 lineages | [63] |
| Ci-FoxF | A | Trunk ventral cells at tailbud stage | [64] |
| Ci-FoxN2/3 | A | Mesenchyme, tail muscles | [55]+ |
| B | CNS, mesenchyme, epidermis | ||
| Ci-orphanFox1 | A | Mesenchyme | [55]+ |
| Ci-GATAb | A | a-line ectoderm | [55]+ |
| B | Mesenchyme | ||
| Ci-Hes-a | A | Head endoderm, endodermal strand, epidermis | [55]+ |
| Ci-Hndx | A | Endoderm, trunk lateral & trunk ventral cells | [65] |
| Ci-Hox1 | A | Epidermis, neural tube | [66] |
| B | RA response element | ||
| Ci-Hox3 | A | Brain | [67] |
| Ci-Irx-B | A | Endoderm, palps, tail epidermis | [55]+ |
| Ci-KLF1/2/4 | A | Endodermal strand, mesenchyme, tail muscles | [55]+ |
| Ci-Lhx3 | A | Mesenchyme (B8.5 & B7.7 lines), muscle | [55]+ |
| Ci-meis | A | Sensory vesicle, tail muscle | [57] |
| Ci-mesp | A | B7.5 cells at 112-cell stage | [68] |
| Ci-Msxb | A | CNS | [69] |
| B | Pharynx | ||
| C | Ventral epidermis | ||
| Ci-neurogenin | A | Nerve cord, muscle, mesenchyme | [55]+ |
| Ci-Nodal | A | Notochord, tail epidermis, ventral head epidermis | [55]+ |
| B | b8.17, b8.18, b8.19, b8.20 at 112-cell stage | ||
| Ci-NPP | A | Head endoderm. TVCs | [65] |
| Ci-Otx | A | Neuroectoderm from 32-cell stage (“a-element”) | [41] |
| Ci-paraxis | A | Primary muscle lineage | [70] |
| Ci-Pax6 | UB | Nerve cord & sensory vesicle amplifier | [44] |
| UA | Sensory vesicle | ||
| I1 | Photoreceptors & nerve cord amplifier | ||
| I4 | Ectopic repression | ||
| Ci-Pitx | A | Neurohypophysis | [71] |
| B | Epidermis | ||
| Ci-RAR | A | Muscle | [72] |
| B | CNS | ||
| C | Epidermis (“E element”) | ||
| Ci-Rora | A | a-line ectoderm at gastrula stage (“AS2” | [73] |
| Ci-Rorb | A | a-line ectoderm at gastrula stage (“BS1”) | [73] |
| B | a-line neural ectoderm gastrula stage (“BS4”) | ||
| C | Neural gland of adult & early repressors (“BL”) | ||
| Ci-Rx | A | CNS | [74] |
| Ci-sFRP1/5 | A | Anterior gastrula ectoderm | [75] |
| Ci-sna | A | B4.1 lineage | [76] |
| Ci-SoxB1 | A | Ectoderm | [55]+ |
| B | Mesenchyme | ||
| Ci-SoxC | A | Mesenchyme, muscle, epidermis (only midline), epidermal neruons, palps and a part of brain | [55]+ |
| Ci-Tbx6b | A | Muscle?? | [70] |
| Ci-TTF1 (Nkx2-1) | A | Endoderm | [77] |
| Ci-Trim2/3 | A | Anterior neural precursors (weak) | [55]+ |
| Ci-Unc4A | A | Notochord | [55]+ |
| Ci-Wnt5 | A | Muscle precursors | [55]+ |
| Ci-ZicL-B | A | A6.2 & A6.4 lineages | [78] |
| B | B6.2 & B6.4 lineages & later A-line notochord, nerve cord & muscle | ||
| Ci-Znf(C2H2)-24 | A | Mesenchyme, endodermal strand, muscle | [55]+ |
| Ci-Znf(C3H) | A | CNS, mesenchyme, notochord, tail muscles, palps | [55]+ |
This element not shown on alignment.
Data from Y. Ohtsuka, unpublished, accessed on www.aniseed.cnrs.fr.
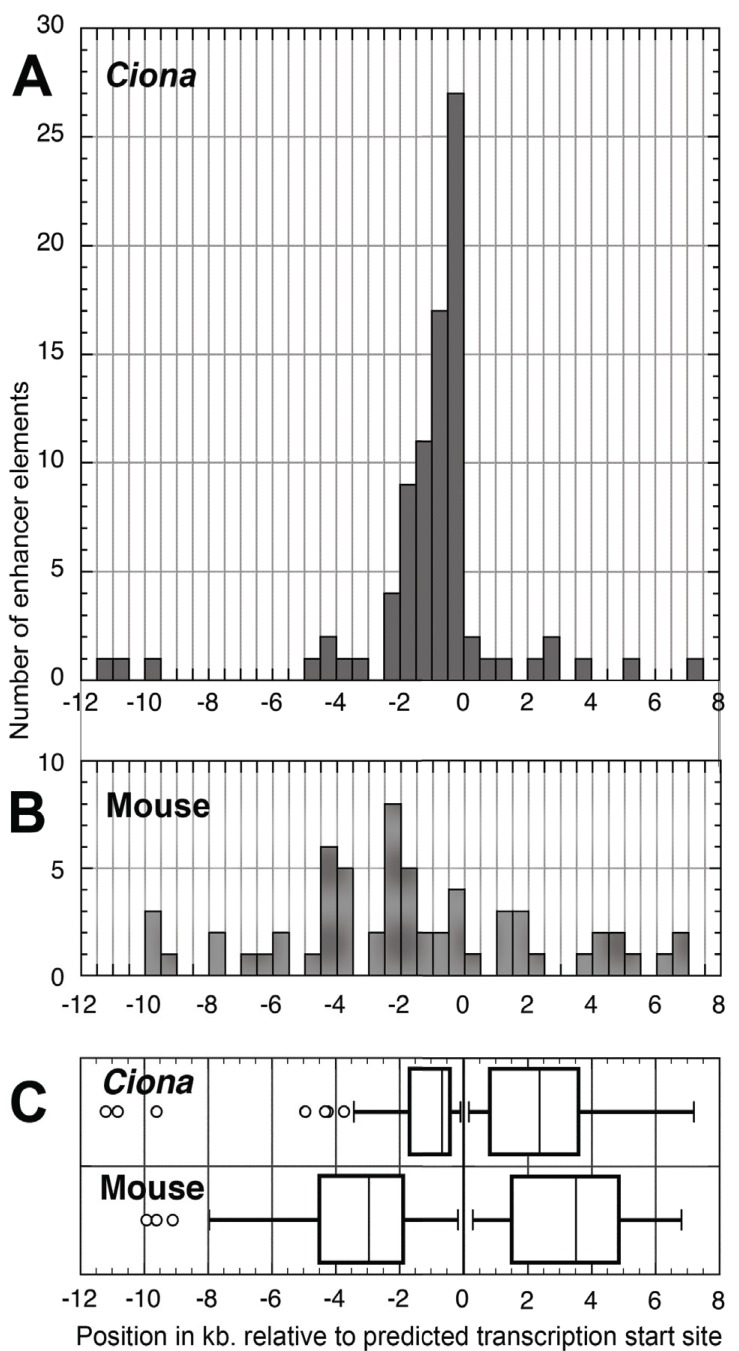
The 83 Ciona enhancers listed in Table 2 were all found within 12 kb. upstream or 7.5 kb. downstream of their respective estimated transcription start sites (TSSs). Consistent with the compact genome, most enhancers are found within 1.5 kb. upstream of the TSS [22], as shown in Fig. 2A. Fig. 2B shows the contrasting condition in the mouse. Out of a random selection of 79 experimentally verified mouse enhancers from 30 different transcription factor and cell-cell signaling genes, those within the total range of the Ciona enhancers were much less concentrated close to the TSS, and more likely to be located downstream. In fact, in this sample, mouse enhancers ranged as far as 123 kb downstream and 93 kb upstream. (Refer to Supplemental Material Table S1 (1.7MB, pdf) for Ciona enhancer locations, and Table S2 (1.7MB, pdf) for the list of mouse genes and enhancer locations).
Fig. (2).
Genomic positions of experimentally localized cis-regulatory elements (CREs) in C. intestinalis as compared with those in mouse. A) Histogram depicting frequencies of positions of CREs for 62 C. intestinalis transcription factor and cell-cell signaling genes. Positions based on 5' or 3' mean distance of experimentally determined cis-regulatory elements from the TSS. B) Mouse CRE positions for 30 mouse transcription factor and cell-cell signaling genes, depicted as in A). Only CREs between -12 and +8 kb relative to the TSS are included. C) Distribution of mean positions of CREs compared between C. intestinalis and mouse. For C. intestinalis and mouse data and references refer to Supplemental Tables 1 (1.7MB, pdf) and 2 (1.7MB, pdf) .
There is clearly a much greater spread in the locations of mouse CREs as compared with those of C. intestinalis (Fig. 2C). In this sample the median locations of C. intestinalis CREs was 630 bp upstream from the TSS and 2.4 kb downstream, while in the mouse the medians were 4.1 kb upstream from the TSS, and 6.3 kb downstream. Because CREs more than 12 kb upstream or 8 kb downstream of the mouse TSSs were ignored in constructing Fig. 3C, the actual median distance to CREs in the mouse is even further than shown here.
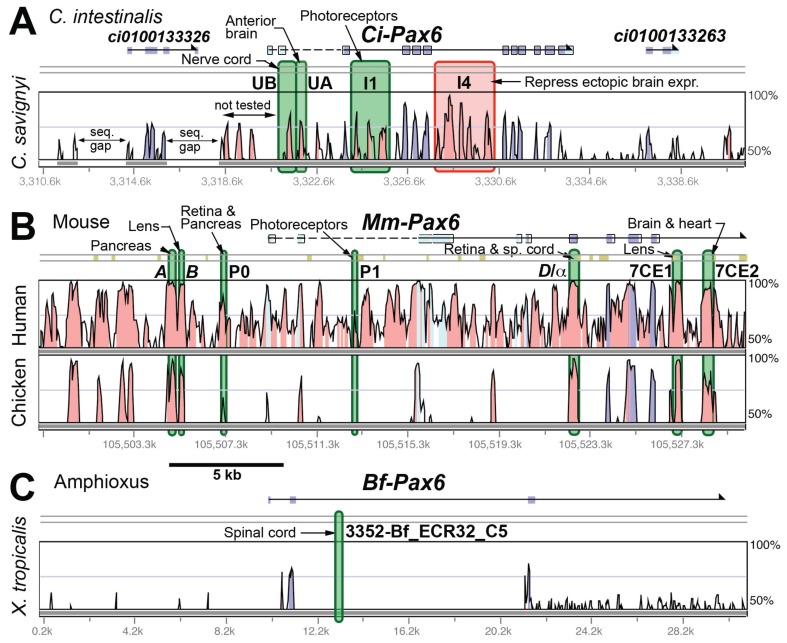
Fig. (3).
Graphical representations of sequence conservation and CRE locations for Pax6 genes in C. intestinalis, mouse and amphioxus at the same genomic scale. Alignments were generated by AVID or LAGAN using the VISTA web application [43]. Experimentally verified CREs are denoted by colored overlays - green for enhancers, red for repressors, blue for CREs that modulate expression in some other way. Regions of expression for each CRE are noted. A) Plot of sequence conservation between C. intestinalis and C. savignyi for the ascidian Pax6 gene. B) Mouse vs. chicken. C) Amphioxus vs. Xenopus. No more conservation was seen in alignments of amphioxus with human, zebrafish, or lamprey DNA. (CREs: [44-47, 29]).
The close position to the TSS of most C. intestinalis CREs means that investigators have a good chance of finding major CREs by examining only a few kb upstream of the TSS. However, an important caveat to this axiom is that it is possible that searches for CREs in C. intestinalis are biased towards elements proximal to the TSS. The kinds of long-range CRE searches that have been done in mouse, using BAC reporter transgenes, or Drosophila using P-element transgenes, for example, have not been done in C. intestinalis, so the presence of CREs acting at more than a few kb cannot be ruled out.
This difference in the distances to CREs between the mammal and C. intestinalis reflect the extreme compactness of the Ciona haploid genome, which is only 160 Mb, as opposed to 2700 Mb in the mouse. However, genome size doesn't necessarily correlate with compactness of CRE architecture, as seen in the large distances to some CREs found in Drosophila, which has a similar genome size to C. intestinalis[23]. For example, in a very incomplete look at D. melanogaster CREs in the Redfly database [24], the most proximal enhancers for the Hox genes lab and Scr are 1-2 kb 5' of the TSS, and for Dfd about 4 kb 5'. However, numerous other enhancers for each gene are found further from the promoter, as much as 40 kb 5' and 25 kb 3' of the TSS for Scr. For the fly Dll gene, the most proximal enhancer found is about 5 kb 5' of the TSS, with longer range enhancers up to 15 kb 5' and 40 kb 3'. In C. intestinalis, on the other hand, the enhancers for Ci-Hox1 and Ci-Hox3 are within 2 kb 5' and 2.5 kb 3' of the TSS, and for Ci-DllA and DllB, major enhancers are within 0.5 kb 5', without longer range enhancers having been found. Thus, the ascidian may have a constraint on CRE distance that is not present in some other metazoans with small genomes, such as Drosophila.
While this apparent constraint on distance of enhancers from transcription start sites may be an artifact of the lack of longer range enhancer searches, if the constraint turns out to be real it may be related to the apparent prevalence of rearrangements in the Ciona genome. For example, the Hox and ParaHox genes, which have conserved cluster arrangements in many other animals, are dispersed and rearranged in C. intestinalis [25, 26]. If the C. intestinalis genome is subject to frequent rearrangement, there may be a selective advantage to having enhancers located close to TSSs, since there would be a lower probability of a rearrangement event between the enhancer and basal promoter that might render the enhancer non-functional. In this scenario, enhancers located more distally would be lost to purifying selection if rearranged away from their appropriate basal promoter.
CONSERVED SEQUENCES AND CIS-REGULATORY MODULES
Another advantage of C. intestinalis for CRE study is the fortuitous genetic distance to its congener C. savignyi, also sequenced, which allows for reliable prediction of CRE positions by finding conserved non-coding elements (CNEs) in alignments of the genomes [27]. Even though C. savignyi and C. intestinalis are capable of producing viable hybrid offspring by in-vitro fertilization, they exhibit very low similarity in most non-coding DNA sequences. Genomic comparisons indicate that the genetic distance between the two congeneric ascidians is in the range of human-chicken or human-frog [27, 28]. The fact that the two ascidians can be hybridized suggests that the coding and non-coding DNA sequences that are highly conserved represent genuine functional sequences. This observation has been borne out by many investigations, and allows for reliable prediction of putative CREs for experimental verification.
In order to visualize the relationship between CNEs and CREs, Figs. 3-5 map experimentally determined CRE locations onto same-scale plots of sequence conservation for 3 transcription factor genes in C. intestinalis, mouse, and certain other taxa. These comparisons were chosen because of the availability of published experimental CRE data along with complete genome sequences. Each comparison provides different insights into CRE characteristics.
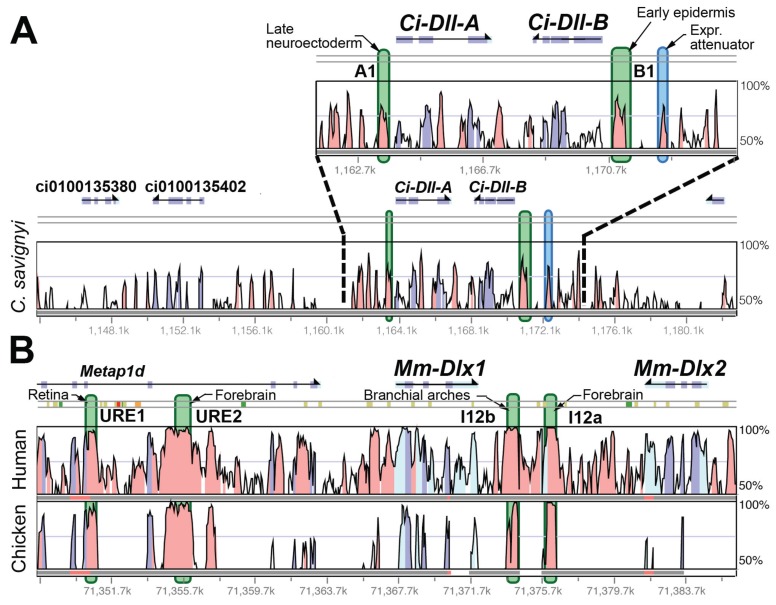
Fig. (5).
Graphical representations of sequence conservation and CRE locations for Dlx cluster genes in C. intestinalis and mouse. Plots constructed as in Fig. 3. The mouse Dlx1-2 cluster is used in comparison with the C. intestinalis DllA-B cluster, since there are more CRE data available for that cluster, although the orthology of the C. intestinalis cluster with respect to the three mouse Dlx clusters has not been able to be determined by phylogenetic means [31]. A) C. intestinalis Pax6 vs. C. savignyi genome. The region proximal to the TSSs is shown enlarged. B) Mouse Dlx1-2 cluster vs. chicken. (CREs: [37, 51, 52]).
Case 1 - Pax6
In Fig. 3, data for Pax6 are plotted. Here an alignment for amphioxus vs. frog is also included, as a CRE has been reported for this other invertebrate chordate. In the alignment of C. intestinalis vs. C. savignyi (Fig. 3A), each of the four CNEs tested by reporter gene assay had some cis-regulatory activity, although the I1 and I4 regions were not dissected in enough detail to exclude the possibility that some individual CNE peaks within those regions are not involved in cis-regulation. The mouse Pax6 gene is clearly orthologous to Pax6 in C. intestinalis. The alignment shown (Fig. 3B) compares the mouse genome with that of chicken, as the chicken-mouse overall genetic distance is similar to C. intestinalis-C. savignyi. More individual CREs have been found in Mm-Pax6, than in Ci-Pax6, as would be expected given the much greater anatomical complexity of the mammal. (In addition, long-range downstream CREs have been found in the mouse, which are not shown here.) Interestingly, although 5 of the 7 CREs identified map to mouse-chicken CNEs, 2 of the CREs (P0 and P1) do not. In fact, even in the mouse-human comparison, these CREs correspond to regions much less conserved than several others in this region that were not found to have CRE activity. Included in this catagory are 3 upstream and two intronic CNEs conserved with chicken. An unusual case of an active CRE is the "D/α(" element, which is coincident with one of the protein coding exons.
In the case of amphioxus, a survey of CNEs found in an exhaustive and sensitive search [29] located a short (48 bp) CNE in the Bf-Pax6 gene Fig. (3C). The less sensitive AVID or LAGAN alignment programs implemented in VISTA, and used for the Fig. (3-5) plots, failed to show an amphioxus CNE shared with frog, lamprey, zebrafish or human genomes at the empirically identified CRE. For amphioxus, either alignment algorithms more sophisticated than those commonly used, or a more closely related comparison genome, will be required to discover more CREs using sequence analysis.
Case 2 - FoxA
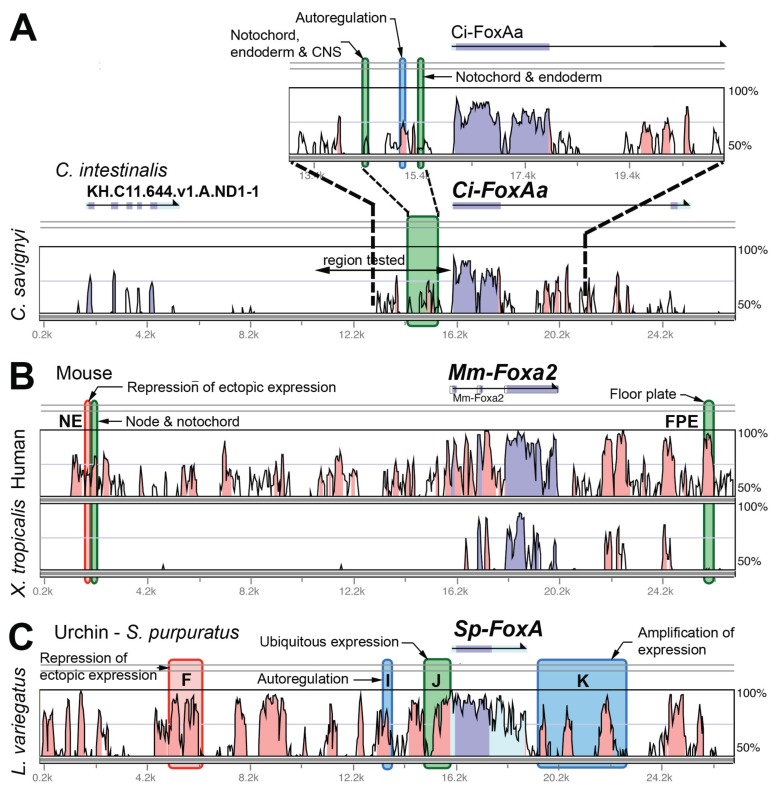
Empirical data on FoxA CREs are available for C. intestinalis, mouse, and sea urchin. In the case of C. intestinalis, CREs were carefully mapped on a VISTA plot of CNEs. This mapping shows that 3 of the experimentally identified CREs correspond with CNEs, but two significant CREs do not (the notochord, endoderm, and CNS enhancers in Fig. 4A). In fact, two CNEs fall just adjacent to CREs. For the mouse Foxa2 gene 2 of 3 CREs coincide with CNEs in the mouse-human comparison. However, once again there are equally or more conserved sequence elements in the mouse-human comparison that failed to show CRE activity in the published studies. As for a more distant comparison, most of the mouse Foxa2 locus fails to align with the chicken genome, so the comparison here is with the frog, X. tropicalis. None of the 3 identified CREs show up as CNEs in the frog alignment.
Fig. (4).
Graphical representations of sequence conservation and CRE locations for FoxA genes in C. intestinalis, mouse and sea urchin. Plots constructed as in Fig. 3. While these genes may not be strictly orthologous, they belong to the same gene subfamily (FoxA. A) Plot of sequence conservation between C. intestinalis and C. savignyi for the ascidian FoxAa (a.k.a. forkhead) gene. The region proximal to the TSS is shown enlarged. B) Mouse Foxa2 vs. human and chicken genomes. Note that there are extensive gaps in the mouse vs. chicken alignment. C) S. purpuratus vs. L. variegatus FoxA (two sea urchins). (CREs: [48-50, 30]).
A study has also been published of the sea urchin Strogylocentrotus purpuratus FoxA gene, which revealed 4 CREs (Fig. 4C). All 4 correspond with CNEs showing up in an alignment with the urchin Lytechinus variegatus, the species the study authors used for comparison [30]. While 98 kb of upstream and 50 kb of downstream sequence was tested for CRE activity, the acting CREs were found within 12 kb upstream and 7 kb downstream of the TSS. Within the 26 kb of sequence shown in Fig. (4C), on the other hand, only about half of the CNEs correspond with CREs that show up in the reporter assays. Interestingly, from the work published on these gene loci to date, it appears that the complexity in terms of numbers of CREs regulating each gene is similar in the three taxa, even though anatomical complexity varies widely from C. intestinalis to urchin to mouse, and the genes are expressed both early and late in development.
Case 3 - Dlx Clusters
Both vertebrates and C. intestinalis have Dlx homeobox genes arranged in convergently transcribed 2-gene clusters, which are linked to Hox clusters on the same chromosomes [31]. Mammals have 3 such clusters - the relationships of which to the single C. intestinalis cluster have not been able to be robustly determined. Because the most CRE data are available for the mouse Dlx1-2 cluster, that locus has been used here (Fig. 5) for comparison. For C. intestinalis the major enhancer elements are located proximal to the respective TSSs and correspond with the largest regions of conserved sequence with C. savignyi (Fig. 5A). For Ci-DllB, additional upstream sequence in which four CNEs are located was tested, only one of which had an effect (attenuation) on expression.
For the mouse Dlx1-2 cluster, the most broadly acting CREs are located in the large intergenic region. Other CREs activating Mm-Dlx1 (URE1 and 2) have been found far upstream in the introns of the Metap1d gene model. The number of CREs found to date, 3 in C. intestinalis and 4 in mouse, suggest a comparable level of cis-regulatory complexity. However, it is possible that more mouse CREs would be found with more extensive searching, and that the individual mouse cis-regulatory modules have more internal complexity than those in the ascidian.
For the Dlx clusters the C. intestinalis-C. savignyi and mouse-chicken genomic comparisons do a good job of correlating CNEs with CREs. Once again though, several CNEs are found that do not correspond with the CREs uncovered in reporter gene assays.
Correspondance Between Conserved Non-Coding Elements and cis-regulatory Elements
As found by Johnson et al. (2004) [27], in many cases verified C. intestinalis CREs correspond to non-coding sequences conserved with C. savignyi. (In the present analysis at >70% identity over 75 bp scanned with a 75 bp window in VISTA.) However, even in the small sample of 3 genes examined here, one case - Ci-FoxAa - has limited conserved sequence showing up in the commonly used VISTA analysis. Here, a more sensitive conservation-finding method might have located more of the discovered CREs. On the other hand, it may be the case that there is too much rearrangement or reorganization of the CREs since the split of C. intestinalis and C. savignyi to be able to use sequence analysis as a CRE finding tool. It is likely that this would be the case for a significant minority of both individual CREs, or the complement of CREs for particular genes.
Interestingly, the mouse Foxa2 gene exhibits a somewhat similar situation to that of Ci-FoxAa. In the intermammalian comparison, there are many CNEs with the same or greater degree of conservation to the few that have been empirically shown to have CRE activity, while no sequence conservation with the phylogenetically closest non-mammalian sequenced genome, X. tropicalis, was found. In other words, for this gene the consensus of mammalian alignments have too much sequence conservation to efficiently identify CREs, but not enough sequence conservation to the next closest taxon.
Alternative approaches for identifying CREs rely on various ways of searching for shared non-coding motifs, in, for example, coexpressed genes, or searching for combinations of known transcription factor binding sites (TFBSs) (reviewed in [12]). Even these sophisticated sequence analytical approaches can still be quite error prone as shown by Halfon et al. [32].
Taken together, these comparative observations point out 2 major advantages of the C. intestinalis system for cis-regulatory study. First, non-coding sequence conservation comparisons with C. savignyi, are at a fortuitous genetic distance to have a high probability of locating functional CREs. These C. intestinalis-C. savignyi comparisons consistently identify CNEs that correspond with CREs. Mammals and other chordates, such as amphioxus, do not have such phylogenetically well-positioned sequenced comparison taxa for this purpose. Second, the close proximity of C. intestinalis CREs to the transcription start site means that only a few kb of DNA must be scanned experimentally to have a high probability of recovering the major CREs for any gene. This is certainly not the case for the mouse, and to some extent even for the sea urchin.
Conserved Non-Coding Sequences That Do Not Show cis--regulatory Activity
Even though phylogenetic or binding site sequence analysis can point the way to locating CREs, many, if not most, CNEs do not correspond with CREs identified in empirical experiments, such as reporter gene assays, or ChIP-Seq. In a study of human conserved sequence elements tested in mouse [33], only 29% of DNA sequences conserved between human and Fugu had positive enhancer activity when placed upstream of a heat shock promoter and tested at stage e11.5. Another approach in the same study scanned for possible forebrain specific enhancer sequences. Only 17% of the new elements found had reporter gene forebrain expression. In the small sample of C. intestinalis genes shown here (Figs. 3-5) the correspondance is much better, 67%, although there are many distal CNEs that were not tested for cis-regulatory activity. In addition, two Ci-FoxAa CREs did not align with a CNE, and would have been missed if only testing conserved sequence elements.
There are, of course, a number of reasons that a conserved non-coding sequence might be functional, and therefore constrained by purifying selection, but not show CRE activity in a reporter assay [12]. One set of possibilities is that the CNE is actually involved in cis-regulation, but does not have activation activity that would drive reporter gene expression. These elements could be repressors, insulators or modifiers that have negative regulatory function, and can only be detected by their effect on another cis-regulatory element. A CRE may only have activity in concert with another element, so the individual element shows no activity on its own eg. [34]. The CNE could also be a positive acting CRE, but just operate at a developmental stage not tested. Another possibility is "shadow enhancers" - remote activating elements redundant with other CREs but further from the gene that they act upon [35, 36]. Characterization of these cryptic CREs will depend on more sensitive reporter assays, or more exhaustive assays such as ChIP-Seq, that reveal protein-DNA binding without regard to effects on a promoter.
Apart from these genuine but undetected CREs, are other non-coding functional elements, such as matrix attachment regions, or transcribed non-coding RNAs, such as micro RNAs, or long non-coding RNAs. Identification of the function of these CNEs will rely on completely different types of experimental and bioinformatic approaches than those used for CRE discovery.
Not All cis-regulatory Activity is Confined to Conserved Sequences
As mentioned above, some CREs found through reporter gene assays do not correspond with CNEs (at least at the same level of sensitivity and genetic distance that reveals other CREs). In addition, in cases where CRE dissections have been done in some detail, even if major CRE activity is found in conserved sequences there are sequences outside those conserved that contribute to the expression pattern. For example, in a detailed study of the Troponin I gene in C. intestinalis [27], 4 conserved sequences were located within a 400 bp region 5' of the TSS, all of which were required to recapitulate the endogenous expression pattern. However, for maximal expression, intervening less conserved sequence was also required. In another case, Ci-DllB, a CNE was identified that accounted for the majority of the endogenous expression pattern ("B1" in Fig. 5A), but an adjacent non-conserved sequence was capable of acting with a portion of the conserved region to activate a similar expression pattern [37].
THE CIONA MODEL AS A MEANS TO STUDY DETAILS OF CIS-REGULATORY ARCHITECTURE AND FUNCTION
Apart from the dissection of the cis-regulatory architecture of individual genes, C. intestinalis lends itself to both broader and deeper investigations of cis-regulation. Two studies may serve to demonstrate the particular advantages of reporter gene assays in this animal. Brown and colleagues [38] produced hundreds of reporter gene constructs and produced thousands of transgenic embryos to dissect the cis-regulation of co-expressed muscle-specific genes in C. intestinalis. They found that though similar DNA-binding proteins cluster in muscle gene CREs, their arrangement and relative contributions to activation of expression vary greatly. The authors also compared the cis-regulation of six orthologous muscle-specific genes in C. intestinalis vs. C. savignyi, and found large variations between the relative activity conferred by individual binding sites. The simplicity and rapidity of transient in vivo transgenic reporter experiments in Ciona contributed to the feasibility of performing the large number of assays required for this study. Other workers have also used the C. intestinalis system for study of groups of co-expressed genes e.g. [39, 40].
Another study by Lemaire and colleagues built on a previous finding of paired ETS and GATA binding sites in an Otx enhancer that confers early neural expression [41]. They then used in silico analysis to find 55 occurances of this motif in the C. intestinalis genome, and then tested each of these in in vivo reporter assays. 19 of the sequences showed enhancer activity, and examination of these CREs revealed a nucleosome exclusion motif that was found to be conserved between Ciona and Drosophila [42]. Once again, the ability to test a large number of reporter transgenes rapidly, along with the compactness of the C. intestinalis genome and CREs, enhanced the practicality of this study.
CONCLUSION
Cis-regulatory elements in C. intestinalis tend to be located relatively close to the transcription start sites of genes, which may be associated with the small size of its genome. This fact combined with the ease and economy of producing transient transgenic embryos in large numbers, have made it an attractive model system for the study of cis-regulation. Comparison of experimentally verified CREs with those found in other animals, such as the mouse, suggests that C. intestinalis transcriptional regulation relies on a similar complexity of CREs given its anatomical simplicity. Recent work touched on here makes increasingly sophisticated use of the extensive bioinformatic resources for Ciona, coupled with analysis of large numbers of in vivo reporter gene experiments, to study fundamental aspects of transcriptional regulation. Future work promises to further exploit the C. intestinalis model to advance understanding of the function and evolution of cis-regulation.
ACKNOWLEDGEMENTS
The author acknowledges the constructive critique of two anonymous reviewers which helped to improve the manuscript, and Amardeep Gill for help with the literature search. Analysis of Ciona cis-regulation was greatly facilitated by the extensive work put into the ANISEED database by members of the Patrick Lemaire lab and others. Thanks also to Anna DiGregorio for insights on Ci-FoxAa cis-regulation. This work was supported by a University of Rhode Island Council for Research grant, and NSF Epscor grant No. 0554548 to URI.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers Web site along with the published article.
REFERENCES
- 1.Satoh N. Developmental Biology of Ascidians. Cambridge: Cambridge Univ. Press; 1994. [Google Scholar]
- 2.Lemaire P. Unfolding a chorate developmental program, one cell at a time: Invariant cell lineages, short-range inductions and evolutionary plasticity in ascidians. Dev. Biol. 2009;332(1 ): 48–60. doi: 10.1016/j.ydbio.2009.05.540. [DOI] [PubMed] [Google Scholar]
- 3.Satoh N, Levine M. Surfing with the tunicates into the post-genome era. Genes & Development. 2005;19(20 ): 2407–2411. doi: 10.1101/gad.1365805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dehal P, Satou Y, Campbell R K, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein D M, Harafuji N, Hastings K E M, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen I A, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang H G, Awazu S, Azumi K, Boore J, Branno M, Chin-bow S, DeSantis R, Doyle S, Francino P, Keys D N, Haga S, Hayashi H, Hino K, Imai K S, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee B I, Makabe K W, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki M M, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt P D, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar D S. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298(5601 ): 2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 5.Imai K S, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- 6.Christiaen L, Davidson B, Kawashima T, Powell W, Nolla H, Vranizan K, Levine M. The tran-scription/migration interface in heart precursors of Ciona intestinalis. Science. 2008;320:1349–1352. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- 7.Jose-Edwards D S, Kerner P, Kugler J E, Deng W, Jiang D, Di Gregorio A. The Identification of Transcription Factors Expressed in the Notochord of Ciona intestinalis Adds New Potential Players to the Brachyury Gene Regulatory Network. Dev. Dyn. 240(7 ): 1793–1805. doi: 10.1002/dvdy.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbo J C, Levine M, Zeller R W. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- 9.Zeller R W, Virata M J, Cone A C. Predictable mosaic transgene expression in ascidian embryos produced with a simple electroporation device. Dev. Dyn. 2006;235:1921–1932. doi: 10.1002/dvdy.20815. [DOI] [PubMed] [Google Scholar]
- 10.Vierra D A, Irvine S Q. Optimized conditions for transgenesis of the ascidian Ciona using square wave electroporation. Dev. Genes Evol. 2012;222(1 ): 55–61. doi: 10.1007/s00427-011-0386-0. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Christiaen L. Transcriptional enhancers in ascidian development. In: Plaza S P F, editor. Transcriptional Switches During Development. Vol 98. 2012. pp. 147–172. [DOI] [PubMed] [Google Scholar]
- 12.Haeussler M, Joly J S. hen needles look like hay: How to find tissue-specific enhancers in model organism genomes. Dev. Biol. 2011;350(2 ): 239–254. doi: 10.1016/j.ydbio.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 13.Simmen M W, Bird A. Sequence Analysis of Transposable Elements in the Sea Squirt Ciona intestinalis. Mol. Biol. Evol. 2000;17:1685–1694. doi: 10.1093/oxfordjournals.molbev.a026267. [DOI] [PubMed] [Google Scholar]
- 14.Feng S H, Cokus S J, Zhang X Y, Chen P Y, Bostick M, Goll M G, Hetzel J, Jain J, Strauss S H, Halpern M E, Ukomadu C, Sadler K C, Pradhan S, Pellegrini M, Jacobsen S E. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. USA. 2010;107(19 ): 8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small K S, Brudno M, Hill M M, Sidow A. Extreme genomic variation in a natural population. Proc. Natl. Acad. Sci. USA. 2007;104:5698–5703. doi: 10.1073/pnas.0700890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 17.Dunn C W, Hejnol A, Matus D Q, Pang K, Browne W E, Smith S A, Seaver E C, Rouse G W, Obst M, Edgecombe G D, Sorenson M V, Haddock S H D, Schmidt-Rhaesa A, Okusu A, Kristensen R M, Wheeler W C, Martindale M Q, Giribet G. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–750. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 18.Lemaire P. Evolutionary crossroads in developmental biology: the tunicates. Development. 2011;138(11 ): 2143–2152. doi: 10.1242/dev.048975. [DOI] [PubMed] [Google Scholar]
- 19.Natale A, Sims C, Chiusano M L, Amoroso A, D'Aniello E, Fucci L, Krumlauf R, Branno M, Locascio A. Evolution of anterior Hox regulatory elements among chordates. BMC Evol. Biol. 2011;11 doi: 10.1186/1471-2148-11-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaster-Jones L. Cis-regulation and conserved non-coding elements in amphioxus. Brief. Funct. Genomics. 2012;11(2 ): 118–130. doi: 10.1093/bfgp/els006. [DOI] [PubMed] [Google Scholar]
- 21.Kusakabe T. Decoding cis-regulatory systems in ascidians. Zool. Sci. 2005;22:129–146. doi: 10.2108/zsj.22.129. [DOI] [PubMed] [Google Scholar]
- 22.Satoh N, Satou Y, Davidson B, Levine M. Ciona intestinalis: an emerging model for whole-genome analyses. Trends Genet. 2003;19(7 ): 376–381. doi: 10.1016/S0168-9525(03)00144-6. [DOI] [PubMed] [Google Scholar]
- 23.Arnosti D N. Analysis and function of transcriptional regulatory elements: insights from Drosophila. Annu. Rev. Entomol. 2003;48:579–602. doi: 10.1146/annurev.ento.48.091801.112749. [DOI] [PubMed] [Google Scholar]
- 24.Gallo S M, Gerrard D T, Miner D, Simich M, Des Soye B, Bergman C M, Halfon M S. REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res. 39:D118–D123. doi: 10.1093/nar/gkq999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrier D, Holland P. Ciona intestinalis ParaHox genes: evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Mol. Phyl. Evol. 2002:412–417. doi: 10.1016/s1055-7903(02)00204-x. [DOI] [PubMed] [Google Scholar]
- 26.Ikuta T, Yoshida N, Satoh N, Saiga H. Ciona intestinalis Hox gene cluster: Its dispersed structure and residual colinear expression in development. Proc. Natl. Acad. Sci. USA . 2004;101(42 ): 15118–15123. doi: 10.1073/pnas.0401389101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson D S, Davidson B, Brown C D, Smith W C, Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 2004;14(12 ): 2448–2456. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berna L, Alvarez-Valin F, D'Onofrio G. How fast is the sessile Ciona? Comp. Funct. Genomics. 2009;2009:875901. doi: 10.1155/2009/875901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hufton A L, Mathia S, Braun H, Georgi U, Lehrach H, Vingron M, Poustka A J, Panopoulou G. Deeply conserved chordate noncoding sequences preserve genome synteny but do not drive gene duplicate retention. Genome Res. 2009;19(11 ): 2036–2051. doi: 10.1101/gr.093237.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De-Leon S B T, Davidson E H. Information processing at the foxa node of the sea urchin endomesoderm specification network. Proc. Natl. Acad. Sci. USA. 2010;107(22 ): 10103–10108. doi: 10.1073/pnas.1004824107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irvine S Q, Cangiano M C, Millette B J, Gutter E S. Non-overlapping expression patterns of the clustered DllA/B genes in the ascidian Ciona intestinalis. J. Exp. Zool. (Mol. Dev. Evol.) 2007;308B:428–441. doi: 10.1002/jez.b.21169. [DOI] [PubMed] [Google Scholar]
- 32.Halfon M S, Zhu Q, Brennan E R, Xhou Y. Erroneous attribution of relevant transcription factor binding sites despite successful prediction of cis- regulatory modules. BMC Genomics . 2011;12:578. doi: 10.1186/1471-2164-12-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pennacchio L A, Ahituv N, Moses A M, Prabhakar S, Nobrega M A, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis K D, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black B L, Couronne O, Eisen M B, Visel A, Rubin E M. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444(7118 ): 499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 34.Guerrero L, Marco-Ferreres R, Serrano A L, Arredondo J J, Cervera M. Secondary enhancers synergise with primary enhancers to guarantee fine-tuned muscle gene expression. Dev. Biol. 0000;337(1 ): 16–28. doi: 10.1016/j.ydbio.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Hong J W, Hendrix D A, Levine M S. Shadow enhancers as a source of evolutionary novelty. Science . 2008;321(5894 ): 1314–1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barolo S. Shadow enhancers: Frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays. 2011;34(2 ): 135–141. doi: 10.1002/bies.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irvine S Q, Vierra D A, Millette B J, Blanchette M D, Holbert R E. Expression of the Distalless-B gene in Ciona is regulated by a pan-ectodermal enhancer module. Dev. Biol. 2011;353:432–439. doi: 10.1016/j.ydbio.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown C D, Johnson D S, Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science . 2007;317:1557–1560. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- 39.Kusakabe T, Yoshida R, Ikeda Y, Tsuda M. Computational discovery of DNA motifs associated with cell type-specfic gene expression in Ciona. Dev. Biol. 2004;276(2 ):563–580. doi: 10.1016/j.ydbio.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 40.Haeussler M, Jaszczyszyn Y, Christiaen L, Joly J S. A cis-Regulatory Signature for Chordate Anterior Neuroectodermal Genes. PLoS Genet. 2010;6(4 ) doi: 10.1371/journal.pgen.1000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell. 2003;115(5 ): 615–627. doi: 10.1016/s0092-8674(03)00928-0. [DOI] [PubMed] [Google Scholar]
- 42.Khoueiry P, Rothbacher U, Ohtsuka Y, Daian F, Frangulian E, Roure A, Dubchak I, Lemaire P. A cis-Regulatory Signature in Ascidians and Flies, Independent of Transcription Factor Binding Sites. Curr Biol. 2010;20(9 ): 792–802. doi: 10.1016/j.cub.2010.03.063. [DOI] [PubMed] [Google Scholar]
- 43.Mayor C, Brudno M, Schwartz J R, Poliakov A, Rubin E M, Frazer K A, Pachter L S, Dubchak I. VISTA: Visualizing Global DNA Sequence Alignments of Arbitrary Length. Bioinformatics. 2000;(16): 1046. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 44.Irvine S Q, Fonseca V C, Zompa M A, Antony R. Cis-regulatory organization of the Pax6 gene in the ascidian Ciona intestinalis. Dev. Biol. 2008;317(2 ): 649–659. doi: 10.1016/j.ydbio.2008.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev. Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 46.Xu P X, Zhang X, Heaney S, Yoon A, Michelson A M, Maas R L. Regulation of Pax6 expression is conserved between mice and flies. Development. 1999;126(2 ): 383–95. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- 47.Kleinjan D A, Seawright A, Childs A J, van Heyningen V. Conserved elements in Pax6 intron 7 involved in (auto)regulation and alternative transcription. Dev. Biol. 2004;265(2 ): 462–77. doi: 10.1016/j.ydbio.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 48.Di Gregorio A, Corbo J, Levine M. The regulation of forkhead/HNf-3 beta expression in the Ciona embryo. Dev. Biol. 2001;229:31–43. doi: 10.1006/dbio.2000.9964. [DOI] [PubMed] [Google Scholar]
- 49.Sasaki H, Hogan B L M. Enhancer analysis of the mouse HNF-3beta gene: regulatory elements for node/notchord and floor plate are independent and consist of mutiple sub-elements. Genes Cells. 1996;1:59–72. doi: 10.1046/j.1365-2443.1996.04004.x. [DOI] [PubMed] [Google Scholar]
- 50.Nishizaki Y, Shimazu K, Kondoh H, Sasaki H. Identification of essential sequence motifs in the node/notochord enhancer of Foxa2 (Hnf3 beta) gene that are conserved across vertebrate species. Mech. Dev. 2001;102(1-2 ): 57–66. doi: 10.1016/s0925-4773(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 51.Ghanem N, Jarinova O, Amores A, Long Q, Hatch G, Park B K, Rubenstein J L R, Ekker M. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 2003;13:533–543. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghanem N, Yu M, Long J, Hatch G, Rubenstein J L R, Ekker M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. Journal of Neuroscience. 2007;27(19 ): 5012–5022. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satou Y, Kawashima T, Shoguchi E, Nakayama A, Satoh N. An integrated database of the ascidian, Ciona intestinalis: Towards functional genomics. Zool. Sci. 2005;22(8 ): 837–843. doi: 10.2108/zsj.22.837. [DOI] [PubMed] [Google Scholar]
- 54.Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, Yamada L, Matsumoto J, Wasserscheid J, Dewar K, Wiley G B, Macmil S L, Roe B A, Zeller R W, Hastings K E M, Lemaire P, Lindquist E, Endo T, Hotta K, Inaba K. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into intron and operon populations. Genome Biol. 2008;9(10 ) doi: 10.1186/gb-2008-9-10-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tassy O, Dauga D, Daian F, Sobral D, Robin F, Khoueiry P, Salgado D, Fox V, Caillol D, Schiappa R, Laporte B, Rios A, Luxardi G, Kusakabe T, Joly J S, Darras S, Christiaen L, Contensin M, Auger H, Lamy C, Hudson C, Rothbacher U, Gilchrist M J, Makabe K W, Hotta K, Fujiwara S, Satoh N, Satou Y, Lemaire P. The ANISEED database: Digital representation, formalization, and elucidation of a chordate developmental program. Genome Res. 2010;20(10 ): 1459–1468. doi: 10.1101/gr.108175.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiba S, Sasaki A, Nakayama A, Takamura K, Satoh N. Development of Ciona intestinalis juveniles (Through 2nd ascidian stage) Zool. Sci. 2004;21(3 ): 285–298. doi: 10.2108/zsj.21.285. [DOI] [PubMed] [Google Scholar]
- 57.Sierro N, Kusakabe T, Park K J, Yamashita R, Kinoshita K, Nakai K. DBTGR: a database of tunicate promoters and their regulatory elements. Nucleic Acids Res. 2006;34:D552–D555. doi: 10.1093/nar/gkj064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Endo T, Ueno K, Yonezawa K, Mineta K, Hotta K, Satou Y, Yamada L, Ogasawara M, Takahashi H, Nakajima A, Nakachi M, Nomura M, Yaguchi J, Sasakura Y, Yamasaki C, Sera M, Yoshizawa A C, Imanishi T, Taniguchi H, Inaba K. CIPRO 2.5: Ciona intestinalis protein database, a unique integrated repository of large-scale omics data, bioinformatic analyses and curated annotation, with user rating and reviewing functionality. Nucleic Acids Res. 2011;39:D807–D814. doi: 10.1093/nar/gkq1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hotta K. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev. Dyn. 2007;236:1790–1805. doi: 10.1002/dvdy.21188. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida R, Sasakura Y. Establishment of Enhancer Detection Lines Expressing GFP in the Gut of the Ascidian Ciona intestinalis. Zoological Science. 2012;29(1 ): 11–20. doi: 10.2108/zsj.29.11. [DOI] [PubMed] [Google Scholar]
- 61.Harafuji N, Keys D N, Levine M. Genome-wide identification of tissue-specific enhancers in the Ciona tadpole. Proc. Natl. Acad. Sci. USA. 2002;99:6802–6805. doi: 10.1073/pnas.052024999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rothbacher U, Bertrand V, Lamy C, Lemaire P. A combinatorial code of maternal GATA, Ets and beta-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development. 2007;134(22 ): 4023–4032. doi: 10.1242/dev.010850. [DOI] [PubMed] [Google Scholar]
- 63.Mita K, Fujiwara S. Nodal regulates neural tube formation in the Ciona intestinalis embryo. Dev. Genes Evol. 2007;217(8 ): 593–601. doi: 10.1007/s00427-007-0168-x. [DOI] [PubMed] [Google Scholar]
- 64.Beh J, Shi W, Levine M, Davidson B, Christiaen L. FoxF is essential for FGF- induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134(18 ): 3297–3305. doi: 10.1242/dev.010140. [DOI] [PubMed] [Google Scholar]
- 65.Davidson B, Levine M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc. Natl. Acad. Sci. USA. 2003;100(20 ): 11469–11473. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanda M, Wada H, Fujiwara S. Epidermal expression of Hox1 is directly activated by retinoic acid in the Ciona intestinalis embryo. Dev. Biol. 2009;335(2 ): 454–463. doi: 10.1016/j.ydbio.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 67.Locascio A, Aniello F, Amoroso A, Manzanares M, Krumlauf R, Branno M. Patterning the ascidian nervous system: structure, expression and transgenic analysis of the CiHox3 gene. Development. 1999;126:4737–4748. doi: 10.1242/dev.126.21.4737. [DOI] [PubMed] [Google Scholar]
- 68.Christiaen L, Stolfi A, Davidson B, Levine M. Spatiotemporal intersection of Lhx3 and Tbx6 defines the cardiac field through synergistic activation of Mesp. Dev. Biol. 2009;328(2 ): 552–560. doi: 10.1016/j.ydbio.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 69.Russo M, Donizetti A, Locascio A, D'Aniello S, Amoroso A, Aniello F, Fucci L, Branno M. Regulatory elements controlling Ci-msxb tissue-specific expression during Ciona intestinalis embryonic development. Dev. Biol. 2004;267:517–528. doi: 10.1016/j.ydbio.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 70.Erives A. Non-Homologous Structured CRMs from the Ciona Genome. Journal of Computational Biology . 2009;16(2 ): 369–377. doi: 10.1089/cmb.2008.20TT. [DOI] [PubMed] [Google Scholar]
- 71.Christiaen L, Bourrat F, Joly J S. A modular cis-regulatory system controls isoform-specific pitx expression in ascidian stomodaeum. Dev. Biol. 2005;277(2 ): 557–566. doi: 10.1016/j.ydbio.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 72.Fujiwara S. Promoter activity of the retinoic acid receptor gene in the Ciona intestinalis embryo. Dev. Dyn. 2005;232(4 ): 1124–1130. doi: 10.1002/dvdy.20265. [DOI] [PubMed] [Google Scholar]
- 73.Auger H, Lamy C, Haeussler M, Khoueiry P, Lemaire P, Joly J S. Similar regulatory logic in Ciona intestinalis for two Wnt pathway modulators, ROR and SFRP-1/5. Dev. Biol. 2009;329(2 ): 364–373. doi: 10.1016/j.ydbio.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 74.D'Aniello E, Pezzotti M R, Locascio A, Branno M. Onecut is a direct neural-specific transcriptional activator of Rx in Ciona intestinalis. Dev. Biol. 2011;355(2 ): 358–371. doi: 10.1016/j.ydbio.2011.05.584. [DOI] [PubMed] [Google Scholar]
- 75.Lamy C, Rothbacher U, Caillol D, Lemaire P. Ci-FoxA-a is the earliest zygotic determinant of the ascidian anterior ectoderm and directly activates Ci-sFRP1/5. Development. 2006;133(15 ): 2835–2844. doi: 10.1242/dev.02448. [DOI] [PubMed] [Google Scholar]
- 76.Erives A, Corbo J C, Levine M. Lineage-specific regulation of the Ciona snail gene in the embryonic mesoderm and neuroectoderm. Dev. Biol. 1998;194(2 ): 213–225. doi: 10.1006/dbio.1997.8810. [DOI] [PubMed] [Google Scholar]
- 77.Fanelli A, Lania G, Spagnuolo A, Di Lauro R. Interplay of negative and positive signals controls endoderm-specific expression of the ascidian Cititf1 gene promoter. Dev. Biol. 2003;263:12–23. doi: 10.1016/s0012-1606(03)00397-x. [DOI] [PubMed] [Google Scholar]
- 78.Anno C, Satou A, Fujiwara S. Transcriptional regulation of ZicL in the Ciona intestinalis embryo. Dev. Genes Evol. 2006;216(10 ): 597–605. doi: 10.1007/s00427-006-0080-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publishers Web site along with the published article.