Abstract
Glucosinolates are a major class of sulphur-containing secondary metabolites involved in plant defence against pathogens. Recently many regulatory links between glucosinolate biosynthesis and sulphate assimilation were established. Since sulphate assimilation undergoes diurnal rhythm and is light regulated, this study analysed whether the same is true for glucosinolate biosynthesis. The levels of glucosinolates and glutathione were found to be higher during the day than during the night. This agreed with variation in sulphate uptake as well as activity of the key enzyme of the sulphate assimilation pathway, adenosine 5’-phosphosulphate reductase. Correspondingly, the flux through sulphate assimilation was higher during the day than during the night, with the maximum flux through primary assimilation preceding maximal incorporation into glucosinolates. Prolonged darkness resulted in a strong reduction in glucosinolate content. Re-illumination of such dark-adapted plants induced accumulation of mRNA for many genes of glucosinolate biosynthesis, leading to increased glucosinolate biosynthesis. The light regulation of the glucosinolate synthesis genes as well as many genes of primary sulphate assimilation was controlled at least partly by the LONG HYPOCOTYL5 (HY5) transcription regulator. Thus, glucosinolate biosynthesis is highly co-regulated with sulphate assimilation.
Key words: Diurnal, glucosinolates, HY5, light, primary metabolism, secondary metabolism, sulphate assimilation
Introduction
Glucosinolates (GLS) are sulphur-rich compounds found in the Brassicaceae family. GLS and their breakdown products (thiocyanates, isothiocyanates, and nitriles) play an important role in plant defence against pathogens and herbivores, while humans may gain protection against cancer through the consumption of vegetables containing GLS, such as broccoli, kale, and Brussels sprouts (Traka and Mithen, 2009).
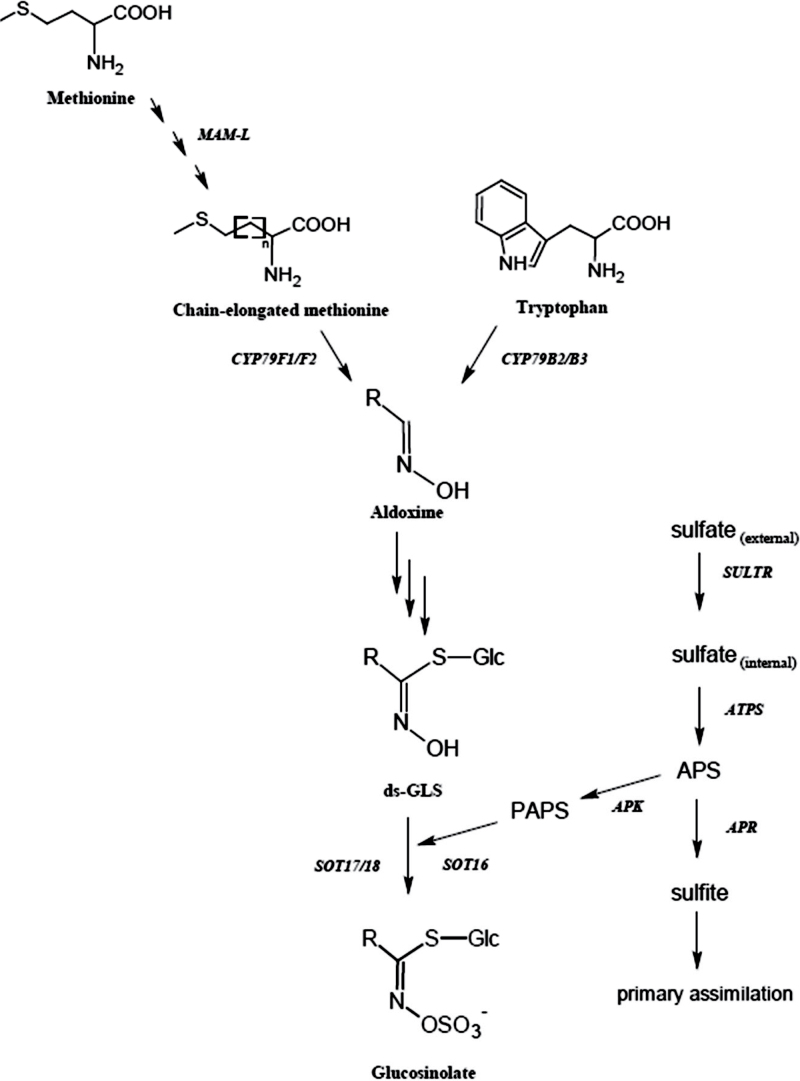
The GLS core structure is an S-glucose moiety and a sulphoxime group with an attached amino acid-derived side chain (Fig. 1). GLS are classified based on the origin of their side chain, and the main groups are aliphatic, indolic, and benzenic GLS derived from the amino acids methionine, tryptophan, and phenylalanine, respectively (Brown et al., 2003; Agerbirk and Olsen, 2012). The biosynthesis of GLS starts with chain elongation of the amino acids followed by oxidation, to form an aldoxime (Fig. 1). Glutathione donates a sulphur atom to the aldoxime, resulting in a thiohydroxymate, which quickly undergoes glycosylation by UDP-glucose to give desulpho-GLS. GLS are formed when the desulpho-GLS are sulphated by sulphotransferases using 3’-phosphoadenosine 3’-phosphosulphate (PAPS), linking GLS biosynthesis to primary sulphate assimilation (Mugford et al., 2009). The GLS can further undergo secondary modifications, such as hydroxylations, methylations, or oxidations, which give rise to a large variation of structures (Grubb and Abel, 2006; Sønderby et al., 2010).
Fig. 1.
The biosynthesis of glucosinolates linked with the sulphate assimilation pathway. APS, adenosine 5’-phosphosulphate; PAPS, 3’-phosphoadenosine 3’-phosphosulphate.
Glucosinolates contain at least two sulphur atoms in their core structure, and aliphatic GLS may have additional sulphur incorporated in their side chains (Fig. 1). The sulphur status of the plant therefore affects the biosynthesis of GLS (Falk et al., 2007), which is co-regulated with sulphate assimilation, both positively controlled by the same group of MYB transcription factors (Hirai et al., 2004, 2005; Yatusevich et al., 2010). The availability of other nutrients such as nitrogen and potassium can also affect production of GLS (Omirou et al., 2009; Troufflard et al., 2010).
GLS content and the expression of GLS biosynthetic genes are regulated by a range of environmental factors. In line with their function in biotic stress defence, the genes are inducible by wounding, jasmonate, or pathogens (Mikkelsen et al., 2003; Jost et al., 2005; Kusnierczyk et al., 2007). In addition, GLS and the mRNA levels of biosynthetic genes have been shown to change during a 24h light/dark cycle; the metabolic fluctuations are affected by temperature in cultivars of Brassica oleracea (Rosa et al., 1994; Rosa, 1997; Schuster et al., 2006). Branched-chain aminotransferase (BCAT4) and methylthioalkylmalate synthase (MAM1), both involved in the early side chain elongation process, show higher expression levels under light compared with darkness, and the expression levels remain high when the plants are kept under continuous light (Schuster et al., 2006). The three sulphotransferases involved in core GLS biosynthesis, SOT17 in particular, show higher expression levels during the light period followed by a gradual decrease during the dark period (Klein et al., 2006). AOP2, involved in secondary modification of the side chain, has a high expression level under continuous light, while it is not detectable in continuous darkness (Neal et al., 2010).
Despite the link that seems to exist between light and the biosynthesis of GLS, their total content often fluctuates more than the gene expression, and elevated levels can be seen during the dark period when the genes have low expression levels (Rosa, 1997; Klein et al., 2006; Schuster et al., 2006). In addition, the closely related sulphate assimilation is also regulated by light. The key enzyme of sulphate assimilation, adenosine 5’-phosphosulphate reductase (APR), undergoes diurnal rhythm in Arabidopsis and maize, with a maximum during the light period (Kocsy et al., 1997; Kopriva et al., 1999). On the other hand, no diurnal changes in cysteine or glutathione contents were observed in poplar (Noctor et al., 1997), although the capacity for glutathione synthesis is higher in light than in the dark (Buwalda et al., 1988). Other components of sulphate assimilation are regulated by light: ATP-sulphurylase activity was higher in irradiated oat, barley, and maize (Passera et al., 1989). In Arabidopsis, mRNA levels of genes of sulphate assimilation were several times higher in green leaves than in etiolated tissues (Hell et al., 1997). APR activity is repressed in dark-adapted plants, and induced rapidly by re-illumination (Neuenschwander et al., 1991; Kopriva et al., 1999). The transcription factor LONG HYPOCOTYL5 (HY5) has been shown to be involved in the regulation of APR light response (Lee et al., 2011). HY5 is a bZIP transcription factor and a positive regulator of photomorphogenesis (Ang et al., 1998). Mutation in HY5 causes defects in the inhibition of hypocotyl elongation in all light conditions, suggesting that HY5 acts downstream of phytochromes A and B, cryptochromes, and UV-B (Ang et al., 1998; Ulm et al., 2004). HY5 directly binds to the promoters of >1000 light-inducible genes, including APR (Lee et al., 2007, 2011).
However, while the light and diurnal regulation of sulphate assimilation is well established, the data have been obtained in different species in various growth conditions. In addition, particularly little is known about such regulation of GLS biosynthesis and the coordination with sulphate assimilation in Arabidopsis. The aims of this study were, therefore, to analyse light induction and diurnal regulation of GLS biosynthesis genes and to provide a comprehensive study of diurnal variation of sulphur-containing metabolites, sulphate uptake, flux through sulphate assimilation, and the rate of GLS biosynthesis, in the same plant material.
Materials and methods
Plant material and growth conditions
For all experiments, Arabidopsis thaliana ecotype Col-0 or the hy5 mutant [T-DNA insertion line SALK_056405 (Lee et al., 2011)] were grown on vertical plates with Murashige and Skoog (MS) medium, without sucrose supplement, and 0.5% phytagel. The seeds were stratified for 72h at 4 °C before being transferred to a controlled environment room at 20 °C and with a 16h light/8h dark cycle. For the diurnal experiments, samples of 15-day-old seedlings were collected every fourth hour, for 24h. For the light induction experiments, 21-day-old seedlings were either kept under a normal 16h/8h day–night rhythm or transferred to continuous darkness. After 44h, half of the dark-treated plants were re-illuminated while the remainder were kept in the dark. Sampling was done 3h (metabolites and RNA) or 4h (35S incorporation) after re-illumination. For the experiments with hy5, the seedlings were grown for 7 d, transferred to dark for 38h, and re-illuminated for 90min before sampling. The control group were kept in the dark. This allowed direct comparison of these results with the previous analysis of hy5 (Lee et al., 2011) and of regulation of the GLS synthesis genes at different developmental stages.
GLS analysis
The content of GLS and desulpho-GLS was determined using ~50mg of frozen plant material. The extraction and quantification of GLS followed the protocol described in Mugford et al. (2009). Quantification was based on UV absorption at 229nm and response factors relating to the internal standard. Identification was done by liquid chromatography–mass spectrometry (LC-MS), using atmospheric pressure chemical ionization and the +H+ molecular ion. The following GLS were identified: 3MSOP (3-methylsulphinylpropyl GLS), 4MSOB (4-methylsulphinylbutyl GLS), I3M (indol-3-ylmethyl GLS), 4OHI3M (4-hydroxy-indolyl-3-methyl GLS), 4MOI3M (4-methoxyindol-3-methyl GLS), and 1MOI3M (1-methoxyindol-3-ylmethyl GLS).
RNA extraction and expression analysis
RNA was isolated by phenol–chloroform–isoamyl mixture (25:24:1) extraction and LiCl precipitation. cDNA was synthesized from 1 µg of total RNA with a QuantiTect Reverse Transcription Kit (Qiagen, Crawley, UK), which includes a DNase step to remove possible DNA contamination. Quantitative real-time PCR was performed using gene-specific primers (Supplementary Table S1 available at JXB online) and the fluorescent intercalating dye SYBR Green (Sigma, Dorset, UK) as described by Lee et al. (2011). All quantifications were normalized to the TIP41 gene, and relative quantification was performed using the comparative Ct-method. The real-time PCRs were performed in duplicate for each of the three independent biological replicates.
Sulphate uptake
Every fourth hour plants were placed in 24-well plates with 1ml of MS nutrient solution {adjusted to contain 0.2mM sulphate and supplemented with 5.6 µCi of [35S]sulphate (Hartmann Analytic, Braunschweig, Germany)}. Plants were incubated in light or dark according to the light period previously described. When collecting samples in the dark, the plants were transferred using green light. After 4h incubation the plants were washed in sterile water, blotted in paper tissue, weighed, and placed in scintillation vials. A 4ml aliquot of tissue solubilizer (Soluene-350, Perkin Elmer) was added and the samples were left overnight to dissolve. After addition of 10ml of Optiphase HiSafe3 scintillation cocktail (Perkin Elmer), the radioactivity of the samples was measured in a scintillation counter (Beckman, High Wycombe, UK).
Determination of flux through sulphate assimilation and rate of GLS biosynthesis
The flux through primary sulphate assimilation, measured as incorporation of 35S from [35S]sulphate to thiols and proteins, and the GLS biosynthesis rate were determined essentially as described by Mugford et al. (2011). The plants were transferred into 24-well plates containing 1ml of MS nutrient solution adjusted to a sulphate concentration of 0.2mM and supplemented with 5.6 µCi of [35S]sulphate (Hartmann Analytic), and incubated for 4h in the corresponding light/dark regime. After the incubation, the seedlings were extensively washed in water, carefully blotted in paper tissue, and the shoots were cut, weighed, transferred, into 1.5ml tubes, and frozen in liquid nitrogen. The quantification of 35S in proteins and thiols followed extraction in 0.1M HCl, while for the [35S]GLS analysis the shoots were extracted in 70% methanol. The incorporation of 35S in desulpho-GLS was determined by scintillation counting of the flow-through and wash of the DEAE-Sephadex used to isolate GLS.
APR activity
APR activity was determined as the production of [35S]sulphite, assayed as acid volatile radioactivity formed in the presence of [35S]APS and dithioerythritol as reductant (Lee et al., 2011). The protein concentrations were determined with a Bio-Rad protein kit (Bio-Rad, Hemel Hempstead, UK) with bovine serum albumin as a standard.
Glutathione measurements
Total glutathione levels were determined by HPLC as described by Lee et al. (2011) from 20–30mg of plant material.
Statistical analysis
The results were analysed for variance by the Genstat software using ANOVA and a significance level of P=0.05. Where only two genotypes were compared, Student’s t-test was used.
Results
Diurnal variation in sulphur metabolism and GLS biosynthesis
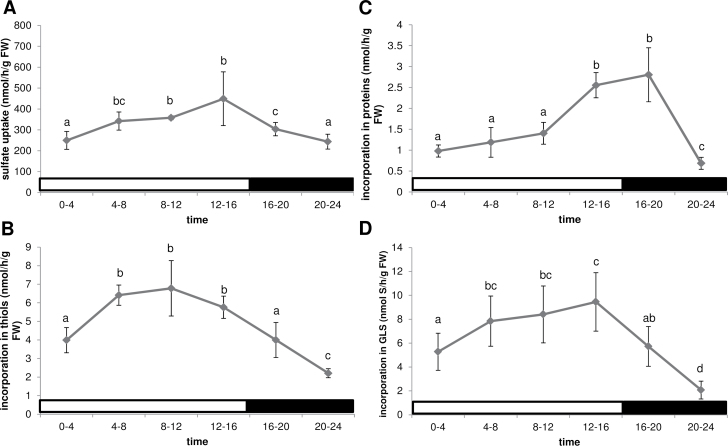
To obtain a more detailed understanding of the diurnal variation of sulphur metabolism, [35S]sulphate was used to determine sulphate uptake and fluxes in both primary and secondary sulphate assimilation. Sulphate uptake was measured in six 4h intervals over a 24h period, starting at light onset. The uptake increased from the start of the day to a higher level during the rest of the light period, followed by a decrease, until reaching the same level as at the beginning of the light period (Fig. 2A). The flux through primary assimilation was determined as incorporation of radioactivity into thiols (cysteine and glutathione) and proteins, while GLS represented the secondary metabolites (Mugford et al., 2011). The incorporation of 35S into thiols was higher in the light than in the dark, with maximal values between 8h and 12h (Fig. 2B). The incorporation of 35S into proteins, however, had a different pattern, showing an increase only late in the day and in the first part of the dark period (Fig. 2C). There was a clear minimum in incorporation of sulphate into thiols and proteins in the second part of the night. The GLS biosynthesis rate showed a very similar pattern, with a minimum at the end of the night (Fig. 2D). Thus, sulphur metabolism seems to be well coordinated throughout the day, with higher activity during the light period than during the night.
Fig. 2.
Diurnal variation in sulphate uptake and fluxes through primary and secondary sulphate assimilation in Arabidopsis (Col-0) grown on MS-agarose plates for 2 weeks in long days (16h light/8h dark). Sulphate uptake (A) and incorporation of 35S into thiols (B), proteins (C), and GLS (D) was determined. The light regime is indicated by white or black bars. Data are presented as means ±SD. Different letters mark significantly different values from four biological replicates (P < 0.05).
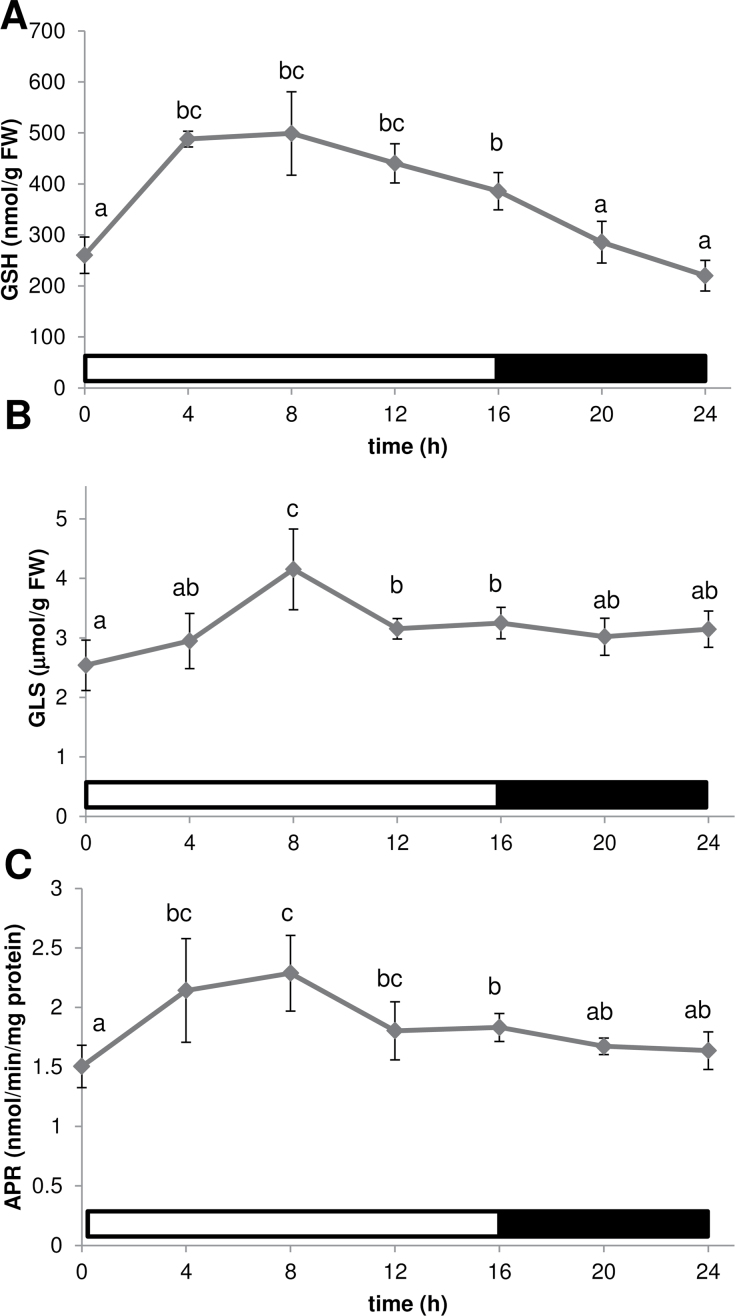
The levels of two major sulphur-containing metabolites, glutathione and GLS, were compared as representatives of primary and secondary sulphate assimilation pathways. Glutathione levels were higher during the light period than in the dark, without a clear maximum (Fig. 3A). GLS levels were relatively stable except for a peak 8h after light onset (Fig. 3B; Supplementary Table S2 at JXB online). APR, which controls flux through the sulphate assimilation pathway (Vauclare et al., 2002), had previously only been shown to undergo diurnal rhythm in plants adapted to short days (Kopriva et al., 2009); therefore, it was necessary to confirm the same regulation in plants that were grown in long days. APR activity was again higher during the light period than in the dark, but without the strong maximum observed under short days (Fig. 3C).
Fig. 3.
Diurnal variation in contents of glutathione (GSH) (A), total GLS (B), and APS reductase activity (C). The light regime is indicated by white or black bars. Data are presented as means ±SD. Different letters mark significantly different values from four biological replicates (P < 0.05).
Regulation of GLS biosynthesis by light
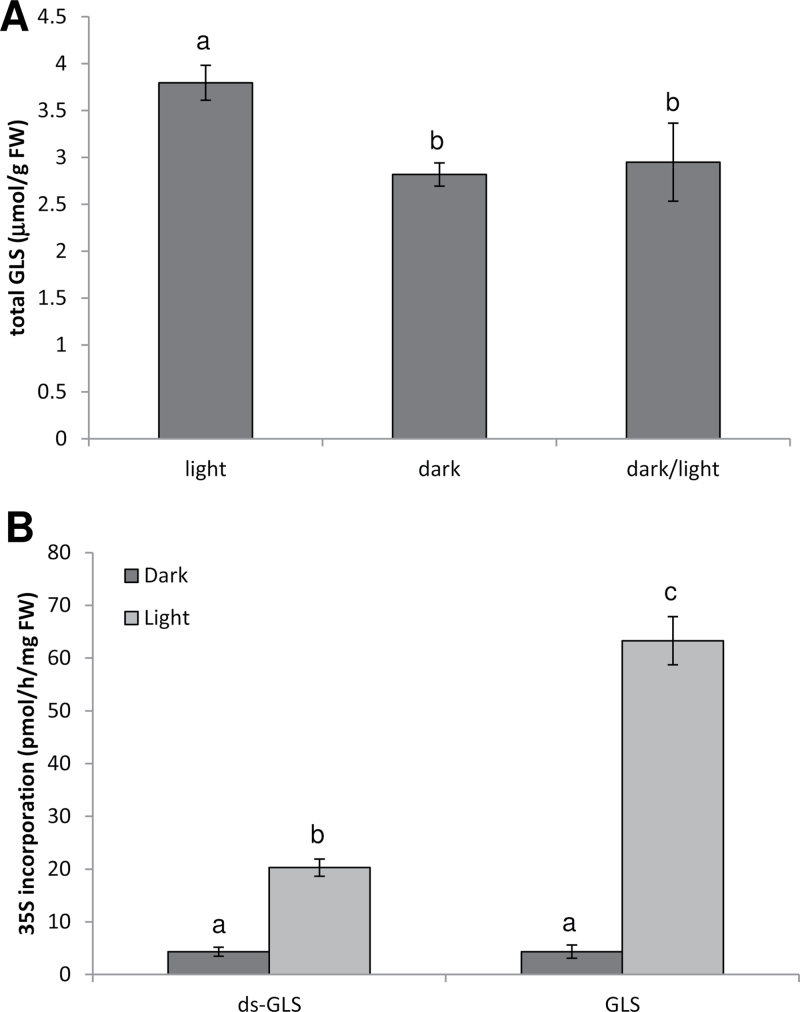
The direct effect of dark–light transitions on GLS biosynthesis was analysed in more detail through a light induction experiment testing both the biosynthetic rate and gene expression. Plants kept under the normal light/dark rhythm contained the highest GLS levels (Fig. 4A; Supplementary Table S3 at JXB online). Forty-four hours of darkness resulted in an ~25% decrease in GLS levels. Re-illumination for 3h was not sufficient to increase the GLS content significantly, and this stayed significantly lower than in control plants (Fig. 4A). However, 44h of darkness affect many processes in plants, and the lack of difference in total GLS content between the dark-treated and re-illuminated plants does not preclude that light directly regulates GLS biosynthesis.
Fig. 4.
GLS synthesis in Arabidopsis seedlings under different light regimes. Three-week-old Arabidopsis seedlings on MS-agarose plates were transferred to darkness for 44h. One part of these seedlings was re-illuminated by white light for 3h (dark/light), while the other part remained in darkness (dark). (A) Total GLS levels in these plants were compared with those of plants continuing to grow in the normal day/night cycle (light). (B) 35S incorporation in desulpho-GLS and GLS during 4h was determined. Data are presented as means ±SD from four pools of three seedlings. Different letters mark significantly different values from four biological replicates (P < 0.05).
To measure the effect of light on the GLS biosynthesis rate, the incorporation rate of 35S into GLS and desulpho-GLS was determined for the different light regimes. While there was some basal incorporation of 35S in dark-incubated plants, the rate of biosynthesis was greatly increased by light (Fig. 4B). Labelling was found in both GLS and desulpho-GLS, with the former being labelled to a higher degree, reflecting the nature of desulpho-GLS as pathway intermediates. Thus, biosynthesis of GLS is indeed a light-regulated process.
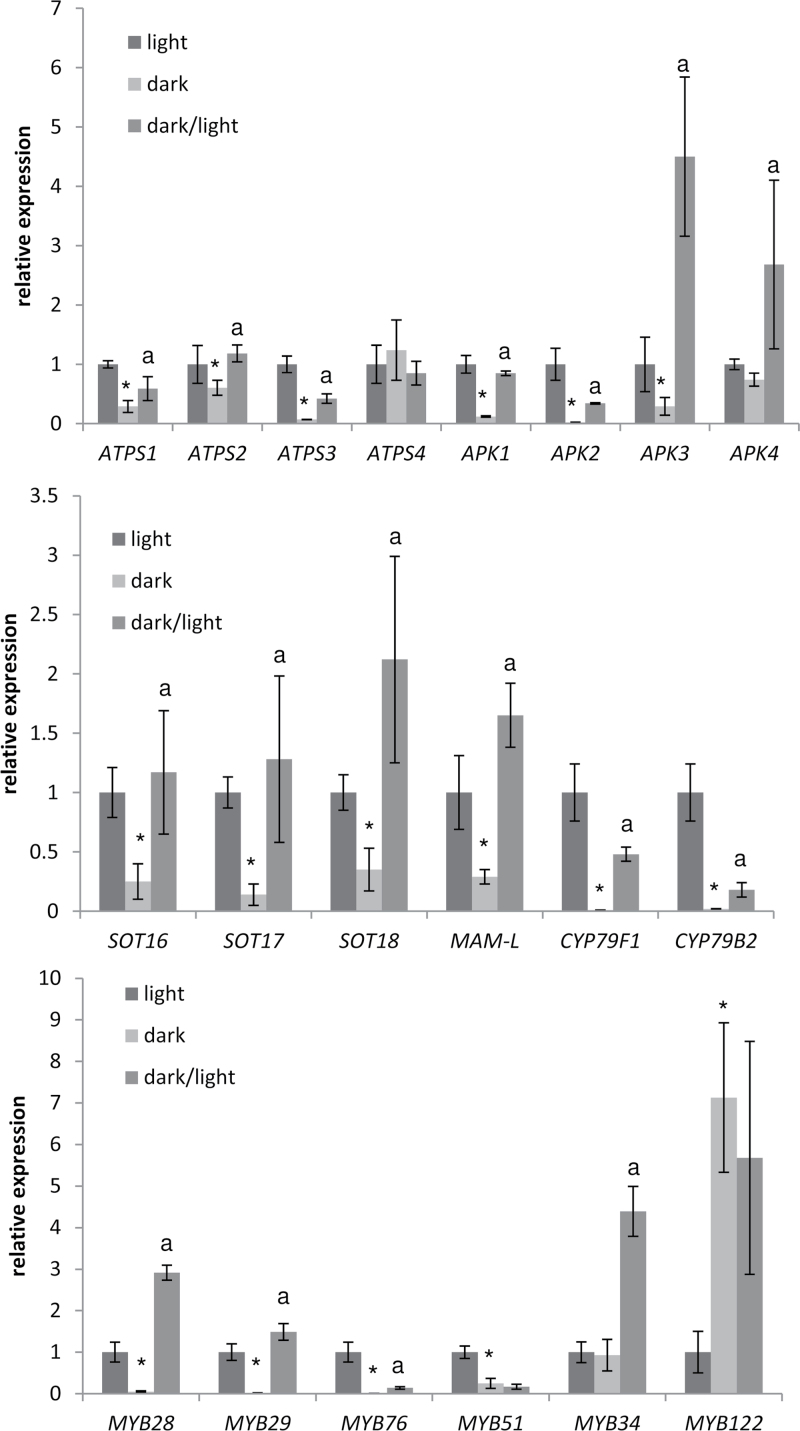
Among the sulphate assimilation genes, ATPS1–ATPS3 isoforms of ATP sulphurylase and APK1–APK3 genes encoding APS kinase were down-regulated after 44h in the dark, while ATPS4 and APK4 were not affected (Fig. 5). The three ATPS genes were induced by light, but only ATPS2 reached the levels in control plants after 3h re-illumination. All APK isoforms were up-regulated by light; in particular, APK1 and APK4 transcript levels were increased very strongly, ~15-fold compared with the levels in dark-adapted plants. The increase led to mRNA levels for APK3 and APK4 being higher than in control plants, but not for APK2 which was most strongly repressed (Fig. 5). The six genes directly associated with the GLS biosynthesis pathway (SOT16, SOT17, SOT18, MAM-L, CYP79F1, and CYP79B2) all followed the same pattern of down-regulation in the dark and up-regulation after light induction, with both CYP genes being repressed to the highest degree. The same expression pattern can be seen in regulation of mRNAs for transcription factors MYB28, MYB29, and MYB76, which have been shown to control biosynthesis of aliphatic GLS (Hirai et al., 2007). For the transcription factors associated with indolic GLS (MYB51, MYB34, and MYB122), the results are more varied. MYB51 was down-regulated in the dark, but showed no response to light induction. MYB34 was regulated in an opposite manner, namely it did not respond to dark treatment but was still induced by light. MYB122, on the other hand, was the only gene that showed a strong up-regulation under dark conditions and no response after light induction (Fig. 5). Thus, the genes of GLS metabolism are highly regulated by light but not in a completely coordinated manner.
Fig. 5.
Light regulation of genes of sulphate assimilation and GLS biosynthesis. Three-week-old Arabidopsis seedlings on MS-agarose plates were transferred to darkness for 44h. One part of these seedlings was re-illuminated by white light for 3h (dark/light), while the other part remained in darkness (dark). These plants were compared with plants continuing to grow in the normal day/night cycle (light). Transcript levels of genes involved in sulphur metabolism and GLS biosynthesis were determined by quantitative real-time PCR. The values for control plants (light) were set to 1. Data are presented as means ±SD from three pools of three seedlings. Asterisks mark significant differences between dark and light transcript levels, while the letter ‘a’ marks values significantly different between dark/light and dark samples (P < 0.05).
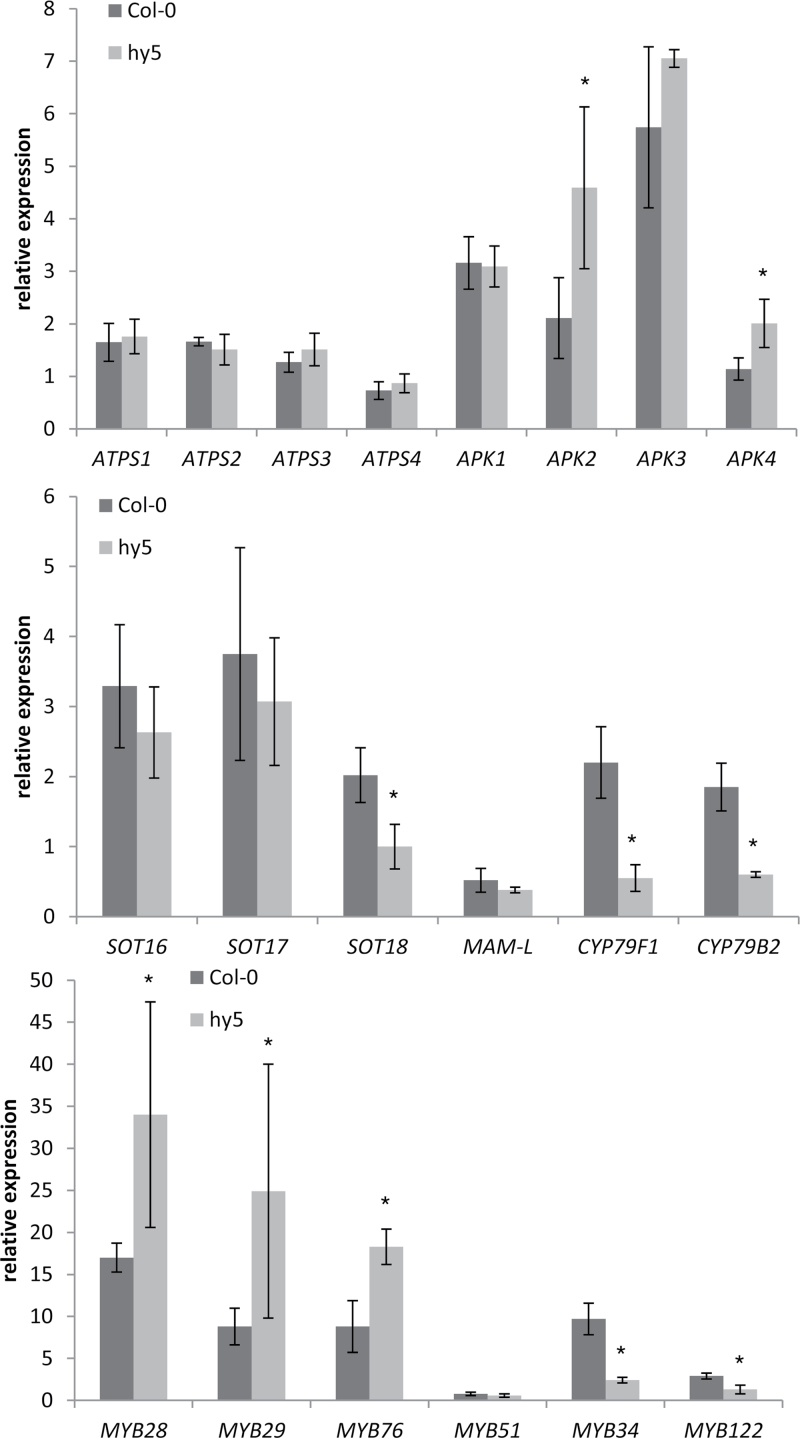
Having established that the GLS biosynthetic genes are light regulated, tests were conducted to determine whether they may also be under HY5 control (Fig. 6). Although 1-week-old plants were analysed, compared with 3-week-old plants in a previous experiment, in Col-0 most genes showed the same response to re-illumination. The major difference seems to be the regulation of MAM-L, which was repressed by 90min re-illumination of 1-week-old plants but induced in 3-week-old plants. The regulation of several genes of GLS biosynthesis was strongly affected in the hy5 mutant. The most pronounced difference was the opposite effect of light on the two genes involved in core GLS biosynthesis (CYP79F1 and CYP79B2), which were induced in Col-0 but repressed in hy5 (Fig. 6). Also SOT18 was up-regulated to a much lower degree in hy5 compared with Col-0, whereas the other SOT genes and MAM-L were regulated in the same way in the two genotypes. The regulation of all ATPS isoforms and APK1 and APK3 was not affected by HY5 disruption. However, strikingly, the MYB factors that are associated with aliphatic GLS biosynthesis were more strongly up-regulated in hy5 than in Col-0. The same was true for APK2 and APK4, indicating that HY5 may act as a transcription repressor as well as an activator. MYB51 was not regulated by light in either genotype, whereas both MYB34 and MYB122 were induced to a lower degree in hy5. Thus, most genes of the GLS biosynthetic network, but not genes of sulphate assimilation involved in the biosynthesis of PAPS, are light regulated in a HY5-dependent manner.
Fig. 6.
HY5 controls light regulation of genes of sulphate assimilation and GLS biosynthesis. One-week-old Arabidopsis Col-0 and hy5 seedlings on MS-agarose plates were transferred to darkness for 38h. One part of these seedlings was re-illuminated by white light for 90min, while the other part remained in darkness. Transcript levels of genes involved in sulphur metabolism and GLS biosynthesis were determined by quantitative real-time PCR. The values in dark-grown plants were set to 1. Data are presented as means ±SD of the relative expression in re-illuminated plants compared with dark-grown plants determined in three pools of three seedlings. Asterisks mark values significantly different between the two genotypes (P < 0.05).
Discussion
Diurnal variation of sulphur metabolism and GLS biosynthesis in Arabidopsis
Plant physiology and the life cycle are under strong control of diurnal and circadian rhythms. For example, more than a third of Arabidopsis genes are under circadian control (Pruneda-Paz and Kay, 2010). Key genes of the major pathways of primary metabolism, carbon, nitrate, and sulphate assimilation show clear diurnal and/or circadian rhythmicity (Pilgrim et al., 1993; Kopriva et al., 1999; Zeeman et al., 2007). Much less is known about circadian regulation of secondary metabolism, even though a large number of cytochrome P-450 genes were found to be controlled by the clock (Pan et al., 2009). There is, however, evidence for diurnal variation in levels of GLS in field-grown Brassica oleracea plants (Rosa et al., 1994) which was confirmed under controlled conditions (Rosa, 1997). In the results presented here, a variation in GLS content in Arabidopsis grown under control conditions has been detected, with greater accumulation in the day than in the night. This is surprisingly in contrast to the results with B. oleracea, which showed a significant decrease of GLS content during the day both in field and in controlled conditions (Rosa et al., 1994; Rosa, 1997). On the other hand, the GLS levels agree well with the variation in sulphate uptake, APR activity, sulphate reduction rate, and, most importantly, the GLS biosynthesis rate measured in the same plants. The difference compared with B. oleracea may thus be due to the different developmental stages of plants analysed or due to species-specific variation, possibly linked to the different nature of the major herbivores associated with these species. Diurnal rhythms for a large range of secondary compounds involved in plant–herbivore interactions have been described, some showing higher foliar accumulation in the light, but others, importantly for nocturnal insects, more abundant in the night (De Moraes et al., 2001; Kim et al., 2011).
The various components of sulphur metabolism were to a large extent coordinated throughout the 24h day cycle. Only subtle differences were found between, for example, accumulation of glutathione or GLS and their rates of synthesis (Figs 2, 3). This is confirmed by a high level of correlation between individual components, particularly APR activity and incorporation into thiols, with glutathione and GLS levels (Supplementary Table S4 at JXB online). Only the incorporation in proteins shows a very different pattern and only a weak correlation with sulphate uptake. Interestingly, for both GLS and glutathione, the highest metabolite accumulation seems to be achieved before the period of the highest synthesis rate. This may seem contra-intuitive; however, metabolite levels are not dependent solely on the biosynthesis rate but also on the breakdown of the compound. This has been demonstrated, for example, in the apr2 mutant lacking the major isoform of APR, which shows lower flux through sulphate assimilation without affecting glutathione levels or in apk1 apk2 mutants which showed an increased rate of incorporation into GLS despite much lower GLS levels (Mugford et al., 2011). Interestingly, reduced sulphur seems to be utilized first for synthesis of glutathione and GLS and only in later stages incorporated into proteins (Fig. 2). This corresponds to the results of Koprivova et al. (2000) who showed that after resupply of nitrogen to N-starved plants the glutathione pool was filled before proteins. The diurnal variation of APR (Fig. 3C) agrees with a previous report (Kopriva et al., 1999); however, the amplitude of the cycle was much less in the present study. This is probably caused by the difference in light regime between the studies, short days (10h light) in Kopriva et al. (1999) versus long days (16h light) reported here. Thus, although the transcript level of APR2 is under circadian control (Harmer et al., 2000), the control of APR variation also has to include a component dependent on daylength. The daylength also seem to affect the variation in fluxes, as the significant drop at the end of the night (Fig. 2) was not detected in plants grown in short days (Kopriva et al., 1999).
Sulphate uptake showed a diurnal variation, with the highest rate measured in the second half of the day (Fig. 2A). The maximum rate of sulphate uptake seems to follow after the maximal flux through the pathway. The trigger for such regulation thus could be depletion of sulphate due to increased reduction, as the maximum APR activity was measured in the first half of the day. Indeed, low internal sulphate levels in mutants of the sulphate transporter SULTR1;2 and a FIERY1 gene trigger sulphate deficiency responses, which include induction of sulphate transporters (Lee et al., 2012; Matthewman et al., 2012). The gradual increase in sulphate uptake rate during the light period is very similar to the diurnal regulation of nitrate uptake (Lejay et al., 1999). The variation in nitrate uptake can be explained by changes in expression of the nitrate transporters Nrt1 and Nrt2;1 (Lejay et al., 1999). Also the transcripts of the PHT4 group of phosphate transporters undergo circadian regulation, with a maximum during the day (Guo et al., 2008). Whether the same is true for sulphate transporters still has to be established.
GLS biosynthesis is controlled by light
Diurnal regulation is often connected with light regulation. In sulphur metabolism, APR activity and mRNA levels were shown to decrease strongly when plants were kept in continuous darkness and increased rapidly after re-illumination (Kopriva et al., 1999). The present results show that other components of the pathway are also light regulated. With the exception of ATPS4 and APK4, genes encoding the two enzymes necessary for PAPS biosynthesis (ATP sulphurylase and APS kinase) were down-regulated upon incubation of plants in prolonged darkness. ATPS4 is the major isoform of ATPS in the roots, while APK4 is a minor plastidic form of APS kinase, so that the lack of light regulation is not entirely surprising (Kopriva et al., 2009). The ATPS and APK genes, including APK4 but not ATPS4, were induced upon re-illumination of dark-adapted plants. This regulation is consistent with regulation of APR and shows a well-coordinated response of the whole pathway to changes in light regime.
Similarly coordinated was the regulation of GLS biosynthetic genes (Fig. 5). It has been shown previously that the expression of genes of GLS biosynthesis is well coordinated, for example repressed by sulphur deficiency (Hirai et al., 2005) or induced in apk1 apk2 mutants (Mugford et al., 2009). Similar coordination of the transcript accumulation is seen in plants with modulated expression of the two groups of MYB factors (Gigolashvili et al., 2007a , b , 2008; Sønderby et al., 2007; Malitsky et al., 2008). The MYB factors themselves are regulated in the same way (Hirai et al., 2005; Mugford et al., 2009), so it appears to be the changes in the expression of MYB factors driving the regulation of the downstream transcript abundance. This seems to be true for the light regulation of the aliphatic GLS subset of the network, as the biosynthetic genes were regulated in the same way as the genes for MYB28, MYB29, and MYB76 factors. The genes involved in biosynthesis of indolic GLS, CYP79B2 and SOT16, were co-regulated with the other biosynthetic genes; however, the MYB factors responded to light in a different manner. Nevertheless, mRNA for the main indolic MYB factor, MYB51, was reduced in dark-grown plants, resulting in repression of CYP79B2 and SOT16. This decrease was probably not compensated by the presence of MYB34 and MYB122, which were not affected or were even induced by darkness, respectively, which agrees with the finding that disruption of MYB51 severely reduces transcription of genes for biosynthesis of the indolic GLS (Gigolashvili et al., 2007a). On the other hand, MYB34 was induced by re-illumination and shown previously to induce expression of genes of biosynthesis of indolic GLS (Celanza et al., 2005). It seems, therefore, that unlike the aliphatic group of MYBs, the MYB factors controlling indolic GLS biosynthesis have a specific function in light regulation of their target genes.
The effect of light on GLS biosynthesis, however, was not confined to regulation of gene expression. In agreement with the low transcript levels of GLS biosynthetic genes, the total GLS contents were reduced in plants kept in darkness. The decrease in GLS was not as strong as the reduction in mRNA levels, probably since the turnover of GLS is slower. In line with the induction of gene expression, re-illumination induced GLS biosynthesis that was very low in the dark-incubated plants. The down-regulation of GLS biosynthesis in prolonged darkness is consistent with a decrease in the synthesis rate in the night, particularly in the second half (Fig. 2D). The light induction also seems to be similar to that seen during the day; however, it has to be noted that in the first 4h of re-illumination, the GLS biosynthesis rate was still almost 100-fold lower than during the day (cf. Figs 2D and 4). This is similar to the light regulation of the primary sulphate assimilation. The sulphate reduction rate also decreases in prolonged darkness and is induced by light (Lee et al., 2011). The flux during the first 4h of re-illumination (Lee et al., 2011) was ~8-fold lower than the flux during the day determined in this study. This is partly caused by averaging the flux over the 4h experimental period, with initial rates expected to be particularly low because the necessary enzymes need to be synthesized. The lower rate of GLS biosynthesis compared with sulphate reduction presumably reflects the plants’ needs for cysteine for protein biosynthesis and thus represents further evidence that primary and secondary sulphur metabolism are highly coordinated.
HY5 contributes to regulation of GLS synthesis by light
The induction of APR mRNA by light is at least partly dependent on HY5. HY5 also controls the regulation of SULTR1;2, the major sulphate transporter in the roots. Thus, it was intriguing to test whether the pathway of sulphate assimilation is regulated by HY5 coordinately, particularly as the regulation of ATPS1–ATPS3 (Fig. 5) was very similar to light regulation of APR (Lee et al., 2011). This, however, was not the case; ATPS was regulated by light in the same way in Col-0 and the hy5 mutant (Fig. 6). Different mechanisms of regulation between ATPS and APR, despite catalysing the subsequent steps in the pathway, are not surprising. For example, APR is highly induced by sulphate deficiency but ATPS is repressed (Takahashi et al., 1997; Kawashima et al., 2011). Also the mechanisms of this regulation are different. APR is transcriptionally regulated, whereas ATPS is a target of a sulphur limitation-inducible microRNA, miR395, and is thus down-regulated post-transcriptionally (Kawashima et al., 2011). Given the different mechanisms of regulation of APR and ATPS by sulphate deficiency, it is not surprising that the mechanism of their light regulation differs.
On the other hand, many genes of the GLS biosynthesis network are regulated in a HY5-dependent manner and remarkably several of the MYB factors, too (Fig. 6). Again, the aliphatic and indolic MYB groups were regulated differently; whereas all three aliphatic MYB factors were more strongly induced by light in hy5, the light induction of MYB34 and MYB122 was almost abolished in the mutant. Thus, HY5 acts as a repressor of the aliphatic MYB factors and an activator for the two indolic ones. This is consistent with the influence of HY5 on many regulatory networks not through direct binding but through controlling other transcriptional regulators (Zhang et al., 2011). It seems that GLS biosynthesis is another example of such networks, even though the mechanism of HY5 action is less than clear. While the MYB28, MYB29, and MYB76 genes are induced by light to a higher degree in the hy5 mutant than in Col-0, the target genes SOT18 and CYP79F1 are less strongly up-regulated. The hierarchy of HY5 and the MYB factors in regulation of GLS biosynthesis by light thus still needs to be established.
Altogether, these results show that GLS biosynthesis is regulated by light and shows diurnal variation that is well coordinated with general sulphur metabolism. The transcription factor HY5 seems to be involved in regulating the GLS biosynthetic network. These new data complement and expand the available knowledge on the coordination of primary and secondary sulphur metabolism in Arabidopsis.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primer sequences used for quantitative real-time PCR.
Table S2. Diurnal variations of individual GLS.
Table S3. Light regulation of individual GLS.
Table S4. Correlation analysis of diurnal variations in levels of sulphur-containing metabolites, fluxes, and APR activity.
Acknowledgements
The work of SHs is supported by the Research Council of Norway (project no. 185017/O10). The work of SK is supported by British Biotechnology and Biological Sciences Research Council (BBSRC).
References
- Agerbirk N, Olsen CE. 2012. Glucosinolate structures in evolution. Phytochemistry 77, 16–45 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. 1998. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Molecular Cell 1, 213–222 [DOI] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J. 2003. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana . Phytochemistry 62, 471–481 [DOI] [PubMed] [Google Scholar]
- Buwalda F, De Kok LJ, Stulen I, Kuiper PJC. 1988. Cysteine, γ-glutamylcysteine and glutathione contents of spinach leaves as affected by darkness and application of excess sulfur. Physiologia Plantarum 74, 663–668 [Google Scholar]
- Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J. 2005. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiology 137, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moraes CM, Mescher MC, Tumlinson JH. 2001. Caterpillar-induced nocturnal plant volatiles repel conspecific females. Nature 410, 577–80 [DOI] [PubMed] [Google Scholar]
- Falk KL, Tokuhisa JG, Gershenzon J. 2007. The effect of sulfur nutrition on plant glucosinolate content: physiology and molecular mechanisms. Plant Biology 9, 573–581 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Muller C, Weisshaar B, Fluegge UI. 2007. a .The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana . The Plant Journal 50, 886–901 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Engqvist M, Yatusevich R, Muller C, Flugge UI. 2008. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana . New Phytologist 177, 627–642 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Yatusevich R, Berger B, Muller C, Flugge UI. 2007. b The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. The Plant Journal 51, 247–261 [DOI] [PubMed] [Google Scholar]
- Grubb CD, Abel S. 2006. Glucosinolate metabolism and its control. Trends in Plant Science 11, 89–100 [DOI] [PubMed] [Google Scholar]
- Guo B, Irigoyen S, Fowler TB, Versaw WK. 2008. Differential expression and phylogenetic analysis suggest specialization of plastid-localized members of the PHT4 phosphate transporter family for photosynthetic and heterotrophic tissues. Plant Signaling and Behaviour 3, 784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch LB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. 2000. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hell R, Schwenn JD, Bork C. 1997. Light and sulfur sources modulate mRNA levels of several genes of sulfate assimilation. In: Cram J, ed. Sulfur metabolism in higher plants. Leiden, The Netherlands: Backhuys Publishers, 181–185 [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, et al. 2005. Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. Journal of Biological Chemistry 280, 25590–25595 [DOI] [PubMed] [Google Scholar]
- Hirai M, Sugiyama K, Sawada Y, et al. 2007. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proceedings of the National Academy of Sciences, USA 48, S67–S67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. 2004. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 101, 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost R, Altschmied L, Bloem E, et al. 2005. Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynthesis Research 86, 491–508 [DOI] [PubMed] [Google Scholar]
- Kawashima CG, Matthewman CA, Huang SQ, et al. 2011. Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis . The Plant Journal 66m, 863–876 [DOI] [PubMed] [Google Scholar]
- Kim SG, Yon F, Gaquerel E, Gulati J, Baldwin IT. 2011. Tissue specific diurnal rhythms of metabolites and their regulation during herbivore attack in a native tobacco, Nicotiana attenuata. PLoS One 6, e26214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Reichelt M, Gershenzo J, Papenbrock J. 2006. The three desulfoglucosinolate sulfotransferase proteins in Arabidopsis have different substrate specificities and are differentially expressed. FEBS Journal 273, 122–136 [DOI] [PubMed] [Google Scholar]
- Kocsy G, Owttrim G, Brander K, Brunold C. 1997. Effect of chilling on the diurnal rhythm of enzymes involved in protection against oxidative stress in a chilling-tolerant and a chilling-sensitive maize genotype. Physiologia Plantarum 99, 249–254 [Google Scholar]
- Kopriva S, Mugford SG, Matthewman C, Koprivova A. 2009. Plant sulfate assimilation genes: redundancy versus specialization. Plant Cell Reports 28, 1769–1780 [DOI] [PubMed] [Google Scholar]
- Kopriva S, Muheim R, Koprivova A, Trachsel N, Catalano C, Suter M, Brunold C. 1999. Light regulation of assimilatory sulphate reduction in Arabidopsis thaliana. The Plant Journal 20, 37–44 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Suter M, den Camp RO, Brunold C, Kopriva S. 2000. Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiology 122, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnierczyk A, Winge P, Midelfart H, Armbruster WS, Rossiter JT, Bones AM. 2007. Transcriptional responses of Arabidopsis thaliana ecotypes with different glucosinolate profiles after attack by polyphagous Myzus persicale and oligophagous Brevicoryne brassicae. Journal of Experimental Botany 58, 2537–2552 [DOI] [PubMed] [Google Scholar]
- Lee BR, Huseby S, Koprivova A, et al. 2012. Effects of fou8/fry1 mutation on sulfur metabolism: is decreased internal sulfate the trigger of sulfate starvation response?. PLoS One 7, e39425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BR, Koprivova A, Kopriva S. 2011. The key enzyme of sulfate assimilation, adenosine 5’-phosphosulfate reductase, is regulated by HY5 in Arabidopsis. The Plant Journal 67, 1042–1054 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao HY, Lee I, Deng X. 2007. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. The Plant Cell 19, 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive FD, Filleur S, Daniel-Vedele F, Gojon A. 1999. Molecular and functional regulation of two NO3– uptake systems by N- and C-status of Arabidopsis plants. The Plant Journal 18, 509–519 [DOI] [PubMed] [Google Scholar]
- Malitsky S, Blum E, Less H, Venger I, Elbaz M, Morin S, Eshed Y, Aharoni A. 2008. The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiology 148, 2021–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthewman CA, Kawashima CG, Húska D, Csorba T, Dalmay T, Kopriva S. 2012. miR395 is a general component of sulfate assimilation regulatory network in Arabidopsis. FEBS Letters 586, 3242–3248 [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA. 2003. Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiology 131, 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford SG, Lee BR, Koprivova A, Matthewman C, Kopriva S. 2011. Control of sulfur partitioning between primary and secondary metabolism. The Plant Journal 65, 96–105 [DOI] [PubMed] [Google Scholar]
- Mugford SG, Yoshimoto N, Reichelt M, et al. 2009. Disruption of adenosine-5’-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. The Plant Cell 21, 910–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal CS, Fredericks DP, Griffiths CA, Neale AD. 2010. The characterisation of AOP2: a gene associated with the biosynthesis of aliphatic alkenyl glucosinolates in Arabidopsis thaliana. BMC Plant Biology 10, 170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenschwander U, Suter M, Brunold C. 1991. Regulation of sulfate assimilation by light and O-acetyl-l-serine in Lemna minor L. Plant Physiology 97, 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Arisi ACM, Jouanin L, Valadier MH, Roux Y, Foyer CH. 1997. Light-dependent modulation of foliar glutathione synthesis and associated amino acid metabolism in poplar overexpressing γ-glutamylcysteine synthetase. Planta 202, 357–369 [Google Scholar]
- Omirou MD, Papadopoulou KK, Papastylianou I, Constantinou M, Karpouzas DG, Asimakopoulos I, Ehaliotis C. 2009. Impact of nitrogen and sulfur fertilization on the composition of glucosinolates in relation to sulfur assimilation in different plant organs of broccoli. Journal of Agricultural and Food Chemistry 57, 9408–9417 [DOI] [PubMed] [Google Scholar]
- Pan YH, Michael TP, Hudson ME, Kay SA, Chory J, Schuler MA. 2009. Cytochrome P450 monooxygenases as reporters for circadian-regulated pathways. Plant Physiology 150, 858–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passera C, Ghisi R, Ferretti M. 1989. Light-activation of ATP-sulfurylase in leaves and chloroplasts of Zea mays. Photosynthetica 23, 166–172 [Google Scholar]
- Pilgrim ML, Caspar T, Quail PH, McClung CR. 1993. Circadian and light-regulated expression of nitrate reductase in Arabidopsis. Plant Molecular Biology 23, 349–364 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Kay SA. 2010. An expanding universe of circadian networks in higher plants. Trends in Plant Science 15, 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa EAS. 1997. Daily variation in glucosinolate concentrations in the leaves and roots of cabbage seedlings in two constant temperature regimes. Journal of the Science of Food and Agriculture 73, 364–368 [Google Scholar]
- Rosa EAS, Heaney RK, Rego FC, Fenwick GR. 1994. The variation of glucosinolate concentration during a single day in young plants of Brassica oleracea var. Acephala and Capitata. Journal of the Science of Food and Agriculture 66, 457–463 [Google Scholar]
- Schuster J, Knill T, Reichelt M, Gershenzon J, Binder S. 2006. BRANCHED-CHAIN AMINOTRANSFERASE4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. The Plant Cell 18, 2664–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Geu-Flores F, Halkier BA. 2010. Biosynthesis of glucosinolates—gene discovery and beyond. Trends in Plant Science 15, 283–290 [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ. 2007. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS One 2, e1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, Engler JD, Engler G, VanMontagu M, Saito K. 1997. Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 94, 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M, Mithen R. 2009. Glucosinolates, isothiocyanates and human health. Phytochemistry Reviews 8, 269–282 [Google Scholar]
- Troufflard S, Mullen W, Larson TR, Graham IA, Crozier A, Amtmann A, Armengaud P. 2010. Potassium deficiency induces the biosynthesis of oxylipins and glucosinolates in Arabidopsis thaliana. BMC Plant Biology 10, 172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F. 2004. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, von Ballmoos P, Krahenbuhl U, den Camp RO, Brunold C. 2002. Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5’-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. The Plant Journal 31, 729–740 [DOI] [PubMed] [Google Scholar]
- Yatusevich R, Mugford SG, Matthewman C, Gigolashvili T, Frerigmann H, Delaney S, Koprivova A, Flugge UI, Kopriva S. 2010. Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. The Plant Journal 62, 1–11 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. 2007. The diurnal metabolism of leaf starch. Biochemical Journal 401, 13–28 [DOI] [PubMed] [Google Scholar]
- Zhang HY, He H, Wang XC, Wang XF, Yang XZ, Li L, Deng XW. 2011. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. The Plant Journal 65, 346–358 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.