Abstract
The TetL antiporter from the Bacillus subtilis inner membrane is a tetracycline-divalent cation efflux protein that is energized by the electrochemical proton gradient across the membrane. In this study we expressed tetL in Escherichia coli and investigated the oligomeric state of TetL in the membrane and in detergent solution. Evidence for an oligomeric state of TetL emerged from SDS-PAGE and Western blot analysis of membrane samples as well as purified protein samples from cells that expressed two differently tagged TetL species. Furthermore, no formation or restoration of TetL oligomers occurred upon detergent solubilization of the membrane. Rather, oligomeric forms established in vivo persisted after solubilization. Mass spectrometry of the purified protein showed the absence of proteolysis and post-translational modifications. Analytical size-exclusion chromatography of the purified protein revealed a dimeric TetL in dodecyl-maltoside solution. In addition, TetL dimers were found in a number of other detergents and over a wide pH range. It is therefore likely that the oligomeric form of the protein in the membrane is also a dimer.
The TetL protein from the Bacillus subtilis inner membrane is a tetracycline (Tc) efflux protein that catalyzes the exchange of a tetracycline-chelated metal complex for protons in an electrogenic exchange (H+ > Tc-Me2+) that is energized by the transmembrane electrochemical proton gradient. Sodium and potassium ions can replace the singly charged Tc-Me2+ as efflux substrate (i.e. the transporter can function as a low affinity monovalent cation/proton antiporter) and potassium ions can replace protons at least in part as coupling ions. The assays of these modes have been conducted in defined strains of Escherichia coli or in proteoliposomes, but physiological studies on wild type and mutant strains of Bacillus subtilis support functions for all the modes in the native host (1). A member of the major facilitator superfamily (MFS) (2–4), the protein consists of 458 amino acids. It is predicted to possess 14 transmembrane α-helices, a characteristic shared with the closely related tetracycline transporter TetK (5, 6). TetL has been overexpressed in E. coli, and the purified protein shows transport activity following reconstitution into proteoliposomes (7). The oligomeric state of the protein in the membrane, however, remains unclear.
Molecular mechanisms of substrate translocation across the cell membrane for secondary active transporters are incompletely understood. High-resolution structural information that is beginning to emerge for some of these proteins (8, 9) will greatly facilitate progress toward increasingly detailed transport models. Among the characteristics that need to be carefully assessed, as part of structural work and even before detailed structural models can be made, is the association state of the particular transporter. This information is essential for the complete understanding of the substrate translocation process and, especially, for cooperative and regulatory aspects of transporter function (10). There is evidence that many secondary transporters form oligomers in the membrane. Therefore, intensive recent efforts have been made to determine the oligomeric state of transporter proteins in the early stages of structure-function studies using various biochemical and biophysical techniques. Whereas electron microscopy of freeze-fracture samples suggests that the human neuronal glutamate transporter (EAAT3) has a pentameric structure (11), the serotonin transporter (SERT) is shown to form a dimeric functional unit, which may further form a tetramer (12, 13). On the other hand, the sodium/proton antiporter NhaA from E. coli is a dimer in solution and in reconstituted two-dimensional crystals as well (14, 15). The newly-determined crystal structure of AcrB, a multidrug exporter of the Resistance-Nodulation-Cell Division family from E. coli, shows a trimer, with a single substrate translocation pathway thought to be located in the center of the trimer (16, 17). In the major facilitator superfamily, many members purify as monomers and are also believed to function as monomers, including the human erythrocyte glucose transporter (Glut1) (18, 19) and three proteins from the E. coli inner membrane, the lactose permease (LacY) (20, 21), the hexose-6-phosphate transporter (UhpT) (22) and the glycerol-3-phosphate transporter (GlpT) (23). However, some other members of the superfamily form various types of oligomers. TetA from E. coli, a tetracycline transporter predicted to have 12 transmembrane α- helices, has been shown to be a trimer in solution and in reconstituted two-dimensional crystals (24), even though genetic and biochemical data suggest a dimeric structure (25). On the other hand, the lactose transporter (LacS) from Streptococcus thermophilus works as a dimer in the membrane with two sugar translocation pathways that exhibit cooperativity. This transporter protein also purifies as a dimer in detergent solution (26–28).
In the current work we investigated the association state of the TetL protein in the native membrane using both genetic and biochemical approaches. The results were compared to the oligomeric state determined in detergent solution. From these experiments we conclude that the state of TetL in the membrane is an oligomer that is most likely a dimer.
Materials and Methods
Unless otherwise mentioned, standard reagents were purchased from Sigma (St. Louis, MO), and cloning kits from Qiagen (Chatsworth, CA). Protein concentration was determined using the Micro-BCA assay (Pierce, Rockford, IL).
Cloning of TetL constructs
C-terminally His-tagged TetL (TetL-His) was subcloned in pET23b(+) (Novagen, Madison, WI), which contained pBR322 origin of replication and encoded ampicillin resistance; N-terminally T7-tagged TetL (T7-TetL) was subcloned in a pACYC184-derived vector, which contained p15A origin of replication and encoded chloramphenicol resistance. Hence these two plasmids were compatible and we were able to maintain them within the same cell using a combination of different antibiotics. Both constructs conferred tetracycline resistance in E. coli cells. The sequence of the T7 tag is Met-Ala-Ser-Met-Thr-Gly-Gly-Gln-Gln-Met-Gly.
The construct for TetL-His expression was subcloned as follows: A 1.4 kb tetL fragment was amplified by oligonucleotides 5′-GGAATTCCATATGAATACGTCTATATCACAG - 3′ and 5′-CCGGATCCCTCGAGGCCATGTCTCCGCGAACGTTT -3′, digested with Nde I and Xho I and ligated with pET23b(+) which was linearized with these two restriction enzymes. The whole TetL-encoding ORF, in frame with the C-terminal His-tag, was confirmed by DNA sequencing. The construct for T7-TetL expression was generated in two steps as follows. A 300 bp T7 promoter containing fragment was first amplified from pET23b(+) (Novagen) by oligonucleotides 5′-CCATCGATAGATCTCGATCCCGCGAAAT - 3′ and 5′-TGACGATATGATCACAAAAAACCCCTCAAGACCC - 3′, digested with Cla I and Bcl I, and ligated with a 2.2 kb Cla I/Bcl I fragment of pACYC184. The resulting construct contained the replication origin of pACYC184 and the chloramphenicol resistance gene but not the original tetracycline resistance element, and the plasmid was designated pJST7. This construct was then linearized by Bam HI digestion and ligated to a 1.4 kb Bam HI tetL fragment which was amplified by oligonucleotides 5′-CCCGGATCCGAATACGTCTTATTCACAGTCA-3′ and 5′-CCCGGATCCTTTCACTCATTTA-3′. Only plasmids with the correct orientation of insert were selected. The whole ORF encoding TetL, in frame with the N-terminal T7-tag, was confirmed by DNA sequencing.
Analysis of TetL association in the membrane
The association state of TetL in the native E. coli membrane was analyzed by co-expressing the protein tagged with different markers, essentially according to the approach by Gerchman et al. in their study of NhaA protein (15). Briefly, E. coli strain BL21(DE3) was either transformed with TetL-His/pET23b(+) and T7-TetL/pJST7 plasmids separately, or co-transformed with both plasmids simultaneously. For overexpression, 1 L LB containing 100 μg/ml ampicillin and 35 μg/ml chloramphenicol in a 4 L flask was inoculated with 30 ml of overnight culture. After growth at 30 °C for 2–3 hours until optical density reached 0.6–0.7, the temperature was reduced to 20 °C. Expression of TetL was induced with 1 mM IPTG at a cell density of 0.9–1.0. Following growth for an additional 1 hour and 45 minutes, cells were harvested by centrifugation at 15000g for 15 minutes. The pellet was washed briefly in 50 ml cold TBS buffer (100 mM NaCl, 50 Tris, pH 7.5) and resuspended in lysis buffer (20 mM Tris/HCl pH 8, 100 mM NaCl, 1 mM PMSF, a protease inhibitor cocktail and 1 μg/ml DNaseI), at 5 ml buffer per gram cells. Cells were broken with two passes of French Press at 20,000 psi. After the removal of unbroken cells at 15000g for 20 minutes, the membranes were collected by ultracentrifugation at 100000g for 2.5 hrs. The membrane sample obtained from cells co-expressing TetL-His and T7-TetL was designated Co-expr, and the membranes from cells expressing TetL-His and cells expressing T7-TetL were designated His-only and T7-only, respectively.
Following membrane isolation, equal parts of the His-only and T7-only membrane samples were mixed. This membrane sample was designated Mixed. The remaining samples were distributed in equal amounts so that all four samples, Co-expr, His-only, T7-only and Mixed, contained the same amounts of membrane protein. These membranes were then solubilized and subjected to metal affinity chromatography purification as described in the following section. Western blot analysis of membrane vesicles and partially purified samples were performed using two types of polyclonal antibodies, one against the N-terminus of TetL (7) and the other against the T7 peptide (Novagen).
TetL purification
Before developing a strategy for affinity-based purification, we examined the effect of a single modification, addition of hexahistidine to the C-terminus vs. the N-terminus of TetL on its tetracycline efflux capacity. For the N-terminal modification, a 1.4 kb tetL fragment was amplified by oligonucleotides 5′-GGAATTCCATATGAATACGTCTATATCACAG-3′ and 5′-CCCGGATCCTTTCACTCATTTA-3′. The fragment was digested with Nde I and Bam HI and subcloned in pET15b (Novagen). The construct encodes a TetL protein that is in frame with a N-terminal His-tag and a thrombin site. For C-terminal modification, the Nde I-containing primer was again used, this time together with 5′-CCGGATCCCTCGAGGCCATBTCTCCGCGAACGTTT-3′. The PCR product was digested with Nde I and Xho I and subcloned into pET-23b or pET-29b (Novagen). In both vectors, which conferred different resistances, the encoded TetL protein was now in-frame with a C-terminal His-tag. The two modified TetL proteins were compared with a wild type TetL that was expressed comparably after having been cloned into pET23b(+) (Novagen). For that cloning, an Nde I/Bam HI fragment of TetL was subcloned in pET23(+) by primers 5′-GGAATTCCATATGAATACGTCTATATCACAG - 3′ and 5′-CCCGGATCCTTTCACTCATTTA - 3′ so that the endogenous stop codon of TetL was retained. The three constructs were transformed into E. coli DH5α. Tetracycline efflux was assayed in everted vesicles as described previously (29). In three independent experiments, the C-terminally modified TetL exhibited very low activity relative to the un-modified TetL while the N-terminally modified TetL exhibited activity that was slightly higher than that of unmodified TetL (data not shown). For subsequent purification, N-terminal modification was chosen. Although ultimate removal of the tag was envisioned, it was prudent to have the more active form of the transporter assembled in the membrane in case the poorer activity reflected a problem in an oligomerization that influences activity. The 1.4TetL protein was cloned in pET15b as described above, and overexpressed in E. coli BL21 (DE3) pLysS cells. The presence of the pLysS plasmid facilitated the lysis process and therefore only one pass of French Press was required for complete cell breakage, all other steps for membrane preparation were similar to those described in the previous section. The membrane was solubilized in (20 mM Tris/HCl pH 8, 400 mM NaCl, 20 % glycerol, 1 mM PMSF, protease inhibitor cocktail, and 1 % DDM) at 10 ml per gram of membrane for 30 min. Unsolubilized debris was removed by centrifugation at 100000g for 30 min. The resulting clear supernatant was incubated overnight with Co2+-TALON (Clontech, Mountain View, CA) at 0.25 ml per gram of membrane. Three wash steps were performed with buffer A (20 mM Tris/HCl pH 8, 200 mM NaCl, 20 % glycerol, 0.1 % DDM) at increasing concentrations of imidazole: 10 mM, 20 mM and 50 mM, respectively. TetL was eluted in two steps with buffer A containing 200 mM and 500 mM imidazole, respectively.
Purified TetL was incubated with thrombin (ICN, Costa Mesa, CA) at 22 °C for 16 hrs at 4–5 units protease per mg of protein. The protein sample was concentrated with a 50 kDa cut-off concentrator (Millipore, Bedford, MA), and loaded onto a preparative size-exclusion Superdex200 column on FPLC (Amersham-Pharmacia, Piscataway, NJ), in 50 mM imidazole pH 7.0, 100 mM NaCl, 20 % Glycerol, 0.5 mM EDTA, 0.075 % DDM. Elutes were collected in 0.3 ml fractions at a flow rate of 0.1 ml/min. Protein purity and completeness of His-tag removal were analyzed by Coomassie Blue- and silver-stained SDS-PAGE and by Western blot using polyclonal anti-TetL antibodies and the India His-Probe-HRP conjugate (Pierce, Rockford, IL).
Determination of Stokes radius of TetL by size-exclusion chromatography
The oligomeric state of TetL in DDM was determined by analytical size-exclusion chromatography (19, 30). Purified TetL was loaded onto a Shodex KW804 or KW803 size-exclusion column on HPLC (Waters, Milford, MA), in buffer containing (50 mM Tris pH 8.0, 200 mM Na2SO4, 3 mM NaN3 and 0.05 % DDM). The Stokes radius of the TetL-DDM complex was determined using soluble proteins as references (31–33). The association state of TetL was derived by comparing its Stokes radius with those of membrane transporters with known oligomeric states (19, 23, 34–36). The TetL association state in other detergents was analyzed at a concentration of 0.2 % above their corresponding Critical Micellar Concentrations (CMC), using protein sample purified in 0.075 % DDM. Approximately 50 μg of protein was required for each experiment.
Mass spectrometry
The mass of the purified TetL was determined using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry in the laboratory of Dr. T. Neubert following published protocols (23, 37).
Assessment of functional complementation of co-expressed TetL mutant proteins
We tested whether mutant TetL proteins could form an active transporter when co-expressed from compatible plasmids in the same E. coli DH5α cells, using the following combinations of mutants that had been generated and characterized earlier (38): G70R and E397N; E152Q and E397N; and R110C and E397N. E397N cloned in pJST7, a vector derived from pACYC184 (New England BioLabs) as follows. The E397N construct cloned earlier in pBK15 was digested with Nde I and Bam HI; the resulting fragment was cloned into similarly digested pJST7. The E152Q and R110C mutants were used in their original pBK15 vector (38). Tetracycline antiport activity was assayed in everted membrane vesicles from single and double transformants and vector control transformants as described previously (29). In order to detect whether recombination occurred among the co-expressed plasmids, PCR primers were designed that were specific for each individual mutant and a third set was designed for each pair that would detect a full length recombinant product.
Results
Association state of TetL in the membrane
To investigate the oligomeric state of TetL in the cell membrane we co-expressed two TetL constructs, containing either a His- or a T7-tag, in E. coli. TetL was then purified, and analyzed in comparison with protein derived from cells expressing each of the constructs separately. As part of the purification, membrane vesicles were prepared from all three samples. In addition to purification starting from these membrane types, another experiment involved mixing vesicles containing the two types of singly-tagged species prior to solubilization. Solubilized protein from all four samples, T7-Tet/Tet-His co-expressed, T7-Tet, Tet-His and T7-Tet/Tet-His mixed after membrane preparation, was further purified by Co2+-TALON affinity chromatography and analyzed by Western blotting using two different antibodies, one directed against the T7-tag, and the other the N-terminus of TetL.
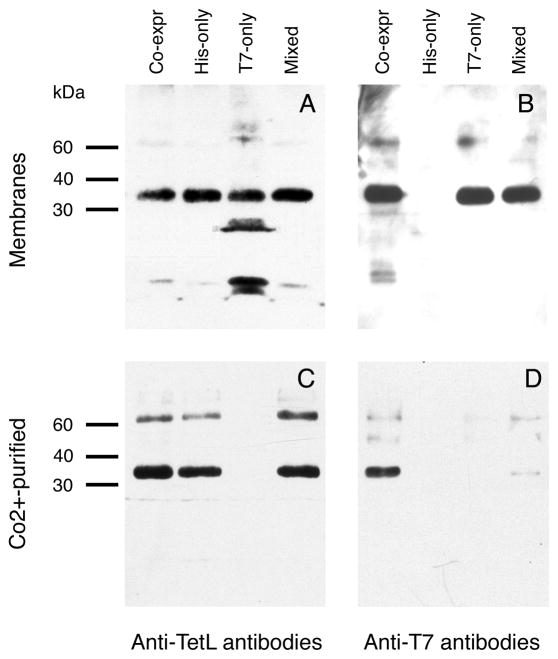
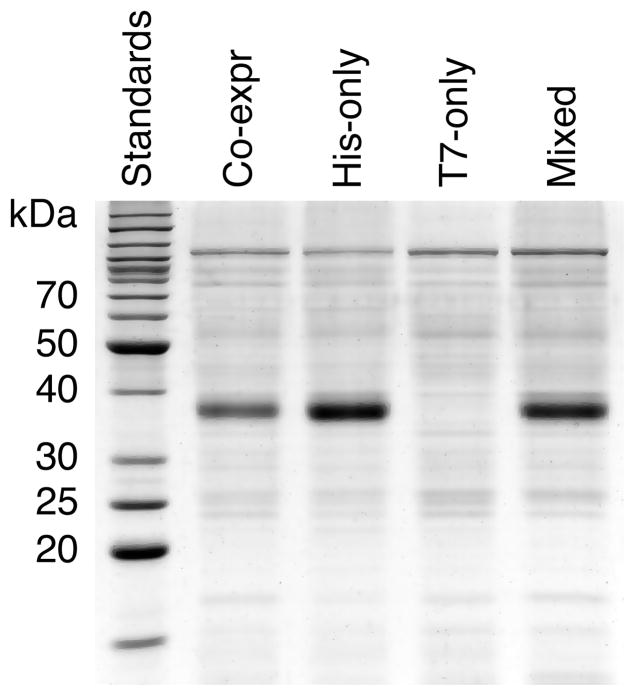
As expected, Western blot analysis using anti-TetL antibodies detected the TetL protein in all four samples (Figure 1A), and the anti-T7 antibodies detected positive signals in all but the His-only sample (Figure 1B). After Co2+-TALON purification, TetL was detected with anti-His probe in the Co-expr, His-only and Mixed sample, but not in the T7-only sample (Figure 1C) which lacked a His-tag. In the same panel of four Co2+ column-purified samples, however, a positive Western blot reaction was detected only in the Co-expr sample when anti-T7 antibodies were used (Figure 1D). This showed co-purification of His- and T7-tagged TetL protein in the cells of the co-expression experiment; no co-purification occurred when individual TetL-His and T7-TetL samples were mixed prior to solubilization. These results strongly suggested that (1) TetL-His and T7-TetL formed oligomers in the membrane of the co-expression system and (2) TetL oligomers did not derive from association of differently tagged monomers during membrane solubilization. The second hypothesis was further supported by the pattern of Coomassie Blue-stained SDS-PAGE of the four purified samples (Figure 2), which showed that equal amounts of TetL were purified from the His-only membrane and Mixed membranes, again indicating the absence of re-association of TetL oligomers during solubilization. Therefore, it is likely that TetL oligomerized in the membrane and, once formed, the oligomers were stable during solubilization and purification.
Figure 1.
Western blot analysis of membrane and Co2+-TALON purified samples from membranes with co-expressed or individually expressed TetL-His and T7-TetL protein constructs. A. Membrane samples probed with anti-TetL antibodies. B. Membrane samples probed with anti-T7 antibodies. C. Co2+-TALON purified samples probed with anti-TetL antibodies. D. Co2+-TALON purified samples probed with anti-T7 antibodies. Four samples were examined in each experiment: co-expressed TetL-His and T7-TetL, expressed TetL-His only, expressed T7-TetL only, and singly-expressed TetL-His and T7-TetL samples mixed prior to solubilization.
Figure 2.
Coomassie Blue-stained SDS-PAGE of co-expressed and individually expressed TetL-His and T7-TetL. The four lanes are co-expressed TetL-His and T7-TetL, TetL-His expressed alone, T7-TetL expressed alone, and individual TetL-His and T7-TetL samples mixed prior to solubilization. After purification with Co2+-TALON, the same volume of sample was loaded onto first three lanes, but twice the volume was loaded onto the Mixed-sample lane.
TetL purification
E. coli BL21(DE3)pLysS cells transformed with a pET vector containing the TetL gene produced tightly-controlled TetL protein synthesis. The pLysS strain was particularly suitable for large-scale protein expression and purification due to the convenience in breaking the cells. Prior to induction of tetL expression, the rates of cell growth were indistinguishable from non-transformed cells. Reducing the temperature from 30 to 20 °C prior to the induction of TetL overexpression minimized endogenous proteolytic digestion of the protein. Western blotting of total membrane protein showed only small amounts of fragmented target protein (Figure 3B), with the full-length TetL as the dominant species.
Figure 3.
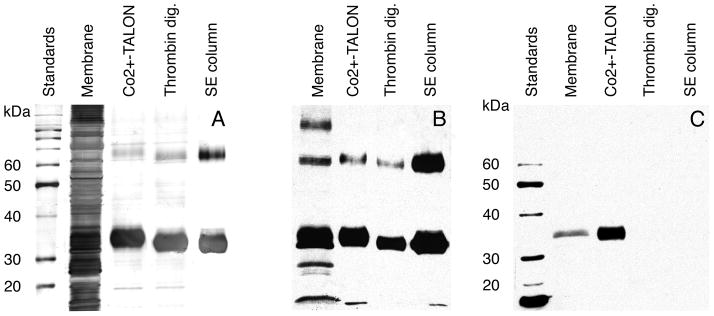
Expression and purification of TetL. A. Silver-stained SDS-PAGE. B. Western blot analysis using polyclonal anti-TetL antibodies. C. Western blot analysis using India His-Probe-HRP. Dimers are detected in purified TetL samples by both Coomassie-blue stained SDS-PAGE and Western blot. Low molecular weight fragments of TetL and high-molecular weight aggregates can be seen in B. They did not co-purify with intact transporter protein. Four samples were shown in each panel: solubilized membrane, Co2+-TALON-purified sample, thrombin-digested sample and SE-column purified sample.
During purification, very little proteolytic digestion TetL occurred as shown by silver-stained SDS-PAGE and Western blot analysis (Figures 3A,B). The monomeric protein migrated as a fuzzy band in SDS-PAGE, with an apparent mass of 34 kDa. This is smaller than the 52 kDa as calculated from the protein sequence, a phenomenon often observed with hydrophobic membrane proteins (30). Interestingly, even under reducing conditions, a band at precisely twice the monomeric mass was detected in total membranes as well as in purified samples (Figure 3A,B). Western blot detection of this band, presumably a TetL dimer, was possible with an antibody directed against the 14 N-terminal amino acids of TetL, but not with the INDIA probe against the His-tag (Figure 3C). This suggests a possible masking of the His-tag in the dimeric state, which would also explain the slow binding of His-tagged TetL to Co2+-TALON during purification. Co2+-TALON purification produced a TetL purity of above 80 % and a protein yield of 10–15 mg per 12 L culture (Figure 3A). The purity is significantly higher than the 50 % observed when Ni2+-NTA resin was used (data not shown). Therefore, Co2+-TALON resin was used for all subsequent purifications.
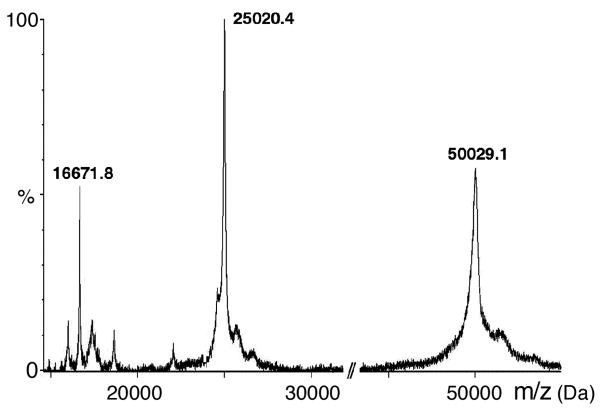
Complete removal of the His-tag was achieved by overnight incubation with thrombin (Figure 3). TetL was further purified by preparative size exclusion chromatography, yielding a protein purity of higher than 95%. MALDI-TOF mass spectrometry of TetL produced a sharp peak, corresponding to 50038.8 Da (Figure 4). This value is within 1.2 Da of 50037.6 Da as the predicted mass from the wild-type TetL sequence with the N-terminal Gly-Ser-His sequence of the thrombin cleavage site. This indicated the presence of a single polypeptide species and absence of post-translational modification, such as N-terminal methionine cleavage or in vivo proteolysis. The protein was also found to be resistant to further digestion against a range of proteases (data not shown). The final yield of TetL purification was 3–4 mg from 12 liters of bacterial culture.
Figure 4.
MALDI-TOF mass spectrometry of purified TetL. The observed m/z values for single, double and triple charged TetL protein molecules are indicated. Due to the standardization used in this experiment, mass calculation is most accurate at m/z = 15000–30000 (± 0.002 %). The subunits mass calculated from the doubly charged species is 50038.8 Da, in agreement with the value of 50037.6 Da predicted from protein sequence. The sharpness of the peaks indicates homogeneity of the sample.
Oligomeric state of TetL in DDM solution
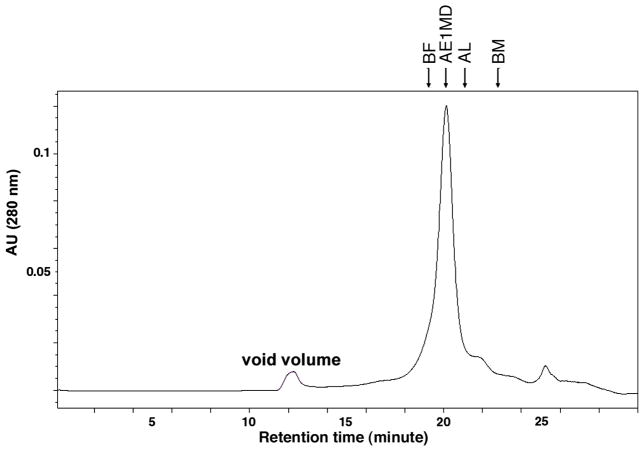
To further characterize the nature of the TetL oligomer, we examined its oligomeric state in DDM solution after purification. The Stokes radius of TetL in DDM was determined using analytical size exclusion chromatography. TetL eluted as a single, sharp and symmetrical peak at 20.2 min, with very little aggregation or secondary peaks (Figure 5). Its Stokes radius was determined to be 55±3 Å. This is significantly larger than the Stokes radii of 50 Å for glycosylated Glut1 (19) and 46 Å for GlpT (23), two monomeric transporter proteins with 12 transmembrane α-helices. Importantly, the dimeric membrane domain of the human erythrocyte anion exchanger (AE1MD) (34–36), as an internal standard with a similar molecular weight and number of transmembrane α-helices per monomer, eluted with the same retention time as TetL. We therefore conclude that TetL in DDM solution is a dimer.
Figure 5.
Determination of Stokes radius of purified TetL in DDM solution at pH 7.5 by analytical size exclusion chromatography. Protein standards are indicated:β-ferritin (BF, 63 Å), aldolase (AL, 46 Å), and bumin (BM, 35 Å), as well as the dimeric membrane domain of AE1 (AE1MD).
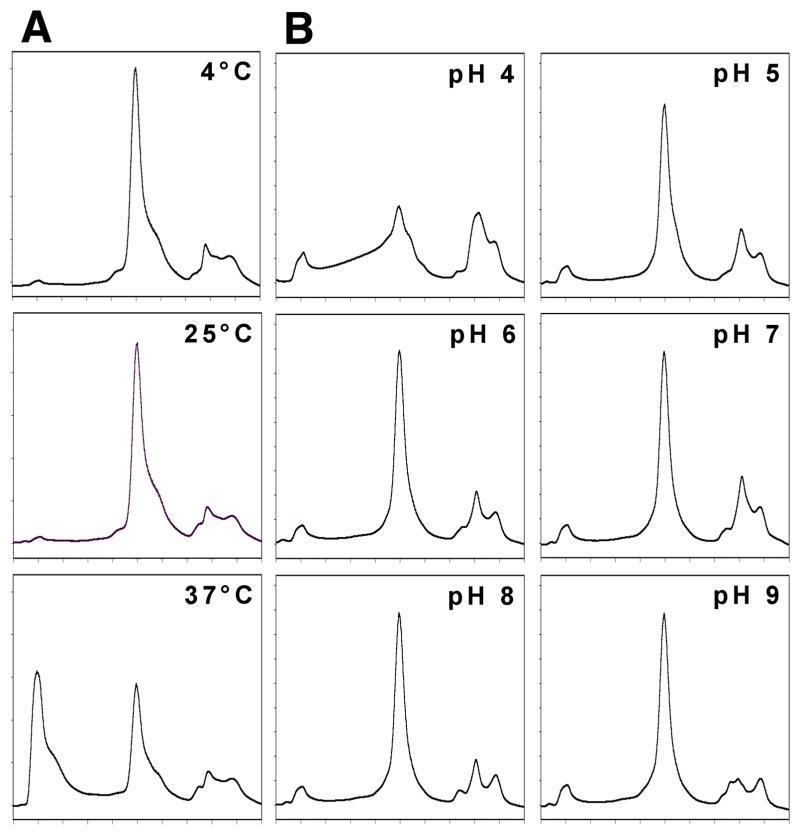
Oligomeric state of TetL at different temperatures and pH ranges
TetL remained as a dimer in DDM over a broad pH and temperature range, as shown by analytical size-exclusion chromatography (Figure 6). Little aggregation or dimer disintegration occurred at pH 5–9. Only at pH 4 did we observe non-specific association of TetL subunits and significant aggregation. Similar results were observed at elevated temperatures: at 25 °C, the structural integrity of TetL dimers was maintained, but at 37 °C TetL aggregated significantly as it mostly eluted in the void volume. However, even under these conditions the TetL remained in solution was still a dimer. Finally, TetL preserved its dimeric state after several cycles of freezing and thawing. Therefore, we conclude that the dimeric form is indeed the favored oligomeric state of TetL in DDM.
Figure 6.
Analysis of the oligomeric state of purified TetL in DDM at different temperatures and pH ranges using analytical size-exclusion chromatography. Identical amounts of TetL were injected for each experiment. For simplicity, profiles are shown only between retention time 8 and 25 min in this and the next figure.
Oligomeric state of TetL in other detergents
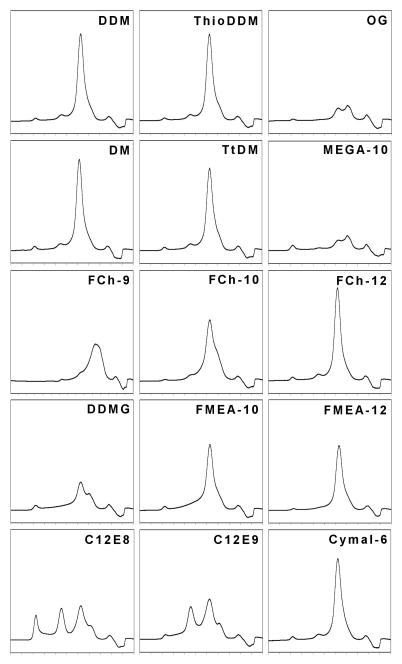
The oligomeric states of TetL in 16 additional detergents were examined (Figure 7). These detergents cover a broad range of chemical properties: non-ionic vs. zwitterionic head-groups, and long vs. short acyl-chain lengths. In half of the detergents tested, TetL showed a retention time and elution profile virtually identical to that seen in DDM; in the rest of the detergents TetL precipitated out of solution. These observations further support our suggestion that TetL is a dimeric membrane protein, since it is highly unlikely that the association state is an artifact due to the diversity in chemical properties and characteristics of the detergents examined.
Figure 7.
Analysis of TetL oligomeric state in different detergents. TetL was purified in 0.075 % DDM, A second detergent was added to a concentration of 0.2 % above its corresponding critical micellar concentrations, followed by size-exclusion chromatography analysis on HPLC. Approximately 50 μg of protein was used for each experiment.
Assessment of complementation by co-expression of TetL mutant proteins
Partial functional complementation of defects in the catalytic activity of inactive mutants of closely related 12-transmembrane spanning Tet proteins has been reported to occur when two mutants with defects in different halves of the molecule are co-expressed (39, 40). In a different antiporter, NhaA from E. coli, co-expression of different mutants with regulatory phenotypes indicated an interaction that supported an oligomeric state (15). We therefore tested whether mutant TetL proteins could form an active transporter when co-expressed from compatible plasmids in the same E. coli cells. In initial experiments, the cells for vesicle preparation, that expressing the plasmids with the two test mutant TetL versions, were grown in the presence of tetracycline. In every combination, but in no single transformant controls, significant tetracycline uptake was observed. However, in all instances, PCR analysis of the cells from which the vesicles had been prepared showed that detectable recombination had occurred between the two plasmids. Thereafter, tetracycline was omitted from the growth medium. PCR analysis indicated that the two plasmids were retained with their inserts during the growth period and that no recombination was detectable. Expression levels were comparable to those in which activity at 15% or more of wild type TetL would have been detected. No tetracycline uptake activity was observed in vesicles in which mutant pairs were co-expressed in the absence of recombination (data not shown).
Discussion
In this work, TetL was shown to exist as an oligomer rather than a monomer in the membrane. The major piece of evidence came from tests of the expectation that if TetL exists as an oligomer in the membrane, some hetero-oligomers containing both His- and T7- tags would be found in membranes of cells expressing TetL proteins labeled with these two different tags (Figure 8 and Table 1). Consequently, we would be able to identify T7-tags in protein samples that were purified by His-tag directed Co2+-TALON affinity chromatography. Thus, the starting criterion was fulfilled for positing a TetL oligomeric state in the membrane (Figures 1&2).
Figure 8.
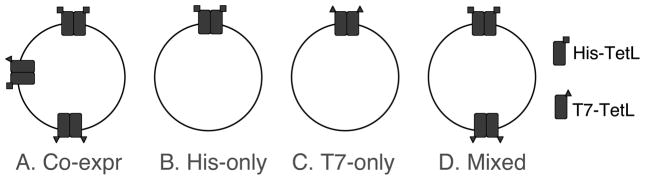
Illustration showing dimeric organizations of cells or mixed membrane vesicles that overexpressed combinations of TetL-His and T7-TetL.
Table 1.
Possibilities of TetL oligomeric states in the membrane
| In membrane | In solution | Yes/No | Reason | Comment |
|---|---|---|---|---|
| monomer | monomer | No | A, C | |
| monomer × N | Nmer | No | B | N > 1 |
| dimer | dimer | Yes | C | |
| dimer | monomer + monomer | No | C | |
| dimer | Nmer | No | B | N > 2 |
| trimer | trimer | No | C | |
| trimer | monomer + dimer | No | C | |
| trimer | monomer × 3 | No | C | |
| trimer × 2 | trimer × 3 | No | B | |
| tetramer | tetramer | No | C | |
| tetramer | monomer + trimer | No | C | |
| tetramer | monomer + monomer + dimer | No | C | |
| tetramer | dimer × 2 | ? | ||
| tetramer | monome × 4 | No | C |
Notes: A. TetL was oligomeric in the membrane as determined by the co-expression experiment. B. TetL did not re-associate upon solubilization as shown by the mixed sample experiment. C. TetL was dimeric in detergent solution and no other forms were detected, as shown by analytical size-exclusion chromatography.
Evidence for a dimeric TetL complex comes from a number of observations (Table 1). First, association was seen between differently tagged TetL subunits only when they were co-expressed in the same cell (Figures 1&2). When subunits in membranes of different cells were mixed prior to solubilization, no such interaction was observed, indicating that transporter complexes formed in the membrane and were stable enough to withstand detergent extraction from the membrane. This is very similar to the results obtained for NhaA (15). Second, a TetL dimer band in denaturing PAGE experiments was seen during protein purification, even at low protein concentrations (Figure 3), indicating a strong interaction of TetL subunits within the dimer. No higher oligomers were observed under these conditions. The behavior of TetL is thus similar to those of other stable oligomeric membrane proteins like KscA K+-channel (41). Third, evidence for a TetL dimer was obtained from analytical size exclusion chromatography of purified protein, where purified TetL eluted as a dimer in detergent solution (Figures 5,6&7). The existence of TetL dimmer in solution was observed across broad pH and temperature ranges, and in a variety of different detergents, making it unlikely to be an artifact caused by specific protein-detergent interactions.
We cannot rule out the possibility that higher oligomers, such as tetramers, exist in the membranes (Table 1). It is possible that tetramers in the membrane dissociated into dimers upon solubilization. Whereas the monomer-monomer interactions within a dimer are clearly very strong, dimer-dimer interactions, if any, must be relatively weak and thus less important for proper functioning of the protein. Given the prevalence of a dimer under diverse conditions in solution, we hypothesize that a dimer is the in vivo structure as well.
It remains unclear whether the dimeric association of TetL is necessary for substrate translocation. As noted, several MFS proteins have been shown to function as monomers, including Glut1 (18, 19), LacY (20, 21) and UhpT (22). On the other hand, although the functional unit of LacS is a monomer, it forms a dimer in the membrane with two sugar translocation pathways that exhibit cooperative behavior (26–28). Most importantly, the recently determined structures of glycerol-3-phosphate transporter and the lactose permease, both MFS proteins, reveal that the substrate translocation pore is located between the N- and C-terminal halves of the proteins (8, 9). Since all MFS proteins share a similar topology and even some degree of sequence homology, it is likely that they all work as monomers.
The complementation observed by Rubin and Levy with Gram-negative Tet proteins (39, 40) and additional properties have led to a proposal for TetB in which a dimeric Tet might be able to form a translocation pathway within a monomer but could also form a dimer in which two translocation pathways were formed through interactions between the two complementing domains of the protein (25). This latter option probably does not apply to the larger TetL, inasmuch as complementation among mutations in different regions was not detected. Large-scale conformational changes between the two halves of monomeric GlpT are thought to be necessary for substrate translocation. In an MFS oligomer, such gross conformational changes are likely to affect the transport activity of the other subunit(s). This means that various subunits would be expected function cooperatively. Therefore, oligomerization, as observed with the TetL protein, provides another dimension of regulation, e.g. by allosteric interactions, which may be particularly important for this multifunctional antiporter. During earlier kinetic experiments (42), Jin1 observed stimulation of tetracycline efflux by very low concentrations of a competitive efflux substrate, Na+; such stimulation at low concentrations and competitive inhibition at higher concentrations is most consistent with a cooperative interaction that would be best accommodated by an oligomeric transporter. Once the appropriate mutants of TetL are available for complementation experiments, it is anticipated that the physiologically-related functions of the TetL dimer will be shown to relate to elements of regulation, such as the different pH dependence of its different catalytic modes (1), analogous to the findings with NhaA (15). The challenge of demonstrating the functional implications of transporter oligomerization are not, however, trivial and there are only a few clear examples thus far (10).
Acknowledgments
The research was financially supported by NIH (RO1 GM052837) and the Robert Leet and Clara Guthrie Patterson Trust.
We are grateful to Ms. Y. Lu and Dr. T. Neubert for mass spectrometry measurements, and to Drs. R. Goetz and M.J. Lemieux for helpful discussions.
Abbreviations
- CMC
critical micellar concentration
- DDM
dodecyl-maltoside
- HPLC
high-performance liquid chromatography
- PAGE
polyacrylamide gel electrophoresis
- PMSF
phenyl methyl sulfonyl fluoride
- SDS
sodium dodecyl sulfate
Footnotes
Jin, J., unpublished data.
References
- 1.Krulwich TA, Jin J, Guffanti AA, Bechhofer H. J Mol Microbiol Biotechnol. 2001;3:237–46. [PubMed] [Google Scholar]
- 2.Reizer J, Finley K, Kakuda D, MacLeod CL, Reizer A, Saier MH., Jr Prot Sci. 1993;2:20–30. doi: 10.1002/pro.5560020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson PJF. Curr Opin Cell Biol. 1993;5:708–712. doi: 10.1016/0955-0674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 4.Pao SS, Paulsen IT, Saier MH., Jr Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginn SL, Brown MH, Skurray RA. J Bacteriol. 1997;179:3786–9. doi: 10.1128/jb.179.11.3786-3789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirata T, Fujihira E, Kimura-Someya T, Yamaguchi A. J Biochem (Tokyo) 1998;124:1206–11. doi: 10.1093/oxfordjournals.jbchem.a022239. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J, Hicks DB, Krulwich TA. Proc Natl Acad Sci U S A. 1996;93:14446–51. doi: 10.1073/pnas.93.25.14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 9.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Science. 2003;301:610–5. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 10.Veenhoff LM, Heuberger EH, Poolman B. Trends Biochem Sci. 2002;27:242–9. doi: 10.1016/s0968-0004(02)02077-7. [DOI] [PubMed] [Google Scholar]
- 11.Eskandari S, Kreman M, Kavanaugh MP, Wright EM, Zampighi GA. Proc Natl Acad Sci U S A. 2000;97:8641–6. doi: 10.1073/pnas.97.15.8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilic F, Rudnick G. Proc Natl Acad Sci U S A. 2000;97:3106–11. doi: 10.1073/pnas.060408997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid JA, Scholze P, Kudlacek O, Freissmuth M, Singer EA, Sitte HH. J Biol Chem. 2001;276:3805–10. doi: 10.1074/jbc.M007357200. [DOI] [PubMed] [Google Scholar]
- 14.Williams KA. Nature. 2000;403:112–5. doi: 10.1038/47534. [DOI] [PubMed] [Google Scholar]
- 15.Gerchman Y, Rimon A, Venturi M, Padan E. Biochemistry. 2001;40:3403–12. doi: 10.1021/bi002669o. [DOI] [PubMed] [Google Scholar]
- 16.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Nature. 2002;419:587–93. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 17.Yu CA, Xia JZ, Kachurin AM, Yu L, Xia D, Kim H, Deisenhofer J. Biochimica et Biophysica Acta. 1996;1275:47–53. doi: 10.1016/0005-2728(96)00049-7. [DOI] [PubMed] [Google Scholar]
- 18.Burant CF, Bell GI. Biochemistry. 1992;31:10414–20. doi: 10.1021/bi00157a032. [DOI] [PubMed] [Google Scholar]
- 19.Boulter JM, Wang DN. Prot Expr Purif. 2001;22:337–348. doi: 10.1006/prep.2001.1440. [DOI] [PubMed] [Google Scholar]
- 20.Wright JK, Weigel U, Lustig A, Bocklage H, Mieschendahl M, Muller-Hill B, Overath P. FEBS Lett. 1983;162:11–5. doi: 10.1016/0014-5793(83)81039-4. [DOI] [PubMed] [Google Scholar]
- 21.Roepe PD, Kaback HR. Proc Natl Acad Sci U S A. 1989;86:6087–91. doi: 10.1073/pnas.86.16.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambudkar SV, Anantharam V, Maloney PC. J Biol Chem. 1990;265:12287–92. [PubMed] [Google Scholar]
- 23.Auer M, Kim MJ, Lemieux MJ, Villa A, Song J, Li XD, Wang DN. Biochemistry. 2001;40:6628–35. doi: 10.1021/bi010138+. [DOI] [PubMed] [Google Scholar]
- 24.Yin CC, Aldema-Ramos ML, Borges-Walmsley MI, Taylor RW, Walmsley AR, Levy SB, Bullough PA. Mol Microbiol. 2000;38:482–92. doi: 10.1046/j.1365-2958.2000.02149.x. [DOI] [PubMed] [Google Scholar]
- 25.McMurry LM, Levy SB. J Biol Chem. 1995;270:22752–7. doi: 10.1074/jbc.270.39.22752. [DOI] [PubMed] [Google Scholar]
- 26.Friesen RH, Knol J, Poolman B. J Biol Chem. 2000;275:33527–35. doi: 10.1074/jbc.M004066200. [DOI] [PubMed] [Google Scholar]
- 27.Veenhoff LM, Heuberger EH, Poolman B. Embo J. 2001;20:3056–62. doi: 10.1093/emboj/20.12.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heuberger EH, Veenhoff LM, Duurkens RH, Friesen RH, Poolman B. J Mol Biol. 2002;317:591–600. doi: 10.1006/jmbi.2002.5416. [DOI] [PubMed] [Google Scholar]
- 29.Jin J, Guffanti AA, Beck C, Krulwich TA. J Bacteriol. 2001;183:2667–71. doi: 10.1128/JB.183.8.2667-2671.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang DN, Lemieux MJ, Boulter JM. In: Membrane Protein Protocols: Expression, Purification and Characterization. Selinsky B, editor. Humana Press; Totowa, New Jersey: 2003. pp. 239–256. [Google Scholar]
- 31.Le Maire M, Aggerbeck LP, Monteilhet C, Andersen JP, Moller JV. Anal Biochem. 1986;154:525–35. doi: 10.1016/0003-2697(86)90025-4. [DOI] [PubMed] [Google Scholar]
- 32.Harlan JE, Picot D, Loll PJ, Garavito RM. Anal Biochem. 1995;224:557–63. doi: 10.1006/abio.1995.1087. [DOI] [PubMed] [Google Scholar]
- 33.de Haen C. Anal Biochem. 1987;166:235–45. doi: 10.1016/0003-2697(87)90570-7. [DOI] [PubMed] [Google Scholar]
- 34.Lemieux MJ, Reithmeier RA, Wang DN. J Struct Biol. 2002;137:322–32. doi: 10.1016/s1047-8477(02)00010-2. [DOI] [PubMed] [Google Scholar]
- 35.Casey JR, Reithmeier RA. J Biol Chem. 1991;266:15726–37. [PubMed] [Google Scholar]
- 36.Casey JR, Reithmeier RA. Biochemistry. 1993;32:1172–9. doi: 10.1021/bi00055a023. [DOI] [PubMed] [Google Scholar]
- 37.Cadene M, Chait B. Anal Chem. 2000;72:5655–58. doi: 10.1021/ac000811l. [DOI] [PubMed] [Google Scholar]
- 38.Jin J, Krulwich TA. J Bacteriol. 2002;184:1796–800. doi: 10.1128/JB.184.6.1796-1800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubin RA, Levy SB. J Bacteriol. 1990;172:2303–12. doi: 10.1128/jb.172.5.2303-2312.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubin RA, Levy SB. J Bacteriol. 1991;173:4503–9. doi: 10.1128/jb.173.14.4503-4509.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valiyaveetil FI, Zhou Y, MacKinnon R. Biochemistry. 2002;41:10771–7. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 42.Jin J, Guffanti AA, Bechhofer DH, Krulwich TA. J Bacteriol. 2002;184:4722–32. doi: 10.1128/JB.184.17.4722-4732.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]