Abstract
The genus Cuscuta (Convolvulaceae, the morning glory family) is one of the most intensely studied lineages of parasitic plants. Whole plastome sequencing of four Cuscuta species has demonstrated changes to both plastid gene content and structure. The presence of photosynthetic genes under purifying selection indicates that Cuscuta is cryptically photosynthetic. However, the tempo and mode of plastid genome evolution across the diversity of this group (~200 species) remain largely unknown. A comparative investigation of plastid genome content, grounded within a phylogenetic framework, was conducted using a slot-blot Southern hybridization approach. Cuscuta was extensively sampled (~56% of species), including groups previously suggested to possess more altered plastomes compared with other members of this genus. A total of 56 probes derived from all categories of protein-coding genes, typically found within the plastomes of flowering plants, were used. The results indicate that two clades within subgenus Grammica (clades ‘O’ and ‘K’) exhibit substantially more plastid gene loss relative to other members of Cuscuta. All surveyed members of the ‘O’ clade show extensive losses of plastid genes from every category of genes typically found in the plastome, including otherwise highly conserved small and large ribosomal subunits. The extent of plastid gene losses within this clade is similar in magnitude to that observed previously in some non-asterid holoparasites, in which the very presence of a plastome has been questioned. The ‘K’ clade also exhibits considerable loss of plastid genes. Unlike in the ‘O’ clade, in which all species seem to be affected, the losses in clade ‘K’ progress phylogenetically, following a pattern consistent with the Evolutionary Transition Series hypothesis. This clade presents an ideal opportunity to study the reduction of the plastome of parasites ‘in action’. The widespread plastid gene loss in these two clades is hypothesized to be a consequence of the complete loss of photosynthesis. Additionally, taxa that would be the best candidates for entire plastome sequencing are identified in order to investigate further the loss of photosynthesis and reduction of the plastome within Cuscuta.
Key words: Dodders, heterotroph, plastid genome, slot-blot hybridization.
Introduction
Heterotrophic plants show a wide range of degradation of photosynthetic capability. These plants are generally categorized either as parasites or as mycoheterotrophs (MHTs). Parasitic plants attach directly to their hosts through a haustorial connection (specialized organ allowing transfer of nutrients and water from host to parasite; Kujit, 1969), whereas MHTs acquire nutrients via a mycorrhizal intermediate (Leake, 1994). Obligate heterotrophy often coincides with the loss or impairment of the photosynthetic apparatus, and obligate heterotrophs rely on their autotrophic hosts for nutrition and water (Krause, 2008). The nutritional shift to obligate heterotrophy, or holoparasitism, is frequently accompanied by the loss or near loss of chlorophyll and reduced stem, root, and leaf morphology, resulting in a condition referred to as the ‘parasitic reduction syndrome’ (Colwell, 1994). Overall, the evolution of heterotrophy has been inferred to have occurred a minimum of 22 times independently within flowering plants (Nickrent, 2002; Nickrent et al., 2004; Bidartondo, 2005; Barkman et al., 2007; Merckx and Freudenstein, 2010).
One of the most studied groups of heterotrophic plants is Cuscuta (dodders), the sole parasitic genus of Convolvulaceae (reviewed in Stefanović and Olmstead, 2004, 2005). Species of Cuscuta are characterized by long slender stems, with scale-like leaves and no roots. They twine counter-clockwise and attach to their hosts via direct interplant haustorial connection. Once attached, dodders depend entirely or almost entirely upon a host to meet their carbon budget, water, and other nutrient demands (Kujit, 1969; Dawson et al., 1994). This genus is cosmopolitan in its distribution, but the majority of the species diversity (~140 out of 200 species) is encountered in the Americas (Yuncker, 1932; Stefanović et al., 2007). Cuscuta is considered economically important because several species can cause significant losses to agricultural crops (Parker and Riches, 1993; Costea and Tardiff, 2006). However, many Cuscuta species are also ecologically important, acting as keystone species in their natural ecosystems (Press and Phoenix, 2005), and some dodders are in need of conservation (Costea and Stefanović, 2009). Based on cytological, morphological, and anatomical evidence, Cuscuta is traditionally subdivided into three subgenera, Cuscuta, Grammica, and Monogynella (Yuncker, 1932). Recent molecular data (McNeal et al., 2007a; Stefanović et al., 2007) revealed a fourth major clade, consisting of Cuscuta species native to South Africa, and we refer to it here as ‘Pachystigma’ while awaiting formal classification.
Photosynthetic ability is variable across Cuscuta; it contains both hemi- and holoparasitic species. Some dodders produce significant amounts of chlorophylls in the tips of unattached seedlings as well as in fruiting sepals and ovaries (Panda and Choudhury, 1992; Dawson et al., 1994). This diversity of photosynthetic ability among Cuscuta species prompted several anatomical, physiological, and molecular evolutionary studies. Despite having plastids with no visible grana and a reduced number of thylakoids, C. reflexa (subgenus Monogynella) possesses chlorophylls a and b, and is capable of performing photosynthesis, albeit at a very reduced rate (Machado and Zetche, 1990; Hibberd et al., 1998). In contrast, C. europaea (subgenus Cuscuta) lacks chlorophyll, grana, and thylakoids, and appears incapable of fixing CO2 (Machado and Zetche, 1990). Within subgenus Grammica, in most cases, thylakoids, chlorophylls, and low quantities of the large subunit of Rubisco could be detected (van der Kooij et al., 2000). However, two species within subgenus Grammica (C. odorata and C. grandiflora) were found to lack not only chlorophyll and thylakoids but also the rbcL gene and its protein product (van der Kooij et al., 2000). Initial investigations into the plastid genome structure of C. reflexa indicated the loss of many plastid genes (Bommer et al., 1993; Haberhausen and Zetsche, 1994). The parallel loss of ribosomal polymerase (rpo) genes in three holoparasitic species indicates a transition from plastid-encoded polymerase (PEP) to nuclear-encoded polymerase (NEP) in subgenus Grammica (Krause et al., 2003). Subsequently, the loss of the rpo genes has been demonstrated to be shared by all of subgenus Grammica (Stefanović and Olmstead, 2005).
This early body of work on physiology and plastid molecular evolution in Cuscuta culminated with sequencing of four entire plastomes, two from subgenus Monogynella (C. reflexa and C. exaltata) and two from subgenus Grammica (C. campestris and C. obtusiflora; Funk et al., 2007; McNeal et al., 2007b). Both C. reflexa and C. exaltata retained much of their plastid genomes (~121–125 kbp), with losses being restricted primarily to the chlororespiratory (ndh) genes and non-coding regions, such as intergenic spacers and introns (Funk et al., 2007; McNeal et al., 2007b). C. campestris and C. obtusiflora, two very closely related species from clade ‘B’ of subgenus Grammica (Stefanović et al., 2007), have substantially smaller plastomes (~85–87 kbp). In addition to losses shared with C. reflexa and C. exaltata, they also lack a suite of rpo and some other ‘housekeeping’ genes (Funk et al., 2007; McNeal et al., 2007b). Both C. campestris and C. obtusiflora also share the loss of the group IIA introns, correlated with the loss of the intron maturase gene, matK (McNeal et al., 2009). This loss is rather unique in plants and is currently known to be shared only by an MHT orchid, Rhizanthella gardneri (Delannoy et al., 2011). Surprisingly, despite all these changes, the plastomes of Cuscuta retain many plastid genes required for photosynthesis, such as rbcL, psa, psb, pet, and atp genes, and appear generally unaffected compared with some other heterotrophs. For example, this is in contrast to the much reduced plastomes of Epifagus virginiana (~70 kbp) and R. gardneri (~59 kbp), which have retained only a few intact protein-coding genes, related to functions other than photosynthesis (Wolfe et al., 1992; Delannoy et al., 2011). Some MHT species, such as Neottia nidus-avis and Aneura mirabilis, also retain larger plastomes (~92 kbp and ~108 kbp, respectively), and many photosynthetic genes are still present either as open reading frames or as pseudogenes (Wickett et al., 2008; Logacheva et al., 2011).
Despite previous investigations into plastid genome evolution in Cuscuta, the extent of loss of plastid genes and its phylogenetic distribution within this genus remain largely unknown. A comparison of the plastid gene content across the diversity of Cuscuta would allow: (i) assessment of the degree to which the plastomes have been affected in various lineages; (ii) elucidatation of the fine-scale tempo, pattern, and limits of plastome gene loss; and (iii) dissection of the evolutionary constraints imposed on plastid genomes by non-photosynthetic metabolic functions, such as fatty acid biosynthesis (Krause, 2008). In this investigation, using slot-blot Southern hybridization, the presence of plastid genes is surveyed across an extensive sample of Cuscuta species. These data are interpreted within a rigorous phylogenetic framework, and in comparison with previously sequenced dodders and other heterotrophs. Finally, these results ase used to identify the most interesting species from the molecular evolution point of view, those that possess highly modified plastomes, thus representing prime candidates for targeted entire plastome sequencing.
Materials and methods
Taxon sampling
The sampling (Supplementary Table S1 available at JXB online) encompasses all major groups/subgenera of Cuscuta, as defined by several broad-scale molecular treatments (García and Martín, 2007; McNeal et al., 2007a; Stefanović et al., 2007). Out of ~200 species described for this genus, 149 accessions of Cuscuta representing 112 species (~56% of diversity) were included. Subgenus Grammica, the largest and most diverse lineage of Cuscuta, is represented by 93 species (124 accessions), with multiple samples from each of 15 clades (A–O) circumscribed in Stefanović et al. (2007). In particular, sampling was concentrated extensively on clades ‘K’ and ‘O’, two groups in which the presence of plastomes has been previously questioned (van der Kooij et al., 2000; McNeal et al., 2007a; Stefanović et al., 2007; Costea et al., 2011). The three remaining subgenera were also sampled in proportion to their diversity: four species from ‘Pachystigma’, eight from subgenus Cuscuta, and seven from subgenus Monogynella. As representatives of autotrophic lineages, 23 species representing nine out of 11 photosynthetic tribes within Convolvulaceae (Stefanović et al., 2003) were included. Taken together, the sampling strategy provides a broad phylogenetic background in which to compare the plastid gene content of Cuscuta with that of their autotrophic relatives (Supplementary Table S1).
DNA extraction and hybridization
Total genomic DNA was isolated from fresh, silica gel-dried, and herbarium tissue using the modified 2× cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987) and quantified using a UV spectrophotometer (BioPhotometer®; Eppendorf AG). To survey for the presence/absence of plastid genes of interest, the slot-blot hybridization method was used. A detailed description and rationale for this approach is provided in Doyle et al. (1995), Braukmann et al. (2009), and Braukmann and Stefanović (2012). In brief, a slot-blot apparatus (Bio-Rad) was used to make five sets of pseudoreplicate filter-blots, following the manufacturer’s protocol. Approximately 500–800ng of total DNA (per sample and per set) was bound to Immobilon-Ny+ nylon membrane (Millipore). DNAs from four Cuscuta species (C. obtusiflora, C. campestris, C. exaltata, and C. reflexa) whose entire plastomes were recently sequenced (Funk et al., 2007; McNeal et al., 2007b) were included on the membranes as known controls. Membranes were pre-hybridized and hybridized at 60–62 °C in 5× standard saline citrate (SSC), 0.1% SDS, 50mM TRIS (pH 8.0), 10mM EDTA, 2× Denhardt’s solution, and 5% dextran sulphate. After hybridization, filters were washed twice for 30–45min in 0.5% SDS and 2× SSC at the hybridization temperature. Probes were labelled with 32P using random oligonucleotide primers (Invitrogen). Autoradiography was carried out using intensifying screens at –80 °C for 18–48h. Filters were stripped of probe between hybridizations by boiling twice for 5–10min in 0.1% SSC. Prior to subsequent rounds of hybridization, the absence of carry-over signal was assured by an overexposure of decayed blots on a phosphor imaging screen for 6–8h (Personal Molecular Imager™; Bio-Rad) or autoradiography using intensifying screens at –80 °C for 72h. Hybridization probes for 48 plastid protein-coding genes (Supplementary Table S1 at JXB online) as well as controls from both plastid (16S and 23S rDNA) and mitochondrial (ATP synthase subunit 1) genomes were derived from tobacco (Nicotiana tabacum L.) via polymerase chain reaction (PCR). For the most part, two probes were used to survey genes interrupted by an intron, with each probe covering an exon. Exceptions to this approach were made for several genes that contain a very short exon (e.g. rps16, petB, and petD), as well as rpoC1, for which only the larger of the exons was probed. An additional exception was made for clpP, a gene that contains three short exons; only a single probe was constructed spanning the largest of the exons. Also, longer genes were surveyed using two probes situated at the 5’ and 3’ ends, respectively. A total of 56 probes were used, sampling every major functional category of protein-coding genes typically observed in green plant plastomes (refer to Wicke et al., 2011 for a detailed review). Primer names and sequences used to construct the probes are provided in Supplementary Table S2. For each probe, their length, GC content, and the structural location within the plastome of tobacco are provided in Supplementary Table S3. In addition, to estimate the unspecific background hybridization levels, an initial negative hybridization control was performed under the same stringency conditions (see above) and the same amount of 32P, but without probe added.
Results and Discussion
Interpretation of slot blots
The presence or absence of plastid protein-coding genes was determined by eye, by comparison of hybridization signal with the corresponding plastid and mitochondrial controls. Given the conserved nature of the genes encoding the plastid small (16S) and large (23S) ribosomal subunits and their near ubiquitous presence among plants (Bendich, 1987; Wicke et al., 2011), these two probes were used as controls to establish the presence of significant amounts of plastid DNA (ptDNA). However, because of weak to absent signal for 16S and 23S across clade ‘O’ in subgenus Grammica, a mitochondrial ATP synthase subunit 1 (atp1) probe was introduced as an additional control to verify the presence of organellar DNA. This ensures that a lack of hybridization signal is not due to insufficient quantities of DNA on the membrane, but is an indication of a significantly altered or absent plastid gene. These probes also serve as a baseline measure against which the presence or absence of other plastid genes was estimated. Additionally, a diverse sample of green Convolvulaceae was included to compare Cuscuta with more closely related autotrophic taxa to differentiate between losses common to the family and those common to Cuscuta.
A representative example of hybridization results, arranged phylogenetically, is depicted in Fig. 1, and the scores for all of the surveyed accessions and probes are listed in Supplementary Table S1 at JXB online. For all probes, the relative absence or presence of signal was scored for each taxon as indicating either full (++), diminished (+), absent (–), or unknown (?) in comparison with both the plastid (16S and 23) and mitochondrial (atp1) controls. For genes assayed with two probes (two exons or 5’ and 3’ end), a full hybridization signal to both probes is necessary to indicate that a functional copy of the gene is present. A full hybridization signal is assumed to indicate that the surveyed gene is present and putatively functional. Diminished signals, where hybridization is weaker than the controls but there is definite signal presence, can be interpreted in two different ways. It can indicate that the gene is present and functional but divergent with respect to tobacco or, alternatively, that the homologous region is rendered non-functional (pseudogene). Absence was scored if no detectable hybridization to a probe was observed. Given the experimental conditions used here, a gene transferred to the nucleus would not produce a hybridization signal when compared with a gene copy retained in the plastid genome. Transferred genes are significantly reduced in copy number and have accelerated substitution rates relative to the plastid (Wolfe et al., 1987). Given the typically low substitution rates for functional genes in ptDNA, a lack of signal suggests either loss of the gene or intracellular gene transfer (IGT) to the nucleus, rather than a highly divergent yet functional gene. Another potential destination for plastid genes includes mitochondria. Similarly to plastids, mitochondria are present in cells in high copy number, and have substantially lower rates of substitution compared with the plastid (Wolfe et al. 1987). In those rare cases where defunct copies of plastid genes (pseudogenes) are present in the mitochondrial genome due to IGT, they could result in a hybridization signal. However, this outcome is unlikely to affect the majority of hybridizations, as evidenced by the general absence of plastid gene signal in holoheterotrophs (see Supplementary Table S1; Fig. 1; and Braukmann et al., 2012 for the proof of principle). In certain cases, some taxa were scored as unknown (‘?’; see Supplementary Table S1). These ambiguities are a consequence of insufficient amounts or poor quality DNA for a given pseudoreplicate.
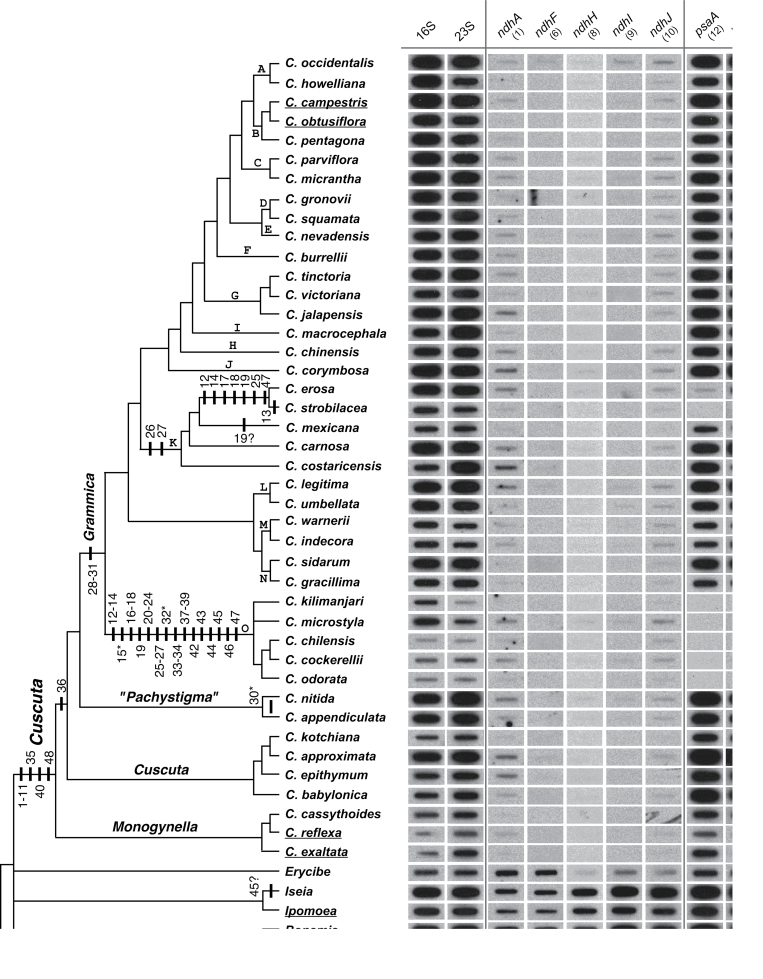
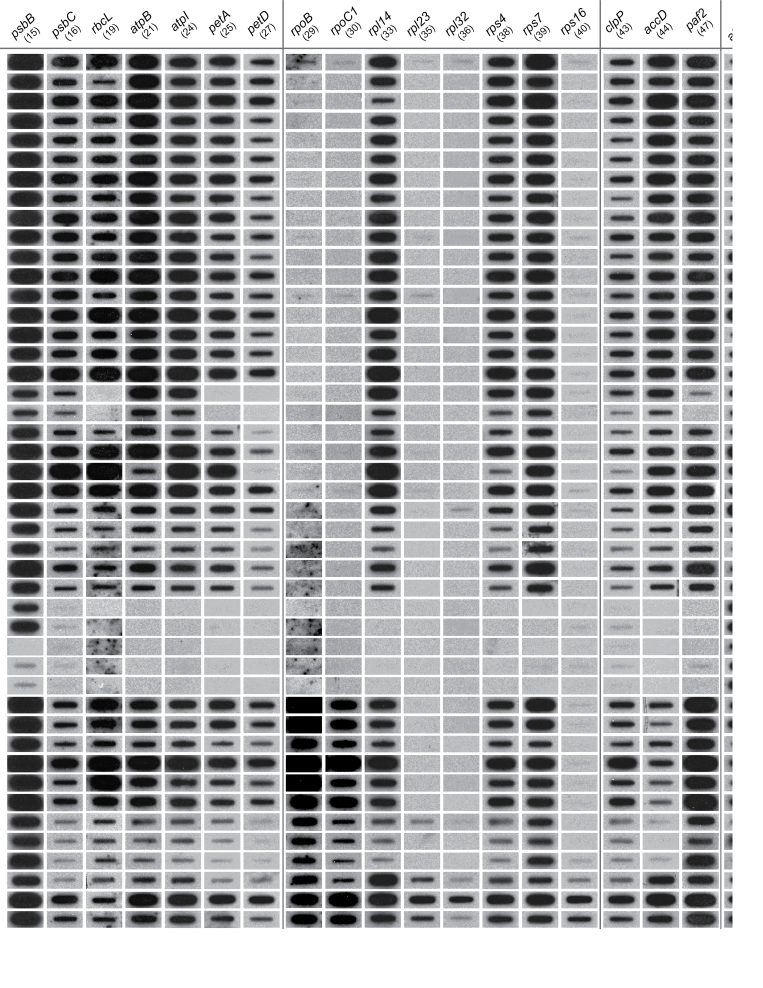
Fig. 1.
Autoradiographs representing a subset of slot-blot hybridization results for the presence/absence of 48 plastid protein-coding genes in Cuscuta and its close outgroups presented in a phylogenetic context. The topology shown is a composite tree depicting current understanding of relationships within Cuscuta derived from several published phylogenetic analyses (see text for references). Taxa with sequenced plastomes (Funk et al., 2007; McNeal et al., 2007a) are underlined. Parsimony reconstruction of plastid gene losses within Cuscuta under the assumption of irreversibility are mapped (bars) on the composite tree (depicted left). The numbers below each gene and above/below the bars refer to probes used in the hybridizations (see Supplmentary Table S1 at JXB online). Genes that are followed by a ‘?’ indicate potentially divergent copies of a plastid genes, whereas those followed by ‘*’ refer to genes that are present or absent (polymorphic) within the indicated clades. The plastid small (16S) and large (23S) rRNA subunits and mitochondrial ATP synthase subunit 1 (atp1) were used as positive controls (shown here is one representative out of five sets). Clades ‘K’ and ‘O’ show the greatest number of absences or near absences of hybridization signal for the plastid genes. Note that diminution of hybridization signal for clade ‘O’ extends to plastid positive controls (16S and 23S rDNA) compared with mitochondrial atp1. For full details, see Supplementary Table S1.
Altogether, these assumptions on the presence or absence of genes can lead to potential underestimates or overestimates of gene losses. For example, signals that appear present could potentially represent relatively recent pseudogenized genes, while significantly diminished signals might be due to divergent but functional genes. Despite these potential difficulties, Southern hybridization allows for the evaluation of the gene content of a broad and diverse set of taxa in an efficient and cost-effective manner (Doyle et al., 1995; Braukmann et al., 2009; Braukmann and Stefanović, 2012).
Plastid gene losses common to Cuscuta
Overall, the present hybridization results are consistent with a number of gene losses that were previously associated with the transition to parasitism in Cuscuta (Stefanović and Olmstead, 2005; McNeal et al., 2007a). Similarly to previous studies (Funk et al., 2007; McNeal et al., 2007b), the genes encoding the plastid NADH dehydrogenase complex (ndh genes) have been found to be lost across Cuscuta, as indicated by the general absence of hybridization signal (Fig. 1; Supplementary Table S1 at JXB online). The loss of these genes is a common feature amongst heterotrophic plants, whether parasitic or mycoheterotrophic, and whether hemi- or holoheterotrophic (Wolfe et al., 1992; Funk et al., 2007; McNeal et al., 2007b; Krause, 2008; Wickett et al., 2008; Logacheva et al., 2011; for comparison, see Table 1). The loss of the ndh genes has been observed in a limited number of autotrophic seed plant lineages as well (Braukmann et al., 2009; Wu et al., 2010; Blazier et al., 2011; Wicke et al., 2011; see Jansen et al., 2007 for a detailed review). The complex is thought to enable cyclic electron flow around photosystem I, by adjusting the ratio between ATP and NADPH, thereby helping protect a plant from photo-oxidative stress (Casano et al., 2000; Krause, 2011). The NDH complex is thought to be dispensable under conditions of low environmental stress (Martin and Sabater, 2010). Only under low CO2 conditions do ndh mutants exhibit a different phenotype from that of plants with a functional set of ndh genes (Horvath et al., 2000). Many dodders have limited gas exchange, which can lead to high internal levels of CO2 (Hibberd et al., 1998). Hence, the loss of the NDH complex can be viewed as selectively neutral, even potentially advantageous for a parasite (Krause, 2011).
Table 1.
Comparison of the 48 plastid protein-coding genes surveyed across Cuscuta with selected sequenced plastid genomes of heterotrophs and their respective autotrophic outgroups. Gene losses and pseudogenes are indicated for each protein-coding gene category. Taxa with fully sequenced plastid genomes are indicated by an asterisk, and achlorophyllous holoparasitic species are indicated in bold.
| Family subgenus* species | ||||
| NADH dehydrogenase | Photosystem I and II | Cytochrome b 6/f complex | ATP synthase | |
| SOLANACEAE | ||||
| Nicotiana tabacum* | ||||
| CONVOLVULACEAE | ||||
| Ipomoea purpurea* | ||||
| Monogynella | ||||
| C. exaltata* (S) | ndhA, ψndhB, ndhC, ψndhD, ndhE–K | |||
| C. reflexa* (S) | ndhA, ψndhB, ndhC–K | |||
| Cuscuta | ||||
| C. approximata (Q) | ψndhA–C, ndhD–K | |||
| Pachystigma | ||||
| C. nitida (P) | ψndhA–C, ndhD–I, ψndhJ, ψndhK | |||
| Grammica | ||||
| C. chilensis (O) | ψndhA, ndhB–I, ψndhJ, ndhK | psaA–C, psbB, ψpsbC, psbD, psbE | petA, petB, petD | atpA, atpB, atpF, atpH, atpI |
| C. kilimanjari (O) | ndhA, ψndhB, ndhC–K | psaA–C, psbB–E | petA, petB, petD | atpA, atpB, atpF, atpH, atpI |
| C. microstyla (O) | ψndhA–C, ndhD–I, ψndhJ, ndhK | psaB, psaC, ψpsbC, psbD, psbE | petA, petB, petD | atpA, atpB, atpF, atpH, atpI |
| C. sidarum (N) | ψndhA, ψndhB, ndhC–I, ψndhJ, ndhK | ψpetB | ψatpA, atpF | |
| C. costaricensis (K) | ψndhA–C, ndhD–K | ψpetB, ψpetD | ψatpB, atpF | |
| C. mexicana (K) | ndhA–K | ψpetB, ψpetD | atpF | |
| C. strobilacea (K) | ψndhA, ndhB, ψndhC, ndhD–I, ψndhJ, ndhK | psaA–C, ψpsbB, ψpsbD, psbE | petA, petB, petD | atpF |
| C. gronovii (D) | ψndhA–C, ndhD–I, ψndhJ, ndhK | atpF | ||
| C. campestris* (B) | ndhA–K | |||
| C. obtusiflora* (B) | ndhA–K | |||
| OROBANCHACEAE | ||||
| Epifagus virginiana* | ndhA, ψndhB, ndhC–K | psaA–C, psaI, ψpsbA, ψpsbB, psbC, psbD, psbE | petA, petB, petD | ψatpA, ψatpB, atpF, atpH, atpI |
| ORCHIDACEAE | ||||
| Phalaenopsis aphrodite * | ndhA, ψndhB–C, ψndhD, ψndhE, ndhF, ψndhG, ndhH, ψndhI, ψndhJ, ψndhK | |||
| Neottia nidus-avis* | ψndhA–C, ndhD–I, ψndhJ, ndhK | ψpsaA, ψpsaB, psaC, psbB ψpsbC, ψpsbD, psbE | ψpetA, ψpetB, petD | ψatpA, ψatpB, atpF, atpH, ψatpI |
| Rhizanthella gardneri* | ndhA–J, ψndhK | psaA, ψpsaB, psaC, psbA-E | petA, petB, petD | atpA, atpB, atpF, atpH, atpI |
| ANEURACEAE | ||||
| Aneura mirabilis* | ndhA, ψndhB–F, ndhG, ndhH, ndhI, ψndhJ, ndhK | ψpsaA, ψpsaB, ψpsbB-E | ψpetA, ψpetB | |
| CO2 fixation | RNA synthesis | Large and small ribosomal proteins | Genes with other function | |
| SOLANACEAE | ||||
| Nicotiana tabacum* | ||||
| CONVOLVULACEAE | ||||
| Ipomoea purpurea* | ψrpl23 | ycf15 | ||
| Monogynella | ||||
| C. exaltata* (S) | rps16, ψrpl23 | ψycf15 | ||
| C. reflexa* (S) | ψrps16, ψrpl23 | ψycf15 | ||
| Family subgenus* species | ||||
| CO2 fixation | RNA synthesis | Large and small ribosomal proteins | Genes with other function | |
| Cuscuta | ||||
| C. approximata (Q) | rps16, ψrpl23, rpl32 | ψaccD?, ycf15 | ||
| Pachystigma | ||||
| C. nitida (P) | rpl23, rpl32, rps16 | ψclpP, ψaccD, ψycf15 | ||
| Grammica | ||||
| C. chilensis (O) | rbcL | ψrpoA, rpoB, rpoC1, rpoC2 | ψrpl2, rpl14, rpl20, rpl23, rpl32, rps2, rps4, ψrps7, rps16 | ψmatK, ccsA, clpP, accD, ycf1, ycf2, paf2, ycf15 |
| C. kilimanjari (O) | rbcL | ψrpoA, rpoB, rpoC1, rpoC2 | rpl2, rpl14, rpl20, rpl23, rpl32, rps2, rps4, rps7, rps16 | ψmatK, ccsA, clpP, accD, ycf1, ycf2, paf2, ycf15 |
| C. microstyla (O) | rbcL | ψrpoA, rpoB, rpoC1, rpoC2 | ψrpl2, rpl14, rpl20, rpl23, rpl32, rps2, rps4, rps7, rps16 | ψmatK, ccsA, ψclpP, accD, ycf1, ψycf2, paf2, ycf15 |
| C. sidarum (N) | ψrbcL | ψrpoA, rpoB, rpoC1, rpoC2 | ψrpl2, rpl23, rpl32, ψrps2, rps16 | ψmatK, ψaccD, ψycf1, ycf15 |
| C. costaricensis (K) | ψrpoA, rpoB, rpoC1, ψrpoC2 | rpl23, rpl32, rps16 | ψmatK, ψclpP, ycf1, ψycf15 | |
| C. mexicana (K) | ψrpoA, rpoB, rpoC1, ψrpoC2 | ψrpl2, rpl23, rpl32, rps16 | ψmatK, ψclpP, ψycf1, ycf15 | |
| C. stobilacea (K) | rbcL | ψrpoA, rpoB, rpoC1, ψrpoC2 | ψrpl2, rpl23, rpl32, rps16 | ψmatK, ψclpP, ycf1, paf2, ψycf15 |
| C. gronovii (D) | ψrpoA, rpoB, rpoC1, rpoC2 | ψrpl2, rpl23, rpl32, rps16 | ψmatK, ψclpP, ψycf1, ψycf15 | |
| C. campestris* (B) | ψrpoA, rpoB, rpoC1, rpoC2 | rps16, rpl23, rpl32 | matK, ψycf2, ψycf15 | |
| C. obtusiflora* (B) | ψrpoA, rpoB–C2 | rps16, rpl23, rpl32 | matK, ycf15 | |
| OROBANCHACEAE | ||||
| Epifagus virginiana * | ψrbcL | ψrpoA, rpoB–C2 | rps16, ψrpl14, ψrpl23, rpl32 | cemA, ccsA, paf2 |
| ORCHIDACEAE | ||||
| Phalaenopsis aphrodite* | ||||
| Neottia nidus-avis* | ψrbcL | rpoA, ψrpoB, rpoC1, ψrpoC2 | rps16, rpl23 | ψmatK, ccsA, cemA, paf2, ycf15 |
| Rhizanthella gardneri* | rbcL | rpoA–C2 | rps16, rpl32 | ccsA, cemA, matK, paf2, ycf15 |
| ANEURACEAE | ||||
| Aneura mirabilis* | ψccsA | |||
In addition to the loss of the NDH complex, a number of other shared losses are indicated by the present results, common to the entire genus (Fig. 1). For example, there was no hybridization signal for rpl32 and rps16 across all Cuscuta species. Also, hybridization for rpl23 typically exhibited a complete absence of signal, but a weak signal was detected in a number of species (Supplementary Table S1 at JXB online). At the same time, all three of these genes had a full hybridization signal in autotrophic taxa, indicating that their loss is confined to Cuscuta. Both rpl23 and rps16 are known to be present only as pseudogenes in C. reflexa and C. exaltata and completely absent in C. campestris and C. obtusiflora (Funk et al., 2007; McNeal et al., 2007b). The hybridization approach used here confirms these findings and extends them to the entire genus; that is, the absence of hybridization signal is consistent with the functional loss of these genes from the plastome of all Cuscuta species. Loss of a gene from the plastome can either result from a complete loss of the gene from the cell or be a product of functional transfer to the nucleus. In several angiosperms, rps16 is encoded in the nucleus and targeted to both the chloroplast and mitochondria (Ueda et al. 2008), as are many other proteins and tRNAs (Carrie et al., 2009). The loss of large (rpl) and small (rps) ribosomal protein genes from the plastome does not necessarily represent loss of these genes from the cell but a shift to an increased reliance on nuclear-encoded products for plastid gene expression (Krause, 2011). In contrast to this, in both C. exaltata and C. reflexa, rpl32 is present within the plastome (Funk et al., 2007; McNeal et al., 2007b); however, no hybridization signal was observed for rpl32 in any Monogynella species sampled in this study. This is likely to be a consequence of high sequence divergence in comparison with tobacco (McNeal et al., 2007b), compounded by the large genome sizes observed for subgenus Monogynella (McNeal et al., 2006).
Subgenus Monogynella exhibited a full hybridization signal to ycf15 but, despite this strength, ycf15 is known to exist as a pseudogene in both C. reflexa and C. exaltata based on sequencing of the entire plastome (Funk et al., 2007; McNeal et al., 2007b). Hence, the relative strength of signal for the ycf15 pseudogene in Monogynella species is probably a result of the differential age of pseudogenes and/or rates of plastome decay within various lineages of Cuscuta. Only a weak to absent signal for this gene was observed for all other Cuscuta species (Fig. 1; Supplementary Table S1). This is consistent with the complete loss of ycf15 from plastomes of C. campestris and C. obtusiflora (Funk et al., 2007; McNeal et al., 2007b) and the functional loss of ycf15 is thought to be shared by all Cuscuta species (McNeal et al., 2007a).
Subgenus Grammica
The largest and most diverse subgenus of Cuscuta, Grammica, has the most variable plastid gene content (Stefanović and Olmstead, 2005; McNeal et al., 2007a; Stefanović et al., 2007). Clades ‘O’ and ‘K’ exhibit substantially more plastid gene loss compared with any other Cuscuta investigated to date, and will be discussed in detail below. However, there are a few common gene losses associated with the entire subgenus Grammica. Chief among these, and consistent with previous investigations, all rpo genes have been lost from the plastomes of species belonging to this group (Stefanović and Olmstead, 2005; Funk et al., 2007; McNeal et al., 2007b). The general absence of hybridization for any rpo gene (Supplementary Table S1 at JXB online; Fig. 1) is consistent with the view that there has been a shift from a PEP to a NEP. The transition to an NEP in subgenus Grammica is accompanied by the corresponding shift in promoters (Krause et al., 2003; Berg et al., 2004; Funk et al., 2007; McNeal et al., 2007b; Krause 2008, 2011). The loss of rpo genes from the plastome has previously been linked to the loss of photosynthetic ability (deSantis-Maciossek et al., 1999). However, despite a conversion to NEP for plastid expression (Krause et al., 2000; Berg et al., 2003; Krause, 2008), many Cuscuta species are thought to be cryptically photosynthetic.
To date, the loss of matK, a maturase for splicing group IIA introns in the plastome (Zoschke et al., 2010), has only been observed for Cuscuta subgenus Grammica and the mycoheterotrophic orchid R. gardneri (Funk et al., 2007; McNeal et al., 2007b, 2009; Delannoy et al., 2011; Krause, 2011). However, the present hybridization results indicate that there are a number of clades within Grammica that show a strong hybridization signal to a matK probe constructed from C. pedicillata, a species belonging to subgenus Cuscuta. For example, within clade ‘A’, C. salina had a full hybridization signal for matK. Additionally, most species within clades ‘H’ and ‘M’ (except C. azteca and C. coryli, respectively) also showed a full hybridization signal for this matK probe. These results suggest that matK might still be present in some members of Grammica or, more probably, that matK has decayed at very different rates within this subgenus, following its functional loss.
Overall, the present results indicate that the majority of subgenus Grammica species are similar to C. obtusiflora and C. campestris regarding their photosynthetic potential. However, it was also observed that the coding content, and correspondingly the photosynthetic ability, is highly variable across the breadth of subgenus Grammica, even more so than previously reported (van der Kooij et al., 2000). For example, two species of clade ‘L’ (C. odontolepis and C. hyalina) and all species belonging to clades ‘M’ and ‘N’ show only a weak hybridization signal for rbcL, petB, psaC, and psbD. This suggests that a number of species within these closely related clades have functionally lost more plastid genes or possess a more divergent plastome than C. campestris and C. obtusiflora (Supplementary Table S1 at JXB online).
The ‘O’ clade
The hybridization results for the ‘O’ clade (i.e. the C. grandiflora species complex) revealed a drastic reduction of plastid gene content, compared with other Cuscuta species or any other highly reduced holoheterotrophs, such as E. virginiana and R. gardneri (Table 1; Wolfe et al., 1992; Delannoy et al., 2011). The signal is completely absent across all gene categories probed and for all protein-coding genes, with only one exception, namely the full signal observed for psbB in C. microstyla, C. kilimanjari, and C. purpurata. These three species are not immediately related to each other and hence there is no phylogenetic pattern for the presence of signal for psbB in the ‘O’ clade. This again hints at a diverse tempo of pseudogene decay in different lineages of Cuscuta and the random nature of this process. Alternatively, it may indicate that some fragments of ptDNA have been transferred to the mitochondrial chromosome in the past and are now ‘frozen’ in this genome due to its very low mutation rates (Wolfe et al., 1987).
Another surprising result from the clade ‘O’ data is the weak hybridization signal observed for both the 16S and 23S controls, prompting the need for a mitochondrion-derived probe (atp1) to verify the presence of sufficient quantities of organellar DNA on the blots. Typically, these ribosomal RNA (rrn) genes are highly conserved elements of plastomes across plants, including in the vast majority of heterotrophs (Krause, 2011; Wicke et al., 2011). Absence of Southern hybridization has been reported previously only from some non-asterid holoparasites, such as Corynaea (Balanophoraceae), Hydnora (Hydnoraceae), and Rafflesiaceae (Nickrent et al., 1997a; Nickrent, 2008). Lack of hybridization signal for 16S in these parasites raised the possibility for the first time for the wholesale loss of the plastid genome in some angiosperms (Nickrent et al., 1997a), a situation analogous to that of hydrogenosomes (i.e. hydroxy somes; de Paula et al., 2012). These double-membrane bounded organelles are probably lacking DNA in some lineages [e.g. Trichomonas (Clemens and Johnson, 2000), Entamoeba (León-Avila and Tovar, 2004)] and are thought to have evolved from mitochondria in several lineages of anaerobic eukaryote parasites (Bui et al., 1996; Hackstein et al., 2001). In aggregate, the comprehensive hybridization investigation in Cuscuta presented here, taken together with previously published clues from a small number of species (van der Kooij et al., 2000; McNeal et al., 2006) or limited sequencing efforts (McNeal et al., 2007a; Stefanović et al., 2007), suggests that the plastomes within the ‘O’ clade may have reached the same or a similar evolutionary endpoint, where the very presence of a plastid genome is questionable (Nickrent et al., 1997a).
Alternatively, it has also been hypothesized that the plastomes in higher plants cannot actually be completely lost. The presence of 16S sequences derived via PCR amplification in most surveyed haustorial parasite lineages except Rafflesiaceae indicated that this evolutionary reduction has not yet gone to completion (Nickrent et al., 1997b). Instead, it has been proposed that plastomes of holoparasites can at most be reduced to mini-circles containing the plastid glutamyl-tRNA, encoded by trnE (Barbrook et al., 2006). In plants, the trnE has an essential role in tetrapyrole synthesis in both mitochondria and plastids, and, hence, this gene cannot functionally be replaced by the nuclear-encoded glutamyl-tRNA, because it cannot interact in haem synthesis (Howe and Smith, 1991; Barbrook et al., 2006). Given this essential role, the plastid-encoded trnE must remain separated from the rest of the cell (i.e. compartmentalized). Thus, it is predicted that even those heterotrophic plants that have lost most of their plastomes, including rrn genes, would still retain a residual plastome containing trnE, a suggestion known as the ‘essential tRNA’ hypothesis (Barbrook et al., 2006). Finding examples in which plastomes have been completely lost would tell us that these ‘essential’ genes are merely very difficult, but not impossible, to relocate functionally to the nucleus, and therefore that the plastome in heterotrophic plants is not fundamentally indispensable but should best be viewed as a partially or completely frozen product of evolutionary inertia.
The ‘K’ clade
Within Grammica, the plastid gene content of clade ‘K’ (i.e. the C. chapalana species complex) indicates a more gradual, stepwise degradation of plastome content across the clade. Overall, the entire clade shares the losses of petB and petD (Supplementary Table S1 at JXB online; Fig. 1). Otherwise, the plastomes of C. costaricensis, C. carnosa, and C. mexicana are relatively unaffected, indicating full hybridization for most genes (except petB and petD). However, it is clear from sequence data that rbcL is present as a pseudogene in C. mexicana (GenBank KC013278), despite its strong hybridization signal. The extent of plastid gene loss is greatest in C. erosa and C. strobilacea, compared with the rest of the ‘K’ clade (Fig. 1). The photosystem genes psaA, psaC, psbD, and psbE are the most affected, having only weak or completely absent signal. In addition to these photosystem genes, C. erosa and C. strobilacea also share the functional losses of rbcL, petA, and paf2 (ycf4), as evidenced by a substantially diminished signal. Finally, unique to C. strobilacea is the further absence of signal for psaB (Supplementary Table S1 at JXB online). Altogether, the progression of plastome degradation starts with the least affected C. costaricensis and C. carnosa, followed by C. mexicana, and then the most affected plastomes of C. erosa and especially C. strobilacea.
The Evolutionary Transition Series (ETS) hypothesis posits that changes associated with the evolution of parasitic plants are expected to be phylogenetically progressive (Boeshore, 1920; Young et al., 1999). The pattern of plastid gene loss observed within the ‘K’ clade appears to be stepwise and consistent with the ETS hypothesis. Alternatively, plastome evolution in Cuscuta is thought to be more consistent with the punctuated equilibrium hypothesis, which states that modifications at various evolutionary time points are followed by long periods of stasis, during which time no, or relatively few, events are inferred (Young et al., 1999; McNeal et al., 2007a). Albeit these two hypotheses are not necessarily mutually exclusive, the exact mechanism of plastome reduction is difficult to elucidate because of the lack of recent and intermediate transitional cases. Plastomes of most of the heterotrophic plants sequenced to date are found either to retain a large complement of plastid genes, as observed in all of the investigated Cuscuta species (Funk et al., 2007; McNeal et al., 2007b), or to be highly reduced and lacking most of the photosynthetic apparatus, as reported for Epifagus, Neottia, and Rhizanthella (Wolfe et al., 1992; Delannoy et al., 2011). The more recent the shift to holoparasitism is in this kind of comparative molecular endeavours, the more likely we are to discover clues about underlying mechanisms as well as capture certain processes ‘in action’ such as intermediate stages in gene transfer to the nucleus, pseudogenization, etc.
Subgenera Cuscuta, ‘Pachystigma’, and Monogynella
Excluding gene losses common to the entire genus, the plastid gene content of subgenera Cuscuta, ‘Pachystigma’, and Monogynella appears relatively unaffected. Genes involved in photosynthesis, transcription, and translation, and genes with other and unknown function gave full hybridization to most probes with only a few exceptions. The one exception for all non-Grammica Cuscuta species is the weak hybridization signal for accD to both tobacco- and C. obtusiflora-derived probes. Whole plastome analyses of both C. obtusiflora and C. campestris reveal that accD is divergent in C. obtusiflora and C. campestris (Funk et al., 2007; McNeal et al., 2007b). The weak hybridization signal for accD within these three groups suggests that this gene is not absent but rather divergent from the probes used in this study. However, the approach used here is unable to differentiate between divergent gene sequences and pseudogenes present in the plastome, and this issue will be elucidated only through further sequencing efforts.
Results presented here indicate that the plastid gene content is not as conserved in ‘Pachystigma’ as it is in subgenus Cuscuta. Notably, a weak signal was observed for psbC, psbD, ccsA, and rbcL for all members of ‘Pachystigma’ investigated. The hybridization signal for clpP was diminished in C. natalensis, C. nitida, and C. appendiculata, but remains relatively strong in C. angulata. Also, C. natalensis indicated weak hybridization for atpB, while C. angulata showed a unique absence of hybridization for rpoC1. This indicates that there is potentially another shift from a PEP into a NEP in ‘Pachystigma’. Overall, this clade of Cuscuta retains most photosynthetic genes, suggesting that there is still some photosynthetic ability.
A full complement of genes is generally retained in subgenus Monogynella as well, consistent with early physiological studies demonstrating that selected species within this group contain chlorophyll and are capable of performing photosynthesis at a very reduced rate (Machado and Zetche, 1990; Hibberd et al., 1998). The present results across this subgenus are also consistent with what is known from the fully sequenced plastomes of C. reflexa and C. exaltata. A few notable exceptions include the weak to absent signals for petD and psaC (Supplementary Table S1 at JXB online). Both petD and psaC are present in C. reflexa and C. exaltata (Funk et al., 2007; McNeal et al., 2007b), but full signal was detected only for psaC in C. exaltata. Previously, C. exaltata was found to have a significantly higher non-synonymous to synonymous (Dn/Ds) mutation rate for photosystem genes, ATP synthase, and phytochrome oxidase genes (McNeal et al., 2007b), which can in part explain the observed reduction in the hybridization signal strength. In addition, the hybridization signal can be diminished by the large nuclear genome size in Monogynella species (McNeal et al., 2006), because very large nuclear genomes substantially reduce the relative quantity of ptDNA loaded on the membranes.
Green Convolvulaceae
As expected, there is no diminution of hybridization signal for genes involved directly in photosynthesis among fully photosynthetic members of the family. Genes encoding the photosynthetic machinery, ATP synthase, phytochrome oxidase, and the large subunit of Rubisco, as well as those for the NADH dehydrogenase complex are present across autotrophic Convolvulaceae. However, hypothetical chloroplast open reading frames 1 and 15 (ycf1 and ycf15) have weak to absent signal for a number of autotrophic Convolvulaceae (see Supplementary Table S1 at JXB online). These results are consistent with previous Southern hybridization data, suggesting that ycf1 is lost or altered among some Convolvulaceae (Downie and Palmer, 1992). Among other green flowering plants, ycf1 has been reported to be absent from the plastid genomes of Passiflora and in some monocots (Jansen et al. 2007).
In addition to ycf1, seven species of green Convolvulaceae show a loss of ycf15 and an additional four indicate that ycf15 is present as a pseudogene or is very divergent from tobacco (Supplementary Table S1 at JXB online; Fig. 1). The ycf15 gene does not encode a protein but is instead hypothesized to act as a regulatory sequence or as a structural RNA (Schmitz-Linneweber et al., 2001). Previous work has shown that ycf15 is not under purifying selection and tends to be highly divergent across a wide range of taxa (Raubeson et al., 2007; Wicke et al., 2011). Overall, the losses of ycf1 and ycf15 amongst green Convolvulaceae exhibit no phylogenetic pattern and further support the idea that the losses of some plastid genes and non-coding regions in Convolvulaceae are not necessarily associated with parasitism (Stefanović and Olmstead, 2005; McNeal et al., 2007b).
Conclusions
This study provides a comprehensive investigation of the plastid genome content across the phylogenetic breadth and depth of Cuscuta. Most Cuscuta species retain plastid genome content similar to the previously published plastomes of C. reflexa, C. exaltata, C. campestris, and C. obtusiflora (Funk et al., 2007; McNeal et al., 2007b); however, the results clearly indicate that clades ‘K’ and ‘O’ within subgenus Grammica are divergent compared with the rest of Cuscuta. Generally, the most affected within subgenus Grammica is clade ‘O’, a group that has lost all the plastid protein-encoding genes probed in this study. In addition, within this clade, the rrn genes are also substantially affected, a condition reported previously only for some non-asterid holoparasites (Nickrent et al., 1997b). A representative species from the ‘O’ clade is currently a target for whole plastome sequencing, to explore further the limits of reduction and the ultimate fate of the plastome in holoparasitic plants. In the ‘K’ clade, the pattern of plastid gene loss is phylogenetically progressive and is consistent with the ETS hypothesis. Sequencing the entire plastomes in a number of species of the ‘K’ clade along the gradient of plastome degradation, from relatively unaffected (e.g. C. costaricensis or C. carnosa) to highly modified (e.g. C. strobilacea or C. erosa) via apparent intermediate cases (e.g. C. mexicana), will provide a good opportunity to capture plastome evolutionary processes ‘in action’ following the transition to holoparasitism amongst recently diverged, closely related species.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Cuscuta and its autotrophic outgroups from Convolvulaceae surveyed for the presence/absence of 48 plastid protein-coding genes.
Table S2. Oligonucleotides used in this study.
Table S3. List of probes used in the hybridizations.
Acknowledgements
The authors warmly thank A. Colwell, T. Van Devender, T. Deroin, M. García, R. Olmstead, and D. Tank, as well as the curators/directors of A, AAU, ALTA, ARIZ, ASU, CANB, CHR, CIMI, DAO, F, GH, IND, J, JEPS, LL, K, MEL, MEXU, MICH, NMC, NY, OKLA, PRE, QCNE, P, RSA, SD, SGO, TEX, TRT, UBC, UNB, UNM, UPRRP, US, USAS, WTU, and XAL for supplying plant material. We would also like to thank two anonymous reviewers whose suggestions greatly improved our manuscript. Financial support from the Natural Sciences and Engineering Research Council of Canada (grant no. 326439), the Canada Foundation for Innovation (grant no. 12810), and the Ontario Research Funds to SS is gratefully acknow ledged. We also thank the Natural Sciences and Engineering Research Council of Canada for the scholarship award provided to TB.
References
- Barbrook AC, Howe CJ, Purton S. 2006. Why are plastid genomes retained in non-photosynthetic organisms?. Trends in Plant Science 11, 101–108 [DOI] [PubMed] [Google Scholar]
- Barkman TJ, McNeal JR, Lim S-H, Coat G, Croom HB, Young ND, dePamphilis CW. 2007. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evolutionary Biology 7, 248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich AJ. 1987. Why do chloroplasts and mitochondria contain so many copies of their genome. BioEssays 6, 279–282 [DOI] [PubMed] [Google Scholar]
- Berg S, Krupinska K, Krause K. 2003. Plastids of three Cuscuta species differing in plastid coding capacity have a common parasite-specific RNA composition. Planta 218, 135–142 [DOI] [PubMed] [Google Scholar]
- Berg S, Krause K, Krupinska K. 2004. The rbcL genes of two Cuscuta species, C. gronovii and C. subinclusa, are transcribed by the nuclear-encoded plastid RNA polymerase (NEP). Planta 219, 541–546 [DOI] [PubMed] [Google Scholar]
- Bidartondo MI. 2005. The evolutionary ecology of myco-heterotrophy. New Phytologist 167, 335–352 [DOI] [PubMed] [Google Scholar]
- Blazier JC, Gusinger MM, Jansen RK. 2011. Recent loss of plastid encoded ndh genes within Erodium (Gerianaceae). Plant Molecular Biology 76, 263–272 [DOI] [PubMed] [Google Scholar]
- Boeshore I. 1920. The morphological continuity of Scrophulariaceae and Orobanchaceae. Contributions from the Botanical Laboratory of the University of Pennsylvania 5, 139–177 [Google Scholar]
- Bömmer D, Haberhausen G, Zetsche K. 1993. A large deletion in the plastid DNA of the holoparasitic flowering plant Cuscuta reflexa concerning two ribosomal proteins (rpl2, rpl23), one transfer RNA (trnI) and an ORF 2280 homologue. Current Genetics 24, 171–176 [DOI] [PubMed] [Google Scholar]
- Braukmann TWA, Kuzmina M, Stefanović S. 2009. Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Current Genetics 55, 323–337 [DOI] [PubMed] [Google Scholar]
- Braukmann TWA, Stefanović S. 2012. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Molecular Biology 79, 5–20 [DOI] [PubMed] [Google Scholar]
- Bui ET, Bradley PJ, Johnson PJ. 1996. A common evolutionary origin for mitochondria and hydrogenosomes. Proceedings of the National Academy of Sciences, USA 93, 9651–9656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casano LM, Zapata JM, Martin M, Sabater B. 2000. Chlororespiration and poising of cyclic electron transport: plastoquinone as electron transporter between thylakoid NADH dehydrogenase and peroxidase. Journal of Biological Chemistry 275, 942–948 [DOI] [PubMed] [Google Scholar]
- Carrie C, Giraud E, Whelan J. 2009. Protein transport in organelles: dual targeting of proteins to mitochondria and chloroplasts. FEBS Journal 276, 1187–1195 [DOI] [PubMed] [Google Scholar]
- Clemens DL, Johnson PJ. 2000. Failure to detect DNA in hydrogenosomes of Trichomonas vaginalis by nick translation and immunomicroscopy. Molecular and Biochemical Parasitology 106, 307–313 [DOI] [PubMed] [Google Scholar]
- Colwell AE. 1994. Genome evolution in a non-photosynthetic plant, Conopholis americana. PhD dissertation, Washington University; St Louis, MO, USA: [Google Scholar]
- Costea M, García IR, Stefanović S. 2011. ‘Horned’ dodders: phylogenetic relationships and two new species within Cuscuta chapalana complex (Convolvulaceae). Botany 89, 715–730 [Google Scholar]
- Costea M, Stefanović S. 2009. Cuscuta jepsonii (Convolvulaceae), an invasive weed or an extinct endemic?. American Journal of Botany 96, 1744–1750 [DOI] [PubMed] [Google Scholar]
- Costea M, Tardif FJ. 2006. Biology of Canadian weeds: Cuscuta campestris Yuncker, C. gronovii Willd. ex Schult., C. umbrosa Beyr. ex Hook., C. epithymum (L.) L. and C. epilinum Weihe. Canadian Journal of Plant Science 86, 293–316 [Google Scholar]
- Dawson JH, Musselman LJ, Wolswinkel P, Dorr I. 1994. Biology and control of Cuscuta . Reviews of Weed Science 6, 265–317 [Google Scholar]
- Delannoy E, Fijii S, Colas des Francs C, Brundett M, Small I. 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Molecular Biology and Evolution 28, 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paula WBM, Allen JF, van der Giezen M. 2012. Mitochondria, hydrogenosomes, and mitosomes in relation to the CoRR hypothesis for genome function and evolution. In: Bullerwell CE, ed. Organelle genetics: evolution of organelle genomes and gene expression Berlin: Springer; 105–119 [Google Scholar]
- deSantis-Maciossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, Rüdiger W, Koop HU, Herrmann RG. 1999. Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. The Plant Journal 18, 477–489 [DOI] [PubMed] [Google Scholar]
- Downie SR, Palmer JD. 1992. Use of chloroplast DNA rearrangements in reconstructing plant phylogeny. In: Soltis PS, Soltis DE, Doyle JA, eds. Molecular systematics of plants New York: Chapman and Hall; 14–35 [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19, 11–15 [Google Scholar]
- Doyle JJ, Doyle JL, Palmer JD. 1995. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Molecular Phylogenetics and Evolution 5, 429–438 [DOI] [PubMed] [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K. 2007. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii . BMC Plant Biology 7, 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García MA, Martín MP. 2007. Phylogeny of Cuscuta subgenus Cuscuta (Convolvulaceae) based on nrDNA ITS and chloroplast trnL intron sequences. Systematic Botany 32, 899–916 [Google Scholar]
- Haberhausen G, Zetsche K. 1994. Functional loss of all ndh genes in an otherwise relatively unaltered plastid genome of the holoparasitic flowering plant Cuscuta reflexa . Plant Molecular Biology 24, 217–222 [DOI] [PubMed] [Google Scholar]
- Hackstein JH, Akhmanova A, Voncken F, et al. 2001. Hydrogenosomes: convergent adaptations of mitochondria to anaerobic environments. Zoology 104, 290–302 [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Bungard RA, Press MC, Jeschke WD, Scholes JD, Quick WP. 1998. Localization of photosynthetic metabolism in the parasitic angiosperm Cuscuta reflexa . Planta 205, 506–513 [Google Scholar]
- Horvath EM, Peter SO, Joet T, Rumeau D, Cournac L, Horvath GV, Kavanagh TA, Schafer C, Peltier G, Medgyesy P. 2000. Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiology 123, 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe CJ, Smith AG. 1991. Plants without chlorophyll. Nature 349, 109 1986302 [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proceedings of the National Academy of Sciences, USA 104, 1369–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K, Berg S, Krupinska K. 2003. Plastid transcription in the holoparasitic plant genus Cuscuta: parallel loss of the rrn16 PEP promoter and of the rpoA and rpoB genes coding for the plastid encoded RNA polymerase. Planta 216, 815–823 [DOI] [PubMed] [Google Scholar]
- Krause K. 2008. From chloroplasts to ‘cryptic’ plastids: evolution of plastid genomes in parasitic plants. Current Genetics 54, 111–121 [DOI] [PubMed] [Google Scholar]
- Krause K. 2011. Piecing together the puzzle of parasitic plant plastome evolution. Planta 234, 647–656 [DOI] [PubMed] [Google Scholar]
- Kuijt J. The biology of parasitic flowering plants. Berkeley, CA: University of California Press; 1969. [Google Scholar]
- Leake JR. 1994. The biology of mycoheterotrophic (‘saprophytic’) plants. New Phytologist 127, 171–216 [DOI] [PubMed] [Google Scholar]
- Leon-Avila G, Tovar J. 2004. Mitosomes of Entamoeba histolytica are abundant mitochondrion-related remnant organelles that lack a detectable organellar genome. Microbiology 150, 1245–1250 [DOI] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Penin AA. 2011. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis . Genome Biology and Evolution 3, 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado MA, Zetsche K. 1990. A structural, functional, and molecular analysis of plastids of the holoparasites Cuscuta reflexa and Cuscuta europaea . Planta 181, 91–96 [DOI] [PubMed] [Google Scholar]
- Martin M, Sabater B. 2010. Plastid ndh genes in plant evolution. Plant Physiology and Biochemistry 48, 636–645 [DOI] [PubMed] [Google Scholar]
- McNeal JR, Arumugunathan K, Kuehl JV, Boore JL, dePamphilis CW. 2007. a Systematics and plastid genome evolution of the cryptically photosynthetic parasitic plant genus Cuscuta (Convolvulaceae). BMC Biology 5, 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, dePamphilis CW. 2007. b Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta . BMC Plant Biology 7, 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, Leebens-Mack J, dePamphilis CW. 2009. Parallel loss of plastid introns and their maturase in the genus Cuscuta . PLoS One 4, e59825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Leebens-Mack JH, Arumuganathan K, Kuehl JV, Boore JL, dePamphilis CW. 2006. Using partial genomic fosmid libraries for sequencing complete organellar genomes. BioTechniques. 41, 69–73 [DOI] [PubMed] [Google Scholar]
- Merckx V, Freudenstein JV. 2010. Evolution of mycoheterotrophy in plants: a phylogenetic perspective. New Phytologist 185, 605–609 [DOI] [PubMed] [Google Scholar]
- Nickrent DL. 2002. Origenes filosgeneticos de las plantas parasitas. In: Lopez-Saez JA, Catalan P, Saez L, eds. Plantas parasitas de la Penisula Iberica Islas Baleares Madrid: Mundi-Prensa Libras; 29–56 [Google Scholar]
- Nickrent DL. 2008. Parasitic plants. In: McGraw Hill yearbook of science and technology New York: McGraw Hill Book Co; 251–253 [Google Scholar]
- Nickrent DL, Blarer A, Qiu Y-L, Vidal-Russel R, Anderson FE. 2004. Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer. BMC Evolutionary Biology 4, 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent DL, Duff RJ, Konings DAM. 1997. b Structural analyses of plastid-derived 16S rRNAs of holoparasitic angiosperms. Plant Molecular Biology 34, 717–729 [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Ouyang Y, Duff RJ, dePamphilis CW. 1997. a Do nonasterid holoparasitic flowering plants have plastid genomes?. Plant Molecular Biology 34, 731–743 [DOI] [PubMed] [Google Scholar]
- Panda MM, Choudhury NK. 1992. Effect of irradiance and nutrients on chlorophyll and carotenoid content and Hill reaction activity of Cuscuta reflexa . Photosynthetica 26, 585–592 [Google Scholar]
- Parker C, Riches CR. Parasitic weeds of the world. Biology and control. Wallingford, UK: CAB International; 1993. [Google Scholar]
- Press MC, Phoenix GK. 2005. Impacts of parasitic plants on natural communities. New Phytologist 166, 737–751 [DOI] [PubMed] [Google Scholar]
- Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK. 2007. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus . BMC Genomics 8, 174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Maier RM, Alcaraz J, Cottet A, Herrmann RG, Mache R. 2001. The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Molecular Biology 45, 307–315 [DOI] [PubMed] [Google Scholar]
- Stefanović S, Austin DF, Olmstead RG. 2003. Classification of the Convolvulaceae: a phylogenetic approach. Systematic Botany 28, 791–806 [Google Scholar]
- Stefanović S, Kuzmina M, Costea M. 2007. Delimitation of major lineages within Cuscuta subgenus Grammica (Convolvulaceae) using plastid and nuclear DNA sequences. American Journal of Botany 94, 568–589 [DOI] [PubMed] [Google Scholar]
- Stefanović S, Olmstead RG. 2004. Testing the phylogenetic position of a parasitic plant (Cuscuta, Convolvulaceae, Asteridae): Bayesian inference and the parametric bootstrap on data drawn from three genomes. Systematic Biology 53, 384–399 [DOI] [PubMed] [Google Scholar]
- Stefanović S, Olmstead RG. 2005. Down the slippery slope: plastid genome evolution in Convolvulaceae. Journal of Molecular Evolution 61, 292–305 [DOI] [PubMed] [Google Scholar]
- Ueda M, Nishikawa T, Fujimoto M, Takanashi H, Arimura S, Tsutsumi N, Kadowaki K. 2008. Substitution of the gene for chloroplast RPS16 was assisted by generation of dual targeting signal. Molecular Biology and Evolution 25, 1566–1575 [DOI] [PubMed] [Google Scholar]
- van der Kooij TAW, Krause K, Dorr I, Krupinska K. 2000. Molecular, functional and ultrastructural characterization of plastids from six species of the parasitic flowering plant genus Cuscuta . Planta 210, 701–707 [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Muller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Molecular Biology 76, 273–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Zhang Y, Hansen SK, Roper JM, Kuehl JV, Plock SA, Wolf PG, dePamphilis CW, Boore JL, Goffinet B. 2008. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis . Molecular Biology and Evolution 25, 393–401 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences, USA 84, 9054–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Ems SC, Palmer JD. 1992. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. Journal of Molecular Evolution 35, 304–317 [DOI] [PubMed] [Google Scholar]
- Wu F, Chan M, Liao D, Hsu C, Lee Y, Daniell H, Duvall MR, Lin C. 2010. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding of Oncidiinae. BMC Plant Biology 10, 68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Steiner KE, dePamphilis CW. 1999. The evolution of parasitism in Scrophulariaceae and Orobanchaceae: plastid gene sequences refute an Evolutionary Transition Series. Annals of the Missouri Botanical Garden 86, 876–893 [Google Scholar]
- Yuncker TG. 1932. The genus Cuscuta . Memoirs of the Torrey Botanical Club 18, 113–331 [Google Scholar]
- Zoschke R, Nakamura M, Liere K, Sugiura M, Börner T, Schmitz-Linneweber C. 2010. An organellar maturase associates with multiple group II introns. Proceedings of the National Academy of Sciences, USA 107, 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.